SUMMARY
Autophagy has been implicated as a component of host defense, but the significance of antimicrobial autophagy in vivo and the mechanism by which it is regulated during infection are poorly defined. Here we found that antiviral autophagy was conserved in flies and mammals during infection with Rift Valley fever virus (RVFV), a mosquito-borne virus that causes disease in humans and livestock. In Drosophila, Toll-7 limited RVFV replication and mortality through activation of autophagy. RVFV infection also elicited autophagy in mouse and human cells, and viral replication was increased in the absence of autophagy genes. The mammalian Toll-like receptor adaptor, MyD88, was required for anti-RVFV autophagy, revealing an evolutionarily conserved requirement for pattern-recognition receptors in antiviral autophagy. Pharmacologic activation of autophagy inhibited RVFV infection in mammalian cells, including primary hepatocytes and neurons. Thus, autophagy modulation might be an effective strategy for treating RVFV infection, which lacks approved vaccines and therapeutics.
INTRODUCTION
Autophagy is a fundamental process that mediates the breakdown of cytoplasmic material and is conserved from yeast to humans (Mizushima and Komatsu, 2011). As an adaptive response to cellular stress such as starvation, autophagy involves the formation of double-membrane vesicles that degrade the engulfed content to recycle nutrients (Lum et al., 2005). In addition to controlling this bulk degradation program, autophagy genes have also been co-opted by the innate immune system to orchestrate cell-autonomous antimicrobial responses against diverse pathogens, including viruses (Deretic and Levine, 2009; Levine et al., 2011). For example, in mammals, autophagy restricts the replication or pathogenesis of Herpes simplex virus type 1 (HSV-1), Sindbis virus (SINV), and Chikungunya virus (CHIKV) (Joubert et al., 2012; Leib et al., 2009; Orvedahl et al., 2010). Thus, pharmacological autophagy manipulation has been proposed as an attractive strategy for treating viral infection, as well as other diseases including neurodegenerative disorders in which pathogenic protein aggregates are cleared by autophagy (Rubinsztein et al., 2012; Shoji-Kawata et al., 2013). Nevertheless, a better understanding of how autophagy is regulated during infection is critical to harness this pathway for therapeutic purposes.
Emerging data suggest that pattern-recognition receptors (PRRs) can regulate autophagy activation. These receptors detect conserved microbial signatures known as pathogen-associated molecular patterns (PAMPs) to elicit protective effector responses (Akira et al., 2006). Viruses can be recognized at the cell surface or in endosomal compartments by Toll-like receptors (TLRs), which were originally identified through homology to Drosophila Toll (Lemaitre et al., 1996; Uematsu and Akira, 2006). Intracellular sensors such as retinoic acid-inducible gene-1 (RIG-I) and melanoma differentiation-associated protein 5 (MDA-5), collectively termed RIG-I-like receptors (RLRs), can also detect viral nucleic acids in the cytoplasm (Loo and Gale, 2011). Interestingly, canonical ligands for several TLRs induce autophagy in macrophages (Campbell and Spector, 2012; Delgado et al., 2008; Shi and Kehrl, 2008; Xu et al., 2007). However, the role of PRR-mediated autophagy in mammals, across diverse cell types and during viral infection, has remained largely unexplored.
Taking advantage of Drosophila, recent studies provided critical insights into the significance of this PRR-autophagy axis in host defense (Nakamoto et al., 2012; Yano et al., 2008). In flies, autophagy is essential for controlling Vesicular stomatitis virus (VSV) replication and limiting host lethality (Shelly et al., 2009). Toll-7 is the PRR that recognizes VSV to trigger antiviral autophagy and is rapidly induced transcriptionally by infection, along with several other Toll receptors (Nakamoto et al., 2012; Xu et al., 2012). Yet it remains unclear whether Toll receptors and autophagy restrict other arthropod-borne viruses (arboviruses), especially those that are more pathogenic in humans, and whether this TLR-autophagy axis is conserved in mammals during infection.
By screening Toll receptor mutant flies, we identified a role for Toll-7 in controlling the bunyavirus Rift Valley fever virus (RVFV), a mosquito-transmitted pathogen that causes disease in humans and livestock (Walter and Barr, 2011). Though RVFV infection typically causes a self-limiting febrile illness, the liver is a major target of infection, with ~50% of infected humans presenting with jaundice. Some patients develop more severe manifestations such as hemorrhagic fever and encephalitis, and recent outbreaks have been particularly deadly (Ikegami and Makino, 2011). Livestock are exceptionally susceptible to infection, and mortality rates approach 100% in newborn animals. Furthermore, although historically RVFV has been limited to Sub-Saharan Africa, the virus has recently spread and can be weaponized. Vaccines and therapeutics are unavailable for RVFV, necessitating additional insight into the pathogenesis and immunologic control of the virus.
We found that Toll-7 mutant flies are more susceptible to RVFV infection as a result of a defect in virus-triggered antiviral autophagy, which is essential for controlling viral replication and survival. The role of antiviral autophagy is conserved in mammalian cells, because RVFV infection activated autophagy in murine and human cells, and loss of autophagy genes enhanced viral replication. The mammalian TLR signaling-adaptor myeloid-differentiation primary response 88 (MyD88) restricted RVFV infection and was required for anti-RVFV autophagy, suggesting a clear parallel whereby both Drosophila Tolls and mammalian TLRs direct antiviral autophagy. Pharmacologic autophagy activation potently inhibited RVFV infection in mammalian cells, including primary hepatocytes and neurons, cell types targeted during pathogenic human RVFV infection. Therefore, autophagy activation might be an effective strategy for treating RVFV and other viral infections, especially neurotropic and hepatotropic pathogens that target tissues in which autophagy is highly manipulable.
RESULTS
Toll-7 Restricts RVFV Replication in Adult Flies
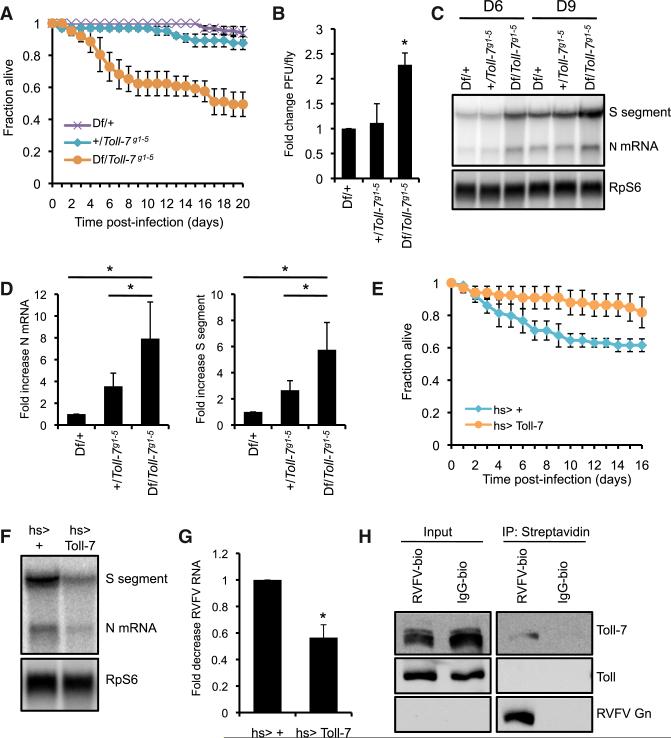
Flies encode nine Toll receptors, but their functions during viral infection have not been comprehensively evaluated. Therefore, we screened a subset of these receptors (Toll, Toll-2, Toll-6, Toll-7, Toll-8, Toll-9) for antiviral functions by challenging mutant flies with RVFV. From these studies, we identified a critical role for Toll-7 in limiting susceptibility to RVFV infection. Compared to sibling controls, heteroallelic Toll-7 mutant flies (Df(2R) BSC22/Toll-7g1-5) that express no Toll-7 (Yagi et al., 2010) exhibited diminished survival and elevated viral titers after infection with the attenuated MP12 strain of RVFV (Figures 1A and 1B). Furthermore, loss of Toll-7 increased levels of RVFV RNA (S segment genomic RNA and N mRNA) (Figures 1C and 1D). These data suggest that Toll-7 is required for resistance to RVFV infection in vivo.
Figure 1. Toll-7 Restricts RVFV Infection in Adult Flies.
(A) Survival of RVFV-infected Toll-7 mutant flies (Toll-7 null: Df(2R)BSC22/Toll-7g1-5) or sibling controls (Toll-7 heterozygous: Df(2R)BSC22/+, +/Toll-7g1-5). Mean ± SE; p < 0.001, log-rank test.
(B) Viral titers from RVFV-infected flies as measured by plaque assay 3 dpi. Mean ± SD; *p < 0.05, Student's t test.
(C) RNA blot for RVFV RNA 6 dpi.
(D) Fold increase in RVFV S segment genomic RNA and N mRNA 6 dpi normalized to Df(2R)BSC22/+ control. Mean ± SE; *p < 0.01, Student's t test.
