Abstract
Cutting edge immune monitoring techniques increasingly measure multiple functional outputs for various cell types, such as intracellular cytokine staining (ICS) assays that measure cytokines expressed by T cells. To date, however, there is no precise method to measure virus-specific cytokine production by both T cells as well as NK cells in the same well, which is important to a greater extent given recent identification of NK cells expressing a memory phenotype. This study describes an adaptable and efficient ICS assay platform that can be used to detect antigen-driven cytokine production by human T cells and NK cells, termed “viral ICS”. Importantly, this assay uses limited amount of cryopreserved PBMCs along with autologous heat-inactivated serum, thereby allowing for this assay to be performed when sample is scarce as well as geographically distant from the laboratory. Compared to a standard ICS assay that detects antigen-specific T cell cytokine expression alone, the viral ICS assay is comparable in terms of both HIV-specific CD4 and CD8 T cell cytokine response rates and magnitude of response, with the added advantage of ability to detect virus-specific NK cell responses.
Keywords: Intracellular cytokine staining, Flow cytometry, Cellular immunity, Viral infection, Immune monitoring
1. Introduction
A robust and reliable method of assessing cellular immunity is vital for several key areas of investigation, including infectious disease, vaccine, and cancer research. A limitation to the study of human immunology as opposed to use of murine or other animal model systems lies in the relative scarcity of human samples, both generally as well as from a particular individual. A lack of unlimited samples thus leads to prioritization of research questions such that only a limited set of assays can be performed on precious vials of PBMCs, thereby limiting the types of cells and immune outputs that can be studied.
The standard assays used to examine antigen-specific immune cell responses, ELISpot and intracellular cytokine staining (ICS), have long been used to measure antigen-driven human T cell responses in the context of many types of research, such as HIV, tumor immunology, and to evaluate vaccine immunogenicity. Both of these immune monitoring approaches, however, have limitations. Quantitation of antigen-specific T cell responses elicited by virus infection or vaccination have relied largely on the ELISpot assay, which yields a measurement on the quantity of a single cytokine produced in response to the antigen in question. Thus, while this assay provides a precise measurement of a single cytokine produced in response to antigen stimulation, it yields no information on what kind of cells produced this protein, nor does it allow us to assess the immune phenotype of the cellular sources. While FACS-sorting or magnetic bead enrichment of various cell populations of PBMCs prior to in vitro stimulation can allow for cytokine measurement by defined sub-populations of cells, this requires a larger number of cells, which are often not available in many research settings. ICS improves upon this information by allowing for further immunophenotyping of the cells responding to antigen, including multiple cytokine readouts as well as activation and phenotyping markers, since current technologies allow for more than 15 color flow cytometry. Thus, ICS is used frequently to quantify both the CD4+ and CD8+ T cell responses to various virus infections as well as vaccines.
Recently, NK cells in the context of virus infection were found to generate an “immune memory”, thus highlighting an unappreciated role for NK cells in immune control of virus infection (Foley et al., 2012; Sun, Beilke, and Lanier, 2009; Zhang et al., 2013). Due to these recent findings, there is renewed focus on this cell subset in the context of infectious disease research and in particular anti-viral immunity. Because both NK and T cells play a role in the immune response to a variety of viral infections, the goal of this assay is to be able to measure the antigen-specific immune response from both of these cell types using a single assay and a limited amount of PBMCs sample. Previous studies examining the HIV-driven cytokine expression by NK cells have used ICS of fresh whole blood (Meddows-Taylor et al., 2007; Stratov, Chung, and Kent, 2008; Tiemessen et al., 2009), which limits the researcher to use of local study cohorts. For studies including vaccine clinical trials and HIV research in areas that are sometimes geographically distant from the laboratory, logistics necessitate the use of cryopreserved samples, thus pointing to the need for an alternative assay.
Therefore, a standard T cell ICS assay was altered to include the ability to measure virus-driven cytokine production by NK cells within the same assay while using cryopreserved PBMCs, termed “viral ICS”. Addition of autologous serum to the well, as well as the use of phenotypic markers capable of identifying NK cells, resulted in a new ICS assay able to measure virus-driven cytokine production by T cells and NK cells for improved detection of immune responses vital to clearance of common viral infections in humans.
2. Methods
2.1 Study participants
Cryopreserved PBMCs and serum were obtained from 20 HIV-1 positive subjects from an HIV-1 prevention study in East Africa (Baeten et al., 2012). In addition, PBMCs from 20 HIV-1 seronegative subjects from the US with no known exposure to HIV were obtained. The study protocol was approved by the institutional review board at the University of Washington and African sites; all participants provided written informed consent.
