It has been more than 20 years since the first micro total analysis systems (μTAS) papers were published. Initial reports of these devices, which are also commonly referred to as Labs-on-a-Chip (LOC), LabChips, microchips or microfluidic devices, generally focused on separations and the development of a variety of functional elements for sample manipulation and handling. One of the greatest potentials of μTAS, however, has always been in the integration of multiple functional elements to produce truly sample-in/answer-out systems. In the last decade, the march toward developing such integrated devices has accelerated significantly. Many μTAS reported now are quite sophisticated with multiple sample handling and processing steps that are highly integrated and often automated. While most of these devices are not yet strictly sample-in/answer-out several come quite close. There are, however, some significant hurdles still facing the development of true sample-in/answer-out systems especially in the areas of sample preparation, chip-to-real-world interfacing and detection. Additionally, further progress is needed in the miniaturization or elimination of external fluidic control elements.
μTAS have found a major niche in the areas of biological and biomedical analyses, especially cellular and nucleic acid analysis. This focus on biological applications reflects the capabilities of these devices to precisely and accurately handle picoliter volumes of materials and to integrate cell transport, culturing or trapping with reagent delivery, and on-chip detection. Significant progress has been made in the development of a variety of cellular analysis systems; this field, however, is still rapidly growing and many papers focused on the expansion of such capabilities continue to be seen. Areas of focus remain the development of substrate materials and culturing conditions that do not unnaturally perturb or stress cells and that allow for extended culturing so that changes in cell physiology over time can be monitored. In addition, a significant amount of work has been directed to developing cell co-cultures on μTAS to mimic tissues, organs, and organ systems. μTAS can create unique, controlled environments to study cell-cell interactions that can not be replicated in any other way. For cellular assays substantial increases in throughput are also a focus. While significant development toward completely integrated cell assays has occurred and even some clinical demonstrations of such assays have been reported, the availability of commercial, fully integrated devices, however, has lagged.
In addition to biological assays, the creative expansion of the basic μTAS toolkit with centrifugal platforms, digital microfluidics, and paper-based devices has substantially expanded its potential application base. Interest in these devices is generally more clinical in nature and again focused on generating sample-in/answer-out analyses. Significant work, however, is still needed for most of these platforms in terms of substrate materials, fluid control, sample handling, integration and throughput. Finally, the development of label-free detection technologies remains of interest.
This review focuses on recent advances in μTAS technology in the areas of integrated biological assays and diagnostics with an analytical focus. We have also tried to highlight some material, fabrication, coating, separation, and detection advances with more general applicability. We have not included, for the most part, papers on synthesis, biosensors, theory, simulations or reviews. The papers included in this review were published between September 2012 and September 2013. The material was compiled using several strategies including extensive searches using Scifinder, Web of Science, PubMed, and Google Scholar. The contents of high impact journals were also scanned, including Analytical Chemistry, Lab-on-a-Chip, Nature, PNS, Appl. Phys., Letters, and Langmuir. Almost 2000 papers relating in some way to microfluidics were examined. We have done our best to try to identify some of the most interesting and promising papers and to report on them in this review. Without a doubt we have missed a few excellent papers and had to eliminate others based on space constraints and readability. For those papers that we have failed to include, we apologize in advance and welcome comments regarding any oversight that we have made.
Fundamentals
Fabrication
Materials
Poly(dimethylsiloxane) (PDMS) is by far the most popular material for the fabrication of microfluidic devices due in most part to easy fabrication and low cost. Most of the devices discussed in this review were made using PDMS. PDMS does, however, have its limitations as a substrate material. It is quite hydrophobic and difficult to wet, will absorb hydrophobic analytes, can be toxic to some cell types1 and generates a low electroosmotic flow. As such, considerable effort has been invested in developing coatings for PDMS to modify its surface properties and these will be discussed under the surface modification section of this review. While PDMS molding techniques are quite mature, ongoing interest in this material is focused on modifications to the fabrication process or to the chemical composition of PDMS itself. For example, novel 3-D PDMS structures, i.e. tubes, were fabricated through the inhomogeneous swelling of thin films of PDMS in chloroform vapor (Figure 1A).2 Silver nanoparticles embedded in the film were then used as 3-D heaters or solenoids. PDMS was also modified to create long-pass filters for fluorescent emission.3 The addition of a UV-absorbing chromophore to a ∼5μm thick film of PDMS rejected UV light with an efficiency of -40dB at 342 nm making this material potentially suitable as a long pass filter for laser induced fluorescence (LIF) applications.
Figure 1.
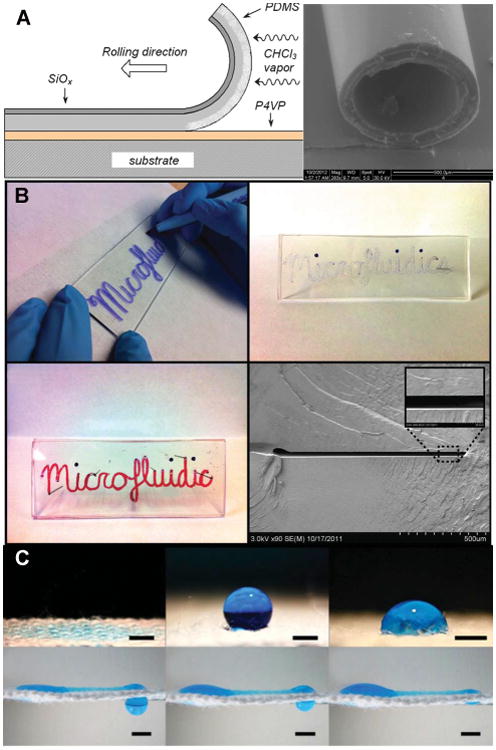
(a) Innovative fabrication methods and materials. (a) The production of self-rolled PDMS microcapillaries. The rolling process and a SEM image are shown. Reprinted with permission from Chia, G. L. P.; Bollgruen, P.; Egunov, A. I.; Mager, D.; Malloggi, F.; Korvink, J. G.; Luchnikov, V. A. Lab Chip 2013, 13, 3827-3831. Copyright 2013 The Royal Society of Chemistry. (b) COC device fabrication procedure uses a wet-erase pen to define a channel pattern on a COC chip followed by swelling and solvent bonding. SEM image shows the cross-section of a sealed microchannel. Reprinted with permission from Rahmanian, O.; DeVoe, D. L. Lab Chip 2013, 13, 1102-1108. Copyright 2013 The Royal Society of Chemistry. (c) The wetting and transport by surface tension driven flow of a 5uL droplet upon micropatterned superhydrophobic textile. Reprinted with permission from Xing, S.; Jiang, J.; Pan, T. Lab Chip 2013, 13, 1937-1947. Copyright 2013 The Royal Society of Chemistry.
While PDMS is the most common microfluidic substrate material, a variety of other materials are also used. Glass, while more expensive than PDMS in both materials and processing costs, still has some advantages in terms of known surface chemistries, transparency and high thermal conductivities. In fact many different commercial glass microfluidic devices are readily available. Glass devices are generally fabricated using a combination of photolithographic and wet-etching processes followed by thermal annealing to form closed channels. This is a time consuming process. A novel Ag particle masking agent, however, was shown to speed the fabrication process and to make it less expensive.4 The Ag film serves as both the photoresist and etch mask. It was vapor deposited in a variety of thicknesses and allowed etching to 300+ μm. An alternative to masking involves the use of a programmable proximity lithography (PPAL) system. An improved PPAL system using a six MeV ion beam was used to create sub 100μm features in soda-lime glass.5 Importantly, after developing and cleaning, these open-face features were successfully annealed to create enclosed channels.
The use of PDMS is popular in academic labs, however, other polymers, such as poly(methylmethacrylate) (PMMA) and cyclic olefin copolymers (COCs), are more amenable to high volume manufacturing techniques such as hot embossing and injection molding. Devices seeking to take advantage of these properties included an integrated PMMA device with single use valves for generating a sample-in-to-PCR-result-out in 45 min,6 and a COC device for the analysis of banned aromatic amines.7 In another COC device, the channels were created using a unique solvent swelling and sealing technique.8 This device is further discussed in the bonding section below. Polystyrene is of particular interest in the area of cell culturing, as most standard cell culture flasks are polystyrene. Polystyrene was molded against a PDMS master to create a channel manifold that integrated multiple sample handling, processing and electrochemical detection functions.9 The device was successfully used to culture endothelial and PC12 cells, and to monitor the release of endogenous species upon external stimulation.10 Polystyrene devices are likely to become more popular in the near future for cell-based assays because the interactions between polystyrene and cells are well understood and biologists are more comfortable using the material.11 μTAS can also be fabricated directly from a laser printer using polyester transparencies providing another avenue for high throughput production. Channels from 5 to 10 μm deep can be printed and then sealed using a laminator. This technique affords a very cheap and readily available fabrication source. Recent efforts in this area have focused on increasing the chip lifetime and on improved analytical performance.12 For highly corrosive materials, special polymers such as poly(vinylidene fluoride) (PVDF) must be used. A novel PVDF device with embedded gold electrodes was fabricated via embossing and welding to monitor droplets.13 Channel manifolds have also been recently created using UV-curable adhesives.14 After an initial curing to form the channel manifold, the adhesive substrate with the patterned channels in it was then sealed against a glass slide.
Hydrogels are suitable materials for cell culturing, but they are not generally used as a μTAS substrate material as they actively absorb water and swell. Cross-linked cellulose, however, was shown to possess an excellent structural replication ability, good mechanical properties and cell compatibility.15 Such a device was used to culture endothelial cells. The porous nature of the substrate allowed for the generation of chemical gradients between closely spaced parallel channels that would otherwise have been impossible. These gradients were used to investigate cellular responses. Microfluidic channels were also formed in collagen. These collagen devices were then seeded with endothelial cells to study vascular growth in response to gradients in the extracellular (hydrogel) matrix.16 The ability to use these biocompatible materials with microfluidic channels should lead to further improvements in the ability of μTAS to realistically mimic in vitro environments.
PDMS-glass hybrid devices are common, but other hybrid-type devices have potential advantages in terms of integrating detection and control electronics with fluidic channels. One popular approach to fabricating such devices is the integration of printed circuit boards (PCB)s with fluidic layers.17 A hybrid PCB-polyurethane device was used for on-chip mixing, cell lysis and nucleic acid extraction (Figure 9E). This device included integrated heating elements.18 Silicon-based CMOS devices can also be integrated with microfluidics. A silicon-based CMOS potentiostat was combined with an SU-8 channel manifold to create system with a miniaturized electrochemical detector.19 Another small CMOS device was completely embedded in PDMS with unique liquid metal interconnects to create a flexible sensor system that was used to detect magnetic nanoparticles.20 This device could be bent with a 1 cm radius of curvature allowing it to conform to the human body, thus potentially making future monitoring devices wearable.
Figure 9.