(E) Survival of RVFV-infected Toll-7-overexpressing flies (hs-Gal4 > UAS-Toll-7) and control flies (hs-Gal4 > +). Mean ± SE; p < 0.02, log-rank test.
(F) RNA blot for RVFV RNA 6 dpi.
(G) Fold decrease in RVFV N mRNA levels 6 dpi. Mean ± SE; *p < 0.05, Student's t test.
(H) Biotinylated RVFV virions or IgG were incubated with Drosophila cells at 4°C for 1 hr and precipitated with streptavidin from cell lysates. Bound proteins were monitored by immunoblot. All data are representative of three independent experiments. Figure 1, see also Figure S1.
Restriction of RVFV replication is specific for Toll-7. Flies lacking Toll-8 showed no change in lifespan, RVFV titers or viral RNA compared to control flies (see Figures S1A–S1C available online). Likewise, mutants in Toll, Toll-2, Toll-6, and Toll-9 were not more susceptible to RVFV challenge (Figures S1D–S1G). Moreover, we observed no change in survival for Toll-7 mutants infected with Drosophila C virus (DCV), Flockhouse virus (FHV), or SINV (Figure S1H–S1J), and Toll-7 mutant flies harbored equivalent amounts of DCV protein compared to sibling controls (Figure S1K). Toll-7 is also dispensable for antimicrobial peptide induction after bacterial infection (Yagi et al., 2010). Taken together, these data reveal that Toll-7 specifically limits viral replication and mortality in RVFV-challenged flies.
To complement these loss-of-function studies, we next determined whether Toll-7 overexpression impacts RVFV infection by using previously characterized flies that express Toll-7 with heat-shock induction (Nakamoto et al., 2012). Compared to control flies, Toll-7-overexpressing flies showed modestly enhanced survival (Figure 1E) and decreased viral RNA (Figures 1F and 1G). In contrast, overexpression of other Toll receptors did not reduce mortality during RVFV infection (Figure S1L). Furthermore, Toll-7 overexpression did not limit lethality with DCV, SINV, and FHV infection (Figure S1M; data not shown). Thus, both loss-of-function and gain-of-function studies demonstrate that Toll-7 is an essential facet of defense against RVFV infection.
To determine whether Toll-7 functions as a PRR for RVFV, we prebound biotinylated purified RVFV virions to Drosophila cells in the cold to allow surface binding but no internalization, and precipitated the virus using streptavidin. Biotinylated RVFV but not control immunoglobulin G (IgG) precipitated endogenous Toll-7, suggesting that RVFV and Toll-7 physically interact at the cell surface (Figure 1H). Moreover, biotinylated RVFV did not precipitate Toll, demonstrating specificity (Figure 1H). Therefore, RVFV particles, which only contain the viral glycoproteins on the surface, act as functional ligands for Toll-7, and this recognition is critical for restricting RVFV infection.
Toll-7-Dependent Antiviral Autophagy Is Required for Host Defense in Flies
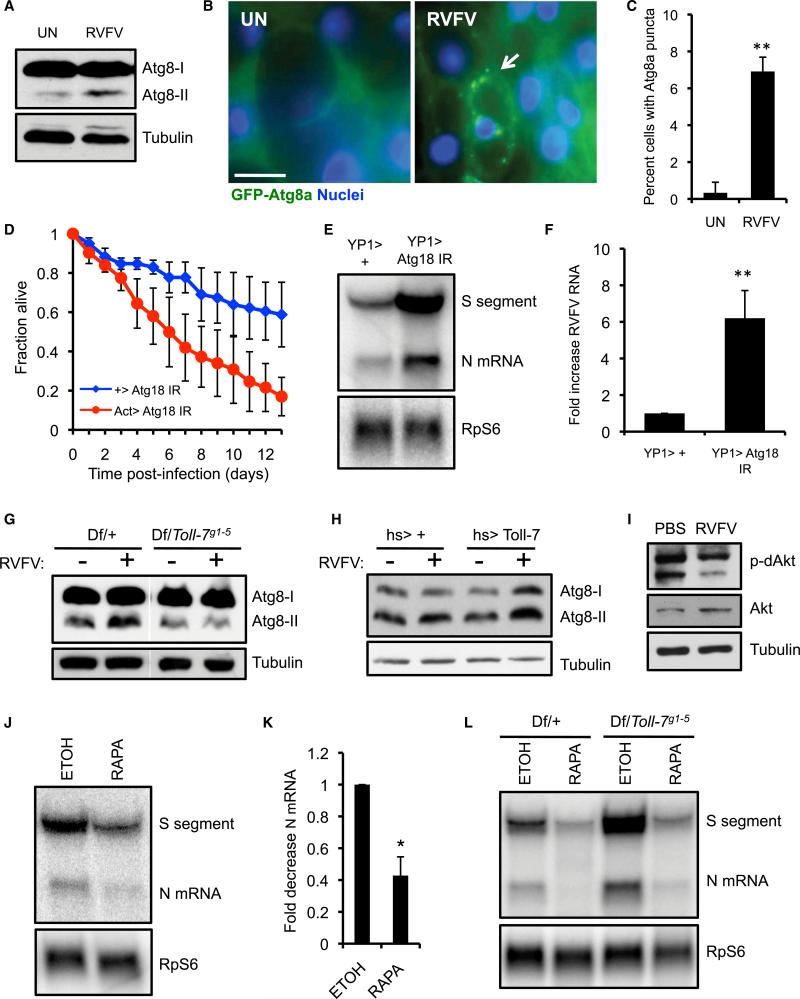
We next examined the mechanism by which Toll-7 controls antiviral immunity during RVFV infection. Because Toll-7 is required for VSV-induced antiviral autophagy (Nakamoto et al., 2012), we hypothesized that RVFV infection engages a similar autophagic pathway through Toll-7. During autophagy, Atg8-I is conjugated to a lipid moiety, and this form (Atg8-II) is recruited to autophagosomal membranes (Shelly et al., 2009). An elevation in Atg8-II expression consequently reflects autophagy activation. Compared to uninfected flies, RVFV-infected flies showed increased Atg8-II, suggesting that RVFV infection induces autophagy (Figure 2A). In contrast, DCV and SINV infection did not promote Atg8-II conversion (Figures S2A and S2B). We also monitored flies that express green fluorescent protein (GFP)-tagged Atg8a in the adult female fat body, the functional equivalent of the mammalian liver and a site of arboviral replication, including RVFV (Deddouche et al., 2008; Shelly et al., 2009) (data not shown). GFP-Atg8a forms discrete puncta that correspond to autophagosomes (Scott et al., 2004). While GFP+ puncta were largely absent in fat bodies from uninfected flies, fat bodies from RVFV-infected flies showed an increase in cells containing GFP+ foci (Figures 2B and 2C). Collectively, these results suggest that RVFV infection elicits autophagy in vivo.
Figure 2. Toll-7 Is Required for RVFV-Induced Autophagy.
(A) WT flies (w1118) were infected with RVFV, and Atg8 expression was monitored by immunoblot 3 dpi.
(B) Fat body images from uninfected or RVFV-challenged flies expressing GFP-Atg8a (YP1-Gal4 > UAS-GFP-Atg8a). The arrow indicates a cell with GFP-Atg8a puncta in an infected animal. Scale bar represents 10 μM.
(C) Percent of fat body cells containing GFP-Atg8a puncta from uninfected or RVFV-infected flies. Mean ± SD; **p < 0.01, Student's t test.
(D) Survival of RVFV-infected Atg18-silenced flies (Act-Gal4 > UAS-Atg18 IR) or control flies (+ > UAS-Atg18 IR). Mean ± SE; p < 0.001, log-rank test.
(E) Representative RNA blot of RVFV RNA from flies depleted of Atg18 in the fat body (YP1-Gal4 > Atg18 IR) or control flies 6 dpi.
(F) Fold increase in RVFV RNA in Atg18-silenced flies. Mean ± SE; **p < 0.01, Student's t test.
(G) Atg8 immunoblot from uninfected or RVFV-infected Toll-7 mutant or control flies.
(H) Atg8 immunoblot from uninfected or RVFV-challenged Toll-7-overexpressing or control flies. I. Flies (hs > +) were challenged with insulin and either RVFV or PBS, and Akt activity was monitored by immunoblot 3 dpi.
(J) RNA blot for RVFV RNA from flies fed either rapamycin (100 μM) (RAPA) or vehicle control (ethanol, ETOH) 6 dpi.
(K) Fold decrease in viral RNA with rapamycin treatment. Mean ± SE; *p < 0.05, Student's t test.
(L) RNA blot for RVFV RNA 6 dpi from Toll-7 mutant or control flies fed either rapamycin or ethanol. All data are representative of at least three independent experiments. Figure 2, see also Figure S2.
To determine whether RVFV-induced autophagy restricts viral infection, we silenced a core autophagy gene (Atg18) by using established in vivo RNAi transgenic flies (Shelly et al., 2009). Compared to sibling controls, Atg18-depleted flies showed greatly enhanced lethality after RVFV infection (Figure 2D). Moreover, silencing Atg18 and two other core autophagy genes (Atg5 and Atg7) specifically in the fat body significantly increased amounts of RVFV RNA (Figures 2E and 2F; Figure S2C). Therefore, autophagy genes are critical for resistance to RVFV infection in adult flies.
