Abstract
Here we describe Protein Interaction Quantitation (PIQ), a computational method that models the magnitude and shape of genome-wide DNase profiles to facilitate the identification of transcription factor (TF) binding sites. Through the use of machine learning techniques, PIQ identified binding sites for >700 TFs from one DNase-seq experiment with accuracy comparable to ChIP-seq for motif-associated TFs (median AUC=0.93 across 303 TFs). We applied PIQ to analyze DNase-seq data from mouse embryonic stem cells differentiating into pre-pancreatic and intestinal endoderm. We identified (n=120) and experimentally validated eight ‘pioneer’ TF families that dynamically open chromatin, enabling other TFs to bind to adjacent DNA. Four pioneer TF families only open chromatin in one direction from their motifs. Furthermore, we identified a class of ‘settler’ TFs whose genomic binding is principally governed by proximity to open chromatin. Our results support a model of hierarchical TF binding in which directional and non-directional pioneer activity shapes the chromatin landscape for population by settler TFs.
Manipulation of TFs can reprogram cellular identity1, 2 and re-wire intercellular signaling pathways3, 4. Efforts to predict TF binding patterns have been hampered by incomplete understanding of the rules governing TF binding site choice. Highly accurate genome-wide methods have been developed to localize the condition-specific binding of TFs to the genome, facilitating the elucidation of genome regulatory elements and gene regulatory networks5, 6. Chromatin immunoprecipitation of selected protein-DNA complexes followed by high-throughput sequencing and mapping of the immunoprecipitated DNA (ChIP-seq)7 has become a valued method for TF location analysis and can reliably identify where TFs bind genome-wide within 10 bp8, 9. Each ChIP-seq experiment profiles a single TF and requires either an antibody specific to the TF or the incorporation of a tag into the TF being profiled. DNase-seq10 is an assay that takes advantage of the preferential cutting of DNase I in open chromatin11 and the steric blockage of DNase I by tightly-bound TFs that protect associated genomic DNA sequences12. After deep sequencing of DNase-digested genomic DNA from intact nuclei, genome-wide data on chromatin accessibility as well as TF-specific DNase-protection profiles revealing the genomic binding locations of a majority of TFs are obtained13-16. These TF signature “DNase profiles” reflect the TF's effect on DNA shape and local chromatin architecture, extending hundreds of base pairs from a TF binding site, and they are centered on “DNase footprints” at the binding motif itself that reflect the biophysics of protein-DNA binding15, 17, 18. As DNase-seq experiments are TF-independent and do not require antibodies, it is possible to predict the binding of hundreds of different TFs to their genomic motifs from a single DNase-seq experiment. Several groups have developed algorithms to infer TF binding from DNase-seq data13, 15, 17-19, but these existing methods do not model TF-dependent chromatin accessibility well.
Here we aimed to improve upon these methods conceptually in two ways. First, we take into account how individual TFs contribute to both the magnitude and spatial pattern of DNase hypersensitivity. Not only does this improve our ability to identify binding of all TFs regardless of their DNase profiles, it also allows us to probe whether a factor increases local hypersensitivity. Second, we carefully integrate prior information, such as the quality of a motif match, so that the method behaves robustly even with weak motifs or low coverage data.
RESULTS
Protein Interaction Quantitation
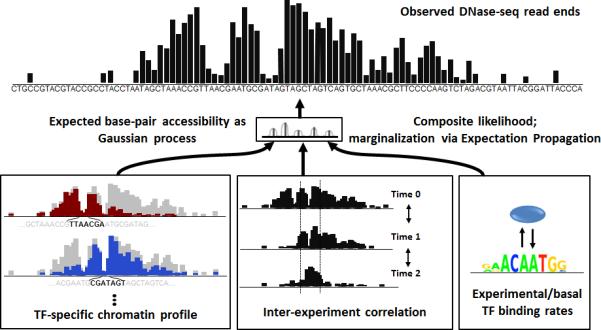
PIQ is a method for analyzing genome-wide DNaseI hypersensitivity data. The input of PIQ is one or more DNase-seq experiments, the genome sequence of the organism assayed and a list of motifs represented as position weight matrices (PWMs) that describe candidate TF binding sites. PIQ uses machine learning methods to normalize input DNase-seq data and then predicts TF binding by detecting both the shape and magnitude of DNase profiles15 specific to each TF (Fig. 1). The output of PIQ is the probability of occupancy for each candidate binding site in the genome, along with aggregate TF-specific scores (e.g. metrics for TF-specific chromatin opening). For the results in this paper, PIQ outputs protein binding at the locations of 733 TF motifs.
Figure 1.
Accurate detection of dynamic TF binding using DNase-seq and PIQ. Schematic outlining the PIQ algorithm. See text and Supplementary Information for details.
The PIQ algorithm consists of three steps: candidate site identification, background model computation and TF binding estimation (Fig. 1).
In the first step, PIQ scans for DNase profiles at PWM motifs for 1,331 TFs derived from the JASPAR, UniPROBE and TRANSFAC databases9-11 (see Supplementary Information for further explanation of motif choice). We choose to scan potentially bound motifs from databases and subsequently determine whether each site has a profile8, instead of detecting genome-wide footprints de novo and subsequently matching them to underlying motifs4-7, because motif-centered searching can take into account each TF's unique signature DNase profile information that is learned in subsequent steps of PIQ (Supplementary Fig. 1). This motif-specific information about the expected hypersensitivity profile surrounding a bound site improves individual binding prediction and allows complex enhancer and promoter profile clusters to more easily be deconvolved into a set of bound motifs each imparting its signature profile on the chromatin.
In the second step, PIQ smooths the raw reads from each DNase-seq experiment to produce a robust foundation for profile detection. PIQ models raw DNase-seq reads as arising from a Gaussian process, which is a statistical model that removes noise by adaptively smoothing the reads from neighboring bases (see Supplementary Information for details on how reads are combined). Optionally, reads from multiple experiments, whether replicates or time-series data, are integrated and collectively smoothed using the same Gaussian process framework, which serves to maximize consistent signal while minimizing stochastic noise.