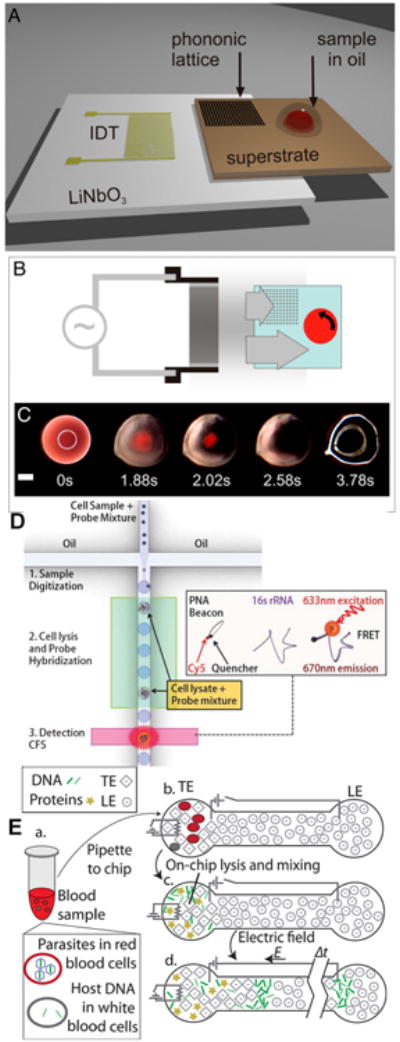
DNA. (a) SAW device for lysis and PCR of whole blood. (b) Transmission of ultrasonic wave on-chip for cell lysis. (c) Lysis of whole blood sample. Reprinted with permission from Reboud, J.; Bourquin, Y.; Wilson, R.; Pall, G. S.; Jiwaji, M.; Pitt, A. R.; Graham, A.; Waters, A. R.; Cooper, J. M. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 15162-15167, S15162/1-S15162/7. Copyright 2012 National Academy of Sciences. (d) μTAS for detecting DNA from encapsulated cells using PNA beacons coupled with FRET. Reprinted with permission from Rane, T. D.; Zec, H. C.; Puleo, C.; Lee, A. P.; Wang, T.-H. Lab Chip 2012, 12, 3341-3347. Copyright 2012 The Royal Society of Chemistry. (e) Separation of Plasmodium DNA using isotachophoresis on a printed circuit board (PCB) in polyurethane stamped channels. Reproduced from Marshall, L. A.; Wu, L. L.; Babikian, S.; Bachman, M.; Santiago, J. G. Anal. Chem. 2012, 84, 9640-9645. Copyright 2012 American Chemical Society.
Paper microfluidics has become a popular area of research because of the potential to make cheap devices for resource-poor situations. Paper itself has some limitations in its use as a substrate material so a variety of other substrates that support capillary wetting are being explored. One very interesting material is electroflocked nylon microfibers. These microfibers were deposited on an adhesive-based substrate, and specific patterns of different types of biofunctionalized fibers were created through shadow masking. 21 Layers of these fiber sheets were used to create 3-D channel networks without the need for hydrophobic wall patterning. Another approach to improving paper devices focused on the limitations of capillary flow through the creation of a hollow channel above the paper surface.22 Flow in this device was based on the pressure difference created between reservoirs and was seven times faster than flow in paper capillary driven devices. The use of superhydrophobic polymers integrated with superhydrophilic yarn micropatterns was also reported to overcome the limitations of flows driven by capillary action (Figure 1C).23 In this case, the flow in the channels continued even when they were completely wetted. The creation of these superhydrophilic and superhydrophobic regions can be a time consuming process. In order to decrease fabrication time, adhesive tape was used to replicate superhydrophilic/phobic patterns on paper.24 Channel patterns were replicated from a single master up to 12 times significantly decreasing the time needed to make multiple devices.
Bonding
Most microchip fabrication methods require that the substrate containing the channel manifold be bonded to a non-patterned flat substrate in order to form enclosed channels. There are a variety of issues with reproducible bonding, especially between heterogeneous substrates that still need to be addressed. In addition to the photoresist25 bonding described above in the materials section, a doubly cross-linked nano-adhesive was demonstrated to improve the bonding between any combination of PDMS, glass, silicon, polyimide and poly(ethylene) terephthalate (PET).26 This promising coating was vapor deposited in a 200nm layer and the bond strength was at least 2.5 MPa in all cases. PDMS was also bonded to a biologically friendly photoresist poly(2,2-dimethoxy nitrobenzyl methacrylate-r-methyl methacrylate-r-poly(ethylene glycol) methacrylate) (PDMP) using a mussel inspired poly(dopamine) adhesion layer. This allowed for multiprotein patterning against the PDMP in the channels.25 For COC polymers, a very simple and fast method for both creating channels and then solvent bonding them was reported (Figure 1B).8 In this paper, a marking pen was used to define a channel pattern on a COC substrate. The marked substrate was exposed to solvent and the non-marked area swelled. A flat substrate with access holes was then placed on top of the swelled substrate and pressure was applied to form an enclosed channel manifold. In some cases, the adhesive itself can be used to form the sidewalls of the channels simplifying the fabrication process. For example, a UV-active bisphenol A acrylate polymer was sandwiched between two quartz plates to define both the channel pattern in the polymer and to bond the two glass slides together.27 Dry film photoresist on glass substrates can similarly be employed to create channels through photolithographic processes and then used to bond the photoresist patterned glass to oxidized PDMS to form an enclosed channel manifold.28
One issue of particular importance when bonding multiple substrates with channel patterns that must interconnect is alignment. While most alignment is performed optically, a new method embedded micromagnets in patterned PDMS sheets to improve alignment.29 Linear and angular errors using the magnets were three times smaller than using optical alignment methods.
Finally, when channels have to be patterned with biologically active substrates prior to bonding, the bonding process must retain the activity of the biological materials. Retention of biological activity during bonding was accomplished using low melting point (mp < 40°C) wax30 for glass devices, methanol solvent bonding at 35°C with pressure for PMMA devices,31 or poly(dopamine) for polymer-glass hybrid devices.25 The wax spacer was patterned on a glass substrate and channels 25μm deep were easily formed.30
Surface Modification
The large surface area-to-volume ratios in microfluidic channels make interfacial chemistry critically important especially in regard to analyte adsorption and cell adhesion. Many materials from which microfluidic channels are fabricated generate unwanted or detrimental interactions with analytes and cells making surface modifications necessary. Coatings, however, often require a surface activation step. When channels have low aspect ratios in relation to a photoactivating light source surface activation is generally not an issue, but surface activation in high aspect channels can be problematic. A recent report indicated that COC-based polymers with high aspect ratio channels were easier to activate than similar aspect ratio PMMA channels.32 In addition to surface activation, the stability of coatings is always an issue, especially when they are biologically based. In situ coating just prior to detection is a potential method for solving this problem. An in situ coating for gold electrodes was successfully demonstrated using self-assembled monolayer (SAM) chemistry and biotin-streptavidin complexation chemistry. 33 Impedance-based sensing of a variety of biomolecules was successfully demonstrated on this device.
A wide range of materials can be used to coat or modify a surface. For the purposes of this review we have chosen to categorize such materials into one of three broad classes: chemically generated thin films, physical texturing of the surface including the fabrication of pillars, and biologically active films.
Chemically Patterned Films
The high surface area-to-volume ratio in microfluidic channels often creates unwanted interactions between analytes or cells and the walls. In order to minimize or moderate these interactions, surface coatings are necessary. A wide variety of effective surface modifications have been reported previously, and modifications to, or applications of, such surfaces continue to be investigated. For example, an allyl-polyethylene glycol (PEG) coating was applied to a PDMS device to generate a stable environmentally friendly coating that significantly improved protein separation efficiency.34 A poly(dopamine)-coated channel for the electrochromatographic separation of amino acid enantiomers showed good resolution between d and l enantiomers of several amino acids even though the channel was 18μm deep.35 A hydrophilic quaternized poly(dimethylaminoethyl methacrylate) coating for PDMS channels was applied using a surface-initiated atom transfer radical polymerization (SI-ATRP).36 The coating significantly reduced nonspecific protein adsorption and cell and bacterial adhesion. All of the above the coatings increased the hydrophilicity of the surface to mediate analyte-wall interactions. Conversely, an interesting hydrophobic fluoropolymer was selectively coated on only pre-roughened PDMS surfaces.37 This coating greatly reduced PDMS swelling when exposed to organic solutions and fluorescent dye adsorption. Importantly, it did not interfere with standard PDMS-glass bonding.
Coatings have also been applied to paper devices to improve their separation capabilities. A paper device coated with grafted poly(methacrylic acid- co-ethylene glycol dimethacrylate)-g-poly(methacrylic acid) (gPMAA) and poly(dimethylaminoethyl methacrylate-co-ethyl-ene glycol dimethacrylate)-g-poly(dimethylaminoethyl meth-acrylate) (gPDMAEMA) was shown to substantially improve the separation of mixtures of organic compounds.38 Significantly, the polymers were also coated with a hydrophobic poly(o-nitrobenzyl methacrylate) (oNBMA) that could be converted to a hydrophilic methacrylate using UV light. This conversion was used to create a flow switch.
Physically Textured Films and Posts
The development of surface features in microfluidic channels can be used to mediate cell attachment and migration behavior or to move particles selectively through multiple flowing streams thus simplifying many types of sample handling processes in μTAS. For example, cells are very sensitive to surface stiffness. Recently a device was reported with a surface consisting of long rows of PDMS hemicylinders on a glass substrate. The changes in surface PDMS thickness were related to the stiffness of the substrate, and cells were shown to move along these surface stiffness gradients.39 While substrate stiffness seems to be an important parameter for some cell types, for other cell types the chemical nature of the surface coating is more important. The effects of fibronectin, bovine serum albumin (BSA) and collagen on both native and plasma oxidized PDMS surfaces were examined.40 The coatings using collagen and fibronectin gave cell phenotypes that were indistinguishable from standard polystyrene petri dishes. This is a critically important point for those developing cellular assays using μTAS.
The inclusion of topographical features in a channel can be used to steer liquids and particles in microfluidic devices without the need for active fluid control elements. This can make the design and fabrication of devices integrating multiple sample handling and processing steps easier. For example, liquid crystals have been used to form soft “rails” in microfluidic devices to guide particles.41 The soft rails were created from disclination lines and positioned in a well-controlled manner that allowed the subsequent control of particles through a variety of interconnected channels. Micropost arrays have also been used for the passive guiding of particles and cells into adjacent, but distinct, fluidic streams.42 These arrays allowed the automation of reaction and washing steps for bead-based chemistries and significantly simplified device design and fabrication.
Biologically Active Films
Many coatings are used to control the interactions of cells, particles or biomolecules with the surface. These surface modifications provide powerful tools for the development of bio-based assays in μTAS. For example, a graphene oxide (GO)-silica composite material was used to immobilize trypsin on the surface of a PMMA microchannel bioreactor. The reactor demonstrated on-chip digestion efficiencies in 5 s that were similar to that of conventional techniques taking 12 hrs as measured by MS sequence coverages for the digested proteins.43 In a significant development, a nanofiber coating consisting of poly(lactic-co-glycolic acid) (PLGA) was used to sort circulating tumor cells (CTCs).44 Antibodies to specific receptors on CTCs were attached to the PLGA surface to create a “nanovelcro” chip. This chip was used with real clinical samples to successfully monitor changes in CTC concentrations in patients undergoing cancer therapy. In addition to simply capturing cells, stimuli-sensitive materials can also release them in a controlled manner potentially allowing for the collection or manipulation of purified cells downstream. For example, a stimuli-responsive smart interfacial polymer (poly(N-isopropylacrylamide) was coated onto a polystyrene device with an integrated heater to control the selective capture of CD4+ T-lymphocyte cells both spatially and temporally.45 Aptamer-coated channels were also used for the controlled capture and release of cells through temperature modulation.46 Specific capture of CCRF-CEM cells on this device was realized, and the released cells remained viable.
Channel Layout/Patterning and Molding
The ability to create patterned microstructures of varying depth in a one step process in μTAS increases the range of applications for these devices and can lead to cheaper fabrication. The fabrication of such structures, however, has been a difficult problem to solve. It is important to note, therefore, that an especially interesting single mask process technique was developed to create 3-D structures in PDMS microfluidic devices.47 The process used a mask with varying opacity that could be used in an ion etching process. This resulted in the fabrication of glass relief molds that allowed channels of continuously varying depths to be produced, as well as weirs and pillars. In some molding techniques adhesion of the PDMS to the mold can be problematic. This is especially the case for PDMS on PDMS molding. In order to overcome this problem and create high quality PDMS master molds through double casting, perylene C was used as a demolding and anti-adhesion layer.48 Microstructures with aspect ratios of 4:1 to 20:1 and angles from 5° to 40° were successfully replicated using this process. While PDMS devices are commonly molded against an SU-8 template, more exotic materials such as mammalian hairs can be used to create multichannel interconnecting structures with channel surface features reflective of the hair surface topology.49
Lengthy fabrication times, from concept design to the complete μTAS device chip, significantly slow the engineering design cycle and hinders rapid research progress. A proximity aperature lithography technique can be used to decrease cycle time and was shown to be capable of writing and etching complex channel patterns in PMMA.50 Fabrication time from channel layout to completed device took only a few hours and channel dimensions from 1 to 500μm were fabricated.
