Abstract
The role of RNA interference (RNAi) in post-transcriptional regulation of complementary targets is well known. However, less is known about transcriptional silencing mechanisms mediated by RNAi. Such mechanisms have been characterized in yeast and plants, which suggests that similar RNA silencing mechanisms might operate in animals. A growing amount of experimental evidence indicates that short RNAs and their co-factor Argonaute proteins can regulate many nuclear processes in metazoans. PIWI-interacting RNAs (piRNAs) initiate transcriptional silencing of transposable elements, which leads to heterochromatin formation and/or DNA methylation. In addition, Argonaute proteins and short RNAs directly regulate Pol II transcription and splicing of euchromatic protein-coding genes and also affect genome architecture. Therefore, RNAi pathways can have a profound global impact on the transcriptional programs in cells during animal development. This article is part of a special issue entitled: Chromatin and epigenetic regulation of animal development.
1. Introduction
The discovery of RNA interference (RNAi) [1] and other related RNA silencing phenomena [2–7] as well as the characterization of different classes of small non-coding RNAs has radically changed the way we think about the role of RNA in gene regulation. Although the idea of non-coding RNAs as regulators of gene expression is not recent [8], appreciation of their fundamental role in many biological processes was only achieved after the emergence of new RNA-based silencing phenomena and rigorous biochemical studies. Most of the RNA-mediated gene silencing phenomena include a double-stranded RNA (dsRNA) intermediate, which is processed by the RNase III enzyme Dicer into short interfering RNAs (siRNAs) ranging from 18 to 32 nucleotides (nt). These siRNAs are loaded onto Argonaute proteins, which serve as the catalytic component of the RNA-induced silencing complex (RISC). The catalytic domain of the Argonaute proteins allows them to cleave an RNA target (slicer activity) in cases when there is perfect complementarity between the loaded siRNA and their RNA target (reviewed in [9]). siRNAs can be directly transfected into the cell, produced by Dicer processing of exogenous dsRNA, or even generated endogenously.
RNA silencing processes are widely conserved in eukaryotes. In animals, there are three major endogenous classes of short RNAs: microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs) and endogenous short interfering RNAs (endo-siRNAs) (Fig. 1). miRNAs are generated from longer primary precursor molecules that are sequentially processed by the RNase III enzymes Drosha and Dicer and are involved in a wide variety of biological processes (reviewed in [10]). miRNAs are mostly known to act at the post-transcriptional level and to negatively regulate their messenger RNA (mRNA) targets (reviewed in [11]). piRNAs are short RNAs associated with the PIWI subfamily of Argonaute proteins; they are mostly expressed in the germline and mainly act to repress parasitic elements, such as transposons (reviewed in [12, 13]). The endo-siRNAs are a recently discovered class of short RNAs; they can be antisense to protein-coding transcripts or can be derived from pseudogenes and intergenic regions. endo-siRNAs can regulate the expression level of protein-coding genes and transposons [14–22]. The mechanisms used by these three classes of short RNA to regulate their targets are still being investigated, although they are mostly known to act at the post-transcriptional level. However, a growing amount of experimental evidence is changing this view, and one of the most intriguing possibilities is that short RNAs and their co-factor Argonaute proteins also act in the nucleus and induce epigenetic chromatin changes. Previous studies in yeast (reviewed in [23]) and plants (reviewed in [24]) have already characterized the connection between the RNA silencing machinery and chromatin, opening the possibility that similar mechanisms exist in the animal kingdom.
Figure 1. Biogenesis of the three endogenous classes of short RNAs.
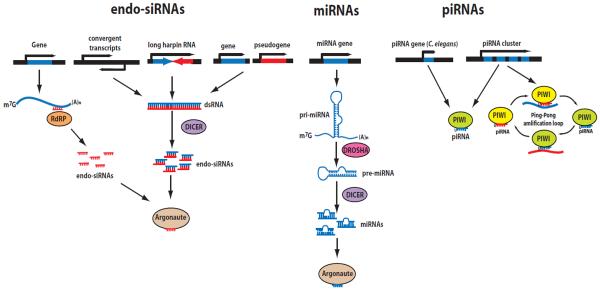
(left) endo-siRNAs can be produced by RNA-dependent RNA polymerases (RdRPs) using mature mRNAs as templates (in C. elegans), or generated by Dicer from long hairpin RNAs or from double-stranded RNAs that arise from convergent transcription and hybridization between spliced protein-coding transcripts and homologous pseudogenes. (center) miRNAs are transcribed from miRNA genes as long hairpin-structured RNA precursors, which are sequentially processed by RNase III enzymes Drosha and Dicer and loaded onto Argonaute proteins. (right) piRNAs are generated in a Dicer-independent manner from long piRNA precursors, which include multiple piRNAs, or from single piRNA transcriptional units (in C. elegans) and loaded onto PIWI proteins. They can be amplified by PIWI family Argonautes in a ping-pong amplification loop.
In this Review, we discuss the main findings that highlight the new roles of short RNAs in different nuclear processes, such as transcriptional gene silencing (TGS), induction of epigenetic chromatin modifications, and regulation of RNA Polymerase II (Pol II) transcription and splicing. We first describe studies in which exogenous RNAi has been used for the investigation of nuclear functions of Argonaute/siRNA complexes. Next, we discuss the endogenous nuclear functions of the three major short RNA classes: the role of miRNAs in inducing transcriptional gene silencing, the role of piRNAs in transcriptional silencing and heterochromatin formation, and possible new functions of endo-siRNA/Argonaute complexes in different nuclear and chromatin processes. Discussions of the nuclear function of specific short RNAs in different animal models, such as Caenorhabditis elegans, Drosophila and mouse, are presented separately in order to provide a better comparison of similar mechanisms in different species.
2. Exogenous short RNAs in TGS
2.1 siRNA-induced TGS in mammalian cells
The best known examples of siRNA-induced TGS come from yeast and plants, where it has been shown that siRNAs associated with Argonaute proteins can recruit chromatin modifying enzymes or DNA methyltransferases and contribute to the formation of heterochromatin and/or repression of transposable elements (reviewed in [23, 24]). The molecular mechanisms of siRNA-induced TGS in these systems are well characterized and have encouraged many researchers to find parallel RNAi mechanisms in metazoans. The experimental approach, which has been extensively used in mammalian cell culture to prove the existence of siRNA-induced TGS, is the use of exogenous siRNAs directed against specific regulatory DNA regions, such as promoters.
In general, either activation or repression can occur in response to siRNAs matching the promoter and repression occurs in response to siRNAs targeting the 3' end of a gene. Morris and colleagues first tested whether TGS can be induced in mammalian cells by using siRNAs complementary to the promoter region of a transgene [25]. An integrated transcriptional reporter with an endogenous promoter fused to a Green fluorescent protein (GFP) sequence was used, and after introducing siRNAs targeting promoter sequences, the authors observed a decrease in the fluorescence signal from the reporter gene [25]. The level of the reduction in the GFP signal was similar to the reduction obtained using siRNAs targeting the GFP coding sequence. Nuclear run-on assays demonstrated that silencing occurred at the transcriptional level and appeared to depend on DNA methylation [25], a known epigenetic modification associated with transcriptional silencing. A similar effect was also observed using siRNAs targeting the promoter of an endogenous gene [25]. In a different study transfection of siRNAs complementary to promoter regions also induced an increase in epigenetic chromatin marks associated with gene silencing, such as methylation of lysine 27 of histone H3 (H3K27me) and methylation of lysine 9 of histone H3 (H3K9me) [26]. However, these initial studies did not show a direct connection between the exo-siRNAs and the RNAi silencing machinery. Subsequent works have demonstrated the involvement of Argonaute proteins in this process [27, 28]. In particular, only two of the four human Argonaute proteins (AGO1 and AGO2) were shown to induce TGS [27, 28]. Moreover, Chromatin Immunoprecipitation (ChIP) experiments have shown that AGO proteins and Polycomb group complex components, which promote H3K27me, bind to the promoter of the silenced reporter or to the endogenous genes targeted by siRNAs [27, 28]. In another case, siRNAs targeting the promoter of the endogenous gene c-myc were shown to recruit AGO2 and to silence the expression of the gene without inducing H3K27me and H3K9me [29]. The silencing of c-myc was associated with a reduction in the formation of the pre-initiation complex (PIC). Therefore, the reports described above suggest that siRNAs targeting promoter sequences can induce TGS through multiple mechanisms. Surprisingly, two other studies have shown that siRNAs targeting different endogenous promoters were able to activate the corresponding genes after recruitment of AGO2 [30, 31]. The work by Schwartz et al. provided a plausible explanation of these contradictory results by showing that transfection of siRNAs targeting promoter regions leads to an activating or repressive effect on gene expression depending on the basal transcription level and on the factors bound to the promoter of the targeted gene [32]. Moreover, it was shown that Argonautes localize to the promoter regions through interaction with antisense transcripts, which extend to the promoter regions of the target genes [32]. The binding of the Argonaute/siRNA complex to the promoter antisense RNA leads to the displacement of factors already recruited to the promoter [32]. Therefore, depending on the factors that are already present at the promoter, siRNAs can induce different effects on transcription of the target gene. Finally, the use of siRNAs targeting the 3' end of the gene can also induce TGS [33].
