Abstract
Absent in melanoma 2 (AIM2) is a cytoplasmic dsDNA sensor involved in innate immunity. It uses its C-terminal HIN domain for recognizing dsDNA and its N-terminal Pyrin domain (PYD) for eliciting downstream effects through recruitment and activation of Apoptosis-associated Speck-like protein containing CARD (ASC). ASC in turn recruits caspase-1 and/or caspase-11 to form the AIM2 inflammasome. The activated caspases process proinflammatory cytokines IL-1β and IL-18, and induce the inflammatory form of cell death pyroptosis. Here we show that AIM PYD (AIM2PYD) self-oligomerizes. We notice significant sequence homology of AIM2PYD with the hydrophobic patches of death effector domain (DED)-containing proteins, and confirm that mutations on these residues disrupt AIM2PYD self-association. The crystal structure at 1.82 Å resolution of such a mutant, F27G of AIM2PYD, shows the canonical six-helix (H1-H6) bundle fold in the death domain (DD) superfamily. In contrast to the wild-type (WT) AIM2PYD structure crystallized in fusion with the large maltose-binding protein tag, the H2-H3 region of the AIM2PYD F27G is well defined with low B-factors. Structural analysis shows that the conserved hydrophobic patches engage in a type I interaction that has been observed in DED/DED and other DD superfamily interactions. While previous mutagenesis studies of PYDs point to the involvement of charged interactions, our results reveal the importance of hydrophobic interactions in the same interfaces. These centrally localized hydrophobic residues within fairly charged patches may form the hot spots in AIM2PYD self-association, and may represent a common mode of PYD/PYD interactions in general.
Inflammasomes are large supramolecular complexes responsible for sensing cytosolic danger signals associated with microbial infection or endogenous perturbations in the cell. The assembly of the inflammasomes is important for eliciting innate immune responses through the maturation and secretion of two inflammatory cytokines, IL-1β and IL-18, and may lead to an inflammatory form of cell death known as pyroptosis 1; 2; 3. Inflammasomes are typically composed of an upstream sensor molecule, an adaptor protein known as Apoptosis-associated Speck-like protein containing CARD (ASC), and an effector molecule caspase-1 (and/or caspase-11). Upon recruitment into a full inflammasome, caspase-1 activates through dimerization and auto-proteolysis in order to process pro-inflammatory cytokines.
The sensor molecules, after which the inflammasomes are named, can be grouped into two families – NOD-like receptors (NLRs) and AIM2-like receptors (ALRs), based on their distinct domain architecture. There are a total of 23 NLRs in the human genome 2. Most NLRs contain an N-terminal interaction domain (PYD, CARD, or BIR), followed by a nucleotide binding and oligomeriziation domain (NACHT or NBD), along with a C-terminal leucine-rich repeat (LRR) responsible for auto-inhibition 4. Only a handful of these NLRs are found to assemble into functional inflammasomes, including NLRP1, 6, 7, 12, and NLRC4, while the functions of the other NLRs remain to be elucidated. ALRs, which include absent in melanoma 2 (AIM2) and interferon inducible protein 16, also assemble functional inflammasomes 5; 6; 7; 8; 9. They lack the NACHT domain found in NLRs and directly bind to dsDNA, a trigger associated with microbial invasion, though a C-terminal HIN domain 10.
The NLRP3, 6, 7, and 12 inflammasomes and the AIM2 and IFI16 inflammasomes require the bipartite adaptor molecule, ASC, which contains an N-terminal pyrin domain (PYD) followed by a C-terminal caspase recruitment and activation domain (CARD). ASC is recruited by homotypic PYD/PYD interaction with the activated upstream sensor. Oligomerization of the sensor molecules, either through oligomerization of the NACHT domain in NLRs 2; 11 or the release of an auto-inhibition mechanism in ALRs 10; 12, initiates inflammasome assembly. To date, several PYD monomeric structures have been solved using crystallography and NMR. These include the solution structure of ASCPYD 13, full-length (ASCFL) 14, NALP1PYD 15, ASC2 16, NLRP10PYD 17, NLRP7PYD 18, NLRP12PYD 19, NALP3PYD 20, and MNDAPYD. While our paper was in preparation, the crystal structure of WT AIM2PYD as a fusion protein to maltose binding protein (MBP) was also reported 12.