The ability to mold 3-D surfaces, especially in the sidewalls of channels, is an extreme challenge due to mold release issues. A novel 3-D nanopatterning technique for PMMA was used to partially solve this problem and to mold nanostructures in the sidewalls of channels using a two-step molding technique. 51 In the first step, a thin PDMS layer was applied to a nanomolded PMMA substrate. A second molding step was then used to create micron-sized features on the already nanomolded PMMA. Subsequent removal of the two stamps resulted in devices with both nano and micron-sized features. In another approach, free-standing PDMS microstructures were formed using two photon laser sculpting after the addition of a photoinitiator to the uncured PDMS.52 The accuracy of the structuring was ∼5μm. Such techniques could provide the ability to create very sophisticated structures within μTAS channels in the future to specifically capture or filter particles or cells.
Finally, a miniature and inexpensive CO2 laser-based cutting tool was used to create novel flow barriers, i.e. side walls, in paper devices in < 20s.53 The channel sidewalls were thus defined by the thin lines of material removed by the cutting process. This process may be an effective alternative to present wax printing methods.
Functional Elements
One of the key advantages of μTAS is the ability to integrate multiple functional (sample processing) elements onto a single device with a small footprint. Most of the devices discussed in this review integrate several functional elements that have been reported previously, and it is the combination of elements or application of the device that is novel. In this section we will concentrate on reports of novel or significantly improved functional elements, whether integrated with other elements or not.
Fluid Flow
Fluid flow and control in microfluidic channels is of particular importance. The ability to visualize this flow with high resolution is often critical in validating device design. Particle imaging velocimetry (PIV) is commonly used in this visualization process. The ability to monitor flow profiles without having to resort to expensive fluorescence-based PIV would have important advantages. Recently, spatiotemporal correlation spectroscopy was used to monitor velocity flow fields in 2-D. This technique required only the use of a bright field microscope.54 Flows up to 10 mm/s could be visualized with a resolution of 5 μm. In contrast to optical velocimetry methods, electrochemical velocimetry has also been demonstrated.55 This technique was able to monitor flow rates of 200-1800μl/min without the need for fluorescent particles.
There are many methods to generate fluid flow in μTAS. For the purposes of this review, however, we have tried to categorize them into three major areas - peristaltic pneumatic pumping, active pumping by means other than peristalsis, and passive pumping methods. Interesting papers advancing each area have been reported especially in terms of minimizing pumping components external to the microfluidic device itself.
Pneumatically Controlled Peristaltic Pumping
At present, most pneumatic pumping schemes require the use of off-chip pressure sources and solenoid valves. Moving some of that equipment on-chip has significant advantages in terms of finer flow control and better multiplexing. The ability to control 31 valves and 4 liquid handling operations using only 4 inputs and a vacuum line was demonstrated through the development of an on-chip pneumatic digital logic circuit (Figure 2F).56 This circuit eliminated a significant amount of off-chip machinery normally necessary for individual valve control. A similar report extended the potential of digital logic circuits to include the concatenated operation of normally closed pneumatic valves to create on-chip oscillators, flip-flops and shift registers.57 In addition to digital logic circuits, a serial digital-to-analog pressure convertor allowed for the on-chip control of fluidic resistances, and therefore, the relative flow of fluids at channel intersections.58
Figure 2.
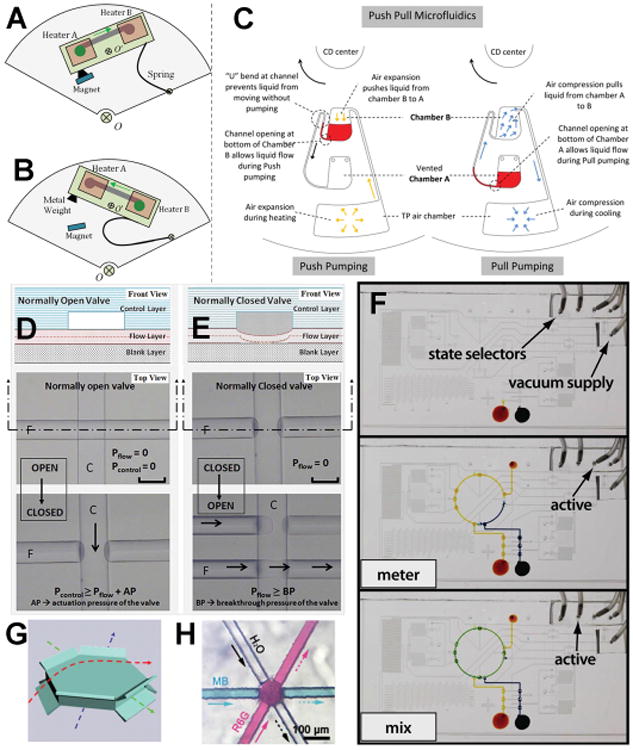
Flow and Valving. (a,b) Centrifugal device design using magnetic actuation for forward (a) and (b) reverse flow. Reprinted with permission from Wang, G.; Ho, H.-P.; Chen, Q.; Yang, A. K.-L.; Kwok, H.-C.; Wu, S.-Y.; Kong, S.-K.; Kwan, Y.-W.; Zhang, X. Lab Chip 2013, 13, 3698-3706. Copyright 2013 The Royal Society of Chemistry. (c) Thermopneumatic pumping design for a centrifugal μTAS. Reprinted with permission from Thio, T. H. G.; Ibrahim, F.; Al-Faqheri, W.; Moebius, J.; Khalid, N. S.; Soin, N.; Kahar, M. K. B. A.; Madou, M. Lab Chip 2013, 13, 3199-3209. Copyright 2013 The Royal Society of Chemistry. (d,e) Pneumatically actuated microfluidic valves in PDMS. (d) A normally open valve. (e) A normally closed valve. Reprinted with permission from Devaraju, N. S. G. K.; Unger, M. A. Lab Chip 2012, 12, 4809-4815. Copyright 2012 The Royal Society of Chemistry. (f) On-chip pneumatic digital logic circuit. Reprinted with permission from Nguyen, T. V.; Duncan, P. N.; Ahrar, S.; Hui, E. E. Lab Chip 2012, 12, 3991-3994. Copyright 2012 The Royal Society of Chemistry. (g, h) “Overpass” structures for fluid flow. Reprinted with permission from He, Y.; Huang, B.-L.; Lu, D.-X.; Zhao, J.; Xu, B.-B.; Zhang, R.; Lin, X.-F.; Chen, Q.-D.; Wang, J.; Zhang, Y.-L.; Sun, H.-B. Lab Chip 2012, 12, 3866-3869. Copyright 2012 The Royal Society of Chemistry.
Active Pumping Methods
Redox-magnetohydrodynamics (MHD) is a particularly interesting method of generating pumping on-chip. However, it generally requires the use of high concentrations of redox species in order to work well and this can limit the applications for which it can be used. A novel approach to redox-MHD overcomes this limitation by allowing the depleted redox species time to recover thus eliminating the high concentration limitation.59 Bead velocities of 0.1 mm/s were generated using this pumping method. The use of molecular motors to drive flow on nano- and microfluidic devices is also of significant interest due to their small size. There is concern, however, about how different types of buffers or sample matrices might interfere with the operation of these motors. A recent paper, however, demonstrated that these motors worked well in a range of biologically based buffers and could potentially be used for pumping in biological assays.60 Finally, a clever bio-inspired pump was fabricated using agarose gel, a micro-perforated silicon sheet, and a micro-heater to mimic the stomata in plant leaves.61 The device worked by simulating transpiration and could be attached directly to a small planar microfluidic device with only a small external power supply. The micro-pump was able to generate a water potential of 72.5KPa raising the water upwards of 7m.
Power-Free Passive Pumping Methods
In order to make μTAS inexpensive and portable with low power requirements, the minimization of external components are of high interest. One avenue to eliminate pump-related external components is to rely on passive pumping to control fluids. For example, passive flow was generated in channels molded in degassed PDMS.62 Pumping was initiated by exposing the PDMS to air. As air diffused back into the PDMS the pressure in the waste reservoir, which was covered with adhesive tape, was lower than ambient air. Droplets containing reagents and analytes placed at the entrances to other channels were drawn into the device due to this pressure difference. The ability to label and detect MicroRNA was demonstrated using this passive pumping mechanism. There is some skepticism, however, that passive capillary flow can actually be used for well-controlled assays involving the use of multiple channels and sequential processing steps. Such capabilities, however, were recently demonstrated successfully.63 Additionally, a second passive capillary pumping device was used to automate the sequential addition of four reagents and to perform a model immunoassay.64 These three passive pumping papers indicate a promising future for the implementation of more sophisticated multi-step assays using this flow generation method. Finally, a unique diaphragm pump was reported that consisted of a one way diaphragm valve and chamber.65 The chamber inflated to 240uL upon filling and through subsequent deflation provided a 125μL/min pressurized flow to downstream elements.
Pumping on Inertial (Centrifugal) Flow Microfluidic Devices
Fluid flow in centrifugal microfluidic devices is generated by inertial forces when the device is spun. The generallimitation with such devices is that the fluid must always flow out toward the edge of the disk. A couple of new design modifications were reported that make it significantly easier to implement bidirectional flows (Figure 2A,B).66,67 Bidirectional flow between two temperature zones was generated on a pseudo-centrifugal platform by physically altering the channel alignment on the device using magnetic actuation.66 These alterations to the standard centrifugal pumping mechanism allowed, for the first time, real time droplet PCR on this type of device. Reversible pumping was implemented using a thermopneumatic pump consisting of an air chamber and the use of localized heating to pump fluid back toward the interior of a multi-level 3-D compact disk (CD)(Figure 2C).67
Flow Control – Other
A variety of other methods to control flow rates independent of external pumps have also been reported. A very interesting and potentially important paper described the control of electroosmotic flow (EOF) using a flow field effect transistor (FET).68 The transistor was created by coating an Au electrode in the channel with a SAM. Voltage applied to this electrode provided excellent, even reversible, control of the EOF. An electroactive polymer (EAP)69 and a thermally responsive phospholipid preparation70 were also used to control fluid flow in μTAS channels. Actuation of the EAP altered the fluidic resistance in the channel by compressing it.69 Temperature control of the phospholipid in various channels generated viscosity differences that could be used to generate injections.70 Additionally, microstructures can be fabricated and inserted into channel intersections to control flow (Figure 2G,H).71 These microstructures literally created small over/underpasses at an intersection to direct fluid flows past one another. It remains to be seen how practical this approach to controlling flow will be.
Several analytical techniques including cytometry require the use of flow focusing at channel intersections to reduce coincidental detection. While 2-D flow focusing is quite straightforward to accomplish 3-D control is much more difficult due to the multiple photolithography, molding, and alignment steps involved. Nevertheless, a 3-D focusing device was recently constructed that produced a well-focused stream in the middle of the channel. This design substantially reduced focused flow velocity variations compared to some previous 3-D focusing schemes.72 Another 3-D flow focusing device was used to study protein folding dynamics by diffusional mixing in under 80 microseconds.73
Valves
Valves are used to control or gate the transport of material throughout a channel manifold (Figure 2D,E). Two particularly interesting magnetically actuated valves have been demonstrated recently. One valve consisted simply of a gas or paraffin spacer between 15μL segments of liquid.74 The static liquids contained magnetic particles upon which analytes were attached. The valves were “actuated” when a magnet was used to move the particles from one liquid to another through the air gap. Sample processing and PCR were carried out on this device with results that were comparable to commercial kits, but without any fluid movement. Another device used magnetic timing valves to control flow on a paper device.75 An ionic sensor detected flow through specific channels and actuated an electromagnet to open or close a paper cantilever valve. With this device, an enzyme-based colorimetric assay was performed. Another creative valving method relied on modulating the fluidic resistances in multiple channels via frequency tuning of an acoustic pump.76 The pump operating frequency was used to control the selective actuation of diaphragm valves in different channels, thus determining into which channel the fluid flowed.
