Abstract
Propofol is the most important intravenous general anesthetic in current clinical use. It acts by potentiating GABAA receptors, but where it binds to this receptor is not known and has been a matter of some controversy. We have synthesized a novel propofol analogue photolabeling reagent that has a biological activity very similar to that of propofol. We confirmed that this reagent labeled known propofol binding sites in human serum albumin which have been identified using X-ray crystallography. Using a combination of the protiated label and a deuterated version, and mammalian receptors labeled in intact membranes, we have identified a novel binding site for propofol in GABAA receptors consisting of both β3 homopentamers and α1β3 heteropentamers. The binding site is located within the β subunit, at the interface between the transmembrane domains and the extracellular domain, and lies close to known determinants of anesthetic sensitivity in transmembrane segments TM1 and TM2.
Propofol is the world’s most widely used intravenous general anesthetic, but how it acts at the molecular level is unknown. A key role for the GABAA receptor in the actions of propofol seemed likely ever since the demonstration1 that clinically relevant concentrations of the drug markedly enhance the GABA-induced chloride current. That this receptor is the most important target for propofol is now beyond doubt following the work of Jurd et al.2 who showed that a point mutation in the β3 subunit of the receptor in a knock-in mouse is sufficient to abolish the actions of propofol in preventing a response to a painful stimulus, and to greatly reduce its effects in causing a loss of righting reflex (a surrogate for loss of consciousness). Although it is generally agreed that the principal target for propofol is the GABAA receptor, where it binds to this receptor, and how binding is translated into an enhanced chloride current is not clear.
A complete picture of how propofol acts requires a knowledge of where it binds to its targets. Because no high-resolution X-ray crystal structure yet exists for a mammalian GABAA receptor, most work that has aimed at identifying the binding site or sites has been indirect. Work on subunit selectivity shows that homomeric β3 receptors will form a complex that provides a binding site for propofol3,4 with an affinity that is little different from that of the intact αβγ receptor3. While the presence, or isoform, of the α, γ and δ subunits have all been shown to affect propofol modulation of the GABAA receptor5–8, it seems likely that the propofol binding site involves the β subunit.
This conclusion is strengthened by site-directed mutagenesis which has provided a wealth of information on how mutations in specific subunits can affect anesthetic action9. While mutations in the α10 and γ11 subunits have been shown to change propofol’s modulation of the GABAA receptor, most work has focused on mutations in the β subunit. Amino acids in the β112, β213–17 and β318 subunits have been found that, when mutated, influence propofol’s actions. These mutations, however, are distributed throughout the membrane-spanning region of the receptor, being found in TM114, TM215,18, TM312,13,15,18 and TM416 as well as in the extracellular domain17. The question then remains, which if any of these amino acids form part of a binding site? A crystal structure of a proton-activated bacterial homologue of the GABAA receptor, GLIC, shows a propofol binding site buried between these four helices19. This channel, however, is inhibited by propofol, so the relationship between this site and the potentiating site in mammalian channels remains unclear.
One approach to this problem involves mutating amino acids at a putative propofol binding site to cysteine residues, and then probing that site using chemical reagents that bind to cysteine irreversibly20. This is an elegant approach and has been used to argue that a propofol binding site must exist close to the middle of TM3. Unfortunately, it suffers from the same problem that bedevils the interpretation of site-directed mutagenesis - are the observed effects a consequence of allosteric conformational changes? For example, while propofol does indeed block the chemical modification of a cysteine mutated into the middle of TM320, is this due to a direct steric interaction or an effect of propofol binding elsewhere21?
An alternative, and direct, method of identifying a drug binding site is to use an analogue of the drug that contains a photoaffinity group whose activation by light results in irreversible covalent binding. This requires, of course, that the photoaffinity analogue of the drug resembles the parent compound sufficiently closely so that it binds in exactly the same location. Additionally, the photolabeling reagent must preferentially participate in inter-molecular crosslinking rather than intra-molecular rearrangement; this is a difficult condition to achieve for propofol whose simple aromatic structure makes internal rearrangement following photoactivation highly likely. In addition to these problems, separating peptides from a multimeric transmembrane protein such as the GABAA receptor in sufficient quantities to enable a specific labeled amino acid to be identified is very challenging22. In this paper we show how we have solved these problems by synthesizing novel protiated and deuterated versions of a propofol diazirine with properties very similar to propofol, using them to photolabel GABAA receptors expressed in intact membranes and utilizing high-resolution mass spectroscopy23 to identify sites of photolabel incorporation. Using this approach we have identified a novel binding site for propofol.
RESULTS
Synthesis and characterization of propofol photolabels
We synthesized four different putative propofol photolabels, each of which incorporated a trifluoromethyl diazirine group in either the ortho (1), meta (2) or para (3, 4) positions (Supplementary Results, Supplementary Fig. 1). Only the analogue with a trifluoromethyl diazirine group at the ortho position (1) replacing one of the two isopropyl groups (Fig. 1a) proved to be an effective photolabel, with essentially 100% incorporation into ethanol, as well as being very similar to the parent compound propofol in its ability to directly activate and potentiate GABAA receptors (Fig. 1b). The other analogues were either very poor photolabels (the para-substituted derivatives; 3, 4) or were virtually inactive in potentiating GABAA receptors (the meta-substituted derivative; 2).
Figure 1. Characterization of ortho-propofol diazirine.
