Abstract
TrkA is a tyrosine kinase receptor required for development and survival of the peripheral nervous system. In the adult, TrkA and its ligand NGF are peripheral pain mediators, particularly in inflammatory pain states. However, how TrkA regulates the function of nociceptive neurons and whether its activity levels may lead to sensory abnormalities is still unclear. Here we report the characterization of a 3 aa (KFG) domain that negatively regulates TrkA level and function in response to NGF. Deletion of this domain in mouse causes a reduction of TrkA ubiquitination leading to an increase in TrkA protein levels and activity. The number of dorsal root ganglia neurons is not affected by the mutation. However, mutant mice have enhanced thermal sensitivity and inflammatory pain. Together, these data suggest that ubiquitination is a mechanism used in nociceptive neurons to regulate TrkA level and function. Our results may enhance our understanding of how ubiquitination affects TrkA activation following noxious thermal stimulation and inflammatory pain.
Introduction
The Trk neurotrophin tyrosine kinase receptors are key regulators of the mammalian nervous system and impairments in their function have been implicated in a number of neurodegenerative disorders in humans (Bibel and Barde, 2000; Chao et al., 2006). Efforts to modulate the activity of these receptors have been focused on the development of Trk receptor agonists or antagonists (Longo and Massa, 2004). So far, this effort has not been successful partly due to the still insufficient knowledge of the mechanisms controlling the activity of these ligand/receptor systems (Thoenen and Sendtner, 2002). This family of receptors includes TrkA, whose ligand is NGF; TrkB, which is activated by BDNF and NT-4; and TrkC activated by NT3 (Tessarollo, 1998). TrkA was the first identified Trk receptor and its downstream signaling pathways have been extensively characterized including the mitogen-activated protein kinase (MAPK), phospholipase C gamma (PLC-γ) and phosphatidylinositol-3 kinase (PI3-K) intracellular signaling cascades (Reichardt, 2006). Although these pathways are common to other tyrosine kinase receptors, it has been shown that in PC12 cells, TrkA activation of MAPK leads to a different cellular outcome compared with the activation of this pathway by the epidermal growth factor (EGF) receptor. In this case, the EGF receptor causes a transient activation of MAPK leading to cell proliferation, whereas TrkA signaling causes sustained activation of MAPK, inducing cell differentiation (Marshall, 1995). Overall, it appears that the engagement of positive and negative feedback mechanisms by different receptors determines the duration of MAPK activation. Of course, the cellular context including the presence of different cellular signaling networks is key to determining how tyrosine kinase receptors signal after binding to their ligands (Santos et al., 2007; Lemmon and Schlessinger, 2010). Moreover, in vivo, this scenario can be further complicated by extrinsic factors such as the simultaneous engagement of different receptors by gradients of growth factors (Coppola et al., 2001).
Ubiquitination has recently emerged as a new mechanism controlling tyrosine kinase receptor signaling (Acconcia et al., 2009; Haglund and Dikic, 2012). So far, there has been very limited information on how ubiquitination affects Trk signaling and virtually no reports are available on how this mechanism controls Trk function in vivo (Geetha et al., 2005; Makkerh et al., 2005; Arévalo et al., 2006; Georgieva et al., 2011; Takahashi et al., 2011; Yu et al., 2011). Here we investigate the role of a 3 aa (KFG) domain on TrkA function in vivo. This domain is conserved among all Trk receptors and previous experiments have suggested a role in cell differentiation, at least in vitro (Peng et al., 1995). Surprisingly, we found that in vivo this domain negatively regulates TrkA function. This activity is mediated by a lysine that is responsible for a change in TrkA ubiquitination in response to NGF. In peripheral sensory neurons, the KFG deletion causes an increase in the levels of TrkA that does not affect the number of dorsal root ganglia (DRG) neurons but increases thermal and mechanical sensitivity. These data establish ubiquitination as a physiologically important mechanism regulating TrkA function.
Materials and Methods
Mice
TrkAΔKFG mice were generated by a conventional gene-targeting approach using the CJ7 (129/SV1 strain) embryonic stem (ES) cell line. Electroporation and selection were performed as described previously (Southon and Tessarollo, 2009). DNAs derived from G418/FIAU-resistant ES clones were screened by the diagnostic BamHI restriction enzyme digestions using, respectively, a 5′ and 3′ probe external to the targeting vector sequence. Recombinant clones containing the predicted rearranged band were injected into C57BL/6 blastocysts to generate chimeras that transmitted the mutated allele to the progeny (Reid and Tessarollo, 2009). Following germline transmission of the targeted ES cell clones, BamHI digestion was used again for screening as it allowed us to distinguish between the wild-type (WT) and all the targeted alleles including those generated after cre-recombination.
Mutant mice were backcrossed for at least 10 generations onto the C57BL/6 background. Male mice were used for behavioral analysis and mice of either sex were used for biochemical studies. Animals were bred in a specific, pathogen-free facility with food and water ad libitum. All experimental procedures followed the National Institutes of Health Guidelines for Animal Care and Use (ACUC), and were approved by the National Cancer Institute–Frederick ACUC committee.
DRG neuronal culture
DRG neurons were isolated from E13.5 (6–8 embryos per genotype) and cultured for 3 d in minimal essential medium (MEM), B27 (Invitrogen) serum supplement, and 10 ng/ml NGF (Alomone Labs) to promote survival of TrkA-expressing neurons. On the third day in vitro (DIV), the cells were incubated in MEM for 4 h followed by MEM with or without 100 ng/ml NGF for 5 min. Cells were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (Millipore) and subjected to Western blot analysis using an antibody specific for TrkA (1 μg/ml; Advanced Targeting Systems), AKT (1:1000; Cell Signaling Technology), MEK (1:1000; Cell Signaling Technology), and their phosphorylated forms (pTrkA, pAKT, and pMEK all at 1:1000 dilution; Cell Signaling Technology). For quantification, band intensity from the blots was quantified using National Institutes of Health (NIH) Image software and the data were expressed as the mean ± SEM the WT control and subjected to Student's t test.
Generation and analysis of HEK293 cell lines with inducible expression of TrkA.