Mixing
Mixing can be an issue in microfluidic devices due to the low Reynolds numbers. A considerable amount of effort has been directed to developing both passive and active methods to increase the speed of mixing. For instance, a creative bio-mimetic mixer used artificial cilia with embedded magnetic particles.77 The cilia were fabricated in microfluidic channels and an applied magnetic field induced figure-eight oscillations that improved mixing rates over simple diffusion (Figure 3A).77 A digital logic circuit for combinatorial mixing, similar to those discussed above under peristaltic pumping, was also demonstrated (Figure 3B).78 The 6-sample processor was shown to be able to mix 26 unique combinations in < 1 s. This mixing approach has significant implications for the development of high throughput clinical applications in microfluidics.
Figure 3.
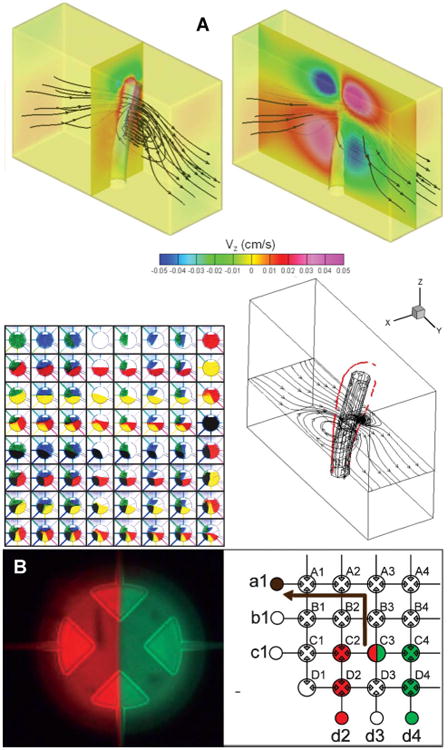
On-chip Mixing. (a) Magnetically actuated artificial cilia induce a mixing vortex; the in-plane fluid path is denoted by black lines. Reprinted with permission from Chen, C.-Y.; Chen, C.-Y.; Lin, C.-Y.; Hu, Y.-T. Lab Chip 2013, 13, 2834-2839. Copyright 2013 The Royal Society of Chemistry. (b) Mixing using pneumatically actuated microvalves allows multiple reagents to be loaded into the combining valve. Reprinted with permission from Jensen, E. C.; Stockton, A. M.; Chiesl, T. N.; Kim, J.; Bera, A.; Mathies, R. A. Lab Chip 2013, 13, 288-296. Copyright 2013 The Royal Society of Chemistry.
Gradient Formation
The generation of a physical gradient78 has already been discussed, but most gradients in microfluidic devices are chemical in nature. The ability to generate stable chemical gradients is critical to a variety of biological analyses where concentration effects on cell physiology need to be investigated In an interesting and novel approach to gradient formation, a series of trapped bubbles was used to form both stable and pulsatile chemical gradients (Figure 4A).79 The bubbles were staggered across the channel width in a ladder arrangement and acoustically activated at 30kHz to generate the gradient. In another paper the formation of stable oxygen gradients across a microfluidic device was reported to study aerotaxis in bacteria.80 A two-layer PDMS device integrated with a computer-controlled gas mixer was used to generate the gradients. One problem with gradient generation is that the gradient can get blurred when moving across the uneven surface presented by cultured cells. A method to overcome this problem inserted a porous track-etched polycarbonate sheet to separate the cultured cells from the gradient.81 Importantly, the cells were cultured on the membrane allowing them to be exposed to different chemical environments from above and below as typical in vivo.
Figure 4.
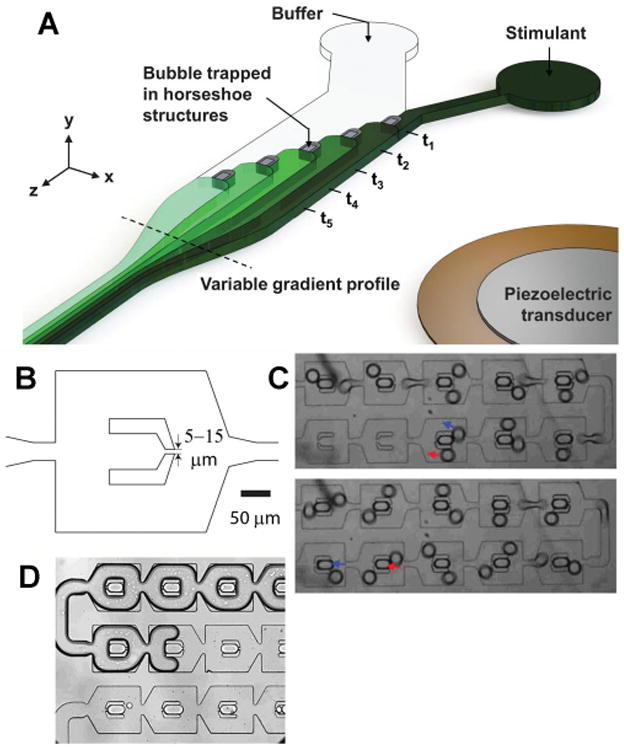
Gradients and Concentration. (a) Gradient generation using a piezoelectric transducer to manipulate trapped bubbles within PDMS. Reprinted with permission from Ahmed, D.; Chan, C. Y.; Lin, S.-C. S.; Muddana, H. S.; Nama, N.; Benkovic, S. J.; Huang, T. J. Lab Chip 2013, 13, 328-31. Copyright 2013 The Royal Society of Chemistry. (b, c) Analyte concentration through evaporation is implemented by trapping droplets (b,c) in a sequential manner followed by (d) gas infusion into the device. Reprinted with permission from Casadevall, i. S. X.; Turek, V.; Prodromakis, T.; Edel, J. B. Lab Chip 2012, 12, 4049-4054. Copyright 2012 The Royal Society of Chemistry.
Analyte Concentration and Filtering
The ability to concentrate an analyte in a controlled manner is difficult to implement on microfluidic devices. A novel approach to concentration, trapped droplets of analyte-containing solutions in arrays of obstacles molded in a channel (Figure 4B, C, D).82 Air was then flushed through the channel, partially evaporating the trapped droplets and concentrating the solutes. Fluorescence detection enhancement and crystallization experiments demonstrated the utility of the device. Filtering can be used to concentrate particles, and the ability to select particles based upon size is crucial to many biologically based μTAS experiments. Such sorting thus far on planar devices, however, has been slow. A method to significantly increase particle sorting speed using inertial microfluidics within a four or sixteen parallel-channel device has been demonstrated.83 The filtration efficiency for 10μm particles was >95% at throughput several orders of magnitude higher than previously shown through straight channel inertial flow devices.
Droplet-Based Microfluidic Elements
An especially productive area of high interest is the use of segmented flows in microfluidic devices for a variety of chemical analyses. Much of this research has focused on the active control of droplet formation or the development of droplet-on-demand techniques. Surface acoustical wave (SAW) structures, for example, were integrated into microfluidic devices in order to generate on-demand droplets to encapsulate 10μm particles.84 Pneumatically controlled PDMS diaphragm valves were also used to promote on-demand mixing, dosing, and sizing of droplets.85-87 One of these devices integrated a pneumatic pump to actively generate droplets of various radii allowing for multivolume digital PCR.85 In a second device, pneumatic valves were used to open and close different side channels that were used for droplet-generation.86 Droplets from different channels were selectively mixed in 0.5 s, incubated, and then extracted from the oil. The extracted samples contained protein digests that were then analyzed using ESI-MS. The automation and integration displayed in this device demonstrate several of the important strengths of μTAS for performing true total analyses. In a third device, large sample plugs were generated off-chip and brought on-chip through a capillary where they were converted into smaller daughter droplets. Four sequential reagents were then injected into the droplets, followed by incubation under the control of pneumatically-operated valves (Figure 5G).87
Figure 5.

Droplet manipulation. (a) Use of a surface acoustic wave device to sort droplets. Reproduced from Li, S.; Ding, X.; Guo, F.; Chen, Y.; Lapsley, M. I.; Lin, S.-C. S.; Wang, L.; McCoy, J. P.; Cameron, C. E.; Huang, T. J. Anal. Chem. 2013, 85, 5468-5474. Copyright 2013 American Chemical Society. (b) Droplet size control using side channels to decrease volume. Reprinted with permission from Schoeman, R. M.; Kemna, E. W. M.; Wolbers, F.; van, d. B. A. Electrophoresis 2013. Copyright 2013 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (c) Parallel high-throughput design for PCR microreactors. (d) The use of rail architectures to move and mix droplets. Reprinted with permission from Dangla, R.; Kayi, S. C.; Baroud, C. N. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 853-858, S853/1-S853/6. Copyright 2012 National Academy of Sciences. (e) Suspended microfluidic screening array using microDots which are accessible from above and below channels for cell assays. Reprinted with permission from Casavant, B. P.; Berthier, E.; Theberge, A. B.; Berthier, J.; Montanez-Sauri, S. I.; Bischel, L. L.; Brakke, K.; Hedman, C. J.; Bushman, W.; Keller, N. P.; Beebe, D. J. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 10111-10116, S10111/1-S10111/10. Copyright 2013 National Academy of Sciences. (f) Protein-protein reactions in droplets can be measured multiple times in one long mixing channel using fluorescent lifetime measurements. Reproduced with permission from Benz, C.; Retzbach, H.; Nagl, S.; Belder, D. Lab Chip 2013, 13, 2808-2814. Copyright 2013 The Royal Society of Chemistry. (g) Droplets can be produced on-demand, injected with reagents, and incubated using valving mechanisms. Reproduced with permission from Zec, H.; Rane, T. D.; Wang, T.-H. Lab Chip 2012, 12, 3055-3062. Copyright 2012 The Royal Society of Chemistry.
Part of the interest in droplet production is high throughput analysis. Several new methods to increase droplet production, registration, and interrogation have been recently demonstrated. A high-throughput parallel droplet generator that integrated controlled droplet mixing used 256 nozzles to produce ∼226 pL droplets at a frequency of 1.5 kHz (Figure 5C, D).88 Droplet volume was controlled through a combination of nozzle design and surface energy gradients. The production of droplets on a second device used a sipper capillary to imbibe samples and spacer plugs from a multiwell plate.89 The droplets were mixed and then interrogated on-chip at rates of 1-5 droplets/s. While droplet generation on this device was slower than on the previous device,88 the drops in the second device were registered. In addition, the droplet analysis frequency could be increased by increasing flow velocity. This might allow higher throughputs in the future. On a more fundamental level, the ability to both sense and heat droplets in a high-throughput fashion was demonstrated using microwave radiation.90 Importantly, both the excitation loop and resonator were integrated onto the chip.
Bacteria and cell encapsulation in emulsions and droplets on microfluidic devices can be used to mimic micro-scale environments to study biological events. Two especially interesting devices incorporating cells in emulsions and droplets have been demonstrated. In the first device, bacteria were encapsulated in water-in-oil-in-water double emulsions to separate cells, yet provide them with a source of nutrients and to allow the addition of reagents to affect gene expression.91 In the second device, cells were encapsulated at rates of ∼1500Hz and with efficiencies of 40% (Figure 5B).92 The drops were paired with a second drop at 100% efficiency, after which the volume of the drop was selectively reduced in volume by 75%. These devices show great potential for automating a variety of biological assays in the future. One important point that needs to be considered for these cell encapsulation strategies is that in order to ensure that only one cell is in each droplet, low cell densities are used. This leads to many droplets, as in the last example, not having any cells. To overcome this problem, a fluorescently activated cell sorting system was integrated after the encapsulation step.93 Droplets without cells were removed in the sorting process so that 94.1% of the droplets continuing for downstream processing contained cells.
