Abstract
Cohesins are conserved and essential Structural Maintenance of Chromosomes (SMC) protein-containing complexes that physically interact with chromatin and modulate higher-order chromatin organization. Cohesins mediate sister chromatid cohesion and cellular long-distance chromatin interactions affecting genome maintenance and gene expression. Discoveries of mutations in cohesin’s subunits and its regulator proteins in human developmental disorders, so-called “cohesinopathies,” reveal crucial roles for cohesins in development and cellular growth and differentiation. In this review, we discuss the latest findings concerning cohesin’s functions in higher-order chromatin architecture organization and gene regulation and new insight gained from studies of cohesinopathies.
Keywords: cohesin, cohesinopathies, NIPBL, Cornelia de Lange Syndrome, Roberts' Syndrome
1. Introduction
Chromosomes undergo both global and local structural changes during the cell cycle and cellular differentiation. Accumulating evidence indicates that proper structural organization of chromosomes is critical for genome maintenance and functions, including proper chromosome segregation during cell division, DNA replication and repair, and gene expression. Structural Maintenance of Chromosomes (SMC) protein-containing complexes are a unique class of conserved and essential factors that control these processes by altering chromatin structural organization.
The first SMC gene, Smc1, was identified in yeast as being essential for mitotic chromosome segregation [1]. SMC proteins have conserved ATPase motifs, and ATP binding and hydrolysis by SMC proteins were shown to be important for the complexes’ functions [2–4]. SMC proteins are folded in half at the hinge domain, which brings the conserved head and tail globular domains with divided ATPase motifs together. They form highly stable heterodimers in specific combinations in eukaryotes (SMC1-SMC3, SMC2-SMC4, and SMC5-SMC6) that further interact with specific sets of non-SMC subunits to assemble three major complexes: cohesin, condensin and the SMC5-SMC6 complex, respectively.
The common feature of SMC complexes is that they physically associate with chromatin and regulate higher-order chromatin structure. Early studies of condensin and cohesin in a Xenopus in vitro system and in yeast established solid biochemical and cell biological grounds to appreciate the intricate cell cycle-specific regulation and essential mitotic function of these two complexes [5, 6]. SMC complexes organize mitotic chromosomes to ensure proper segregation during cell division: cohesin through sister chromatid cohesion and metaphase chromosome congression, and condensin through orderly chromatin compaction and chromosome resolution. Studies in multiple organisms including S. cerevisiae, S. pombe, Drosophila, human and chicken cells, C. elegans, and more recently zebrafish and mice, have provided further insight into both conserved and species-specific functions of SMC complexes in genome regulation. Comprehensive reviews of condensin [7–9] and the SMC5-SMC6 complex [10, 11] have been recently published and they will not be discussed in detail here.
We now understand that cohesin has pivotal roles in mitosis, DNA replication, DNA repair, and gene expression, though the underlying molecular mechanisms and implications for development and disease are still under active investigation [12–14]. In somatic vertebrate cells, there are two different cohesin complexes (Fig. 1), and their functional redundancies and distinctions have just begun to be uncovered (see below). In this review, we mainly discuss cohesin’s functions and regulation in mammalian cells, but we will not address its role in meiosis.
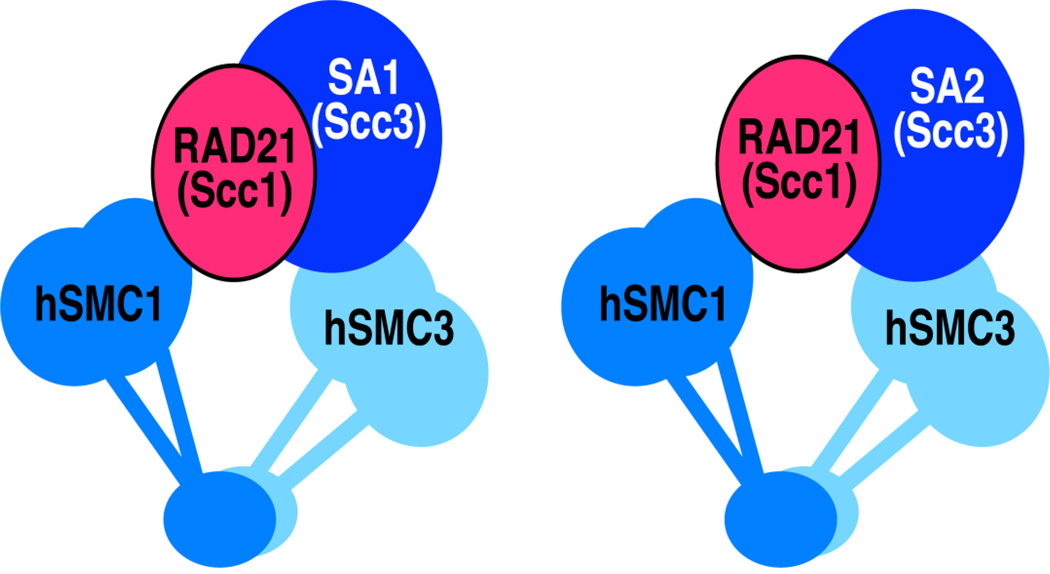
Figure 1.
Schematic diagrams of cohesin-SA1 and cohesin-SA2.
1.1. Structural features of cohesin
Cohesin consists of the SMC family proteins SMC1 (also known as SMC1A) and SMC3 as a heterodimer with the two non-SMC components Rad21 (also called Mcd1 or Scc1) and Scc3 (also called SA or STAG) [5]. SMC1 and SMC3 interact through their central hinge regions, while their respective paired amino- and carboxyl-terminal globular domains are further bridged by the kleisin family component Rad21 (or Scc1) (Fig. 1) [6, 15]. The primary function of cohesin is to mediate genome-wide sister chromatid cohesion in a cell cycle-regulated manner to ensure proper segregation of chromosomes in mitosis [16–18]. High-resolution microscopy and biochemical studies revealed that cohesin forms a ring structure [19–22]. Further analyses of purified cohesin-circular minichromosome complexes assembled in vivo, in conjunction with various mutational manipulations of cohesin subunits, supports the notion that the cohesin ring traps sister chromatids inside to mediate sister chromatid cohesion with distinct chromatin entry and exit mechanisms [20, 23–26]. However, alternative models of DNA trapping and cohesion by cohesin are still being discussed [27], and the exact mechanism is not yet fully resolved.
1.2. Two cohesin complexes in vertebrates
While a single Scc3 is present in yeast, two SA proteins, SA1 and SA2 (STAG1 and STAG2 in mice), are found in higher eukaryotes to form two distinct cohesin complexes in somatic cells: cohesin-SA1 and cohesin-SA2 (Fig. 1) [28, 29]. Both cohesin-SA1 and cohesin-SA2 contribute to genome-wide sister chromatid cohesion, with SA1 being particularly important for telomeric sister chromatid cohesion in mammalian cells [28, 30–32]. This appears to be determined by the specific interactions of SA1, but not SA2, with the telomere binding proteins TRF1 and TIN2 [30]. Thus, while the exact function of the SA/Scc3 subunit in yeast cohesin function remains elusive, SA proteins appear to dictate the recruitment specificity of cohesins through protein: protein interactions in mammalian cells. More recently, knockout and depletion experiments revealed that SA1 and SA2 have non-redundant functions in the transcriptional regulation of certain, if not all, genes [32]. Mutations in SA2, with an intact SA1, are associated with aneuploidy in a diverse range of human cancers [33], and SA1 knockout caused aneuploidy and increased cancer risk despite the presence of an intact SA2 [34]. These studies suggest both redundant and distinct functions of the two cohesin complexes that are likely to have emerged to manage the increased complexity of chromosome organization and functions in higher eukaryotes.
2. Cell cycle-specific cohesin regulation in chromatin loading and cohesion
2.1. Cohesin loading onto chromatin by Scc2-Scc4 (NIPBL-MAU2) in telophase
In S. cerevisiae, cohesin is loaded onto chromosomes during G1 phase, which requires the heterodimeric cohesin loading factor Scc2-Scc4 [35]. Chromatin loading, but not establishment of cohesion, requires ATP hydrolysis [2, 3, 36]. Despite the early discovery of this cohesin loading factor, the exact loading mechanism remains enigmatic. Human cohesin also requires NIPBL (or delangin, yeast Scc2 homolog) and its partner MAU2 (yeast Scc4 homolog) for chromatin loading (Fig. 2) [37, 38]. Cohesin loading takes place in telophase in higher eukaryotes (see also 4.1.). A recent study suggests that this requires the opening of the SMC dimer at the hinge region, though how Scc2-Scc4 mediates this process is not understood [39].
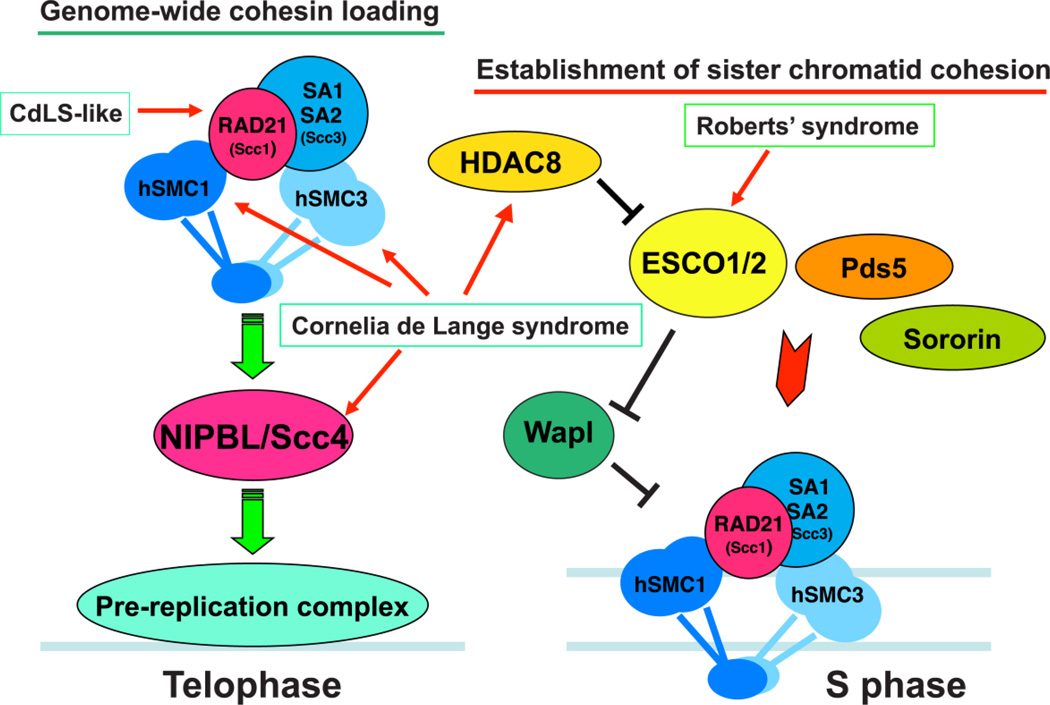
Figure 2.
Regulators of cohesin loading and establishment of sister chromatid cohesion. The cohesin loading factor, NIPBL-MAU2 (Scc2-Scc4), is required for cohesin loading onto chromatin in telophase in mammalian cells. The initial loading of NIPBL-MAU2 is dependent on the pre-replication machinery. In S phase, the establishment of sister chromatid cohesion requires sororin and Pds5A/B as well as the ESCO1/2 (Eco) acetyltransferases that coordinately antagonize the activity of the cohesin destabilizing factor Wapl. ESCO-mediated acetylation of the cohesin subunit SMC3 must be reversed by histone deacetylase HDAC8 in order to refresh and recycle cohesin for the subsequent cell cycle. Mutations associated with the cohesinopathies RBS and CdLS are indicated.
2.2. Establishment of sister chromatid cohesion in S phase
ESCO1/2 (Eco1 in yeast), sororin and Pds5 are additionally needed to antagonize the cohesin destabilizing factor Wapl and establish sister chromatid cohesion in S phase (Fig. 2) [40, 41]. ESCO1 and ESCO2 (and Eco1p in yeast) are acetyltransferases, and their acetylation of SMC3 is required for antagonizing Wapl and establishment of sister chromatid cohesion [41–46]. Wapl appears to release cohesin from chromatin by opening the gate between SMC3 and Scc1 (Rad21) [26, 47, 48]. A recent structural study suggests that the binding of Wapl to the ATPase head domain of Smc3 may regulate its activity, though the detailed gate opening mechanism is unclear [49]. Interestingly, a sororin homolog has not been found in yeast, and although interfering with Wapl activity is critical for sister chromatid cohesion, how it leads to cohesion of the two sister chromatids is not well understood. Interestingly and somewhat counterintuitively, SMC3 acetylation also facilitates DNA replication fork progression, suggesting that this cohesin modification is also important to switch cohesin to a configuration that does not obstruct fork advancement [50]. ESCO-mediated acetylation of SMC3 is reversed by the deacetylase Hos1 in S. cerevisiae and HDAC8 in human cells, which is required for the next cycle of cohesion establishment [51–54].