(a) The chemical structures of propofol (left) and ortho-propofol diazirine (1). (b) The percentage potentiation of GABA-evoked currents by propofol (3 μM; n=12), ortho-propofol diazirine (5 μM: n=12), para-methylene-propofol diazirine (4; 50 μM; n=7) and meta-propofol diazirine (2; 200 μM; n=3). GABA (3 μM; EC7) was applied to HEK293 cells expressing α1β2γ2s GABAA receptors, and the current was recorded under whole-cell patch clamp. The inset shows a typical current trace during switches into GABA, in the presence and absence of 5 μM ortho-propofol diazirine. (c) UV absorption spectrum of ortho-propofol diazirine in ethanol (0.2 mg ml−1). (d) Inhibition of [35S]-TBPS binding to GABAA β3 homomers by ortho-propofol diazirine. The IC50 concentration for inhibition of TBPS binding was 2.9 ± 0.4 μM (n=3). (e) The percentage potentiation of GABA-evoked currents by ortho-propofol diazirine acting on α1β3 GABAA receptor heteromers. The EC50 concentration for potentiation was 1.7 ± 0.7 μM (n=10) with a Hill coefficient of 0.8±0.2 and the maximum potentiation was 151 ± 18%. (The point at the highest drug concentration was excluded from the fit to the Hill equation.) (f) Quantal dose-response curves for propofol and ortho-propofol diazirine for loss of righting reflex in rats. The ED50 concentrations for propofol and ortho-propofol diazirine were 4.7 ± 0.8 mg kg−1 (n=12) and 14.7 ± 0.2 mg kg−1 (n=13) respectively. All error bars are standard errors of the mean and if not shown, were smaller than the size of the symbols.
The peak absorbance of ortho-propofol diazirine (λmax=283 nm; ε =1970 ± 20 M−1 cm−1) was very similar to that of propofol (λmax=275 nm; ε =2490 ± 90 M−1 cm−1) but, in addition, there was an absorption band between 320 and 360 nm due to the diazirine group (Fig. 1c), with an extinction coefficient at 350 nm of 40.5 ± 2.5 M−1 cm−1. Following exposure of an ethanolic solution of ortho-propofol diazirine to light (>320 nm) from a mercury-xenon arc lamp, changes in the spectrum occurred with a halftime of 23.8 ± 1.0 s. In the absence of light, ortho-propofol diazirine was stable over several hours at room temperature, and several months at −20°C.
As a prelude to our attempts to label GABAA receptors, we examined the concentration-dependent modulation of [35S]TBPS binding to GABAA β3 homomeric receptors expressed in Sf9 cells, and the EC50 concentration for potentiation of currents elicited by an EC60 concentration of GABA in α1β3 heteromers expressed in HEK cells. ortho-Propofol diazirine inhibited TBPS binding to β3 homomers with an IC50 of 2.9 ± 0.4 μM (Fig. 1d), similar to previous reports with propofol3,4. The EC50 for the potentiation of GABA-induced currents mediated by α1β3 heteromers (Fig. 1e) was comparable, at 1.7 ± 0.7 μM.
In order to compare the anesthetic potency of propofol and ortho-propofol diazirine in animals, we determined the ED50 doses needed to induce loss of righting reflex (LORR) in Sprague-Dawley rats. Various doses of both compounds were administered by tail-vein injection and dose-response curves constructed using the method of Waud24. Both propofol and ortho-propofol diazirine were able to cause reversible loss of righting reflex at comparable concentrations (Fig. 1f)
Labeling human serum albumin
To further validate the use of ortho-propofol diazirine to identify propofol binding sites in GABAA receptors, we tested its ability to label known propofol binding sites that have been identified in the high-resolution crystal structure25 of the soluble protein human serum albumin (HSA). The structure of HSA with propofol bound was solved at a resolution of 2.4 Å, and showed two propofol binding sites: propofol site 1 in subdomain IIIA of HSA and propofol site 2 in subdomain IIIB. We carried out labeling experiments with HSA in the presence of either the protiated or the deuterated forms of ortho-propofol diazirine to see if these known sites could be identified.
We exposed HSA to light with wavelengths >320 nm in the absence (control) or presence of either the protiated or deuterated forms of ortho-propofol diazirine, then analyzed tryptic digests of the samples using LC-MS26. In the control samples, tandem spectra (MS2) unambiguously identified peptides that contained 569 of the 585 amino acids in HSA (97% coverage; Supplementary Fig. 2). In some cases, labeling of peptides could be detected by changes in absorbance at 214 and 280 nm in the HPLC traces. Our search of the mass spectrometric data for labeled peptides was greatly facilitated by the combined use of the protiated and deuterated forms of the photolabel. This is because, while the isotopic distributions of most peptides would be expected to be very similar, whether or not they labeled with the protiated ortho-propofol diazirine (see e.g. Figs. 2a,b), a peptide labeled with our deuterated version (which contains a known distribution of d0, d1 and d2 species) is characteristically different and immediately recognizable (see e.g. Fig. 2c). Hence it was relatively easy to identify the peptides that had been labeled.
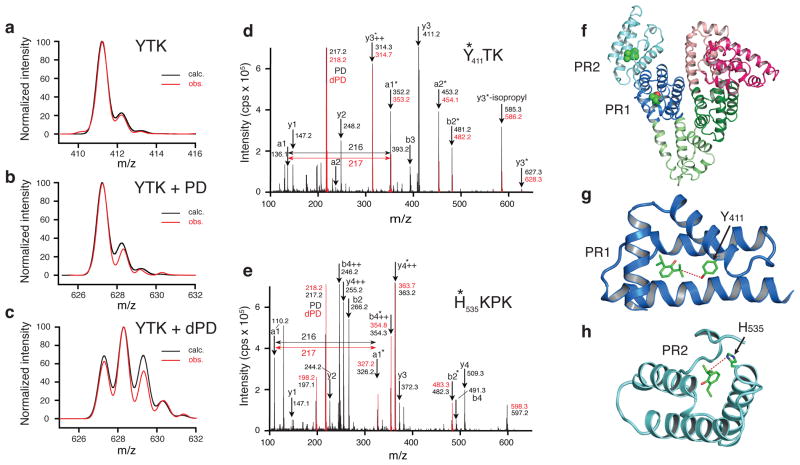
Figure 2. Labeling human serum albumin with ortho-propofol diazirine.