HEK293 cells with the Flp-In T-Rex system (Life Technologies) were used to create cell lines that express WT, KFG-deleted TrkA, AFG (single lysine to alanine mutation), and KAA (F and G amino acids converted to alanine) mutated TrkA in a stable and inducible manner. Cell lines were generated as suggested by the manufacturer, by inserting a single copy cDNA into an inducible locus within the genome by frt site-specific recombination. To induce expression of the specific TrkA cDNA, cells were treated overnight with doxycycline (0.005 μg/ml) and serum starved in DMEM for 10 h before NGF treatment or biotinylation as needed. Very low levels of doxycycline were used to prevent excessive expression of TrkA and, consequently, its auto-activation in the absence of NGF. After a specific treatment, cells were rinsed thoroughly with DMEM and PBS before lysis at the indicated time points for immunoprecipitation and Western blot analysis. For ubiquitination experiments cells were treated for 5 min with 100 ng/ml NGF and lysed at varying time points, ranging from 5 to 90 min, in a lysis buffer composed of 30 mm Tris, pH.8, 75 mm NaCl, 10% glycerol, and 1%-Triton X-100. Equal amounts of total protein (determined using a BCA Assay Kit; Thermo Scientific) were immunoprecipitated with Pan Trk (C-15; 2 μg/ 500 μg of lysate protein; Santa Cruz Biotechnology) antibodies. Samples were subjected to Western blot analysis to measure the extent of ubiquitination with an anti-Ubiquitin (P4D1; 1:500; Santa Cruz Biotechnology) antibody that detects both poly- and mono-ubiquitinated proteins. The FK1 antibody (Enzo Life Science) was used to detect poly-ubiquitinated proteins. The same blots were stripped and re-probed with Pan Trk (C-15) antibodies. The immunoblotting results were visualized and quantified with a Syngene gel documentation and analysis system. Quantification of ubiquitination levels was performed by normalizing the ubiquitin band intensity values to corresponding Pan Trk (C-15) levels relative to non-NGF-treated WT conditions. The values are given as the mean ± SEM and blots are representative of at least three independent experiments. Statistical significance (P) was calculated by Student's t test using GraphPad Prism Software. Additionally, input samples were subjected to Western blot analysis, assessing phospho-Erk1/2 (1:1000; Cell Signaling Technology) levels and identical gels were run and probed with total-Erk1/2. The blots are representative of at least three independent experiments.
For biotinylation assays to assess the level of TrkA present at the membrane, HEK293T cells were serum starved, washed, and treated with or without NGF (100 ng/ml) at 37°C for the indicated time points. The cells were then biotinylated with 0.3 mg/ml Sulfo-NHS-SS-Biotin as indicated by the manufacturer (Pierce) for 30 min at 4°C. Nonreacted biotinylation reagent was removed by three washes with PBS after which the cells were lysed in 10 mm Tris, pH7.4, 150 mm NaCl, 2 mm EDTA, and 1% Nonidet P-40. For biotinylation assays to assess the level of recycling TrkA, HEK293T cells were serum starved, biotinylated as above, washed to remove nonreacted biotin, and treated with or without NGF (100 ng/ml) at 37°C for 5 min to induce internalization. After NGF treatment, cells were rinsed three times in PBS and biotin attached to noninternalized proteins was cleaved by 50 mm dithiothreitol (DTT) treatment for 15 min at 4°C. Cells were then rinsed in PBS and prewarmed DMEM media and incubated at 37°C for the indicated time points before lysis as above. Biotinylated proteins were immunoprecipitated with Streptavidin beads (Pierce). Samples were then subjected to Western blotting using the Pan Trk, actin (1:500; Santa Cruz Biotechnology), phospho-Erk1/2, and GAPDH (1:1000; Millipore) antibodies. For the input samples, Pan Trk and phospho-Erk1/2 antibodies were simultaneously applied to the membrane as they have different molecular weights. GAPDH was used as a loading control for inputs and actin was the negative control for biotinylated pull-down samples. Quantification of surface TrkA levels was performed by determining the biotinylated TrkA band intensity values normalized with the GAPDH band relative to the non-NGF-treated WT condition. Quantification of biotinylated TrkA in the cleaved biotin assay was performed by determining the band intensity of the TrkA immunoprecipitated samples normalized with the GAPDH input band relative to the TrkA WT sample value at 5 min NGF treatment. The values are given as the mean ± SEM and blots are representative of three independent experiments. Statistical significance (p) was calculated by Student's t test using GraphPad Prism.
Immunohistochemistry and neuronal cell counts
Heterozygous (+/−) TrkAΔKFG mice were intercrossed by brother/sister matings for histological analysis of pups as described previously (Tessarollo et al., 1997). Spinal cords with attached DRGs from P0 mice were embedded in the same block, sectioned sagittally at 5 μm, and stained with 0.1% cresyl violet (Nissl). DRGs at P0 and lumbar level L4 from mutant embryos and WT littermates were counted. Neurons having a clear nucleus and nucleolus were counted in every sixth section and the sum of the counts was multiplied by six. To assure unbiased analysis, neuronal counts were performed in a blinded fashion. Differences were evaluated using a one-tailed Student's t test. For adult DRG analysis, animals were perfused with 4% paraformaldehyde and L4 DRGs were dissected and cryoprotected overnight in 30% sucrose. Serial sections (10 μm) were mounted and one in every six sections was stained with the NeuN antibody and positive cells were counted. The total number of DRG neurons was estimated as six times the total number of NeuN-positive neurons. The statistical significance was determined by Student's t test analysis.
The following antibodies have been used for immunohistochemistry staining: NeuN-488 (1:50; Millipore), rabbit anti-TrkA antibody (1:500; Advanced Targeting Systems), and IB4-568 (1:100; Life Technologies).
Behavioral tests
Open field test.
For the assessment of general locomotor and open field activity, eight WT and nine TrkAΔKFG male mice were subjected to the open field test (Omnitech Electronics Digiscan animal activity monitor). Each mouse was placed in the arena (a 16 × 16 inch Plexiglas chamber) for 5 min. Animal movements including assessment of total distance traveled, rearings, ambulations, and time spent in the center part of the field were recorded by infrared sensors mounted on to the walls of the chamber.
Rota rod test.
Evaluation of muscle strength and coordination of male mice was performed with a rota rod apparatus (Ugo Basile) as previously described (Yanpallewar et al., 2012). Briefly, animals (n = 15 WT and n = 13 TrkAΔKFG) were placed on a rotating rod accelerating from 5 to 40 rpm over a 5 min period. Animal performance was recorded as the time spent on the rotating rod, averaged over three trials separated by 1 h intervals.
The Hargreaves test.
This was performed with IITC Life Science apparatus. Briefly, the mice were placed in individual Plexiglas chambers set on a glass plate maintained at 30°C and allowed to acclimate for 1 h. Once acclimated, the response latencies (flinching or paw lifting) to noxious thermal stimulation were measured by applying a radiant heat stimulus to the glabrous skin of each hindpaw. Three measurements from the left paws were averaged to obtain a single score for each animal. The test was repeated for 3 consecutive days. The experimenter was blind to the animal's genotype. Statistical significance was determined by Student's t test analysis.
Formalin test.