Taken together, these studies suggest that some Argonaute proteins and their associated siRNAs may enter into the nucleus and interfere with the transcriptional apparatus and other protein complexes associated with promoters. Given the widespread antisense transcription in mammalian genomes [34, 35] and the peculiar feature of bi-directional Pol II transcription at the promoters of human genes (reviewed in [36]), it is reasonable to expect that endogenous Argonaute/siRNA complexes may be tethered to the antisense transcripts and regulate endogenous genes by mechanisms similar to the those described for siRNA-induced TGS or activation.
2.2 Exogenous RNAi in Caenorhabditis elegans
The first evidence of TGS in C. elegans was reported in worms containing a repetitive transgene carrying an intestinal GFP reporter, which became silenced if nematodes were subjected to RNAi induced by feeding bacteria with a plasmid expressing GFP dsRNA [37]. Surprisingly, the silencing of the GFP reporter occurred even if the plasmid used for RNAi contained only the backbone sequence identical to that of the transgene [37]. The authors demonstrated that the silencing of the somatic transgene occurred at the transcriptional level, since it was associated with a decrease in pre-mRNA levels and a reduction in Pol II occupancy and histone acetylation on the transgenic array [37]. A candidate RNAi screen identified the requirement of chromatin-associated proteins and known RNAi factors for this form of TGS [37]. However, the exact mechanism of dsRNA-induced TGS of repetitive transgenes in somatic tissues of C. elegans is not fully understood.
In order to identify specific nuclear RNAi factors, Kennedy and colleagues performed a forward genetic screen for mutants affecting dsRNA-induced silencing of a nuclear multicistronic RNA [38]. They have identified fifty-five nuclear RNAi defective (nrde) mutant alleles. Forty-six of these alleles matched to the nrde-3 gene, which encodes an Argonaute protein required for nuclear RNAi silencing [38]. The nuclear localization of NRDE-3 was necessary for silencing and required its interaction with siRNAs. Moreover, NRDE-3-associated exo-siRNAs targeted the NRDE-3/siRNA complex to the complementary nuclear pre-mRNA [38]. Therefore, the exo-siRNAs induced NRDE-3 translocation from the cytoplasm to the nucleus, where it silenced pre-mRNA targets in a sequence-specific manner [38]. Notably, NRDE-3 is naturally localized to the somatic nuclei and is bound by endo-siRNAs, which suggests an endogenous nuclear function [38]. Studies of the other nrde genes led to the genetic and molecular dissection of the NRDE pathway [39, 40]. NRDE-2 physically interacts with NRDE-3 and is recruited to the target nuclear pre-mRNA in an NRDE-3/exo-siRNA-dependent manner [40]. The recruitment of NRDE-2 to the target nuclear pre-mRNA induces H3K9me3 and a decrease in Pol II occupancy on the target locus [40]. Moreover, nuclear run-on experiments demonstrated that nuclear RNAi interferes with elongating Pol II on the target gene [40]. NRDE-1 functions downstream of NDRE-3/2 and localizes to the target nuclear pre-mRNA in a NRDE-3/2-exo-siRNA-dependent manner [39]. NRDE-1 also localizes to the chromatin and promotes RNAi-directed H3K9 methylation. NRDE-4 functions downstream of the NRDE-1/2/3 interaction with nuclear mRNA target and is not required for this process [39]. NRDE-4 binds to chromatin and mediates recruitment of NRDE-1 to the target locus [39]. The compelling molecular characterization of the NRDE pathway sheds light on the first step in RNAi-induced TGS. Moreover, it was recently demonstrated that the H3K9me3 signal induced by dsRNA at several loci persisted for two generations after the dsRNA induction [41]. The dsRNA-induced H3K9me3 signal was shown to spread up to 11 kb away from the dsRNA trigger region in an NRDE-2-dependent manner [41]. In addition, the germline-specific nuclear Argonaute HRDE-1/WAGO-9 has been implicated in a multigenerational germline TGS through the NRDE pathway and H3K9me [42] (see also section 4.3).
2.3 Transgene-induced TGS and heterochromatin silencing in Drosophila
Early studies in Drosophila have also suggested an interesting connection between TGS and the RNAi machinery [5, 6, 43]. Introduction of multiple copies of a transgene containing the promoter of the white gene fused to the Alcohol dehydrogenase (Adh) gene (w-Adh) led to the progressive copy number-dependent silencing of the transgene together with the silencing of the corresponding endogenous adh gene [5]. The silencing effect was partially dependent on the chromatin-binding repressive Polycomb Group (PcG) proteins, suggesting the involvement of a transcriptional silencing mechanism [5]. Indeed, the use of nuclear run-on assay demonstrated that the silencing occurred at the transcriptional level [6]. Interestingly, loss-of-function of the Piwi Argonaute protein impaired the transcriptional silencing effect [6]. The involvement of Piwi suggested a possible role for RNA silencing in other known TGS phenomena, such as position effect variegation (PEV) (reviewed in [44]). PEV occurs when a euchromatic gene is silenced after having been moved next to heterochromatin. Several factors associated with heterochromatin, including heterochromatin proteins 1 and 2 (HP1 and HP2) and a H3K9 methyltransferase, are required for PEV [44]. As suspected, Piwi and other proteins involved in RNA-mediated silencing, such as Aubergine and Spindle-E, were also implicated in PEV [43]. Moreover, using immunofluorescence staining of methylated H3K9 and HP1 on polytene chromosomes it was shown that inactivation of piwi, aubergine or spindle-E caused a reduction in H3K9 methylation level and a redistribution of HP1 protein [43]. It is notable that the RNAi components described in the above studies were later associated with the endogenous piRNA pathway and therefore suggest a possible link between piRNAs and TGS (see section 4.4). A further connection between the RNAi machinery and transgene-induced TGS has been revealed by the Cavalli group [45]. This study used transgenes containing two copies of the fab-7 boundary element, which contains an insulator and a core Polycomb Response Element (PRE), fused to a reporter gene. Upon integration into the Drosophila genome, the transgene was silenced by the PcG proteins [45]. Surprisingly, this silencing also required the activity of RNAi components, such as Dicer2, Piwi and Argonaute1 [45]. Mutations of these RNAi genes did not affect the recruitment of PcG proteins to the fab-7 boundary elements but compromised the maintenance of long-distance chromosomal interactions between transgenic and endogenous fab-7 [45]. Moreover, another study also showed that RNAi factors, and particularly AGO2, contribute to the interaction between the two transgenic mcp boundary elements [46]. The effect of RNAi factors on chromosomal pairing has also been observed for some other known endogenous PRE sites, which suggests a possible role for the RNAi pathway in mediating long-distance chromosomal interactions between PcG targets [45]. A compelling review on the topic of chromatin insulators is presented in this issue (Matzat, L.H. and Lei, E.P., Surviving an Identity Crisis: A Revised View of Chromatin Insulators in the Genomics Era).
3. miRNAs in TGS
miRNAs are a well characterized class of short RNA molecules involved in the regulation of mRNA targets at the post transcriptional level [11]. However, we cannot exclude the possibility that miRNAs also regulate their targets at the transcriptional level. Indeed, Kim et al. [47] described such a role for miR-320, which is encoded within the promoter region of its proximal gene POLR3D. Inhibition of this miRNA in tissue culture led to the upregulation of the POLR3D gene, and silencing of POLR3D by miR-320 likely occurred at the transcriptional level [47].