Calculated and observed isotopic distributions for (a) unlabeled YTK peptide, (b) YTK peptide labeled with protiated ortho-propofol diazirine and (c) YTK peptide labeled with partially deuterated ortho-propofol diazirine. MS2 data for peptides YTK (d) and HKPK (e) with protiated ortho-propofol diazirine (black) and deuterated ortho-propofol diazirine (red). The asterisks denote the presence of photolabel bound to the peptide fragment. The two sites are illustrated in (f) with PR1 being in subdomain IIIA (dark blue) and PR2 being in subdomain IIIB (light blue). More detailed views are shown in (g) for PR1 where the labeled Y411 is shown and in (h) for PR2 where the labeled H535 is shown. Dashed lines mark the distances between the propofol molecules to Y411 (4.9 A) and to H535 (5.0 A).
A number of peptides were identified whose molecular weights had increased by the expected amount (216.1 Da) on labeling with the protiated ortho-propofol diazirine and which also showed the characteristic isotopic distribution when labeled with the deuterated version of the photolabel. In all cases the labeled peptide adduct eluted later than the parent peptide during HPLC separation, consistent with an increase in hydrophobicity. As expected, we observed specific labeling of amino acids in the two known propofol binding sites 1 and 2 (Figs. 2d–h). In the highest affinity propofol site25, site 1, a single amino acid (Y411) was labeled (Fig. 2d,g), while in the lower affinity site25, propofol site 2, four amino acids (K525, H535, K536, K541) were labeled (Fig. 2e,h). Interestingly, we were not able to show any measurable labeling of HSA by meta-propofol diazirine (2), indicating that meta-substituted derivatives of propofol may not bind at the same sites as the parent compound.
Labeling GABAA receptors
To identify propofol binding sites on GABAA receptors, membranes were prepared from Sf9 cells transfected with epitope-tagged α1strep and β3His subunits27; these membranes contain both β3 homomers and α1β3 heteromers. The membranes (30–100 pmol of receptor) were incubated in the absence (control) or presence of the protiated or deuterated version of ortho-propofol diazirine (3 μM, 30 μM or 100 μM) and irradiated with wavelengths >320 nm for 3 minutes. In the first set of experiments GABAA receptors were extracted in 2% TritonX-100 to preserve pentameric receptor structure and the α1β3 heteromers and β3 homomers were separately purified by sequential affinity chromatography on Strep-tactin superflow plus resin™ (Qiagen) and Ni2+ NTA-agarose (Qiagen) beads. In two subsequent experiments, GABAA receptors were extracted from photolabeled membranes using denaturing conditions (2% SDS) to enhance the protein yield and increase mass spectrometric sequence coverage. The α1 and β3 subunits were separately purified by detergent dilution followed by sequential affinity chromatography with Strep-tactin superflow plus resin™ and Ni2+ NTA-agarose. In all of the experiments, the affinity-purified GABAA receptor subunits were delipidated, reduced and alkylated, and digested in-solution with endoproteases, employing a strategy that generates multiple small overlapping peptides from the TMDs by using multiple-timed chymotryptic digestions23. LC-MS analysis of the protein digests yielded MS2 spectra that provided unambiguous identification of 96% of the amino acids in β3 subunit including 95% of the residues in the TMD regions as well as 95% of the amino acids in the α1 subunit including 88% of the TMD residues (Supplementary Figs. 3 and 4 and Tables 1 and 2). The undetected sequences in the N-terminal domains contain glycosylation sequons (N-X-T) and are probably not detected because they are glycosylated.
MS2 spectra (Supplementary Dataset) from all samples were searched for peptides modified by the added mass of either the protiated or deuterated propofol diazirines. The only candidate peptides that satisfied our criteria for labeling (see Methods) were the doubly charged peptides m/z=632.299 and m/z=632.802 corresponding to the protiated and deuterated ortho-propofol diazirine-adducts of 260-TMoxiTTINTHL-268, a β3 peptide located near the extracellular end of TM2.
Diagnostic MS2 spectra of the protiated and deuterated adducts of TMoxiTTINTHL were identified in both the α1β3 heteromers and the β3 homomers. The MS2 spectra (Fig. 3a) contain fragment ions (y series) that identify the sequence of the peptide and localize the site of photoincorporation to either H267 or L268. The major features in the MS2 spectra (m/z=326.148 and 327.151) correspond to internal ion fragments (H*) that definitively identify the site of protiated and deuterated propofol incorporation as H267 (Fig. 3b). The stable heavy isotope labeling greatly assisted in interpretation of the fragmentation spectra, as fragment ions containing the deuterated or protiated propofol adduct differed by a single mass unit, whereas fragments (e.g. b2 ions) that were not labeled had identical mass. The parent peptide, TMoxiTTINTHL (m/z=524.261, z=2) was also observed at a chromatographic retention time significantly earlier than the propofol adducts. Comparison of the intensities of the parent and adduct MS1 features was used to estimate the efficiency of labeling as 13% for both the protiated and deuterated adduct.
Figure 3. Labeling GABAA receptors with ortho-propofol diazirine.
With both α1β3 heteromers and β3 homomers only a single amino acid, H267, was labeled. (a) MS2 data for the peptide 260-TMTTINTHL-268 labeled with the protiated ortho-propofol diazirine (black) and the deuterated ortho-propofol diazirine (red) superimposed. (b) Fragmentation diagram showing that the MS2 data are consistent with the ortho-propofol diazirine labeling the histidine side chain which subsequently rearranges as internal ions H* and TH* following fragmentation. (c) A view of our proposed propofol binding site seen from the center of the pore. Two neighboring subunits are shown in yellow and blue and a single ortho-propofol diazirine molecule is shown associated with the yellow subunit. (d) A view from the extracellular side of the receptor, but with the extracellular domain removed for clarity. The β3 homopentamer is shown with five equivalent ortho-propofol diazirine molecules bound. (e) A surface representation of our proposed propofol binding pocket with an ortho-propofol diazirine molecule sitting in the pocket close to H267 which is the amino acid that is photolabeled. (f) Electrophysiological data from α1β3 heteromers showing the effects of the mutations β3-H267A and β3-F221W on the GABA apparent affinity (n=21), (g) the extent to which 3 μM ortho-propofol diazirine potentiates a GABA-evoked chloride current (at the GABA EC60) (n=4–10) and (h) the extent to which 10 μM ortho-propofol diazirine directly activates the receptor (n=4). All error bars are standard errors of the mean.