PYD belongs to the death domain (DD) superfamily, which also includes two other subfamilies – CARD and death-effector domain (DED) 21. Many PYDs also oligomerize, in addition to interactions with other PYD-containing proteins. This property is observed in DD signalosomes such as the MyDDosome and PIDDosome complexes 22; 23. In other cases, these homotypic complexes can form extensive cytosolic signaling filaments, such as the DED filaments of caspase-8 22; 23 and the CBM filamentous complex in NF-κB activation 24. Therefore, the oligomerization property of AIM2PYD may represent a suitable activation mechanism of the AIM2 inflammsome and warrants more careful studies. Here we used biochemical and structural studies to deduce the mode of AIM2 oligomerization.
Sequence similarity with DED and mutagenesis of AIM2PYD
As a first step in the elucidation of AIM2 inflammasome formation, we decided to pursue the structure determination of the PYD of AIM2 (AIM2PYD). However, AIM2PYD (residues 1–100) was insoluble when expressed alone. Even when fused with the solubility tag Sumo, AIM2 still formed large aggregates. Upon searching for suitable mutation sites to solubilize and de-aggregate AIM2PYD, we noticed a short, but significant, sequence homology between AIM2PYD and the DEDs of FADD, v-FLIP and caspase-8 centered at the known conserved hydrophobic patches of DEDs (Figure 1(a)). In particular, the sequence homology placed F27 of AIM2 as the homologous residue for the surface exposed F25 of FADD, which has been shown to be important for apoptosis induction 25. While F25W and F25Y essentially maintained the activity of WT FADD, F25G and F25V are almost completely compromised in the cell death induction function. The mutations on F25 disrupted FADD self-association 26 and rendered the proteins monomeric 25. They also compromised the ability of FADD to interact with the FLICE-inhibitory protein c-FLIP, a caspase-8-like protein 27. In the tandem DED structure of a viral caspase-8-like protein known as the v-FLIP MC159 from the poxvirus Molluscum contagiosum virus, the F25-analagous residue in the similar hydrophobic patch of the first DED (DED1, F30) interacts with the second DED (DED2) to form a rigid dumbbell shaped structure 28; 29. In caspase-8, the analogous single site F122G mutant exhibited weakened interaction with FADD, while the double mutant, F122G and L123G, completely abolished its interaction with FADD 28.
Figure 1.
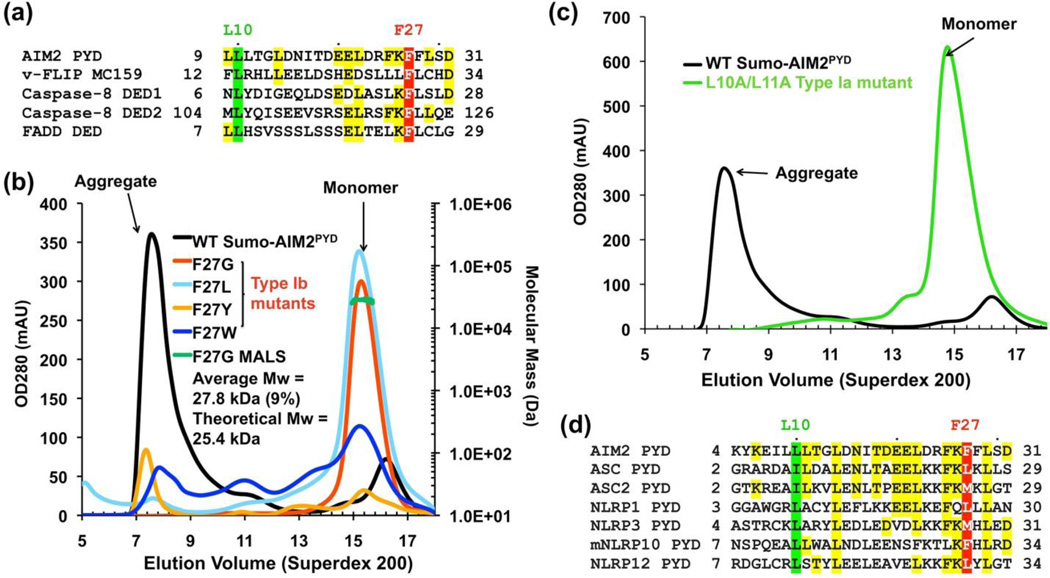
Sequence similarity between PYDs and DEDs, and identification of monomeric AIM2 PYD mutants. (a) Sequence alignment of human AIM2 PYD with human caspase-8 DED1 and DED2, and human FADD DED. The conserved hydrophobic positions are highlighted in red for that corresponding to type Ib and in green for that corresponding to type Ia interactions. F25 of FADD is analogous to F27 of AIM2, and has been shown to be important for apoptosis induction, self-association, and c-FLIP and caspase-8 interaction 25; 26; 27. Additional residues that are identical with the AIM2 sequence are highlighted in yellow. Residue numbers are shown. (b) Gel filtration profiles of His-Sumo-AIM2PYD WT and F25 mutant proteins expressed in E. coli using the pSMT3 vector. The proteins were purified using Ni-NTA columns. WT: mostly in the aggregation fraction; F27W, F27Y: in both aggregate and monomeric fractions; F27L: mostly in the monomeric fraction; F27G: only in the monomeric fraction. (c) Gel filtration profiles of His-Sumo-AIM2PYD WT and the L10A/L11A mutant protein expressed in E. coli using the pSMT3 vector. (d) Sequence alignment among PYDs in the region around L10 and F27 of AIM2.
