Abstract
Piwi-interacting RNAs (piRNAs) are small RNAs required to maintain germline integrity and fertility but their mechanism of action is poorly understood. Here we demonstrate that C. elegans piRNAs silence transcripts in trans through imperfectly complementary sites. Target silencing is independent of Piwi endonuclease activity or “slicing”. Instead, piRNAs initiate a localized secondary endogenous small interfering RNA (endo-siRNA) response. Endogenous protein-coding gene and transposon transcripts exhibit Piwi-dependent endo-siRNAs at sites complementary to piRNAs and are de-repressed in Piwi mutants. Genomic loci of piRNA biogenesis are depleted of protein-coding genes and tend to overlap the start and end of transposons in sense and antisense, respectively. Our data suggest that nematode piRNA clusters are evolving to generate piRNAs against active mobile elements. Thus, piRNAs provide heritable, sequence-specific triggers for RNAi in C. elegans.
The Piwi/piRNA pathway has an evolutionarily conserved role in germline transposon silencing in animals. C. elegans encodes two Piwi family proteins, PRG-1 and PRG-2, although PRG-2 has likely little or no function (1, 2). PRG-1 and piRNA expression is restricted to the male and female germline. The piRNAs of C. elegans are 21 nucleotides in length with a 5′ uracil (21U-RNAs) (1-4). In C. elegans piRNAs have a sequence motif, situated ~40 bp upstream of each piRNA locus, that is thought to be required for piRNA biogenesis (2, 3). A challenge in the field is to understand the mechanism(s) by which piRNAs act on their targets. Proposed functions for Piwi/piRNA complexes include the RNAi-like slicing of RNA transcripts (5-7), transcript deadenylation (8) and de novo DNA methylation (9, 10). Here we identify the targeting mechanism and the endogenous targets of C. elegans piRNAs.
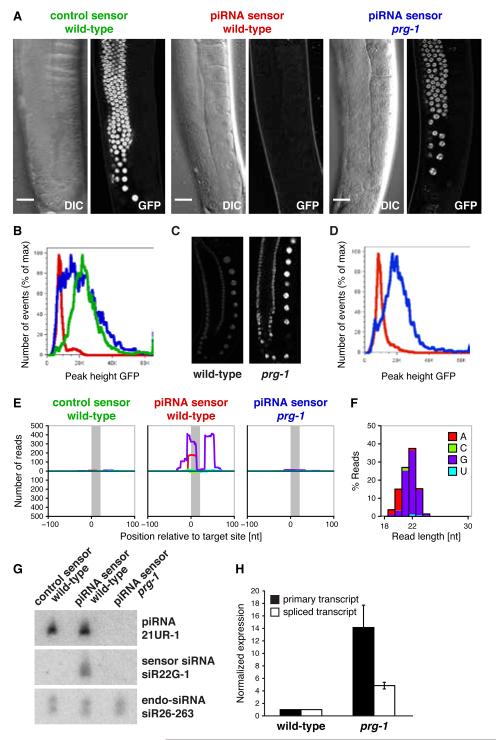
We generated C. elegans strains carrying GFP-histone H2B fusion transgenes into which we inserted a short sequence complementary to an endogenous piRNA (21UR-1) or its reverse complement, hereafter referred to as the piRNA sensor and control sensor, respectively (see fig. S1, supplementary online text) (11). While the control sensor expressed nuclear, chromatin-associated GFP throughout germline development, GFP expression was silenced in animals carrying the piRNA sensor transgene (Fig. 1A,B). piRNA sensor silencing was dependent on prg-1. In contrast, prg-2 did not show an effect (fig. S2, supplementary online text). An independent sensor for the unrelated endogenous piRNA 21UR-1349 confirmed these results (Fig. 1C,D).
Fig. 1.
A single antisense piRNA site is sufficient for target silencing in vivo. (A) Fluorescence microscopy (GFP-H2B) and differential interference contrast (DIC) images of adult hermaphrodites. Scale bar 20 μm. (B) Flow cytometry analysis of control sensor strain (green) and piRNA sensor strain in wild-type (red) or prg-1 (n4357) mutant background (blue) as in (A). (C) Germline GFP-H2B expression of the 21UR-1349 piRNA sensor. (D) Flow cytometry analysis of the 21UR-1349 piRNA sensor strain in wild-type (red) and prg-1 mutant (blue). (E) Profiles of small RNA high-throughput sequencing reads with unique match to the sensor relative to the target site (indicated in grey). Colors correspond to 5′ nucleotides as indicated in the color key in (F). Positive and negative y-axes correspond to antisense and sense reads, respectively. (F) Length and 5′ nucleotide identity of small RNAs antisense to the piRNA sensor in wild-type. (G) Northern blot of total RNA. Probes were against piRNA 21UR-1, a piRNA sensor-specific 22G-RNA and the PRG-1-independent endo-siRNA siR26-263. (H) qRT-PCR of primary piRNA sensor transcript and mRNA. Data were normalized to wild-type transcript levels. Error bars are standard errors of the mean.
