Abstract
This review of the exercise genomics literature emphasizes the strongest papers published in 2010 as defined by sample size, quality of phenotype measurements, quality of the exercise program or physical activity exposure, study design, adjustment for multiple testing, quality of genotyping, and other related study characteristics. One study on voluntary running wheel behavior was performed in 448 mice from 41 inbred strains. Several quantitative trait loci for running distance, speed, and duration were identified. Several studies on the alpha-3 actinin (ACTN3) R577X nonsense polymorphism and the angiotensin converting enzyme (ACE) I/D polymorphism were reported with no clear evidence for a joint effect, but the studies were generally underpowered. Skeletal muscle RNA abundance at baseline for 29 transcripts and 11 single nucleotide polymorphisms (SNPs) were both found to be predictive of the VO2max response to exercise training in one report from multiple laboratories. None of the 50 loci associated with adiposity traits is known to influence physical activity behavior. However, physical activity appears to reduce the obesity-promoting effects of at least 12 of these loci. Evidence continues to be strong for a role of gene-exercise interaction effects on the improvement in insulin sensitivity following exposure to regular exercise. SNPs in the cAMP responsive element binding position 1 (CREB1) gene were associated with training-induced heart rate response, in the C-reactive protein (CRP) gene with training-induced changes in left ventricular mass, and in the methylenetetrahydrofolate reductase (MTHFR) gene with carotid stiffness in low-fit individuals. We conclude that progress is being made but that high-quality research designs and replication studies with large sample sizes are urgently needed.
Keywords: Genetics, exercise training, candidate genes, gene-exercise interaction, single nucleotide polymorphism, quantitative trait locus, genomic predictors
Introduction
From 2000 until 2009, our international group of exercise genetics and genomics investigators published an annual comprehensive and cumulative review of the published literature in the area of genomics relative to exercise, fitness, and performance. As discussed in last year’s publication, this annual project eventually became too large of an effort to publish in terms of the number of journal pages it required and the effort necessary from the coauthors to review and summarize the published literature annually and, more importantly, to maintain the gene map and summary tables that were an important part of the publication (45). As a result, the review evolved into a substantially shorter version, with the goal of summarizing the literature published in the preceding year (actually 2008 and 2009 in the initial version) and focusing only on the strongest studies based on design, sample size, phenotypes, novelty, and potential impact. Thus, this review is no longer intended to be a comprehensive and cumulative summary of all published literature addressing genomics relative to exercise, fitness, and performance.
The present paper is the second annual version of the focused review of the “scientifically strongest and substantively most important articles in exercise genomics” and covers the calendar year 2010. As indicated in the introduction to last year’s review (45), this new version intends to be highly selective in choosing articles for presentation in order to provide an ongoing and updated source for exercise and sports medicine scientists to understand the current literature in this area and to better grasp the necessary components of a high-quality exercise genomics study. Also, we hope this effort will help people identify the gaps in our knowledge concerning genomics and genetics relative to exercise, fitness, and performance.
In this review, we have selected papers focusing on (a) physical activity level, (b) muscular strength and power, (c) endurance performance, (d) adiposity, (e) glucose and insulin metabolism, and (f) hemodynamic traits.
Physical Activity Level
During 2010, no major studies on the genetics of physical activity level in humans were published. However, a comprehensive study reported several quantitative trait loci (QTLs) for spontaneous physical activity level in mice (32).
Running wheel behavior (distance, time, speed) was measured in 448 mice (212 females, 236 males) representing 41 inbred strains (4 to 64 animals per strain). Interestingly, there were marked differences in running behavior, with a 27-fold range for the distance run, a 23-fold range for the time the animal spent running, and a 3-fold range for the speed of running. Each mouse was given access to a solid-surface running wheel at 8 weeks of age, and wheel use was monitored for either 7 or 21 consecutive days. Genomic data for each strain were derived from a SNP database containing over 8 million markers developed by Perlegen Sciences, Inc. It should be noted that association analyses were based on strain-specific (in silico) SNP data rather than markers genotyped on individual animals. QTL mapping was done with a Bayesian-imputation-based haplotype association method.
The haplotype association mapping revealed three significant (log10 Bayes Factor ≥ 3.0) QTLs for running distance: rs46860253 on chromosome 12, rs3692956 on chromosome 18, and rs3666297 on chromosome 19 (Table 1). In addition, nine sex-specific QTLs were reported: five for males (chromosomes 5, 6, 8, and 13 for distance and chromosome 6 for running speed) and four for females (chromosomes 8 and 11 for distance, chromosome 11 for speed, and X chromosome for duration). The QTLs for distance and speed on chromosome 11 in females overlapped, which is not surprising given the strong correlation between these traits (r = 0.70).
Table 1.
Summary of the significant QTLs for wheel running phenotypes from haplotype association mapping in mice
| Trait | SNP | BFlog10 | Chr | Map, bp | CI size, Mbp | # of known genes within CI * |
|---|---|---|---|---|---|---|
| All animals: | ||||||
| Distance | rs46860253 | 4.966 | 12 | 89,352,286 | 0.106 | 0 |
| Distance | rs3692956 | 4.195 | 18 | 11,576,341 | 0.173 | 0 |
| Distance | rs36666297 | 3.865 | 19 | 16,111,217 | 0.411 | 2 |
| Males only: | ||||||
| Distance | rs47227633 | 3.12 | 5 | 118,050,446 | 3.085 | 36 |
| Distance | rs30945756 | 4.317 | 6 | 145,453,995 | 0.004 | 0 |
| Distance | rs49863494 | 3.041 | 8 | 61,372,952 | 24.030 | 155 |
| Distance | rs46617906 | 3.074 | 13 | 96,447,450 | 0.386 | 5 (Iqgap2) |
| Speed | rs31691517 | 3.078 | 6 | 119,765,173 | 0.353 | 1 (Erc1) |
| Females only: | ||||||
| Distance | rs32100214 | 3.094 | 8 | 97,083,316 | 0.261 | 4 (Cpne2) |
| Distance | rs26981291 | 3.327 | 11 | 85,044,150 | 2.468 | 24 (Appbp2) |
| Speed | rs28225821 | 3.599 | 11 | 83,840,288 | 2.525 | 23 (Ap1gbp1) |
| Duration | rs33880738 | 3.242 | X | 108,411,954 | 2.348 | 5 |
Number of confirmed, functional genes within the confidence interval (CI). Gene where the SNP is located is given in parentheses (if applicable).
Iqgap2 = IQ motif containing GTPase activating protein 2
Erc1 = ELKS/RAB6-interacting/CAST family member 1
Cpne2 = copine II
Appbp2 = amyloid beta precursor protein (cytoplasmic tail) binding protein 2
Ap1gbp1 = AP1 gamma subunit binding protein 1 [the new name for this gene is Synrg (synergin, gamma)]
Adapted from Lightfoot JT, Leamy L, Pomp D, et al. Strain screen and haplotype association mapping of wheel running in inbred mouse strains. J Appl Physiol. 2010;109:623–34, with permission of the American Physiological Society.