In the final step, PIQ identifies binding sites of each TF in each experiment by iteratively combining direct evidence of binding with indirect analysis of whether the observed DNase-seq data is consistent with a computer-generated model of DNase hypersensitivity that includes that binding event. First, PIQ preliminarily assigns genomic binding events for each TF motif on the basis of whether a profile exists at each putative binding site. Then, PIQ uses TF-signature profile shapes and magnitudes for each TF to build a model of the expected genomic DNase hypersensitivity given the assigned binding events. These TF binding estimation and DNase hypersensitivity model building steps are iteratively performed using a fast approximate machine learning method called expectation propagation20 to arrive at the final binding calls for each motif. PIQ is implemented on the Amazon EC2 cloud server, exploiting parallel computation to substantially speed up run time (see Supplementary Information for a more detailed description). Post-processing to cull motifs without significant binding and merging sets of motifs with >90% overlapping binding sites reduces the number of informative TF motifs in the cell types we examine in this work to 733.
Benchmarking PIQ against DGF, CENTIPEDE and ChIP-seq data
We applied PIQ, as well as two published DNase-seq–based TF binding detection methods, DGF (which uses only DNase-seq data)15 and CENTIPEDE (which, like PIQ, incorporates DNase-seq and motif data)14, to published DNase-seq data from K562 cells and validated these predictions against 303 matched ChIP-seq experiments15 (Supplementary Table 1 and available online, see Supplementary Information). Compared with other methods, PIQ exhibited higher accuracy in the prediction of sequence-specific TF binding events, as determined by ChIP-seq peaks covering factor motifs, while displaying comparable overall coverage of all ChIP-seq peaks (Supplementary Fig. 2 and Supplementary Table 1).
To summarize these accuracy numbers, we used a standard statistical technique to gauge predictive accuracy, the area under the receiver operating characteristic curve (AUC, Supplementary Methods), which represents the probability of correctly ranking a ChIP-seq bound motif above an unbound motif for each method. Corresponding AUC scores revealed that PIQ's predictions were more accurate than those of both other methods at every one of the 303 ChIP-seq experiments (PIQ mean AUC of 0.93, CENTIPEDE AUC of 0.87, and DGF AUC of 0.65 (Fig. 2a and Supplementary Table 1)). A similar comparison on six mouse embryonic stem cell ChIP-seq profiles21 that matched known motifs also finds PIQ highly concordant (AUC minimum 0.86, mean 0.92; Fig. 2b). The median fraction of total ChIP-seq binding sites recapitulated by PIQ predictions was 66% for the 200/303 sequence-specific ChIP-seq experiments with more than half of their sites backed by motifs, and 50% over all 303 experiments (Supplementary Table 1 and Supplementary Fig. 2). Similarly, median positive predicting value (PPV, Supplementary Methods) scores, which reveal the precision of PIQ predictions over the top 500 predictions, were 76% for the top quarter of ChIP-seq experiments, 32% for the 200 motif-enriched experiments noted above and 39.4% over 194 experiments for which any DNase-Seq method achieved >0% PPV, substantially outperforming CENTIPEDE and DGF. Thus, PIQ is consistently highly concordant with ChIP-seq (median AUC of 0.93 over 303 ChIP-seq comparison datasets) and thus is a highly accurate tool to uncover TF-DNA binding.
Figure 2.
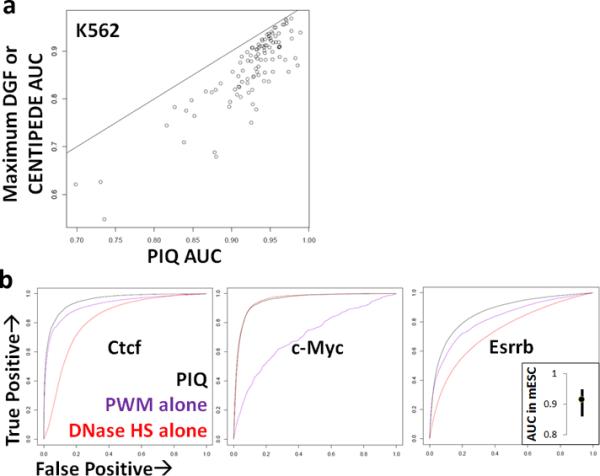
Benchmarking PIQ. (a) Comparison of AUC values (the probability of correctly ranking a bound TF site above an unbound one) comparing PIQ versus ChIP-seq (X-axis) and DGF/CENTIPEDE versus ChIP-seq (Y-axis) for 303 matched ChIP-seq experiments in K562 cells. The Y-axis value plots the higher AUC value between DGF and CENTIPEDE for each experiment. (b) ROC curves (which show the tradeoff between true positives to false positives as the cutoff for defining what is bound is varied) comparing mESC-stage PIQ binding calls for the TFs Ctcf, c-Myc and Esrrb against matched ChIP-seq binding calls. To calculate ROC curves, we ranked all above-threshold genomic motif instances for each TF according to their PWM motif strength (purple), total adjacent DNase hypersensitivity in a 400 bp window (red) or the per-site binding score given by PIQ (black). Plots compare true positives (Y-axis) to false positives (X-axis) at progressively lower ranked sites. Inset plot displays average, minimum and maximum AUC values for six mESC-stage PIQ versus ChIP-seq comparisons.
The high correspondence of PIQ output with ChIP-seq results suggests that PIQ provides a valuable tool for predicting protein regulatory interactions for hundreds of TFs genome wide. PIQ allows TF binding site prediction with similar accuracy to ChIP-seq for motif-supported direct protein-DNA binding events, with a median AUC of 0.93. With a small number of replicate experiments PIQ is able to predict the binding of over 733 factors (Supplementary Information), and can do so in the absence of specific TF antibodies or tagged TFs. However, PIQ cannot detect TF motif-free binding events which are observed in ChIP-seq for certain TFs. Some motif-free ChIP-seq events may be mediated by cofactor proteins with diverse sequence specificities, and PIQ would miss these regulatory interactions, although some motif-free events may also be artifacts.
PIQ identifies pioneer and settler transcription factors
We next used PIQ to explore why ChIP-seq experiments have consistently shown that transcription factors bind to fewer than 5% of their 5–15 base pair thermodynamic high-affinity genomic motifs 22, 23. To explain this disparity, we sought to test the hypothesis that TFs, rather than interacting with the epigenetic environment uniformly, act hierarchically, with some TFs actively manipulating chromatin state and others passively responding to local chromatin architecture. The idea that a subset of TFs, defined as pioneer factors, occupy previously closed chromatin and, once bound, allow other TFs to bind nearby has been proposed previously24-26 but not systematically explored. We decided to test whether PIQ, which directly models TF-dependent chromatin accessibility, could discover pioneer factors de novo and characterize TFs into classes based upon their behavior with respect to chromatin accessibility.