As opposed to some of the droplet merging examples described above, the ability to precisely split droplets and then sort them is important for many types of assays. An interesting device to precisely and actively split charged droplets used a noncontact method to electrostatically charge the drops.94 This device allowed significantly better control over the splitting and sorting of droplets compared to other previously reported methods.94 In addition, a SAW based droplet sorter was shown capable of sorting droplets into one of 5 channels at rates in excess of 200 droplets/s (Figure 5A). 95
Other
Another very creative and unique method to generate “droplets” used small picoliter amphiphilic solvent carriers (i.e. vessels) that were created in situ and arrayed in a microfluidic chamber.96 These vessels, which resemble hexagonal nuts, were fabricated through the photopolymerization of trimethylolpropane triacrylate (TMPTA). They have a hydrophobic exterior and a hydrophilic interior. Each vessel can be uniquely encoded for identification providing some interesting opportunities for performing high-throughput assays that require registration. Finally, a new class of microfluidic system was reported in which fluids were suspended between multiple air or immiscible fluid-air interfaces (Figure 5E).97 Flow was generated using spontaneous capillarity. Cell assays on microDots suspended between fluid flows were performed to investigate cell invasion, cell growth, and metabolite extraction. The authors believe that such a design will lower the barrier of adoption of microfluidics in the life sciences due to its simplicity, the ease with which reagents can be added, and the ability to easily retrieve cells from such devices.
Separations
Separations are often included in many microfluidic devices as part of an integrated chemical analysis system. Several significant advancements in separations have been reported especially in the areas of proteins and nucleic acids. Protein separations, in particular, have always been problematic due to analyte-wall interactions. A significant reduction of such interactions in a PDMS device was demonstrated using an environmentally friendly poly(ethylene glycol) coating.34 The coating also generated good separation efficiencies. While high efficiency protein separations are often difficult to implement, the separation of nucleic acids has been one of the most successful application areas for μTAS. Even considering past successes, notable advances in both gel-based and free solution separations have taken place. For example, the free solution conjugate electrophoretic separation of 19 ligase detection products was achieved using drag tags in only 70s.98 This technique potentially provides a viable alternative to gel-based separations for some nucleic acid analyses where single base pair resolution is not necessary. In another device, rapid separations of 100-300bp DNA fragments with resolutions of 10-20 bp in a 2 mm separation distance were accomplished in under a minute.99 The separations were performed using a dynamic coating and a replaceable linear polyacrylamide (LPA) sieving matrix. It is the shortest separation yet reported that meets the meets the high-resolution criterion for the post-amplification analysis of DNA. Finally, a novel coating and sieving polymer composition consisting of 95% w/w hydroxyethylcellulose and 5% w/v of polyvinyl pyrrolidone at a final polymer concentration of 2.5-3.0% generated robust separations with resolutions of 1.2 bp for < 200bp.100 This composition allowed the gel to be vacuum-loaded loaded in a single step without any pretreatment providing a simple, fast, and inexpensive method for DNA separation.
Chromatographic separations in packed columns have lagged behind other separation methods on microfluidic devices due to the difficulty of packing the stationary phase particle support efficiently in the channels. In an attempt to improve the packing efficiency, an innovative column geometry utilizing bypass channels was used to pack microfluidic channels with beads. High efficiency chromatography separations were generated with sub-micron plate heights at low applied pressures.101 Due to the packing difficulty with particles, a popular alternative has been the use of column monoliths. The performance of a variety of pillar designs for electrochromatographic separations on monolith-based chips were investigated.102 A new type of foil-shaped pillar performed better than previously reported diamond and hexagon-shaped pillars. While electrochromatography can be performed on devices with attached stationary phases, it is most often performed using a much easier to implement pseudo-stationary phase. Micellar electrokinetic capillary chromatography (MEKC) separations were demonstrated using unique mixtures of ionic, nonionic and zwitterionic surfactants.103 A nonionic and zwitterionic mixture gave better results than other common surfactant systems.
Gel-based separations of proteins are fundamental to biochemistry and molecular biology. Miniaturization of these techniques could both significantly reduce the sample and reagent volumes needed and decrease analysis times, thus having a significant impact on these fields. One of the most commonly used protein separation techniques is 2-D IEF-SDS PAGE. A miniaturized μTAS version of this technique was introduced.104 For the IEF separation dimension, a resolution of 0.1 pH units in < 20 min was achieved and both second dimension assays - PAGE and pore-limit electrophoresis – were completed in < 15 min. The 2-D peak capacities ranged from 35 to 256. Another 2-D protein separation system incorporating SDS μ-CGE and microemulsion electrokinetic chromatography (MEEKC) was used to generate oxidative-stress induced bio-marker profiling in vitro nitrosylated proteins.105 This device generated “fingerprints” of brain tissues for mice with Alzheimer's disease using LIF detection. Lastly, a miniaturized 48-plex Western blot system was reported in which sample enrichment, protein sizing, protein immobilization (blotting), and in situ antibody probing were carried out in an automated manner.106 Validation of the technique was performed using purified proteins, crude cell lysate and human sera. With the human sera, detection limits of 50 pM with quantitation of 3 orders of magnitude were demonstrated. While these 3 devices, at present, cannot compete with conventional techniques, they point the way for potentially better and more powerful future incarnations that could have a significant impact on how multidimensional protein separations are performed in the future.
Detection
The ability to perform miniaturized chemical analysis is only useful if the analytes can actually be detected at analytically or clinically relevant levels. A wide variety of detection techniques have been reported for analytes in microfluidic devices over the last 20 years. Progress, however, is still being made in improving these techniques or adding additional ones.
Electrochemical
Electrochemical detection is of particular interest with μTAS because electrodes and detection electronics can be miniaturized and have low power requirements, as was discussed above with the integration of CMOS electrochemical detection electronics and fluidics (Figure 6C).19 Amperometric detection is the most popular electrochemical technique integrated with microfluidic devices. For amperometry, better electrode materials and geometries that can be integrated with the microchip fabrication techniques and provide adequate detection limits are being constantly pursued. One novel composite electrode was fabricated from graphite/PMMA (20:1) and used for the detection of dopamine and catechol.107 In another device, enhanced sensitivity for catechol was demonstrated using 3-D gold-coated micro-pillar electrode arrays. 108 Additionally, a 10× detection enhancement for NO was shown using platinized glassy carbon electrodes.108 Selectivity for NO on these electrodes was increased through the application of a Nafion film. Finally, a method to increase detection sensitivity by increasing the electrode surface area was reported by electrodepositing copper nanostructures sheathed with carbon.109
Figure 6.

Integration of electronics and fluidics. (a, b) Electrodes are incorporated into device using a low melting point alloy allowing the detection of analyte without contact. Reprinted with permission from Gaudry, A. J.; Breadmore, M. C.; Guijt, R. M. Electrophoresis 2013. Copyright 2013 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (c) Electrochemical detection with CMOS integrated circuit which includes two mixers and separation channel. Reprinted with permission from Huang, Y.; Mason, A. J. Lab Chip 2013, 13, 3929-3934. Copyright 2013 The Royal Society of Chemistry (d) Integrated planar coil electromagnets for mixing in droplets. Reprinted with permission from Chiou, C. H.; Shin, D. J.; Zhang, Y.; Wang, T. H. Biosens Bioelectron 2013, 50, 91-9. Copyright 2013 Elsevier Inc. (e) Optical detection using a (GO)-based (FRET) detection scheme in a 33-channel device. Reprinted with permission from Cao, L.; Cheng, L.; Zhang, Z.; Wang, Y.; Zhang, X.; Chen, H.; Liu, B.; Zhang, S.; Kong, J. Lab Chip 2012, 12, 4864-4869. Copyright 2012 The Royal Society of Chemistry.
Bonding is always an issue with integrated electrodes, as they are generally not coplanar with one of the substrates. This is especially the case for glass bonding, where even sub-100nm surface variations can cause glass-to-glass bonding to fail. To improve such bonding, Pt electrodes were deposited into channels etched 500nm deep into the substrate. This allowed the fabrication of electrodes nearly flush with the surface.110 An additional limitation of electrochemical detection is that the electrodes must generally be placed at the end of the separation channel where it meets the waste reservoir to minimize the interference of the separation potential with the electrochemical measuring potential. This placement can reduce the usefulness of the detector. To overcome this limitation, an amperometric detection system with an improved electrically isolated potentiostat allowed the use of in-channel electrodes for the detection of hydrogen peroxide.110 Many electrochemical detectors also require the use of potentiostats which are generally the most specialized and expensive component of the system. A cheap, readily available alternative to purchasing dedicated potentiostats has recently been demonstrated using a smart phone's audio jack and video camera.111
Amperometric detection can also now be readily implemented on paper-based microfluidic devices using carbon ink electrodes.112 The electrodes were masked onto paper while microfluidic channels were milled into a layer of PDMS. Afterward the paper and PDMS were sealed together. Other forms of electrochemical detection, besides amperometry, can be integrated with microfluidic devices. For example, cyclic voltammetry was used to detect hydrogen peroxide from oxidatively stressed hepatocytes surrounding a Ag electrode encased in a poly(ethyleneglycol)-horse radish peroxidase (PEG-HRP) coating.113
Finally, integrated prototypes of potential commercial devices have recently appeared. This included a second generation portable microfluidic device with integrated high voltage power supply and potentiostat for electrochemical detection, but the detection limits for most compounds tested on this device were still limited.114
Conductivity
Standoff detectors are always interesting, as they do not have to make physical contact with the analyte in order to detect it. Capacitively coupled contactless conductivity detectors (C4D) are examples of standoff detectors with a variety of potential analyte, cell, particle, and droplet detection applications. The major issue in terms of implementing these detectors in μTAS is the need for a more facile method for integrating the electrodes and optimizing capacitive coupling. A recent implementation of C4D on a PMMA device minimized stray capacitance by placing the 100μm wide electrodes in-plane with but isolated from the separation channel at an effective electrode distance of ∼1mm (Figure 6A,B).115 The electrodes were composed of a low melting point (80°C) alloy that that could be pumped through channels next to the separation channel and promoted quick and simple fabrication. Electrophoretic separations of cations were accomplished in less than 22s with LOD of 1.5-3.5 ppm.
Impedance
Impedance detectors are very commonly used in biosensors. They are also of interest in μTAS because of their compact nature and minimal power requirements. Such detectors can be used for sensing the presence of droplets or cells but the discrimination of live and dead cells can be problematic. This issue, however, has been addressed using a device capable of detecting and discriminating between viable and nonviable cells in droplets at throughput rates of 100Hz.17 Pathogen detection by a bio-recognition array of impedance detectors was also carried out with species specific immobilized antimicrobial peptides coating the microsensors in a μTAS.116 This detector array was used to rapidly detect S. mutans and P. aeruginosa within 25 min.
Surface Enhance Raman Spectroscopy (SERS)
Raman detection schemes are of special interest for development on microfluidic devices because of their capability to detect and differentiate specific chemical species at low levels in real time without the need for labels. SERS, for example, was used to detect methamphetamine in saliva117 and MRSA in fluids.118 For the detection of methamphetamine, the salt-induced aggregation of Ag nanoparticles substantially increased SERS signal.117 Additionally, the reproducible and specific nature of the SERS spectra allowed the differentiation between MRSA and non-MRSA strains in a μTAS with an accuracy of 95%.118 Inter-laboratory comparisons showed the analysis to be robust. A technique was also reported for spray-coating paper microfluidic devices with Ag nanoparticles for SERS detection.119 Nanomolar detection limits were achieved with coating costs < $0.02/chip.