We analyzed small RNA populations in the sensor strains by high-throughput sequencing (Fig. 1E) (11). We detected a set of small RNAs that map unambiguously to the piRNA sensor mRNA, mostly within ~20 bp of the piRNA target site. These small RNAs were predominantly 22 nucleotides in length with a 5′ guanosine, characteristic features of secondary endo-siRNAs, also referred to as 22G-RNAs (Fig. 1F). 22G-RNAs represent the most abundant class of endogenous small RNAs in C. elegans, are RNA-dependent RNA polymerase products and have a 5′ triphosphate (12-14). The set of 22G-RNAs that map in close proximity to the piRNA target site was dependent on prg-1 and absent in the control sensor (Fig. 1E). We validated the high-throughput sequencing data using northern blotting for an abundant 22G-RNA mapping to the piRNA sensor (Fig. 1G). As endo-siRNA silencing can involve post-transcriptional regulation of mRNAs (15) or co-transcriptional gene regulation (16) we tested if the piRNA sensor was regulated at either level. We find that both primary transcript and mRNA are upregulated in prg-1 mutants, consistent with co-transcriptional or a combination of post- and co-transcriptional regulation of the piRNA (Fig. 1H).
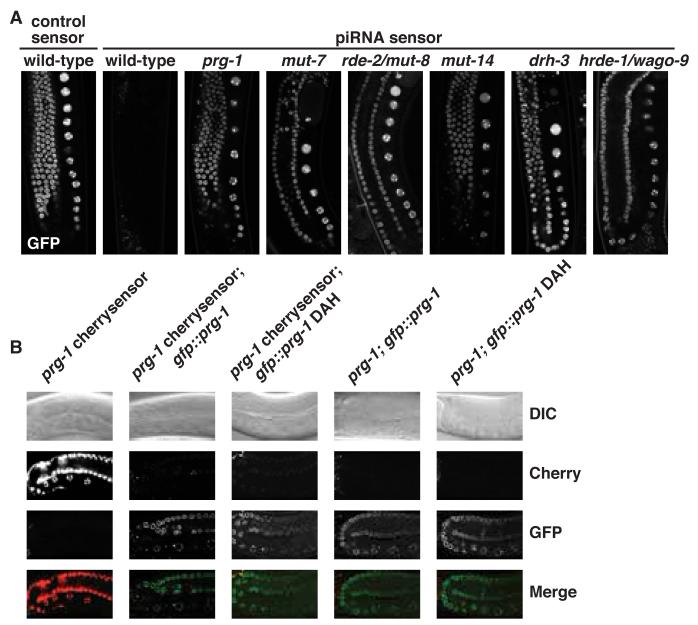
To investigate the role of endo-siRNAs in piRNA-mediated gene silencing we screened a set of 24 siRNA pathway genes. Mutants were crossed into the piRNA sensor and assayed for transgene GFP expression through fluorescent microscopy (Fig. 2A, fig. S3A,B), flow cytometry (fig. S3C,D), northern blotting (fig. S4), or high-throughput sequencing (fig. S5). We found that a specific subset of siRNA pathway genes is required for piRNA-mediated silencing and encodes pathway components including three putative helicases (MUT-7, DRH-3, MUT-14), two RNA-dependent RNA polymerases (EGO-1 and RRF-1), the enzymes that generate secondary siRNAs, and several “worm”-specific Argonaute proteins (WAGOs) (17) (Fig. S6A,B). While several strains with mutations in multiple WAGOs are defective in piRNA sensor silencing, hrde-1/wago-9 is the only single mutant WAGO strain defective in piRNA sensor silencing (Fig. 2A, fig. S3). PRG-1 is necessary for both piRNA expression and sensor-specific 22G-RNA expression. In contrast, the identified siRNA pathway genes act downstream of piRNA expression and are required only for the expression of piRNA sensor-derived 22G-RNAs (fig. S4,S5). We conclude that a specific endo-siRNA pathway acts as a downstream effector of the piRNA pathway.