2.2 In vitro stimulations
To determine virus specific T cell and NK cell responses, cryopreserved PBMCs were thawed and rested overnight in R10 (RPMI Media 1640 (Gibco, NY, USA), containing 10% FBS (Gemini Bio-Products, CA, USA), 2mM L-Glutamine (Gibco, NY, USA), 1X Penicillin Streptomycin (Gibco, NY, USA), 1mM Sodium Pyruvate (Gibco, NY, USA) and 10mM HEPES buffer solution (Gibco, NY, USA)) at 37°C/5% CO2. PMBC were resuspended at 10×106 cells/ml, then plated in a 96-well U-bottom plate at 1×106 cells/well and stimulated with (1) global potential T cell epitope peptides for HIV-1 Gag or Env, each including the 40 most frequent 15-mers among all sequences (Li et al., 2006); or (2) one (or more) of UL39 HSV-2 peptide pools (Posavad et al., 2010) in the presence of 10μg/ml Brefeldin A (Sigma-Aldrich Co., MO, USA), Golgi stop (BD Biosciences, CA, USA), and CD107a-APC (BD Biosciences, CA, USA). Peptide diluent (DMSO) served as a negative control and stimulation with 1μg/ml PMA (Sigma-Aldrich Co., MO, USA) and 1μM Ionomycin (Sigma-Aldrich, MO, USA) served as a positive control. Autologous serum was heat inactivated at 56°C for 30 minutes and 100μl was added to each well. After a 5 hour incubation at 37°C/5% CO2, 2mM EDTA was added to each well and placed at 4°C overnight.
2.3 Viral ICS protocol
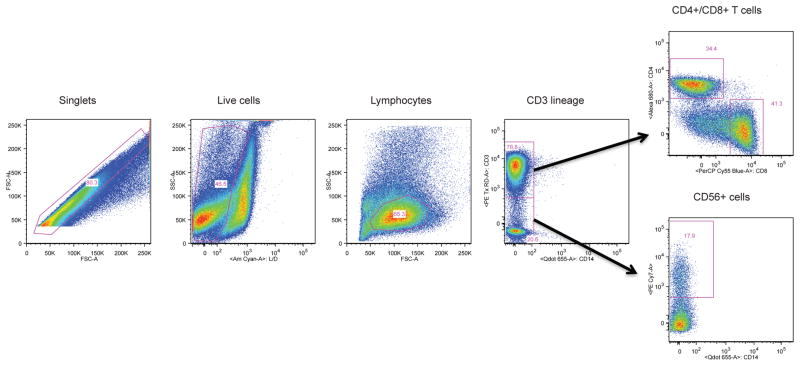
Live/Dead staining was done using a Live/Dead Fixable Aqua Dead Cell Stain Kit from Molecular Probes (OR, USA). All remaining antibodies were purchased through BD Biosciences (CA, USA), with the exceptions of CD3 (Beckman Coulter, Marseille France) and CD14 (Molecular Probes, CA, USA). The complete panel is reported in Table 1. All antibodies were titrated and Fluorescence minus one (FMO) performed to optimize the gating scheme. Samples were collected using a High Throughput Sample device on a LSRII flow cytometer (BD Biosciences, CA, USA) immediately following staining. The LSRII flow cytometer used for this study was configured with four lasers (violet, blue, green, red) and 20 detectors (18 fluorescence detectors) as described previously (De Rosa, Carter, and McElrath, 2012). Fig. 1 shows the gating scheme. Flow cytometry analysis was performed using Flowjo software (Treestar, Inc, OR).
Table 1.
Staining Panel used for two methods of Intracellular Cytokine Staining.
| Format | Standard ICS | Viral ICS |
|---|---|---|
| PE | -- | MIP1β |
| FITC | TNFα | TNFα |
| ECD | CD3 | CD3 |
| Qdot655 | CD14 | CD14 |
| PerCP Cy5.5 | CD8 | CD8 |
| APC | -- | CD107a |
| APC Cy7 | CD4 | -- |
| Alexa700 | MIP1β | CD4 |
| PE Cy7 | CD107a | CD56 |
| V450 | IFNγ | IFNγ |
| Am Cyan | Live/dead | Live/dead |
Figure 1. Staining profile for identification of T cells and NK cells.
The plots show sequentially, left to right, the gating hierarchy for PBMCs from an HIV-positive individual: Single cells, live cells, lymphocytes, CD3+ cells, further gated for CD4+ and CD8+ T cells, and CD3−CD14+ cells, further gated for CD56+ NK cells.
2.4 Standard ICS protocol
Stimulations and staining for the standard ICS were done according to published methods (Horton et al., 2007). The complete antibody panel is reported in Table 1.
2.5 Statistical analysis
The Wilcoxon signed-rank test was used to compare the magnitude of responses across the two ICS assays as shown in Fig. 3. Peptide stimulated wells were classified as having positive or negative virus specific T cell and NK cell responses by comparing the proportion of cells responding in stimulated wells to the proportion responding in the negative control well using MIMOSA (Mixture Models for Single-Cell Assays), a recently published statistical model (Finak et al., 2013). We accepted a false discovery rate of 5%. Spearman’s correlation coefficient was calculated to determine the correlation between results obtained with the two assays. Statistical analyses were performed using SAS 9.3.