There are additional factors that function in sister chromatid cohesion that all relate to DNA replication. These include the Ctf18-RFC complex, the DNA polymerase α-associating Ctf4, Trf4 (DNA polymerase κ PCNA and, more recently, Timeless and Tipin, further suggesting the coupling of DNA replication and cohesion [36, 55–60]. How these factors orchestrate the establishment of sister chromatid cohesion remains obscure. Consistent with the apparent coupling of DNA replication and establishment of sister chromatid cohesion, cohesin newly expressed in G2 phase after the completion of DNA replication fails to establish sister chromatid cohesion despite its loading onto chromatin in S. cerevisiae [36, 61]. This observation has not yet been confirmed in higher eukaryotes.
2.3. Cohesin removal and spindle-associated function in mitosis
In higher eukaryotes, cohesin is removed from chromosomes in a two-step process during mitosis that results in chromosome separation in anaphase [62]. The first step is removal of the majority of cohesin from chromatin in prophase, and the second step is destruction of the residual cohesin remaining primarily at centromeres by separase-mediated Rad21 cleavage at the end of metaphase, which leads to chromosome segregation in anaphase. This mitosis-specific regulation of cohesin was reviewed extensively [63–67] and will not be discussed here in detail. More recent studies indicate that the SMC3-Rad21 gate opening by Wapl is important for cohesin release in prophase [47, 48]. A small population of cohesin associates with centrioles, and a proteolytic cleavage of Rad21 also regulates centriole disengagement [68–70]. In addition, a significant population of cytoplasmic cohesin associates with spindles and spindle poles in a mitosis-specific fashion, contributing to proper spindle assembly and chromosome congression [69, 71]. Thus, cohesin ensures proper congression and segregation of chromosomes during cell division through both chromatin-dependent and -independent actions.
2.4. Non-mitotic functions of cohesin
Cohesin functions in maintaining genome stability through post-replicative DNA double-strand break (DSB) repair, specifically sister chromatid homologous recombination (HR) repair [72, 73]. In mammalian cells, cohesin is also involved in DNA damage checkpoint control [74–77]. An excellent comprehensive review of the regulation and function of cohesin in DSB damage response and repair was recently published [14]. A recent study also indicated that cohesin affects normal DNA replication [78]. In addition, an expanding body of literature is documenting cohesin as a key regulator of gene expression (see below).
3. Mechanism of cohesin-mediated gene regulation
3.1. Long-distance chromatin interactions
3.1.1. CTCF-dependent and -independent long-distance chromatin interactions
Cohesin was shown to mediate chromatin looping at multiple gene loci important for imprinting and differential gene expression during development [79–85]. These interactions include CCCTC-binding factor (CTCF)-dependent insulator interaction, which blocks enhancer activity and/or inhibits the spreading of heterochromatic domains, as well as distal enhancer-promoter interactions important for gene activation (there are a number of comprehensive reviews, including but not limited to [12, 86, 87]). The roles of CTCF and related insulator binding proteins in Drosophila are discussed in a companion article in this issue (Matzat and Lei, this issue). In mammalian cells, the total number of cohesin binding sites vary from ~25,000 to ~120,000 as determined by chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) analyses, depending on the antibody, cell type, experimental conditions, and analytic tools employed [32, 83, 88]. Approximately 50–70% of cohesin sites overlap with CTCF binding sites genome-wide [88–92]. Cohesin mediates chromatin domain organization and insulator functions at many of these CTCF sites, including the H19/IGF2 locus, the IFNG locus, the apolipoprotein gene cluster, the β -globin locus, the Igh locus, the MHC class II gene cluster, the HoxA locus, and the T-cell receptor α locus [79–81, 84, 93–97]. However, a significant number of cohesin sites appear to be CTCF-free and often overlap with binding sites for cell type-specific transcription factors [88, 98, 99]. Thus far, however, no significant DNA sequence preference was observed at cohesin binding sites other than the CTCF binding motif. CTCF-free cohesin binding sites coincide significantly with enhancer elements and genes that exhibit tissue/cell type-specific patterns of expression, and cohesin appears to help stabilize transcription factor binding to these sites [88]. It should be noted that a CTCF-associated function of cohesin has not been observed in Drosophila, in which cohesin mediates gene regulation in an insulator-independent manner (Matzat and Lei, this issue).
At the β-globin locus, both CTCF-dependent insulator interaction and CTCF-independent enhancer-promoter interactions can be observed [82]. Both types of interaction involve cohesin in mouse and human erythroid lineage cells as detected by chromatin conformation capture (3C) and 3C combined with ChIP (ChIP-loop) (Fig. 3) [82]. The distal enhancer in the locus control region (LCR) interacts with the developmental stage-specific globin genes, which correlates with their specific expression [100, 101]. The lineage-specific transcription factors EKLF (Klf1), GATA-1, Fog-1, and Ldb1 are required in this process [102–104]. Both Nipbl and cohesin binding rapidly increases at chromatin loop anchoring sites upon cellular differentiation [82]. Depletion of either cohesin or Nipbl decreased both the insulator interaction and the LCR enhancer-promoter interaction, while CTCF depletion only affected the insulator interaction [82]. Consistent with this, cohesin depletion, but not CTCF depletion, decreased β-globin gene expression [82].
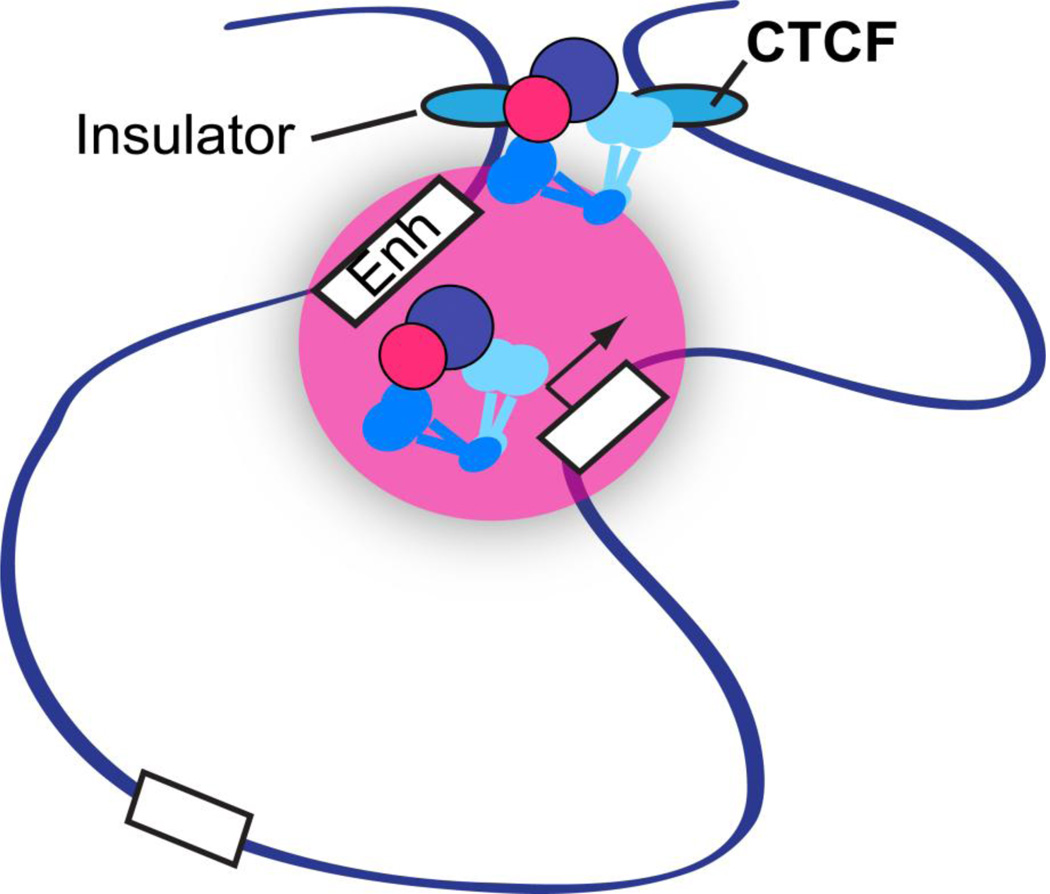
Figure 3.
CTCF-dependent and -independent chromatin loop formation at the β-globin locus. Cohesin binds to and mediates the long-distance interactions of CTCF-bound insulator elements flanking the locus as well as between the distal enhancer (Enh) in the locus control region and the adult globin genes (white box with an arrow) [82]. The pink circle represents the presence of various transcription factors involved in globin gene expression, such as EKLF (Klf1), GATA-1, Fog-1, Ldb1, and NF-E2 [82, 102–104, 147]. A white box without an arrow represents the inactive gene, which is not interacting with the enhancer.
3.1.2. Genome-wide analyses of cohesin-mediated long-distance chromatin interactions
Recent studies examined cohesin-mediated chromatin interactions genome-wide using high-resolution high-throughput 3C-based techniques, circular 3C followed by high-throughput sequencing (4C-seq) with and without ChIP [105, 106], 3C carbon copy (5C) [107], and Chromatin Interaction Analysis by Paired-End Tag sequencing (ChIA-PET) [108] (for experimental details, see a recent review [109]).
An SMC1 ChIA-PET study, in which chromatin interactions involving cohesin were selectively analyzed in developing mouse limb, identified over 2,200 interactions at both CTCF-positive and -negative cohesin binding sites [108]. In either the promoter or intergenic/intronic regions, ~65% of chromatin interaction sites coincided with CTCF occupancy. The study revealed that in addition to tissue-specific promoter-enhancer interactions and constitutive chromatin domain demarcations, a subset of promoter-enhancer interactions reflect the poised state in embryonic stem cells (ESCs) and are maintained in multiple tissues even when the genes are not expressed.
Cohesin plays an important role in the maintenance of pluripotency. Cohesin was found to interact with Mediator and colocalize at the anchoring sites of enhancer-promoter interactions at pluripotency genes in mouse ESCs (mESCs), with its depletion causing spontaneous differentiation [83, 110]. High-resolution 5C analysis of the regions surrounding the major developmentally regulated genes during neuroectoderm differentiation was compared to corresponding ChIP-sequencing data for CTCF, cohesin and Mediator [107]. The results revealed that CTCF/cohesin tends to mediate relatively constant long-range chromatin interactions defining megabase-sized topologically associating domains (TADs), while Mediator and cohesin bridge short-range enhancer-promoter interactions, which are often cell typespecific, both within and between TADs [107]. Both 3C and 4C with or without ChIP revealed that cohesin and Mediator are involved in pluripotency-specific chromatin interactions at the Oct4 and Nanog promoters [83, 105, 106, 111]. The interaction patterns are altered during differentiation and restored in induced pluripotency cells (iPSCs). Cohesin recruitment is induced concomitant with the induction of long-distance chromatin interactions during the iPSC reprogramming process. Cohesin depletion disrupts the enhancer-promoter interaction, blocks self-renewal, induces differentiation in pluripotent cells, and interferes with reprogramming of fibroblasts to iPSCs [105, 106, 111].
3.1.3. Chromatin looping: cause or consequence of gene expression?
The aforementioned studies strongly suggest that cohesin-mediated chromatin interactions are critical for gene expression. Furthermore, a recent study also showed that forced induction of distal enhancer-promoter interaction indeed activates β-globin gene expression (albeit to lesser extent than the full activation), demonstrating the pivotal role of long-distance chromatin interactions in gene regulation [112]. Comparison of mESCs and differentiated cells as well as examination of iPSC reprogramming described above also provided evidence that reorganization of chromatin interactions precedes the actual gene expression changes, supporting the idea that chromatin interactions are causative rather than a consequence of gene expression changes [105, 106, 111]. Whether cohesin is involved in the initiation and/or maintenance of these interactions is unclear.