To determine if F27 of AIM2PYD also impacts its solubility and aggregation solution behavior, we generated the F27Y, F27L, F27W and F27G mutants on the His-Sumo-AIM2PYD construct. All mutations caused the proteins to shift to the monomeric fraction, at least to some extent (Figure 1(b)). The more conserved substitutions F27W and F27Y showed a mixture of aggregated and monomeric proteins, while the F27L mutant is mostly monomeric and the F27G mutant is completely monomeric. Upon removal of the Sumo-tag, the F27G AIM2PYD mutant showed solubility greater than 20 mg/ml. These data suggest that F27 is involved in self-association of AIM2PYD.
Based on the definition of the three types of asymmetric interactions that have been observed in the DD fold superfamily 21; 30, F27 resides on a type Ib surface. Modeling and sequence alignment revealed that the corresponding type Ia surface likely includes residues L10 and L11 on the predicted helix H1 (Figure 1(a)). In previously reported interactions in the DD superfamily, the type Ia and the type Ib surfaces form a conserved asymmetric interaction pair 21. Indeed, the mutant L10A/L11A of AIM2PYD also showed almost all monomeric species (Figure 1(c)), revealing that the type Ia and Ib surfaces are both involved in AIM2 self-association. Both the type Ia and the type Ib residues are also conserved among the different PYDs (Figure 1(d)), suggesting that the type I interaction may be a common feature in PYD/PYD interactions.
AIM2PYD form ordered filaments in vitro and in cells
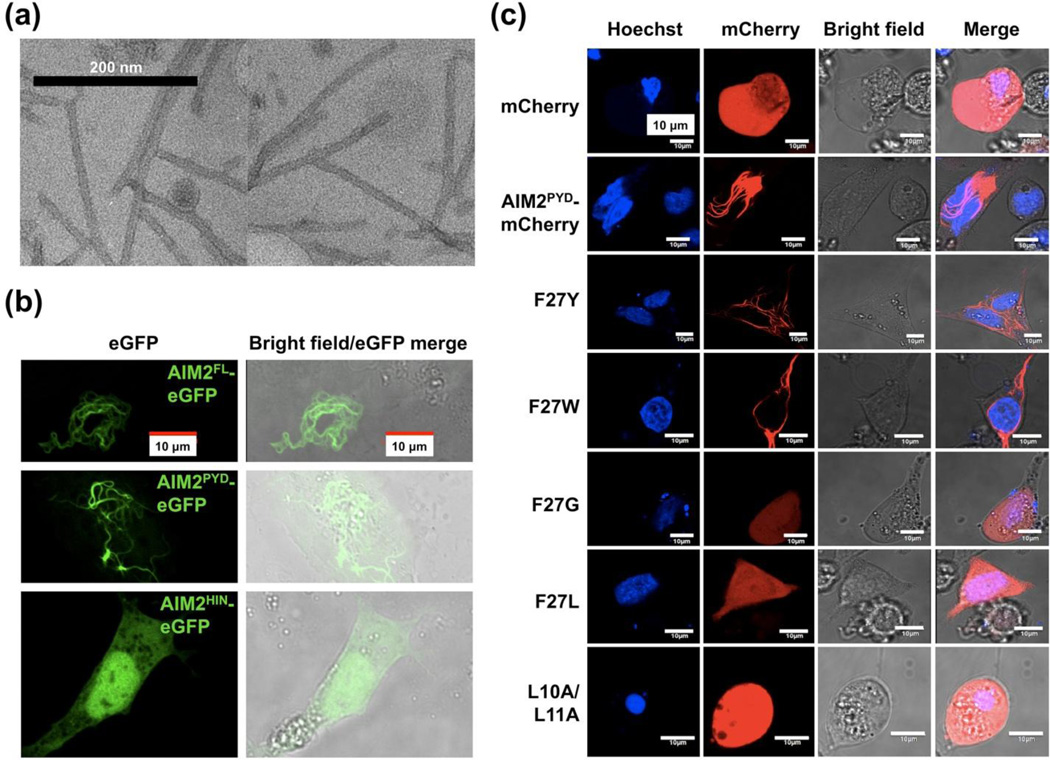