We next tested the role of Toll-7 in controlling RVFV-induced autophagy. While flies heterozygous for Toll-7 exhibited an increase in Atg8-II protein after RVFV infection, Atg8-II induction was abrogated in Toll-7 mutant flies, suggesting that Toll-7 is required for autophagy activation in response to RVFV infection (Figure 2G). Conversely, Toll-7 overexpression enhanced the increase in Atg8-II after infection, demonstrating that Toll-7 expression positively correlates with RVFV-elicited autophagy activation (Figure 2H).
VSV infection in Drosophila attenuates PI3K-Akt signaling, which activates the antiviral autophagy program (Shelly et al., 2009). To evaluate whether RVFV infection represses this pathway, we monitored phospho-Akt expression. Because resting phospho-Akt was not detectable, we pretreated flies with insulin to increase basal phospho-Akt and challenged them with RVFV. Under these conditions, RVFV-infected flies showed reduced phospho-Akt compared to uninfected flies with no decrease in total Akt (Figure 2I), demonstrating that RVFV infection also inhibits Akt signaling to regulate antiviral autophagy in vivo.
Given that Toll-7-dependent autophagy restricts RVFV replication, we next determined whether pharmacologic autophagy activation protects against infection. Because Tor inhibits autophagy, drugs that inhibit Tor (such as rapamycin) are classically used to activate autophagy. We fed flies 100 mM rapamycin, a dose that has been used previously, which activated autophagy as shown by increased Atg8-II conversion (Figure S2D). Lower amounts of viral RNA were detected in rapamycin-fed flies compared to controls (Figures 2J and 2K). We next determined whether pharmacologically-induced autophagy could suppress the increased virus replication in Toll-7 mutant flies. Again, vehicle-fed Toll-7 mutant flies showed increased amounts of viral RNA compared to sibling controls (Figure 2L). Rapamycin treatment of Toll-7 mutants reduced viral RNA to amounts similar to those detected in control flies. Thus, rapamycin treatment protects against RVFV infection and bypasses the requirement for Toll-7 in limiting virus replication. Together, these data suggest that Toll-7 orchestrates a protective antiviral autophagy response during RVFV infection in vivo.
Autophagy-Mediated Antiviral Defense against RVFV Is Conserved in Mammalian Cells
VSV can induce autophagy in murine and human cell lines, as well as in mouse macrophages and plasmacytoid dendritic cells (Lee et al., 2007; Tal et al., 2009). Despite this robust activation, autophagy inhibition does not result in increased VSV replication, which has been attributed to the role of autophagy in attenuating RLR-signaling and thus Type I interferon (IFN-I) production. However, a role for autophagy in RVFV infection of mammalian cells has not been explored.
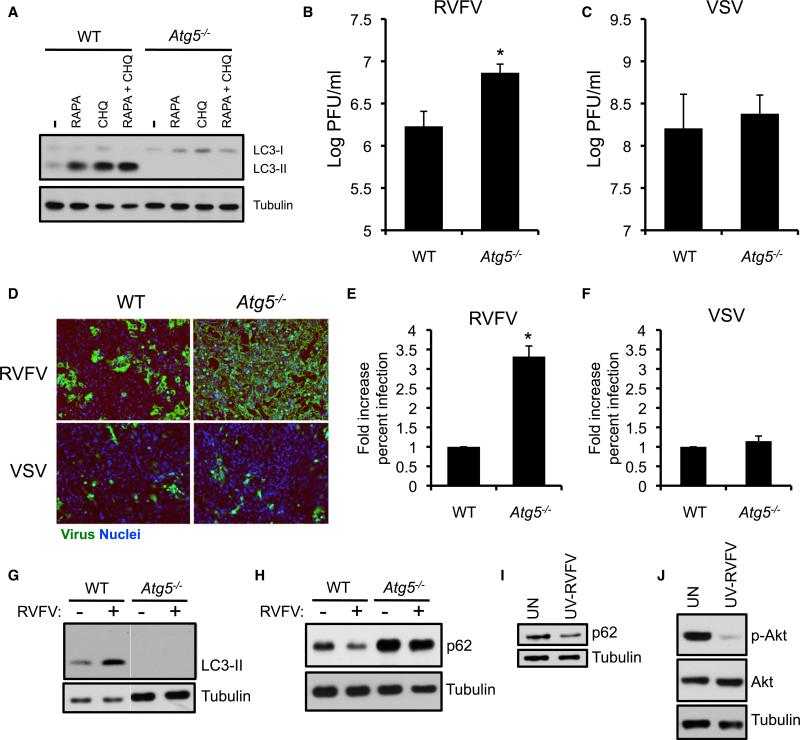
To determine whether autophagy restricts RVFV infection in murine cells, we examined mouse embryonic fibroblasts (MEFs) genetically null for Atg5, an essential and conserved autophagy gene (Kuma et al., 2004). As expected, Atg5−/− MEFs were deficient in processing of LC3 (the mammalian Atg8 homolog) in response to rapamycin and chloroquine, which inhibits late stages of autophagy and allows for LC3-II accumulation (Figure 3A). Compared to wild-type (WT) cells, Atg5−/− MEFs exhibited an increase in RVFV titers by plaque assay (Figure 3B) and RVFV protein by immunofluorescence (Figures 3D and 3F). In contrast, we observed no difference in VSV replication (Figures 3C, 3D, and 3F). Thus, Atg5 specifically controls RVFV infection in murine cells.
Figure 3. Atg5 Limits RVFV Replication in Mammalian Cells.
(A) WT and Atg5−/− MEFs were treated with rapamycin (RAPA, 1 μM) or chloroquine (CHQ, 20 μM) for 6 hr and monitored for LC3 expression by immunoblot.
(B and C) RVFV (B) or VSV (C) was titered on WT or Atg5−/− MEFs by plaque assay. Mean ± SD; *p < 0.05, Student's t test.
(D) Immunofluorescence images of WT or Atg5−/− MEFs infected with RVFV (16 hr) or VSV (12 hr).
(E and F) Quantification of the percentage of RVFV-infected (E) or VSV-infected (F) cells normalized to the WT control. Mean ± SD; *p < 0.05, Student's t test.
(G) Immunoblot for LC3 in uninfected or RVFV-infected WT and Atg5−/− MEFs 8 hpi.
(H) Immunoblot for p62 in WT or Atg5−/− MEFs 12 hpi.
(I) Immunoblot for p62 in WT MEFs incubated with UV-inactivated RVFV for 16 hr.
(J) Immunoblot for phospho-Akt and total Akt from WT MEFs incubated with UV-inactivated RVFV for 8 hr. All data represent three independent experiments.
Because Atg5 is required for canonical autophagy, we assessed whether RVFV infection induces autophagy in MEFs. RVFV infection resulted in the accumulation of processed LC3-II in WT but not Atg5−/− MEFs, suggesting that RVFV infection elicits Atg5-dependent autophagy (Figure 3G). Next, we monitored expression of the autophagy adaptor p62, which recruits cytoplasmic components to autophagosomes and is consequently degraded. Therefore, a decrease in p62 expression denotes flux through the autophagy pathway (Mizushima et al., 2010). We found that RVFV infection reduced p62 protein expression in WT MEFs, and this decrease was not observed in Atg5−/− MEFs (Figure 3H). UV-inactivated virus also decreased p62 protein, demonstrating that RVFV-activated autophagy is stimulated by a component of the incoming virions, and that the induction cannot be attributed to viral replication products or indirect effects due to altered cellular demands during infection (Figure 3I). MEFs incubated with UV-inactivated virus showed substantially reduced phospho-Akt without decreased total Akt, suggesting conserved regulation of antiviral autophagy by the Akt pathway in both flies and mammals (Figure 3J). Together, these data show that autophagy is activated in MEFs to defend against RVFV infection.