Optical
Optical interrogation techniques are frequently used for the detection of analytes on μTAS devices. Of particular interest are fluorescence techniques due to their high selectivity and sensitivity. For example, fluorescent lifetime and FRET approaches were used for the detection of protein-protein interactions within droplets120 and cancer cells.121 Fluorescent lifetime measurements were shown to improve the data quality compared to intensity–based approaches (Figure 5F).120 In an interesting application of graphene oxide (GO), cancer cells were detected when a fluorescently tagged aptamers interacted with the cell causing the release of the GO quencher (Figure 6E).121 Seven samples were analyzed in parallel on this 33-channel device.
For some compounds direct detection is inconvenient and so indirect detection techniques have been developed. One example involved the use of a platinum porphyrin polymer luminescent probe to monitor dissolved oxygen in microfluidic channels.122 The probe was used to follow the oxidation of small inorganic compounds. While fluorescence provides excellent sensitivity and selectivity, most molecules are either not fluorescent or difficult to derivatize, especially on column. Label-free detection techniques, therefore, are highly sought after. One such technique made use of a dual ring resonator for the label-free optical detection of biological molecules in a microfluidic device.123 The gapless light coupling photonic configuration was simple to fabricate and was used to detect both proteins and carbohydrates using visible wavelength light. Simple optical imaging can also be implemented as a detector for μTAS. A nice demonstration of such a system used magnetic particles and magnetic tweezers to perform a one-step, high-throughput, low cost agglutination assay in a droplet device.124 The agglutinated beads were imaged using a low cost USB camera, and hundreds of assays per hour could be performed with detection limits of 100pM. Another unique label-free sensing method was demonstrated using a liquid-crystal-based sensor.125 4-cyano-49-pentylbiphenyl (5CB) microdroplets coated with PAA-b-LCP were functionalized with protein binding moieties. The 5CB microdroplets underwent a configurational change that could be detected using cross polarizers when the proteins bound. The detection limit for this technique, however, was 2-4 μM.
Mass spectrometry
Mass spectrometry (MS) is one of the best and most sensitive methods to identify specific compounds. An area of special interest is the integration of droplet-based devices with MS. A direct method of integration used a micropillar filter to separate aqueous droplets from oil.86 The extracted droplets were then introduced into the MS using an nanoelectrospray interface.86 Droplets from a microfluidic device could also be interfaced to a MALDI-MS by spotting them on a MALDI plate.126 The droplets were spatially confined to hydrophilic spots on the plate.126 Over 26,000 300μm droplets could be registered on the device. A MS-coupled microfluidic device was also used to measure sub second hydrogen/deuterium exchange (HDX).127 The device integrated all of the functions necessary for “bottom-up” HDX labeling experiments and was directly interfaced to the MS through a nanoelectrospray interface. The integration of microfluidics with MS as in the last example provides the ability to examine exchange kinetics at time scales difficult to access with any other method.
Surface Plasmon Resonance (SPR)
SPR is a sensitive label-free detection technique that has shown promise as a microfluidic biosensor. SPR detectors, however, require the use of an Au or Ag film in close proximity to the analyte. This need for the direct integration of the film in the microfluidic channel has hindered its application. Interesting approaches to make such integration more facile have recently been reported. For example, the inexpensive, in situ fabrication of Au nanoparticle films within a PDMS microfluidic channel was accomplished.128 Antibodies were attached to the nanoparticles for the specific detection of growth hormones. To increase throughput, this type of detector was multiplexed.129 The detection system was simplified, however, so that a common UV-Vis detector could be used to detect the spectral shift and intensity of the plasmonic band. Further expanding on the parallel analysis capability of SPR, a nanohole SPR system capable of performing and detecting 50 assays in parallel was demonstrated.130 This system was used to quantify ligand-binding kinetics and affinities in a high throughput fashion.
Other
X-ray detection systems have been used to both detect and monitor the growth of Au particles on microfluidic devices. Small angle X-ray scattering131 and in situ X-ray scattering132 were both utilized to detect the concentration, size and shapes of Au particles synthesized in droplets or aqueous solutions, respectively. Such real time monitoring of the synthesis of such particles is difficult to implement in any other manner. Finally, a novel detection technique using terahertz sensing was reported using a photonic crystal pillar array.133 An initial proof of concept detector was demonstrated and its response was in agreement with simulations.
Chip-to-World Interface
One key issue in the development of μTAS is the integration of these devices with real world samples and other types of laboratory instrumentation. Several novel interfaces or improvements in interfacing have recently been demonstrated including a generic microfluidic chip to liquid-handling-station interface design.134 This interface allowed the simultaneous molding of PDMS ports on a connector apron with the channel layout using a specially designed mold. A more flexible connector based on the standard nine-pin sub-D connector was used to make an instrument independent generic connector.135 The male side consisted of an industry standard connector with the wires replaced with tubes. The female side was micromolded from PDMS and permanently attached to a microfluidic device. A third approach to this interfacing problem presented a methodology for integrating ports into a PDMS device over any part of a 100 cm2 wafer surface.136 The PDMS device consisted of two layers – one ∼10μm thick into which channels were molded and the other 500μm thick into which connectors were molded. The problem with this type of double molding technique is that a thin layer of residual PDMS can block connections between the 2 separately molded layers. To remove any residual PDMS after demolding, a fluorine-based dry etching technique was used. In addition to these interfaces, a novel component platform for μTAS used Lego® Mindstorms® motors, controllers and software.137 The system was robust and inexpensive compared to custom-made actuators. The MainSTREAM platform consisted of a peristaltic pump, 8-channel valve, sample-to-waste liquid management and interconnections to a microfluidic device.
Microfluidic Platforms
Integrated Devices
The ultimate goal of many μTAS projects is the development of an integrated platform with rapid sample-in/answer-out capability. Several devices that satisfy this criterion or come close have been reported and highlight the advantages and strengths of using μTAS for a variety of different applications. For example, an external power-free device for the rapid and sensitive detection of microDNA was developed that integrated sandwich hybridization and dendritic amplification with fluorescent detection to detect products in < 20 min.62 A centrifugal microfluidic device for the analysis of pesticide residue was demonstrated that integrated liquid-solid magnetically actuated extraction, filtration, sedimentation and detection. 138 The detection limits were on the order of 0.1 ppb. A second centrifugal microfluidic device was used for the determination of nutrients in water.139 All of the sample processing steps were integrated onto the device from sample metering to detection. An automated microfluidic device for multiplexed magnetic bead assays integrated both the incubation and washing steps.140 No external controls except a syringe pump to apply pressure were used, and the system was compatible with a variety of commercial immunoassay technologies. Another μTAS integrated and automated immunoassays for the detection of cancer biomarkers using SERS.141 Sandwich immunocomplexes were formed with detection limits in the 0-10 ng/mL range. A cell assay μTAS integrating cell culture, stimulation, and incubation with cytometry permitted the automated and hands-free analysis of cell receptor signaling. Additionally, the sampling of extracellular rat hippocampus fluid was integrated with automated injection, electrokinetic separation and detection.142 In a more universal approach to generating generic μTAS devices, a “Kit-on-a-lid-assay” (KOALA) platform integrated a lid containing reusable microfluidic channels with disposable bases containing cryopreserved cells and reagents.143
Automated sample-in/answer-out PCR analyses of biological samples have received a great deal of attention over the past few years. A new commercial In-Check system was reported that integrated sample preparation, nucleic acid amplification, and DNA microarray detection in < 2 hrs.144 A second system provided the automated analysis of DNA collected on buccal swabs in 45 min.6 Three genes and 15 STR's were selected for amplification and detection. All sample preparation, DNA amplification and DNA separation steps were integrated onto the device.
POC devices
Closely related and overlapping with the integrated devices discussed above are point-of-care (POC) devices. These devices are focused on giving rapid sample-in/answer-out assays for a variety of clinical applications. Many prototypes have been and continue to be reported, but few have been commercialized. The reason for the lack of commercial devices is the process of product qualification.145 An interesting article recently discussed the problems with qualifying POC devices along with some potential solutions.145 Several papers in this area focused on developing the functional elements and packaging necessary to create complete sample-in/answer-out POC devices. For example, a microfluidic biomolecular amplification reader (μBAR) performed rapid, low cost isothermal nucleic acid amplification.146 Sample pretreatment, however, was carried out off-chip for this device. Another simple POC device was designed to incorporate bar-coded beads and magnetic actuation in a microfluidic channel.147 This device was used for the detection of infectious diseases such as HIV and hepatitis B in less than 20 min with a detection limit of ∼ 1 nM. A particularly interesting, simple, and inexpensive μTAS for measuring a variety of interesting biomarkers used a directly read “bar-chart” to report results.148 The assays on this “V-chip” were linked to catalase. The production of oxygen by catalase displaced ink in a channel creating a “bar chart.” The displacement was proportional to the amount of analyte. A comparison of an assay for carcinoembryonic antigen (CEA) performed on the device with a conventional commercial instrument showed that the results were statistically the same.
Digital Microfluidics (DMF)
DMF is a unique branch of microfluidics in which discrete nL-sized droplets are individually addressed and moved around on a dielectric coated electrode array based usually on controlled electrowetting. These devices have shown promise in a variety of clinical application areas where the reduction of reagent volumes and analysis times made possible by the small droplet handling capabilities of DMF could significantly increase throughput and lower cost. Precise and accurate control of droplet volumes, however, is of concern with DMF devices. Typical droplet splitting can vary as much as 10%. Methods to better control droplet splitting, therefore, are an area or active interest. One approach to decreasing this variability to < 1% was recently demonstrated by ramping of voltages rather than simply switching electrodes on and off during the splitting process.149 Another significant limitation of DMF platforms is the number of electrical connections that can be practically made. A new approach to device fabrication using thin-film transistors (TFT) substantially increased the number of individually addressable electrodes.150 The increased addressing capability allowed more flexibility in terms of actively managing droplet operations. A colorimetric assay for glucose on 64 × 64 TFT array demonstrated on this device was shown to give similar results to conventional DMF devices. Most detection on DMF devices is carried out using optical techniques. In order to increase the range of potential applications of DMF additional detection modalities are being integrated. For example the interfacing of an MS detector with a DMF platform was demonstrated using a folded polyimide nanoelectrospray ionization emitter.151 Electrochemical detection in the form of voltammetry has also been integrated with a DMF device.152 Integration of such detectors, especially in situ detectors requires some reorganization of the droplet controlling electrodes which decreases the forces that the electrode can generate on the droplet. Careful design of the electrodes, however, has been shown to be able to reduce this loss of force.153 In addition to the basic instrumental development, new assays are being adapted for this platform. One assay of note was a particle-based immunoassay. 154 One this device the particles could be separated from droplets using magnets and then resuspended in other droplets. This device performed immunoassays 10× faster and used 100× less reagent volume compared to conventional assays. Both basic instrumental development and the development or adaptation of new assays will likely continue at a rapid pace in the near future in DMF. In order to entice other researchers into this area an source instrument called the DropBot has been introduced.155
Applications
Droplet-Based Applications
Droplet-based microfluidic devices provide a unique small volume environment to monitor many different types of chemical processes that might be difficult or impossible to perform on larger volume scales. For example, the process of Ag nanoparticle growth was examined using X-ray detection.131 These droplet-based systems were also used to monitor enzyme kinetics156 and perform heterogeneous enzyme assays, both related to biofuels.157 The heterogeneous assay device was quite unique and examined the enzymatic saccharification of insoluble biomass entrained in droplets.157 Multiplexed protease assays in droplets were also performed to determine the effects of inhibitors on protease activities.158 This device was then used to measure multiple protease activities in clinical samples. In addition, a droplet-based device was used to detect pathogenic cells using a FRET-based, amplification-free detection system (Figure 9D).159
General Analytical Measurements and Sensing
While most μTAS applications have a definite biological slant, there are some interesting non-biological applications. For example, monodisperse polymeric ionic liquid microgel beads were synthesized on a microfluidic device and used as chemical sensors.160 These novel beads were sensitive to redox changes and pH. They could also be used to deliver chemical payloads for controlled release or remove toxic metals from water. Such microgel beads have the potential to be applied to a wide variety of μTAS-based assays in the future.