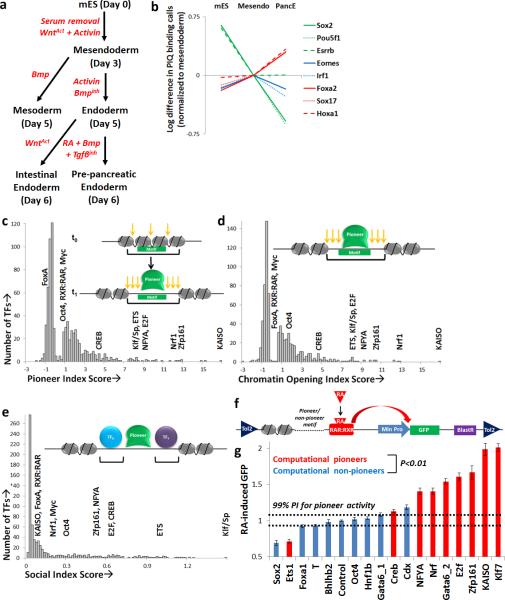
We applied PIQ to data from a developmental lineage paradigm that involves the stepwise differentiation of mouse embryonic stem cells (mESC) to pre-pancreatic and intestinal endoderm (PancE and IntE)27. We induced PancE and IntE differentiation by treating mESC for six days with an in vitro growth factor and small molecule treatment protocol (Fig. 3a). We collected DNase-seq data at two intermediate stages along this stepwise differentiation pathway, mesendoderm (day 3) and endoderm (day 5), as well as from lateral plate mesoderm, which we derived by treating mesendoderm cells with distinct growth factors. This experimental structure yielded a total of six cell states (Fig. 3a) all of which were generated with >90% efficiency (Supplementary Fig. 3), providing relatively homogenous populations. We found that PIQ identifies extensive changes in TF occupancy through differentiation. TFs most strongly expressed in the mESC state such as Pou5f1, Sox2 and Esrrb also bind most often in mESC, and likewise for mesendoderm-enriched TFs, Eomes and Irf1, and PancE-enriched TFs Sox17, Foxa2 and Hoxa1 (Fig. 3b).
Figure 3.
Systematic identification of pioneer TFs. (a) Flow chart outlining mESC-derived populations used for dynamic DNase-seq analysis. (b) Differences in PIQ-detected binding sites for eight selected TFs with strong microarray expression values in mESC (green), mesendoderm (blue) and PancE (red). For each TF at each stage, PIQ calculates a score representing the overall number and strength of binding sites, plotted in natural log PIQ binding strength units normalized to mesendoderm values. (c) Pioneer index log odds scores for all PIQ motifs. (d) Chromatin opening index log odds scores for all PIQ motifs. (e) Social index scores for all PIQ motifs. Scores of selected pioneer and non-pioneer TFs in A-C are noted. (f) Schematic of modular Tol2 transposon-based pioneer reporter system to test pioneer and non-pioneer motifs for chromatin opening ability. Chromatin openness is read out by the level of RA-induced RAR:RXR DNA binding and consequent GFP transcriptional activation, as measured by flow cytometric fluorescence. (g) Average increase in flow cytometric fluorescence after RA addition for 18 pioneer reporter lines grouped as predicted pioneer (red) and non-pioneer (blue) TFs, normalized to RA-induced GFP of the control reporter line. Error bars indicate SEM, and dotted line represents 99% prediction interval based on control RA-induced GFP, indicating lines with RA-induced GFP out of the predicted control range. Predicted pioneers as a group have significantly higher average RA-induced GFP than predicted non-pioneers. n=4, P<0.01 in t-test.
We asked whether PIQ could provide an initial understanding of the rules governing TF binding site choice. We focused first on whether some TFs act as “pioneers24,” shaping the chromatin landscape and the binding of other TFs. Several reports of TFs possessing pioneer activity exist in the literature24, 26, 28-33, but these reports are empirical experimental studies that do not use standard criteria to define pioneer TF activity, are often unconfirmed functionally and to date no systematic attempts have been taken to categorize pioneer TFs. Although pioneer TFs have been defined in various ways, we chose to probe the existence of pioneer TFs capable of binding to closed chromatin and opening nearby chromatin for future occupancy by other TFs. Utilizing our time series, we designed a pioneer index to measure the expected motif-specific local increase in DNase accessibility with respect to baseline at sites whose binding changes between successive timepoints according to PIQ for each of our 733 motifs (Supplementary Information). A higher pioneer index corresponds to higher chromatin opening activity from one timepoint to the next in our developmental timecourse.
We found that most motifs show little appreciable pioneer activity, whereas a small number of motifs appear to open chromatin substantially upon binding (Fig. 3c and Supplementary Table 2). Although there is no clear division between weak pioneers and non-pioneers, a stringent cutoff gives an estimate that 120 of the 733 motifs (16%) show pioneer activity, and the motifs with strongest pioneer activity can be classified into ten TF families (Klf/Sp, NFYA, Nrf, ETS, Creb/ATF, Zfp161, KAISO, Zinc Finger, E2F and CTCF, Supplementary Tables 3 and 4). Of note, previously identified pioneer TFs in the GATA28, Klf26 and NFYA29 families are found to display high pioneer indices whereas FoxA1 (ref. 28), the first identified pioneer, has a low pioneer index.
As binding sites that vary across our observations do not represent a majority of all binding events and are influenced by dynamic TF expression profiles in the particular cell types analyzed, we devised a second metric, the chromatin opening index, to measure the expected static local increase in DNase accessibility attributed to each motif (Supplementary Information). The chromatin opening index is highly concordant with the pioneer index (r2=0.98, Fig. 3d, Supplementary Fig. 4 and Supplementary Table 2), indicating that pioneers can be identified through their static association with open chromatin, thus providing an alternative metric for pioneer TFs that does not require temporal DNase-seq data. TF families with high chromatin opening index scores are conserved in K562 cells (r2=0.84, Supplementary Fig. 4), indicating that chromatin opening is a TF-intrinsic activity consistent across cell type and species.