To assess whether AIM2PYD forms ordered aggregates, we first used negative stain EM to examine purified His-Sumo-AIM2PYD, which elutes in the void position of a Supderdex 200 column (Figure 1(b)). The protein showed filamentous morphology (Figure 2(a)). To confirm that AIM2PYD also forms ordered aggregates in cells, we transiently transfected in 293T cells full-length, PYD domain, and HIN domain of AIM2 in fusion to eGFP at the C-terminus (AIM2FL-eGFP, AIM2PYD-eGFP and AIM2HIN-eGFP). Confocal microscopy showed that while both AIM2FL-eGFP and AIM2PYD-eGFP formed filamentous structures, AIM2HIN-eGFP did not, suggesting that PYD is responsible for the ordered aggregation (Figure 2(b)). We further transiently transfected in HeLa cells AIM2PYD WT and Type I interface mutants identified above, as fusions to C-terminal mCherry. Confocal microscopy images showed filamentous aggregates for WT, F27Y, and F27W AIM2PYD-mCherry, while F27G, F27L, and L10A/L11A AIM2PYD-mCherry distributed throughout the cells (Figure 2(c)). These cellular data correlate well with migration positions in gel filtration chromatography (Figure 1(b)). Together, they suggest that bulky hydrophobic residues at the predicted Type I interface are essential for filament formation.
Figure 2.
AIM2 PYD forms filamentous aggregates. (a) EM images of purified His-Sumo-AIM2PYD, which showed ordered filamentous structures. (b, c) Confocal microscopy images of C-terminal eGFP tagged AIM2 constructs (b) and of mCherry tagged AIMPYD WT and Type I interface mutants (c). 0.8 μg of recombinant DNA was transfected into cells using Lipofectamine 2000 (Invitroggen) accordining to manufacturer’s instructions. Twenty-four hours post transfection, cells were fixed in 0.5% paraformaldehyde followed by nuclei staining using Hoechst 33342 (Molecular probes). The images were collected on an Olympus Fluoview FV1000 confocal microscope.
Crystal structure of F27G of AIM2PYD
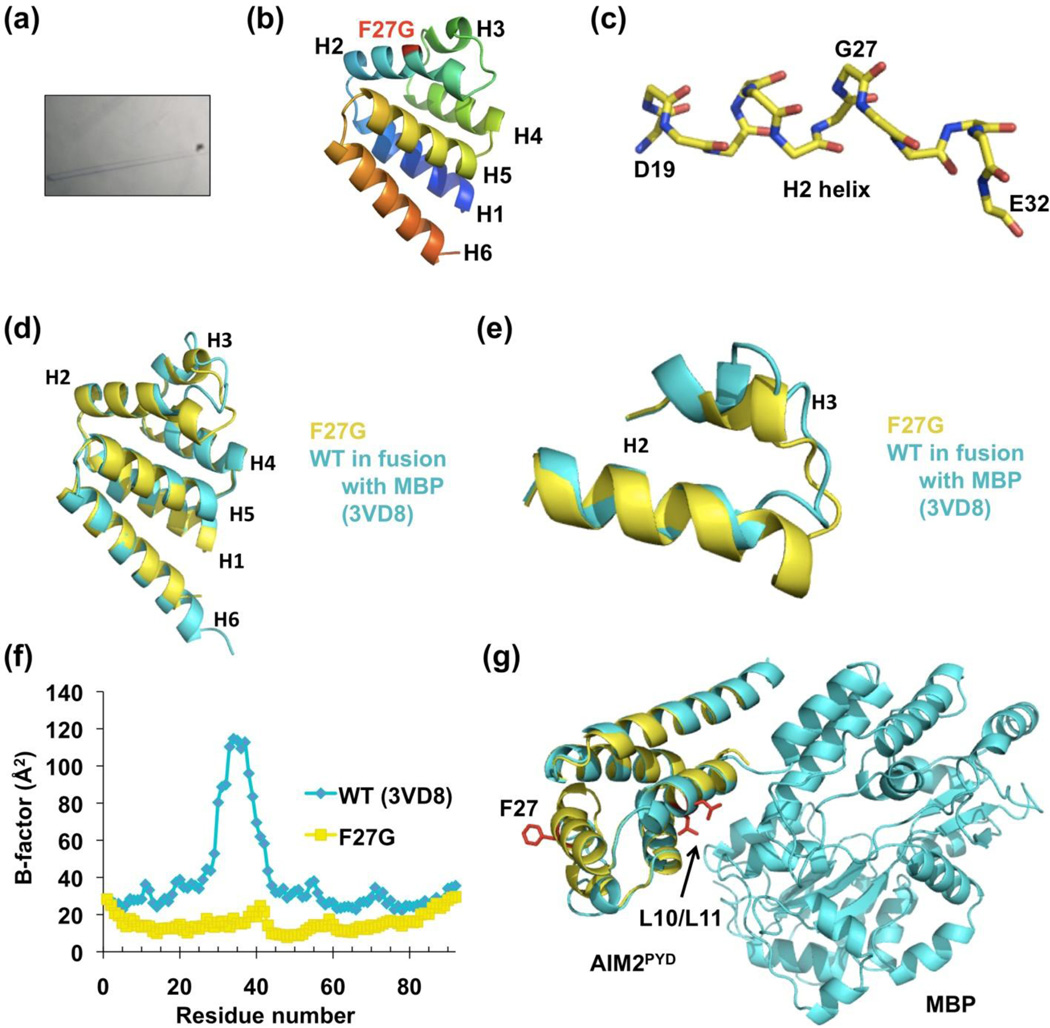