Mice were placed in Plexiglas boxes on top of a glass plate to be acclimated for 30 min in an isolated behavioral room before formalin injection. The Plexiglas chambers were centered over a wide-angle, high-resolution video camera placed 35 cm below the glass plate to record the behavioral responses. For the test, animals were anesthetized briefly by isoflurane vapor (using a nose cone) and subcutaneously injected with 25 μl of 1.85% formaldehyde in 0.9% saline to the right hindpaw, using a 50 μl Hamilton microsyringe attached to a 30 gauge needle. Then, they were immediately replaced into the Plexiglas chamber and video recorded for 60 min. The total amount of licking/biting of the affected hindpaw was measured in 5 min intervals by two separate experimenters who were blind to the genotype of the animals. The acute phase (Phase I) is defined as 0–10 min after injection, and the persistent (tonic) phase (Phase II) is defined as 10–60 min after injection. Statistical significance was determined by two-way ANOVA.
Two-temperature choice test.
Experimental mice were placed into a thermal gradient apparatus (IITC Life Science) and allowed to explore two adjacent areas. One was held at a constant 32°C (side A) while the other side had variable temperatures ranging from 4 to 50°C (side B). Mice were placed in the apparatus for 10 min, and the time spent in each area was recorded. The percentage of time spent on side B during the last 5 min interval was measured. Statistical significance was determined by two-way ANOVA followed by “t” tests at each temperature using the Holm–Sidak multiple-comparison method (GraphPad Prism).
Results
Deletion of the KFG domain of the TrkA gene causes an increase in DRG TrkA protein level
To investigate the biological significance of the conserved KFG domain in the juxtamembrane region of Trk receptors, we decided to delete it from the TrkA gene. This locus does not generate TrkA receptor isoforms with different intracellular domains as do the TrkB and TrkC genes (Tessarollo, 1998). We reasoned that this would simplify the interpretation of the phenotypic and biochemical characterization. To this end, by conventional gene targeting technology, we first generated an allele with a deletion of the KFG sequence located in exon 11 (Fig. 1). A loxP-flanked neomycin resistance gene was placed in the downstream intron for selection of the targeted ES cells, as described in Figure 1. Following germline transmission of the mutant ES cells we removed the neomycin resistance gene by crossing of heterozygous TrkAΔKFG-neo mice with a ubiquitously expressing β-actin CRE transgenic mouse. TrkAΔKFG mutant mice were obtained by crossing heterozygous mice lacking the neo cassette. TrkAΔKFG−/− mice are viable, develop to adulthood, and are fertile. Moreover, mutants subjected to the open field test to analyze their general exploratory behavior including distance traveled, number of rearings, ambulation, and time spent in the center of the arena showed no impairments compared with WT controls. Likewise, in the rota rod test to evaluate muscle strength and motor coordination ability, mutant mice showed no impairments, again suggesting a lack of major abnormalities (data not shown).
Figure 1.
Generation of the TrkAΔKFG mutant mice. A, Schematic of the vector and strategy to target the TrkA locus. The replacement-type targeting vector consists of an ∼9.0 kb 129/SV mouse genomic fragment. Exon 11 was modified by removal of the 9 nt encoding the KFG domain by PCR (asterisk). The pGKneobpA cassette flanked by loxP sites was placed in the intron downstream of exon 11. The pGK-thymidine kinase (TK) cassette was introduced as a negative selectable marker. ED, extracellular domain; TM, transmembrane domain; JM, juxtamembrane domain; TK tyrosine kinase domain. B, Southern blot analysis of tail DNA from 3-week-old mice with a 5′ external probe shows the switch of the WT BamHI fragment from 12 kb (WT) to a 7 kb restriction fragment (TrkAneo) after neo insertion due to the targeting of the TrkA locus. Removal of the neo gene causes a further decrease of the BamHI-generated restriction fragment to 5.5 Kb. The presence of the KFG deletion was verified by PCR analysis using one primer upstream and downstream of the KFG domain. C, Northern blot analysis of total RNA extracted from DRG neurons of E13.5 embryos either WT(+/+) or TrkA ΔKFG mutants after germline removal of the neo cassette and hybridized with a TrkA-kinase domain-specific probe (exon 14–17). Bottom, Shows the ethidium bromide stained gel at the 28S ribosomal RNA level to confirm RNA loading and integrity. D, Western blot analysis of total protein lysates from E13.5 DRG neurons from WT (+/+) heterozygous (+/−) and mutant (−/−) animals. The lysates were probed with an antibody specific for TrkA (top) and GAPDH (bottom) as a control for loading. E, Quantification of mutant TrkAΔKFG (−/− and +/−) protein levels relative to WT TrkA (+/+). TrkA protein was normalized relative to GAPDH used as loading control. Values are the mean ± SEM; N = 5; *p < 0.01 (t test).
To test whether the residual loxP site in the TrkA locus affects trkA mRNA levels we isolated DRG RNA from E13.5 embryos and analyzed it by Northern blotting. DRGs from TrkAΔKFG and control embryos had similar levels of trkA mRNA, suggesting that the presence of the loxP site has no influence on TrkA splicing or mRNA stability (Fig. 1C). Next, we investigated the TrkA protein levels from E13.5 DRG neurons. To our surprise, we found that mutant DRGs have higher levels of TrkA protein (Fig. 1D,E). Together these data suggested that deletion of the KFG domain of TrkA does not affect the transcription level or quality of the transcripts produced by this locus but it does affect TrkA protein levels.
TrkAΔKFG mutant mice have normal DRG neuron numbers
The evidence that TrkA and its ligand NGF are critical for survival of neurons of the peripheral nervous system prompted us to investigate whether the increased level of TrkA affected DRG neuron numbers (Crowley et al., 1994; Smeyne et al., 1994). L4 DRG neurons were counted from TrkA and control mice at P0. No significant changes were found in the total number of neurons at this developmental stage (WT 6564 ± 437; TrkAΔKFG 6696 ± 379; n = 3 mice per genotype). We also extended our analysis to adult DRGs to investigate whether the change in the overall level of TrkA protein results in long-term consequences on the neuronal composition of this structure. Again, as in P0 animals, no changes were found in the total number of neurons from adult L4 DRG (WT 3904 ± 36; TrkAΔKFG 3954 ± 57; n = 3 mice per genotype) and, surprisingly, the percentage of TrkA-positive neurons was also unaffected (WT, 25.39 ± 1.49%; TrkAΔKFG, 27.17 ± 2.01%; n = 3 per genotype). Similarly, double staining with the IB4 and NeuN markers to examine the percentage of small diameter TrkA-negative neurons was not altered by the mutation. The percentage of IB4 binding neurons was 25.39 ± 1.49 in control versus 27.17 ± 2.01 in mutant mice (n = 3 per genotype, p = 0.42) again suggesting no overall change in the composition of the small diameter nociceptive neurons. Together, these data suggest that increased expression of TrkA protein during embryonic development does not affect the proliferation of precursors or the fate and survival of mature DRG neurons. These results coupled with previous data from animals lacking one copy of TrkA that show no DRG defects suggest that this neuronal population is not sensitive to either an increase or a decrease of TrkA for its anatomical development (Silos-Santiago et al., 1995).