Protein Analysis
Protein-protein interactions are key to understanding many biochemical functions. The ability to quickly and precisely control fluid flow and mixing gives these platforms unique capabilities to measure protein-protein interactions161 and protein folding kinetics.73 For example, a μTAS was used to study the aggregation of amyloid fibrils into spherical aggregates as a function of flow rate. The use of this device provided novel insights into the aggregation process.161
Cell Analysis
Cytometry
One of the most common functions of μTAS in the area of cellular analysis is cytometry. The ability to interrogate cells quickly with additional sorting capabilities on μTAS has been widely reported. Rapid advances, however, are being made in the area to broaden the applications, increase the throughout and integrate both upstream and downstream components.17,162-164 Two recent devices consisted of simple on-chip cytometers. In one device, the cells were not focused but rather detected using a line-confocal detection scheme.162 In the second device, the cells were focused using a unique self-focusing design and then detected using epifluorescence.163 Both detection approaches mark significant advances in the development of faster and inexpensive automated cytometric methods. A third device integrated cell culturing, stimulation and incubation upstream of the cytometer.164 While these systems used fluorescence markers162-164 and/or forward scattering164 for detection, another system implemented impedance to detect and differentiate between viable and nonviable cells in droplets.17
Cell Sorting
Cell sorting is an integral part of cellular analysis and a wide variety of new or improved methods to sort and enrich cells using μTAS devices are being pursued. For example, optoelectronic tweezers were used to select and isolate specific cells from a suspension of cells and transfer them to a tube where RT-qPCR was performed (Figure 7F).165 Another more automated method of sorting used isoelectric focusing to separate yeast deletion strains (Figure 7A, B).166 Only about 10% of genetic deletions, however, were found to affect the electrical properties of yeast cells in specific ways. Higher throughput sorting was accomplished in a droplet-based device where cells were first encapsulated, then incubated off-chip, and finally sorted using dielectrophoresis on a separate microfluidic device. This system was capable of screening and sorting up to 300,000 hybridomas cell clones in < 1 day.167 Using a different, more passive separation approach, several inertial microfluidic devices implemented contraction-expansion channel designs for the separation of cancer cells with high efficiencies (Figure 7C, H).168-170 Another inertial microfluidics design was based upon trapezoidal cross-sectional spiral channels and was used for the separation of leukocytes from blood (Figure 7E).171 Cell sorting can also be performed based upon a cell's physical deformability. The incorporation of micropillar arrays into microfluidic channels enabled the enrichment of mechanically deformable tumor-initiating cells.172 This device provided a novel method with which to isolate such cells. Other reported sorting methods used ferrohydrodynamics,173 acoustics,174 acoustophoretics (Figure 7D),175 affinity flow fractionation (Figure 8E,F,G),176 and a fluorescently triggered solenoid valve(Figure 8A,B,C,D).177
Figure 7.
Cell sorting and assays. (a,b) Isodielectric separation of yeast deletion strains. Reprinted with permission from Vahey, M. D.; Quiros, P. L.; Svensson, J. P.; Samson, L. D.; Voldman, J. Lab Chip 2013, 13, 2754-2763. Copyright 2013 Lab Chip. (c) Inertial focusing of living cell clumps in microscale flow. Reprinted with permission from Hur, S. C.; Brinckerhoff, T. Z.; Walthers, C. M.; Dunn, J. C. Y.; Di, C. D. PLoS One 2012, 7, e46550. Copyright 2012 PLoS One. (d) Separation of viable (white) and nonviable (black) mammalian cells by piezoelectric actuation. Reproduced from Yang, A. H. J.; Soh, H. T. Anal. Chem. 2012, 84, 10756-10762. Copyright 2012 American Chemical Society. (e) Inertial focusing in rectangular vs. trapezoidal shaped channels. Reproduced from Wu, L.; Guan, G.; Hou, H. W.; Bhagat, A. A. S.; Han, J. Anal. Chem. 2012, 84, 9324-9331. Copyright 2012 American Chemical Society. (f) Target cells are identified and selectively transported into branch channels using optical tweezers. Reprinted with permission from Huang, K.-W.; Wu, Y.-C.; Lee, J.-A.; Chiou, P.-Y. Lab Chip 2013, 13, 3721-3727. Copyright 2013 Lab Chip. (g) CTC “nanovelcro”capture device using SiNW. Reprinted with permission from Lu, Y.-T.; Zhao, L.; Shen, Q.; Garcia, M. A.; Wu, D.; Hou, S.; Song, M.; Xu, X.; Ouyang, W.-H.; Ouyang, W. W. L.; Lichterman, J.; Luo, Z.; Xuan, X.; Huang, J.; Chung, L. W. K.; Rettig, M.; Tseng, H.-R.; Shao, C.; Posadas, E. M. Methods 2013. Copyright 2013 Methods. (h) A high aspect ratio channel (segment 1) was used to focus the molecules at their equilibrium positions then a low aspect ratio channel (segment 2) was used to focus the larger particles to the center of the channel. Reprinted with permission from Zhou, J.; Giridhar, P. V.; Kasper, S.; Papautsky, I. Lab Chip 2013, 13, 1919-1929. Copyright 2013 Lab Chip.
Figure 8.
Cell sorting and capture. (a) Droplet sorter using a solenoid valve. Reprinted with permission from Cao, Z.; Chen, F.; Bao, N.; He, H.; Xu, P.; Jana, S.; Jung, S.; Lian, H.; Lu, C. Lab Chip 2013, 13, 171-178. Copyright 2013 Lab Chip. (b) Sorting of cells using multiple P-selectin gold strips. Target cells interacted with strips which altered their flow trajectories. Reprinted with permission from Bose, S.; Singh, R.; Hanewich-Hollatz, M.; Shen, C.; Lee, C.-H.; Dorfman, D. M.; Karp, J. M.; Karnik, R. Sci Rep 2013, 3, 2329. Copyright 2013 Lab Chip. (c) Microfluidic device for cell-to-cell communication study. Reproduced from Mao, S.; Zhang, J.; Li, H.; Lin, J. M. Anal Chem 2013, 85, 868-76. Copyright 2013 American Chemical Society.
Capturing Circulating Tumor Cells (CTCs)
Ultra-high efficiency cell sorting conditions are required when attempting to isolate circulating tumor cells (CTCs) as these cells can be at densities as low as 1/109 cells. Several devices have been described which attempt to sort CTCs mostly using affinity capture techniques.32,44,162,178-181 In one case, the capture efficiency of antibodies attached to PMMA and COC walls was examined. The COC μTAS devices were found to exhibit higher capture efficiencies.32 The potential of μTAS devices to make significant contributions to this area was shown in the ability of one device to successfully isolate CTCs in clinical samples and to monitor changes in CTCs over time as a function of treatment regime (Figure 7G).44 While efficiently capturing CTCs has become more routine, the collection of isolated tumor cells attached to the capturing surface can be challenging. The development, however, of a transparent nanovelcro chip was reported that allowed for selected individual cells to be microdissected and collected.178 One final method to simplify the collection of the isolated cells is to attach the affinity ligands to magnetic nanocarriers. These particles can be easily manipulated, and the cells attached to them easily collected.180,181
Non-CTC Cell and Organelle Capture
Beyond CTCs, there are many reasons to use microfluidic devices for capturing cells, particles and other biological molecules. Several devices have recently been reported that are capable of controlled capture and release of cells based upon thermoresponsive polymers,45 aptamers,46 multivalent DNA182,183 or optical waveguide loops.184 In addition to cells, a microfluidic device was demonstrated for the capture of subcellular organelles such as mitochrondria.185 The mitochondria were physically entrapped in sub-μm channels and interrogated using fluorescence microscopy.
Cell Culturing
Many assays require the culturing of cells. Microfluidic devices have the ability to culture small numbers of cells under very precisely controlled conditions and consume only very small volumes of reagents. The large surface area-to-volume ratio and small total volume of cell culture chambers on these devices, however, can potentially induce cell stress.186 Such stresses could be partially compensated for by increasing the buffer capacity of the media and the frequency of buffer replacement.186 Cell culturing assays can require the precise positioning of cells. The capture and positioning of such cells may also generate cell stress. It is important, therefore, to ensure cell viability after capture. Many different passive and active methods to position cells on microfluidic devices are available, and improvements to existing methods, or the reporting of new methods, occur frequently. For example, a device for trapping E. coli cells used dielectrophoresis. Trapped cells on this device were shown to retain viability under a variety of trapping conditions.187 Contactless negative dielectrophoresis was also demonstrated to trap individual bacterial cells using a 3-D octupole geometry.188 Single trapped bacterium could be cultured without contact with any surface for > 3 hours. An alternative physical trapping strategy for E. coli used sub-micron electron-beam fabricated channels.189 Cell viability is also a concern for 3-D cultures, as the center of the culture can be anoxic. A method to solve this problem through the creation of celloidosomes involved the encapsulation of yeast into shells of alginate on liquid cores.190 These celloidosomes were made by double emulsion technology. This process enabled precise control of the size of the celloidosome, the thickness of the outer shell and the celloidosome density. Because the cells were cultured on a liquid core, oxygen supply was not a concern.
Cell Assays
The ability to develop generic methods to perform cells assays would broaden microfluidic technology's appeal to the biomedical field. In an effort to make such generic assay development easier, a KOALA design was shown to be able to perform a variety of assays.143 In addition, a generic microfluidic design for performing ELISAs on arrays of single cells was demonstrated with detection limits at the attomole to zeptomole levels.191 Single cells were arrayed in a 2-D set of single cell wells, stimulated, and lysed. ELISAs were performed on the lysates with all assay and wash steps performed on the device.
Many recent microfluidic-based cell assays have used a variety of fluorescent markers to examine cell signaling81,192,193 and cytotoxicity. 194-196 In addition to the advantages of small scale culturing and the low reagent consumption mentioned above, most of these systems made use of the ability of microfluidic devices to generate precisely controlled spatial and temporal gradients. For example, a μTAS was reported that generated stable gradients in morphogen concentrations, e.g. bromoindirubin-39-oxime (BIO) to examine their effects on cell fate through the activation of Wnt/β-catenin signaling. Another device examined cell response to gradients of chloride channel-modulating compounds.193 The IC50 for known activators and inhibitors were in agreement with conventional plate reader measurements, but only a single culture chamber needed to be used. A device for performing whole cell dynamic mass redistribution (DMR) assays allowed estimation of residence antagonist times that had not been available with conventional assays 192 The ability to make such a measurement was the key factor in determining blockage efficiency. Measurement of cytokines (TNF-α) from THP-1 cells stimulated by lipopolysaccharide (LPS) was also demonstrated.197 Finally, the intracellular calcium response of suspension leukemic cells to mechanical stimulation was examined by designing a device in which the cells were compressed under controlled conditions within a channel manifold.198
Novel cell cytotoxicity assays using μTAS were well represented in a series of interesting reports seeking to measure IC50 values. For example, a microfluidic device integrating cell culturing and gradient generation was used to determine the IC50s for a variety of toxins, including Triton X-100m H2O2, and cadmium chloride.194 The IC50s compared well with standard assays. Another automated gradient-generating device was used to determine the cytotoxic effects of celecoxib and 5-fluorouracil on normal and cancer cells.195 Such automation can avoid operator error and improve reliability of the results. Finally, the cytotoxicity of drugs under step gradients in oxygen tensions in a microfluidic cell culture chamber was reported.196 In disease states, drugs may be introduced to hypoxic locations (e.g. interiors of tumors) and may work differently in such environments. This device allowed a better assessment of true drug cytotoxicity under realistic conditions in a cost effective manner
Gene Expression Assays
The ability to monitor and follow gene expression through several generations of cells provides extremely valuable insights into cell differentiation and mutations. Microfluidic devices, with their ability to precisely position, trap and culture cells over several days are a potentially very valuable platform to study such processes. Several devices have been reported to study gene expression over time through on-chip manipulation of the cell environment.199-204 Of course, one does need to make certain that the microfluidic environment itself does not perturb the system as pointed out in a recent report examining the effects of culturing cells on PDMS and PMMA chips. These devices were shown to affect gene expression in PC12 cells.1 In some of these μTAS, gene expression was tracked in individual lineages,202 while in other cases the overall expression of the culture was monitored in response to external stimuli.203 The ability to follow gene expression changes is critically important in stem cell biology. Recently, the differentiation of pluripotent stem cells was followed in response to gradients of molecular factors and inhibitors of mesodermal commitment.205 Eight concentration levels and 15 replicates were examined.