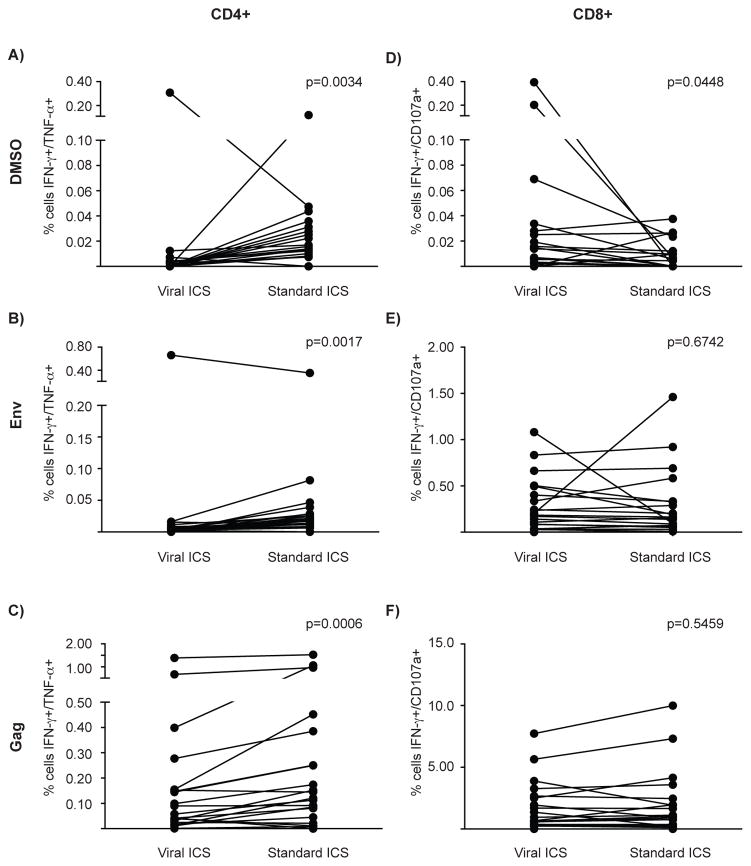
Figure 3. Magnitude of the responses detected by the two different assays.
After five hours of stimulation with DMSO (top row), Env peptide pool (middle row) or Gag peptide pool (bottom row), the responses from 20 HIV-positive individuals were evaluated for CD4+ T cells (IFNγ and TNFα) and for CD8+ T cells (IFNγ and CD107a). Individual plots show the frequency of cytokine producing/degranulating cells detected by viral and standard ICS. The Wilcoxon signed-rank test was used to generate p values as reported in the figure.
3. Results
3.1 Generation of the Viral ICS assay
In order to create the viral ICS flow cytometry panel, 10-color flow was used to maximize the immune parameter outputs. Cells and cytokines critical to combat viral infection were the chosen as the focus: NK cells and CD4+ and CD8+ T cells producing MIP1β, TNFα, and IFNγ, with the inclusion of CD107a (LAMP1) to assess degranulation of these effector cells (Table 1). The benefit of this viral ICS method lies in the ability to analyze NK cell responses in a single well along with the T cell responses, whereas the “standard ICS” method includes the same cytokines but does not include NK cell markers.
To identify these anti-viral immune effector cell populations, single live cells within the lymphocyte size and granularity gate were plotted for CD3 versus CD14 to enable further gating of CD3+ T cells into CD4 and CD8 populations. Cells double negative for CD14 and CD3 were examined for CD56+ NK cells (Fig. 1). These three effector populations were next examined for cytokine production and gating was achieved using fluorescence minus one (FMO) controls.