We set up the F27G mutant of AIM2PYD (residues 1–100) and obtained initial crystals that diffracted to ~4Å resolution. Using in situ proteolysis with trypsin, needle clusters grew within 3 days at 4 °C in the same condition with improved resolution to ~1.8 Å (Figure 3(a)). The structure was determined using molecular replacement in the program MOLREP 31. A composite of several PYD structures was used as the search model because of the low sequence identity of AIM2PYD to any known PYD structures. Model building and refinement at 1.82 Å was carried out using Coot 32 and PHENIX 33 (Supplementary Table 1). The final atomic coordinates comprise one residue from the vector, and AIM2PYD residues M1 to K93. It is likely that trypsin cleaved the C-terminal tail off at residue K93. This C-terminal region is involved in crystal packing and the cleavage must have facilitated crystal growth.
Figure 3.
Crystal structure of the AIM2PYD F27G mutant. (a) Photograph of a crystal under a transmission light microscope. (b) A ribbon diagram of the structure in rainbow colors. Helices 1–6 are labeled and the location of F27G is shown in red. (c) A stick model of helix H2 showing the kink at G27. (d) Superposition of the crystal structures of F27G (yellow) with the WT in fusion with the MBP tag (cyan). (e) Zoom-up of the superposition in the H2-H3 region, showing the longer H2 in the F27G structure and the different conformations in this region. (f) B-factors along the protein sequence for both the F27G (yellow) and the WT (Cyan) structures, showing the low values in F27G even in the H2-H3 region. (g) Superposition of the WT AIM2-MBP structure (cyan) with the F27G structure (yellow). L10/L11 and F27 residues are shown as red sticks, showing that L10/L11 are packed against MBP to avoid aggregation.