Fig. 2.
A specific endo-siRNA pathway acts downstream of and is required for piRNA-mediated silencing. (A) piRNA sensor expression in siRNA pathway mutants as in Fig. 1A. (B) A second 21UR-1 piRNA sensor strain (cherrysensor) expressing mCherry-H2B in prg-1 (n4357).
The Ping-Pong piRNA amplification loop in insects (18) and vertebrates requires endonuclease or slicing activity for biogenesis of at least a subset of piRNAs (7, 19). PRG-1 contains an evolutionarily conserved DDH motif (catalytic triad) that confers endonuclease or slicing activity to some Argonaute superfamily proteins (20) and recombinant PRG-1 appears to have some slicing activity in vitro (fig. S7, S8, supplementary online text). To test the requirement for PRG-1 catalytic activity in vivo, we generated two transgenes expressing wild-type GFP-PRG-1 or GFP-PRG-1 DAH mutant fusion proteins (fig. S9). In prg-1 mutants both wild-type and DAH mutant GFP-PRG-1 were sufficient to rescue piRNA expression (fig. S4). To address whether the catalytic triad of PRG-1 is required for piRNA-mediated silencing of targets we generated a second 21UR-1 piRNA sensor strain (cherrysensor) expressing mCherry-H2B. While the cherrysensor was de-silenced in a prg-1 mutant background, both wild-type GFP-PRG-1 and GFP-PRG-1 DAH restored silencing of the cherrysensor to the same extent (Fig. 2B). This was also true in the prg-1;prg-2 double mutants (fig. S10). In addition, both wild-type and DAH mutant GFP-PRG-1 rescue the fertility defects of prg-1 mutants (fig. S11). Finally, we generated a panel of mutated piRNA sensors and find that two mismatches are tolerated throughout the target sequence for PRG-1-dependent sensor silencing, including mismatches at positions 10 and 11 that are required for slicing (fig. S12). We conclude that PRG-1 slicing is not required in vivo.
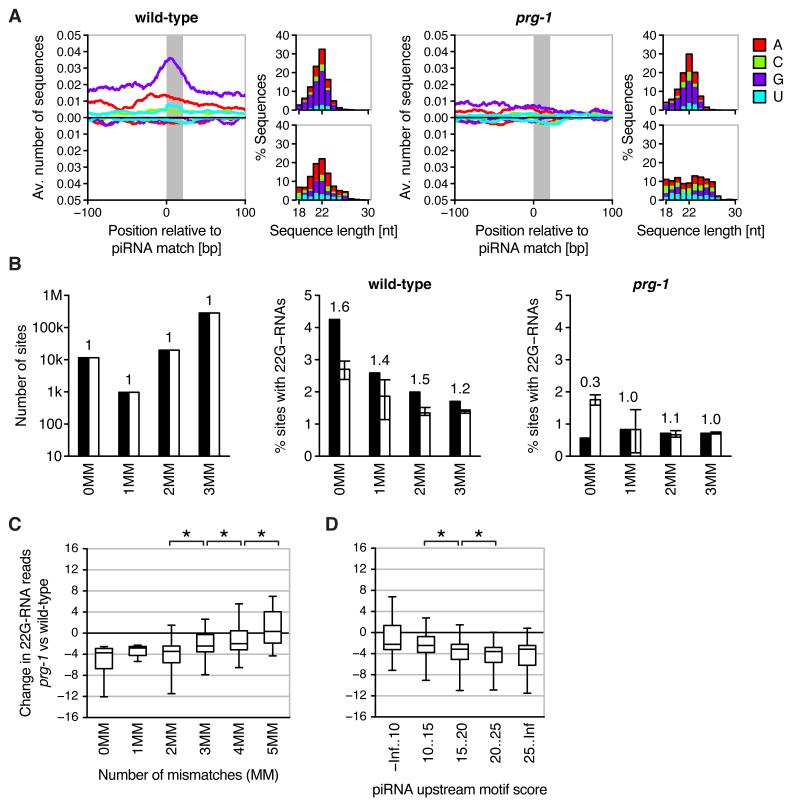
To investigate whether piRNAs target endogenous transcripts, we considered piRNA matches in the C. elegans genome allowing for up to 3 mismatches. For 16,003 piRNAs we identified a total of 681,746 sites (Additional Data Table S1, S2). We found that PRG-1-dependent 22G-RNAs localize in close proximity to imperfect piRNA matches, recapitulating our observations for the piRNA sensor (Fig. 3A, fig. S13). To assess how many of these represent functional target sites, we compared genomic matches of piRNAs to those of matched control sequences. In wild-type animals, approximately 4.2%, 2.6%, 2.0% and 1.7% of the 0, 1, 2 and 3 mismatch sites exhibit unambiguously mapping 22G-RNAs, corresponding to an enrichment of 1.6, 1.4, 1.5 and 1.2 compared to control sites, respectively (Fig. 3B). In prg-1 mutants the percentage of genomic piRNA matches with 22G-RNAs was comparable to control sites. The distribution of piRNA matches (and those with 22G-RNAs) across the genome resembled that of controls, except for an enrichment of perfect piRNA matches in the two piRNA clusters on chromosome IV (fig. S14, S15).