To determine whether pioneer motifs facilitate binding of other TFs in addition to governing chromatin structure, we devised the social index, the mean number of PIQ-identified binding sites within 200 bp of PIQ-called binding events for a given TF (Supplementary Information) and found that pioneer TFs tend to have more neighbors than non-pioneer TFs (Fig. 3e and Supplementary Table 2). In all analyses, sites adjacent to annotated TSS were excluded to avoid artifacts associated with the strong nucleosome depletion at promoters15, 16, and the results remained consistent after a more stringent removal of unannotated promoters detected through GRO-seq, RNA-seq and histone marks characteristic of promoters (Supplementary Fig. 4).
We experimentally tested the ability of a variety of predicted pioneer and control motifs to open up surrounding chromatin and allow other TFs to bind. To evaluate these criteria in a high-throughput, functional assay, we designed 18 versions of a reporter vector driven by a strong RXR:RAR motif directly adjacent to a pioneer or non-pioneer motif at a locus >1 kb from a minimal promoter and GFP reporter gene (Fig. 3f). We chose the RXR:RAR motif for three reasons. First, RXR:RAR binding shows no effect on surrounding chromatin in the computational analysis (Supplementary Table 2). Second, nuclear hormone receptors, which bind the RXR:RAR motif, respond primarily to surrounding chromatin state rather than specific cofactor interactions34 (also see later text). Third, the RXR:RAR motif allows strong inducible expression of GFP upon addition of retinoic acid (RA), allowing a straightforward quantitative readout of cellular fluorescence intensity. We inserted this vector into the genome of mESC by means of Tol2 transposition35 followed by antibiotic selection, allowing for random genomic integration in a highly polyclonal fashion (>1,000 distinct clones per reporter line), thus controlling for site-specific effects. Consistent with this idea, biological replicates of several lines produced from distinct rounds of Tol2 transposition yielded highly reproducible results (Supplementary Fig. 5). We then used flow cytometry to measure cellular GFP levels in mESC after 24 hours in the presence or absence of RA, interpreting the RA-induced increase in GFP as a correlate of the accessibility of the RXR:RAR site (Fig. 3g).
The pioneer reporter assay data support the computational pioneer TF predictions. Eight of nine predicted pioneer motifs showed significantly above-control RA-induced GFP as compared with only one of eight non-pioneer motifs (Fig. 3g), and pioneer TFs on average promoted significantly higher RA-induced GFP than did controls (P<0.01 in t-test). None of the 18 tested motifs showed significant GFP induction in the absence of RA as compared to the control line (Supplementary Fig. 5), indicating that pioneer and non-pioneer motifs alike do not activate significant gene expression on their own. RT-qPCR analyses also confirmed that RA-induced transcripts do not span the promoter region and pioneer sequences still increase RA-induced GFP when the enhancer is 3 kb from the minimal promoter, confirming that the reporter constructs act as distal enhancers (Supplementary Fig. 5). Lastly, to control for the relative expression of TFs, we performed the reporter assays in mesendoderm and in the presence of ectopically expressed pioneer and non-pioneer TFs, obtaining consistent results (Supplementary Fig. 5).
Asymmetrical opening of chromatin by directional pioneer TFs
Evidence exists that TFs deposit histone marks asymmetrically36. We identified a subset of pioneer TF families that open chromatin more significantly on one side of their motif than on the other (Supplementary Fig. 6 and Fig. 4a). We call factors that possess this novel asymmetrical chromatin opening ability ‘directional pioneers.’ To quantify directional pioneer activity, we measured the expected difference in chromatin opening on either side of each pioneer motif (Supplementary Table 3), identifying strong directional pioneer activity in the Klf/Sp, NFYA, Creb/ATF and Zfp161 pioneer TF families. As we cannot observe directional pioneer activity at palindromic motifs because PIQ cannot orient them, we note that the directional pioneer TF Creb/ATF has multiple PWMs, one of which is non-palindromic. Although directional motifs are known to be important at promoters37, our analyses exclude TSS-adjacent regions and we do not find appreciable transcript production or promoter-characteristic histone marks at distal pioneer sites (Supplementary Fig. 4). Thus, the unidirectional opening of chromatin relative to pioneer TF motif appears to represent a property of certain TFs that to our knowledge has not been described.
Figure 4.
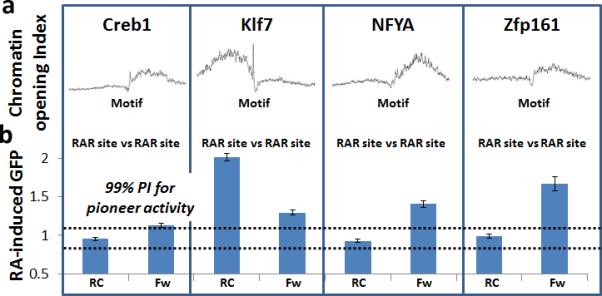
Asymmetrical chromatin opening by directional pioneers. (a) Per-base chromatin opening index log odds scores, which represent expected local increase in hypersensitivity induced by TF binding at all above-threshold genomic motifs for Creb1, Klf7, NFYA, and Zfp161. X-axis for each plot is +/−200 bp from motif center. (b) Experimental validation of directional pioneers. Average increase in flow cytometric fluorescence after RA addition for pioneer reporter lines for the stated motifs. For each TF noted, the left plot (labeled as RC for reverse complement) shows reporter results when the motif orientation is such that the RAR site is on the left of the motif with respect to the plot in a, and the right plot (labeled as Fw for forward) shows results when the RAR site is on the right of the motif with respect to a. All plots are normalized to control line RA-induced GFP as in Figure 3f, error bars indicate SEM, and a 99% prediction interval is shown as in Figure 3f.
To experimentally assess directional pioneer activity, we performed reporter analysis on four motifs displaying strongly directional pioneer activity (Fig. 4b), placing both motif orientations relative to the RXR:RAR site. In all four cases, RA-induced GFP was significantly stronger in the direction predicted to have higher pioneer activity (Fig. 4b), and as predicted, NFYA, Creb and Zfp161 only open chromatin in a single direction from their motif. Directional pioneer activity does not occur during transient transfection (Supplementary Fig. 5), suggesting that this activity occurs through interaction with the local chromatin state.