Autophagy Genes Restrict RVFV Replication in Human Cells
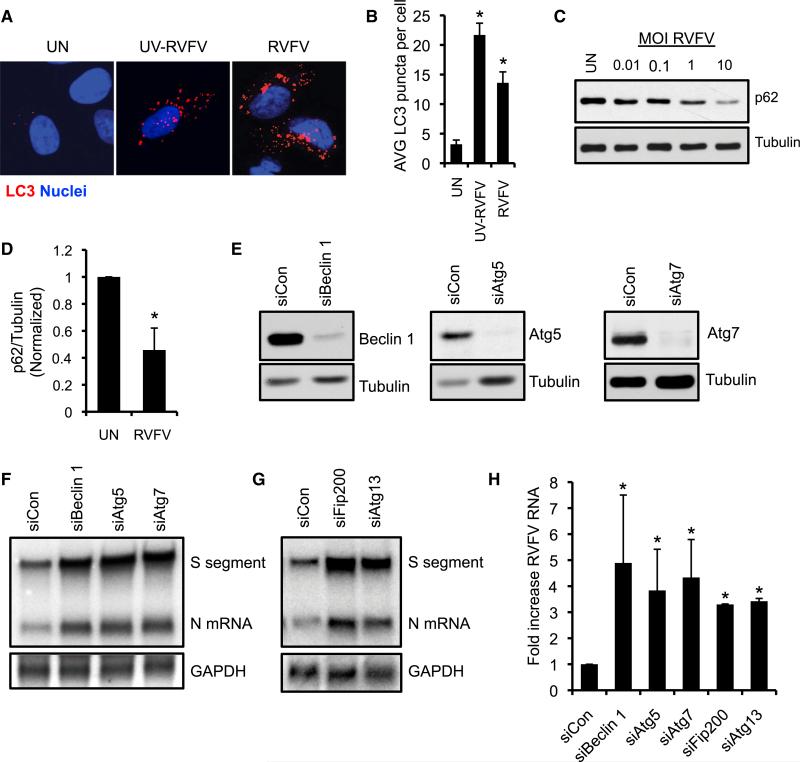
We next assessed whether RVFV infection activates autophagy in a human osteosarcoma cell line (U2OS cells). We monitored autophagy activation by examining U2OS cells stably expressing a mCherry-GFP-tagged LC3 reporter; the GFP tag is acid-sensitive, whereas the mCherry tag is acid-insensitive, and thus mCherry+GFP+ puncta represent early autophagosomes, whereas mCherry+GFP− puncta represent autolysosomes. LC3 puncta accumulate upon treatment with rapamycin and chloroquine, with chloroquine primarily leading to mCherry+ GFP+ puncta due to a block of autophagosomal acidification (Figures S3A and S3B). We observed an increase in LC3 puncta (mCherry+) after RVFV infection (Figures 4A and 4B), suggesting that RVFV infection induces autophagy. Furthermore, UV-inactivated RVFV also elicited an increase an LC3 puncta, which were preferentially mCherry+GFP−, consistent with mature autolysosomes (Figures 4A and 4B; Figure S3B). Moreover, RVFV infection decreased p62 protein (Figures 4C and 4D), further indicating that RVFV activates autophagy in human cells.
Figure 4. Autophagy Genes Restrict RVFV Infection in Human Cells.
(A) Representative image of human U2OS cells stably expressing a mCherry-GFP-LC3 reporter either mock infected or infected with RVFV (MOI 10) or UV-inactivated RVFV at 12 hpi. mCherry puncta represent total autophagosomes.
(B) Quantification of the average mCherry+ LC3 puncta per cell. Mean ± SE; *p < 0.05, Student's t test.
(C) U2OS cells were infected with RVFV at the indicated MOI and p62 expression was assessed by immunoblot 18 hpi.
(D) Average p62 protein levels normalized to tubulin from uninfected or RVFV-infected U2OS cells. Mean ± SE; *p < 0.05, Student's t test.
(E) Immunoblot from U2OS cells transfected with the indicated siRNAs.
(F and G) RNA blot for RVFV RNA from U2OS cells transfected with the indicated siRNAs 16 hpi.
(H) Average fold increase in RVFV RNA by RNA quantification. Mean ± SE; *p < 0.05, Student's t test. All data represent three independent experiments. Figure 4, see also Figure S3.
To define the importance of autophagy activation during infection, we silenced the autophagy genes Atg5, Atg7, and Beclin 1 by using small interfering RNA (siRNA), which reduced protein expression (Figure 4E) and inhibited LC3-II accumulation with rapamycin or chloroquine treatment (Figures S3C and S3D). Cells treated with siRNAs against Atg5, Atg7, and Beclin 1 exhibited elevated viral RNA levels after RVFV infection (Figures 4F and 4H). siRNA-mediated silencing of Atg13 and Fip200, members of the autophagic preinitiation complex, also increased viral RNA (Figures 4G and 4H). These data show that multiple stages of the core autophagy pathway are required for antiviral defense against RVFV from flies to humans.
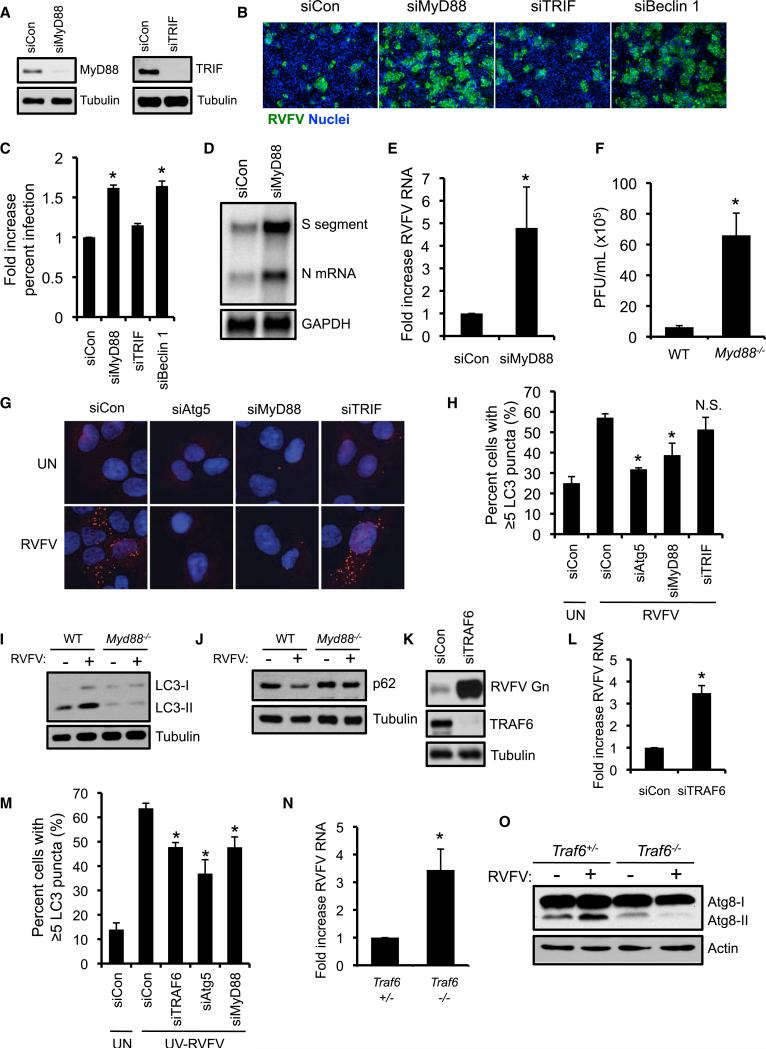
TRAF6 and MyD88 Restrict RVFV Infection and Promote Antiviral Autophagy
The pathways that control autophagy downstream of viral recognition are not fully characterized. Because Drosophila Toll-7 recognizes RVFV to activate antiviral autophagy, we postulated that a PRR-linked signaling pathway is required for anti-RVFV autophagy in mammals. We first tested the role of cytosolic RLRs, which detect viral RNA species such as the 5′-triphosphate on RVFV genomic RNAs (Habjan et al., 2008), by examining cells lacking the common signaling adaptor mitochondrial antiviral-signaling protein (MAVS) (Dixit et al., 2010). Consistent with a known role for RIG-I in sensing VSV (Kato et al., 2005), MAVS−/− MEFs demonstrated increased VSV titers (Figure S4A). Perhaps surprisingly, we found no difference in RVFV titers between MAVS−/− and WT MEFs, suggesting that RLRs are dispensable for restricting RVFV infection in this cell type (Figure S4B). Similar results were found when monitoring VSV and RVFV infection by immunofluorescence (Figures S4C–S4E). In addition, MAVS was dispensable for RVFV-induced p62 degradation (Figure S4F). We also examined the role of TANK-binding kinase-1 (TBK1), which functions downstream of RLRs (and certain endosomal TLRs such as TLR3) and is essential for antimicrobial autophagy against Mycobacterium tuberculosis (Fitzgerald et al., 2003; Watson et al., 2012). TBK1−/− MEFs exhibited increased infection with VSV but not RVFV, as well as normal RVFV-associated p62 degradation (Figures S4G–S4L). Consequently, neither TBK1 nor MAVS is required for RVFV-induced antiviral autophagy in MEFs.
TLRs recognize viruses at the plasma membrane or in endosomes and TLR ligands activate autophagy in macrophages (Delgado et al., 2008; Xu et al., 2007). Because Toll-7 is required for RVFV-induced autophagy in flies, we examined whether TLRs regulate RVFV-induced autophagy. TLRs signal through two Toll-IL-1 receptor (TIR) domain-containing adapters, MyD88 and TRIF (O'Neill and Bowie, 2007). To determine whether either TIR adaptor restricts RVFV infection, we silenced MyD88 and TRIF in U2OS cells by using siRNAs and verified knockdown by immunoblot (Figure 5A). MyD88 silencing increased RVFV infection by immunofluorescence similar to Beclin 1 knockdown (Figures 5B and 5C). In contrast, we found no change in infection with TRIF depletion, suggesting that MyD88, but not TRIF, mediates cell-intrinsic immunity to RVFV (Figures 5B and 5C). MyD88-silenced cells also demonstrated increased RVFV RNA (Figures 5D and 5E). Consistent with our results in human cells, RVFV-challenged Myd88−/− MEFs (Tal et al., 2009) showed a significant increase in viral titers, similar to the increase observed in Atg5−/− MEFs (~10-fold, Figure 5F). These data implicate MyD88 as a critical component in mammalian host defense against RVFV.