Mutant TrkAΔKFG signals more robustly in response to NGF
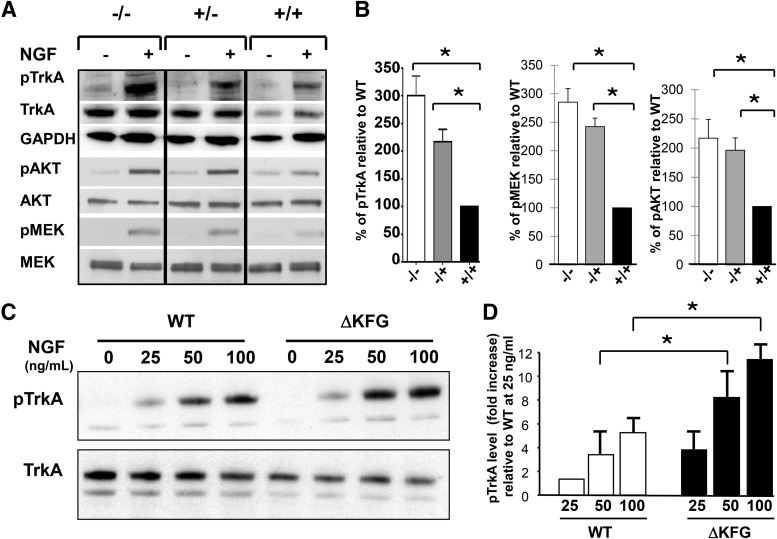
Although the increased level of TrkA apparently does not affect the anatomical development of the DRG we decided to test whether NGF-induced signaling was altered in the mutant neurons. For this, we isolated E13.5 DRG neurons, and after 3 d in culture we treated them with NGF to investigate the signaling intensity elicited by TrkA as a measure of its own phosphorylation and phosphorylation of the downstream Ras-MAP kinase and the PI3/Akt pathways. As shown in Figure 2, A and B, in line with the increased level of activated TrkA, both MAP kinase and Akt were significantly more phosphorylated in response to NGF treatment in DRG neurons from the TrkAΔKFG-null and heterozygous mice.
Figure 2.
NGF leads to increased TrkA activation and higher AKT and MEK phosphorylation in TrkAΔKFG mutant neurons. A, DRG neurons were isolated from E13.5 WT (+/+), heterozygous (+/−), or TrkAΔKFG mutant (−/−) embryos (6–8 embryos per genotype) and cultured for 3 d, as described in Materials and Methods with 10 ng/ml NGF to promote survival of TrkA-expressing neurons. On the third DIV, the cells were starved for 4 h and treated with or without 100 ng/ml NGF for 5 min. Cell lysates were subjected to Western blot analysis using antibodies specific for TrkA, AKT, and MEK (for loading) and their phosphorylated forms (pTrkA, pAKT, and pMEK). A GAPDH-specific antibody was used to control for loading in the blots probed for TrkA. The data shown are representative of four independent experiments. B, Phosphorylation of TrkA, AKT, and MEK is significantly higher in NGF-treated DRG neurons. Band intensity from blots as shown in A was quantified using NIH ImageJ software. TrkA, AKT, or MEK phosphorylation of +/− and −/− neurons in response to NGF is expressed as percentage increase over, respectively, the ratio of WT pTrkA/GAPDH, and the ratio of AKT or MEK phosphorylation over total AKT or MEK level. Mean ± SEM (n = 4), *p < 0.03 (t test). C, TrkAΔKFG is more sensitive to NGF activation. HEK293 cells expressing WT or ΔKFG TrkA were treated with increasing amounts of NGF (0, 25, 50, and 100 ng/ml) for 5 min and analyzed by Western blotting with an antibody recognizing the phosphorylated Shc binding site of TrkA (top). Total TrkA level was used as control (bottom). D, Quantification of results from C. The data represent the fold increase relative to the band intensity of the WT samples treated with 25 ng/ml and normalized by the total level of TrkA. The results represent the average from three different experiments and are the mean ± SD, *p < 0.05 (t test).
Since the increased signaling could have been caused by the increased number of receptors present in the mutant DRG neurons we decided to employ an in vitro system where we could manipulate TrkA protein levels to study signaling of this mutant Trk receptor compared with WT. For this we used an HEK293 cell line containing an flp site that allows site-specific recombination of a single cDNA copy into the genome where it is driven by a tetracycline-inducible promoter (Flp-In T-Rex system;Life Technologies, see Materials and Methods for details). We generated two cells lines, one expressing a single copy of WT TrkA and another with TrkA lacking the KFG domain. Following induction with doxycycline to allow for comparable expression of TrkA, cells were treated with varying amounts of NGF ranging from 25 to 100 ng/ml. We found that TrkAΔKFG receptors, expressed at comparable levels to that of WT TrkA, were phosphorylated more effectively in response to NGF (Fig. 2C,D). These data suggest that the deletion of the KFG domain leads not only to an increase in total TrkA protein level in DRG, but has also a potentiating role on TrkA signaling since its removal increases TrkA phosphorylation in response to NGF.