Previous studies based on site-directed mutagenesis or molecular modeling have suggested that M286 in β3-TM3 and M236 in α1-TM1 constitute two sides of a possible propofol binding site between the α1 and β3 subunits12,13,20. Unambiguous MS2 spectra of peptides containing both M286 and M236 were identified in our analyses. MS1 features with an m/z corresponding to the calculated propofol adduct of these peptides were not observed. Based on the intensities of the observed peptides (β3- 285-LMoxiGCF-289, β3-278-VKAIDMoxiYLMoxiGCF-289, α1-227-IQTYLPcIMT-236) and the signal-to-noise ratio of our spectra, MS1 features of the propofol adducts of these peptides should have been observed if they were photolabeled with efficiencies of more than 3%. Given that the efficiency might vary between sites, we cannot rule out these residues as candidates for additional sites of propofol binding.
We explored the location of the labeled H267 in the structure of the GABAA receptor using the homology model based (Supplementary Fig. 5) on the open state of an homologous glutamate-gated chloride channel28 that was described previously22. On inspection of the model it was clear that, immediately adjacent to H267 is a predominantly hydrophobic cleft opening out to a larger cavity at the top of, and between, the first and second transmembrane domains TM1 and TM2 (Fig. 3 c–e and Supplementary Fig. 6). This cavity essentially lies within a single β subunit, although there is also an interaction with the main-chain at the top of TM2 in the neighboring subunit. This cleft is open to the central ion pore, allowing drug access. Figure 3c shows a side view of a putative propofol binding site seen from the center of the pore, and Figure 3d shows a top view (with the extracellular domain removed for clarity) illustrating the position of the ortho-propofol diazirine molecules with respect to the four transmembrane domains. Five equivalent drug molecules are shown in this model of a β3 pentamer, but given that the binding site is essentially composed of amino acid side chains from a single β subunit, we would predict only two or three drug molecules would bind to the αβ or αβγ heteromers, depending on the number of β subunits present. Figure 3e illustrates the binding cavity as a surface, with the location of the labeled histidine indicated at the bottom of the pocket.
We next mutated the labeled H267 to an alanine to test the effects of this mutation on propofol modulation of the α1β3 receptor. The H267A mutation caused a significant reduction (p<0.01) in the GABA apparent affinity (Fig. 3f) as previously reported29, a significant reduction (p<0.05) in the extent to which ortho-propofol diazirine potentiated (Fig. 3g) the receptor (at a GABA concentration equivalent to the GABA EC60 for both the wild-type and mutant receptors), and a comparable reduction (Fig. 3h) in the ability of the drug to directly activate the receptor (p<0.01). We also mutated F221 to the more bulky amino acid, tryptophan, because the model predicted that this should partially occlude drug binding (Supplementary Fig. 6). The F221W mutation had only a small effect on the GABA EC50 (Fig. 3f), although there was a measureable picrotoxin-sensitive increase in baseline current, indicating some constitutive activity (16 ± 4% of the maximum GABA-induced current; n=6). This mutation, however, virtually eliminated (Fig. 3g) anesthetic-induced potentiation (P<0.01) and significantly (P<0.01) reduced direct anesthetic activation (Fig. 3h).
DISCUSSION
If the binding site of a drug is to be determined using photolabeling, then one of the most important problems to overcome is the attachment of a photoreactive group that does not significantly alter the interaction of the drug with its targets. With the simple general anesthetic propofol, this is particularly challenging because few, if any, modifications are known which do not significantly reduce its anesthetic potency30. Furthermore, the GABAA receptor, to which propofol binds, is affected by an extraordinarily diverse range of chemical compounds that almost certainly bind to a variety of distinct sites9. Thus, it is particularly important that any photoreactive propofol analogue is as similar as possible to propofol itself. That said, designing an analogue that is chemically stable and is capable of covalently labeling a protein binding site before an unproductive internal rearrangement occurs, is a challenge in itself.
A widely used photoreactive group is the trifluoromethyl diazirine moiety, which is a chemically stable group with an absorbance band centered around 350 nm, well away from the principal absorbance band of proteins (280 nm). Propofol analogues with this group substituted at the para and meta positions were either inefficient photolabels (with both solvents and human serum albumin), or did not closely mimic the effects of the parent compound propofol on GABAA receptors. An analogue with this group replacing one of the two isopropyl side chains at the ortho position, however, worked remarkably well, being both a highly efficient photolabel, as well as retaining the ability to directly activate and potentiate the GABAA receptor almost as well as propofol. We further validated the ortho-propofol diazirine analogue as a propofol photolabel by showing that it labeled the two propofol binding sites that had been identified using X-ray crystallography25 (Fig. 2). Our results with human serum albumin, together with the functional and binding data on GABAA receptors, strongly suggest that any binding sites identified using the ortho-propofol diazirine photolabel are likely to represent sites where propofol itself will bind.
Our labeling experiments with GABAA receptors, whether α1β3 heteromers or β3 homomers, identified a single peptide (260-TMoxiTTINTHL-268) as being labeled, with the ortho-propofol diazirine photolabel attached to histidine 267. A binding site involving the β subunit might have been expected, given the comparable apparent affinities of the α1β3 heteromers or β3 homomers to propofol3,4, although the absence of labeling in the α subunit cannot be taken as definitive proof that another site involving this subunit might not exist. If, for example, the diazirine group is surrounded by solvent, the protein might not be measurably labeled. Also, our label might react more efficiently with some amino acids than with others. The labeling of H267 in the β3 subunit, however, was robust and reproducible.