3.2. Role of cohesin in gene repression
Cohesin was found to repress gene expression by enhancer blocking, for example, at the cut gene in Drosophila [113] and the IGF2-H19 locus in mammalian cells [89, 91]. Although cohesin is also known to bind to centromeric and non-centromeric heterochromatin repeats [114–116], only a limited number of examples of cohesin’s involvement in heterochromatin-mediated gene silencing have been documented. In Drosophila, both cohesin and Nipped-B (Nipbl homolog) bind to the Enhancer of split and invected-engrailed gene complexes coinciding with histone H3 lysine 27 trimethylation (H3K27me3), the repressive histone modification associated with the polycomb silencing pathway [117, 118]. Depletion of cohesin resulted in upregulation of these genes [115]. More recent studies provided additional evidence for the functional interaction between cohesin and polycomb proteins and the effect of cohesin on polycomb silencing in Drosophila [119, 120]. In S. pombe, cohesin binds to subtelomeric heterochromatin regions harboring H3 lysine 9 methylation (H3K9me) [114]. Cohesin is co-recruited with Swi6, a heterochromatin binding protein 1 (HP1) homolog that recognizes methylated lysine 9 residues, and they function together in gene silencing [114] (see 4.3.3). Similar co-recruitment of cohesin and HP1γ is observed at subtelomeric heterochromatin repeats in human cells, whose loss is associated with a specific muscular dystrophy (see 7.1).
3.3. RNA polymerase II (RNAPII) occupancy and transition from pausing to elongation
In Drosophila, cohesin and Nipped-B bind to a subset of active genes, in particular to genes with a paused RNAPII [121–123]. Cohesin or Nipped-B depletion results in increased RNAPII pausing at cohesin-bound genes, suggesting that cohesin facilitates RNAPII transition to elongation [123]. Whether this is a consequence of cohesin’s function in enhancer-promoter bridging or by cohesin’s direct effect on RNAPII is currently unclear. Interestingly, cohesin depletion also results in a general decrease of RNAPII pausing and transcription of noncohesin- bound genes [123]. Whether a similar effect of cohesin depletion on non-cohesin-bound genes exists in other organisms is currently unknown, and whether cohesin facilitates RNAPII transition from pausing to elongation in mammalian cells remains to be determined.
3.4. Intragenic cohesin binding and RNA transcription
In contrast to the studies in Drosophila, intragenic binding of cohesin together with CTCF appears to cause RNAPII pausing in mammalian cells, resulting in alternative mRNA products. In human cells, cohesin/CTCF binding in intragenic regions functions as a chromatin boundary to block transcriptional read-through of the full-length PUMA gene [124]. In addition, RNAPII complexes accumulate at the CTCF-cohesin binding site within the first intron of the latency transcript of Kaposi's sarcoma-associated herpesvirus [125]. This pausing, which also involves the binding of pausing factors SPT5 and NELF-A at the intragenic CTCF-cohesin binding site, appears to be important for proper mRNA processing and production. Although the presence of cohesin was not tested, intragenic binding of CTCF also dictates alternative mRNA splicing of the CD45 gene [126], and the presence of CTCF at promoter proximal sites was shown to be associated with RNAPII pausing in mammalian cells [127]. Since no significant overlap between cohesin and CTCF binding is seen in Drosophila [128] (Matzat and Lei, this issue), how this relates to the observations in Drosophila (see 3.3.) is currently unclear.
4. Cohesin recruitment mechanisms
4.1. The NIPBL-Mau2 (SCC2-SCC4) cohesin loading factor
Cohesin is not a canonical sequence-specific DNA binding factor, and how it is recruited to chromatin is critical for both its cell cycle- and differentiation stage-specific functions. In metazoans, genome-wide cohesin loading occurs at the end of mitosis during telophase, which also requires their Scc2 and Scc4 homologs (NIPBL (human)/Nipbl (mouse) and MAU2 (human)/Mau2 (mouse), respectively) [38, 129, 130]. In Xenopus and human cells, pre-replication complex components, including ORC, Cdc6, Cdt1, and MCM2-7, were shown to be required for loading of Scc2-Scc4 (NIPBL-MAU2) and subsequent cohesin binding to chromatin [129–131]. This suggests that the initial loading sites for cohesin are at pre-replication complex assembly sites (i.e. replication origins) in higher eukaryotes. In contrast, no obvious relationship between the replication origin (Autonomously replicating sequence (ARS)) and Scc2-Scc4 binding sites has been demonstrated in S. cerevisiae. Interestingly, all three SMC complexes (cohesin, condensin, and the SMC5-SMC6 complex) independently require Scc2 in S. cerevisiae [132, 133]. Condensin additionally requires RNA polymerase (RNAP) III transcription factor TFIIIC and is preferentially recruited to RNAPIII genes, such as tRNA genes [133]. In C. elegans, loading of condensins and the SMC5-SMC6 complex appears to be Scc2-independent despite the partial overlap of Scc2 and condensin binding sites [134, 135]. The relationship between other SMC complexes and,NIPBL-MAU2 is unclear in mammalian cells.
4.2. Cohesin sliding?
In S. cerevisiae, cohesin binding appears to be affected by the transcriptional status of nearby genes, and cohesin tends to accumulate at sites of transcriptional convergence [136–138]. Interestingly, ChIP analyses using antibody specific for Scc2 has revealed that the peaks associated with Scc2 binding often do not coincide with cohesin peaks, suggesting that cohesin may “slide” from its initial loading sites marked by Scc2-Scc4 [137, 139]. However, another study using FLAG-tagged Scc2 revealed the presence of Scc2 at all cohesin binding peaks, arguing that the loading factor functions at all cohesin binding sites [140]. It should be noted, however, that even in the latter study, the peak signals for Scc2 binding are not always proportional to cohesin peaks, suggesting that an additional factor(s) impacts cohesin accumulation (e.g., the transcriptional status of the neighboring genes) [140]. If the Scc2 ChIP efficiency is low, these weak sites may be considered negative and give the impression that cohesin binds to Scc2-free regions. Interestingly, the binding of ATP hydrolysis-defective cohesin appears to be more restricted and more closely correlates with the major Scc2 binding peaks [141]. This suggests that sliding, but not initial loading, of cohesin requires ATP hydrolysis. However, whether cohesin can change its binding sites in the absence of Scc2-Scc4 has not been explicitly tested.
In higher eukaryotes, there is thus far no clear evidence for cohesin sliding and accumulation at transcriptional convergence sites. In Drosophila, Nipped-B and cohesin binding sites virtually overlap and are associated with active genes, often with paused RNAPII [121, 122]. Cohesin was found to be significantly enriched at the promoters and gene regions in mammalian cells [89, 91]. Furthermore, the increase of Nipbl binding closely accompanies the increase of cohesin binding at the adult globin enhancer and promoter regions upon β-globin gene activation [82]. A study in mESCs identified two different populations of cohesin binding sites, one overlapping with CTCF with no apparent Nipbl peaks, and the other coinciding with Nipbl and Mediator [83]. This led to the notion that Nipbl may not load cohesin at CTCF sites. However, specific Nipbl binding peaks can be identified at cohesin-bound CTCF insulator sites by manual ChIP-PCR, and depletion of Nipbl also affects cohesin binding at these regions, suggesting that Nipbl also loads cohesin at CTCF sites [82]. The difficulty in detecting Nipbl peaks consistently by ChIP-seq at all cohesin binding sites may be due to the fact that Nipbl binds chromatin less stably than cohesin [131]. Recent attempts to reconstitute Scc2-Scc4-dependent cohesin loading in vitro in yeast and human cells are an important first step towards addressing this issue [131, 142].
4.3. Cohesin recruitment through protein and RNA interactions
4.3.1. Modulation of cohesin recruitment to CTCF sites
The majority of cohesin binding sites contain the CTCF motif in mammalian cells [89–92], which appears to be sufficient to recruit cohesin [91]. The SA proteins (both SA1 and SA2) interact with CTCF [143]. CTCF depletion decreases cohesin binding to some of these sites, suggesting that cohesin is recruited to these sites by CTCF though this relationship is not observed in Drosophila (Matzat and Lei, this issue). However, not all the CTCF sites in mammalian cells are co-occupied with cohesin [96, 144], suggesting that an additional factor(s) dictates cohesin binding at CTCF sites. Indeed, the cohesin and CTCF interaction is modulated by the DEAD-box RNA binding protein p68, together with its associated non-coding RNA (ncRNA) called steroid receptor RNA activator (SRA), and promotes insulator function, for example, at the Igf2/H19 locus [145]. ATR-X, mutated in the Alpha-Thalassemia mental Retardation, X-linked (ATR-X) syndrome, together with methyl-CpG binding protein 2 (MeCP2), also interact with cohesin and CTCF in the brain, affecting their binding and postnatal imprinting function at the Igf2/H19 and Gtl2/Dlk1 loci [146].
4.3.2. Other factors that dictate cohesin recruitment
Cohesin was shown to interact with Mediator, Nanog and Klf4, suggesting that these interactions may mediate the specific recruitment of cohesin [83, 105, 110]. Cohesin was also found to interact with NF-E2, which is specifically recruited to the LCR enhancer and the promoter regions of the adult β-globin locus coinciding with cohesin [82, 147]. In addition, cohesin was found to be part of the human ISWI (SNF2h)-containing chromatin remodeling complex together with the Mi2/NuRD complex, and bind chromatin together in an SNF2h ATPase activity-dependent manner in human cells [148]. Rad21 directly interacts with SNF2h [148]. Recently, Drosophila Mi-2 was also found to recruit cohesin to polytene chromosomes in salivary grands [149]. In addition, cohesin was reported to bind to the non-coding RNAs (ncRNAs) transcribed on enhancer regions, termed enhancer RNAs (eRNAs) [150]. Ligand-activated estrogen receptor (ER) upregulates transcription of eRNAs, which act in cis to promote upregulation of nearby ER target genes. The eRNAs bind to cohesin and increase cohesin recruitment to the enhancer regions in response to the ER ligand estradiol, and stimulate the enhancer-promoter interactions in MCF7 breast cancer cells [150]. Though the exact mechanism is unclear, this raises the intriguing possibility that other ncRNAs may also affect long-distance chromatin interactions through recruitment of cohesin.
4.3.3. Cohesin recruitment to heterochromatin repeats
In S. pombe, cohesin is recruited to both pericentromeric and subtelomeric heterochromatin via the H3K9me-Swi6 (HP1) pathway, except that the recruitment of cohesin and Swi6 is mutually dependent at subtelomeric heterochromatin [114, 151, 152]. While cohesin is recruited by Swi6 to mediate centromeric sister chromatid cohesion with no role in gene silencing at pericentromeric heterochromatin [151, 152], cohesin co-recruited with Swi6 to the subtelomeric heterochromatin participates in gene regulation [114]. Interestingly, similar co-recruitment of cohesin and one of the HP1 variants, HP1γ, was observed at subtelomeric heterochromatic D4Z4 macrosatellite repeat regions marked by H3K9me3 in human cells, whose loss is closely associated with a muscular dystrophy (see 7.1) (Fig. 4A). Though it was controversial whether the H3K9me-HP1-cohesin pathway is conserved at mammalian centromeres [153, 154], a recent study demonstrated that cohesin recruitment to pericentromeric heterochromatin indeed involves HP1 in human cells [155]. While mainly HP1α and also HP1γ are involved in pericentromeric heterochromatin recruitment of cohesin, HP1γ is specifically involved in cohesin co-recruitment at subtelomeric D4Z4 heterochromatin. NIPBL, but not cohesin, was shown to directly bind to all three HP1 variants [116, 154, 156]. More recently, the Suv4-20h histone methyltransferase that specifically mediates H4K20 trimethylation (H4K20me3) was shown to interact with cohesin and functions in cohesin recruitment to pericentromeric heterochromatin in mouse cells in a catalytic activity-independent manner, which is important for centromeric sister chromatid cohesion and proper segregation of chromosomes in mitosis [157]. Suv4-20h recruitment to pericentromeric heterochromatin itself is dependent on H3K9me3 and HP1. Thus, cohesin recruitment to heterochromatin appears to be more complex than previously thought (Fig. 4B).
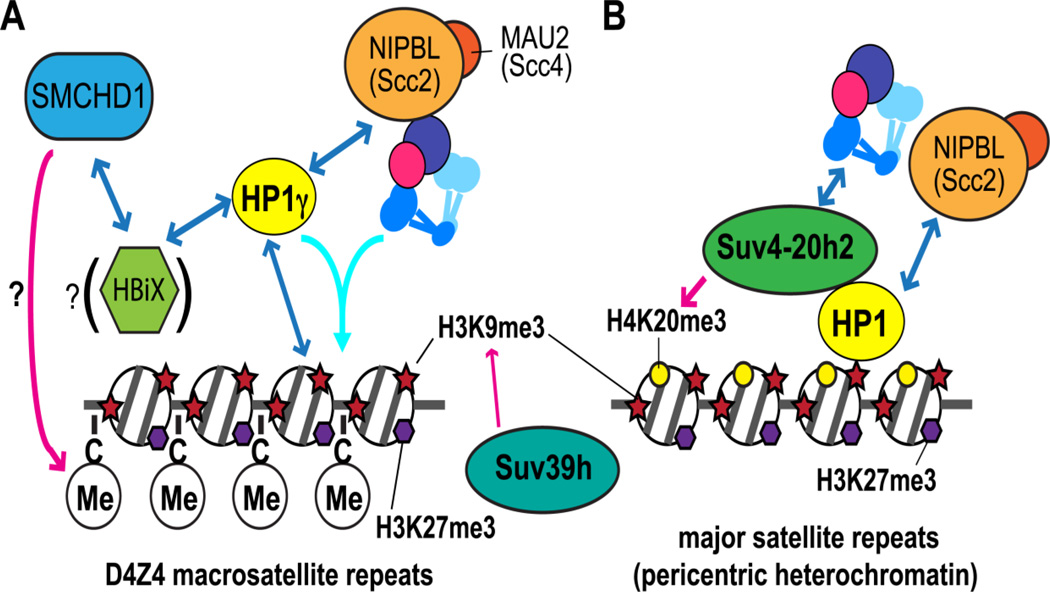
Figure 4.