Deletion of the KFG domain causes a reduction in the ubiquitination of TrkA receptors
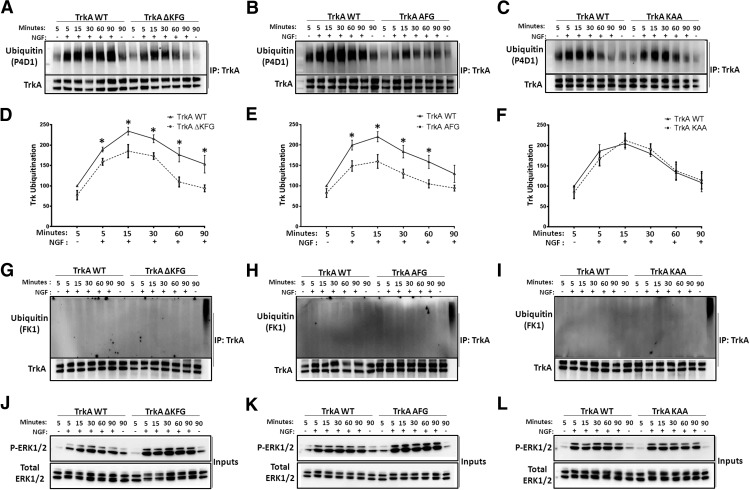
The increased level of TrkA in the mutant animals and the presence of a lysine in the KFG domain prompted us to investigate whether the mutation of this domain influenced TrkA ubiquitination. We again used the HEK293 cell lines that allowed us to control for TrkA expression level. Treatment of both WT-TrkA and TrkAΔKFG lines with NGF caused rapid ubiquitination of the receptor, as previously reported (Arévalo et al., 2006). However, the ubiquitination level of the mutant TrkAΔKFG was significantly reduced compared with the WT TrkA (Fig. 3A,D). To test whether deletion of the lysine in the KFG domain was responsible for the reduced ubiquitination, we generated two additional cell lines: one with an alanine replacing the lysine (TrkA-AFG) and one with a replacement of the phenylalanine and the glycine with two alanines (TrkA-KAA). Analysis of TrkA ubiquitination of these lines showed that while the KAA mutation did not have any effect, the AFG mutation caused a reduction in TrkA ubiquitination comparable to that of the ΔKFG mutant (Fig. 3A–F). These data suggest that the lysine in the KFG domain is important for ubiquitination of TrkA in response to NGF. Moreover, to gain insight as to whether the KFG domain was affecting mono- or poly-ubiquitination we tested the same protein lysates with an antibody that recognizes poly-ubiquitinated but not mono-ubiquitinated proteins. Surprisingly, we found that none of the protein lysates from the NGF-treated cells expressing TrkA was reactive to this antibody strongly suggesting that TrkA is multi-mono-ubiquitinated, as previously suggested (Arévalo et al., 2006; Figure 3G–I). Interestingly, when we examined the downstream signaling of the two new mutant cell lines, i.e., the TrkA-AFG and the TrkA-KAA, we found that the TrkA-AFG line elicited increased Erk phosphorylation in response to NGF similar to the TrkAΔKFG line suggesting that even this phenotype was linked to the lysine of the KFG domain (Fig. 3 J–L).
Figure 3.
Lys450 in the KFG motif is important for the ubiquitination of TrkA receptors in response to NGF. A–C, HEK293T cell lines stably expressing a single copy of WT, KFG deleted (ΔKFG), AFG (single lysine to alanine mutation), or KAA (phenylalanine and glycine converted to two alanines) mutated TrkA were generated as described in Materials and Methods. Cells treated for 5 min with 100 ng/ml NGF and lysed at time points ranging from 5 to 90 min were immunoprecipitated (IP) and blotted with an anti pan-TrkA antibody as a control (bottom) or the anti-ubiquitin antibody P4D1 (top). D–F, Graphs showing the quantification of ubiquitination levels relative to total TrkA in A–C were performed by normalizing the ubiquitin band intensity values to corresponding TrkA bands relative to TrkA WT samples without NGF. The values are given as mean ± SEM and blots are representative of at least three independent experiments; *p < 0.05 (t test). G–I, Blots as in A–C were probed with the FK1 antibody that detects poly-ubiquitinated but not mono-ubiquitinated proteins. Previously characterized poly-ubiquitinated mouse embryo fibroblasts transfected with an HA-ubiquitin expression vector and treated with MG132 were used as controls to exhibit the specificity of the antibody. J–L, Input lysates were blotted with anti-phospho-Erk (as a control of NGF activity) and Erk (as a loading control) antibodies.
TrkAΔKFG receptors are more resistant to downregulation after NGF activation
Since the TrkAΔKFG receptor protein level is increased in DRG neurons and we found that this receptor is less mono-ubiquitinated in response to NGF, we next performed biotinylation experiments to determine the fate of these receptors after NGF activation. It has been proposed that mono-ubiquitin carries both internalization and degradation signals suggesting that a change in the level of ubiquitination of a specific receptor may affect its degradation or turnover (Haglund et al., 2003). HEK293 cells expressing either the WT or ΔKFG TrkA were treated with NGF for a period ranging from 0 to 6 h. For each time point we biotinylated and immunoprecipitated the membrane-bound TrkA receptors to determine the level of internalization and recycling of the receptor. We found that contrary to the WT TrkA that was gradually internalized and most likely degraded within 6 h from the NGF treatment, the mutant receptor was present at a higher level at the cell membrane suggesting that it was more resistant to internalization or more prone to recycling (Fig. 4A,C). Interestingly, again we found that Erk was activated more robustly by NGF as observed in DRG neurons and other in vitro experiments (Figs. 1, 2). To further investigate the fate of activated mutant and WT TrkA receptors, we used an alternative approach in which cells expressing TrkA receptors were biotinylated and, following NGF treatment, were treated with DTT to cleave residual membrane-bound biotin. This procedure allows us to investigate over time the level of TrkA receptors that have been internalized by NGF activation. We found that TrkA receptors lacking the KFG domain are present in higher amounts after internalization for a period up to 3 h (Fig. 4 B,D). Together, these data suggest that reduction in mono-ubiquitination of TrkA facilitates its recycling and possibly makes it more resistant to internalization, providing an explanation for why TrkA receptor levels are augmented in the DRGs of TrkAΔKFG mutant mice.
Figure 4.
Deletion of the KFG domain from TrkA alters NGF-induced trafficking of TrkA. A, HEK293T cells stably expressing WT or KFG deleted TrkA as in Figure 3 were treated with 100 ng/ml NGF for the indicated time (1–6 h) followed by biotinylation. Cell lysates were immunoprecipitated (IP) with Streptavidin beads (Strept) and blotted with an anti-TrkA and actin (used as a negative control) antibodies. Note the increased levels of TrkAΔKFG suggesting reduced degradation and/or internalization of the mutant receptors. B, Recycling of TrkA receptors was analyzed by cleavable biotinylation assay as described in Materials and Methods and immunoblotted as in A. Note the higher level of mutant TrkAΔKFG receptors following internalization suggesting increased recycling. Input lysates in A and B were blotted with anti-TrkA, phospho-Erk (as a control of NGF activity), and GAPDH (as a loading control) antibodies. C, Quantification of surface TrkA levels in A was performed by determining the band intensity of the different biotinylated TrkA samples relative to the non-NGF-treated WT sample. D, Quantification of biotinylated TrkA levels in IP samples in B was determined by comparing the GAPDH-normalized band intensities of the IP TrkA samples at various time points relative to the value of the TrkA WT sample treated with NGF for 5 min. All values are the mean ± SEM from three independent experiments. Statistical significance was calculated with the Student's t test using GraphPad Prism; *p < 0.05.