Settler TFs depend on open chromatin for binding
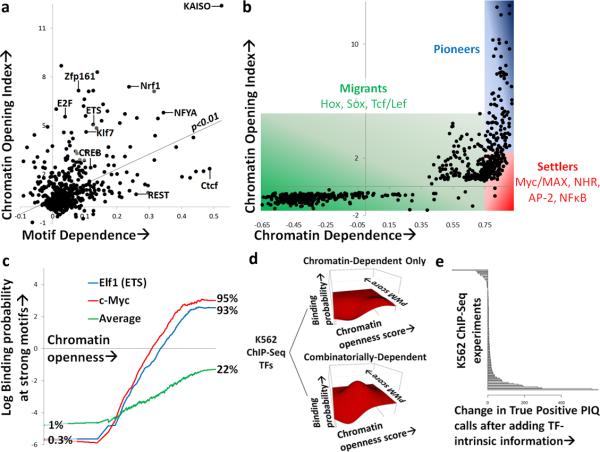
Next we reasoned that classifying TFs by their interactions with chromatin might reveal distinctions in how TFs choose binding sites. As pioneers have been shown to scan nucleosomal DNA for their motifs38, we reasoned that they may be more likely than other TFs to bind to their motif wherever it occurs. To assess this idea, we devised a metric to indicate the likelihood of a TF to bind to an instance of its motif, the correlation of PWM score and binding probability (referred to hereafter as motif dependence). Plotting motif dependence against the chromatin opening index, we find a statistically significant (P<0.01 in t-test) but imperfect positive correlation between motif dependence and chromatin opening (Fig. 5a and Supplementary Table 4), suggesting that pioneer TFs generally do not bind to a high fraction of their genomic motif candidates. Several non-pioneer TFs, including REST, also display strong motif dependence (Fig. 5a and Supplementary Table 4). Motif dependence is uncorrelated with motif information content, suggesting that it is not an artifact of database PWM quality (Supplementary Fig. 7). Thus, although pioneers TFs are more likely to bind their motifs than are non-pioneers, they still rely on facets other than their motif in a majority of their binding decisions.
Figure 5.
Binding of settler TFs is governed by underlying chromatin state. (a) Comparing motif dependence (X-axis) versus chromatin opening index (Y-axis) for all 733 motifs in mouse lineage. Positions of select TFs are denoted and a linear trendline is displayed which shows imperfect but statistically significant positive correlation. (b) Comparing chromatin dependence (X-axis) versus chromatin opening index (Y-axis) for all 733 motifs in mouse lineage. Classes of pioneer TFs (blue), settler TFs (red), and migrant TFs (green) as defined by their chromatin opening and dependence properties are shaded, and select members of each class are listed. (c) Comparing K562 DNase-seq chromatin openness score (X-axis) vs. binned K562 ChIP-seq binding probability at strong motifs (Y-axis) for Elf1 (ETS family, pioneer), c-Myc (settler), and the average of all ChIP-seq experiments. (d) Contour plots showing log odds binding probability (contour) for bins of strong motifs at varying chromatin openness scores (X-axis) and PWM scores (Y-axis) for the K562 ChIP-seq TF clusters displaying chromatin-dependence only (left) or combinatorial motif-dependence and chromatin-dependence (right). For chromatin-dependent TFs, binding probability is predominantly dependent on chromatin openness score, whereas binding probability scores of combinatorially-dependent TFs increase as both chromatin openness and PWM score are increased. (e) Change in number of true positive PIQ calls per TF motif at a 10% false discovery rate as a result of incorporating motif-dependence and chromatin-dependence as prior information for all K562 ChIP-seq motif comparisons. Prior information improves PIQ accuracy for most TFs.
Among non-pioneer TFs, we reasoned that some TFs might be disproportionately dependent on the pre-existing chromatin state as established by pioneer TFs. We explored this possibility computationally by measuring the correlation between DNase accessibility surrounding high-confidence TF motifs and binding probability (Supplementary Table 4). Plotting this metric against the chromatin opening index, which controls for TF-intrinsic chromatin opening, we found that TFs vary substantially in their dependence on chromatin openness in order to bind genomic DNA (Fig. 5b). A subset of TFs were highly likely to bind wherever their motif occurs in an open chromatin landscape but do not open chromatin themselves.
We coin the term “settler” TFs to define the set of TFs whose binding is predominantly dependent on the openness of chromatin at their motifs. Chromatin dependence of TFs is graded, but a stringent cutoff gives an estimate that 131 of the 733 motifs (18%) act as settler TFs (Supplementary Table 4). The majority of non-pioneer TFs, which we term “migrant” TFs, bind only sporadically even when chromatin at their motifs is open and are presumably more heavily dependent on specific cofactor interactions (see Supplementary Table 4 for factor-specific classifications in the mESC pancreatic lineage). Accurate a priori prediction (AUC>0.9) of ChIP-seq genomic binding of “settler” TFs, such as members of the Myc/MAX, nuclear hormone receptor (i.e. RXR:RAR), Ap-2 and NF-κB families, can be obtained simply by measuring DNase accessibility surrounding their motifs (Figs. 2b, 5c), so settler TF binding can be accurately determined solely based on chromatin accessibility in the absence of ChIP or DNase profile information. Pioneer TF binding can also be predicted a priori by local DNase accessibility (Fig. 5c), presumably a result of pioneer-induced chromatin opening at binding sites either in the profiled developmental stage or at a prior timepoint. Thus, we have identified a class of settler TFs that to our knowledge has not been described that obey one simple rule, binding DNA when chromatin is open, establishing settler TFs as a class whose binding is directly dependent on the chromatin opening ability of pioneer TFs.
Although pioneers and settlers typify chromatin opening and chromatin dependence, respectively, we reasoned that the motif-dependence and chromatin-dependence properties of migrants might also contribute to their binding decisions. To test this hypothesis, we clustered TFs possessing matched ChIP-seq and DNase-seq experiments in K562 cells39 by their combination of motif-dependence and chromatin-dependence. We found that TFs broadly fall into two categories: those in which ChIP-seq binding probability increases only with chromatin openness, and those in which binding probability is combinatorially linked to motif score and chromatin openness (Fig. 5d and Supplementary Fig. 7). Modifying PIQ to incorporate these TF-intrinsic binding dependencies into its binding calls improves predictive accuracy for a majority of TFs with matched ChIP-seq data (Fig. 5e), indicating that TF-intrinsic chromatin interaction can be exploited to improve binding prediction. Although we have not included data on histone modification or DNA methylation status in PIQ, we find that DNase hypersensitive regions and PIQ-identified TF binding sites have low levels of DNA methylation in mESC (Supplementary Fig. 7). This suggests that future addition of data types may further improve binding prediction.