3.2 Detection of HIV-specific CD4 T cell responses
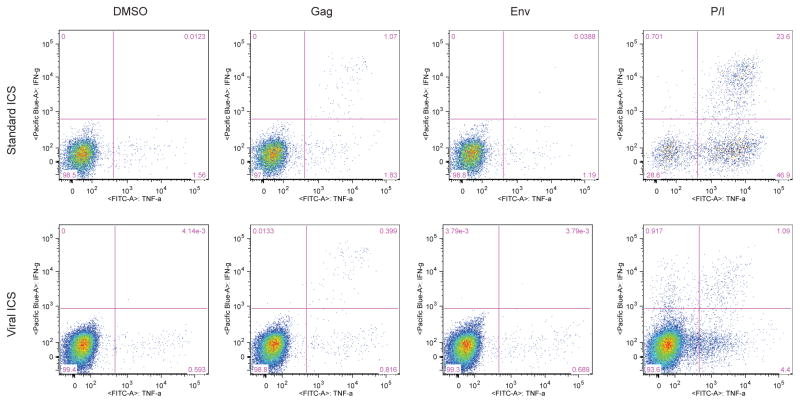
The viral ICS was compared to a standard T cell ICS assay in order to determine if addition of serum to PBMCs compromised the ability to detect virus-specific T cell responses. Specifically, the HIV-specific T cell response was examined in 20 HIV-positive individuals using stimulation with HIV peptide pools from Env and Gag. Positive responses were calculated using the statistical package MIMOSA with a false discovery rate of 5% applied to the values obtained for CD4+ T cells producing both IFNγ and TNFα (representative flow plots shown in Fig. 2). Most HIV-specific CD4 T cell responses were to Gag using either method, and more responses were detected using the viral ICS assay as compared to the standard ICS (95% versus 75% of individuals, respectively; Table 2). When the frequency of Gag-stimulated CD4 T cells producing IFNγ and TNFα was examined, the standard ICS assay revealed a higher frequency of cytokine producing cells (p=0.0006; Fig. 3C), despite the viral ICS assay yielding a higher response rate for CD4 T cell responses to Gag (Tables 2). Although the focus was on the double positive cells to identify the positive responses in order to decrease the chance of false discovery, the percentages of single cytokine-secreting cells was also examined. Table 3 reports the medians and interquartile ranges (IQR) of single and double cytokine-secreting cell frequencies. Significant correlations were observed between the two tests following Gag stimulation for single and double cytokine producers, with spearman rho values of 0.7821 for IFNγ and TNFα producing cells, 0.7616 for IFNγ- and 0.8396 for TNFα-producing cells (p<0.001). There was no statistically significant correlation for the negative control, DMSO-treated PBMCs; in fact, although with both methods few CD4 T cells expressed cytokines, their frequencies were lower using viral ICS (p=0.0034; Fig. 3A), demonstrating lower background for viral ICS. Only one individual was found to have an Env-specific CD4 T cell response, by both viral and standard ICS (Fig. 3B).
Figure 2. Representative flow plots of ICS results for CD4+ T cells by two methods.
The plots show the gating for a representative sample from an HIV-positive individual stimulated for five hours with DMSO, PTE peptide pools for Gag and Env and PMA/Ionomycin. Plots show the co-expression of IFNγ and TNFα by CD4+ T cell as detected by standard ICS (top row) and viral ICS (bottom row).
Table 2. Rate of HIV-specific response for CD4 T, CD8 T, and NK cells in HIV-positive and HIV-negative individuals.
A positive response was considered to be double positive for TNFα and IFNγ (CD4 T cells) or for CD107a and IFNγ (CD8 T cells and NK cells).
| HIV-Positive | HIV-Negative | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| N tested | Standard ICS | Viral ICS | N tested | Standard ICS | Viral ICS | |
|
| ||||||
| CD4 T cell | 20 | 20 | ||||
| Env | 1 (5.0) | 1 (5.0) | 0 | 0 | ||
| Gag | 15 (75.0) | 19 (95.0) | 0 | 0 | ||
|
| ||||||
| CD8 T cell | 20 | 20 | ||||
| Env | 18 (90.0) | 18 (90.0) | 0 | 0 | ||
| Gag | 19 (95.0) | 20 (100.0) | 0 | 0 | ||
|
| ||||||
| CD56+ NK cell | 20 | 20 | ||||
| Env | -- | 14 (70.0) | -- | 0 | ||
| Gag | -- | 20 (100.0) | -- | 0 | ||
|
| ||||||
| CD56− NK cell | 20 | 20 | ||||
| Env | -- | 10 (50.0) | -- | 0 | ||
| Gag | -- | 0 (0.0) | -- | 0 | ||
Table 3. Magnitude of CD4+ T cell responses by viral and standard ICS.
Medians and interquartile ranges (IQR) were calculated for the frequencies of CD4+ T cells secreting both IFN-γ and TNF-α or only one of either cytokine.
| Stim | Median (IQR) | Wilcoxon signed rank p value | ||
|---|---|---|---|---|
| Viral ICS | Traditional ICS | |||
| IFNγ/TNFα | DMSO | 0.0020 (0.0000, 0.0030) | 0.0090 (0.0050, 0.0150) | 0.0034 |
| Env | 0.0040 (0.0010, 0.0070) | 0.0060 (0.0000, 0.0180) | 0.0017 | |
| Gag | 0.0100 (0.0030, 0.0740) | 0.0090 (0.0030, 0.1330) | 0.0006 | |
| IFNγ | DMSO | 0.0030 (0.0000, 0.0070) | 0.0110 (0.0070, 0.0190) | 0.0095 |
| Env | 0.0070 (0.0040, 0.0110) | 0.0090 (0.0040, 0.0220) | 0.0897 | |
| Gag | 0.0140 (0.0040, 0.0920) | 0.0140 (0.0070, 0.1560) | 0.0007 | |
| TNFα | DMSO | 0.5520 (0.3300, 0.6880) | 0.7050 (0.2670, 1.5220) | <.0001 |
| Env | 0.6300 (0.2990, 0.7460) | 0.4490 (0.1420, 1.2150) | 0.0027 | |
| Gag | 0.6540 (0.3330, 0.8670) | 0.5520 (0.1200, 1.3260) | 0.0006 | |
Finally, to test the specificity of the viral ICS assay, the two ICS assays were also compared using 20 HIV-unexposed and −negative individuals. Both the viral as well as the standard ICS assays found zero responses for CD4 T cells stimulated with Env or Gag peptide pools (Table 2).