TrkAΔKFG mutant mice show increased sensitivity to thermal stimuli and are more sensitive to inflammatory pain
Next, we wanted to investigate whether increased TrkA levels have physiological consequences in the mutant mouse. Since NGF/TrkA signaling is important for nociceptor function (Ugolini et al., 2007; McNamee et al., 2010) we used a series of tests aimed at evaluating the ability of the mice to detect noxious stimuli. Testing of mutants and controls with the Hargreaves test showed that TrkAΔKFG mice had significantly lower withdrawal latency to a noxious thermal stimulus when compared with WT mice (WT 9.39 ± 0.64 s; TrkAΔKFG, 7.06 ± 0.58 s, n = 11 per genotype, p < 0.05; Fig. 5A) suggesting increased sensitivity. Moreover, in the two-temperature choice test, in which mice can choose to spend time on a plate at either 32°C or a plate at different temperatures, mutant mice spent significantly less time at either cold (14°C, 20°C) or hot temperatures (41°C, 45°C; Fig. 5B) suggesting increased sensitivity to thermal stimulation. Next, we used the formalin test as a model of inflammatory pain. We measured spontaneous pain behavior elicited by the injection of a diluted solution of formalin in the plantar surface by scoring both licking and lifting/favoring of the injected paw (Fig. 5). In Phase I, corresponding to acute nociception immediately after the injection and lasting ∼10 min, we found no significant difference between control and TrkAΔKFG mice. However, in Phase II, during the central sensitization and inflammation period mutant mice displayed significantly higher spontaneous pain behavior as compared with their WT littermates. Together, these data strongly suggest that deletion of the TrkA KFG domain enhances the pain response in mice and points to an inhibitory activity of this domain on the nociception mediated function of TrkA.
Figure 5.
TrkAΔKFG mice have increased sensitivities to thermal and chemical stimuli. A, TrkAΔKFG mice have a lowered threshold in the Hargreaves test (Student's t test, p < 0.05). Graph showing the average time (in seconds) by which the mice responded to the thermal stimuli. B, TrkAΔKFG mice spend less time than controls in the chamber with variable temperature in the two-temperature choice test. Side A of the chamber was kept at the constant temperature of 32°C whereas side B of the chamber had variable temperature ranging from 4 to 50°C as described in Materials and Methods. TrkAΔKFG mice spent significantly less time at 14, 20, and 45°C (two-way ANOVA, p < 0.05). C, TrkAΔKFG mice have increased sensitivity in Phase II formalin test (two-way ANOVA, p < 0.05); n = 11 for each genotype in each test.
Discussion
In this study, we have investigated the physiological significance of a 3 aa domain (KFG) in the juxtamembrane region of the TrkA tyrosine kinase receptor. This domain (mouse residues 450–452 corresponding to human residues 441–443) is conserved among all Trk genes suggesting an important biological function (Peng et al., 1995). Our analysis has revealed two interesting features of this domain. It controls TrkA protein level and it has an inhibitory role on TrkA signaling. Physiologically, KFG deletion increases the sensitivity to pain while leaving DRG number and cell-type composition unaffected. Mechanistically, it controls the level of TrkA ubiquitination, with lysine 450 being critical for this function. Together, our data establish the in vivo relevance of TrkA ubiquitination and underscore the importance of ubiquitination in the physiology of nociceptive neurons.
The in vitro mechanistic dissection of KFG function suggests that lysine 450 is necessary and sufficient for the change in TrkA ubiquitination and increased signaling. This result was somewhat surprising because all three KFG amino acids are conserved among Trk receptors. Thus, it is conceivable that, despite the data from the in vitro analysis, in a more complex in vivo scenario the mutation of lysine 450 or the mutation/deletion of all three KFG amino acids may have different penetrance on the phenotype as the physiological cellular context is more complex. Nevertheless, our data clearly identify this domain as a critical modulator of TrkA ubiquitination. They also provide evidence that ubiquitination is an important mechanism to control TrkA function in vivo. In fact, to date, such a role was only postulated based on in vitro or ex vivo experiments and its physiological significance has remained untested (Geetha et al., 2005; Arévalo et al., 2006; Yu et al., 2011).
The increase in TrkA protein level caused by KFG deletion suggests that this domain may affect recycling and/or internalization of this receptor. Indeed, in biotinylation experiments we have found that, over time, there are more TrkAΔKFG mutant receptors at the membrane and more receptors are recycled after NGF-induced internalization (Fig. 4). This is in agreement with other data showing that depletion of the E3 ligase NEDD4-2 results in a reduction of TrkA mono-ubiquitination and an increase in membrane bound TrkA receptor due to more efficient recycling (Yu et al., 2011). Conversely, TrkA mutants with stronger binding affinity for NEDD4-2 are more ubiquitinated and have a higher degradation rate with lower recycling to the membrane (Georgieva et al., 2011). The report that a less ubiquitinated TrkA mutant (i.e., TrkA K485; Geetha et al., 2005) does not get internalized efficiently and has reduced signaling ability suggests that the TrkAΔKFG mutant that is also less ubiquitinated but is activated more by NGF, probably has an increased recycling rate rather than a defect in internalization (Fig. 4). However, the K485 and KFG domains of TrkA most likely control different mechanisms of ubiquitination with separate outcomes for the receptor. One is dependent on p75 recruiting E2 and E3 ligases, which in turn ubiquitinate TrkA receptors, to internalize and direct them to the endosomal compartment (Geetha et al., 2005). Whereas in the KFG mutant the mechanism is p75 independent since we were able to reproduce the initial results from mutant DRG neurons in HEK293 cells that do not express p75 (Fig. 4). It should also be noted that the role of p75 in promoting TrkA ubiquitination is still not clear as another study has reported that it may prolong TrkA signaling by suppressing its ubiquitination and thereby delaying its internalization and degradation rate (Makkerh et al., 2005). Nevertheless, our data suggest that a change in TrkA interaction with p75 is unlikely to be the cause of the TrkAΔKFG phenotype (E. Kiris and L. Tessarollo, unpublished observations) and future studies should also include, for example, the analysis of the interaction of TrkAΔKFG mutants with ubiquitin ligases that have been shown to mediate the ubiquitination and degradation of other receptor tyrosine kinases. Indeed, c-Cbl, a ubiquitin ligase that promotes the ubiquitination and degradation of hepatocyte growth factor, EGF, and platelet-derived growth factor receptors recently has been shown to mediate the NGF-induced ubiquitination and degradation of TrkA as well (Takahashi et al., 2011). Regardless, despite the different mechanisms leading to changes in ubiquitination of TrkA receptors in all models investigated so far–including the TrkA K485 depending on TrkA/p75 interaction for ubiquitination, TrkA P782S disrupting the binding of the Nedd4-2 ubiquitin ligase to TrkA, and the TrkAΔKFG described in this study–suggest the importance of ubiquitination in controlling TrkA signaling.