Hierarchical binding of pioneer and settler TFs
Our hierarchical binding model predicts that loss of pioneer TF binding should result in closing of chromatin and loss of settler TF binding, at times directionally. Sites at which pioneer TF binding is lost during mESC differentiation do in fact show dramatic loss of DNase hypersensitivity and of adjacent TF binding (Fig. 6a, b). To address this idea mechanistically, we constructed mESC with Doxycycline-inducible dominant negative (DN) alleles for two pioneer TFs, NFYA and Nrf1, that consist solely of DNA-binding domains (Fig. 6c). These DN proteins should bind to their cognate motifs and compete with their native counterparts, blocking pioneer-induced increase in chromatin accessibility. Creation of Doxycycline-inducible lines avoids the lethality associated with knockouts of these TFs40, 41. DNase-qPCR analysis at a set of strongly bound sites revealed that both DN NFYA and DN Nrf1 significantly reduced hypersensitivity at their respective binding sites (Fig. 6d). Furthermore, impairing NFYA and Nrf1 binding also impaired adjacent binding of the settler TF c-Myc at several genomic loci (Fig. 6e). Consistent with our prediction of NFYA's directional pioneer activity (Fig. 4), impairing NFYA binding diminished c-Myc binding when the c-Myc site was downstream of the NFYA site but not upstream (Fig. 6e). Thus, pioneer TF binding is required to maintain open chromatin and to allow nearby settler TF binding, confirming that pioneer TFs sit atop a TF binding hierarchy.
Figure 6.
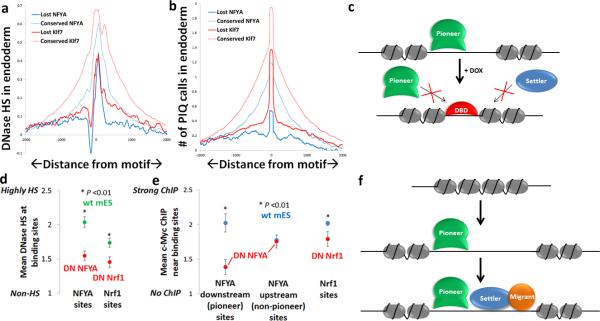
Pioneer TFs control chromatin state and settler TF binding. (a–b) Per-base average DNase hypersensitivity (HS) (a) and number of PIQ binding sites (b) within 4 kb of Klf7 and NFYA motifs for sites conserved (dotted lines) or lost (solid lines) between mesendoderm and endoderm stages. Both DNase HS and adjacent TF binding are diminished when Klf7 and NFYA binding are lost between successive stages. (c) Schematic of pioneer dominant negative (DN) competition experiments in which Doxycycline (Dox) induces DN pioneer TF expression (DBD), which should block pioneer-induced chromatin opening and prevent settler binding to opened chromatin. (d) Mean DNase hypersensitivity at several strong binding sites for NFYA (left) and Nrf1 (right) in wildtype (wt) (green) or DN NFYA/DN Nrf1 (red) mES, normalized to background DNase activity at non-hypersensitive sites. Asterisk indicates statistically significant difference between average DNase HS between wt and DN (n=4, P<0.01) using t-test. (e) Mean ChIP enrichment for four c-Myc sites downstream (in direction of predicted pioneer activity) of NFYA (left column), upstream (in direction of predicted non-pioneer activity) of NFYA (middle column) or adjacent to Nrf1 (right column) in wt (blue) or DN NFYA/DN Nrf1 (red) mES, normalized to positive and negative control genomic c-Myc sites. N=3, P<0.01 using t-test. (f) Model of TF binding hierarchy. Pioneers open chromatin, some directionally, and open chromatin is populated by settler TFs and by certain combinations of migrant TFs.
DISCUSSION
We conclude that PIQ offers a valuable window into TF binding and behavior and has facilitated the elucidation of pioneer TFs that represent a mechanistically diverse set of TFs that play a disproportionately large role in organizing chromatin structure. In a chromatin-based view of TF binding, pioneer TFs shape the chromatin landscape, allowing settler TFs and specific combinations of migrant TFs to populate open chromatin (Fig. 6f). We have shown both computationally and experimentally that through mESC differentiation, gain of pioneer TF binding opens chromatin and that loss of pioneer TF binding closes chromatin, and so we posit that pioneer TFs play an important role in controlling the TF binding dynamics that control cell fate acquisition.
PIQ was designed to model factors that directly modulate chromatin accessibility and is thus uniquely capable of identifying pioneer factors from DNase-seq experiments. PIQ fits a background read model over the entire genome, which allows us to precisely quantify how much a transcription factor opens chromatin relative to both other factors and genomic background. Prior methods such as CENTIPEDE model TF binding on a factor-to-factor basis and therefore would normalize out cross-factor effects. In addition, the chromatin opening index is a natural extension of a TF's profile in PIQ, whereas in DGF or CENTIPEDE profiles are by definition normalized to a mean of zero and do not indicate chromatin opening. We have found in practice that this more detailed model of chromatin accessibility has made it possible to detect TFs with indistinct footprints but large chromatin effects. In some of our identified pioneers such as Gata6, PIQ detects distinct binding sites whereas CENTIPEDE fails to do so (Supplementary Fig. 8).
Recent work42, 43 has suggested that DNase sequence bias may add noise to narrow DNase-seq footprints. In PIQ, TF binding detection is performed on a TF-specific profile, extending 400 bp from each motif and thus not limited to the 5-10 bp footprint itself (Supplementary Fig. 9). PIQ performs a profile-level significance test for whether or not an estimated TF profile is significant outside its motif match region, and all identified pioneer TFs are highly significant.
Our identification of pioneer and settler TFs is limited by the breadth of the motifs used in PIQ, by the degrees of expression and dynamic binding of TFs in the cell types analyzed in this dataset, and by the focus on single motifs which may exclude emergent chromatin opening of TF combinations. Thus the list of pioneer and settler TF families should expand with the collection of more DNase-seq data and TF motifs15. We further note that TFs that do not open chromatin but still facilitate the binding of other factors and those that induce chromatin repression are not captured by our DNase-based assay. Notably, the most well studied pioneer TF, Foxa1, has a relatively low score in all indices (Fig. 3c–e). This may result from the dual role of Foxa1 as a chromatin opening and compacting agent44, 45, its dependence on prior binding of Foxd346 whose strong expression in mESC could obscure its pioneer activity in this lineage, or its minimal role in coordinating chromatin structure as determined by knockout studies in mouse liver47. In any case, this result exemplifies that the computational approach taken here focuses on pioneer TFs that increase DNase hypersensitivity when they bind and thus does not exhaustively identify pioneer TFs.