Using an homology model based on the crystal structure28 of the open state of the glutamate-gated chloride channel22, we have identified a plausible binding site for propofol, in a cavity close to the interface between the transmembrane region and the ligand-binding extracellular domain. Our only constraints were the requirement for an amphiphilic cavity large enough to accommodate the drug, and the necessity for the diazirine moiety to be in proximity to histidine 267. Other than this, we cannot be definitive about the detailed molecular architecture of the binding site, although we note that the amino acid side chains in the vicinity of our postulated binding site are similar between the GABAA receptor and the glutamate-gated chloride channel, indicating that the homology model in this region should be relatively reliable.
The mutation of H267 to an alanine caused changes in both GABA activation and drug modulation of the receptor (Fig. 3f–h), and this is certainly consistent with this amino acid forming part of an anesthetic binding site. More persuasively, our model predicted that the mutation of F221, which lies at the back of the cavity, to a more bulky tryptophan would partially block the binding of propofol. Indeed, this mutation caused some constituitive activity, as if the residue were mimicking the anesthetic, but also virtually eliminated anesthetic potentiation, as well as reducing direct anesthetic activation (Fig. 3f–h). The histidine that we have labeled is on the opposite side of the helix from an amino acid N2652,12,18,31 in TM2 that is known to be a key determinant of anesthetic action. The drug also lies equally close to another anesthetic determinant, G21914,32 at the top of TM1. Having said this, however, given that multiple mutations throughout the receptor can affect its modulation by anesthetics and other drugs, we consider our mutagenesis results are consistent with our model, but not definitive. An additional possibility that is difficult to rule out completely is that the labeling of H267 reflects an inhibitory site in the channel pore. This would seem to be very unlikely because we observe robust labeling at concentrations of the photolabel (3 μM) which do not cause measurable channel inhibition.
The site we have identified does not overlap with the propofol site19 in GLIC (which is inhibited, rather than activated by propofol). Nor does it overlap with an inter-subunit site (that lies between TM3 in a β subunit and TM1 in an adjacent α subunit) that has been shown to bind etomidate by photolabeling33, and which has also been proposed as a likely propofol binding site12,13,20. As mentioned above, although we did not label this site, we cannot rule it out, and further experiments with other propofol photolabels34,35 may provide a complete picture.
The approach that we have employed to identify propofol binding sites in GABAA receptors has several advantages. Most importantly, photolabeling is performed in intact membranes, in which the receptor is in a native lipid environment. While the ideal condition might be to photolabel in an intact cell, GABAA receptors are more likely to be in a native conformation in membranes than in the detergent environment used in most anesthetic photolabeling studies. The binding sites that we have identified are thus unlikely to be artifacts of unnatural or denaturing conditions.
In summary, we have identified a novel binding site for the widely used intravenous general anesthetic propofol on its major target, the GABAA receptor expressed in intact membranes. We have shown the utility of using a combination of deuterated and protiated variants of ortho-propofol diazirine, combined with mass spectrometry, to identify unknown propofol binding sites on membrane receptors.
METHODS
Synthesis of ortho-propofol diazirine and its deuterated analogue
The synthesis of ortho-propofol diazirine is outlined in Supplementary Note. ortho-Propofol diazirine was stable for hours at room temperature in organic solvents. It showed no signs of decomposition by 19F NMR spectroscopy over a period of six months as a 250 mM solution in ethanol at −18°C. The deuterated analogue of ortho-propofol diazirine was prepared in a like manner from deuterated ortho-isopropylphenol (see Supplementary Note). The penultimate, methylated diazirine had the following distribution of deuterium by mass spectrometry: 34:43:23 d0:d1:d2.
Electrophysiology
GABAA receptors formed from either α1β2γ2s or α1β3 subunits were transiently expressed in HEK 293 cells and currents were recorded using whole-cell patch clamping as described previously26. Extracellular solution (140 mM NaCl, 2.5 mM KCl, 2 mM MgCl2, 10 mM HEPES, 10 mM D-glucose, 1 mM CaCl2, pH 7.4) was used to perfuse the cells. Currents were recorded using borosilicate glass electrodes filled with intracellular solution (140 mM CsCl, 4 mM NaCl, 10 mM HEPES, 10 mM D-glucose, 5 mM EGTA, 0.5 mM CaCl2, 2 mM Mg-ATP, pH 7.3) with cells voltage clamped at −60 mV.
[35S]-TBPS binding to GABAA receptor β3 homomers
[35S]-TBPS binding was performed as previously described22,36. Briefly, aliquots of membranes prepared from SF9 cells expressing β3 homomeric GABAA receptors (final protein concentration, 20 μg ml−1) were resuspended in 100 mM KCl, 10 mM potassium phosphate, pH 7.5, containing 1 to 2 nM [35S]-TBPS (60–100 Ci mmol−1) and 2-μl aliquots of ortho-propofol diazirine in dimethylsulfoxide (final concentration 10 nM–100 μM), in a total assay volume of 1 ml. Values for the maximum binding level, Bmax, were determined by using 30 nM [35S]-TBPS. Control binding was defined as the binding observed in the absence of ortho-propofol diazirine. All assays contained 0.2% ethanol. Nonspecific binding was defined as binding observed in the presence of 200 μM picrotoxin. The [35S]-TBPS binding data were fitted to a Hill equation.
Anesthetic potency in rats
ED50 concentrations for propofol and ortho-propofol diazirine causing loss of righting reflex in male Sprague-Dawley rats (average weight 498 ± 33 g; mean ± sd) were determined. The anesthetic, dissolved in Intralipid (Sigma-Aldrich), was introduced via a 26-gauge tail-vein cannula and the animals were then placed in a cylinder rotating at 3 rpm. Loss of righting reflex was scored within one minute of injection and the data analyzed using the method of Waud24. All experiments were carried out in accordance with the United Kingdom Animals (Scientific Procedures) Act of 1986 and have been approved by the Ethical Review Committee of Imperial College London.