3.3 Detection of HIV-specific CD8 T cell responses
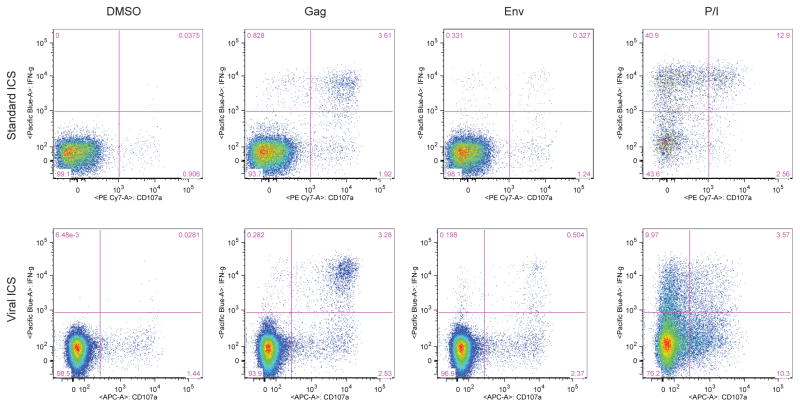
The HIV-specific CD8 T cell response was examined next. For both ICS assays, IFNγ and CD107a were considered for positivity (Fig. 4). Nearly all of the 20 HIV-positive individuals had detectable Gag- and Env-specific CD8 T cell responses, though there were differences depending on the assay used. Both ICS assays yielded a 90% response rate using an Env peptide pool (Table 2), though each assay failed to detect a single response that the other did. Viral ICS detected 100% Gag-specific CD8 T cell responses as compared to 95% for standard ICS (Table 2).
Figure 4. Representative flow plots of ICS results for CD8+ T cells by two methods.
The plots show the gating for a representative HIV-positive sample stimulated for five hours with DMSO, PTE peptide pools for Gag and Env and PMA/Ionomycin. Plots show the co-expression of CD107a and IFNγ by CD8+ T cells as detected by standard ICS (top row) and viral ICS (bottom row).
Next, the magnitude of HIV-specific CD8 T cell responses was compared between the two ICS method. The negative control, DMSO-treated PBMCs, generally resulted in low frequencies of CD8 T cells producing cytokine, although there were a few instances of high levels of background with viral ICS (p=0.0448 vs. standard ICS; Fig. 3D). Despite this difference, however, the frequency of Env- or Gag-stimulated CD8 T cells double positive for IFNγ and CD107a was similar regardless of the assay used (p=0.6742 and p=0.5459, respectively; Fig. 3E–F and Table 4). Accordingly, the Spearman’s rho was 0.81129 for Gag and 0.8832 for Env stimulation (p<0.001). The same trend was observed when the responses using a single marker were analyzed (Table 4).
Table 4. Magnitude of the CD8+ T cell responses by viral and standard ICS.
Medians and IQR were calculated for the frequencies of CD8+ T cells secreting both IFN-γ and TNF-α or only one of either cytokine.
| Stim | Median (IQR) | Wilcoxon signed rank p value | ||
|---|---|---|---|---|
| Viral ICS | Traditional ICS | |||
| IFNγ/CD107a | DMSO | 0.0030 (0.0000, 0.0120) | 0.0000 (0.0000, 0.0100) | 0.0448 |
| Env | 0.0180 (0.0000, 0.1940) | 0.0100 (0.0000, 0.1600) | 0.6742 | |
| Gag | 0.0160 (0.0020, 0.8040) | 0.0100 (0.0000, 0.1600) | 0.5459 | |
| IFNγ | DMSO | 0.8270 (0.5340, 1.2500) | 0.2340 (0.1070, 0.5190) | <.0001 |
| Env | 1.0130 (0.5800, 1.7680) | 0.4280 (0.2480, 0.8460) | <.0001 | |
| Gag | 1.2850 (0.6530, 2.7810) | 0.7330 (0.1930, 1.6390) | 0.114 | |
| CD107a | DMSO | 0.0090 (0.0030, 0.0300) | 0.0150 (0.0060, 0.0360) | 0.5949 |
| Env | 0.0390 (0.0050, 0.2750) | 0.0260 (0.0040, 0.2120) | 0.2455 | |
| Gag | 0.0660 (0.0060, 0.9940) | 0.0540 (0.0090, 1.4300) | 0.0532 | |
Finally, the two ICS assays were compared using 20 HIV-unexposed and −negative individuals. Both the viral as well as the standard ICS assays found zero responses for CD8 T cells stimulated with Env or Gag peptide pools (Table 2).