A more compelling study investigated the role of AGO2 and miRNAs in TGS during cellular senescence [48]. The authors compared genome-wide chromatin localization of Argonaute proteins in pre-senescent and senescent cells. The genomic regions bound by Argonaute proteins in senescent cells corresponded to the promoters of genes negatively regulated by the retinoblastoma complex during cellular senescence [48]. It was shown that during cellular senescence AGO2 was localized to the nucleus and its binding to some retinoblastoma-repressed promoters correlated with an increase in H3K9me and H3K27me levels and a reduction in methylation of lysine 4 on histone H3 (H3K4me) at these promoters [48]. Conversely, upon AGO2 knock-down a reduction in H3K27me levels and the reactivation of the target genes was observed [48]. Interestingly, the overexpression of AGO2 in pre-senescent cells induced a proliferative arrest with features of senescence [48]. Notably, deep sequencing of miRNAs associated with Argonaute during cellular senescence has revealed some candidate miRNAs that may act to recruit AGO2 to the promoters of the target genes [48]. One AGO2 target gene appears to be repressed by let-7 miRNA family targeting the promoter sequence, which opens the possibility that other miRNAs may regulate target genes at the transcriptional level at specific physiological conditions [48]. Lastly, a recent paper demonstrated that a human paralog of GW182 proteins, which play an important role in miRNA-mediated gene silencing, can translocate to the nucleus where it can interact with AGO2 and miRNAs [49]. Therefore, there is a growing experimental validation for the possibility of miRNA function in the nucleus.
4. The role of piRNAs in the nucleus
piRNAs are short RNAs physically associated with the PIWI subfamily of Argonaute proteins [13]. They range in size from 21nt to 33nt with a strong preference for 5' uridine [50]. piRNAs are produced from a discrete limited number of genomic clusters and their biogenesis is Dicer-independent [50]. Most of the PIWI proteins are involved in germline development [51] where they also act to repress transposable elements [50]. Here, we mainly discuss the nuclear role of piRNAs in transcriptional silencing and highlight the differences and similarities in the silencing mechanisms in different animal species (Fig. 2).
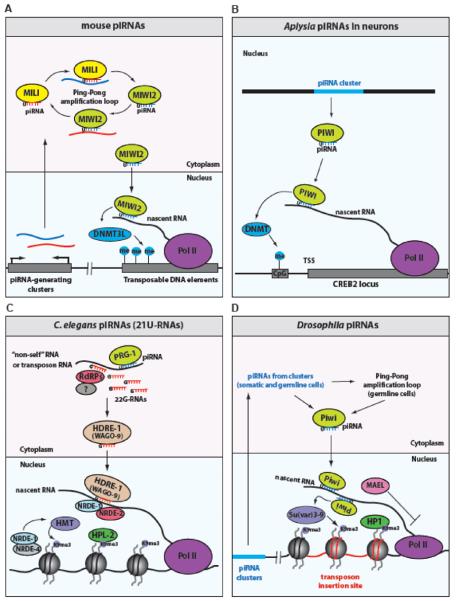
Figure 2. Models of piRNA-mediated transcriptional silencing in different species.
(A) Mouse pre-pachytene piRNAs derived from piRNA-generating clusters are amplified by MILI and MIWI2 proteins in a ping-pong amplification loop in the cytoplasm. MIWI2 translocates to the nucleus where it binds transposon target RNAs and induces de novo DNA methylation of transposable elements by DNA methyltransferase 3-like protein DNMT3L. (B) Aplysia piRNAs are loaded onto PIWI protein in neurons. PIWI is recruited to the transcript derived from the CREB2 locus in a piRNA-dependent manner and silences the expression of CREB2 through the methylation of the CpG island at the promoter, possibly through a DNA methyltransferase protein (DNMT). (C) piRNAs in C. elegans can bind transposon RNAs or “non-self” transgenic RNAs and initiate the production of endogenous antisense 22G-RNAs that are loaded onto HRDE-1 Argonaute protein. HRDE-1 translocates to the nucleus where it binds target RNAs and recruits the NRDE proteins to induce transcriptional silencing through H3K9 methylation mediated by H3K9 methyltransferases (HMT) and HPL-2 (HP1-homologue protein) binding to the target locus. (D) Drosophila piRNAs are produced from piRNA-generating clusters, which are enriched in transposon sequences, and are loaded onto Piwi protein to silence transposable elements in trans. Piwi translocates to the nucleus where it binds nascent RNA transcripts derived from active transposons and induces transcriptional silencing through H3K9 methylation, catalyzed by the H3K9 methyltransferase protein Su(var)3–9, and by direct or indirect recruitment of HP1 to the transposon insertion sites. The piRNA pathway component MAEL translocates to the nucleus and inhibits Pol II transcription independently (or downstream) of H3K9 methylation.
4.1 Mouse piRNAs and DNA methylation
There are three PIWI family proteins in mouse: MIWI, MILI and MIWI2. They are developmentally regulated during spermatogenesis and bind distinct piRNA populations at different stages of germ cell development [50]. MILI binds both pre-pachytene piRNAs and the meiotic pachytene piRNAs [52, 53]; MIWI2 is specifically loaded with pre-pachytene piRNAs [52]; MIWI is loaded with meiotic pachytene piRNAs [54, 55]. The pachytene piRNAs are largely depleted of transposon sequences and are produced from distinct intergenic genomic clusters [53, 54]. Their molecular functions and the biologically relevant targets have not been elucidated. However, a small fraction of pachytene piRNAs targeting repetitive elements can guide MIWI to silence transposons at the post-transcriptional level [56]. A piRNA-independent function for MIWI has also been proposed [57].
The majority of the pre-pachytene piRNAs derive from repetitive elements [52, 53], and MIWI2 and MILI function with pre-pachytene piRNAs to repress transposons during spermatogenesis [53, 54]. In mammals, silencing of transposons during spermatogenesis occurs via de novo DNA methylation generated by the DNA methyltransferase 3-like protein DNMT3L [58]. Deficiencies in either MIWI2 or MILI proteins correlate with a decrease in DNA methylation of transposable elements [53, 54, 59]. Moreover, miwi2 and mili mutants display a phenotype similar to dnmt3L mutant mice [53, 54, 59]. These lines of evidence led to the hypothesis that pre-pachytene piRNAs associated with MIWI2 and MILI serve as sequence-specific guides, which direct the DNA methylation machinery to the transposable elements [53, 59]. MIWI2 is localized to the nucleus during the time when DNA methylation is established and sequencing of piRNAs isolated from MILI complexes in the dnmt3L mutant mice has not revealed depletion of piRNAs, which suggests that both MIWI2 and MILI act upstream of DNMT3L to promote de novo DNA methylation of transposable elements [52]. Therefore, one of the roles of the piRNA pathway in mice is to silence transposons via de novo DNA methylation. Nonetheless, the molecular connection between the piRNAs and the DNA methylation machinery has not been elucidated yet.
4.2 Connection between neuronal piRNAs and DNA methylation in Aplysia
A recent work by Kandel, Tuschl and co-workers provides evidence that the Piwi protein and piRNAs are expressed in the central nervous system (CNS) and other somatic tissues of Aplysia [60]. Piwi is predominantly localized to the nuclei of Aplysia neurons, and deep sequencing of piRNAs showed their distinct genomic clusters [60]. Importantly, the authors identified abundant piRNAs upregulated in response to the exposure to the neurotransmitter serotonin (5HT), which is essential for learning and memory processes [60]. The knockdown (KD) of piwi revealed the upregulation of the transcription factor CREB2, which is involved in the regulation of synaptic plasticity [60]. The promoter of CREB2 becomes methylated at a CpG island upon exposure to 5HT. Strikingly, the 5HT-induced CpG methylation of the CREB2 promoter was impaired upon piwi KD, which suggested a role for Aplysia Piwi in directing DNA methylation similarly to the piRNAs in mouse [60]. The authors found several putative piRNA binding sites at the CREB2 promoter and demonstrated that inhibition of the piRNA corresponding to one of the sites by 2'-O-methyl oligoribonucleotides resulted in an upregulation of CREB2 transcripts [60]. These findings suggest a role for the Piwi/piRNA complex in transcriptional regulation of neuronal genes affecting long-term memory storage [60]. The function of the Piwi/piRNA complex in a tissue other than germline raises the possibility that a similar piRNA-dependent epigenetic mechanism regulates gene expression in neurons and contributes to learning and memory processes in other animals. Indeed, a recent report demonstrated the expression of the PIWI proteins Aubergine (Aub) and Argonaute3 (AGO3) in Drosophila brain [61]. Strikingly, the authors observed that the level of transposon expression anti-correlated with the expression of PIWI proteins and that the transposition events occurred more frequently in mashroom bodies where PIWI proteins are expressed at a lower level [61]. This suggests that transposons and piRNAs can contribute to the generation of genomic heterogeneity in the Drosophila brain.