Cohesin recruitment to heterochromatin. Blue double-sided arrows indicate the interactions reported. Pink arrows indicate downstream effects. A. Cohesin and HP1γ require each other to bind to the D4Z4 subtelomeric heterochromatin in a SUV39H-mediated H3K9me3-dependent manner [116]. Direct interaction of NIPBL with HP1 may contribute [116, 156]. The light blue arrow indicates co-recruitment of cohesin and HP1γ to D4Z4 [116]. In addition, an SMC homolog, SMCHD1, binds to D4Z4 [215]. Whether this binding is mediated by HBiX1, an HP1-interacting protein, as observed at the inactive X chromosome [198] is currently unclear. SMCHD1 was shown to be important for the maintenance of DNA methylation [214]. Whether it contributes to DNA hypermethylation at D4Z4 has not been determined. B. Cohesin is recruited to pericentromeric heterochromatin via interaction with histone methyltransferase Suv4-20h2, which mediates H4K20me3. Suv4-20h2 localization is dependent on HP1 bound to methylated H3K9 mediated by SUV39h [157]. The relevance of the NIPBL-HP1 interaction [156] to pericentromeric cohesin recruitment is unclear.
4.3.4. Cohesin, NIPBL and MAU2 can each specify cohesin recruitment sites
Artificial centromeric tethering of an Scc4 fusion protein is sufficient for the recruitment of Scc2 as well as cohesin in budding yeast, indicating that Scc4 can also be a determinant for binding site specificity [158]. Although the pre-replication complex-dependent loading of NIPBL in telophase is cohesin-independent in human cells [131], cohesin is reciprocally required for Scc2/Scc4 recruitment to centromeres in yeast, supporting the notion that cohesin can dictate its loading site [158].
Collectively, these results suggest that increased binding of either cohesin or NIPBL or MAU2 can trigger cohesin’s accumulation at specific genomic regions. With many potential interaction surfaces available on subunits of cohesin, NIPBL, and MAU2, differential targeting of cohesin may be achieved by interactions with sequence-specific transcription factors, chromatin remodelers, specific histone mark readers, and even with RNA. Many of these interactions may occur at a specific subcellular and/or genomic location and often in a cell cycle- or differentiation stage-specific manner. These differential interactions may be regulated by post-translational modifications or availability of the interacting proteins. This allows cohesin to be recruited to multiple sites in different cell types and contexts, providing further versatility to its actions.
5. Cohesinopathies
Human syndromes caused by cohesin and cohesin-associated factor mutations, resulting in cohesin dysfunction, are called “cohesinopathies” (Fig. 2) [159, 160]. The two classic examples are Roberts’ Syndrome (RBS) and Cornelia de Lange Syndrome (CdLS).
5.1. Roberts’ Syndrome
RBS (OMIM 268300) (more recently, Roberts’ Syndrome/SC phocomelia) is caused by mutations of both alleles of ESCO2 (Fig. 2) [161]. RBS patients have a wide range of clinical phenotypes that include upper and lower limb defects, growth retardation, craniofacial anomalies, and mental retardation with limited similarity to the CdLS phenotype [161, 162]. Importantly, RBS chromosomes exhibit premature centromere separation and heterochromatin puffing, indicative of a sister chromatid cohesion defect [163]. Centromeric cohesion defects and cell cycle aberrations are observed in ESCO2 knockout mice and zebrafish [164, 165].
5.2. Cornelia de Lange Syndrome (CdLS)
CdLS (OMIM 122470, 300590, 610759) is a dominant multisystem developmental disorder characterized by facial dysmorphism, hirsutism, upper limb abnormalities, cognitive retardation, and growth abnormalities [166, 167]. Mutations in the NIPBL gene on chromosome 5p13 are linked to more than 55% of CdLS cases (Fig. 2) [168, 169]. Frameshift or nonsense mutations of NIPBL that result in NIPBL haploinsufficiency often exhibit more severe phenotypes compared to missense mutations [170]. Mutations in the cohesin subunits SMC1 (human SMC1 (hSMC1), SMC1A) and hSMC3 were also found in a minor subset of clinically milder CdLS cases (~5% and <1%, respectively) [171, 172]. SMC1 or SMC3 mutations are always missense mutations and patients often show mental retardation as the primary symptom, with other abnormalities being fewer and/or milder [172]. More recently, mutations in HDAC8, which regulates cohesin dissociation from chromatin in mitosis, were also found in a subset of CdLS patients (OMIM 300882) [53]. HDAC8 functions to deacetylate SMC3 and therefore facilitates cohesin displacement from chromatin during mitotic progression (also see 2.2) [53]. Nonsense or missense mutations that cause loss of HDAC8 activity resulted in SMC3 hyperacetylation and chromatin retention of the cohesin complex during mitosis [53]. CdLS patients with HDAC8 mutations display similar phenotypes as the patients with NIPBL mutations [53]. Furthermore, cohesin component Rad21 mutations were found in patients with a CdLS-like phenotype (OMIM 614701) [173]. In contrast to SMC1 and SMC3 mutations, patients with RAD21 mutations exhibit classical CdLS physical phenotypic characteristics (growth retardation, minor skeletal anomalies, and facial features) but have mild or no cognitive impairment [173]. Taken together, mutations of cohesin subunits and the regulators of cohesin loading to chromatin cause phenotypically related developmental disorders [167, 174].
5.3. Mutations of additional genes in the cohesin pathway?
While mutations in these proteins (NIPBL, HDAC8, SMC1A, SMC3, and possibly RAD21) may explain approximately 65% of CdLS patients, the cause of the remaining 35% remains unclear. For example, mutations in Pds5A and Pds5B, additional factors important for proper cohesin function in sister chromatid cohesion, also result in phenotypes in mouse models reminiscent of those observed in CdLS patients. However, no significant association of Pds5A or Pds5B mutations with CdLS has been observed [175, 176]. Nevertheless, mutations in additional genes involved in the cohesin pathway are expected to contribute to CdLS’ pathogenesis.
6. Mechanism of cohesinopathies
6.1. NIPBL haploinsufficiency causes CdLS
NIPBL haploinsufficiency is the major cause of CdLS (see above) [167, 177, 178]. Nipbl heterozygous mutant (Nipbl+/−) mice exhibit wide-ranging defects characteristic of CdLS, including small size, craniofacial anomalies, microbrachycephaly, heart defects, hearing abnormalities, low body fat, and delayed bone maturation, confirming that partial reduction of Nipbl is sufficient to cause a CdLS-like phenotype [179]. The mutant mice demonstrated only a 25–30% decrease in Nipbl transcripts, suggesting compensatory upregulation of the intact allele, which apparently is not sufficient to block development of the phenotype. Consistent with this, as little as a 15% decrease in NIPBL expression was shown to cause CdLS, though mild, in patients [180, 181]. These observations indicate the extreme sensitivity of mammalian development to NIPBL/Nipbl gene dosage.
6.2. NIPBL haploinsufficiency exhibits no significant sister chromatid cohesion defect
There appears to be a functional hierarchy for cohesin in which the most essential function, which is resistant to partial reduction of cohesin, is its role in sister chromatid cohesion and proper segregation of chromosomes (reviewed in [12]). The differential sensitivities of cohesin functions to cohesin depletion were most systematically demonstrated in yeast with different degrees of cohesin protein reduction [182]. Namely, mitotic sister chromatid cohesion is most resistant to partial reduction of cohesin. Similar observations were made in Drosophila and in human cells, in which partial depletion of cohesin by siRNA does not lead to any significant sister chromatid cohesion defect [69, 115]. Consistent with these findings, CdLS patient cells do not exhibit any obvious sister chromatid cohesion abnormalities [183–186]. This is in contrast to RBS, in which premature sister chromatid separation serves as a prototypical cellular phenotype for the disorder [163]. Though it is currently unclear how sister chromatid cohesion defects specifically contribute to the pathogenesis of RBS, distinct mechanisms are likely involved in the development of this cohesinopathy as opposed to CdLS.
6.3. DNA repair
Increased DNA damage sensitivity appears to be a general feature of cohesinopathies, as it has been reported in RBS, CdLS, and CdLS-like disorder patient cells [173, 184, 186–188]. A study in yeast suggested that an RBS-associated ESCO2 catalytic mutation impairs HR repair [189]. While no obvious HR repair defect was detected in NIPBL-mutated CdLS patient cells, increased chromosome aberrations indicative of a DNA repair defect were observed in SMC1- and SMC3-mutant CdLS patient cells [184, 186]. Cells with the Rad21 mutation found in the CdLS-like disorder also exhibited a repair defect, although impairment of the HR repair pathway was not specifically confirmed [173]. Nevertheless, the defect does not appear to result in prominent genome instability and/or increased cancer incidence [190]. Thus, how the increased DNA damage sensitivity contributes to the disorder’s pathogenesis is currently unclear.
6.4. Nipbl reduction results in decreased cohesin binding and gene expression changes
As discussed above, NIPBL mutations in both CdLS patient cells and in mouse models cause little or no chromatid cohesion defect, suggesting that the developmental abnormalities are a result of defective cohesin-mediated gene regulation [179, 181]. In both patient lymphoblasts and Nipbl-mutant mouse tissues and cells, the partial decrease of Nipbl expression is associated with pervasive, though small, alterations in gene expression. It was proposed, therefore, that diffuse, relatively mild expression perturbations collectively contribute to the developmental defect phenotype. Supporting this model, combined depletion of Nipbl target genes indeed recapitulates the Nipbl depletion phenotype in zebrafish [191]. As noted above, it was shown that Nipbl haploinsufficiency causes both decreased cohesin binding at the β-globin locus in embryonic liver as well as decreased long-distance chromatin interactions (involving both CTCF sites and non-CTCF sites). In particular, reduced chromatin interactions between the enhancer and adult globin genes appear to contribute to decreased globin gene expression [82]. One can envision that diminished cohesin-mediated long-distance chromatin interactions could affect gene regulation genome-wide, resulting in widespread disruption of normal gene expression in a cell type- and differentiation stage-specific manner.
6.5. Cohesinopathy may be a ribosomopathy?
Mutations of genes that impair ribosomal RNA (rRNA) transcription or ribosome biogenesis were found to be associated with various human genetic disorders, many of which are accompanied by growth and mental retardation. These include Treacher Collins Syndrome, Bloom’s and Werner Syndromes, Cockayne Syndrome, and Shwachman–Diamond Syndrome, which can all be considered to be “ribosomopathies” [192].
Recently, the effects of cohesinopathy disorder mutations of ESCO2 (RBS), NIPBL (severe CdLS) and SMC1 (mild CdLS) genes were evaluated by introducing analogous mutations in the corresponding homolog genes Eco1, Scc2, and Smc1 in S. cerevisiae [193]. It was found that Eco1 and, to lesser extent, Smc1 mutations (but not Scc2 mutation), caused decreased rRNA production and ribosomal biogenesis resulting in translational defects [193]. Similar defects were observed in RBS patient cells in which ESCO2 (the Eco1 homolog) is mutated, raising the intriguing possibility that RBS is in fact a ribosomopathy [193]. Although cohesin is known to bind to ribosomal DNA [116, 194], the underlying mechanism of the defect caused by ESCO2 mutation is currently unknown. Whether similar defects contribute to CdLS with NIPBL haploinsufficiency has not been addressed. However, since growth and mental retardation appear to be common phenotypes shared between various ribosomopathies and CdLS, it is possible that defective nucleolar/ribosomal function significantly contributes to CdLS and CdLSlike disorders as well.