While most of the detected QTLs were statistically robust, with well-defined, narrow confidence intervals, interpretation of the results is challenging due to the fact that the most significant QTLs were located in intergenic regions, with no or a very limited number of known genes located within the confidence interval area (Table 1). Thus, it remains to be explored whether these QTLs contain yet unindentified genes, such as microRNA genes, or whether linkage disequilibrium within these genomic regions spans greater distances than suggested by the estimated confidence intervals. On the other hand, the sex-specific QTLs were more promising: five of the nine QTLs were detected with SNPs located within a known characterized gene locus. In addition, confidence intervals for most of the sex-specific QTLs included multiple genes. While the study did not provide any direct candidate genes for wheel running phenotypes in mice, the observed QTLs clearly deserve more detailed follow-up studies.
Muscular Strength and Power
The literature base in the area of genomics and muscular strength and power was expanded only modestly in 2010. The gene of focus for most groups remains alpha-3 actinin (ACTN3) and its nonsense polymorphism, R577X, though the expanding literature in this area is providing less consistent evidence of association with specific muscle traits of strength, mass, or power (5, 16, 49, 63). The primary theme in 2010 was the investigation of ACTN3 in conjunction with the angiotensin converting enzyme (ACE) I/D polymorphism as a gene-by-gene interaction. The hypothesis across most studies was that carriers of ACTN3 R/R or R/X plus ACE D/D genotypes would demonstrate higher levels of muscle strength, mass, or power compared with carriers of X/X and I/I genotypes. That hypothesis was consistently refuted across a number of investigations (5, 14, 33, 39, 46, 51), though the studies were generally poorly powered for such analyses with small sample sizes. The studies are difficult to compare because of differences in the ages, training statuses, etc., of the subjects. However, a number of studies over the past few years have provided evidence that ACTN3 acts as a contributor to muscular trait variation, but the importance of this association, especially in untrained individuals, remains unclear.
There is a growing interest in finding combinations of genes and alleles that more meaningfully reflect the underlying genetic influence of several small-effect genes or larger effect alleles found more rarely in the population. This idea was expanded in a 2010 paper by Ruiz and colleagues (48) on the potential for identifying a “power-oriented polygenic profile.” Ruiz et al. modeled their study on the concept of a “total genotype score” or TGS as developed by Williams and Folland (61). In the TGS, individuals are scored for the number of performance-related genotypes carried across multiple genes, and then that score is transformed across the range from 0 to 100, with 100 representing someone carrying the maximum number of beneficial alleles. Williams and Folland showed how rare in a population such a perfect score would be across 23 polymorphisms in 19 genes, with 99% of the population exhibiting scores in the range of 37 to 65. The use of a genotype score is of course not new, as it has been and continues to be used extensively in displaying the magnitude of associations between SNPs identified in genome-wide association studies (GWASs) and the trait or disease of interest.
In their 2010 study, Ruiz and colleagues (48) examined 53 elite Spanish power athletes compared to 100 controls and 100 endurance athletes. This study was clearly underpowered to achieve its objective. However, we are citing it because it allows us to illustrate a use of the TGS approach. Only six polymorphisms were included in the TGS calculation. Nonetheless, Ruiz et al. (48) reported that the elite power athletes exhibited a higher average TGS of 71 compared to 63 in controls and 60 in the endurance athletes, providing partial support for the idea that a TGS can distinguish different athlete groups. That said, fully 60% of the elite power athletes had three or fewer optimal genotypes (of six possible) and 20% of elite endurance athletes had four or five optimal power genotypes, so the approach isn’t perfect at the individual level. This high false-negative rate reflects both the relatively small number of polymorphisms included in the TGS and the small number of polymorphisms identified confidently in the literature as important to muscle power-related traits, but even more importantly the low statistical power of the study.
For practical reasons, sample sizes tend to be limited in studies of elite-level athletes, but efforts must be made to expand sample sizes quite substantially while maintaining high-quality phenotyping to make genotype score analyses valid and robust, as is the case in common GWAS reports (see last section of this paper). Our understanding of the genetic factors underlying muscular strength and mass traits is clearly in its early stages, and further advances in this area will necessarily require broadened collaboration across groups.
Endurance Performance
Over the past year, a number of reports on genetic markers and endurance performance were published. However, the study of the genomic basis of endurance performance continues to be plagued by small sample sizes and by a lack of hard performance criteria defining endurance performance or elite athlete status. This was the case for many case-control studies (8, 13, 19, 38, 41, 59), and they will not be reviewed here. Other case-control study reports were characterized by a performance surrogate measurement and a clear elite athlete status but were based on rather moderate sample sizes (e.g., (12)).
Experimental intervention studies of relevance to the genomics of endurance performance, cardiorespiratory fitness, or exercise tolerance are not reported frequently. One example of such studies was reported by Timmons et al. (58). They relied on three exercise training studies: one training experiment with 24 subjects to derive a baseline RNA profile predicting the response of VO2max to endurance training, a second group (n = 17) to independently validate the molecular predictor, and a third training group (n = 473 whites from the HERITAGE Family Study) to generate a panel of SNPs predicting the gains in VO2max (58). Skeletal muscle gene expression profiling generated a 29-mRNA signature strongly correlated with the gains in VO2max (r = 0.76). The replication arm also yielded a rather strong correlation between the baseline 29-transcript abundance predictor and the VO2max changes (r = 0.60). Next, haplotype tagging SNPs in the 29 predictor genes were identified and genotyped in the HERITAGE Family Study sample of whites. A multivariable regression analysis using the predictor gene tagSNPs and a set of SNPs previously identified from positional cloning and candidate gene studies of the HERITAGE Family Study yielded a set of 11 SNPs that explained about 23% of the variance in VO2max response to training. Seven of the SNPs were from the RNA predictor gene set, and four were from the HERITAGE QTL projects. Interestingly, when incorporated into the original RNA transcript prediction model, three of the four QTL-derived genes improved the performance of the model. Table 2, abstracted from the Timmons et al. paper (58), defines the SNPs, nearest genes, p values, and r squared contributions to the variance in VO2max changes with training.
Table 2.
Stepwise regression model for standardized residuals of maximal oxygen consumption training response in the HERITAGE Family Study
| Gene (SNP)* | Genomic Location | Partial r2 | Model r2 | P |
|---|---|---|---|---|
| SVIL (rs6481619) | 10p11.2 | 0.0411 | 0.0411 | <.0001 |
| SLC22A3 (rs2457571) | 6q26-q27 | 0.0307 | 0.0718 | 0.0003 |
| NRP2 (rs3770991) | 2q33.3 | 0.0224 | 0.0942 | 0.0017 |
| TTN (rs10497520) | 2q31 | 0.0204 | 0.1146 | 0.0025 |
| H19 (rs2251375) | 11p15.5 | 0.0268 | 0.1414 | 0.0004 |
| ID3 (rs11574) | 1p36.13-p36.12 | 0.02 | 0.1615 | 0.0021 |
| MIPEP (rs7324557) | 13q12 | 0.0163 | 0.1778 | 0.0051 |
| CPVL (rs4257918) | 7p15-p14 | 0.0179 | 0.1957 | 0.0031 |
| DEPDC6 (rs7386139)** | 8q24.12 | 0.0112 | 0.2069 | 0.0185 |
| BTAF1 (rs2792022) | 10q22-q23 | 0.0125 | 0.2194 | 0.0122 |
| DIS3L (rs1546570) | 15q22.31 | 0.0095 | 0.2289 | 0.0279 |
Residual maximal oxygen consumption response values were adjusted for age, sex, baseline body weight, and baseline maximal oxygen consumption (n = 473).
The gene listed is the one nearest to the SNP.
The new symbol is DEPTOR.