4.3 C. elegans piRNAs and H3K9me
The C. elegans piRNAs, also known as 21U-RNAs, are 21nt short RNAs with a strong preference for 5' uridine [62–64]. They differ from piRNAs of other animals in that each of them has an independent identical promoter [62–64]. Thousands of different piRNAs exist in the worm, and while their sequences are not enriched for transposable elements, they are significantly depleted of protein-coding genes. Nonetheless, a single class of transposons showed elevated expression in the C. elegans PIWI mutant prg-1 [65, 66]. Since the sequences of piRNAs do not have obvious complementary targets, several groups utilized single-copy reporter transgenes with or without a complementary site to an endogenous piRNA to investigate their molecular functions [67–69]. Surprisingly, a single piRNA complementary site in the 3'UTR of a reporter transgene was sufficient for silencing. Importantly, the silencing was not dependent on the catalytic domain of PRG-1, and perfect complementarity between the target site and the piRNA was not required [67, 69]. It was discovered that PRG-1/piRNAs initiate the production of secondary siRNAs by RNA-dependent RNA polymerases. These siRNAs mediate post-transcriptional silencing and also induce transcriptional silencing through H3K9me [67, 68, 70]. Although the piRNA-induced transcriptional silencing persists for multiple generations, the PRG-1/piRNA complex is only required for the initiation but not for the maintenance of silencing. The factors involved in piRNA-induced TGS that were identified in candidate screens include: components of the NRDE pathway (NRDE-4/1/2); germline-specific nuclear Argonaute HRDE-1/WAGO-9; a putative histone H3 lysine-9 methyltransferase protein; a C. elegans homologue of the HP-1 protein; and also Polycomb and Trithorax complex-related factors [67, 68, 70, 71].
A similar piRNA-induced silencing effect was also discovered with single-copy transgenes that were not designed to be silenced by piRNAs [70]. This silencing occurs when a transgene contains a sequence that is not present in the C. elegans genome, such as the GFP sequence, and such foreign sequences appear to be fortuitously targeted by imperfectly complementary piRNAs. Consistently, computationally-predicted endogenous piRNA targets were found to be depleted of protein-coding genes expressed in the germline [67, 70]. Therefore, Mello and colleagues proposed a model where thousands of diverse piRNA sequences act to target and silence any “non-self” RNA in C. elegans germline [70]. The existence of such a genome surveillance system suggests a presence of a parallel mechanism that protects “self”, e.g. germline expressed transcripts, from being silenced [70]. It has been proposed that the CSR-1 endo-siRNA pathway could serve this role [70] (see section 5.1).
Collectively, the studies described above identified a surveillance mechanism where the C. elegans piRNAs initiate production of secondary endogenous siRNAs, which in turn associate with the nuclear NRDE pathway to induce TGS through the deposition of H3K9me. Once established, this TGS can be maintained by the endo-siRNA pathways through multiple generations in a PRG-1/piRNA-independent manner.
4.4 piRNA-mediated TGS in Drosophila germline
The Drosophila piRNAs constitute an RNA surveillance system that provides a defense against transposable elements through post-transcriptional and transcriptional silencing [12, 50] and also contributes to global chromatin organization in an indirect manner. There are discrete genomic loci in pericentromeric and telomeric heterochromatin that are enriched in transposon sequences and generate piRNAs [72–74]. piRNAs from those discrete clusters can suppress in trans potentially active transposable elements scattered throughout the genome [72–74]. Drosohila has three PIWI (P-element-induced wimpy testes) family proteins: Piwi, Aubergine (Aub) and Argonaute3 (AGO3). Aub and AGO3 are involved in the piRNA amplification cycle (ping-pong amplification loop) [50, 73], and work together to repress transposable elements through the piRNA-guided cleavage of transposon transcripts [75, 76]. Piwi is predominantly nuclear in germline cells [77] and, unlike AGO3 and Aub, it is also localized to the nuclei of somatic gonad cells, which surround germline cells [73]. The somatic cells surrounding the germline rely only on Piwi for silencing of transposable elements [76]. Nuclear localization of Piwi depends on the loading of primary piRNAs [78], and the catalytic domain, e.g. slicer activity, is not required for its silencing function [79]. The presence of the Piwi/piRNA complex in the nucleus together with the role of Piwi in transgene-induced TGS (discussed in section 2.3) suggested a possibility of piRNA-initiated TGS in transposon control. The first study that evaluated this possibility showed the effect of spindle-E (spn-E) mutation, which reduces piRNA accumulation, on chromatin at different transposable elements [80]. ChIP experiments performed in the spn-E mutant flies revealed a reduction in H3K9me and HP1 binding and an increase in H3K4me at the transposon loci in the mutant compared to wild type [80]. The connection between the PIWI/piRNA complex and heterochromatin silencing was reinforced by in vitro binding and co-immunoprecipitation experiments, which showed a direct interaction between the N-terminal domain of Piwi and the HP1a protein [81]. Staining of polytene chromosomes revealed that HP1 and Piwi partially overlapped in their localization on chromosomes that this co-localization was RNA-dependent [81]. These results suggested that piRNAs may silence transposons through the recruitment of HP1 to the target loci.
However, a different study showed a curious gene-activating role of piRNAs [82]. The Piwi protein was shown to localize to the telomere-associated sequence (TAS), which produces some piRNAs (3R-TAS), and the ChIP assay in piwi mutant detected an increase in HP1 and H3K9me levels and a decrease in H3K4me on the 3R-TAS sequence [82]. Consistently, the expression of a reporter transgene integrated next to the 3R-TAS site was reduced in the piwi mutant [82]. These surprising results can be explained by a competition between the HP1 and Piwi proteins for binding at the 3R-TAS chromatin site. Moreover, ChIP analysis of the two other piRNA-generating clusters determined that the association of HP1 to the clusters was not dependent on the Piwi protein, although Piwi was also localized to these clusters [83]. In addition, deletion of one cluster caused the redistribution of the HP1 protein on polytene chromosomes [84], which suggests an indirect relationship between Piwi and HP1. However, another study performed ChIP analysis on different transposon elements in the germline and showed a significant decrease in HP1 binding and H3K9me upon germline-specific piwi knockdown (KD) [85]. Also, nuclear run-on assay revealed a different degree of transcriptional activation of telomeric transposable elements in the spn-E mutant or upon piwi KD, which demonstrates a clear role for Piwi in suppression of some transposable elements at the transcriptional level [86].
The apparent contradictory results described above can be explained by the fact that different research groups analyze different genomic loci that include either the piRNA-producing clusters or transposable elements that are subject to piRNA regulation in trans. Moreover, the existence of multiple direct mechanisms of regulation and significant indirect effects can potentially explain different results. Finally, a decrease or increase in some chromatin marks in different mutants does not always correlate with changes in transcriptional activity at the piRNA target sites (see [87] and discussion below).