6.6. NIPBL function beyond cohesin loading?
The wide range of defects observed in CdLS shows that abnormalities of cohesin-related functions have significant impact throughout development and on multiple cellular differentiation processes. Mutation of the genes involved in cohesin function and regulation can result in overlapping but not identical phenotypes. Zebrafish mutant analyses of ESCO2, Nipbl, SMC1, and Rad21 revealed only a modest overlap of affected genes [164, 195]. Despite the evidence that cohesin function is affected by NIPBL haploinsufficiency, CdLS cases with NIPBL mutations/haploinsufficiency tend to have a more severe phenotype compared to those with cohesin mutations (increased severity of mental retardation, growth impairment, or structural abnormalities of the limbs and other organ systems) [171–173]. This raises the possibility that NIPBL may in fact govern other pathways in addition to cohesin loading. For example, NIPBL may dictate the chromatin loading of other SMC complexes as seen in yeast [132, 133], though, unlike in yeast [133, 196], no obvious chromosome condensation defect (indicative of condensin dysfunction) was reported to be associated with Nipbl mutation in mammalian cells.
7. Facioscapulohumeral dystrophy (FSHD) as a new cohesinopathy disorder?
FSHD is the third most common heritable muscular dystrophy in the U.S. It is characterized by progressive wasting of facial, shoulder, and upper arm musculature, which can spread to the abdominal and foot-extensor muscles [197–199]. The genetics underlying FSHD are highly unusual; the majority of FSHD cases (>95%) are associated with monoallelic deletion of D4Z4 macrosatellite repeat sequences clustered at the subtelomeric region of chromosome 4q (4qter D4Z4) (FSHD1 (MIM 158900)) [197, 200]. There are between one and ten repeats in the contracted 4qter allele in FSHD1 patient cells, in contrast to 11~150 copies in normal cells. In the more rare form of FSHD (<5% of cases) (FSHD2) there is no D4Z4 repeat contraction, though phenotypically FSHD1 and FSHD2 are largely identical [201].
D4Z4 is a 3.3 kb repeat that contains an open reading frame (ORF) for the double-homeobox transcription factor DUX4 retrogene [202–204]. Artificial overexpression of the fulllength DUX4 (DUX4fl) protein caused a myoblast differentiation defect in human myoblasts and mouse C2C12 cells [205, 206]. Only those individuals with a 4qA haplotype with specific single-nucleotide polymorphisms in the region distal to the last D4Z4 repeat (creating a canonical polyadenylation signal for the DUX4 transcript) develop FSHD, strongly suggesting that DUX4fl mRNA expression is critical for FSHD pathogenesis [207].
7.1. FSHD is associated with disruption of transcriptionally repressive chromatin organization at 4qD4Z4
D4Z4 chromatin normally harbors the transcriptionally repressive histone modification marks histone H3 lysine 9 trimethylation (H3K9me3) and H3K27me3 (Fig. 4A) [116]. Interestingly, H3K9me3 is significantly diminished at D4Z4 repeat regions in both FSHD1 and FSHD2 patient cells, but not in other muscular dystrophies [116, 208]. This change is also found in FSHD patient lymphoblasts, indicating that the loss is not an epiphenomenon of the dystrophic phenotype and suggesting that it occurs early in development before lineage separation [116].
D4Z4 DNA was also shown to be hypermethylated in normal cells (Fig. 4A). Intriguingly, D4Z4 DNA is hypomethylated in both FSHD1 and FSHD2 [209]. D4Z4 DNA, however, is also severely hypomethylated in the Immunodeficiency–Centromeric instability–Facial anomalies (ICF) syndrome, which is phenotypically distinct from FSHD [209, 210]. Thus, the loss of DNA methylation alone is insufficient to cause FSHD. It should also be noted that H3K9me3 is intact in ICF cells or 5-AzaC-treated (DNA methylation-inhibited) cells, indicating that the H3K9me3 loss is not a downstream consequence of DNA hypomethylation [116]. Whether DNA hypomethylation is triggered by H3K9me3 loss is not known. Nevertheless, concomitant loss of H3K9me3 and DNA methylation at D4Z4 indicates that FSHD is an epigenetic abnormality disease.
Although the molecular basis for heterochromatin loss is unclear, the concomitant loss of DNA methylation and H3K9me3 may be directly related to the etiology of FSHD. Cohesin and HP1γ are recruited to D4Z4 in an H3K9me3-dependent manner and are therefore lost in FSHD cells [116]. Interestingly, the two factors require each other for D4Z4 binding, demonstrating the active role of cohesin in heterochromatin organization in human cells (Fig. 4A) [116]. This is analogous to the subtelomeric heterochromatin repeats in S. pombe in which cohesin and Swi6 are recruited in a mutually dependent manner and function in gene silencing [114]. Thus, FSHD can also be considered to be a cohesinopathy, in which D4Z4 heterochromatin-associated cohesin function is specifically disrupted. It is speculated that the loss of heterochromatin contributes to the expression of DUX4fl in FSHD. However, this has not been explicitly demonstrated.
A single copy of D4Z4 repeat sequence was shown to recruit CTCF and A-type lamins and to function as an insulator [211]. However, this binding was lost as the D4Z4 repeats were multimerized, simulating normal non-contracted D4Z4 alleles. Furthermore, CTCF binding is known to be DNA methylation-sensitive [212]. Thus, CTCF binding may be induced in FSHD cells in which D4Z4 heterochromatin, including DNA methylation as well as cohesin binding, is largely lost. Cohesin binding to D4Z4 heterochromatin, therefore, is through an H3K9me3-HP1γ pathway and is independent of CTCF.
7.2. An SMC homolog, SMCHD1, is mutated in FSHD2 and in severe cases of FSHD1
A recent study found that SMCHD1, an epigenetic gene silencer involved in the maintenance of DNA methylation and X inactivation [213, 214], binds to D4Z4 and plays a role in DUX4 gene repression (Fig. 4A) [215]. Importantly, this gene is mutated in many FSHD2 patients (OMIM158901) as well as in severe cases of FSHD1 in conjunction with D4Z4 contraction [215]. How SMCHD1 is recruited to D4Z4 remains unclear, but a recent study indicated that SMCHD1 is recruited to H3K9me3 domains through interaction with HBiX1, an HP1 binding protein, and contributes to the compaction of the inactive X chromosome [198]. This raises the intriguing possibility that in normal cells SMCHD1 is recruited to D4Z4 by the H3K9me3/HP1γ/cohesin heterochromatin, the loss of which in FSHD results in decreased binding of SMCHD1 and subsequent derepression of the DUX4 gene.
8. Concluding remarks
The recent explosion of genome-wide analyses as well as mechanistic studies have established cohesin as a key chromatin organizer in both mitosis and in interphase, influencing virtually all aspects of genomic function. Studies of cohesinopathies highlight the exquisite sensitivity of developmental processes in multiple cell types to subtle dysfunction of cohesin and its associated factors. Further interrogation of mechanistic details (e.g., in vitro reconstitution of cohesin-mediated processes) and how cohesin-mediated genomic organization is integrated into and controlled by other biological processes and signaling pathways, are two exciting areas for further investigation. This will provide significant insight into both basic questions concerning chromosome dynamics and the mechanisms underlying cohesinopathies.
Highlights.
Cohesin is a key chromatin organizer in both mitosis and in interphase.
Cohesin regulates gene expression through different mechanisms and chromatin contexts.
Differential protein and RNA interactions dictate cohesin targeting and function.
Dysfunction of cohesin and its regulators causes related developmental disorders.
Facioscapulohumeral dystrophy (FSHD) may be a new cohesinopathy disorder.
Acknowledgment
The work in the Yokomori laboratory described was supported in part by NIH AR058548, HD062951, MDA4026, and the David and Helen Younger Research Fellowship Research Grant from the FSH Society (FSHS-DHY-001). We apologize to the researchers whose work we could not cite due to space limitations.
List of abbreviations
- ChIP-loop
3C combined with ChIP
- ATR-X
Alpha-Thalassemia mental Retardation, X-linked
- ARS
Autonomously replicating sequence
- CTCF
CCCTC-binding factor
- ChIP
Chromatin immunoprecipitation
- ChIA-PET
Chromatin Interaction Analysis by Paired-End Tag sequencing
- 3C
Chromosome conformation capture
- 5C
Chromosome conformation capture carbon copy
- 4C-seq
Circular chromosome conformation capture followed by high-throughput sequencing
- CdLS
Cornelia de Lange Syndrome
- DSB
DNA double-strand break repair
- eRNAs
Enhancer RNAs
- ER
Estrogen receptor
- H3K9me
H3 lysine 9 methylation
- H3K9me3
H3 lysine 9 trimethylation
- H3K27me3
H3 lysine 27 trimethylation
- H4K20me3
H4 lysine 20 trimethylation
- HR
Homologous recombination repair
- iPSCs
Induced pluripotency cells
- LCR
Locus control region
- mESCs
Mouse embryonic stem cells
- ncRNA
Non-coding RNA
- rRNA
Ribosomal RNA
- RNAPII
RNA polymerase II
- RNAPIII
RNA polymerase III
- RBS
Roberts’ Syndrome
- SRA
Steroid receptor RNA activator
- SMC
Structural Maintenance of Chromosomes
- TADs
Topologically associating domains
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Strunnikov AV, Larionov VL, Koshland D. SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J. Cell Biol. 1993;123:1635–1648. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr. Biol. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 3.Weitzer S, Lehane C, Uhlmann F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr. Biol. 2003;13:1930–1940. doi: 10.1016/j.cub.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 4.Lieb JD, Albrecht MR, Chuang P-T, Meyer BJ. MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell. 1998;92:265–277. doi: 10.1016/s0092-8674(00)80920-4. [DOI] [PubMed] [Google Scholar]
- 5.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell. Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 6.Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 7.Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 2012;26:1659–1678. doi: 10.1101/gad.194746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piazza I, Haering CH, Rutkowska A. Condensin: crafting the chromosome landscape. Chromosoma. 2013;122:175–190. doi: 10.1007/s00412-013-0405-1. [DOI] [PubMed] [Google Scholar]
- 9.Thadani R, Uhlmann F, Heeger S. Condensin, chromatin crossbarring and chromosome condensation. Curr. Biol. 2012;22:R1012–R1021. doi: 10.1016/j.cub.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 10.De Piccoli G, Torres-Rosell J, Aragón L. The unnamed complex: what do we know about Smc5-Smc6? Chrom. Res. 2009;17:251–263. doi: 10.1007/s10577-008-9016-8. [DOI] [PubMed] [Google Scholar]
- 11.Stephan AK, Kliszczak M, Morrison CG. The Nse2/Mms21 SUMO ligase of the Smc5/6 complex in the maintenance of genome stability. FEBS Lett. 2011;585:2907–2913. doi: 10.1016/j.febslet.2011.04.067. [DOI] [PubMed] [Google Scholar]
- 12.Chien R, Zeng W, Ball AR, Yokomori K. Cohesin: a critical chromatin organizer in mammalian gene regulation. Biochem. Cell Biol. 2011;89:445–458. doi: 10.1139/o11-039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood AJ, Severson AF, Meyer BJ. Condensin and cohesin complexity: the expanding repertoire of functions. Nat. Rev. Genet. 2010;11:391–404. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu N, Yu H. The Smc complexes in DNA damage response. Cell Biosci. 2012;2:5. doi: 10.1186/2045-3701-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Losada A, Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 2005;19:1269–1287. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- 16.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S cerevisiae . Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 19.Anderson DE, Losada A, Erickson HP, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 21.Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 22.Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Lowe J. Structure and stability of cohesin's Smc1-kleisin interaction. Mol Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 23.Ivanov D, Nasmyth K. A topological interaction between cohesin rings and a circular minichromosome. Cell. 2005;122:849–860. doi: 10.1016/j.cell.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 24.Ivanov D, Nasmyth K. A physical assay for sister chromatid cohesion in vitro. Mol. Cell. 2007;27:300–310. doi: 10.1016/j.molcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 26.Chan KL, Roig MB, Hu B, Beckouët F, Metson J, Nasmyth K. Cohesin's DNA exit gate is distinct from its entrance gate and is regulated by acetylation. Cell. 2012;150:961–974. doi: 10.1016/j.cell.2012.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nasmyth K. Cohesin: a catenase with separate entry and exit gates? Nat. Cell Biol. 2011;13:1170–1177. doi: 10.1038/ncb2349. [DOI] [PubMed] [Google Scholar]