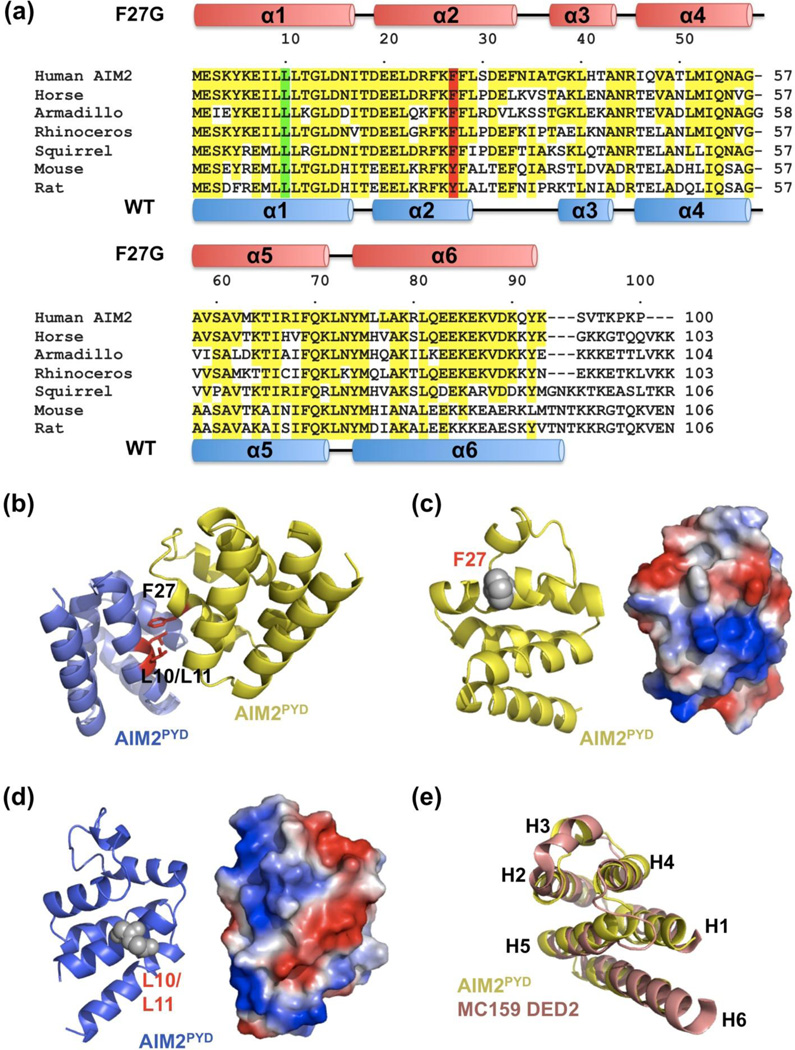
The structure revealed a six-helical bundle structure comprised of helices H1 to H6, which is conserved in the DD fold superfamily 21 (Figure 3(b)). Mutation to Gly at F27 did not dramatically alter the structure; it only created a minor kink in helix H2 (Figure 3(c)). In comparison with the WT AIM2PYD structure that was crystallized as a fusion to N-terminal maltose binding protein (MBP) 12, helix H2 in the F27G mutant is in fact longer (Figure 3(d) and 3(e)). Most conspicuously, in the published WT AIM2PYD structure, the H2-H3 region is highly disordered with high B-factors that go up to ~120 Å2 while the remainder of the residues show B-factors of ~20–40 Å2 (Figure 3(f)). In contrast, in our high resolution F27G mutant AIM2PYD structure, this region is ordered and well defined; the B-factors of residues in this region fall within the average B-factor of 18.7 Å2 in the whole protein chain (Figure 3(f), Supplementary Table 1). The protein conformation in the H2-H3 region is quite different in the WT and the F27G AIM2PYD (Figure 3(e)), suggesting that the ordered F27G mutant provides a clearer structure of this region of AIM2. In comparison with the WT structure, the F27G AIM2PYD structure shows differences in secondary structure boundaries (Figure 4(a)), offering an additional view on the intrinsic conformational flexibility of the molecule.
Figure 4.
Sequence and structural comparisons of AIM2PYD. (a) Sequence alignment among AIM2PYD from different species, with secondary structures from the F27G structure (pink) and the WT structure (blue). (b) An AIM2PYD/AIM2PYD type I interaction model based on the Myddosome structure. The interaction residues F27, L10 and L11 are shown as red sticks. (c, d) Ribbon and electrostatic diagrams of the AIM2PYD surfaces containing F27 (c) and L10/L11 (d), respectively. (e) Superposition between AIM2PYD (yellow) and MC159 DED2 (pink).
Modeled AIM2 PYD/PYD interaction: hydrophobic interactions may comprise the functional epitope
The locations of F27 on a type Ib surface and L10 on a type Ia surface, and the monomeric phenotypes of mutations on these residues, suggest that AIM2PYD oligomerizes through a type I interface. Consistent with this observation, residues L10 and L11 are positioned adjacent to the fusion partner MBP in the WT AIM2PYD structure 12 (Figure 3(g)). Therefore MBP would inhibit the type I interaction of AIM2PYD in the fusion protein, explaining how MBP inhibits AIM2PYD aggregation.