A recent availability of genome-wide approaches to investigate chromatin and transcriptional changes has allowed a more complete and systematic dissection of the role of Piwi in the nucleus. To analyze the consequences of piwi KD on transcription, the Hannon group [88] and the Brennecke group [87] have utilized Global Run-On sequencing (GRO-seq) [89], which allows the precise mapping of the position, amount, and orientation of transcriptionally engaged Pol II molecules in a genome-wide fashion. Rozhkov et al. [88] used a cell-type specific piwi KD to study the function of Piwi proteins in the germline and in the somatic compartment. These authors also performed total RNA sequencing (RNA-seq) to distinguish between transcriptional and post-transcriptional regulation and ChIP assay combined with high-throughput sequencing (ChIP-seq) to detect changes in H3K9me genome-wide. The GRO-seq analysis upon somatic piwi KD identified many upregulated somatic transposable elements and the comparison between the GRO-seq and RNA-seq data showed that Piwi regulates somatic transposons mainly at the transcriptional level [88]. Deep sequencing of piRNAs demonstrated that the majority of somatic piRNAs are antisense to transposable elements and are reduced upon somatic piwi KD [88]. Therefore, these results are consistent with the model where somatic cells repress transposons mainly by the activity of the Piwi protein. In the germline, piwi KD revealed only a modest increase in transcription of transposable elements and showed a greater increase in their steady-state RNA levels, which indicates that both transcriptional and post-transcriptional mechanisms are active in suppressing transposons in germline cells [88]. Deep sequencing of piRNAs from the piwi mutant germlines showed that germline Piwi contributes to the production of both primary and secondary piRNAs, and the latter ones are loaded onto AGO3 and Aub to silence transposons at the post-transcriptional level [88]. The authors also conducted H3K9me3 ChIP-seq experiments using the germline tissue of piwi KD. Consistent with the derepression of transposons, H3K9me3 was decreased over many transposable elements upon piwi KD [88]. Another study by Le Thomas et al. [90] arrived at the same conclusions as Rozhkov et al. [88] by using Pol II ChIP-seq. Transposons showed an increased Pol II occupancy and decreased H3K9me3 upon germline piwi KD, which is consistent with the role of germline Piwi in silencing transposons at the transcriptional level [90]. Curiously, Piwi immunoprecipitation and mass-spectrometry analysis of putative interacting partners mostly identified RNA-binding proteins and just a few chromatin factors [90], which did not include the previously identified HP1 protein [81].
Brennecke and co-workers investigated the role of Piwi in TGS using an established stable cell line derived from Drosophila ovarian somatic cells (OSCs) [87]. The use of a homogeneous cell line allowed the authors to perform several genomic analyses, including GRO-seq, RNA-seq, Pol II ChIP-seq and H3K9me3 ChIP-seq, and to obtain data lacking the background signals that come from other cell types, such as germline cells. In addition to piwi KD they also analyzed the effect of maelstrom (mael) KD; Maelstrom is a protein involved in silencing of transposons in follicle cells, ovaries and OSC cells [87]. It is localized to the nucleus and does not contribute to the piRNA biogenesis in OSC cells, yet its nuclear localization is required for transposon silencing [87]. The RNA-seq profiles of piwi and mael KD showed the upregulation of many families of transposable elements. The upregulation of transposons at the mRNA level correlated with an increase in Pol II occupancy (by Pol II ChIP-seq) and an increase in nascent RNA levels (by GRO-seq) in the piwi and mael KD cells [87]. Interestingly, not all transposons were de-repressed upon piwi or mael KD, but only the ones that were targeted by numerous piRNAs. This argues against a possible general increase in heterochromatin transcription upon piwi and mael KD due to indirect effects. In addition, transcription of piRNA-producing clusters, such as flamenco, was not significantly affected by the piwi or mael KD, which suggests that Piwi/piRNAs induce TGS in trans [87]. The genome-wide analysis of H3K9me3 in OSCs showed only a modest decrease in H3K9me3 at the upregulated transposons in piwi KD and even a smaller decrease in H3K9me3 upon mael KD [87]. The small magnitude of the effects could be due to the fact that the authors considered the average signal of H3K9me3 over all the transposons, irrespective of their genomic positions or transcriptional state [87]. To address this issue they quantified the H3K9me3 signal considering only unique genomic sequences surrounding each transposon insertion of the piwi KD-upregulated transposon families [87]. When quantified in this manner, the H3K9me3 signal was significantly decreased downstream of individual euchromatic transposon insertion sites upon piwi KD. Surprisingly, mael KD did not show a decrease in H3K9me3, instead, a spreading of the H3K9me3 signal downstream of transposon insertions was observed [87]. A spreading of Pol II transcription was also observed downstream of transposon insertions in both piwi KD and mael KD, which indicates that H3K9me3 does not always correlate with the transcriptional status of the locus [87]. Importantly, the ability of piRNAs to induce TGS of transposons can influence the transcriptional state of euchromatic regions surrounding transposon insertions, and therefore may have an impact on the expression of protein-coding genes in close proximity to the insertion site [87].
There is a striking coincidence between the genome-wide profile of Piwi localization to the chromatin, which was identified in a recent ChIP-seq study, and the location of piRNAs, which indicates that piRNAs can guide Piwi to the target genomic sites [91]. Moreover, the derepression of transposons in the piwi mutant leads to a redistribution of Pol II from protein-coding genes to transposable elements, which is likely to have a global effect on cellular transcription [91]. A closer look at a heterochromatic centromeric region in the piwi mutant revealed an increase in H3K27me and in Pol II occupancy. Conversely, the euchromatic chromosomal arm 2L (Chr2L) showed a decrease in the H3K27me level and Pol II occupancy. The abundances of the HP1 protein and H3K9me were also decreased at the euchromatic Chr2L region in the piwi mutant, and a decrease in HP1 was shown for the heterochromatic centromeric region [91]. The euchromatic Chr2L region in the piwi mutant was also characterized by spreading of H3K4me and a decrease in H3K9me around the transcription start sites (TSS) of protein-coding genes, possibly as a consequence of activation of transposons next to the genes [91]. Importantly, the authors used a reporter with an ectopic site complementary to a piRNA to demonstrate the ability of Piwi to localize to the ectopic piRNA site and to recruit HP1 and the H3K9 methyltransferase Su(var)3–9 for inducing transcriptional repression [91]. Collectively, the genome-wide studies described above have proven the fundamental role of Piwi and piRNAs in transcriptional silencing at transposons and their profound indirect impact on transcription of protein-coding genes.
5. endo-siRNAs and Argonaute proteins in nuclear processes
5.1 endo-siRNAs in C. elegans
endo-siRNAs antisense to protein-coding genes and transposons were first discovered in C. elegans by Ambros and colleagues [20, 21]. Subsequent deep sequencing uncovered two classes of endo-siRNAs: 22G-RNAs and 26G-RNAs [62, 92–99]. They have a size of 22nt and 26nt, respectively, and both have a strong preference for 5' guanine [62, 92–99]. Many endo-siRNAs map to exon-exon junctions [62, 99], which is consistent with their synthesis by RNA-dependent RNA polymerases (RdRPs) using mature mRNAs as templates [99–101]. There are two major endo-siRNA pathways in C. elegans, which are defined by the Argonaute proteins to which they bind: the worm-specific AGO (WAGO) pathway [92] and the CSR-1 pathway [93]. The WAGO pathway silences transposable elements, pseudogenes, and cryptic loci providing genome surveillance [92]. Some WAGO Argonautes, such as NRDE-3 and HRDE-1, act in the nucleus to induce TGS, while other WAGOs mediate post-transcriptional silencing [42, 67, 69]. Importantly, mutations in the hrde-1 gene cause progressive sterility in generations, although the underlying cause of this phenotype is not known [42]. The WAGO pathway has also been connected to the piRNA pathway (discussed in section 4.3). Interestingly, a recent study showed that NRDE-3 acts in olfactory neurons to promote odor adaptation by repressing odr-1, a gene required for adaptation [102]. These results suggest an endogenous role for the NRDE pathway in inducing TGS in neurons.
The 22G-RNAs that belong to the CSR-1 pathway are mainly antisense to germline-enriched protein-coding transcripts [93]. Notably, the Argonaute protein CSR-1 binds to the chromatin at the target genes in an endo-siRNA-dependent manner and is required for proper chromosome segregation during mitosis [93]. However, the exact molecular function of CSR-1 in the nucleus is not clear. A recent study by our group revealed a new role for CSR-1 in positively regulating histone mRNA biogenesis [103]. Depletion of CSR-1 and other CSR-1 pathway components led to a severe reduction in histone proteins accompanied by the accumulation of unprocessed histone pre-mRNAs [103]. Moreover, this reduction in histone proteins significantly contributed to the lethal and sterile phenotypes observed upon CSR-1 depletion by RNAi [103]. It is notable that many target genes are downregulated in the csr-1 mutant [93, 103]. Therefore, it is possible that the CSR-1 pathway has a global impact on transcription of its target genes.
5.1 endo-siRNAs in mouse
endo-siRNAs have also been discovered in mouse oocytes and embryonic stem cells [18, 19, 22]. These endo-siRNAs are generated by Dicer from double-stranded RNAs that arise from convergent transcription, hybridization between spliced protein-coding transcripts and homologous pseudogenes, and long hairpin RNAs [18, 19, 22]. It appears that mammalian endo-siRNAs can repress complementary mRNAs, although it is not known if they regulate transcription [18, 19, 22]. endo-siRNAs are also produced from the transcripts of transposable elements and contribute to transposon silencing in mouse oocytes [18, 19, 22]. However, nuclear roles for this class of mouse endo-siRNAs have not been described either.