TrkA receptors lacking KFG have an increased ability to stimulate downstream signaling in response to NGF. Depletion of NEDD4-2 also causes an increase in NGF signaling in isolated DRG neurons. However, the increased level of TrkA caused by NEDD4-2 depletion made it unclear as to whether the underlying cause was an increase in the number of TrkA receptors present at the membrane (Yu et al., 2011). Nevertheless, recent data obtained from primary neurons of mutant mice with TrkA receptors unable to bind NEDD4-2, have also reported reduced levels of ubiquitination and increased signaling efficiency in response to NGF similar to that found in the TrkAΔKFG mutant (J.C. Arévalo, unpublished results). The mechanism for the increased signaling is still not fully elucidated. However, for the TrkAΔKFG mutant we hypothesize that since this domain lies in a region where macromolecular complexes are recruited after TrkA activation, a change in ubiquitination of this domain may alter the dynamics by which specific proteins are recruited and transduce signaling. For example, the juxtamembrane region of phosphorylated TrkA binds two key adapter proteins in a competitive way at Tyr 490, namely Shc and FRS-2/SNT, which activate other signaling proteins after phosphorylation (Meakin et al., 1999; Lemmon and Schlessinger, 2010). Moreover, FRS-2/SNT binds to TrkA in the KFG domain as well (Peng et al., 1995). Thus, one hypothesis is that deletion of this site alters the equilibrium of binding of Shc and FRS-2/SNT to TrkA Tyr490. If mono-ubiquitination is used to displace FRS-2/SNT, removal of the KFG domain could favor FRS-2 interaction with TrkA.
Of course a mechanism using ubiquitination to control the recruitment and interactions of proteins to active TrkA may be more general and be used to modulate the activity of TrkA in response to NGF stimulation. For example, the multimodular guanine nucleotide exchange factor Kalirin-5 has been reported to bind to TrkA and potentiate its autophosphorylation (Chakrabarti et al., 2005). However, TrkA ubiquitination could favor Kalirin-5 displacement to control TrkA activation levels. Although the precise binding site of Kalirin has not yet been determined, in the future it will be of interest to determine whether proteins such as FRS-2 or Kalirin can bind more efficiently to TrkAΔKFG or TrkAP782S, both of which are more resistant to ubiquitination (Arévalo et al., 2006; Yu et al., 2011). In this regard, since FRS-2 is also a substrate for TrkB (Easton et al., 1999) and it has been suggested that Kalirin-5, in addition to TrkA, may affect TrkB and TrkC signaling as well (Chakrabarti et al., 2005), it is tempting to speculate that the conserved Trk receptor KFG domain may have a more general role in controlling all Trk receptors signaling. Deletion of KFG in TrkB and TrkC receptors should help address this issue and should provide information as to whether pharmacologically targeting this domain to boost Trk receptor signaling could have a general effect on the function of all Trk genes.
Very recently, it has been reported that deletion of NEDD4-2 in nociceptive neurons causes thermal hypersensitivity and enhances the reaction in the second phase after formalin injection most likely due to alteration in voltage-gated sodium channel function and distribution (Laedermann et al., 2013). Although they limited their analysis to sodium channels function, it is conceivable that TrkA level or localization has also been affected by NEDD4-2 deletion. In fact, TrkA is a direct target of this E3 ligase and abrogating a NEDD4-2 binding site in TrkA increases pain sensitivity in mice (J.C. Arévalo, unpublished observations). Our finding that the TrkAΔKFG mutants have increased pain sensitivity without causing a change in neuronal number or composition of the DRGs adds a new dimension to the critical role of ubiquitination in nociceptive function. Moreover, it suggests that ubiquitination is critical to the fine-tuning of the function of DRG nociceptors with significant implications to the genesis of neuropathic pain. In the future, it will be of interest to determine, by array or proteomic analysis, which changes are downstream of alterations in TrkA ubiquitination to test whether similar abnormalities are present in patients with pathological pain.
In summary, our data show that ubiquitination of TrkA is critical to the normal function of nociceptors and support the notion that alterations in TrkA ubiquitination can influence pain sensitivity and may be one of the causes of inflammatory pain.
Footnotes
This research was supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, National Institutes of Health. We thank Eileen Southon and Susan Reid for technical help in generating the TrkA mutant mouse model, Colleen Barrick for technical help, and Allan Weissman for discussion and suggestions. We are grateful to Robert Stephens for the discussions at the start of the project and Juan Carlos Arévalo for sharing his unpublished results.
References
- Acconcia F, Sigismund S, Polo S. Ubiquitin in trafficking: the network at work. Exp Cell Res. 2009;315:1610–1618. doi: 10.1016/j.yexcr.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Arévalo JC, Waite J, Rajagopal R, Beyna M, Chen ZY, Lee FS, Chao MV. Cell survival through Trk neurotrophin receptors is differentially regulated by ubiquitination. Neuron. 2006;50:549–559. doi: 10.1016/j.neuron.2006.03.044. [DOI] [PubMed] [Google Scholar]
- Bibel M, Barde YA. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000;14:2919–2937. doi: 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- Chakrabarti K, Lin R, Schiller NI, Wang Y, Koubi D, Fan YX, Rudkin BB, Johnson GR, Schiller MR. Critical role for Kalirin in nerve growth factor signaling through TrkA. Mol Cell Biol. 2005;25:5106–5118. doi: 10.1128/MCB.25.12.5106-5118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MV, Rajagopal R, Lee FS. Neurotrophin signalling in health and disease. Clin Sci. 2006;110:167–173. doi: 10.1042/CS20050163. [DOI] [PubMed] [Google Scholar]
- Coppola V, Kucera J, Palko ME, Martinez-De Velasco J, Lyons WE, Fritzsch B, Tessarollo L. Dissection of NT3 functions in vivo by gene replacement strategy. Development. 2001;128:4315–4327. doi: 10.1242/dev.128.21.4315. [DOI] [PubMed] [Google Scholar]