3.4 Detection of HIV-driven NK cell responses
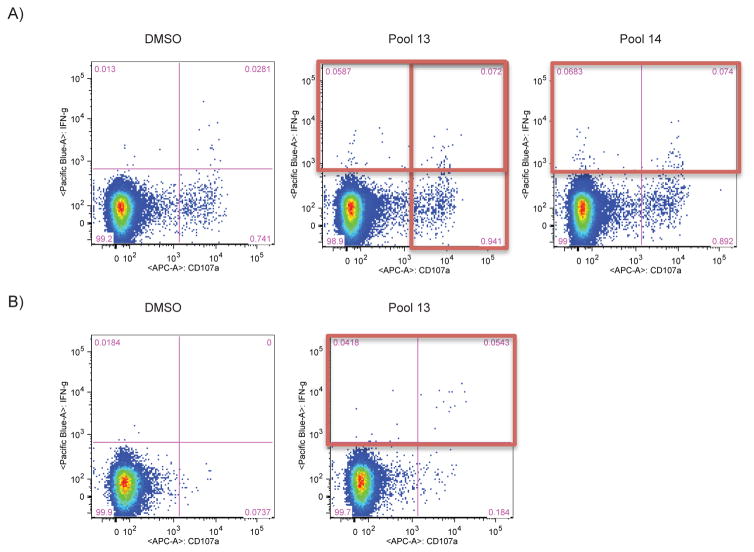
The viral ICS assay allows for the detection of antigen-specific NK cell responses along with T cell responses using a single well of PBMCs together with heat-inactivated autologous serum. Both CD56+ and CD56− NK cells were analyzed, after excluding CD3+ T cells and CD14+ monocytes, as it was reported previously that CD56− NK cells from HIV+ subjects could produce IFNγ as well (Stratov et al., 2008). To be considered a positive response, it was determined that NK cells must test positive for both IFNγ as well as CD107a (Fig. 5). Use of the viral ICS assay with 20 HIV-positive individuals revealed a 70% Env-specific NK cell response rate and a 10% Gag-specific NK cell response (Table 2). The magnitude of the response, measured as median and interquartile range, increased from 0.1060 (0.0570–0.3760) for the negative control to 0.6600 (0.2240–1.9640) with Env stimulation. A lower response rate was observed for the CD56− NK cells: the response rate was 50% with Env stimulation and 0% upon Gag stimulation. These responses are likely HIV-specific, as there were no positive responses found when 20 HIV-negative individuals were tested using viral ICS (Table 2).
Figure 5. Representative flow plots of ICS results for CD56+ NK cells by viral ICS.
The plots show the gating for a representative HIV-positive sample stimulated for five hours with DMSO, PTE peptide pools for Gag and Env and PMA/Ionomycin. Plots show IFNγ secretion and CD107a externalization by CD56+ cells as detected by viral ICS.
3.5 Detection of HSV-specific responses
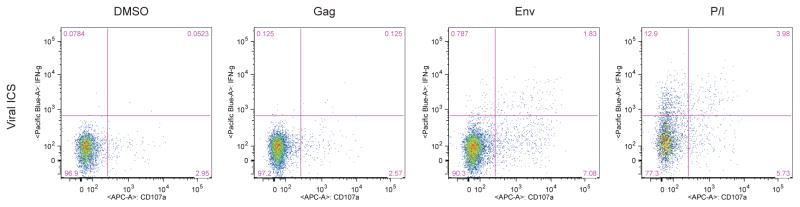
In addition to testing HIV-specific immune effector cell responses, a goal was to determine if the viral ICS assay could be useful for detecting virus-specific immune responses in general. Therefore, viral ICS was performed on a subset of the HIV-positive individuals that were also seropositive for HSV-2 (n=10). Specifically, viral ICS was performed using peptide pools spanning HSV-2 viral protein UL39, as it was found to stimulate CD8 T cell responses from a majority of HSV-2-positive individuals (Hosken et al., 2006). Although screening with these limited peptide pools identified only a few positive responses, representative CD8 T cell responses specific to HSV-2 are depicted in Fig. 6 when either IFNγ or CD107a positivity alone are used as an indication of a positive response. This demonstrates that viral ICS may be a useful tool to examine frequency and magnitude of various virally-induced immune responses.
Figure 6. Detection of HSV-specific CD8+ T cell responses using viral ICS.
After stimulation with four different HSV-2 UL39 peptide pools, CD8+ T cell responses were detected by production of IFNγ or externalization of CD107a by CD8+ T cells upon stimulation with two peptide pools in one sample (ID 3) (A) and by production of IFNγ upon stimulation with one peptide pool in another sample (ID 4) (B). PBMCs were from HIV- and HSV-2-positive individuals.
4. Discussion
The ICS assay described in this study allows for the quantitation of the virus-specific T cell as well as the NK cell response using a single aliquot of cryopreserved PBMCs along with autologous serum. With the advent of multi-color flow cytometry, standard immune monitoring techniques to a greater extent include analysis of multiple cell types and functional outcomes. To demonstrate that this test yields results comparable to a standard ICS assay without addition of autologous serum, both assays were performed on 20 samples from HIV-1-seropositive subjects and 20 from seronegatives with no known exposure to HIV. Importantly, addition of autologous serum to the stimulation cocktail did not lead to an increased background; in fact, the DMSO stimulation resulted in a very low background in both assays, as shown in Figure 3. Furthermore, neither ICS assay uncovered any HIV-1-specific T cell responses in these HIV-negative and unexposed individuals, indicating the specificity of the assays.