Comparing mechanisms by which pioneer TFs function will be a fertile area for future research. Codifying TF properties is a step on the road to a priori TF binding prediction and gene network modeling. And as recent work has implicated pioneer TFs in cellular reprogramming26, categorizing pioneer and settler TFs could lead to principled manipulation of cell fate.
Online methods
PIQ algorithm
Mathematical rationale, principles and implementation of PIQ are described in the Supplementary Information.
Mouse embryonic stem cell line generation, culture and differentiation
Mouse embryonic stem cell culture and endoderm differentiation was modified slightly from previously published protocols27. Undifferentiated 129P2/OlaHsd mouse ES cells were maintained on gelatin-coated plates with mouse embryonic fibroblast (MEF) feeders in mES media composed of Knockout DMEM (Life Technologies) supplemented with 15% defined fetal bovine serum (FBS) (HyClone), 0.1mM nonessential amino acids (Life Technologies), Glutamax (Life Technologies), 0.55mM 2-mercaptoethanol (Sigma), and 1X ESGRO LIF (Millipore).
Prior to differentiation, ES cells were passaged onto gelatin-coated plates for 25 minutes to deplete MEFs. MEF-depleted ES cells were then seeded at 1 *10^4 cells/cm2 onto gelatin-coated dishes in mES media. After 12-24 hours, media was changed to Advanced DMEM (Life Technologies) supplemented with N-2 (Life Technologies), B27 Supplement without vitamin A (Life Technologies), and Glutamax. After 44-48 hours, media was changed to Advanced DMEM with 2% FBS, Glutamax, 5 nM GSK-3 inhibitor XV and 50 ng/mL E. coli-derived Activin A (Peprotech) for 24 hours to produce mesendoderm. For endoderm differentiation, cells were then fed with Advanced DMEM with 2% FBS, Glutamax, 50 ng/mL Activin A and 1 μM Dorsomorphin (Sigma) for 48 hours. For intestinal endoderm differentiation, cells at the endoderm stage were fed for 24 hours with Advanced DMEM with B-27 supplement without vitamin A, Glutamax, and 100 nM GSK-3 inhibitor XV. For pre-pancreatic endoderm differentiation, cells at the endoderm stage were fed for 24 hours with Advanced DMEM with B-27 supplement without vitamin A, Glutamax, 500 nM retinoic acid (Calbiochem), 50 nM A-83-01 (Calbiochem), and 8 ng/mL Bmp4 (Stemgent). For mesodermal differentiation, cells at the mesendoderm stage were treated for 48 hours with 10 ng/mL Bmp4.
ES cells with doxycycline-inducible alleles for Sox2, Foxa1, Hnf1β, Cdx2, Gata6, Zfp161, and Klf7 in the HPRT locus were created as described 48 and maintained and differentiated as above. For dominant negative lines, DNA-binding domains of NFYA and Nrf1 were used to create doxycycline-inducible HPRT lines as above.
Dominant negative lines were grown for >7 days in mES media supplemented with 5 nM GSK-3 inhibitor XV and 500 nM UO126 to enhance pluripotency49 and 2 μg/mL Doxycycline. Cells were harvested at this stage for DNase-qPCR. For ChIP-qPCR, cells were treated for 6 hours with mES media with 1 μM retinoic acid.
Tol2 GFP reporter transposon construct generation, transfectio, and flow cytometry
PCR-amplified constructs containing pioneer and non-pioneer motif regions and RXR:RAR binding sites were generated from primers listed below and cloned into PacI and AscI sites of p2TAL200R175-minHsp-GFP-BlR (Sherwood et al, manuscript under review). To generate the reporter construct with 2 kb spacer DNA added between the enhancer and promoter, 2 kb of genomic DNA from a consistently DNase-insensitive genomic region (primers included in oligonucleotide section) was cloned into the PacI site of p2TAL200R175-minHsp-GFP-BlR.
Tol2-containing reporter plasmids and transposase-containing pCAGGS-mT2TP (Sherwood et al, manuscript under review) were transfected into the mES lines noted in the text using Xfect for mES cells transfection reagent (Clontech). Blasticidin selection was performed for >7 days in mES media with 5 nM GSK-3 inhibitor XV and 500 nM UO126 added to enhance pluripotency49.
For flow cytometric GFP detection, cells were trypsinized and seeded at 3*104 cells/cm2 onto 96-well plates. Cells were treated with mES media alone or supplemented with 1 μM retinoic acid and/or 2 μg/mL Doxycycline or differentiated into mesendoderm prior to treatment. After 24 hours, cells were trypsinized, quenched, and fluorescence of 5-20*103 cells was measured using a BD Accuri C6 flow cytometer and accompanying software (BD Biosciences).
Antibodies and immunofluorescence
For cell immunofluorescence analysis, tissue culture plates were fixed for 20 minutes in 4% paraformaldehyde (Electron Microscopy Sciences) and washed in PBS with 0.1% Triton X-100 (Sigma). Tissues were blocked by 20 minute incubation at 4 degrees in PBS with 20% donkey serum (Jackson Immunoresearch) and 0.1% Triton X-100. Primary and secondary antibody staining were performed overnight at 4 degrees in PBS with 5% donkey serum and 0.1% Triton X-100, and after primary and secondary antibody staining, washing was performed with PBS with 0.1% Triton X-100. After staining, plates were washed and incubated with 1 μg/mL Hoechst 33342 (Life Technologies). Imaging was performed using a DMI 6000b inverted fluorescence microscope (Leica), and image analysis with the Leica AF6000 software package.
The following primary antibodies were used: goat anti-Foxa2 M-20, rabbit anti-RAR M-454, rabbit anti-cMyc N-262 (Santa Cruz Biotechnology), rabbit anti-Foxa2 (Millipore); goat anti-Sox17, mouse anti-Sox2, (R+D Systems); mouse anti-Hnf1β (BD Biosciences). AlexaFluor488 and AlexaFluor594 conjugates (Jackson Immunoresearch) were used for secondary detection.