Labeling of human serum albumin
Fatty-acid-free human serum albumin was dissolved in 1 ml of phosphate buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, pH 7.4) at 10 mg ml−1 and incubated in a quartz cuvette with protiated or deuterated ortho-propofol diazirine at a concentration of 1.5 mM for 5 minutes in the dark. The sample was then continuously stirred while being irradiated for 120 seconds with light from a mercury-xenon arc lamp whose light had been filtered by reflection from a dichroic mirror (to remove the infrared) and by passing through an interference filter (λ > 320 nm). After photolabeling, the protein was denatured with guanidine hydrochloride, the di-sulfide bonds were reduced using dithiothreitol and their reformation blocked using iodoacetamide, and then the protein was digested with trypsin following dialysis26. The resulting peptides were then separated using HPLC and analyzed using mass spectrometry as previously described26.
Membrane preparation and GABAA receptor photolabeling
Sf9 cells were used27 to express α1-streptag and β3-His8 GABAA receptor subunits, and membranes were prepared as described previously22 and stored at −80°C. For photolabeling, the membranes (0.5 mg protein ml−1) were suspended in 10 mM potassium phosphate buffer, 100 mM KCl, containing a cocktail of protease inhibitors including 4-(2-aminoethyl)benzenesulfonyl fluoride, pepstatin A, N-[N-(L-3-trans-carboxyirane-2-carbonyl)-L-leucyl]-agmatine (E-64), bestatin, leupeptin, and aprotinin. Following incubation with 3 μM, 30 μM or 100 μM protiated or deuterated ortho-propofol diazirine for 60 minutes in the dark, the membrane samples were irradiated in a quartz cuvette for 3 min, using a photoreactor emitting light at >320 nm37.
Purification of GABAA Receptor subunits and preparation of peptides
After UV irradiation, membranes were collected by centrifugation at 130,000g for 45 min and resuspended in lysis buffer containing 1% (w/v) Triton X-100, 150 mM NaCl, 50 mM sodium phosphate, pH 7.5 for 120 min at 4°C. The solubilized proteins were loaded on a Strep-tactin™ superflow plus resin (Qiagen) column. After washing the column with lysis buffer, the proteins were eluted with 25 mM desthiobiotin in the lysis buffer. The lysate that passed through the Strep-tactin column was adjusted to an imidazole concentration of 40 mM and loaded on a Ni2+-NTA agarose (Qiagen) column. The column was washed with lysis buffer containing 40 mM imidazole and proteins were eluted with 0.1% Triton X-100, 500 mM imidazole, 25 mM sodium phosphate, 300 mM NaCl, pH 7.5. All procedures were conducted at 4°C in the presence of a protease inhibitor cocktail containing 4-(2-aminoethyl)benzenesulfonyl fluoride, pepstatin A, E-64, bestatin, leupeptin, and aprotinin. To separately purify α1 and β3 subunits, photolabeled membranes were solubilized in 2% SDS, 100 mM sodium phosphate buffer. The membrane lysate was then diluted to an SDS concentration of 0.2% with Triton X-100 lysis buffer and the α1 subunits were purified using Strep-tactin affinity chromatography. The lysate that passed through the Step-tactin columns was diluted to 0.1 % SDS, adjusted to 40 mM imidazole and applied to a Ni2+-NTA agarose column for purification of β3 subunits as described above.
MS samples were prepared as previously described22. Briefly, purified α1 and β3 receptor subunits were concentrated on a 30,000 MW cutoff concentrator (Centricon) and precipitated using a 2D cleanup kit (GE Lifesciences). The proteins were delipidated, suspended in 100 μl of 8 M urea, 0.1% RapiGest (Waters), 100 mM Tris, pH 8.5, reduced and alkylated as previously described23. The samples were then digested with 1 μg of the endoproteinase Lys-C overnight at 37°C followed by trypsin overnight at 37°C, or endoproteinase chymotrypsin for 2, 6, or 18 h at room temperature. For trypsin digestion, the samples were diluted with 100 mM Tris, pH 8.5, to reduce the concentration of urea to 2 M. For chymotrypsin digestion, the samples were diluted by 100 mM Tris, pH 8.5, to a concentration of 1 M urea, and CaCl2 was added to a final concentration of 10 mM. The digests were terminated by adding formic acid to a final concentration of 1% and incubating at 37°C for 30 min. Solid-phase extraction with NuTip C4 (Glygen) and porous graphite carbon (PGC) tips was used to remove salts from the sample using the previously described methods23. The resulting peptides were analyzed using nano-LC-MS with a hybrid mass spectrometer consisting of a linear quadrupole ion trap and an orbitrap ELITE (Thermo Scientific).
Data Processing and Analysis
The LC-MS data files were processed using MASCOT Distiller (Matrix Science Inc.), with previously described settings23. The resulting MS2 centroid-calculated files were used to search (with MASCOT; Matrix Science Inc.) a customized database containing the sequence of all GABAA receptor subunits. The search was conducted with no enzyme specificity and allowed 5 missed cleavages, oxidation of Met, carbamidomethylation of Cys, and a protiated or deuterated ortho-propofol diazirine adduct at every amino acid as variable modifications, with a parent ion tolerance of 10 ppm and a fragment ion mass tolerance of 0.05 Daltons. Scaffold (Proteome Software Inc.) was used to validate MS2-based peptide identifications with the Protein Prophet algorithm module38. To determine the photolabeled peptide identity, seven criterion were used to verify the search results: 1) peptide prophet probability ≥95%; 2) The photolabeled peptide parent ions were within 3 ppm mass error tolerance according to the instrument setting; 3) De Novo sequencing of fragmentation MS2 spectra, based on the theoretical fragmentation generated by using MS-Product (University of California, San Francisco, CA), support the peptide identity; 4) The same features of peptide parent ions (MS1) and fragmentation ions (MS2) were observed in both protiated and deuterated ortho-propofol diazirine -labeled samples with isotope distribution pattern in agreement with the presence of deuterium; 5) The retention time of protiated ortho-propofol diazirine-labeled peptides is similar to that of the deuterated ortho-propofol diazirine-labeled peptides; 6) The features of the labeled peptides were not observed in the non-labeled control samples at the corresponding retention time and 7) The photolabeled peptide and its non-labeled parent were in the same file and the parent had a shorter chromatographic retention time.