Figure 5. TRAF6 and Mammalian MyD88 Restrict RVFV Replication and Regulate Antiviral Autophagy.
(A) Immunoblot of MyD88 and TRIF from U2OS cells transfected with the indicated siRNAs.
(B) Representative immunofluorescence images of U2OS cells transfected with the indicated siRNAs and infected with RVFV at 16 hpi (MOI 0.1).
(C) Average percentage of infected cells normalized to control. Mean ± SD; *p < 0.05, Student's t test.
(D) RNA blot for RVFV RNA from U2OS cells transfected with MyD88-specific siRNA or control siRNA and infected with RVFV (MOI 0.3) 16 hpi.
(E) Quantification of RVFV RNA from (D). Mean ± SE; *p < 0.05, Student's t test.
(F) Plaque assays with RVFV were performed on Myd88−/− or WT MEFs. Mean ± SD; *p < 0.05, Student's t test.
(G) Representative images of siRNA-transfected U2OS cells expressing a mCherry-GFP-LC3 reporter and left uninfected or infected with RVFV (MOI 10). mCherry puncta are shown.
(H) Quantification of the percentage of cells with at least 5 LC3-positive puncta per cell. Mean ± SE; *p < 0.05 compared to siCon RVFV-infected cells, Student's t test.
(I) LC3 immunoblot of Myd88−/− or WT MEFs infected with RVFV 12 hpi.
(J) p62 immunoblot of Myd88−/− or WT MEFs infected with RVFV 16 hpi.
(K) Immunoblot for RVFV Gn and TRAF6 from siRNA-transfected U2OS infected with RVFV 16 hpi.
(L) Fold increase in RVFV S segment RNA quantified by RNA blot. Mean ± SE; *p < 0.05, Student's t test.
(M) Average percentage of cells with at least 5 LC3 puncta from mCherry-GFP-LC3 U2OS cells transfected with the indicated siRNAs and incubated with UV-inactivated RVFV for 12 hr. Mean ± SE; *p < 0.05 compared to siCon UV-RVFV treated cells, Student's t test.
(N) Fold increase in RVFV S segment RNA from Traf6−/− flies (dTraf6Ex1/dTraf6Ex1) or control Traf6+/− flies as quantified by RNA blot 6 dpi. Mean ± SE; *p < 0.05, Student's t test.
(O) Atg8 immunoblot from uninfected or RVFV-infected flies 3 dpi. One representative blot from two independent experiments is shown. All data are representative of three independent experiments unless otherwise indicated. Figure 5, see also Figure S4.
RVFV infection increased the percentage of U2OS cells containing LC3 puncta, and silencing of Atg5 abrogated this response (Figures 5G and 5H). Whereas TRIF was dispensable for RVFV-triggered autophagy, MyD88-silenced cells showed a decrease in RVFV-induced LC3 puncta compared to control cells (Figure 5H). Moreover, RVFV-infected Myd88−/− MEFs demonstrated impaired LC3-II accumulation and p62 degradation (Figures 5I and 5J). This defect was not due to a general requirement for MyD88 in autophagy because MyD88 silencing had no impact on LC3-II accumulation with rapamycin or chloroquine treatment (Figure S4M). These data suggest that antiviral autophagy against RVFV is MyD88-dependent in both human and mouse cells.
Previous studies have shown that the E3 ubiquitin ligase tumor necrosis factor receptor-associated factor 6 (TRAF6) regulates LPS-induced autophagy, which partly depended on MyD88 (Shi and Kehrl, 2008, 2010). Therefore, we tested the requirement for TRAF6 in RVFV-elicited autophagy by using previously validated reagents (Sun et al., 2004). RNAi-mediated depletion of TRAF6 was efficient in U2OS cells and markedly increased RVFV protein and RNA (Figures 5K and 5L; Figure S4N). TRAF6 knockdown also impaired UV-RVFV-induced LC3 puncta accumulation similar to Atg5 and MyD88 silencing (Figure 5M). Thus, TRAF6, which is known to act downstream of MyD88 in TLR signaling pathways, is necessary for activating autophagy to control RVFV replication in human cells.
To test for evolutionary conservation, we next assessed the role of Drosophila Traf6, which has been implicated in canonical Toll signaling (Shen et al., 2001), although other immune functions remain to be determined. We challenged previously characterized Traf6 mutant flies (dTraf6Ex1/dTraf6Ex1) (Cha et al., 2003) or heterozygous controls with RVFV and found that Traf6 mutant flies showed significantly increased viral RNA levels, indicating that Traf6 controls RVFV infection in vivo (Figure 5N; Figure S4O). Further, Traf6 mutants showed impaired autophagy activation during RVFV infection as assessed by Atg8 immunoblot (Figure 5O). Taken together, these data demonstrate a requirement for TRAF6 in antiviral autophagy in both flies and mammals, revealing additional conserved components of this innate defense pathway.
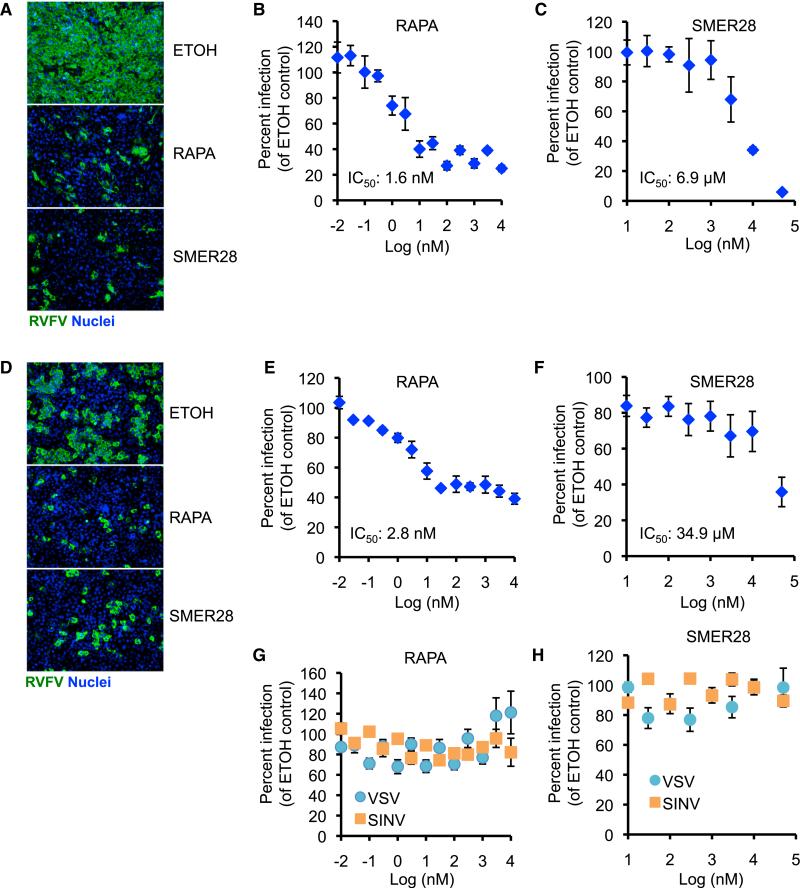
Autophagy-Activating Drugs Protect against RVFV Infection in Mammalian Cells
There are no specific therapeutics against RVFV infection, leading to high mortality during outbreaks (Ikegami and Makino, 2011). Because autophagy genes restrict RVFV replication, drugs that activate this pathway might offer a strategy to treat RVFV infection in humans. In addition to testing rapamycin, we evaluated the protective effect of small molecule enhancer of rapamycin (SMER)28, a recently developed mTor-independent autophagy inducer (Sarkar et al., 2007). As expected, treatment of U2OS cells with rapamycin or SMER28-activated autophagy as measured by increased LC3 puncta formation that was dependent on Atg5 (Figures S5A and S5B).
To determine whether autophagy-activating drugs protect against RVFV infection, we pretreated MEFs with rapamycin or SMER28 and infected with RVFV. Both drugs dramatically reduced the percentage of RVFV-infected cells in a dose-dependent manner (Figure 6A–6C) with IC50 values within previously reported ranges (Noh et al., 2004; Sarkar et al., 2007). Both drugs also protected against RVFV infection in U2OS cells (Figures 6D–6F), and this antiviral effect was specific, because we observed no inhibition of VSV or SINV infection, consistent with previous findings that VSV and SINV replication is insensitive to rapamycin treatment (Figures 6G and 6H) (Connor and Lyles, 2002; Mohankumar et al., 2011). Thus, these data suggest that chemical activation of autophagy limits RVFV in mammalian cells.