Although comparable response rates were observed applying the two ICS methods, some differences were identified. In four samples, there was a detectable CD4+ T cell response by the viral ICS and not with the standard one; in the same way, a CD8+ T cell response was detected only with the viral ICS in two samples. In only one case, the standard ICS showed a response to Env, which was not identified by the viral ICS. Overall, the viral ICS seems to be a more sensitive method as compared to the standard one, demonstrating that addition of serum does not compromise the ability to detect antigen-specific T cell responses, and indeed may lead to enhanced detection of virus-driven cytokine expression.
When comparing the magnitude of the T cell responses, the CD4+ T cell responses varied between the standard and viral ICS methods, with a statistically significant increase resulting when the standard assay was performed. Thus, it appears that the viral ICS is capable of detecting more low-level CD4 T cell responses to Gag (Table 2) and so is perhaps more sensitive, but the standard ICS measures more robust responses (Fig. 3).
The key advantage of the viral ICS assay is that it allows for detection of HIV-specific NK cell responses. By using the assay, in fact, NK cell responses to Env were detectable in 90% of the samples and to Gag in 100% of the samples. Although it is not possible to exclude that these NK cell responses are dependent on T cell activation through cell-to-cell contact, previous findings support the hypothesis that the observed degranulation and IFNγ secretion by NK cells are due to HIV-specific antibodies present in the serum (Stratov et al., 2008). By binding to the HIV peptides, these immunoglobulins induce activation of antibody-dependent cell mediated cytolysis. This hypothesis thus explains the need for addition of heat-inactivated serum to observe the NK cell activity, and would indicate that the NK cells themselves may not be imprinted with “memory”, but rather it is HIV-specific antibodies present in the autologous serum that leads to this antigen-specific memory effect. Alternatively, the assay could be detecting NK cell responses of a heterogeneous nature, in that some of the cytokine-producing cells could be “classical” antigen-specific memory cells as detected in transplant recipients (Foley et al., 2012). However, regardless of whether NK cells are using gene-rearranged Ag-specific receptors or germline-encoded FcR that recognize antibody bound to virally-encoded peptides, this novel assay allows for detection of NK cells that produce cytokine in response to viral antigens, and so can be used to further study the phenomenon of NK cell memory.
Importantly, inclusion of multiple cytokines in this assay allows for customization of the positivity calls. Two parameters for each cell type were chosen based on their functional role, in part to reduce identification of false positive responses. Additionally, measuring polyfunctional cells increases the likelihood that HIV-protective effector cell responses are detected, as it has been shown previously that there are increased frequencies of polyfunctional T cells in LTNP as compared to HIV-1-progressors (Betts et al., 2006). IFNγ and TNFα were used for CD4+ T cells, whereas CD107a, an indicator for degranulation, was included with IFNγ for these cytotoxic CD8+ T and NK cells. However, customized parameters will certainly be appropriate when evaluating immune responses to different antigens, such HSV-2 (Fig. 6).
In conclusion, the viral ICS assay detailed in this study is an adaptable, efficient ICS assay platform that can be used to detect HIV-driven cytokine production by T cell and NK cells, and could be further extended in the future to include even more complex cellular systems, such as dendritic cell-NK cell crosstalk (Gondois-Rey et al., 2012). Further, new methods in mass cytometry such as mass-tag cellular barcoding (Bodenmiller et al., 2012) could further enhance the use of the viral ICS assay in the future, thereby increasing the high-throughput capacity of the approach. Importantly, the viral ICS assay can be performed using cryopreserved PBMCs and heat-inactivated serum, thereby allowing this assay to be used in settings where fresh cells are not possible, such as in the context of a clinical trial or when samples are geographically distant. Finally, the viral ICS assay can be used to detect responses to multiple viral infections as well as to potentially detect antigen-specific cellular responses when studying non-communicable diseases such as cancer.
Highlights.
Our novel method detects anti-viral T and NK cell responses concurrently.
Viral ICS is antigen-specific and has a low background.
Viral ICS detects comparable cytokine response rates as compared to standard ICS.
Acknowledgments
We would like to acknowledge Antje Heit for helpful discussions concerning NK cells, Michael Bolton for technical assistance with setting up the method, Greg Finak for assistance with the MIMOSA package, Christine Posavad for supplying the HSV peptide pools and for insightful discussion and Julie McElrath for helpful discussions and support. We thank the James B. Pendleton Charitable Trust for their generous equipment donation. We are grateful to the research staff and study participants in the Partners PrEP study who made this study possible. Funding for this study was provided by NIAID/NIH 1R01AI096968 (to JMB and JML).