Statistical analysis
For the analysis of the electrophysiological data on the GABAA receptor mutants, one-way ANOVA was used to test for the level of significance between the data from wild-type and mutant receptors.
Supplementary Material
Acknowledgments
We thank the Medical Research Council (UK) (Grant G0901892 to N.P.F), the National Institutes of Health National Institute of General Medical Sciences (Grant PO1-GM47969 to A.S.E) and NIH National Center for Research Resources (Grants P41 RR00954 and UL1 RR024992 to R.R.T)., the Austrian Ministry of Science and Research and the European Seventh Framework Program (Grant HEALTH-F4-2008-202088 to W.S.), for support. We also thank Raquel Yustos for technical assistance, Damian Droste, Mark Bennett, Thom Gent, Kersti Karu, Brad Manion and James Malone for help with the experiments.
Footnotes
Competing financial interests The authors declare no competing financial interests.
Author contributions C.J.E. and E.H.S. synthesized the propofol photolabels and helped with the data analysis, G.M.S.Y. and Z-W.C. carried out the labeling experiments, protein chemistry and mass spectroscopy and helped with the analysis, R.D. made and analyzed the electrophysiological measurements, C.J.E. carried out the animal experiments, E.H. contributed to the molecular modeling, W.S. and K.F. supplied membranes containing expressed receptors, R.R.T. contributed to the design and interpretation of mass spectrometric experiments and A.S.E. and N.P.F. designed and supervised the project, helped with the analysis and wrote the paper.
References
- 1.Hales TG, Lambert JJ. The actions of propofol on inhibitory amino acid receptors of bovine adrenomedullary chromaffin cells and rodent central neurones. British journal of pharmacology. 1991;104:619–628. doi: 10.1111/j.1476-5381.1991.tb12479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jurd R, et al. General anesthetic actions in vivo strongly attenuated by a point mutation in the GABA(A) receptor beta3 subunit. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2003;17:250–252. doi: 10.1096/fj.02-0611fje. [DOI] [PubMed] [Google Scholar]
- 3.Zezula J, Slany A, Sieghart W. Interaction of allosteric ligands with GABAA receptors containing one, two, or three different subunits. European journal of pharmacology. 1996;301:207–214. doi: 10.1016/0014-2999(96)00066-0. [DOI] [PubMed] [Google Scholar]
- 4.Davies PA, Kirkness EF, Hales TG. Modulation by general anaesthetics of rat GABAA receptors comprised of alpha 1 beta 3 and beta 3 subunits expressed in human embryonic kidney 293 cells. British journal of pharmacology. 1997;120:899–909. doi: 10.1038/sj.bjp.0700987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng HJ, Macdonald RL. Multiple actions of propofol on alphabetagamma and alphabetadelta GABAA receptors. Molecular pharmacology. 2004;66:1517–1524. doi: 10.1124/mol.104.003426. [DOI] [PubMed] [Google Scholar]
- 6.Jones MV, Harrison NL, Pritchett DB, Hales TG. Modulation of the GABAA receptor by propofol is independent of the gamma subunit. The Journal of pharmacology and experimental therapeutics. 1995;274:962–968. [PubMed] [Google Scholar]
- 7.Krasowski MD, et al. Alpha subunit isoform influences GABA(A) receptor modulation by propofol. Neuropharmacology. 1997;36:941–949. doi: 10.1016/s0028-3908(97)00074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lam DW, Reynolds JN. Modulatory and direct effects of propofol on recombinant GABAA receptors expressed in xenopus oocytes: influence of alpha-and gamma2-subunits. Brain research. 1998;784:179–187. doi: 10.1016/s0006-8993(97)01334-6. [DOI] [PubMed] [Google Scholar]
- 9.Franks NP. General anaesthesia: from molecular targets to neuronal pathways of sleep and arousal. Nature reviews. Neuroscience. 2008;9:370–386. doi: 10.1038/nrn2372. [DOI] [PubMed] [Google Scholar]
- 10.Moraga-Cid G, Yevenes GE, Schmalzing G, Peoples RW, Aguayo LG. A Single phenylalanine residue in the main intracellular loop of alpha1 gamma-aminobutyric acid type A and glycine receptors influences their sensitivity to propofol. Anesthesiology. 2011;115:464–473. doi: 10.1097/ALN.0b013e31822550f7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Shea SM, Williams CA, Jenkins A. Inverse effects on gating and modulation caused by a mutation in the M2-M3 Linker of the GABA(A) receptor gamma subunit. Molecular pharmacology. 2009;76:641–651. doi: 10.1124/mol.109.055111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krasowski MD, et al. Propofol and other intravenous anesthetics have sites of action on the gamma-aminobutyric acid type A receptor distinct from that for isoflurane. Molecular pharmacology. 1998;53:530–538. doi: 10.1124/mol.53.3.530. [DOI] [PubMed] [Google Scholar]
- 13.Krasowski MD, Nishikawa K, Nikolaeva N, Lin A, Harrison NL. Methionine 286 in transmembrane domain 3 of the GABAA receptor beta subunit controls a binding cavity for propofol and other alkylphenol general anesthetics. Neuropharmacology. 2001;41:952–964. doi: 10.1016/s0028-3908(01)00141-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang CS, Olcese R, Olsen RW. A single M1 residue in the beta2 subunit alters channel gating of GABAA receptor in anesthetic modulation and direct activation. The Journal of biological chemistry. 2003;278:42821–42828. doi: 10.1074/jbc.M306978200. [DOI] [PubMed] [Google Scholar]
- 15.Siegwart R, Krahenbuhl K, Lambert S, Rudolph U. Mutational analysis of molecular requirements for the actions of general anaesthetics at the gamma-aminobutyric acidA receptor subtype, alpha1beta2gamma2. BMC pharmacology. 2003;3:13. doi: 10.1186/1471-2210-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richardson JE, et al. A conserved tyrosine in the beta2 subunit M4 segment is a determinant of gamma-aminobutyric acid type A receptor sensitivity to propofol. Anesthesiology. 2007;107:412–418. doi: 10.1097/01.anes.0000278875.36639.2c. [DOI] [PubMed] [Google Scholar]