From Timmons JA, Knudsen S, Rankinen T, et al. Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. J Appl Physiol. 2010;108:1487–96. Used with permission of the American Physiological Society.
This collaborative effort on the part of several research groups resulted in significant advances in our ability to predict the ability to improve one’s VO2max in response to standardized endurance exercise programs. Using a combination of transcriptomics and genomics, about half of the genetic variance of VO2max trainability was accounted for either by the baseline muscle RNA abundance at 29 transcripts or by 11 SNPs. These results now await large-scale replications in healthy individuals as well as in patients affected by a number of conditions.
Adiposity
The past year has witnessed a flurry of discoveries in the field of adiposity and obesity genetics. This success is fully accounted for by the growing scale of GWASs. While by the end of 2009, GWASs had identified 19 loci unambiguously associated with BMI (9, 15, 36, 52, 57, 60), waist circumference (20, 34), and obesity (40), the number of obesity-susceptibility loci had more than doubled to a total of 50 loci by the end of 2010. The 31 loci confirmed in 2010 were identified by two large-scale genome-wide association meta-analyses that were both performed by the GIANT (Genetic Investigation of ANthropometric Traits) Consortium (21, 54). The first study found 18 loci to be robustly associated with body mass index (BMI) in adults of white European descent using a two-staged design including 249,796 individuals (54). The second study, including 190,803 adults of white European descent, identified 13 loci associated with waist-to-hip ratio adjusted for BMI (21). Despite the enormous progress in the field, little is known about the biological pathways through which these genetic loci confer weight gain and increased obesity risk. None of the 50 obesity-susceptibility loci is unambiguously known to affect exercise behavior or energy expenditure at rest or during physical activity.
Studies to uncover the physiological mechanisms have started to emerge, with the fat mass and obesity associated (FTO) locus being the most intensively examined. FTO was the first obesity-susceptibility locus to be identified, and among the 50 established loci, genetic variation in FTO has the largest effect on BMI and obesity risk. In our previous review (45), we described the apparent contradictory observations from rodent studies, as some suggested that Fto increases obesity susceptibility through the central regulation of food intake, whereas others proposed that Fto affects energy expenditure through peripheral mechanisms. A recent study in mice with systemic Fto overexpression suggests that Fto implicates both peripheral and central physiology (10). Systemic overexpression of Fto reduces leptin expression or secretion from adipose tissue, which in turn affects the central nervous system-mediated control of food intake (10). While there is growing support from studies in rodents for a role of Fto in the regulation of food intake, there has been so far no evidence that Fto affects locomotor activity. This is consistent with the observations of recent studies in humans, which reported no association of genetic variation in FTO with physical activity levels in white Europeans (1, 23), Japanese (28), Korean adults and children (29), and European and African-American youth (35). Although physical activity does not seem to mediate the association between FTO and obesity susceptibility, recent studies continue to show that a physically active lifestyle reduces the effect that FTO has on BMI and obesity risk. Thus, significant FTO-physical activity interactions on BMI and obesity risk have been reported for white European adults and adolescents (1, 25, 50, 53), Japanese adults (28), Korean adults and children (29), and Chinese children and adolescents (62). Four other observational (cross-sectional) studies (22, 27, 31, 35) and one small lifestyle intervention (N = 107) study (11) did not find such an interaction.
Gene-environment interaction studies require on average a sample size four times larger than main effect analyses to provide adequate power. While a meta-analysis of all published data would allow the confirmation or refutation of the presence of an interaction between FTO and physical activity, the large heterogeneity between studies in analyses and measurement of physical activity hamper pooling of such data. Furthermore, as interaction analyses are often secondary to main effect analyses, nonsignificant observations may not be reported, making interaction studies prone to publication bias. Therefore, a meta-analysis based on published and unpublished data that standardizes the analyses and harmonizes the coding of the physical traits across studies is needed to firmly establish whether physical activity indeed attenuates FTO’s BMI-increasing effect. Taken together, genetic variation in FTO does not increase adiposity or obesity risk through reduced physical activity energy expenditure. However, the effect of FTO on adiposity is reduced in those who have a physically active lifestyle. These recent observations further corroborate those summarized in our previous review (45).
The near-melancortin 4 receptor (MC4R) locus was the second locus to be firmly established as an obesity-susceptibility locus with GWASs (9, 36). A genome-wide linkage study had previously found suggestive evidence that this chromosomal region was linked with physical activity levels in Hispanic children (6). However, a recent large-scale population-based study, including more than 12,462 individuals, could not confirm such an association with self-reported physical activity levels (22). Furthermore, five observational studies (sample size ranging from 1,273 to 20,430) found no evidence for interaction between the near-MC4R locus and physical activity in adolescents and adults (7, 22, 30, 31, 64). This is consistent with the results of two intervention studies, a 9-month diet and exercise study (n = 242) (18) and a 12-week resistance training study (n = 785) (17), both showing that weight loss did not depend on the subjects’ genotype at the near-MC4R locus.
So far, only two large-scale population-based studies have examined some of the other obesity-susceptibility loci, but they found no evidence of association or interaction between the individual loci and physical activity (22, 30).
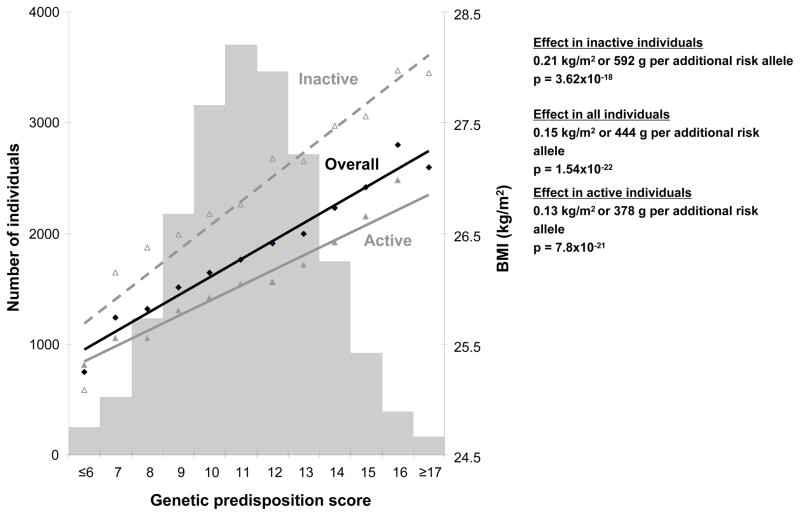
In a large-scale population-based study, including 20,430 white men and women of European descent, each individual’s genetic predisposition to obesity was assessed by adding the number of BMI-increasing alleles across 12 loci to form a score (30). Each additional allele in this genetic predisposition score was associated with a 0.15 kg/m2 (or 444 g for a 1.70 m tall person) increase in BMI, with a 12% increase in the odds of obesity, but not with self-reported physical activity. However, most importantly, the association between the genetic predisposition score and increased BMI was significantly (Pinteraction = 0.005) more pronounced in sedentary individuals (0.21 kg/m2 or 592 g per allele) than in physically active individuals (0.13 kg/m2 or 379 g per allele) (Figure 1) (30). Similar interaction effects were observed between the genetic predisposition score and physical activity for risk of obesity; i.e., 16% per allele in sedentary individuals and 9.5% in physically active individuals. While the main observations were made in cross-sectional analyses, longitudinal analyses confirmed the interaction between the score and physical activity for weight gain (30). These findings convey an important public health message as they challenge the deterministic view of the genetic predisposition to obesity, showing that even the most genetically predisposed individuals could benefit from adopting a physically active lifestyle.