- Crowley C, Spencer SD, Nishimura MC, Chen KS, Pitts-Meek S, Armanini MP, Ling LH, McMahon SB, Shelton DL, Levinson AD. Mice lacking nerve growth factor display perinatal loss of sensory and sympathetic neurons yet develop basal forebrain cholinergic neurons. Cell. 1994;76:1001–1011. doi: 10.1016/0092-8674(94)90378-6. [DOI] [PubMed] [Google Scholar]
- Easton JB, Moody NM, Zhu X, Middlemas DS. Brain-derived neurotrophic factor induces phosphorylation of fibroblast growth factor receptor substrate 2. J Biol Chem. 1999;274:11321–11327. doi: 10.1074/jbc.274.16.11321. [DOI] [PubMed] [Google Scholar]
- Geetha T, Jiang J, Wooten MW. Lysine 63 polyubiquitination of the nerve growth factor receptor TrkA directs internalization and signaling. Mol Cell. 2005;20:301–312. doi: 10.1016/j.molcel.2005.09.014. [DOI] [PubMed] [Google Scholar]
- Georgieva MV, de Pablo Y, Sanchis D, Comella JX, Llovera M. Ubiquitination of TrkA by Nedd4-2 regulates receptor lysosomal targeting and mediates receptor signaling. J Neurochem. 2011;117:479–493. doi: 10.1111/j.1471-4159.2011.07218.x. [DOI] [PubMed] [Google Scholar]
- Haglund K, Dikic I. The role of ubiquitylation in receptor endocytosis and endosomal sorting. J Cell Sci. 2012;125:265–275. doi: 10.1242/jcs.091280. [DOI] [PubMed] [Google Scholar]
- Haglund K, Di Fiore PP, Dikic I. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem Sci. 2003;28:598–603. doi: 10.1016/j.tibs.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Laedermann CJ, Cachemaille M, Kirschmann G, Pertin M, Gosselin RD, Chang I, Albesa M, Towne C, Schneider BL, Kellenberger S, Abriel H, Decosterd I. Dysregulation of voltage-gated sodium channels by ubiquitin ligase NEDD4-2 in neuropathic pain. J Clin Invest. 2013;123:3002–3013. doi: 10.1172/JCI68996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo FM, Massa SM. Neurotrophin-based strategies for neuroprotection. J Alzheimers Dis. 2004;6:S13–S17. doi: 10.3233/jad-2004-6s606. [DOI] [PubMed] [Google Scholar]
- Makkerh JP, Ceni C, Auld DS, Vaillancourt F, Dorval G, Barker PA. p75 neurotrophin receptor reduces ligand-induced Trk receptor ubiquitination and delays Trk receptor internalization and degradation. EMBO Rep. 2005;6:936–941. doi: 10.1038/sj.embor.7400503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- McNamee KE, Burleigh A, Gompels LL, Feldmann M, Allen SJ, Williams RO, Dawbarn D, Vincent TL, Inglis JJ. Treatment of murine osteoarthritis with TrkAd5 reveals a pivotal role for nerve growth factor in non-inflammatory joint pain. Pain. 2010;149:386–392. doi: 10.1016/j.pain.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Meakin SO, MacDonald JI, Gryz EA, Kubu CJ, Verdi JM. The signaling adapter FRS-2 competes with Shc for binding to the nerve growth factor receptor TrkA. A model for discriminating proliferation and differentiation. J Biol Chem. 1999;274:9861–9870. doi: 10.1074/jbc.274.14.9861. [DOI] [PubMed] [Google Scholar]
- Peng X, Greene LA, Kaplan DR, Stephens RM. Deletion of a conserved juxtamembrane sequence in Trk abolishes NGF-promoted neuritogenesis. Neuron. 1995;15:395–406. doi: 10.1016/0896-6273(95)90043-8. [DOI] [PubMed] [Google Scholar]
- Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SW, Tessarollo L. Isolation, microinjection and transfer of mouse blastocysts. Methods Mol Biol. 2009;530:269–285. doi: 10.1007/978-1-59745-471-1_14. [DOI] [PubMed] [Google Scholar]
- Santos SD, Verveer PJ, Bastiaens PI. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol. 2007;9:324–330. doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- Silos-Santiago I, Molliver DC, Ozaki S, Smeyne RJ, Fagan AM, Barbacid M, Snider WD. Non-TrkA-expressing small DRG neurons are lost in TrkA deficient mice. J Neurosci. 1995;15:5929–5942. doi: 10.1523/JNEUROSCI.15-09-05929.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeyne RJ, Klein R, Schnapp A, Long LK, Bryant S, Lewin A, Lira SA, Barbacid M. Severe sensory and sympathetic neuropathies in mice carrying a disrupted Trk/NGF receptor gene. Nature. 1994;368:246–249. doi: 10.1038/368246a0. [DOI] [PubMed] [Google Scholar]
- Southon E, Tessarollo L. Manipulating mouse embryonic stem cells. Methods Mol Biol. 2009;530:165–185. doi: 10.1007/978-1-59745-471-1_9. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Shimokawa N, Esmaeili-Mahani S, Morita A, Masuda H, Iwasaki T, Tamura J, Haglund K, Koibuchi N. Ligand-induced downregulation of TrkA is partly regulated through ubiquitination by Cbl. FEBS Lett. 2011;585:1741–1747. doi: 10.1016/j.febslet.2011.04.056. [DOI] [PubMed] [Google Scholar]
- Tessarollo L. Pleiotropic functions of neurotrophins in development. Cytokine Growth Factor Rev. 1998;9:125–137. doi: 10.1016/S1359-6101(98)00003-3. [DOI] [PubMed] [Google Scholar]
- Tessarollo L, Tsoulfas P, Donovan MJ, Palko ME, Blair-Flynn J, Hempstead BL, Parada LF. Targeted deletion of all isoforms of the trkC gene suggests the use of alternate receptors by its ligand neurotrophin-3 in neuronal development and implicates trkC in normal cardiogenesis. Proc Natl Acad Sci U S A. 1997;94:14776–14781. doi: 10.1073/pnas.94.26.14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoenen H, Sendtner M. Neurotrophins: from enthusiastic expectations through sobering experiences to rational therapeutic approaches. Nat Neurosci. 2002;(5 Suppl):1046–1050. doi: 10.1038/nn938. [DOI] [PubMed] [Google Scholar]
- Ugolini G, Marinelli S, Covaceuszach S, Cattaneo A, Pavone F. The function neutralizing anti-TrkA antibody MNAC13 reduces inflammatory and neuropathic pain. Proc Natl Acad Sci U S A. 2007;104:2985–2990. doi: 10.1073/pnas.0611253104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanpallewar SU, Barrick CA, Buckley H, Becker J, Tessarollo L. Deletion of the BDNF truncated receptor TrkB.T1 delays disease onset in a mouse model of amyotrophic lateral sclerosis. PLoS One. 2012;7:e39946. doi: 10.1371/journal.pone.0039946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Calvo L, Anta B, López-Benito S, Southon E, Chao MV, Tessarollo L, Arévalo JC. Regulation of trafficking of activated TrkA is critical for NGF-mediated functions. Traffic. 2011;12:521–534. doi: 10.1111/j.1600-0854.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]