Fig. 3.
piRNAs initiate a localized secondary siRNA response against endogenous transcripts. (A) Average profiles of collapsed small RNAs mapping uniquely to imperfect genomic piRNA matches (1-3 mismatches) in wild-type (left) and prg-1 mutant (right). Top and bottom panels to the right of each profile illustrate characteristics of antisense and sense small RNAs, respectively. (B) Number of genomic piRNA matches (left) and percentage of matches with uniquely mapping 22G-RNAs in wild-type (middle) and prg-1 (right). Black and white bars correspond to piRNAs and matched controls, respectively. Bars for control sequences indicate medians, error bars the range of values obtained for 20 cohorts of control sequences. Numbers above bars indicate the fold-difference between piRNAs and controls. (C) Difference in 22G-RNAs mapping uniquely within 20 bp of genomic piRNA matches between prg-1 and wild-type. Shown are boxplots of the difference in 22G-RNA reads after square root transformation (box indicates interquartile range, plot extends from 5th to 95th percentile). Asterisks indicate statistical significance (P < 0.001, two-sided Wilcoxon rank-sum test). (D) As in (C) with genomic piRNA matches grouped according to motif score of complementary piRNA (as proxy for abundance).
To further investigate mismatch tolerance of piRNA targeting we considered genomic piRNA matches with up to five mismatches. We observed that levels of PRG-1-dependent 22G-RNAs decrease with increasing number of mismatches (Fig. 3C). Levels of PRG-1-dependent 22G-RNAs were greater at sites with four compared to five mismatches, suggesting that in some cases sites with up to four mismatches can be sufficient for the synthesis of 22G-RNAs. We also observed that levels of 22G-RNAs depend on piRNA abundance (Fig. 3D).
We identified candidate endogenous targets by searching for antisense piRNA matches in annotated protein-coding genes, pseudogenes and transposons allowing for up to three mismatches (Additional Data Table S3, S4, S5). We found that in prg-1 mutant animals 22G-RNAs antisense to candidate targets showed a stronger reduction at target sites (within 20 nt) compared to regions distant from target sites (fig. S16). When analyzing mRNA expression differences between prg-1; prg-2 mutant and wild-type animals by microarray, mRNA expression changes showed stronger correlation with changes in 22G-RNAs at target sites compared to the whole transcript (fig. S16).