Figure 1.
Association between the genetic predisposition score (sum of BMI-increasing alleles from 12 BMI loci) with BMI in all individuals (solid black line), in sedentary individuals (dashed grey line), and in physically active individuals (solid grey line). The bars represent the number of individuals at every unit of the predisposition score. Effect sizes expressed in grams assume a height of 1.70 m.
Adapted from Li S, Zhao JH, Luan Ja, et al. Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from EPIC-Norfolk prospective population study. PLoS Med. 2010;7:e1000332.
In summary, there is so far no evidence that any of the obesity-susceptibility loci, individually or combined, confer weight gain or obesity risk through an effect on physical activity levels. A growing number of studies, however, reported that physical activity reduces the effect of FTO on BMI and obesity risk. While the role of physical activity on the impact of individual obesity susceptibility requires further research, a large-scale population-based study that examined the association of 12 loci in one analysis found that physical activity reduces the overall genetic predisposition to obesity by ~40% (30).
Glucose and Insulin Metabolism
Only two papers were published in the past year analyzing associations between genetic markers and glucose and insulin metabolism phenotypes in response to exercise (26, 47). A summary of these two papers is presented below. Both studies are of interest because they used standardized endurance exercise training programs long enough (> 20 weeks) to produce significant physiological adaptations and relied on measures of glucose metabolism derived from an oral glucose tolerance test or intravenous glucose tolerance test.
Jenkins et al. (26) investigated the associations of common sequence variants in the perilipin (PLIN) gene with body fatness, aerobic fitness, and related cardiovascular disease risk factors before and after a 24-week standardized exercise training program in sedentary men (n = 46) and women (n = 55) aged 50 to 75 years. Plasma samples obtained during a 3-hour oral glucose tolerance test performed before and after training were analyzed for glucose and insulin concentrations, and the Matsuda insulin sensitivity index was calculated. Carriers of the common AA haplotype of two PLIN SNPs, 13041 A>G (rs2304795) and 14995 A>T (rs1052700), were found to have a higher insulin sensitivity and more favorable insulin and glucose responses to oral glucose compared to noncarriers, both in the untrained state and after the endurance exercise program. A significant training × genotype × sex interaction was observed, as noncarrier men had the largest baseline insulin area under the curve (AUC) but were the only group to significantly improve insulin AUC with exercise training. Replication with a substantially larger sample size is clearly warranted.
In the second study, eight polymorphisms in seven type-2 diabetes genes identified through GWASs were tested for associations with changes in glucose and insulin metabolism phenotypes in response to a 20-week endurance exercise training program (47). The study was undertaken in 481 sedentary white adults from 98 families of the HERITAGE Family Study who completed the 20-week standardized and supervised exercise program and in whom an intravenous glucose tolerance test was administered before and after the training. Among the seven candidate genes investigated (CDKAL1, CDKN2A/B, HHEX, IGF2BP2, KCNJ11, PPARG, TCF7L2), the only associations with training responses that remained significant after adjustment for multiple testing were with the PPARG Pro12Ala polymorphism (rs1801282). The results showed that Ala carriers experienced greater increases in overall glucose tolerance (p = 0.0008), glucose effectiveness (p = 0.004), insulin response to glucose (p = 0.03), and disposition index (p = 0.003), a measure of the ability of the pancreatic beta cell to compensate for changes in insulin sensitivity. This study stands out among exercise training studies reporting evidence of gene-exercise interactions for insulin and glucose metabolism phenotypes because of its relatively large sample size, the highly standardized and fully monitored exercise intervention, the extremely high degree of compliance to the program by all completers, and the reliance on a frequently sampled intravenous glucose tolerance test and MINMOD (Minimal Model) to investigate glucose and insulin metabolism in vivo.
Hemodynamic Traits
In 2010, a total of 12 studies were published addressing the effect of genetics on cardiovascular responses to exercise or exercise training. The three studies reviewed below all have large sample sizes and used unique analytic and design strategies or have unique findings that should be considered as models for future studies assessing exercise physiology genotype- phenotype interactions.
An earlier study from the HERITAGE Family Study assessed genome-wide linkage for submaximal exercise heart rate (HR) responses to exercise training and found the strongest linkage on chromosome 2q34 (55). In 2010, Rankinen and HERITAGE colleagues fine-mapped this region by typing 1450 tagSNPs distributed across 2q34 to identify loci underlying submaximal exercise HR responses to training (43). In their total SNP association model, two SNPs near the cAMP responsive element binding position 1 (CREB1) gene exhibited the strongest associations. The effect was sizable, with G homozygotes and heterozygotes having ~60 and ~20% greater training-induced HR reductions during submaximal exercise, respectively, than A homozygotes. This variant accounted for ~5% of the interindividual variance in this response to training. Also, promoter activity varied among C2C12 cell lines transfected with the different CREB1 genotypes. In addition, each of eight other SNPs accounted for >1% of the interindividual variation in this response. After adding these SNPs to the regression model, these genetic variants accounted for 20% of the variance in the response of submaximal exercise HR to training.
This appears to be the third HERITAGE study to use this approach wherein researchers (a) first found significant phenotypic variability and heritability, (b) used genome-wide linkage analyses to identify loci potentially underlying this phenotype, (c) fine-mapped the locus with the highest linkage, and (d) performed cell culture studies to assess the molecular effects of their SNPs. CREB1, found to be so important in this study, would not have been on any list of candidate genes to affect the training-induced change in submaximal exercise HR. Thus, on a larger scale, the use of genome-wide linkage and association studies to identify novel mechanisms involved in the regulation of heritable exercise training response phenotypes has clearly been validated by these HERITAGE studies (2, 43, 44, 55).
In 2010, the Humphries and Montgomery group from the UK published another report from their Litchfield Army Recruit Growth in Exercise Heart Study (LARGE Heart) (37). A major strength of this study is the sample size of men (n = 301) undergoing rigorous and standardized exercise training. Researchers selected the C-reactive protein (CRP) gene as a candidate because of its known role in cardiomyocyte growth. Within this gene, they selected the C1444T variant for study because of its known effects on plasma CRP levels. In the total group, 12 weeks of training increased left ventricular (LV) mass significantly (+3.8 ± 10.8 g, P < 0.0001). However, CRP genotype also significantly affected LV mass change with training, with T homozygotes more than doubling the increase observed in C allele carriers (+8.2 ± 12.1 vs. +3.4 ± 10.6 g, P = 0.033). These results indicate that young male T homozygotes respond better to exercise training in terms of LV mass than men with at least one C allele at the CRP locus.
Then the Humphries and Montgomery group took these results a step further. They had previously found that ACE I/D genotype significantly affected the degree of LV hypertrophy that occurred with exercise training (42). They assessed the impact of the CRP genotype on LV mass changes after accounting for the effect of the ACE genotype. The differential CRP genotype effect on LV hypertrophy remained significant, indicating that this CRP genotype-dependent training effect was independent of the already known effect of ACE genotype.