Figure 6. Autophagy-Activating Drugs Protect against RVFV Infection in Mammalian Cells.
(A) MEFs were pretreated with rapamycin (1 μM), SMER28 (50 μM), or vehicle control (ethanol) for 1 hr and infected with RVFV (MOI 1). Infection was monitored by immunofluorescence.
(B and C) Dose-response curve for RVFV infection of MEFs treated with the indicated concentrations of rapamycin (B) or SMER28 (C). Percent infection was normalized to vehicle control-treated cells.
(D) Representative immunofluorescence image of U2OS cells pretreated with rapamycin or SMER28 and infected with RVFV at 16 hpi.
(E and F) Dose-response curve for RVFV infection of U2OS treated with the indicated concentrations of rapamycin (E) or SMER28 (F).
(G and H) Dose-response curve for VSV and SINV infection of U2OS treated with the indicated concentrations of rapamycin (G) or SMER28 (H). Data show mean ± SE for three independent experiments. Figure 6, see also Figure S5.
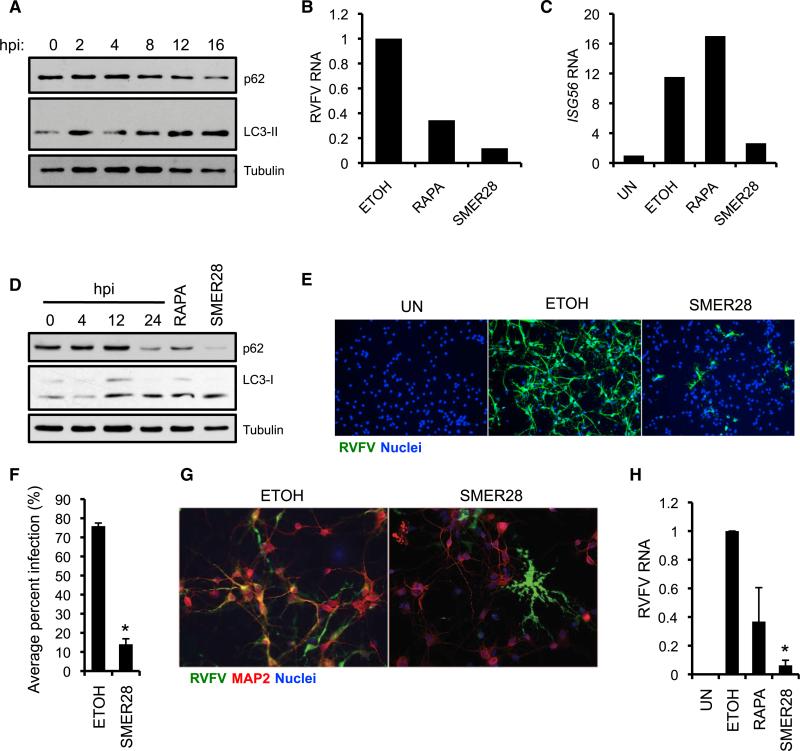
We next asked whether these drugs play a protective role in primary cells from tissues targeted by RVFV during natural infection. RVFV is highly hepatotropic in humans and mice, and infection can progress to fulminant hepatitis (Ikegami and Makino, 2011). RVFV infection induced autophagy in primary mouse hepatocytes as measured by increased LC3-II and decreased p62 expression (Figure 7A). Strikingly, both rapamycin and SMER28 treatment dramatically reduced viral RNA (Figure 7B), indicating that autophagy inducers have powerful antiviral activity in hepatocytes. We observed no increase in induction of the IFN-stimulated gene Isg56 in rapamycin or SMER28 treated cells; thus, the protective effect of autophagy-activating drugs likely cannot be explained by an exaggerated IFN-I response (Figure 7C).
Figure 7. Autophagy Inducers Inhibit RVFV Infection in Primary Mouse Hepatocytes and Rat Neurons.
(A) Primary mouse hepatocytes were infected with RVFV (MOI 5) and protein was isolated at the indicated time points after infection to assess p62 and LC3 expression by immunoblot. Data are representative of two independent experiments.
(B) RVFV RNA levels from primary mouse hepatocytes pretreated with SMER28 or rapamycin and infected with RVFV (MOI 1) for 20 hr as determined by qRTPCR. Data are representative of two independent experiments.
(C) Isg56 mRNA expression quantified by qRT-PCR from RVFV-infected mouse hepatocytes treated with the indicated drugs or vehicle control at 20 hpi.
(D) Primary rat neuronal cultures were infected with RVFV for the indicated times (MOI 1) or treated with rapamycin (1 μM) or SMER28 (50 μM) for 16 hr. Immunoblot analysis of LC3 and p62 expression is shown. Data are representative of two independent experiments.
(E) Primary rat neuronal cultures were pretreated with SMER28 or vehicle control for 1 hr and infected with RVFV (MOI 0.1) for 22 hr. Infection was assessed by immunofluorescence.
(F) Fold decrease in the percent of RVFV-infected rat neuronal cells with SMER28 treatment. Mean ± SE for three independent experiments; *p < 0.05, Student's t test.
(G) Immunofluorescence of RVFV and neurons (MAP2-positive cells) from RVFV-infected neuronal cultures pretreated with SMER28 or vehicle control showing preferential protection of neurons with SMER28 treatment.
(H) RVFV RNA was quantified by qRT-PCR from infected cells pretreated with SMER28 or rapamycin at 22 hpi. Mean ± SE for three independent experiments normalized to ETOH control; *p < 0.05, Student's t test.
Some RVFV-infected humans develop severe neurological disorders and encephalitis, suggesting that RVFV is neuroinvasive (Ikegami and Makino, 2011). Moreover, mice that survive acute RVFV infection can develop encephalitis similar to humans, and viral antigen can be detected in a wide variety of neuronal cell types in the brain (Ross et al., 2012). We consequently asked whether autophagy activation inhibits RVFV infection in primary rat mixed neuroglial cultures. Both SMER28 and rapamycin induced autophagy in neuronal cultures as shown by increased LC3-II accumulation and p62 degradation (Figure 7D). Whereas the majority of neuronal cells were infected in control cultures, SMER28-treated cells were largely protected (Figures 7E and 7F). Costaining for microtubule-associated protein 2 (MAP2), a neuronal differentiation marker, revealed that whereas neurons were efficiently infected by RVFV in control cultures, nonneuronal cells were disproportionately infected in SMER28 cultures, indicating that the SMER28 is particularly protective in neurons (Figure 7G). This inhibition was also observed when monitoring viral RNA, because SMER28 treatment resulted in a 95% reduction in RVFV RNA (Figure 7H). We also observed inhibition of RVFV infection with rapamycin treatment, albeit more modestly (Figure 7H). Notably, RVFV infection activated autophagy in neuroglial cultures as shown by robust p62 degradation and elevated LC3-II levels (Figure 7D), suggesting that autophagy activation is a normal response to RVFV infection in neuronal cells. Collectively, these data suggest that pharmacologic activation of autophagy protects hepatocytes and neurons against RVFV infection and might be a promising treatment strategy for RVFV-induced hepatitis and encephalitis.
DISCUSSION
Autophagy is an ancient host response to starvation that has emerged as an important facet of the innate immune arsenal. We found that in both flies and mammals, autophagy genes restrict the replication of RVFV, a medically and agriculturally relevant arbovirus for which there is no effective treatment or vaccine. Intervention with autophagy-activating drugs potently blocks infection. Therefore, this study provides insight into the mechanisms regulating antiviral autophagy and offers a potential avenue for developing RVFV-directed treatments.
By screening the Drosophila Tolls, we have extended the antiviral Toll receptor-autophagy axis to RVFV. Toll-7 mutant flies succumb to RVFV infection, whereas flies that overexpress Toll-7 are protected. Moreover, Toll-7 interacted with purified RVFV virions, suggesting that Toll-7 likely recognizes surface-exposed RVFV glycoproteins, similar to the autophagy-inducing glycoprotein PAMP on VSV (Nakamoto et al., 2012). Although we found no role for other Toll receptors in RVFV infection, they might function in unknown contexts, perhaps with distinct pathogens. Indeed, a recent study showed that Toll-8 negatively regulates antimicrobial peptide gene expression in larval respiratory epithelia (Akhouayri et al., 2011). Together, these findings suggest that Toll receptors evolved as critical components of host defense and immune regulatory pathways.
We found that Toll-7 restricts RVFV by activating autophagy, because Toll-7 is necessary for RVFV-induced autophagy in vivo, and silencing of autophagy genes leads to increased mortality and viral replication. Furthermore, antiviral autophagy against RVFV infection is conserved in mammals. RVFV triggered autophagy in mouse, rat, and human cells, and its replication was enhanced by deficiency of core autophagy genes in MEFs and U2OS cells. Although diverse viruses can trigger autophagy, the ability of autophagy to limit viral replication is complicated. For example, neuron-specific deletion of Atg5 increases lethality after SINV infection, but viral titers are not generally affected, indicating that autophagy does not restrict SINV replication per se (Orvedahl et al., 2010). WT HSV-1 is also not normally restricted by autophagy because it encodes a virulence factor ICP34.5 that inhibits autophagy (Orvedahl et al., 2007; Tallóczy et al., 2002). Proviral and antiviral functions have also been reported for autophagy in CHIKV infection (Joubert et al., 2012; Krejbich-Trotot et al., 2011). Thus, our data suggest that autophagy might play a more direct and evolutionarily conserved role in restricting RVFV replication than other viruses.