- 17.Williams CA, Bell SV, Jenkins A. A residue in loop 9 of the beta2-subunit stabilizes the closed state of the GABAA receptor. The Journal of biological chemistry. 2010;285:7281–7287. doi: 10.1074/jbc.M109.050294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegwart R, Jurd R, Rudolph U. Molecular determinants for the action of general anesthetics at recombinant alpha(2)beta(3)gamma(2)gamma-aminobutyric acid(A) receptors. Journal of neurochemistry. 2002;80:140–148. doi: 10.1046/j.0022-3042.2001.00682.x. [DOI] [PubMed] [Google Scholar]
- 19.Nury H, et al. X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature. 2011;469:428–431. doi: 10.1038/nature09647. [DOI] [PubMed] [Google Scholar]
- 20.Bali M, Akabas MH. Defining the propofol binding site location on the GABAA receptor. Molecular pharmacology. 2004;65:68–76. doi: 10.1124/mol.65.1.68. [DOI] [PubMed] [Google Scholar]
- 21.Li GD, Chiara DC, Cohen JB, Olsen RW. Numerous classes of general anesthetics inhibit etomidate binding to gamma-aminobutyric acid type A (GABAA) receptors. The Journal of biological chemistry. 2010;285:8615–8620. doi: 10.1074/jbc.M109.074708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen ZW, et al. Neurosteroid analog photolabeling of a site in the third transmembrane domain of the beta3 subunit of the GABA(A) receptor. Molecular pharmacology. 2012;82:408–419. doi: 10.1124/mol.112.078410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen ZW, Fuchs K, Sieghart W, Townsend RR, Evers AS. Deep amino acid sequencing of native brain GABAA receptors using high-resolution mass spectrometry. Molecular & cellular proteomics : MCP. 2012;11:M111 011445. doi: 10.1074/mcp.M111.011445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waud DR. On biological assays involving quantal responses. The Journal of pharmacology and experimental therapeutics. 1972;183:577–607. [PubMed] [Google Scholar]
- 25.Bhattacharya AA, Curry S, Franks NP. Binding of the general anesthetics propofol and halothane to human serum albumin. High resolution crystal structures. The Journal of biological chemistry. 2000;275:38731–38738. doi: 10.1074/jbc.M005460200. [DOI] [PubMed] [Google Scholar]
- 26.Bright DP, et al. Identification of anesthetic binding sites on human serum albumin using a novel etomidate photolabel. The Journal of biological chemistry. 2007;282:12038–12047. doi: 10.1074/jbc.M700479200. [DOI] [PubMed] [Google Scholar]
- 27.Kang SU, Fuchs K, Sieghart W, Lubec G. Gel-based mass spectrometric analysis of recombinant GABA(A) receptor subunits representing strongly hydrophobic transmembrane proteins. Journal of proteome research. 2008;7:3498–3506. doi: 10.1021/pr800236u. [DOI] [PubMed] [Google Scholar]
- 28.Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wooltorton JR, McDonald BJ, Moss SJ, Smart TG. Identification of a Zn2+ binding site on the murine GABAA receptor complex: dependence on the second transmembrane domain of beta subunits. The Journal of physiology. 1997;505 (Pt 3):633–640. doi: 10.1111/j.1469-7793.1997.633ba.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.James R, Glen JB. Synthesis, biological evaluation, and preliminary structure-activity considerations of a series of alkylphenols as intravenous anesthetic agents. Journal of medicinal chemistry. 1980;23:1350–1357. doi: 10.1021/jm00186a013. [DOI] [PubMed] [Google Scholar]
- 31.Cestari IN, Min KT, Kulli JC, Yang J. Identification of an amino acid defining the distinct properties of murine beta1 and beta3 subunit-containing GABA(A) receptors. Journal of neurochemistry. 2000;74:827–838. doi: 10.1046/j.1471-4159.2000.740827.x. [DOI] [PubMed] [Google Scholar]
- 32.Carlson BX, Engblom AC, Kristiansen U, Schousboe A, Olsen RW. A single glycine residue at the entrance to the first membrane-spanning domain of the gamma-aminobutyric acid type A receptor beta(2) subunit affects allosteric sensitivity to GABA and anesthetics. Molecular pharmacology. 2000;57:474–484. doi: 10.1124/mol.57.3.474. [DOI] [PubMed] [Google Scholar]
- 33.Li GD, et al. Identification of a GABAA receptor anesthetic binding site at subunit interfaces by photolabeling with an etomidate analog. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:11599–11605. doi: 10.1523/JNEUROSCI.3467-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall MA, et al. m-Azipropofol (AziPm) a photoactive analogue of the intravenous general anesthetic propofol. Journal of medicinal chemistry. 2010;53:5667–5675. doi: 10.1021/jm1004072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart DS, et al. p-(4-Azipentyl)propofol: a potent photoreactive general anesthetic derivative of propofol. Journal of medicinal chemistry. 2011;54:8124–8135. doi: 10.1021/jm200943f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evers AS, et al. A synthetic 18-norsteroid distinguishes between two neuroactive steroid binding sites on GABAA receptors. The Journal of pharmacology and experimental therapeutics. 2010;333:404–413. doi: 10.1124/jpet.109.164079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darbandi-Tonkabon R, et al. Photoaffinity labeling with a neuroactive steroid analogue. 6-azi-pregnanolone labels voltage-dependent anion channel-1 in rat brain. The Journal of biological chemistry. 2003;278:13196–13206. doi: 10.1074/jbc.M213168200. [DOI] [PubMed] [Google Scholar]
- 38.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical chemistry. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.