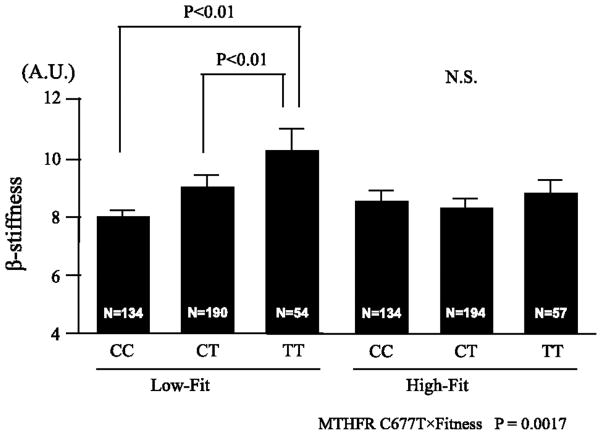
The final study cross-sectionally assessed the role of the methylenetetrahydrofolate reductase (MTHFR) C677T variant on the relationship between cardiovascular fitness and carotid stiffness in 763 Japanese men and women (24). This variant was previously shown to influence plasma homocysteine levels, which can affect arterial stiffness and progression to atherosclerosis. In the total group, there was no effect of MTHFR genotype on carotid stiffness. However, in low-fit individuals, carotid stiffness was ~25% and ~13% higher in TT and CT individuals compared to C homozygotes (Figure 2). On the other hand, there were no differences in carotid stiffness across genotypes in high-fit individuals, with all of them having stiffness values equal to those of the best low-fit genotype group.
Figure 2.
Carotid β-stiffness (β-stiffness) of each fitness group and genotype of MTHFR gene polymorphism (C677T; CC, CT, TT genotypes) of 763 subjects in a cross-sectional study. Subjects were divided into low cardiorespiratory fitness (Low-Fit) and high cardiorespiratory fitness (High-Fit) groups, with the dividing line set at the median value of peak oxygen uptake in each sex and decade of age as a cutoff. Differences in β-stiffness between each fitness group and genotype were assessed by an analysis of covariance (ANCOVA) model that included age as a covariate. Data are expressed as means ± SE for numbers of subjects indicated in the bar. AU, arbitrary units; NS, not significant.
From Iemitsu M, Murakami H, Sanada K, et al. Lack of carotid stiffening associated with MTHFR 677TT genotype in cardiorespiratory fit adults. Physiol Genomics. 2010;42:259–65. Used with permission of the American Physiological Society.
Thus, these results indicate that higher cardiovascular fitness levels protected against the effects of the high-risk MTHFR TT genotype on carotid stiffness. But this would be our conclusion only if we view the low-fit group as the “control” or comparison group. Since humans have evolved progressively from a high level of physical activity to a more inactive state, then perhaps, as was suggested before (3), the high-fit group should be the “control” group. We would then arrive at a completely different conclusion—that the risk genotype expresses itself only when the environmental trigger of low cardiovascular fitness is present. Thus, if this study had been done 50,000 years ago when we were hunters and gatherers on the African plains, we would all have been physically active with reasonably high cardiovascular fitness levels; under these circumstances, there would have been no relationship between MTHFR genotypes and carotid stiffness. Thus, the expression of this “susceptibility” genotype becomes evident only when activated by the environmental trigger of being physically inactive and having low cardiovascular fitness.
Comments and Summary
This review emphasizes the strongest papers in exercise genomics published in 2010. The selection of the papers was made after careful consideration and was based on study design, including sample size and statistical power issues, quality of phenotype measurements, quality of the exercise program or physical activity exposure, adjustment for multiple testing, quality of genotyping, and other related study characteristics. The emphasis on the conditions that are necessary for a valid and conclusive exercise genomics study may seem repetitive to the readership, but we believe that it is absolutely essential considering the publication record of the past decade in this field. Hence, the focus on the highest quality papers accounts for the fact that only a small number of publications are reviewed here.
We believe that the most significant findings of the past calendar year are the following. One study on voluntary running wheel behavior was performed in 448 mice from 41 inbred strains. Several quantitative trait loci for running distance, speed, and duration were identified. As in previous years, several studies on the ACTN3 gene R577X nonsense polymorphism and the ACE gene I/D polymorphism were reported. No clear evidence for a joint effect of these two variants could be found, but the studies were generally underpowered. Skeletal muscle RNA abundance at baseline at 29 transcripts and 11 SNPs were both found to be predictive of the VO2max response to exercise training in one report from multiple laboratories. None of the 50 loci associated with adiposity traits is known to influence physical activity behavior. However, physical activity reduces the obesity-promoting effects of at least 12 of these loci. Evidence continues to be strong for a role of gene-exercise interaction effects on the improvement in insulin sensitivity following exposure to regular exercise. SNPs in the CREB1 gene were associated with training-induced HR response, in the CRP gene with training-induced changes in LV mass, and in the MTHFR gene with carotid stiffness in low-fit individuals.
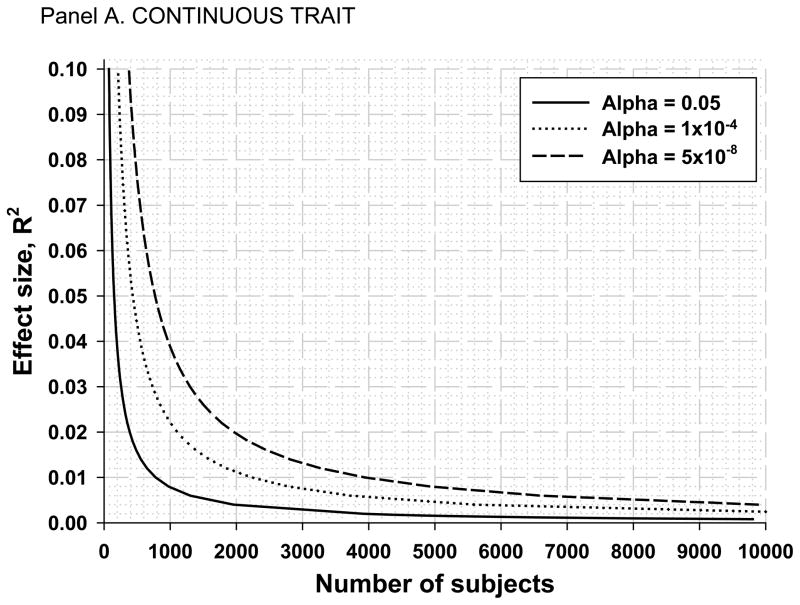
One of the most frequently observed deficiencies of exercise genomics papers is that they are based on small sample sizes and therefore have very limited statistical power to test the hypothesis (or, more commonly, the hypotheses) as defined in their aims statements. In this regard, the relationship between the effect size of a SNP and the sample size required to find a significant association with 80% statistical power under an additive model is shown in Figure 3 for three alpha levels. Panel A summarizes the simulations for a quantitative phenotype. Because the SNP effect size is expressed as R2, the presented simulations apply to all minor allele frequencies (MAFs). In the first scenario, we assume that only one SNP was tested for association with one quantitative trait (the continuous line in the figure). Note that under these conditions, it takes almost 800 subjects to identify an association that accounts for 1% of the trait variance (a value not unrealistic for complex human traits). If the SNP explained only 0.05% of the variance, a sample size of about 1,500 subjects is needed. In the second scenario, 500 SNPs were genotyped, and since all of them were tested for associations with the trait of interest (assuming only one trait), the multiple testing adjusted p value threshold for significance is now set at 0.0001 (the dotted line in the figure). Under these conditions, a minimum of 2,200 subjects are needed to detect a significant association with a SNP that accounts for 1% of the trait variance. In the third scenario (the dashed line in the figure), we illustrate the case of a GWAS undertaken with 1 million SNPs. Here the commonly accepted convention is that we should set the alpha level at p = 5×10−8, and almost 4,000 subjects are needed to detect a 1% effect size with 80% power.