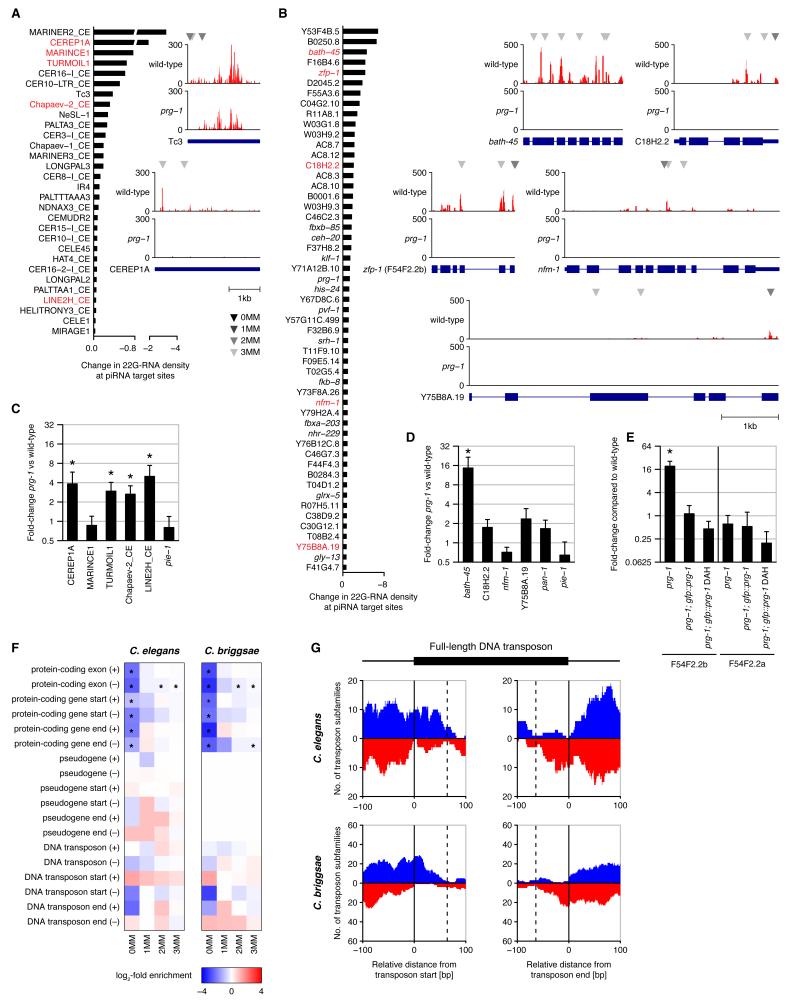
We ranked transposons and protein-coding genes by the decrease in target-site associated 22G-RNA density in prg-1 mutant compared to wild-type animals and examined individual candidate targets (Fig. 4A,B). Transposase mRNA from Tc3 is de-silenced in prg-1 mutants and Tc3 exhibits PRG-1-dependent 22G-RNAs against its terminal inverted repeats (TIRs) but no matching piRNAs (1, 2). We identified three piRNAs with imperfect complementarity to the consensus sequence of Tc3, all of which map to the TIRs, suggesting that these piRNAs target Tc3 elements in trans. We chose five transposable elements (CEREP1A, MARINCE1, TURMOIL1, Chapaev-2_CE, LINE2H_CE) and six protein-coding genes (bath-45, zfp-1, C18H2.2, nfm-1, Y75B8A.19, pan-1) with strong reduction in target-site associated 22G-RNAs for analysis by quantitative RT-PCR (Fig. 4C,D,E). Six of these candidates showed statistically significant increased expression in prg-1 mutants (P < 0.05, two-sided t-test). We tested the requirement of an intact catalytic triad and found that both wild-type and DAH mutant GFP-PRG-1 restored silencing of zfp-1 (F54F2.2b) in prg-1 mutants, while an independent isoform lacking target-site associated 22G-RNAs (F54F2.2a) showed no change in expression (Fig. 4E). In addition, we found that expression of a PRG-1-dependent 22G-RNA against zfp-1 (F54F2.2b) was restored by both wild-type and DAH mutant GFP-PRG-1 (fig. S4). Thus, piRNAs target endogenous transcripts in trans for silencing, independently of slicing activity.
Fig. 4.
Endogenous piRNA targets and piRNA evolution. (A) Candidate transposon targets ranked by change in 22G-RNA density at target sites between prg-1 and wild-type. Transposons selected for qRT-PCR validation are in red. Antisense 22G-RNA profiles are shown for selected elements with target sites indicated above each profile as explained in the color key. (B) Candidate protein-coding targets as in (A). (C) qRT-PCR analysis of candidate transposon targets with fold-changes normalized to actin. Error bars are standard errors of the mean, asterisks denote P < 0.05 (two-sided t-test). (D) qRT-PCR analysis of candidate protein-coding targets. (E) qRT-PCR analysis of targeted (F54F2.2b) and non-targeted (F54F2.2a) transcripts from the zfp-1 locus. Data were normalized to wild-type. (F) Enrichment and depletion of genomic piRNA matches overlapping features of interest. Red and blue indicate increased or reduced number of matches for piRNAs compared to control sequences, respectively. Asterisks indicate statistical significance. Start and end refer to the first and last 50 bp of the annotated feature, respectively. Pseudogene annotation was only available for C. elegans. (G) piRNA matches against start (left) and end (right) of DNA transposons. Profiles indicate the number of transposon subfamilies with perfect piRNA match in at least one full-length genomic copy. Positive (blue) and negative (red) y-axes correspond to sense and antisense matches, respectively. Dashed lines correspond to maximal allowed distance between an upstream sequence motif and piRNA 3′ end.