ChIP-qPCR
ChIP was performed according to the “Mammalian ChIP-on-chip” protocol (Agilent). 1 × 107 - 5 × 107 cells were used for each experiment. qPCR primers are listed in the table of oligonucleotides.
Oligonucleotides
Oligonucleotides used in this work are presented in Supplementary Table 5.
DNase-seq
DNase-seq was performed using adaptations of previous protocols50. A detailed protocol can be found in the Supplementary Information.
DNase-qPCR
DNase-qPCR samples were prepared from the doxycyline-induced dominant-negative cell lines and control cell lines in the absence of doxycyline as per the DNase-seq protocol above. Experimental primers were designed for pioneer transcription factor binding sites and used in conjunction with the positive and negative hypersensitivity control primers described above in quantitative PCR analyses. Hypersensitivity at experimental primers sites was calculated for the dominant negative lines and control lines as follows:
Significance was calculated using Student's t-test.
Supplementary Material
Acknowledgments
The authors thank Grigoriy Losyev, Jennifer Huynh, and the MIT BioMicro Center for technical assistance, Koichi Kawakami and Hynek Wichterle for reagents, and Richard Maas for help with the manuscript. The authors acknowledge funding from The National Institutes of Health Common Fund 5UL1DE019581, RL1DE019021 and 5TL1EB008540; the Harvard Stem Cell Institute's Sternlicht Director's Fund award to R.I.S, and NIH grants 1U01HG007037 and 5P01NS055923 to D.K.G.
Footnotes
Author contributions Experiments were designed by R.I.S., T.H., C.W.O., and D.K.G. DNase-seq experiments were conducted by R.I.S., S.L., A.A.B., and J.P.vH., reporter experiments by R.I.S. and V.K., and dominant negative experiments by R.I.S. and A.A.B. PIQ was designed and implemented by T.H. and C.W.O., DNase-seq and PIQ computational analysis performed by T.H., C.W.O., D.K.G., and T.J. The manuscript was prepared by R.I.S., T.H., C.W.O., and D.K.G.
PIQ implementation and data are available at http://piq.csail.mit.edu The authors declare no competing interests.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mullen AC, et al. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trompouki E, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein- DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 8.Guo Y, et al. Discovering homotypic binding events at high spatial resolution. Bioinformatics. 2010;26:3028–3034. doi: 10.1093/bioinformatics/btq590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Y, Mahony S, Gifford DK. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput Biol. 2012;8:e1002638. doi: 10.1371/journal.pcbi.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weintraub H, Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976;193:848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- 12.Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 13.Boyle AP, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Research. 2011;21:456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pique-Regi R, et al. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neph S, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyle AP, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011;21:456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Hoffman MM, Bilmes JA, Hesselberth JR, Noble WS. A dynamic Bayesian network for identifying protein-binding footprints from single molecule-based sequencing data. Bioinformatics. 2010;26:i334–342. doi: 10.1093/bioinformatics/btq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minka T. Expectation Propagation for Approximate Bayesian Inference.. UAI '01: Proceedings of the 17th Conference in Uncertainty in Artificial Intelligence.2001. pp. 362–369. [Google Scholar]
- 21.Chen X, et al. Integration of External Signaling Pathways with the Core Transcriptional Network in Embryonic Stem Cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 22.Joseph R, et al. Integrative model of genomic factors for determining binding site selection by estrogen receptor-alpha. Mol Syst Biol. 2010;6:456. doi: 10.1038/msb.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplan T, et al. Quantitative models of the mechanisms that control genome-wide patterns of transcription factor binding during early Drosophila development. PLoS Genet. 2011;7:e1001290. doi: 10.1371/journal.pgen.1001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gualdi R, et al. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- 26.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherwood RI, Maehr R, Mazzoni EO, Melton DA. Wnt signaling specifies and patterns intestinal endoderm. Mech Dev. 2011 doi: 10.1016/j.mod.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 29.Nardini M, et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell. 2013;152:132–143. doi: 10.1016/j.cell.2012.11.047. [DOI] [PubMed] [Google Scholar]
- 30.Budry L, et al. The selector gene Pax7 dictates alternate pituitary cell fates through its pioneer action on chromatin remodeling. Genes Dev. 2012;26:2299–2310. doi: 10.1101/gad.200436.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hori S. c-Rel: a pioneer in directing regulatory T-cell lineage commitment? Eur J Immunol. 2010;40:664–667. doi: 10.1002/eji.201040372. [DOI] [PubMed] [Google Scholar]
- 33.Treiber T, et al. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity. 2010;32:714–725. doi: 10.1016/j.immuni.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 34.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawakami K, Noda T. Transposition of the Tol2 element, an Ac-like element from the Japanese medaka fish Oryzias latipes, in mouse embryonic stem cells. Genetics. 2004;166:895–899. doi: 10.1534/genetics.166.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kundaje A, et al. Ubiquitous heterogeneity and asymmetry of the chromatin environment at regulatory elements. Genome Res. 2012;22:1735–1747. doi: 10.1101/gr.136366.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eddy J, et al. G4 motifs correlate with promoter-proximal transcriptional pausing in human genes. Nucleic Acids Res. 2011;39:4975–4983. doi: 10.1093/nar/gkr079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 2009;23:804–809. doi: 10.1101/gad.1775509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattacharya A, et al. The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res. 2003;63:8167–8172. [PubMed] [Google Scholar]
- 41.Huo L, Scarpulla RC. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol Cell Biol. 2001;21:644–654. doi: 10.1128/MCB.21.2.644-654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koohy H, Down TA, Hubbard TJ. Chromatin accessibility data sets show bias due to sequence specificity of the DNase I enzyme. PLoS One. 2013;8:e69853. doi: 10.1371/journal.pone.0069853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He HH, et al. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Methods. 2013 doi: 10.1038/nmeth.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watts JA, et al. Study of FoxA pioneer factor at silent genes reveals Rfx-repressed enhancer at Cdx2 and a potential indicator of esophageal adenocarcinoma development. PLoS Genet. 2011;7:e1002277. doi: 10.1371/journal.pgen.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu J, et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, Schug J, Tuteja G, White P, Kaestner KH. The nucleosome map of the mammalian liver. Nat Struct Mol Biol. 2011;18:742–746. doi: 10.1038/nsmb.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iacovino M, et al. A conserved role for Hox paralog group 4 in regulation of hematopoietic progenitors. Stem Cells Dev. 2009;18:783–792. doi: 10.1089/scd.2008.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5384. pdb prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.