Figure 3.
Panel A. Number of subjects needed for a given effect size for a continuous trait assuming 80% statistical power, an additive model, and three alpha levels: 0.05 (1 SNP), 0.0001 (500 SNPs), and 5×10−8 (1 million SNP GWAS).
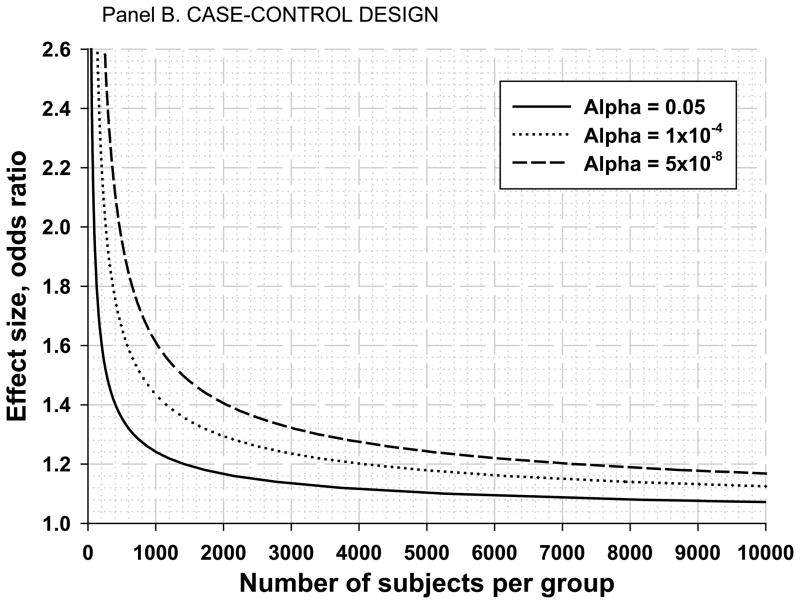
Panel B. Number of subjects needed for a given effect size (measured as an odds ratio [OR]) for a case-control design assuming 80% statistical power, a minor allele frequency (MAF) of 20%, an additive model, and three alpha levels: 0.05 (1 SNP), 0.0001 (500 SNPs), and 5×10−8 (1 million SNP GWAS).
In Panel B, we depict the curves of the odds ratios (ORs) versus the number of subjects per group at three alpha levels for case-control (1:1) designs. The figure is for an additive model and a MAF of 20%. It should be noted that unlike R2 with continuous traits, the sample size needed for a given OR at the same power varies across different MAFs with this design. The majority of the GWAS-derived ORs for common diseases from case-control studies have reached approximately 1.2. If we assume a similar effect size (e.g., comparing athletes and controls) and testing of a single SNP (alpha = 0.05), 1,400 subjects are needed in each of the case and control groups (continuous line). If 500 SNPs were tested (alpha = 1×10−4), we need 4,000 subjects in each group to find an association with an OR of 1.2 with 80% power. Finally, in the context of a GWAS, we can detect a significant difference (p = 5×10−8 or better) at a SNP between cases and controls if we have about 7,100 subjects in each group. If the MAF is less than 20%, we need even larger sample sizes, whereas the sample size requirements decrease by about one-third when MAF approaches 50% (not shown in the figure).
It is important to note that the above estimates apply to a situation where the main effect of a SNP is being tested. The sample size requirements increase four- to fivefold when an interaction term is being investigated for its contribution to the variance of a phenotype. For instance, a test of a SNP–physical activity interaction effect on adiposity would require at least four times more subjects than a test of the main effect of the same SNP.
Based on the above estimates, one can easily see that exercise genomics studies are generally grossly underpowered. For instance, nothing of substance is going to be achieved when comparing athletes and controls with about 50 athletes; even under the most optimistic assumptions (only one SNP tested, alpha = 0.05, MAF = 50%), such a study would have 80% power to detect OR = 2.28 and greater, an effect size that is usually observed only in studies with single-gene disorders, but never with common multifactorial traits. The situation described here is likely to be even less satisfactory, as in many cases the same SNP (or SNPs) is (are) used in different reports but for different traits or the same traits are repeatedly tested against various SNPs or sets of SNPs without ever taking into account the multiple testing effect. Moreover, we should keep in mind that we use a statistical power of only 80% as a target for the above simulations. If we raise the bar to a more conservative 90%, the sample sizes required become markedly larger.
We conclude that progress is being made but that high-quality research designs and replication studies with large sample sizes are urgently needed. It would clearly be more productive for the small community of exercise genomics laboratories to work together in order to be able to undertake collaborative studies that would be statistically adequately powered. There are two examples of multicenter studies in exercise genomics: the FAMUSS project focused on muscle size and strength with about 1,000 subjects at baseline (56) and the HERITAGE Family Study focused primarily on cardiorespiratory fitness and diabetes and cardiovascular risk factors with about 850 subjects at baseline (4). However, even though these studies are substantially larger than the typical exercise genomics experiments, they do not have the statistical power to identify SNPs or genes with small effect sizes. Working collaboratively presents many challenges. For instance, study designs vary, testing methodologies and assays may differ among the units involved, exercise programs may not be comparable across laboratories, recruitment strategies may generate samples of subjects that are not fully comparable, and so on. But there is no other credible solution, particularly in light of the present-day research funding restrictions.
Acknowledgments
CB and TR are funded by the National Institutes of Health (HL-45670). CB is partially funded by the John W. Barton, Sr. in Genetics and Nutrition.
The results of the present study do not constitute endorsement by the American College of Sports Medicine.
Footnotes
Conflict of Interest
The authors have no conflicts of interest to disclose.