Our findings raise the question of how piRNAs and target sites arise during evolution. The majority of the 16,003 C. elegans piRNAs (96%) map to unique locations in the genome and are depleted of protein-coding genes (2). We confirmed that piRNA loci are depleted of protein-coding genes in both C. elegans and the related nematode species C. briggsae when compared to loci of matched control sequences (Fig. 4F). However, we did not observe a depletion of pseudogenes or transposons. piRNA loci showed a trend for depletion at the start and end of full-length DNA transposons in antisense and sense, respectively, and an inverse trend for enrichment at the start and end in sense and antisense, respectively (Fig 4F). This signature suggests recent DNA transposon integrations downstream of instances of the sequence motif thought to be required for piRNA biogenesis. Such integrations may result in the birth of a piRNA either sense to the 5′ end or antisense to the 3′ end of the transposon. In the latter case the new piRNA has the potential to target and silence the mobile element. Indeed, the observed distances between piRNA locus and start or end of the transposon are consistent with transposon insertions downstream of existing sequence motifs (Fig. 4G, fig. S17). A similar signature was observed in C. briggsae (Fig. 4G, fig. S18) despite that neither piRNAs nor transposable elements are conserved between the two species. When considering imperfect piRNA matches we observed a depletion antisense to protein-coding genes (Fig. 4F, fig. S19), suggesting mRNA targeting is often detrimental and piRNAs and sites with potential for mRNA silencing undergo negative selection.
Our data demonstrate that C. elegans piRNAs silence endogenous transcripts by triggering a secondary siRNA response (fig. S20). While we find that individual mRNAs are piRNA targets, the physiological roles of piRNA-mediated gene regulation remain to be explored. RNAi pathways are able to silence repetitive elements regardless of their sequence content but rely on the formation of double-stranded RNA. The piRNA pathway may provide an alternative defense mechanism that is heritable and sequence-specific based on the evolution of new piRNAs against active mobile elements. Secondary siRNA amplification ensures effective silencing of abundant targets, resembling the Ping-pong piRNA amplification cycle in other species.
Supplementary Material
Acknowledgments
We are grateful to Julie Ahringer and Erik Jorgensen for providing reagents. A.S. was supported by the Human Frontier Science Program and the Herchel-Smith Fund. N.J.L. was supported by a PhD studentship from the Wellcome Trust (UK). E.M.W. was supported by a PhD studentship from the Herchel-Smith Fund. S.B. and M.J.S. were funded by the Canadian Institutes of Health Research (CIHR). M.J.S. is a New Investigator from CIHR. This work was supported by Cancer Research UK and the Wellcome Trust. Small RNA sequence data and Affymetrix gene expression data were submitted to the Gene Expression Omnibus (GEO) under accession number GSE37433. Detailed methods can be found in Supplementary Materials.
Footnotes
Materials and Methods
Supplementary Text
Figs. S1 to S21
Tables S1 to S3
References (21-30)
Additional Data Table S1 to S5
References
- 1.Das PP, et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batista PJ, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruby J, et al. Large-Scale Sequencing Reveals 21U-RNAs and Additional MicroRNAs and Endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 4.Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr. Biol. 2008;18:861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 6.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 7.Reuter M, et al. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480:264–267. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- 8.Rouget C, et al. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467:1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aravin AA, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuramochi-Miyagawa S, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Materials and methods are available as supplementary material on Science Online.
- 12.Gu W, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 14.Sijen T, Steiner FA, Thijssen KL, Plasterk RHA. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 15.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 16.Guang S, et al. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1010. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yigit E, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 18.Saito K, et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 19.De Fazio S, et al. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480:259–263. doi: 10.1038/nature10547. [DOI] [PubMed] [Google Scholar]
- 20.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 21.Wood W. The Nematode Caenorhabditis elegans. Cold Spring Harbour Laboratory Press; 1988. [Google Scholar]
- 22.Frøkjaer-Jensen C, et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frøkjaer-Jensen C, Davis MW, Ailion M, Jorgensen EM. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods. 2012;9:117–118. doi: 10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horvitz HR, Sulston JE. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics. 1980;96:435–454. doi: 10.1093/genetics/96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 26.Haley B, Tang G, Zamore PD. In vitro analysis of RNA interference in Drosophila melanogaster. Methods. 2003;30:330–336. doi: 10.1016/s1046-2023(03)00052-5. [DOI] [PubMed] [Google Scholar]
- 27.Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gentleman R. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer Verlag; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.