5.1 endo-siRNAs in Drosophila
Drosophila endo-siRNAs have been cloned from germline and somatic tissues, they are produced by Dicer, which uses dsRNA substrates originating from repetitive transposons, long hairpin RNAs, or overlapping mRNAs predicted to form dsRNAs [14–17, 104]. endo-siRNAs are predominately loaded onto Argonaute2 (AGO2); they contribute to transposon silencing in somatic tissues and also regulate expression of the complementary protein-coding genes, yet the mechanism of this regulation is unknown [14–17, 104, 105]. Intriguingly, based on the phenotypic analysis of the ago2 mutant, it has been proposed that AGO2 is required for the assembly of the pericentromeric heterochromatin, similar to the S. pombe homologue Argonaute [106]. In addition, another study suggested a contribution of AGO2, Dicer2 (DCR2) and endo-siRNAs to heterochromatin formation, which relies on the H3K9 methyltransferase Su(var)3–9 and HP1 [107]. Su(var)3–9 and DCR2 have also been implicated in regulation of nuclear organization and in the control of stability of the ribosomal DNA (rDNA) locus and satellite DNA loci [108]. It was shown that the level of H3K9me2 over the rDNA locus and other DNA repeats was reduced in the Su(var)3–9 and dcr-2 mutants [108]. An increase in the extrachromosomal circular (ecc) repeat DNA was noted in these mutants at the same time, which suggests a possible function for the RNAi pathway and H3K9me2 in regulating the stability and integrity of DNA repeats [108]. Also, another study in mammalian cells showed the association of Dicer with rDNA chromatin, which may indicate a conserved function of RNAi in regulating the stability of rDNA locus in different organisms [109]. However, endo-siRNAs corresponding to genomic DNA repeats have not been detected and an indirect role for the endo-siRNA pathway in this process cannot be excluded.
6. Nuclear Argonaute and transcriptional regulation
The discovery of endo-siRNAs antisense to protein-coding genes and detection of Argonaute proteins in the nucleus suggested a possibility for the Argonaute/endo-siRNA complex in euchromatin regulation. Cernilogar et al. [110] analyzed cellular localization of the key RNAi factors in Drosophila and found that components of the miRNA pathway, Dicer1 (DCR1) and Argonaute1 (AGO1), were equally enriched in the cytoplasm and in the nucleus, whereas DCR2 was greatly enriched in the nucleus. Furthermore, AGO2 and DCR2 were present in the same chromatin fraction as Pol II [110]. Strikingly, staining of polytene chromosomes with AGO2 and DCR2 antibodies showed their co-localization at the transcriptionally active euchromatic regions, including the heat-shock genes [110]. Using the heat-shock genes as a model, the authors showed that AGO2 and DCR2 were localized to heat-shock gene promoters and contributed to the repression of these genes in non-heat-shock conditions, when the genes were not induced [110]. It was shown that AGO2 and DCR2 physically interact with the negative elongation factor (NELF) to maintain Pol II in a paused in absence of heat stress [110]. AGO2 is associated with endo-siRNAs antisense to protein-coding genes and promoters, which suggests a possible role for these endo-siRNAs in guiding AGO2 to the target euchromatic loci [110].
In an earlier study investigating possible nuclear roles of AGO2 at chromatin, Moshkovich et al. [83] performed a genome-wide AGO2 ChIP-seq in S2 and S3 Drosophila embryonic cell lines. In agreement with the AGO2 staining on polytene chromosomes [110], AGO2 was found to be associated with euchromatic sites [83]. Most of the AGO2 binding sites corresponded to known chromatin insulator regions and promoters of active genes and they extensively overlapped with the binding sites of the known insulator proteins CTCF and CP190 [83]. In an earlier study by Lei and Corces [111], a link between the RNAi and the chromatin insulators emerged from an unbiased proteomic approach. A helicase required for the dsRNA-induced gene silencing, Rm62, was found to interact with CP190 in an RNA-dependent manner and to contribute to its insulator function [111]. Interestingly, AGO2 physically interacts with CTCF and CP190, and AGO2 chromatin localization depends on CTCF/CP190 [83]. Using the Adb-B locus as a model to study insulator function, the authors showed the requirement of AGO2 for CTCF/CP190-dependent looping interactions between insulators, promoters and enhancers at the Adb-B locus [83]. AGO2 was also found to be associated with the PRE elements and to co-localize on chromatin with Trithorax group and Polycomb group proteins [83]. Surprisingly, the localization of AGO2 does not correspond to the sites that generate endo-siRNAs, and neither catalytic activity of AGO2 nor DCR2 were required for the insulator function, suggesting an RNAi-independent role of AGO2 in this process [83]. A more recent work by Taliaferro et al. also used AGO2 ChIP-seq to study the function of this protein in transcription and splicing [112]. In agreement with the ChIP-seq data of Moshkovich et al. [83], AGO2 was found to localize to the promoters of euchromatic genes [112]. The authors also performed RNA-seq to measure the changes in mRNA levels in the ago2 mutants compared to wild type and found a modest but significant enrichment of AGO2 ChIP targets among the genes upregulated in ago2 mutant [112]. This result indicates that AGO2 may have a direct negative effect on transcription of some target genes. This effect of AGO2 on transcription is independent of its catalytic domain [112]. A more sensitive approach, such as GRO-seq, can be used in the future to evaluate the global role of AGO2 in transcriptional regulation. Taliaferro et al. also uncovered a possible function of AGO2 in splicing [112]. They used exonic microarray, which covers splice junctions, and RNA-seq to show splicing defects in the ago2 mutant. However, other RNAi factors, including DCR2, DCR1 and AGO1, did not show any splicing defects, which suggests an RNAi-independent function of AGO2 in splicing [112]. Interestingly, the gene set that shows splicing defects in the ago2 mutant is not enriched in AGO2 ChIP-seq targets. Instead, AGO2 was found to bind the mRNAs of these genes, as revealed by AGO2 crosslinking and immunoprecipitation sequencing (CLIP-seq) [112]. Therefore, it appears that Argonaute proteins can regulate multiple nuclear processes by different mechanisms including an RNAi-independent manner.
7. RNAi and alternative splicing
To investigate a possible role of RNAi in alternative splicing, Kornblihtt and colleagues [113] used a well-established system of RNAi-induced TGS in human cell lines (see section 2.1). Instead of using siRNAs against the promoter or the 3'UTR regions, they transfected siRNAs specific to the intronic sequences adjacent to the well-characterized alternative spliced exon (EDI) in the fibronectin gene [113]. AGO1 and AGO2 promoted the inclusion of the EDI exon and the increase in H3K9me at the EDI region upon transfection with intronic siRNAs [113]. RNAi against HP1 by siRNAs introduced together with the intronic siRNAs abolished the inclusion of EDI, indicating the involvement of chromatin mechanisms in this process [113]. Notably, the fibronectin mRNA or pre-mRNA levels were not reduced upon siRNA transfection, which excludes a possible TGS effect. In another similar study it was shown that the transfection of siRNAs targeting intronic sequences caused skipping of the nearby exon, in an AGO2-dependent manner, without any changes in the heterochromatin mark H3K9me [114]. The two opposite effects of siRNAs on alternative splicing demonstrated by these studies might be explained by the involvement of multiple RNAi mechanisms. Interestingly, AGO1 and Dicer knockdowns in mammalian cells affected many cancer-related alternative-splicing events, which suggests an endogenous role for the RNAi pathway in regulating alternative splicing [113].