References
- 1.Ahmad T, Chasman DI, Mora S, et al. The Fat-Mass and Obesity-Associated (FTO) gene, physical activity, and risk of incident cardiovascular events in white women. Am Heart J. 2010;160:1163–9. doi: 10.1016/j.ahj.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argyropoulos G, Stutz AM, Ilnytska O, et al. KIF5B gene sequence variation and response of cardiac stroke volume to regular exercise. Physiol Genomics. 2009;36:79–88. doi: 10.1152/physiolgenomics.00003.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Booth FW, Gordon SE, Carlson CJ, Hamilton MT. Waging war on modern chronic diseases: primary prevention through exercise biology. J Appl Physiol. 2000;88:774–87. doi: 10.1152/jappl.2000.88.2.774. [DOI] [PubMed] [Google Scholar]
- 4.Bouchard C, Leon AS, Rao DC, Skinner JS, Wilmore JH, Gagnon J. The HERITAGE family study. Aims, design, and measurement protocol. Med Sci Sports Exerc. 1995;27:721–9. [PubMed] [Google Scholar]
- 5.Bustamante-Ara N, Santiago C, Verde Z, et al. ACE and ACTN3 genes and muscle phenotypes in nonagenarians. Int J Sports Med. 2010;31:221–4. doi: 10.1055/s-0030-1247529. [DOI] [PubMed] [Google Scholar]
- 6.Cai G, Cole SA, Butte N, et al. A quantitative trait locus on chromosome 18q for physical activity and dietary intake in Hispanic children. Obesity. 2006;14:1596–604. doi: 10.1038/oby.2006.184. [DOI] [PubMed] [Google Scholar]
- 7.Cauchi S, Stutzmann F, Cavalcanti-Proena C, et al. Combined effects of MC4R and FTO common genetic variants on obesity in European general populations. J Mol Med. 2009;87:537–46. doi: 10.1007/s00109-009-0451-6. [DOI] [PubMed] [Google Scholar]
- 8.Cauci S, Di SM, Ryckman KK, Williams SM, Banfi G. Variable number of tandem repeat polymorphisms of the interleukin-1 receptor antagonist gene IL-1RN: a novel association with the athlete status. BMC Med Genet. 2010;11:29. doi: 10.1186/1471-2350-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers JC, Elliott P, Zabaneh D, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40:716–8. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 10.Church C, Moir L, McMurray F, et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet. 2010;42:1086–92. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dlouha D, Suchanek P, Lanska V, Hubacek JA. Body mass index change in females after short-time life style intervention is not dependent on the FTO polymorphisms. Physiol Res. 2010 doi: 10.33549/physiolres.932065. [DOI] [PubMed] [Google Scholar]
- 12.Doring F, Onur S, Fischer A, et al. A common haplotype and the Pro582Ser polymorphism of the hypoxia-inducible factor-1alpha (HIF1A) gene in elite endurance athletes. J Appl Physiol. 2010;108:1497–500. doi: 10.1152/japplphysiol.01165.2009. [DOI] [PubMed] [Google Scholar]
- 13.Eynon N, Alves AJ, Sagiv M, Yamin C, Meckel Y. Interaction between SNPs in the NRF2 gene and elite endurance performance. Physiol Genomics. 2010;41:78–81. doi: 10.1152/physiolgenomics.00199.2009. [DOI] [PubMed] [Google Scholar]
- 14.Eynon N, Alves AJ, Yamin C, et al. Is there an ACE ID - ACTN3 R577X polymorphisms interaction that influences sprint performance? Int J Sports Med. 2009;30:888–91. doi: 10.1055/s-0029-1238291. [DOI] [PubMed] [Google Scholar]
- 15.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson ED, Ludlow AT, Sheaff AK, Park J, Roth SM. ACTN3 genotype does not influence muscle power. Int J Sports Med. 2010;31:834–8. doi: 10.1055/s-0030-1263116. [DOI] [PubMed] [Google Scholar]
- 17.Haupt A, Thamer C, Heni M, et al. Impact of variation near MC4R on whole-body fat distribution, liver fat, and weight loss. Obesity. 2009;17:1942–5. doi: 10.1038/oby.2009.233. [DOI] [PubMed] [Google Scholar]
- 18.Haupt A, Thamer C, Machann J, et al. Impact of variation in the FTO gene on whole body fat distribution, ectopic fat, and weight loss. Obesity. 2008;16:1969–72. doi: 10.1038/oby.2008.283. [DOI] [PubMed] [Google Scholar]
- 19.He ZH, Hu Y, Li YC, Bao DP, Ruiz JR, Lucia A. Polymorphisms in the calcineurin genes are associated with the training responsiveness of cardiac phenotypes in Chinese young adults. Eur J Appl Physiol. 2010;110:761–7. doi: 10.1007/s00421-010-1558-8. [DOI] [PubMed] [Google Scholar]
- 20.Heard-Costa NL, Zillikens MC, Monda KL, et al. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE Consortium. PLoS Genet. 2009;5:e1000539. doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heid IM, Jackson AU, Randall JC, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–60. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holzapfel C, Grallert H, Huth C, et al. Genes and lifestyle factors in obesity: results from 12,462 subjects from MONICA/KORA. Int J Obes. 2010;34:1538–45. doi: 10.1038/ijo.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hubacek JA, Pikhart H, Peasey A, Kubinova R, Bobak M. FTO variant, energy intake, physical activity and basal metabolic rate in Caucasians. The HAPIEE study. Physiol Res. 2010 doi: 10.33549/physiolres.932066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iemitsu M, Murakami H, Sanada K, et al. Lack of carotid stiffening associated with MTHFR 677TT genotype in cardiorespiratory fit adults. Physiol Genomics. 2010;42:259–65. doi: 10.1152/physiolgenomics.00039.2010. [DOI] [PubMed] [Google Scholar]
- 25.Jacobsson J, Riserus U, Axelsson T, Lannfelt L, Schioth H, Fredriksson R. The common FTO variant rs9939609 is not associated with BMI in a longitudinal study on a cohort of Swedish men born 1920–1924. BMC Med Genet. 2009;10:131. doi: 10.1186/1471-2350-10-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins NT, McKenzie JA, Damcott CM, Witkowski S, Hagberg JM. Endurance exercise training effects on body fatness, VO2max, HDL-C subfractions, and glucose tolerance are influenced by a PLIN haplotype in older Caucasians. J Appl Physiol. 2010;108:498–506. doi: 10.1152/japplphysiol.01018.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaakinen M, Laara E, Pouta A, et al. Life-course analysis of a fat mass and obesity-associated (FTO) gene variant and body mass index in the Northern Finland Birth Cohort 1966 using structural equation modeling. Am J Epidemiol. 2010;172:653–65. doi: 10.1093/aje/kwq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karasawa S, Daimon M, Sasaki S, et al. Association of the common fat mass and obesity associated (FTO) gene polymorphism with obesity in a Japanese population. Endocr J. 2010;57:293–301. doi: 10.1507/endocrj.k09e-305. [DOI] [PubMed] [Google Scholar]
- 29.Lee HJ, Kim Ik, Kang JH, et al. Effects of common FTO gene variants associated with BMI on dietary intake and physical activity in Koreans. Clin Chim Acta. 2010;411:1716–22. doi: 10.1016/j.cca.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 30.Li S, Zhao JH, Luan Ja, et al. Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from EPIC-Norfolk prospective population study. PLoS Med. 2010;7:e1000332. doi: 10.1371/journal.pmed.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liem ET, Vonk JM, Sauer PJJ, et al. Influence of common variants near INSIG2, in FTO, and near MC4R genes on overweight and the metabolic profile in adolescence: the TRAILS (TRacking Adolescents’ Individual Lives Survey) Study. Am J Clin Nutr. 2010;91:321–8. doi: 10.3945/ajcn.2009.28186. [DOI] [PubMed] [Google Scholar]