Abbreviations
- ICS
Intracellular cytokine staining
- HIV-1
Human Immunodeficiency Virus
- PBMCs
peripheral blood mononuclear cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baeten JM, Donnell D, Ndase P, Mugo NR, Campbell JD, Wangisi J, Tappero JW, Bukusi EA, Cohen CR, Katabira E, Ronald A, Tumwesigye E, Were E, Fife KH, Kiarie J, Farquhar C, John-Stewart G, Kakia A, Odoyo J, Mucunguzi A, Nakku-Joloba E, Twesigye R, Ngure K, Apaka C, Tamooh H, Gabona F, Mujugira A, Panteleeff D, Thomas KK, Kidoguchi L, Krows M, Revall J, Morrison S, Haugen H, Emmanuel-Ogier M, Ondrejcek L, Coombs RW, Frenkel L, Hendrix C, Bumpus NN, Bangsberg D, Haberer JE, Stevens WS, Lingappa JR, Celum C Partners Pr,EPST. Antiretroviral prophylaxis for HIV prevention in heterosexual men and women. The New England journal of medicine. 2012;367:399–410. doi: 10.1056/NEJMoa1108524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–9. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, Simonds EF, Bendall SC, Sachs K, Krutzik PO, Nolan GP. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nature biotechnology. 2012;30:858–67. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa SC, Carter DK, McElrath MJ. OMIP-014: validated multifunctional characterization of antigen-specific human T cells by intracellular cytokine staining. Cytometry Part A: the journal of the International Society for Analytical Cytology. 2012;81:1019–21. doi: 10.1002/cyto.a.22218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, McDavid A, Chattopadhyay P, Dominguez M, De Rosa S, Roederer M, Gottardo R. Mixture models for single-cell assays with applications to vaccine studies. Biostatistics. 2013 doi: 10.1093/biostatistics/kxt024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley B, Cooley S, Verneris MR, Curtsinger J, Luo X, Waller EK, Anasetti C, Weisdorf D, Miller JS. Human cytomegalovirus (CMV)-induced memory-like NKG2C(+) NK cells are transplantable and expand in vivo in response to recipient CMV antigen. Journal of immunology. 2012;189:5082–8. doi: 10.4049/jimmunol.1201964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondois-Rey F, Granjeaud S, Kieu Sle T, Herrera D, Hirsch I, Olive D. Multiparametric cytometry for exploration of complex cellular dynamics. Cytometry Part A: the journal of the International Society for Analytical Cytology. 2012;81:332–42. doi: 10.1002/cyto.a.22016. [DOI] [PubMed] [Google Scholar]
- Horton H, Thomas EP, Stucky JA, Frank I, Moodie Z, Huang Y, Chiu YL, McElrath MJ, De Rosa SC. Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. Journal of immunological methods. 2007;323:39–54. doi: 10.1016/j.jim.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosken N, McGowan P, Meier A, Koelle DM, Sleath P, Wagener F, Elliott M, Grabstein K, Posavad C, Corey L. Diversity of the CD8+ T-cell response to herpes simplex virus type 2 proteins among persons with genital herpes. Journal of virology. 2006;80:5509–15. doi: 10.1128/JVI.02659-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Malhotra U, Gilbert PB, Hawkins NR, Duerr AC, McElrath JM, Corey L, Self SG. Peptide selection for human immunodeficiency virus type 1 CTL-based vaccine evaluation. Vaccine. 2006;24:6893–904. doi: 10.1016/j.vaccine.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Meddows-Taylor S, Shalekoff S, Kuhn L, Gray GE, Tiemessen CT. Development of a whole blood intracellular cytokine staining assay for mapping CD4(+) and CD8(+) T-cell responses across the HIV-1 genome. Journal of virological methods. 2007;144:115–21. doi: 10.1016/j.jviromet.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posavad CM, Remington M, Mueller DE, Zhao L, Magaret AS, Wald A, Corey L. Detailed characterization of T cell responses to herpes simplex virus-2 in immune seronegative persons. Journal of immunology. 2010;184:3250–9. doi: 10.4049/jimmunol.0900722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratov I, Chung A, Kent SJ. Robust NK cell-mediated human immunodeficiency virus (HIV)-specific antibody-dependent responses in HIV-infected subjects. Journal of virology. 2008;82:5450–9. doi: 10.1128/JVI.01952-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–61. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemessen CT, Shalekoff S, Meddows-Taylor S, Schramm DB, Papathanasopoulos MA, Gray GE, Sherman GG, Coovadia AH, Kuhn L. Cutting Edge: Unusual NK cell responses to HIV-1 peptides are associated with protection against maternal-infant transmission of HIV-1. Journal of immunology. 2009;182:5914–8. doi: 10.4049/jimmunol.0900419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Scott JM, Hwang I, Kim S. Cutting edge: antibody-dependent memory-like NK cells distinguished by FcRgamma deficiency. Journal of immunology. 2013;190:1402–6. doi: 10.4049/jimmunol.1203034. [DOI] [PMC free article] [PubMed] [Google Scholar]