- 28.Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J. Cell Biol. 2000;150:405–416. doi: 10.1083/jcb.150.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters J-M. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 2000;151:749–761. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canudas S, Houghtaling BR, Kim JY, Dynek JN, Chang WG, Smith S. Protein requirements for sister telomere association in human cells. EMBO J. 2007;26:4867–4878. doi: 10.1038/sj.emboj.7601903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canudas S, Smith S. Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J. Cell Biol. 2009;187:165–173. doi: 10.1083/jcb.200903096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Remeseiro S, Cuadrado A, Gómez-López G, Pisano DG, Losada A. A unique role of cohesin-SA1 in gene regulation and development. EMBO J. 2012;31:2090–2102. doi: 10.1038/emboj.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, Toretsky JA, Rosenberg SA, Shukla N, Ladanyi M, Samuels Y, James CD, Yu H, Kim JS, Waldman T. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Remeseiro S, Cuadrado A, Carretero M, Martínez P, Drosopoulos WC, Cañamero M, Schildkraut CL, Blasco MA, Losada A. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 2012;31:2076–2089. doi: 10.1038/emboj.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 36.Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol. Cell. 2006;23:787–799. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 37.Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, Rollins RA, Erdjument-Bromage H, Tempst P, Benard CY, Hekimi S, Newbury SF, Strachan T. Metazoan Scc4 Homologs Link Sister Chromatid Cohesion to Cell and Axon Migration Guidance. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040242. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr. Biol. 2006;16:863–874. doi: 10.1016/j.cub.2006.03.049. [DOI] [PubMed] [Google Scholar]
- 39.Gruber S, Arumugam P, Katou Y, Kuglitsc hD, Helmhart W, Shirahige K, Nasmyth K. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127:523–537. doi: 10.1016/j.cell.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 40.Hou F, Zou H. Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion. Mol. Biol. Cell. 2005;16:3908–3918. doi: 10.1091/mbc.E04-12-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, Peters JM. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell. 2010;143:737–749. doi: 10.1016/j.cell.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 42.Ben-Shahar TR, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 43.Heidinger-Pauli JM, Unal E, Koshland D. Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol. Cell. 2009;34:311–321. doi: 10.1016/j.molcel.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ünal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, Zhang P, Kim ST, Pan X, Qin J. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 46.Higashi TL, Ikeda M, Tanaka H, Nakagawa T, Bando M, Shirahige K, Kubota Y, Takisawa H, Masukata H, Takahashi TS. The prereplication complex recruits XEco2 to chromatin to promote cohesin acetylation in Xenopus egg extracts. Curr. Biol. 2012;22:977–988. doi: 10.1016/j.cub.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 47.Buheitel J, Stemmann O. Prophase pathway-dependent removal of cohesin from human chromosomes requires opening of the Smc3–Scc1 gate. EMBO J. 2013;32:666–676. doi: 10.1038/emboj.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eichinger CS, Kurze A, Oliveira RA, Nasmyth K. Disengaging the Smc3/kleisin interface releases cohesin from Drosophila chro-mosomes during interphase and mitosis. EMBO J. 2013;32:656–665. doi: 10.1038/emboj.2012.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chatterjee A, Zakian S, Hu XW, Singleton MR. Structural insights into the regulation of cohesion establishment by Wpl1. EMBO J. 2013;32:677–687. doi: 10.1038/emboj.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. Cohesin acetylation speeds the replication fork. Nature. 2009;462:231–234. doi: 10.1038/nature08550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beckouët F, Hu B, Roig MB, Sutani T, Komata M, Uluocak P, Katis VL, Shirahige K, Nasmyth K. An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol. Cell. 2010;39:689–699. doi: 10.1016/j.molcel.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borges V, Lehane C, Lopez-Serra L, Flynn H, Skehel M, Rolef Ben-Shahar T, Uhlmann F. Hos1 deacetylates Smc3 to close the cohesin acetylation cycle. Mol. Cell. 2010;39:677–688. doi: 10.1016/j.molcel.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 53.Deardorff MA, Bando M, Nakato R, Watrin E, Itoh T, Minamino M, Saitoh K, Komata M, Katou Y, Clark D, Cole KE, De Baere E, Decroos C, Di Donato N, Ernst S, Francey LJ, Gyftodimou Y, Hirashima K, Hullings M, Ishikawa Y, Jaulin C, Kaur M, Kiyono T, Lombardi PM, Magnaghi-Jaulin L, Mortier GR, Nozaki N, Petersen MB, Seimiya H, Siu VM, Suzuki Y, Takagaki K, Wilde JJ, Willems PJ, Prigent C, Gillessen-Kaesbach G, Christianson DW, Kaiser FJ, Jackson LG, Hirota T, Krantz ID, Shirahige K. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489:313–317. doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiong B, Lu S, Gerton JL. Hos1 is a lysine deacetylase for the Smc3 subunit of cohesin. Curr. Biol. 2010;20:1660–1665. doi: 10.1016/j.cub.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 55.Hanna JS, Kroll ES, Lundblad V, Spencer FA. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 2001;21:3144–3158. doi: 10.1128/MCB.21.9.3144-3158.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mayer ML, Gygi SP, Aebersold R, Hieter P. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae . Mol. cell. 2001;7:959–970. doi: 10.1016/s1097-2765(01)00254-4. [DOI] [PubMed] [Google Scholar]
- 57.Wang Z, Castaño IB, De Las Peñas A, Adams C, Christman MF. Pol κ: a DNA polymerase required for sister chromatid cohesion. Science. 2000;289:774–779. doi: 10.1126/science.289.5480.774. [DOI] [PubMed] [Google Scholar]
- 58.Moldovan GL, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell. 2006;23:723–732. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 59.Leman AR, Noguchi C, Lee CY, Noguchi E. Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion. J. Cell Sci. 2010;123:660–670. doi: 10.1242/jcs.057984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith-Roe SL, Patel SS, Simpson DA, Zhou YC, Rao S, Ibrahim JG, Kaiser-Rogers KA, Cordeiro-Stone M, Kaufmann WK. Timeless functions independently of the Tim-Tipin complex to promote sister chromatid cohesion in normal human fibroblasts. Cell Cycle. 2011;10:1618–1624. doi: 10.4161/cc.10.10.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ström L, Lindroos HB, Shirahige K, Sjögren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 62.Waizenegger IC, Hauf S, Meinke A, Peters J-M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- 63.Carretero M, Remeseiro S, Losada A. Cohesin ties up the genome. Curr. Opin. Cell Biol. 2010;22:781–787. doi: 10.1016/j.ceb.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 64.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 2009;43:525–528. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 65.Shintomi K, Hirano T. Sister chromatid resolution: a cohesin releasing network and beyond. Chromosoma. 2010;119:459–467. doi: 10.1007/s00412-010-0271-z. [DOI] [PubMed] [Google Scholar]
- 66.Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu. Rev. Dev. Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 67.Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- 68.Schöckel L, Möckel M, Mayer B, Boos D, Stemmann O. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat. Cell Biol. 2011;13:966–972. doi: 10.1038/ncb2280. [DOI] [PubMed] [Google Scholar]
- 69.Kong X, Ball ARJ, Sonoda E, Feng J, Takeda S, Fukagawa T, Yen TJ, Yokomori K. Cohesin associates with spindle poles in a mitosis-specific manner and functions in spindle assembly in vertebrate cells. Mol. Biol. Cell. 2009;20:1289–1301. doi: 10.1091/mbc.E08-04-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wong RW, Blobel G. Cohesin subunit SMC1 associates with mitotic microtubules at the spindle pole. Proc. Natl. Acad. Sci. 2008;105:15441–15445. doi: 10.1073/pnas.0807660105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gregson HC, Schmiesing JA, Kim J-S, Kobayashi T, Zhou S, Yokomori K. A potential role for human cohesin in mitotic spindle aster assembly. J. Biol. Chem. 2001;276:47575–47582. doi: 10.1074/jbc.M103364200. [DOI] [PubMed] [Google Scholar]
- 72.Potts PR, Porteus MH, Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377–3388. doi: 10.1038/sj.emboj.7601218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sjogren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol. 2001;11:991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 74.Kim S-T, Xu B, Kastan MB. Involvement of the cohesin protein, Smc1, in Atmdependent and independent responses to DNA damage. Genes & Dev. 2002;16:560–570. doi: 10.1101/gad.970602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM–NBS1–BRCA1 pathway. Genes Dev. 2004;18:1423–1438. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yazdi PT, Wang Y, Zhao S, Patel N, Lee EY-HP, Qin J. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes & Dev. 2002;16:571–582. doi: 10.1101/gad.970702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luo H, Li Y, Mu JJ, Zhang J, Tonaka T, Hamamori Y, Jung SY, Wang Y, Qin J. Regulation of intra-S phase checkpoint by IR-dependent and IR-independent phosphorylation of SMC3. J. Biol. Chem., E pub. 2008 doi: 10.1074/jbc.M802299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guillou E, Ibarra A, Coulon V, Casado-Vela J, Rico D, Casal I, Schwob E, Losada A, Méndez J. Cohesin organizes chromatin loops at DNA replication factories. Genes Dev. 2010;24:2812–2822. doi: 10.1101/gad.608210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc. Natl. Acad. Sci. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chien R, Zeng W, Kawauchi S, Bender MA, Santos R, Gregson HC, Schmiesing JA, Newkirk D, Kong X, Ball ARJ, Calof AL, Lander AD, Groudine MT, Yokomori K. Cohesin mediates chromatin interactions that regulate mammalian β-globin expression. J. Biol. Chem. 2011;286:17870–17878. doi: 10.1074/jbc.M110.207365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo Y, Monahan K, Wu H, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc. Natl. Acad. Sci. 2012;109:21081–21086. doi: 10.1073/pnas.1219280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dorsett D, Merkenschlager M. Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr. Opin. Cell Biol. 2013 doi: 10.1016/j.ceb.2013.02.003. S0955-0674(13)00026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Merkenschlager M, Odom DT. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013;152:1285–1297. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 88.Faure AJ, Schmidt D, Watt S, Schwalie PC, Wilson MD, Xu H, Ramsay RG, Odom DT, Flicek P. Cohesin regulates tissue-specific expression by stabilizing highly occupied cisregulatory modules. Genome Res. 2012;22:2163–2175. doi: 10.1101/gr.136507.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTCbinding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 90.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 92.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Degner SC, Verma-Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, Vettermann C, Lin YC, Ju Z, Schulz D, Murre CS, Birshtein BK, Schork NJ, Schlissel MS, Riblet R, Murre C, Feeney AJ. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc. Natl. Acad. Sci. 2011;108:9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Majumder P, Boss JM. Cohesin regulates MHC class II genes through interactions with MHC class II insulators. J. Immunol. 2011;187:4236–4244. doi: 10.4049/jimmunol.1100688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Majumder P, Gomez JA, Chadwick BP, Boss JM. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J Exp Med. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim YJ, Cecchini KR, Kim TH. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proc. Natl. Acad. Sci. 2011;108:7391–7396. doi: 10.1073/pnas.1018279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, Marks H, Adams DJ, Schatz DG, Aragon, Fisher AG, Krangel MS, Nasmyth K, Merkenschlager M. A role for cohesin in T-cellreceptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schmidt D, Schwalie P, Ross-Innes CS, Hurtado A, Brown G, Carroll J, Flicek P, Odom D. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Prickett AR, Barkas N, McCole RB, Hughes S, Amante SM, Schulz R, Oakey RJ. Genome-wide and parental allele-specific analysis of CTCF and cohesin DNA binding in mouse brain reveals a tissue-specific binding pattern and an association with imprinted differentially methylated regions. Genome Res. 2013;23:1624–1635. doi: 10.1101/gr.150136.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 101.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 102.Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol. Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 105.Wei Z, Gao F, Kim S, Yang H, Lyu J, An W, Wang K, Lu W. Klf4 Organizes Long-Range Chromosomal Interactions with the Oct4 Locus in Reprogramming and Pluripotency. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 106.Apostolou E, Ferrari F, Walsh RM, Bar-Nur O, Stadtfeld M, Cheloufi S, Stuart HT, Polo JM, Ohsumi TK, Borowsky ML, Kharchenko PV, Park PJ, Hochedlinger K. Genome-wide Chromatin Interactions of the Nanog Locus in Pluripotency, Differentiation, and Reprogramming. Cell Stem Cell. 2013:S1934–S5909. doi: 10.1016/j.stem.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, Bland MJ, Wagstaff W, Dalton S, McDevitt TC, Sen R, Dekker J, Taylor J, Corces VG. Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell. 2013 doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Demare LE, Leng J, Cotney J, Reilly SK, Yin J, Sarro R, Noonan JP. The genomic landscape of cohesin-associated chromatin interactions. Genome Res. 2013 doi: 10.1101/gr.156570.113. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zeng W, Mortazavi A. Technical considerations for functional sequencing assays. Nat. Immunol. 2012;13:802–807. doi: 10.1038/ni.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nitzsche A, Paszkowski-Rogacz M, Matarese F, Janssen-Megens EM, Hubner NC, Schulz H, de Vries I, Ding L, Huebner N, Mann M, Stunnenberg HG, Buchholz F. RAD21 Cooperates with Pluripotency Transcription Factors in the Maintenance of Embryonic Stem Cell Identity. PLoS One. 2011;6:e19470. doi: 10.1371/journal.pone.0019470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang H, Jiao W, Sun L, Fan J, Chen M, Wang H, Xu X, Shen A, Li T, Niu B, Ge S, Li W, Cui J, Wang G, Sun J, Fan X, Hu X, Mrsny RJ, Hoffman AR, Hu J-F. Intrachromosomal Looping Is Required for Activation of Endogenous Pluripotency Genes during Reprogramming. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 112.Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila Nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol. Cell. Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dheur S, Saupe SJ, Genier S, Vazquez S, Javerzat JP. Role for cohesin in the formation of a heterochromatic domain at fission yeast subtelomeres. Mol. Cell Biol. 2011;31:1088–1097. doi: 10.1128/MCB.01290-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schaaf CA, Misulovin Z, Sahota G, Siddiqui AM, Schwartz YB, Kahn TG, Pirrotta V, Gause M, Dorsett D. Regulation of the Drosophila Enhancer of split and invectedengrailed gene complexes by sister chromatid cohesion proteins. PLoS One. 2009;4:e6202. doi: 10.1371/journal.pone.0006202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zeng W, de Greef JC, Chen Y-Y, Chien R, Kong X, Gregson HC, Winokur ST, Pyle A, Robertson KD, Schmiesing JA, Kimonis VE, Balog J, R FR, Ball J, R A, Lock LF, Donovan PJ, van der Maarel S, Yokomori K. Specific loss of histone H3 lysine 9 trimethylation and HP1γ/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 118.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 119.Cunningham MD, Gause M, Cheng Y, Noyes A, Dorsett D, Kennison JA, Kassis JA. Wapl antagonizes cohesin binding and promotes Polycomb-group silencing in Drosophila. Development. 2012;139:4172–4179. doi: 10.1242/dev.084566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schaaf CA, Misulovin Z, Gause M, Koenig A, Gohara DW, Watson A, Dorsett D. Cohesin and polycomb proteins functionally interact to control transcription at silenced and active genes. PLoS Genet. 2013;9:e1003560. doi: 10.1371/journal.pgen.1003560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fay A, Misulovin Z, Li J, Schaaf CA, Gause M, Gilmour DS, Dorsett D. Cohesin selectively binds and regulates genes with paused RNA polymerase. Curr. Biol. 2011;21:1624–1634. doi: 10.1016/j.cub.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, Macarthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, Dorsett D. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schaaf CA, Kwak H, Koenig A, Misulovin Z, Gohara DW, Watson A, Zhou Y, Lis JT, Dorsett D. Genome-wide control of RNA polymerase II activity by cohesin. PLoS Genet. 2013;9:e1003382. doi: 10.1371/journal.pgen.1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gomes NP, Espinosa JM. Gene-specific repression of the p53 target gene PUMA via intragenic CTCF-Cohesin binding. Genes Dev. 2010;24:1022–1034. doi: 10.1101/gad.1881010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kang H, Lieberman PM. Mechanism of glycyrrhizic acid inhibition of Kaposi's sarcomaassociated herpesvirus: disruption of CTCF-cohesin-mediated RNA polymerase II pausing and sister chromatid cohesion. J. Virol. 2011;85:11159–11169. doi: 10.1128/JVI.00720-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Paredes SH, Melgar MF, Sethupathy P. Promoter-proximal CCCTC-factor binding is associated with an increase in the transcriptional pausing index. Bioinformatics. 2013;29:1485–1487. doi: 10.1093/bioinformatics/bts596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB, Renkawitz R. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009;28:877–888. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr. Biol. 2004;14:1598–1603. doi: 10.1016/j.cub.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 130.Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat. Cell Biol. 2004;6:991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- 131.Bermudez VP, Farina A, Higashi TL, Du F, Tappin I, Takahashi TS, Hurwitz J. In vitro loading of human cohesin on DNA by the human Scc2-Scc4 loader complex. Proc. Natl. Acad. Sci. 2012;109:9366–9371. doi: 10.1073/pnas.1206840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lindroos HB, Ström L, Itoh T, Katou Y, Shirahige K, Sjögren C. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell. 2006;22:755–767. doi: 10.1016/j.molcel.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 133.D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kranz AL, Jiao CY, Winterkorn LH, Albritton SE, Kramer M, Ercan S. Genomewide analysis of condensin binding in Caenorhabditis elegans. Genome Biol. 2013;14:R112. doi: 10.1186/gb-2013-14-10-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lightfoot J, Testori S, Barroso C, Martinez-Perez E. Loading of Meiotic Cohesin by SCC-2 Is Required for Early Processing of DSBs and for the DNA Damage Checkpoint. Curr. Biol. 2011;21:1421–1430. doi: 10.1016/j.cub.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 136.Bausch C, Noone S, Henry JM, Gaudenz K, Sanderson B, Seidel C, Gerton JL. Transcription alters chromosomal locations of cohesin in Saccharomyces cerevisiae. Mol. Cell Biol. 2007;27:8522–8532. doi: 10.1128/MCB.01007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Glynn EF, Megee PC, Yu HG, Mistrot C, Ünal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ocampo-Hafalla MT, Uhlmann F. Cohesin loading and sliding. J. Cell Sci. 2011;124:685–691. doi: 10.1242/jcs.073866. [DOI] [PubMed] [Google Scholar]
- 140.Kogut I, Wang J, Guacci V, Mistry RK, Megee PC. The Scc2/Scc4 cohesin loader determines the distribution of cohesin on budding yeast chromosomes. Genes Dev. 2009;23:2345–2357. doi: 10.1101/gad.1819409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hu B, Itoh T, Mishra A, Katoh Y, Chan KL, Upcher W, Godlee C, Roig MB, Shirahige K, Nasmyth K. ATP hydrolysis is required for relocating cohesin from sites occupied by its Scc2/4 loading complex. Curr. Biol. 2011;21:12–24. doi: 10.1016/j.cub.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Onn I, Koshland D. In vitro assembly of physiological cohesin/DNA complexes. Proc. Natl. Acad. Sci. 2011;108:12198–12205. doi: 10.1073/pnas.1107504108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xiao T, Wallace J, Felsenfeld G. Specific Sites in the C Terminus of CTCF Interact with the SA2 Subunit of the Cohesin Complex and Are Required for Cohesin-Dependent Insulation Activity. Mol. Cell Biol. 2011;31:2174–2183. doi: 10.1128/MCB.05093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Balakrishnan SK, Witcher M, Berggren TW, Emerson BM. Functional and molecular characterization of the role of CTCF in human embryonic stem cell biology. PLoS One. 2012;7:e42424. doi: 10.1371/journal.pone.0042424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–2555. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kernohan KD, Jiang Y, Tremblay DC, Bonvissuto AC, Eubanks JH, Mann MR, Bérubé NG. ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Dev. Cell. 2010;18:191–202. doi: 10.1016/j.devcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 147.Brand M, Ranish JA, Kummer NT, Hamilton J, Igarashi K, Francastel C, Chi TH, Crabtree GR, Aebersold R, Groudine M. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat. Struc. Mol. Biol. 2004;11:73–80. doi: 10.1038/nsmb713. [DOI] [PubMed] [Google Scholar]
- 148.Hakimi MA, Bochar DA, Schmiesing JA, Dong Y, Barak OG, Speicher DW, Yokomori K, Shiekhattar R. A chromatin remodeling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–998. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- 149.Fasulo B, Deuring R, Murawska M, Gause M, Dorighi KM, Schaaf CA, Dorsett D, Brehm A, Tamkun JW. The Drosophila MI-2 chromatin-remodeling factor regulates higherorder chromatin structure and cohesin dynamics in vivo. PLoS Genet. 2012;8:e1002878. doi: 10.1371/journal.pgen.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, Rosenfeld MG. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013 doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bernard P, Maure J-F, Partridge JF, Genier S, Javerzat J-P, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 152.Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SS, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 153.Koch B, Kueng S, Ruckenbauer C, Wendt KS, Peters JM. The Suv39h-HP1 histone methylation pathway is dispensable for enrichment and protection of cohesin at centromeres in mammalian cells. Chromosoma. 2008;117:199–210. doi: 10.1007/s00412-007-0139-z. [DOI] [PubMed] [Google Scholar]
- 154.Serrano A, Rodríguez-Corsino M, Losada A. Heterochromatin protein 1 (HP1) proteins do not drive pericentromeric cohesin enrichment in human cells. PLoS One. 2009;4:e5118. doi: 10.1371/journal.pone.0005118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shimura M, Toyoda Y, Iijima K, Kinomoto M, Tokunaga K, Yoda K, Yanagida M, Sata T, Ishizaka Y. Epigenetic displacement of HP1 from heterochromatin by HIV-1 Vpr causes premature sister chromatid separation. J. Cell Biol. 2011;194:721–735. doi: 10.1083/jcb.201010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lechner MS, Schultz DC, Negorev D, Maul GG, Rauscher FJr. The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem. Biophys. Res. Commun. 2005;331:929–937. doi: 10.1016/j.bbrc.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 157.Hahn M, Dambacher S, Dulev S, Kuznetsova AY, Eck S, Wörz S, Sadic D, Schulte M, Mallm JP, Maiser A, Debs P, von Melchner H, Leonhardt H, Schermelleh L, Rohr K, Rippe K, Storchova Z, Schotta G. Suv4-20h2 mediates chromatin compaction and is important for cohesin recruitment to heterochromatin. Genes Dev. 2013;27:859–872. doi: 10.1101/gad.210377.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fernius J, Nerusheva OO, Galander S, Alves Fde L, Rappsilber J, Marston AL. Cohesin-dependent association of scc2/4 with the centromere initiates pericentromeric cohesion establishment. Curr. Biol. 2013;23:599–606. doi: 10.1016/j.cub.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bose T, Gerton JL. Cohesinopathies, gene expression, and chromatin organization. J. Cell Biol. 2010;189:201–210. doi: 10.1083/jcb.200912129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liu J, Krantz ID. Cohesin and human disease. Annu. Rev. Genomics Hum. Genet. 2008;9:303–320. doi: 10.1146/annurev.genom.9.081307.164211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Vega H, Waisfisz Q, Gordillo M, Sakai N, Yanagihara I, Yamada M, van Gosliga D, Kayserili H, Xu C, Ozono K, Jabs EW, Inui K, Joenje H. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat. Genet. 2005;37:468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- 162.Dorsett D. Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma. 2007;116:1–13. doi: 10.1007/s00412-006-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tomkins D, Hunter A, Roberts M. Cytogenetic findings in Roberts-SC phocomelia syndrome(s) Am J Med Genet. 1979;4:17–26. doi: 10.1002/ajmg.1320040104. [DOI] [PubMed] [Google Scholar]