- 32.Lightfoot JT, Leamy L, Pomp D, et al. Strain screen and haplotype association mapping of wheel running in inbred mouse strains. J Appl Physiol. 2010;109:623–34. doi: 10.1152/japplphysiol.00525.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lima RM, Leite TK, Pereira RW, Rabelo HT, Roth SM, Oliveira RJ. ACE and ACTN3 genotypes in older women: Muscular phenotypes. Int J Sports Med. 2010 doi: 10.1055/s-0030-1267229. [DOI] [PubMed] [Google Scholar]
- 34.Lindgren CM, Heid IM, Randall JC, et al. Genome-wide association scan meta-analysis identifies three loci influencing adiposity and fat distribution. PLoS Genet. 2009;5:e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu G, Zhu H, Lagou V, et al. FTO variant rs9939609 is associated with body mass index and waist circumference, but not with energy intake or physical activity in European- and African-American youth. BMC Med Genet. 2010;11:57. doi: 10.1186/1471-2350-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loos RJ, Lindgren CM, Li S, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–75. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mann JJ, Payne JR, Shah T, Pennell DJ, Humphries SE, Montgomery HE. C-reactive protein gene variant and the human left ventricular growth response to exercise: data from The LARGE Heart Study. J Cardiovasc Pharmacol. 2010;55:26–9. doi: 10.1097/FJC.0b013e3181c37d2d. [DOI] [PubMed] [Google Scholar]
- 38.Martinez-Redondo D, Marcuello A, Casajus JA, et al. Human mitochondrial haplogroup H: the highest VO2max consumer--is it a paradox? Mitochondrion. 2010;10:102–7. doi: 10.1016/j.mito.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 39.McCauley T, Mastana SS, Folland JP. ACE I/D and ACTN3 R/X polymorphisms and muscle function and muscularity of older Caucasian men. Eur J Appl Physiol. 2010;109:269–77. doi: 10.1007/s00421-009-1340-y. [DOI] [PubMed] [Google Scholar]
- 40.Meyre D, Delplanque J, Chevre JC, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–9. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 41.Muniesa CA, Gonzalez-Freire M, Santiago C, et al. World-class performance in lightweight rowing: is it genetically influenced? A comparison with cyclists, runners and non-athletes. Br J Sports Med. 2010;44:898–901. doi: 10.1136/bjsm.2008.051680. [DOI] [PubMed] [Google Scholar]
- 42.Myerson SG, Montgomery HE, Whittingham M, et al. Left ventricular hypertrophy with exercise and ACE gene insertion/deletion polymorphism: a randomized controlled trial with losartan. Circulation. 2001;103:226–30. doi: 10.1161/01.cir.103.2.226. [DOI] [PubMed] [Google Scholar]
- 43.Rankinen T, Argyropoulos G, Rice T, Rao DC, Bouchard C. CREB1 is a strong genetic predictor of the variation in exercise heart rate response to regular exercise: the HERITAGE Family Study. Circ Cardiovasc Genet. 2010;3:294–9. doi: 10.1161/CIRCGENETICS.109.925644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rankinen T, Rice T, Boudreau A, et al. Titin is a candidate gene for stroke volume response to endurance training: the HERITAGE Family Study. Physiol Genomics. 2003;15:27–33. doi: 10.1152/physiolgenomics.00147.2002. [DOI] [PubMed] [Google Scholar]
- 45.Rankinen T, Roth SM, Bray MS, et al. Advances in exercise, fitness, and performance genomics. Med Sci Sports Exercise. 2010;42:835–46. doi: 10.1249/MSS.0b013e3181d86cec. [DOI] [PubMed] [Google Scholar]
- 46.Rodríguez-Romo G, Ruiz JR, Santiago C, et al. Does the ACE I/D polymorphism, alone or in combination with the ACTN3 R577X polymorphism, influence muscle power phenotypes in young, non-athletic adults? Eur J Appl Physiol. 2010;110:1099–106. doi: 10.1007/s00421-010-1608-2. [DOI] [PubMed] [Google Scholar]
- 47.Ruchat SM, Rankinen T, Weisnagel SJ, et al. Improvements in glucose homeostasis in response to regular exercise are influenced by the PPARG Pro12Ala variant: results from the HERITAGE Family Study. Diabetologia. 2010;53:679–89. doi: 10.1007/s00125-009-1630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruiz JR, Arteta D, Buxens A, et al. Can we identify a power-oriented polygenic profile? J Appl Physiol. 2010;108:561–6. doi: 10.1152/japplphysiol.01242.2009. [DOI] [PubMed] [Google Scholar]
- 49.Ruiz JR, Fernandez Del Valle M, Verde Z, et al. ACTN3 R577X polymorphism does not influence explosive leg muscle power in elite volleyball players. Scand J Med Sci Sports. 2010 doi: 10.1111/j.1600-0838.2010.01134.x. [DOI] [PubMed] [Google Scholar]
- 50.Ruiz JR, Labayen I, Ortega FB, et al. Attenuation of the effect of the FTO rs9939609 polymorphism on total and central body fat by physical activity in adolescents: The HELENA Study. Arch Pediatr Adolesc Med. 2010;164:328–33. doi: 10.1001/archpediatrics.2010.29. [DOI] [PubMed] [Google Scholar]
- 51.Scott RA, Irving R, Irwin L, et al. ACTN3 and ACE genotypes in elite Jamaican and US sprinters. Med Sci Sports Exerc. 2010;42:107–12. doi: 10.1249/MSS.0b013e3181ae2bc0. [DOI] [PubMed] [Google Scholar]
- 52.Scuteri A, Sanna S, Chen W-M, et al. Genome-wide association acan shows genetic variants in the FTO gene are associated with obesity-related traits. PLos Genetics. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sonestedt E, Roos C, Gullberg B, Ericson U, Wirfalt E, Orho-Melander M. Fat and carbohydrate intake modify the association between genetic variation in the FTO genotype and obesity. Am J Clin Nutr. 2009;90:1418–25. doi: 10.3945/ajcn.2009.27958. [DOI] [PubMed] [Google Scholar]
- 54.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spielmann N, Leon AS, Rao DC, et al. Genome-wide linkage scan for submaximal exercise heart rate in the HERITAGE family study. Am J Physiol Heart Circ Physiol. 2007;293:H3366–71. doi: 10.1152/ajpheart.00042.2007. [DOI] [PubMed] [Google Scholar]
- 56.Thompson PD, Moyna N, Seip R, et al. Functional polymorphisms associated with human muscle size and strength. Med Sci Sports Exerc. 2004;36:1132–9. doi: 10.1249/01.mss.0000132274.26612.23. [DOI] [PubMed] [Google Scholar]
- 57.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 58.Timmons JA, Knudsen S, Rankinen T, et al. Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. J Appl Physiol. 2010;108:1487–96. doi: 10.1152/japplphysiol.01295.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsianos GI, Evangelou E, Boot A, et al. Associations of polymorphisms of eight muscle- or metabolism-related genes with performance in Mount Olympus marathon runners. J Appl Physiol. 2010;108:567–74. doi: 10.1152/japplphysiol.00780.2009. [DOI] [PubMed] [Google Scholar]
- 60.Willer CJ, Speliotes EK, Loos RJF, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams AG, Folland JP. Similarity of polygenic profiles limits the potential for elite human physical performance. J Physiol. 2008;586:113–21. doi: 10.1113/jphysiol.2007.141887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xi B, Shen Y, Zhang M, et al. The common rs9939609 variant of the fat mass and obesity-associated gene is associated with obesity risk in children and adolescents of Beijing, China. BMC Med Genet. 2010;11:107. doi: 10.1186/1471-2350-11-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zempo H, Tanabe K, Murakami H, Iemitsu M, Maeda S, Kuno S. ACTN3 polymorphism affects thigh muscle area. Int J Sports Med. 2010;31:138–42. doi: 10.1055/s-0029-1242808. [DOI] [PubMed] [Google Scholar]
- 64.Zobel DP, Andreasen CH, Grarup N, et al. Variants near MC4R are associated with obesity and influence obesity-related quantitative traits in a population of middle-aged people: Studies of 14,940 Danes. Diabetes. 2009;58:757–64. doi: 10.2337/db08-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]