Recent studies propose that autophagy genes can function outside of canonical autophagy. For example, IFN-γ-mediated mouse norovirus restriction requires the Atg5-Atg12-Atg16L1 complex, but not Atg4B or lysosomal degradation, and Atg5 restricts T. gondii replication without the requirement for autophagosome generation (Hwang et al., 2012; Zhao et al., 2008). Because there are no specific genes for these autophagy-related processes, their distinction is in part based on the demonstration that certain “cassettes” of autophagy genes are required while others are dispensable. We show that genes involved in multiple stages of autophagy restrict RVFV replication, from the preinitiation complex (i.e., Atg13 and Fip200) to the elongation phase (i.e., Atg5 and Atg7). Thus, canonical autophagy likely restricts RVFV infection, although the antiviral mechanism remains to be clarified.
Relatively little is known about the PRRs and signaling pathways that activate antiviral autophagy. Although nucleocapsid-associated RVFV RNA activates RIG-I, we found no role for the downstream adaptor MAVS in restricting RVFV infection (Weber et al., 2013). This difference might be related to cell-type-specific functions for autophagy and PRRs. Indeed, MAVS does not restrict RVFV replication in macrophages even though it regulates IFN-I expression, suggesting that IFN-I alone cannot explain RVFV control (Ermler et al., 2013). Moreover, RVFV encodes an IFN antagonist (NSs), and so other restriction mechanisms besides IFN-I must exist (Bouloy et al., 2001). Our data provide evidence that perhaps autophagy is the major antiviral pathway against RVFV in some cell types.
Because MAVS does not restrict RVFV replication in MEFs, we focused on the TLR signaling adapters MyD88 and TRIF. MyD88 but not TRIF was required to restrict RVFV infection and elicit anti-RVFV autophagy. Thus, TLR-mediated signaling has an evolutionarily conserved role in directing antiviral autophagy from flies to mammals. The specific TLR required for RVFV-induced autophagy remains unknown. U2OS cells express all TLRs but TLR8, and MEFs express TLRs 1–9 (Kurt-Jones et al., 2004; Shatz et al., 2012). Given that TRIF and TBK1 do not restrict RVFV infection, we hypothesize that the autophagy-activating receptor is a plasma membrane TLR like TLR2 and TLR4, which have been shown to recognize viral glycoproteins (Barbalat et al., 2009; Georgel et al., 2007). This would parallel Drosophila Toll-7, which binds viral particles at the cell surface. Indeed, the critical role for plasma membrane TLRs in antimicrobial autophagy is supported by recent findings that TLR2 is required for Listeria monocytogenes-induced autophagy (Anand et al., 2011). In addition to revealing a requirement for MyD88 in autophagy activation, our study also suggests an important cell-intrinsic role for MyD88 and TLRs in host defense against viruses. Only a few reports have shown a cell-intrinsic requirement for MyD88 in controlling viral replication, such as in West Nile virus (WNV) infection of macrophages and certain types of neurons (Szretter et al., 2010). We propose that MyD88 has unappreciated functions in cell-intrinsic antiviral immunity, such as by controlling autophagy or other IFN-independent pathways.
Two additional pathways downstream of virus recognition regulate antiviral autophagy in flies and mammals. Because TRAF6 has been associated with LPS-induced autophagy, we investigated the requirement for TRAF6 during RVFV infection. Loss of TRAF6 in mammalian cells or flies resulted in increased viral replication and impaired autophagy activation, revealing a critical role for TRAF6 in anti-RVFV autophagy. Importantly, these data show that Drosophila TRAF6 can restrict pathogen replication in vivo. In Drosophila and mammalian cells, RVFV infection also led to decreased Akt activation, suggesting that this PRR-TRAF6 signaling axis converges on Akt to regulate antiviral autophagy initiation. Taken together, these data demonstrate deep conservation of anti-RVFV autophagy across species.
Autophagy modulation has been proposed as a treatment for diverse human disorders, including cancer, neurodegenerative disease, and infection. We found that treatment with two autophagy inducers, rapamycin and SMER28, greatly reduced viral replication in a variety of human and murine cell types, including primary mouse hepatocytes and rat neurons. Because RVFV targeting of hepatocytes and neurons during natural infection is one of the main causes of morbidity and mortality, autophagy activation might be a viable strategy for RVFV intervention in humans and livestock. These findings are consistent with a recent report identifying an autophagy-inducing peptide (Tat-Beclin 1) that limits susceptibility to WNV, SINV, and CHKV in mice (Shoji-Kawata et al., 2013). Although the host-protective mechanism of Tat-Beclin 1 has not been fully elucidated, the prosurvival effects of autophagy rather than a direct effect on virus growth might be involved because autophagy per se does not normally restrict the replication of these particular viruses (Beatman et al., 2012; Krejbich-Trotot et al., 2011; Orvedahl et al., 2010; Vandergaast and Fredericksen, 2012). Because RVFV replication is normally restricted by autophagy, boosting this process might be a particularly effective therapeutic strategy for RVFV infection.
Most studies on infection and immunity have relied on cell lines or macrophages, but emerging evidence suggests unique functions for autophagy in other cell types. Many of the viruses restricted by autophagy infect neurons, including SINV and HSV-1, as well as RVFV identified in this study. Moreover, some cells rely more heavily on IFN-I-independent mechanisms to counteract virus infection. This was clearly shown for dorsal root ganglion neurons, which are less responsive to IFN-I and use autophagy as a major anti-HSV-1 defense (Yordy and Iwasaki, 2012). We also find that autophagy activation limits RVFV replication in hepatocytes, another cell type that has high basal autophagy and in which autophagy has not been extensively studied for its role in antiviral defense. It will be interesting to determine the relative contribution of IFN-I versus autophagy in viral control in neurons and hepatocytes during RVFV infection and whether other viruses that target these organs are susceptible to antiviral autophagy.
In conclusion, we have taken advantage of the simplified system in Drosophila to discover a conserved role for autophagy genes in restricting RVFV replication in mammals. Toll receptors or signaling molecules downstream of TLRs (i.e., MyD88) are necessary for RVFV-induced autophagy in both flies and mammalian cells, further defining the ancient nature of the antiviral autophagy response. These findings help elucidate the mechanism by which autophagy is triggered during viral infection and add to the mounting evidence that autophagy activation might be beneficial for treating infections and other diseases.
EXPERIMENTAL PROCEDURES
Adult Fly Infections
Four- to seven-day-old flies were challenged as previously described (Shelly et al., 2009). For Toll receptor overexpression experiments, flies were heat shocked at 37° C for 1 hr prior to infection and heat shocked every 2 days. Flies were monitored for mortality or processed at the indicated time points after infection. For rapamycin feeding, flies were starved for 1 hr, placed on food supplemented with 100 μM rapamycin or vehicle, and infected the next day. To monitor Akt activity, we challenged flies with insulin (1 mM) and either PBS or RVFV as previously described (Shelly et al., 2009).
Viral Infections
Cells were infected with RVFV, SINV, and VSV as previously described for the indicated times and MOIs (Moser et al., 2012).
Immunofluorescence
Cells were stained as previously described (Shelly et al., 2009). Automated microscopy (ImageXpress Micro) was used to image cells with at least four sites per well and three wells per condition, and percent infection was calculated with MetaXpress image analysis. Neurons and mCherry-GFP-LC3 U2OS cells grown on coverslips were imaged with a Leica DMI 4000 B fluorescent microscope. MetaXpress software was used to quantify LC3 puncta.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Coyne for mCherry-GFP-LC3 U2OS cells; R. Medzhitov for Myd88−/− MEFs; J. Kagan for MAVS−/− MEFs; T. Neufeld for Atg8-GFP flies; N. Silverman for Traf6 mutant flies; Mahoney Institute of Neurological Sciences Neuron Culture Service Center for neuroglial cell cultures; C. Lopez, A. Yasunaga, T. Nguyen, and D. Schieffer for technical help; Y. Choi and M. Walsh for TRAF6 antibody; and H.W. Virgin for reagents and advice throughout. This work was supported by grants from the National Institutes of Health to S.C. (R01AI074951, U54AI057168, and R01AI095500), B.Z.S. (R01DK083355), and R.H.M. (T32-AI-007324). S.C. is a recipient of the Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2013.10.020.