We generated a PYD/PYD interaction model by superimposing AIM2PYD onto a type I interaction pair in the Myddosome 21; 30 (Figure 4(b)). It has been proposed previously that AIM2PYD interacts with ASCPYD mainly through charge-charge interactions 12. Previous extensive mutagenesis on ASCPYD also suggested the involvement of charged interactions in ASC self-association and interaction with other PYD proteins such as NLRP3 and ASC2 14; 34; 35. In our model of the type I interaction in AIM2PYD oligomerization, both the F27 containing and the L10/L11 containing surfaces are highly charged (Figure 4(c) and 4(d)), consistent with these existing data.
In addition to charge interactions, it has also been proposed that hydrophobic surfaces may be involved in the interactions mediated by AIM2PYD 12. Here we observe that the hydrophobic residues F27 and L10 appear to reside in the center of the charged patches and are likely the major contributors of the interactions. The data provide additional structural and energetic insights because the strength of the hydrophobic interaction may be further enhanced by the local charges surrounding it. In addition, previous detailed mutational studies on protein-protein interfaces have shown that the buried hydrophobic contacts at the center of an interface may be responsible for a majority of the interfacial binding energy 36. Therefore, the hydrophobic residues in AIM2PYD self-association may represent the hot spot or the functional epitope in the interaction, which has been mostly overlooked and not emphasized in the existing literature.
Structural similarities between PYDs and DEDs
In addition to the local sequence homology with DEDs (Figure 1(a)), AIM2PYD was also unexpectedly much more similar in structure to canonical DEDs, such as DED of FADD 25 and DED2 of vFLIP 28, than to CARDs and DDs. A DALI structural homology search 37 found that the top hits are all PYD or DED structures (Supplementary Table 2, Figure 4(e)). This observation is in keeping with a phylogenetic analysis of DD superfamily, which suggests that DED and PYD share common ancestors with PYD derived latest in the evolution of these domains 38. Therefore, the structural and evolutionary features dictate the similarities between DEDs and PYDs.
Supplementary Material
Highlights.
PYD oligomerization plays a pivotal role in inflammasome activation.
Crystal structure of an AIM2 PYD mutant reveals conformational differences from WT.
PYD and DED evolve similar hydrophobic interactions for self-association.
Residues F27 and L10 provide hydrophobic interactions at the Type I interface.
Interaction through oligomerization represents a ubiquitous signaling platform.
Acknowledgements
We thank the staff scientists at beamlines X25 and X29 of National Synchrotron Light Source (NSLS) for assistance with data collection.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lamkanfi M, Dixit VM. Inflammasomes and their roles in health and disease. Annu Rev Cell Dev Biol. 2012;28:137–161. doi: 10.1146/annurev-cellbio-101011-155745. [DOI] [PubMed] [Google Scholar]
- 2.Rathinam VA, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nat Immunol. 2012;13:333–332. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 4.Hu Z, Yan C, Liu P, Huang Z, Ma R, Zhang C, Wang R, Zhang Y, Martinon F, Miao D, Deng H, Wang J, Chang J, Chai J. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341:172–175. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- 5.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 8.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 9.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, Hornung V, Latz E, Bowie AG, Fitzgerald KA, Xiao TS. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan S, Akey CW. Apoptosome structure, assembly, and procaspase activation. Structure. 2013;21:501–515. doi: 10.1016/j.str.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin T, Perry A, Smith PT, Jiang J, Xiao TS. Structure of the AIM2 pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J Biol Chem. 2013 doi: 10.1074/jbc.M113.468033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liepinsh E, Barbals R, Dahl E, Sharipo A, Staub E, Otting G. The death-domain fold of the ASC PYRIN domain, presenting a basis for PYRIN/PYRIN recognition. J Mol Biol. 2003;332:1155–1163. doi: 10.1016/j.jmb.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 14.de Alba E. Structure and interdomain dynamics of apoptosis-associated speck-like protein containing a CARD (ASC) J Biol Chem. 2009;284:32932–32941. doi: 10.1074/jbc.M109.024273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiller S, Kohl A, Fiorito F, Herrmann T, Wider G, Tschopp J, Grutter MG, Wuthrich K. NMR structure of the apoptosis- and inflammation-related NALP1 pyrin domain. Structure (Camb) 2003;11:1199–1205. doi: 10.1016/j.str.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Natarajan A, Ghose R, Hill JM. Structure and dynamics of ASC2, a pyrin domain-only protein that regulates inflammatory signaling. J Biol Chem. 2006;281:31863–31875. doi: 10.1074/jbc.M605458200. [DOI] [PubMed] [Google Scholar]