Another recent work also implicated Argonaute proteins in alternative splicing [115]. To identify putative chromatin interactors of AGO1 and AGO2 the authors used chromatin fraction extracts from human cells for performing immunoprecipitations of these proteins followed by mass-spectrometry analysis [115]. The two Argonaute proteins were found to interact with chromatin-binding and splicing factors, including the HP1 protein, a H3K9 methyltransferase, spliceosomal small nuclear RNP (snRNP) components, and other regulators of alternative splicing [115]. The interaction of AGO1 and AGO2 with chromatin and splicing factors did not depend on RNA, and therefore it may be mediated by protein-protein binding. Sequencing of the siRNAs associated with chromatin-bound Argonaute proteins revealed many endo-siRNAs in the same sense orientation as the corresponding protein-coding RNAs [115]. Notably, some of these endo-siRNAs were enriched at the 3' ends of intronic sequences providing a possible mechanistic link between RNAi pathway and splicing. Next, the authors studied the effect of Argonaute depletion on a well-characterized alternative splicing event at the CD44 gene. They showed that AGO1 and AGO2 promoted the inclusion of the CD44 variant exons [115]. Moreover, AGO1 and AGO2 were detected on the chromatin regions corresponding to the variant exons; they also interacted with the CD44 pre-mRNA in a Dicer-dependent manner [115]. Since HP1 and a H3K9 methyltransferase were found in the AGO1 and AGO2 immunoprecipitates, the authors investigated the possible connection between Argonaute and H3K9me3 in regulation of alternative splicing. AGO1 and AGO2 were shown to promote H3K9me3 at CD44 variant exons, which contributed to slowdown of Pol II and exon inclusion [115]. These results highlight a functional connection between RNAi and H3K9me3 beyond the well-characterized RNAi-induced heterochromatin formation. Finally, the use of genome-wide exon arrays revealed the involvement of Argonaute and Dicer in many alternative splicing events [115], which indicates a more general role of RNAi in alternative splicing. It is notable that most of the nuclear endo-siRNAs associated with Argonaute proteins map to protein-coding genes in the sense orientation [115]. Since the interaction of Argonautes with chromatin is Dicer-dependent [115], the recruitment of Argonaute proteins, if mediated by the RNA-RNA interactions, might occur through the base-pairing with the antisense transcripts that are highly prevalent in mammalian genomes [34, 35].
8. Concluding remarks
In this review, we highlighted the growing number of studies that support new roles of RNA silencing in nuclear processes during animal development. All three major classes of endogenous short RNAs have been shown to be involved in nuclear processes (see Table 1). One of the best-characterized examples of nuclear RNAi function is the control of transposable elements in animal germline (Fig. 2). In different organisms, including Drosophila, C. elegans, and mouse, piRNAs induce sequence-specific transcriptional silencing associated with epigenetic modifications of transposable DNA elements. In mouse, the silencing occurs through the induction of de novo DNA methylation of transposon sequences, whereas in Drosophila and C. elegans, which do not have DNA methylation, an increase in H3K9 methylation is observed. Studies in C. elegans have shown that RNA-induced silencing can persist for multiple generations, which opens the possibility for a more general role of RNA in epigenetic inheritance. Moreover, the role of endo-siRNAs in odor adaptation in olfactory neurons of C. elegans, the neuronal function of Aplysia piRNAs, and the expression of Piwi in Drosophila brain suggest a conserved role for short RNAs animal nervous system.
Table 1.
Endogenous classes of short RNAs associated with Argonaute proteins and their nuclear effects.
| RNA type | Argonaute | Nuclear Targets | Effects | Physiological condition | References | |
|---|---|---|---|---|---|---|
| C. elegans | exo-siRNAs | NDRE-3 | multicistronic RNA | increase in H3K9me3; inhibition of Pol II elongation | soma | [38–41] |
| endo-siRNAs | HRDE-1 (WAGO-9) | transposons; pseudogenes; protein-coding genes; | increase in H3K9me3; decrease in pre-mRNA | germline | [42, 67,69] | |
| endo-siRNAs | CSR-1 | germline genes | associate with chromatin; histone mRNA biogenesis | germline | [93,103] | |
| piRNAs | PRG-1 | “non-self” RNA; transposons | HRDE-1-mediated TGS | germline | [67–71] | |
| D. melanogaster | exo-siRNAs | Piwi; AUB; AGO3 | PEV; transgenes containing PRE | reduction in H3K9me; redistribution of HP1; long-range chromosomal interactions between PcG targets | polytene chromosomes | [5, 6, 43–46] |
| endo-siRNAs | AGO2 | transposons; euchromatic genes; heat-shock genes | regulation of Pol II transcription; three-dimensional genome structure; alternative splicing | germline and somatic cells; polytene chromosomes | [14–17, 105-–107, 110–112] | |
| piRNAs | Piwi | transposons | increase in H3K9me3; inhibition of Pol II transcription; recruitment of HP1 to transposon integration sites | germline cells; somatic gonad cells; OSC cells | [87, 88, 90, 91] | |
| M. musculus | piRNAs | MIWI2 | transposons | CpG methylation; inhibition of transcription | testis | [52–57,59] |
| Aplysia | piRNAs | Piwi | CREB2 | de novo DNA methylation through DNMT3L | neurons | [60] |
| H. sapients | miRNAs | AGO2 | POLR3D; protein-coding genes negatively regulated during cellular senescence | increase in H3K9me and H3K27me; decrease in H3K4me | cell lines | [47,48] |
| exo-siRNAs | AGO1;AGO2 | promoter; UTR; fibronectin intron | TGS; gene activation; alternative splicing of exon (EDI) in the fibronectin gene | cell lines | [25–33, 113, 114] | |
| endo-siRNAs | AGO1;AGO2 | CD44 | alternative splicing; increase in H3K9me; slowdown of Pol II | cell lines | [115] |
The development of genome-wide methods to study transcriptional changes and chromatin modifications has greatly contributed to the better understanding of nuclear processes mediated by RNAi. It was found that Argonaute proteins and siRNAs can regulate nuclear processes that go far beyond the classical RNAi-mediated heterochromatin formation. RNAi-dependent and RNAi-independent functions of nuclear Argonaute proteins have been discovered, which include regulation of Pol II transcription, alternative splicing, and three-dimensional genome structure. The use of different animal models combined with the utilization of the latest genomic, genetic and biochemical approaches will be particularly important for the future characterization of RNAi-mediated nuclear functions in animal development.
Highlights
-
-
piRNAs initiate transcriptional silencing of transposable elements
-
-
piRNAs silence transposons through DNA methylation or H3K9 methylation
-
-
Nuclear Argonaute proteins can associate with euchromatic loci
-
-
Argonaute proteins regulate Pol II transcription, splicing, and genome architecture
Acknowledgments
We thank the members of the Grishok lab for helpful discussions and Dylan Rahe and Lyuda Kovalchuke for editing the manuscript. Our research is supported by NIH Director's New Innovator Award (1 DP2 OD006412-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- [2].Cogoni C, Macino G. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:10233–10238. doi: 10.1073/pnas.94.19.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cogoni C, Romano N, Macino G. Suppression of gene expression by homologous transgenes. Antonie van Leeuwenhoek. 1994;65:205–209. doi: 10.1007/BF00871948. [DOI] [PubMed] [Google Scholar]
- [4].Napoli C, Lemieux C, Jorgensen R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. The Plant cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pal-Bhadra M, Bhadra U, Birchler JA. Cosuppression in Drosophila: gene silencing of Alcohol dehydrogenase by white-Adh transgenes is Polycomb dependent. Cell. 1997;90:479–490. doi: 10.1016/s0092-8674(00)80508-5. [DOI] [PubMed] [Google Scholar]
- [6].Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Molecular cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- [7].van der Krol AR, Mur LA, Beld M, Mol JN, Stuitje AR. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. The Plant cell. 1990;2:291–299. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. Journal of molecular biology. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- [9].Hammond SM. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS letters. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- [10].He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews. Genetics. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- [11].Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature reviews. Genetics. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- [12].Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes & development. 2012;26:2361–2373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & development. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- [14].Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, Hannon GJ, Brennecke J. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- [17].Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- [19].Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Current biology : CB. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- [21].Lee RC, Hammell CM, Ambros V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. Rna. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes & development. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Grewal SI, Jia S. Heterochromatin revisited. Nature reviews. Genetics. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- [24].Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Current opinion in cell biology. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- [25].Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- [26].Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen ZX, Riggs AD, Rossi JJ, Morris KV. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. Rna. 2006;12:256–262. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nature structural & molecular biology. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- [28].Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nature structural & molecular biology. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- [29].Napoli S, Pastori C, Magistri M, Carbone GM, Catapano CV. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. The EMBO journal. 2009;28:1708–1719. doi: 10.1038/emboj.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hu J, Chen Z, Xia D, Wu J, Xu H, Ye ZQ. Promoter-associated small double-stranded RNA interacts with heterogeneous nuclear ribonucleoprotein A2/B1 to induce transcriptional activation. The Biochemical journal. 2012;447:407–416. doi: 10.1042/BJ20120256. [DOI] [PubMed] [Google Scholar]