- 164.Mönnich M, Kuriger Z, Print CG, Horsfield JA. A zebrafish model of Roberts syndrome reveals that Esco2 depletion interferes with development by disrupting the cell cycle. PLoS One. 2011;6:e20051. doi: 10.1371/journal.pone.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Whelan G, Kreidl E, Wutz G, Egner A, Peters JM, G E. Cohesin acetyltransferase Esco2 is a cell viability factor and is required for cohesion in pericentric heterochromatin. EMBO J. 2012;31:71–82. doi: 10.1038/emboj.2011.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.DeScipio C, Kaur M, Yaeger D, Innis JW, Spinner NB, Jackson LG, Krantz ID. Chromosome rearrangements in cornelia de Lange syndrome (CdLS): report of a deR3 t(3;12)(p25.3;p13.3) in two half sibs with features of CdLS and review of reported CdLS cases with chromosome rearrangements. Am. J. Med. Genet. 2005;137:276–282. doi: 10.1002/ajmg.a.30857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liu J, Krantz ID. Cornelia de Lange syndrome, cohesin, and beyond. Clin. Genet. 2009;76:303–314. doi: 10.1111/j.1399-0004.2009.01271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M, Jackson LG. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat. genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat. Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 170.Gillis LA, McCallum J, Kaur M, DeScipio C, Yaeger D, Mariani A, Kline AD, Li HH, Devoto M, Jackson LG, Krantz ID. NIPBL mutational analysis in 120 individuals with Cornelia de Lange syndrome and evaluation of genotype-phenotype correlations. Am J Hum Genet. 2004;75:610–623. doi: 10.1086/424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat. Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- 172.Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodríguez C, Arnedo M, Loeys B, Kline AD, Wilson M, Lillquist K, Siu V, Ramos FJ, Musio A, Jackson LS, Dorsett D, Krantz ID. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am. J. Hum. Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Deardorff MA, Wilde JJ, Albrecht M, Dickinson E, Tennstedt S, Braunholz D, Mönnich M, Yan Y, Xu W, Gil-Rodríguez MC, Clark D, Hakonarson H, Halbach S, Michelis LD, Rampuria A, Rossier E, Spranger S, Van Maldergem L, Lynch SA, Gillessen-Kaesbach G, Lüdecke HJ, Ramsay RG, McKay MJ, Krantz ID, Xu H, Horsfield JA, Kaiser FJ. RAD21 mutations cause a human cohesinopathy. Am. J. Hum. Genet. 2012;90:1014–1027. doi: 10.1016/j.ajhg.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Castronovo P, Delahaye-Duriez A, Gervasini C, Azzollini J, Minier F, Russo S, Masciadri M, Selicorni A, Verloes A, Larizza L. Somatic mosaicism in Cornelia de Lange syndrome: a further contributor to the wide clinical expressivity? Clin. Genet. 2010;78:560–564. doi: 10.1111/j.1399-0004.2010.01408.x. [DOI] [PubMed] [Google Scholar]
- 175.Zhang B, Chang J, Fu M, Huang J, Kashyap R, Salavaggione E, Jain S, Kulkarni S, Deardorff MA, Uzielli ML, Dorsett D, Beebe DC, Jay PY, Heuckeroth RO, Krantz I, Milbrandt J. Dosage effects of cohesin regulatory factor PDS5 on mammalian development: implications for cohesinopathies. PLoS One. 2009;4:e5232. doi: 10.1371/journal.pone.0005232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhang B, Jain S, Song H, Fu M, Heuckeroth RO, Erlich JM, Jay PY, Milbrandt J. Mice lacking sister chromatid cohesion protein PDS5B exhibit developmental abnormalities reminiscent of Cornelia de Lange syndrome. Development. 2007;134:3191–3201. doi: 10.1242/dev.005884. [DOI] [PubMed] [Google Scholar]
- 177.Dorsett D, Krantz ID. On the molecular etiology of Cornelia de Lange syndrome. Ann. N. Y. Acad. Sci. 2009;1151:22–37. doi: 10.1111/j.1749-6632.2008.03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Selicorni A, Russo S, Gervasini C, Castronovo P, Milani D, Cavalleri F, Bentivegna A, Masciadri M, Domi A, Divizia MT, Sforzini C, Tarantino E, Memo L, Scarano G, Larizza L. Clinical score of 62 Italian patients with Cornelia de Lange syndrome and correlations with the presence and type of NIPBL mutation. Clin. Genet. 2007;72:98–108. doi: 10.1111/j.1399-0004.2007.00832.x. [DOI] [PubMed] [Google Scholar]
- 179.Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP, Chua A, Lao T, Lechner MS, Daniel JA, Nussenzweig A, Kitzes L, Yokomori K, Hallgrimsson B, Lander AD. Multiple organ system defects and transcriptional dysregulation in the nipbl mouse, a model of cornelia de lange syndrome. PLoS Genet. 2009;5:e1000650. doi: 10.1371/journal.pgen.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Borck G, Zarhrate M, Cluzeau C, Bal E, Bonnefont JP, Munnich A, Cormier-Daire V, Colleaux L. Father-to-daughter transmission of Cornelia de Lange syndrome caused by a mutation in the 5' untranslated region of the NIPBL Gene. Hum Mutat. 2006;27:731–735. doi: 10.1002/humu.20380. [DOI] [PubMed] [Google Scholar]
- 181.Liu J, Zhang Z, Bando M, Itoh T, Deardorff MA, Clark D, Kaur M, Tandy S, Kondoh T, Rappaport E, Spinner NB, Vega H, Jackson LG, Shirahige K, Krantz ID. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009;7:e1000119. doi: 10.1371/journal.pbio.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Heidinger-Pauli JM, Mert O, Davenport C, Guacci V, Koshland D. Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr Biol. 2010;20:957–963. doi: 10.1016/j.cub.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kaur M, Descipio C, McCallum J, Yaeger D, Devoto M, Jackson LG, Spinner NB, Krantz ID. Precocious sister chromatid separation (PSCS) in Cornelia de Lange syndrome. Am. J. Med. Genet. 2005;138:27–31. doi: 10.1002/ajmg.a.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Revenkova E, Focarelli ML, Susani L, Paulis M, Bassi MT, Mannini L, Frattini A, Delia D, Krantz I, Vezzoni P, Jessberger R, Musio A. Cornelia de Lange syndrome mutations in SMC1A or SMC3 affect binding to DNA. Hum. Mol. Genet. 2009;18:418–427. doi: 10.1093/hmg/ddn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Castronovo P, Gervasini C, Cereda A, Masciadri M, Milani D, Russo S, Selicorni A, Larizza L. Premature chromatid separation is not a useful diagnostic marker for Cornelia de Lange syndrome. Chromosome Res. 2009;17:763–771. doi: 10.1007/s10577-009-9066-6. [DOI] [PubMed] [Google Scholar]
- 186.Vrouwe MG, Elghalbzouri-Maghrani E, Meijers M, Schouten P, Godthelp BC, Bhuiyan ZA, Redeker EJ, Mannens MM, Mullenders LH, Pastink A, Darroudi F. Increased DNA damage sensitivity of Cornelia de Lange syndrome cells: evidence for impaired recombinational repair. Hum. Mol. Genet. 2007;16:1478–1487. doi: 10.1093/hmg/ddm098. [DOI] [PubMed] [Google Scholar]
- 187.van der Lelij P, Godthelp BC, van Zon W, van Gosliga D, Oostra AB, Steltenpool J, de Groot J, Scheper RJ, Wolthuis RM, Waisfisz Q, Darroudi F, Joenje H, de Winter JP. The cellular phenotype of Roberts syndrome fibroblasts as revealed by ectopic expression of ESCO2. PLoS One. 2009;4:e6936. doi: 10.1371/journal.pone.0006936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Mannini L, Menga S, Tonelli A, Zanotti S, Bassi MT, Magnani C, Musio A. SMC1A codon 496 mutations affect the cellular response to genotoxic treatments. Am. J. Med. Genet. 2012;158A:224–228. doi: 10.1002/ajmg.a.34384. [DOI] [PubMed] [Google Scholar]
- 189.Lu S, Goering M, Gard S, Xiong B, McNairn AJ, Jaspersen SL, Gerton JL. Eco1 is important for DNA damage repair in S. cerevisiae. Cell Cycle. 2010;9:3315–3327. doi: 10.4161/cc.9.16.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Dorsett D, Ström L. The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr. Biol. 2012;22:R240–R250. doi: 10.1016/j.cub.2012.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Muto A, Calof AL, Lander AD, Schilling TF. Multifactorial origins of heart and gut defects in nipbl-deficient zebrafish, a model of Cornelia de Lange Syndrome. PLoS Biol. 2011;9:e1001181. doi: 10.1371/journal.pbio.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hannan KM, Sanij E, Rothblum LI, Hannan RD, Pearson RB. Dysregulation of RNA polymerase I transcription during disease. Biochim. Biophys. Acta. 2012 doi: 10.1016/j.bbagrm.2012.10.014. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bose T, Lee KK, Lu S, Xu B, Harris B, Slaughter B, Unruh J, Garrett A, McDowell W, Box A, Li H, Peak A, Ramachandran S, Seidel C, Gerton JL. Cohesin proteins promote ribosomal RNA production and protein translation in yeast and human cells. PLoS Genet. 2012;8:e1002749. doi: 10.1371/journal.pgen.1002749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Laloraya S, Guacci V, Koshland D. Chromosomal addresses of the cohesin component Mcd1p. J. Cell Biol. 2000;151:1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Horsfield JA, Print CG, Mönnich M. Diverse developmental disorders from the one ring: distinct molecular pathways underlie the cohesinopathies. Front. Genet. 2012;3:171. doi: 10.3389/fgene.2012.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Gard S, Light W, Xiong B, Bose T, McNairn AJ, Harris B, Fleharty B, Seidel C, Brickner JH, Gerton JL. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J. Cell Biol. 2009;187:455–462. doi: 10.1083/jcb.200906075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.van der Maarel SM, Frants RR. The D4Z4 repeat-mediated pathogenesis of facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2005;76:375–386. doi: 10.1086/428361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nozawa RS, Nagao K, Igami KT, Shibata S, Shirai N, Nozaki N, Sado T, Kimura H, Obuse C. Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat. Struc. Mol. Biol. 2013;20:566–573. doi: 10.1038/nsmb.2532. [DOI] [PubMed] [Google Scholar]
- 199.Pandya S, King WM, Tawil R. Facioscapulohumeral dystrophy. Phys. Ther. 2008;88:105–113. doi: 10.2522/ptj.20070104. [DOI] [PubMed] [Google Scholar]
- 200.van der Maarel SM, Tawil R, Tapscott SJ. Facioscapulohumeral muscular dystrophy and DUX4: breaking the silence. Trends Mol. Med. 2011;17:252–258. doi: 10.1016/j.molmed.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.de Greef JC, Lemmers RJ, Camaño P, Day JW, Sacconi S, Dunand M, van Engelen BG, Kiuru-Enari S, Padberg GW, Rosa AL, Desnuelle C, Spuler S, Tarnopolsky M, Venance SL, Frants RR, van der Maarel SM, Tawil R. Clinical features of facioscapulohumeral muscular dystrophy 2. Neurol. 2010;75:1548–1554. doi: 10.1212/WNL.0b013e3181f96175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Gabriëls J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, Belayew A. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 203.Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, van der Maarel SM, Ruzzo WL, Gentleman RC, Tawil R, Tapscott SJ. DUX4 Activates Germline Genes, Retroelements, and Immune Mediators: Implications for Facioscapulohumeral Dystrophy. Dev. Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Snider L, Geng LN, Lemmers RJ, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ, Miller DG. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bosnakovski D, Xu Z, Gang EJ, Galindo CL, Liu M, Simsek T, Garner HR, Agha-Mohammadi S, Tassin A, Coppée F, Belayew A, Perlingeiro RR, Kyba M. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 2008;27:2766–2779. doi: 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Vanderplanck C, Ansseau E, Charron S, Stricwant N, Tassin A, Laoudj-Chenivesse D, Wilton SD, Coppée F, Belayew A. The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS One. 2011;6:e26820. doi: 10.1371/journal.pone.0026820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camaño P, Dauwerse JG, Snider L, Straasheijm KR, van Ommen GJ, Padberg GW, Miller DG, Tapscott SJ, Tawil R, Frants RR, van der Maarel SM. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Balog J, Thijssen PE, de Greef JC, Shah B, van Engelen BG, Yokomori K, Tapscott SJ, Tawil R, van der Maarel SM. Correlation analysis of clinical parameters with epigenetic modifications in the DUX4 promoter in FSHD. Epigeneti. 2012;7:579–584. doi: 10.4161/epi.20001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.van Overveld PG, Lemmers RJ, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, Padberg GW, van Ommen G-JB, Frants RR, van der Maarel SM. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- 210.Kondo T, Bobek MP, Kuick R, Lamb B, Zhu X, Narayan A, Bourc’his D, Viegas-Péquignot E, Ehrlich M, Hanash SM. Whole-genome methylation scan in ICF syndrome: hypomethylation of non-satellite DNA repeats D4Z4 and NBL2. Hum. Mol. Genet. 2000;9:597–604. doi: 10.1093/hmg/9.4.597. [DOI] [PubMed] [Google Scholar]
- 211.Ottaviani A, Rival-Gervier S, Boussouar A, Foerster AM, Rondier D, Sacconi S, Desnuelle C, Gilson E, Magdinier F. The D4Z4 macrosatellite repeat acts as a CTCF and A-type lamins-dependent insulator in facio-scapulo-humeral dystrophy. PLoS Genet. 2009;5:e1000394. doi: 10.1371/journal.pgen.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lewis A, Murrell A. Genomic imprinting: CTCF protects the boundaries. Curr. Biol. 2004;14:R284–R286. doi: 10.1016/j.cub.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 213.Ashe A, Morgan DK, Whitelaw NC, Bruxner TJ, Vickaryous NK, Cox LL, Butterfield NC, Wicking C, Blewitt ME, Wilkins SJ, Anderson GJ, Cox TC, Whitelaw E. A genome-wide screen for modifiers of transgene variegation identifies genes with critical roles in development. Genome Biol. 2008;9:R182. doi: 10.1186/gb-2008-9-12-r182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, Apedaile A, Hilton DJ, Dunwoodie SL, Brockdorff N, Kay GF, Whitelaw E. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat. Genet. 2008;40:663–669. doi: 10.1038/ng.142. [DOI] [PubMed] [Google Scholar]
- 215.Lemmers RJLF, Tawil R, Petek LM, Balog J, Block GJ, Santen GWE, Amell AM, van der Vliet PJ, Almomani R, Straasheijm KR, Krom YD, Klooster R, Sun Y, den Dunnen JT, Helmer Q, Donlin-Smith CM, Padberg GW, van Engelen BGM, de Greef JC, Aartsma-Rus AM, Frants RR, de Visser M, Desnuelle C, Sacconi S, Filippova GN, Bakker B, Bamshad MJ, Tapscott SJ, Miller DG, van der Maarel SM. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 2012 doi: 10.1038/ng.2454. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]