REFERENCES
- Akhouayri I, Turc C, Royet J, Charroux B. Toll-8/Tollo negatively regulates antimicrobial response in the Drosophila respiratory epithelium. PLoS Pathog. 2011;7:e1002319. doi: 10.1371/journal.ppat.1002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Anand PK, Tait SW, Lamkanfi M, Amer AO, Nunez G, Pagès G, Pouysségur J, McGargill MA, Green DR, Kanneganti TD. TLR2 and RIP2 pathways mediate autophagy of Listeria monocytogenes via extracellular signal-regulated kinase (ERK) activation. J. Biol. Chem. 2011;286:42981–42991. doi: 10.1074/jbc.M111.310599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbalat R, Lau L, Locksley RM, Barton GM. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat. Immunol. 2009;10:1200–1207. doi: 10.1038/ni.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatman E, Oyer R, Shives KD, Hedman K, Brault AC, Tyler KL, Beckham JD. West Nile virus growth is independent of autophagy activation. Virology. 2012;433:262–272. doi: 10.1016/j.virol.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouloy M, Janzen C, Vialat P, Khun H, Pavlovic J, Huerre M, Haller O. Genetic evidence for an interferon-antagonistic function of rift valley fever virus nonstructural protein NSs. J. Virol. 2001;75:1371–1377. doi: 10.1128/JVI.75.3.1371-1377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell GR, Spector SA. Toll-like receptor 8 ligands activate a vitamin D mediated autophagic response that inhibits human immunodeficiency virus type 1. PLoS Pathog. 2012;8:e1003017. doi: 10.1371/journal.ppat.1003017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha GH, Cho KS, Lee JH, Kim M, Kim E, Park J, Lee SB, Chung J. Discrete functions of TRAF1 and TRAF2 in Drosophila melanogaster mediated by c-Jun N-terminal kinase and NF-kappaB-dependent signaling pathways. Mol. Cell. Biol. 2003;23:7982–7991. doi: 10.1128/MCB.23.22.7982-7991.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor JH, Lyles DS. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J. Virol. 2002;76:10177–10187. doi: 10.1128/JVI.76.20.10177-10187.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deddouche S, Matt N, Budd A, Mueller S, Kemp C, Galiana-Arnoux D, Dostert C, Antoniewski C, Hoffmann JA, Imler JL. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat. Immunol. 2008;9:1425–1432. doi: 10.1038/ni.1664. [DOI] [PubMed] [Google Scholar]
- Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermler ME, Yerukhim E, Schriewer J, Schattgen S, Traylor Z, Wespiser AR, Caffrey DR, Chen ZJ, King CH, Gale M, Jr., et al. RNA heli-case signaling is critical for type i interferon production and protection against Rift Valley fever virus during mucosal challenge. J. Virol. 2013;87:4846–4860. doi: 10.1128/JVI.01997-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Georgel P, Jiang Z, Kunz S, Janssen E, Mols J, Hoebe K, Bahram S, Oldstone MB, Beutler B. Vesicular stomatitis virus glycoprotein G activates a specific antiviral Toll-like receptor 4-dependent pathway. Virology. 2007;362:304–313. doi: 10.1016/j.virol.2006.12.032. [DOI] [PubMed] [Google Scholar]
- Habjan M, Andersson I, Klingström J, Schümann M, Martin A, Zimmermann P, Wagner V, Pichlmair A, Schneider U, Mühlberger E, et al. Processing of genome 50 termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS ONE. 2008;3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Maloney NS, Bruinsma MW, Goel G, Duan E, Zhang L, Shrestha B, Diamond MS, Dani A, Sosnovtsev SV, et al. Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 2012;11:397–409. doi: 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T, Makino S. The pathogenesis of Rift Valley fever. Viruses. 2011;3:493–519. doi: 10.3390/v3050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert PE, Werneke SW, de la Calle C, Guivel-Benhassine F, Giodini A, Peduto L, Levine B, Schwartz O, Lenschow DJ, Albert ML. Chikungunya virus-induced autophagy delays caspase-dependent cell death. J. Exp. Med. 2012;209:1029–1047. doi: 10.1084/jem.20110996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Krejbich-Trotot P, Gay B, Li-Pat-Yuen G, Hoarau JJ, Jaffar-Bandjee MC, Briant L, Gasque P, Denizot M. Chikungunya triggers an autophagic process which promotes viral replication. Virol. J. 2011;8:432. doi: 10.1186/1743-422X-8-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Kurt-Jones EA, Sandor F, Ortiz Y, Bowen GN, Counter SL, Wang TC, Finberg RW. Use of murine embryonic fibroblasts to define Toll-like receptor activation and specificity. J. Endotoxin Res. 2004;10:419–424. doi: 10.1179/096805104225006516. [DOI] [PubMed] [Google Scholar]
- Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- Leib DA, Alexander DE, Cox D, Yin J, Ferguson TA. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J. Virol. 2009;83:12164–12171. doi: 10.1128/JVI.01676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spaätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in meta-zoans: cell survival in the land of plenty. Nat. Rev. Mol. Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohankumar V, Dhanushkodi NR, Raju R. Sindbis virus replication, is insensitive to rapamycin and torin1, and suppresses Akt/mTOR pathway late during infection in HEK cells. Biochem. Biophys. Res. Commun. 2011;406:262–267. doi: 10.1016/j.bbrc.2011.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser TS, Schieffer D, Cherry S. AMP-activated kinase restricts Rift Valley fever virus infection by inhibiting fatty acid synthesis. PLoS Pathog. 2012;8:e1002661. doi: 10.1371/journal.ppat.1002661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto M, Moy RH, Xu J, Bambina S, Yasunaga A, Shelly SS, Gold B, Cherry S. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity. 2012;36:658–667. doi: 10.1016/j.immuni.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh WC, Mondesire WH, Peng J, Jian W, Zhang H, Dong J, Mills GB, Hung MC, Meric-Bernstam F. Determinants of rapamycin sensitivity in breast cancer cells. Clin. Cancer Res. 2004;10:1013–1023. doi: 10.1158/1078-0432.ccr-03-0043. [DOI] [PubMed] [Google Scholar]
- O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- Orvedahl A, Alexander D, Tallóczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Orvedahl A, MacPherson S, Sumpter R, Jr., Tallóczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross TM, Bhardwaj N, Bissel SJ, Hartman AL, Smith DR. Animal models of Rift Valley fever virus infection. Virus Res. 2012;163:417–423. doi: 10.1016/j.virusres.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, Webster JA, Lewis TA, O'Kane CJ, Schreiber SL, Rubinsztein DC. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat. Chem. Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Shatz M, Menendez D, Resnick MA. The human TLR innate immune gene family is differentially influenced by DNA stress and p53 status in cancer cells. Cancer Res. 2012;72:3948–3957. doi: 10.1158/0008-5472.CAN-11-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Liu H, Skolnik EY, Manley JL. Physical and functional interactions between Drosophila TRAF2 and Pelle kinase contribute to Dorsal activation. Proc. Natl. Acad. Sci. USA. 2001;98:8596–8601. doi: 10.1073/pnas.141235698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger auto-phagy in macrophages. J. Biol. Chem. 2008;283:33175–33182. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol. Cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- Szretter KJ, Daffis S, Patel J, Suthar MS, Klein RS, Gale M, Jr., Diamond MS. The innate immune adaptor molecule MyD88 restricts West Nile virus replication and spread in neurons of the central nervous system. J. Virol. 2010;84:12125–12138. doi: 10.1128/JVI.01026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. USA. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallóczy Z, Jiang W, Virgin HW, 4th, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc. Natl. Acad. Sci. USA. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu S, Akira S. Toll-like receptors and innate immunity. J. Mol. Med. 2006;84:712–725. doi: 10.1007/s00109-006-0084-y. [DOI] [PubMed] [Google Scholar]
- Vandergaast R, Fredericksen BL. West Nile virus (WNV) replication is independent of autophagy in mammalian cells. PLoS ONE. 2012;7:e45800. doi: 10.1371/journal.pone.0045800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter CT, Barr JN. Recent advances in the molecular and cellular biology of bunyaviruses. J. Gen. Virol. 2011;92:2467–2484. doi: 10.1099/vir.0.035105-0. [DOI] [PubMed] [Google Scholar]
- Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Gawanbacht A, Habjan M, Rang A, Borner C, Schmidt AM, Veitinger S, Jacob R, Devignot S, Kochs G, et al. Incoming RNA virus nucleocapsids containing a 50-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe. 2013;13:336–346. doi: 10.1016/j.chom.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Grant G, Sabin LR, Gordesky-Gold B, Yasunaga A, Tudor M, Cherry S. Transcriptional pausing controls a rapid antiviral innate immune response in Drosophila. Cell Host Microbe. 2012;12:531–543. doi: 10.1016/j.chom.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y, Nishida Y, Ip YT. Functional analysis of Toll-related genes in Drosophila. Dev. Growth Differ. 2010;52:771–783. doi: 10.1111/j.1440-169X.2010.01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Takada H, Goldman WE, Fukase K, Silverman N, et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat. Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordy B, Iwasaki A. Cell type-dependent requirement of auto-phagy in HSV-1 antiviral defense. Autophagy. 2012;9:236–238. doi: 10.4161/auto.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.