- [31].Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, Janowski BA. Antisense transcripts are targets for activating small RNAs. Nature structural & molecular biology. 2008;15:842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yue X, Schwartz JC, Chu Y, Younger ST, Gagnon KT, Elbashir S, Janowski BA, Corey DR. Transcriptional regulation by small RNAs at sequences downstream from 3' gene termini. Nature chemical biology. 2010;6:621–629. doi: 10.1038/nchembio.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y, F. Consortium. R.G.E.R. Group. G. Genome Science The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- [35].Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C, R.G.E.R. Group. G. Genome Science. F. Consortium Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- [36].Seila AC, Core LJ, Lis JT, Sharp PA. Divergent transcription: a new feature of active promoters. Cell cycle. 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- [37].Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes & development. 2005;19:683–696. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Burkhart KB, Guang S, Buckley BA, Wong L, Bochner AF, Kennedy S. A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS genetics. 2011;7:e1002249. doi: 10.1371/journal.pgen.1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nature genetics. 2012;44:157–164. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- [44].Reuter G, Spierer P. Position effect variegation and chromatin proteins. BioEssays : news and reviews in molecular, cellular and developmental biology. 1992;14:605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- [45].Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- [46].Li HB, Ohno K, Gui H, Pirrotta V. Insulators target active genes to transcription factories and polycomb-repressed genes to polycomb bodies. PLoS genetics. 2013;9:e1003436. doi: 10.1371/journal.pgen.1003436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kim DH, Saetrom P, Snove O, Jr., Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nature cell biology. 2012;14:266–275. doi: 10.1038/ncb2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nishi K, Nishi A, Nagasawa T, Ui-Tei K. Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. Rna. 2013;19:17–35. doi: 10.1261/rna.034769.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- [51].Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & development. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Molecular cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- [54].Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- [55].Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480:264–267. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- [57].Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, Mourelatos Z. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nature structural & molecular biology. 2012;19:773–781. doi: 10.1038/nsmb.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- [59].Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & development. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Perrat PN, DasGupta S, Wang J, Theurkauf W, Weng Z, Rosbash M, Waddell S. Transposition-driven genomic heterogeneity in the Drosophila brain. Science. 2013;340:91–95. doi: 10.1126/science.1231965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- [63].Cecere G, Zheng GX, Mansisidor AR, Klymko KE, Grishok A. Promoters recognized by forkhead proteins exist for individual 21U-RNAs. Molecular cell. 2012;47:734–745. doi: 10.1016/j.molcel.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gu W, Lee HC, Chaves D, Youngman EM, Pazour GJ, Conte D, Jr., Mello CC. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell. 2012;151:1488–1500. doi: 10.1016/j.cell.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, Conte D, Jr., Luo S, Schroth GP, Carrington JC, Bartel DP, Mello CC. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Molecular cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, Matthews N, Berezikov E, Ketting RF, Tavare S, Miska EA. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Molecular cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Luteijn MJ, van Bergeijk P, Kaaij LJ, Almeida MV, Roovers EF, Berezikov E, Ketting RF. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. The EMBO journal. 2012;31:3422–3430. doi: 10.1038/emboj.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lee HC, Gu W, Shirayama M, Youngman E, Conte D, Jr., Mello CC. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr., Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, Pintacuda G, Sakaguchi A, Sarkies P, Ahmed S, Miska EA. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Current biology : CB. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- [73].Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- [74].Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- [75].Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- [76].Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, Kittler EL, Zapp ML, Klattenhoff C, Schulz N, Theurkauf WE, Weng Z, Zamore PD. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- [78].Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes & development. 2010;24:2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Darricarrere N, Liu N, Watanabe T, Lin H. Function of Piwi, a nuclear Piwi/Argonaute protein, is independent of its slicer activity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1297–1302. doi: 10.1073/pnas.1213283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic acids research. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes & development. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- [83].Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes & development. 2011;25:1686–1701. doi: 10.1101/gad.16651211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Moshkovich N, Lei EP. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS genetics. 2010;6:e1000880. doi: 10.1371/journal.pgen.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Wang SH, Elgin SC. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:21164–21169. doi: 10.1073/pnas.1107892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Shpiz S, Olovnikov I, Sergeeva A, Lavrov S, Abramov Y, Savitsky M, Kalmykova A. Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons. Nucleic acids research. 2011;39:8703–8711. doi: 10.1093/nar/gkr552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Sienski G, Donertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Rozhkov NV, Hammell M, Hannon GJ. Multiple roles for Piwi in silencing Drosophila transposons. Genes & development. 2013;27:400–412. doi: 10.1101/gad.209767.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes & development. 2013;27:390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H. A major epigenetic programming mechanism guided by piRNAs. Developmental cell. 2013;24:502–516. doi: 10.1016/j.devcel.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Gu W, Shirayama M, Conte D, Jr., Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, Chen CC, Chaves DA, Duan S, Kasschau KD, Fahlgren N, Yates JR, 3rd, Mitani S, Carrington JC, Mello CC. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Molecular cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, Conte D, Jr., Mello CC. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D., Jr. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3582–3587. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Zhang C, Montgomery TA, Fischer SE, Garcia SM, Riedel CG, Fahlgren N, Sullivan CM, Carrington JC, Ruvkun G. The Caenorhabditis elegans RDE-10/RDE-11 complex regulates RNAi by promoting secondary siRNA amplification. Current biology : CB. 2012;22:881–890. doi: 10.1016/j.cub.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Zhang C, Montgomery TA, Gabel HW, Fischer SE, Phillips CM, Fahlgren N, Sullivan CM, Carrington JC, Ruvkun G. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1201–1208. doi: 10.1073/pnas.1018695108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Molecular cell. 2010;37:679–689. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18674–18679. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- [100].Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- [101].Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. The EMBO journal. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Juang BT, Gu C, Starnes L, Palladino F, Goga A, Kennedy S, L'Etoile ND. Endogenous nuclear RNAi mediates behavioral adaptation to odor. Cell. 2013;154:1010–1022. doi: 10.1016/j.cell.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Avgousti DC, Palani S, Sherman Y, Grishok A. CSR-1 RNAi pathway positively regulates histone expression in C. elegans. The EMBO journal. 2012;31:3821–3832. doi: 10.1038/emboj.2012.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nature structural & molecular biology. 2008;15:581–590. doi: 10.1038/nsmb.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Nishida KM, Miyoshi K, Ogino A, Miyoshi T, Siomi H, Siomi MC. Roles of R2D2, a cytoplasmic D2 body component, in the endogenous siRNA pathway in Drosophila. Molecular cell. 2013;49:680–691. doi: 10.1016/j.molcel.2012.12.024. [DOI] [PubMed] [Google Scholar]
- [106].Deshpande G, Calhoun G, Schedl P. Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes & development. 2005;19:1680–1685. doi: 10.1101/gad.1316805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Fagegaltier D, Bouge AL, Berry B, Poisot E, Sismeiro O, Coppee JY, Theodore L, Voinnet O, Antoniewski C. The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21258–21263. doi: 10.1073/pnas.0809208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nature cell biology. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Sinkkonen L, Hugenschmidt T, Filipowicz W, Svoboda P. Dicer is associated with ribosomal DNA chromatin in mammalian cells. PloS one. 2010;5:e12175. doi: 10.1371/journal.pone.0012175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Lo Sardo F, Saxena A, Miyoshi K, Siomi H, Siomi MC, Carninci P, Gilmour DS, Corona DF, Orlando V. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nature genetics. 2006;38:936–941. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- [112].Taliaferro JM, Aspden JL, Bradley T, Marwha D, Blanchette M, Rio DC. Two new and distinct roles for Drosophila Argonaute-2 in the nucleus: alternative pre-mRNA splicing and transcriptional repression. Genes & development. 2013;27:378–389. doi: 10.1101/gad.210708.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, Klinck R, Chabot B, Kornblihtt AR. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nature structural & molecular biology. 2009;16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- [114].Liu J, Hu J, Corey DR. Expanding the action of duplex RNAs into the nucleus: redirecting alternative splicing. Nucleic acids research. 2012;40:1240–1250. doi: 10.1093/nar/gkr780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, Mathieu J, Hamiche A, Ait-Si-Ali S, Muchardt C, Batsche E, Harel-Bellan A. Argonaute proteins couple chromatin silencing to alternative splicing. Nature structural & molecular biology. 2012;19:998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]