- 17.Su MY, Kuo CI, Chang CF, Chang CI. Three-dimensional structure of human NLRP10/PYNOD pyrin domain reveals a homotypic interaction site distinct from its mouse homologue. PLoS One. 2013;8:e67843. doi: 10.1371/journal.pone.0067843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinheiro AS, Proell M, Eibl C, Page R, Schwarzenbacher R, Peti W. Three-dimensional structure of the NLRP7 pyrin domain: insight into pyrin-pyrin-mediated effector domain signaling in innate immunity. J Biol Chem. 2010;285:27402–27410. doi: 10.1074/jbc.M110.113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinheiro AS, Eibl C, Ekman-Vural Z, Schwarzenbacher R, Peti W. The NLRP12 pyrin domain: structure, dynamics, and functional insights. J Mol Biol. 2011;413:790–803. doi: 10.1016/j.jmb.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bae JY, Park HH. Crystal structure of NALP3 protein pyrin domain (PYD) and its implications in inflammasome assembly. J Biol Chem. 2011;286:39528–39536. doi: 10.1074/jbc.M111.278812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Curr Opin Struct Biol. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegel RM, Martin DA, Zheng L, Ng SY, Bertin J, Cohen J, Lenardo MJ. Death-effector filaments: novel cytoplasmic structures that recruit caspases and trigger apoptosis. J Cell Biol. 1998;141:1243–1253. doi: 10.1083/jcb.141.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan RT, Young S, Liang J, Schmid MC, Mielgo A, Stupack DG. Caspase-8 isoform 6 promotes death effector filament formation independent of microtubules. Apoptosis. 2012;17:229–235. doi: 10.1007/s10495-011-0677-y. [DOI] [PubMed] [Google Scholar]
- 24.Qiao Q, Yang C, Zheng C, Fontan L, David L, Yu X, Bracken C, Rosen M, Melnick A, Egelman EH, Wu H. Structural Architecture of the CARMA1/Bcl10/MALT1 Signalosome: Nucleation-Induced Filamentous Assembly. Mol Cell. 2013;51:766–779. doi: 10.1016/j.molcel.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eberstadt M, Huang B, Chen Z, Meadows RP, Ng S-C, Zheng L, J LM, Fesik SW. NMR structure and mutagenesis of the FADD (Mort1) death-effector domain. Nature. 1998;392:941–945. doi: 10.1038/31972. [DOI] [PubMed] [Google Scholar]
- 26.Carrington PE, Sandu C, Wei Y, Hill JM, Morisawa G, Huang T, Gavathiotis E, Werner MH. The structure of FADD and its mode of interaction with procaspase-8. Mol Cell. 2006;22:599–610. doi: 10.1016/j.molcel.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 27.Kaufmann M, Bozic D, Briand C, Bodmer JL, Zerbe O, Kohl A, Tschopp J, Grutter MG. Identification of a basic surface area of the FADD death effector domain critical for apoptotic signaling. FEBS Lett. 2002;527:250–254. doi: 10.1016/s0014-5793(02)03146-0. [DOI] [PubMed] [Google Scholar]
- 28.Yang JK, Wang L, Zheng L, Wan F, Ahmed M, Lenardo MJ, Wu H. Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition. Mol Cell. 2005;20:939–949. doi: 10.1016/j.molcel.2005.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li FY, Jeffrey PD, Yu JW, Shi Y. Crystal structure of a viral FLIP: insights into FLIP-mediated inhibition of death receptor signaling. J Biol Chem. 2006;281:2960–2968. doi: 10.1074/jbc.M511074200. [DOI] [PubMed] [Google Scholar]
- 30.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vagin AA, Teplyakov A. MOLREP: an Automated Program for Molecular Replacement. J. Appl. Cryst. 1997;30:1022. [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moriya M, Taniguchi S, Wu P, Liepinsh E, Otting G, Sagara J. Role of charged and hydrophobic residues in the oligomerization of the PYRIN domain of ASC. Biochemistry. 2005;44:575–583. doi: 10.1021/bi048374i. [DOI] [PubMed] [Google Scholar]
- 35.Vajjhala PR, Mirams RE, Hill JM. Multiple binding sites on the pyrin domain of ASC protein allow self-association and interaction with NLRP3 protein. J Biol Chem. 2012;287:41732–41743. doi: 10.1074/jbc.M112.381228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 37.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 38.Kersse K, Verspurten J, Vanden Berghe T, Vandenabeele P. The death-fold superfamily of homotypic interaction motifs. Trends Biochem Sci. 2011;36:541–552. doi: 10.1016/j.tibs.2011.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.