Other Assays
The ability to perform fixed cell, in addition to live cell, assays would important consequences to the histological community. Such fixed cell assays on microfluidic devices were recently reported and examined the expression of cancer cell biomarkers that were fluorescently stained.206
Cell-Cell Interaction Measurements
One particular strength of cell culturing on microfluidic devices is the ability to easily generate cell co-cultures in precise spatial arrangements. An interesting device was reported in which endothelial cells were cultured in two different channels separated by a polymerized collagen gel.207 This device attempted to mimic an in vivo cellular environment and encourage vascular anastomosis. Another device made use of microvalves and liquid membranes to examine tumor cell invasion and metastasis at the microscale level.208 The migration, infiltration and coexistence dynamics of different types of breast cancer cells were temporally followed and quantitated. On a third device, channels were patterned to guide the development of neural arrays.209 Multiple cell types were used and all showed differentiation and high interconnection. Finally, a device examining the cell-cell communication through the detection of signaling molecules and metabolites was demonstrated. 210 The cells were cultured in spatially separated but fluidically connected chambers. The effluent from the chambers was interfaced with ESI-Q-TOF-MS (Figure 8H). Changes in the concentrations of epinephrine and glucose were monitored.
Organs/Tissues on a Chip
Microfluidic devices provide the capability of culturing multiple types of cells with tight control over the conditions, as well as the ability to move media from place to place on a chip without significant dilution. This creates intriguing possibilities for building tissue or organ mimics on-chip. Such chips could be used to better understand the metabolism of drugs211 in the body and the effects of primary metabolites on non-target cell types. For example, a synthetic microvasculature model of the blood brain barrier (Sym-BBB) was simulated by designing microfluidic chambers to culture cells in a specified pattern.212 Results indicated that a functional barrier was successfully created. A thick film microfluidic bioreactor was also developed using human mammary epithelial cell tissue to study the effects of drug delivery.211
The most substantial barrier to creating organs on a chip is that organs must be vascularized in order to provide nutrients and remove wastes. To better understand the process of angiogenesis, a microfluidic device was fabricated from cross-linked cellulose and seeded with endothelial cells.15 Heart tissue mimics were created on microfluidic devices to better understand hypoxia-induced myocardial injury.213 Noncellular microfluidic mimics of tissues can also shed light on basic, difficult to study small-scale physiological processes. Both non-cellular stomach214 and lung215,216 mimics have been developed. The stomach mimic device examined the barrier function of stomach mucus in a simple split channel device.214 In one of the lung tissue mimics, channels of varying dimensions were used to examine the pulmonary surfactant layer at the air-liquid interface under oxidative stress conditions.216 In the other lung tissue mimic, the reopening of airways in complex geometries was examined.215
Organisms on a Chip
The ability to create small, enclosed chambers on microfluidic devices creates a unique environment in which to culture not only individual cells, but also small, free living single and multicellular organisms including bacteria, yeast, protozoans, and C. elegans. μTAS have been used to study both physical and chemical changes in unicellular cultures. Vibrio cholera, for example, were shown to exhibit a concentration-dependent avoidance to sub-lethal concentrations of antibiotics in a gradient generating microfluidic device.217 E. coli demonstrated aerotaxis on a microfluidic device in which a gradient of oxygen tension was created.80 E. coli lineages were also tracked over time to study changes in gene expression.200 At the multicellular organism level, C. elegans is a good infection model for humans and is used for drug screening. A μTAS for holding C. elegans examined antimicrobial activity using a whole-animal infection model.218 In addition to drug assays, fundamental developmental studies on C. elegans examining molecular mechanisms of development219 or gene expression were also performed.204
Disease/Pathogen Detection
The rapid and inexpensive detection of potential pathogens using microfluidic devices has many potential public health benefits. Several different types of devices have been reported demonstrating such capabilities. For example, the lysis of cells in a droplet under oil followed by RT-PCR was shown to be a viable method of disease detection(Figure 9A, B, C).220 The cell lysis and temperature cycling required by PCR were both accomplished with the integrated SAW device, removing the need for additional lysing agents or thin film heaters. A droplet-based microfluidic device was used to detect the presence of pathogenic bacteria based upon the amplification-free genetic detection of 16S rRNA.159 Another previously described device also reported the detection of pathogenic bacteria based on their SERS signature.117 Anti-microbial susceptibility testing was performed using AC electrokinetic loading to precisely position the bacteria for observation.221 Results could be obtained in <1 hour, which is a substantial improvement over conventional assays that can take days. In addition to direct pathogenic detection, an interesting study to quantify the adhesion of the bacterial pathogen Pseudomonas aeuginosa was carried out in a 2-D microwell array device with on-chip real-time PCR.222
Physical Assays
All of the assays described above examine the chemical responses of cells to perturbations. Cells also physically respond to stimuli, and several microfluidic devices to measure these physical responses have been reported. For example, a tipchip was reported to measure the penetrative forces of pollen generated by hydrostatic turgor pressure in PDMS microgaps.223 In another approach, cells were rolled across a biofunctionalized surface to remove MHC-I molecules and change the phenotype.224 These cells were shown to be more susceptible to splenocyte-mediated immune response. Similarly, rolling and adhesion assays were performed to examine the leukocyte activation cascade (LAC) under physiologically relevant shear levels.225 Chemotaxis assays were also performed using linear gradients of a chemokine set in a 3-D matrix225 and to examine the effects of controlled hypoxic environments.226 These chips provided robust platforms for studying the effects of gradients that are difficult, if not impossible to produce with conventional cell assay technology.
Nucleic Acid Analysis
Nucleic acid analysis on μTAS is an especially successful area in which μTAS have made significant contributions. The goal in most cases is to develop automated devices capable of sample-in/answer-out analyses, generally of amplified PCR products. In addition, there has been considerable work in moving to mass producible substrates and developing systems capable of parallel analysis. Depending upon the type of analysis one can either detect the PCR products in situ144,227 using fluorescence detection or one can integrate the PCR amplification with separations.6 The development of sample-in/answer-out of in situ methods is more straightforward than with methods which require the integration of a separation step, such as the separation of STR products. So it should be no surprise that more in situ type devices have been developed with complete sample-in/answer-out capabilities. Examples of such devices for the analysis of DNA in biological fluids have been demonstrated for whole blood18,144 using in situ PCR methods and for buccal cells on a swab coupled with separations for STR analysis.6 Also, the direct in situ on-chip extraction and q-RT-PCR amplification of RNA of single bacterial cells was achieved.228 This chip consisted of a 900-microwell array with a microfluidic channel to deliver and remove the appropriate chemical species necessary to perform q-RT-PCR in a massive parallel fashion.
In previous years, many DNA analysis devices have been fabricated from glass or silicon. There has been a significant shift recently to using devices now fabricated from PMMA6,229 or other polymeric materials. The use of polymers and plastics often necessitates re-engineering the various individual functional elements in the device to be material compatible. For example, the use of metal film heaters on plastics can pose several challenges in both the attachment of the film and the temperatures that can be achieved. The incorporation of a thin film aluminum heater on a KMPR photopolymer was shown to be able to reach temperatures of 165°C.230 A second device incorporating a thin film copper heater on a semitransparent poly(ethylene naphthalate) (PEN) foil attached to a co-polymer of N,N-dimethylacrylamide (DMA), N-acryloyloxy-succinimide (NAS) and [3-(methacryloyl-oxy)propyl-trimethoxy-silane] (MAPS) was reported. 231 Channels in this device were cut in a polymer adhesive, and it was used to perform melting curve analyses on PCR products up to 100°C.
A cell-lysing step is often incorporated to release DNA as part of automated nucleic acid analysis. This lysing step generally requires the introduction and removal of chemicals which complicates the process. One method to simplify this step used acoustical waves generated by an on-chip SAW device.220 The SAW both lysed the cell and heated the sample. PCR was then performed directly on the sample without any enrichment steps. Most hybridization control is thermally based. An alternative method to control hybridization was reported using pH.232 Automated DNA sequencing was demonstrated on this bead-based chip without a fluorescent label removal step. Such an approach may allow more selection modules to be incorporated on future devices compared to ones where thermal control is required. In many cases PCR amplification takes place in a single chamber and sample, wash fluids and the PCR mixed are added. In a reversal of this approach, DNA was extracted on magnetic beads, and the beads were moved through static fluid droplets which washed and prepped the DNA for PCR.74,233 These devices are more flexible than the single chamber PCR approach and could be adapted to a broad array of applications. DNA from plasma (Figure 6D)233 and whole blood74 were successfully isolated in an automated manner on these devices. The incorporation of new detection methodologies for DNA can expand the types of analyses that can be performed. Recently, the detection of specific single base extension (SBE) products was reported using MALDI-MS. PCR, allele specific SBE and desalting were all integrated on the device.234 Because of the need for thermal cycling in PCR, other methods of nucleic acid detection are being investigated to allow for the fabrication of simpler chips. Digital loop-mediated amplification (LAMP)146,227 and FRET PNA beacons159 were used to detect nucleic acids on-chip without the need for thermal cycling or amplification, respectively, thus simplifying both the analysis process and the chip design.
Throughput is often an issue, so the development of a parallel analysis system on a single device has garnered some attention. The parallel amplification of STRs was demonstrated in a 5-channel PMMA device.229 In addition, the generation of picoliter droplets of RNA on-chip, followed by coalescence with an RT-PCR droplet, generated 1000s of picoliter PCR reaction vessels that could be used to detect rare cells in a large sample of heterogeneous cells.235 While most nucleic acid analyses are focused on short DNA polymers, e.g. SNPs and STRs, there is also interest in examining much larger megabase segments to map genetic or epigenetic aberrations indicative of disease. Recently, a micropillar array was shown to effectively capture intact chromosomal DNA from single cells.236 DNA capture efficiency was close to 100% and the trapped DNA could be released using endonuclease digestion. In addition, generic cellular damage at the chromosomal level using COMET assays were reported using an agarose-based chip. Lysed DNA from cells was electrophoretically separated and analyzed.237
Forensic Analysis
The interest in developing automated, portable, low cost, robust, single use devices for a variety of forensic analyses is high. The standards for such devices to be accepted for routine forensic use is also high, so it is of importance to note that a system of two microfluidic devices that performed a 27 locus assay in < 1 hr has met all of the U.S. and European law enforcement agency standards.238 A fully integrated device that permitted the sample-in/STR answer-out analysis of buccal cells in 45 min was also demonstrated as discussed under integrated devices above.6 In addition to nucleic acid analysis, other types of forensic analysis (such as for drugs of abuse) may benefit from a μTAS approach. As mentioned previously a microfluidic device was demonstrated for the screening of methamphetamine in saliva.117
Drug Screening
The ability to perform rapid, inexpensive analyses on a variety of samples would greatly enhance the screening of potential drug candidates. μTAS devices show great potential in this area. An agarose-in-oil microfluidic droplet device was described to rapidly screen for novel antibiotics.239 In this device, yeast and E. coli were genetically modified to produce and excrete potential antibiotics. These cells were then encapsulated with S. aureus to examine the secreted drug's efficacy. In another device, bacteria were again encapsulated in droplets to form chemostats.240 These droplets were combined with other droplets to examine the effects of various levels of antibiotic dosing. A third device examined the effects of polychlorinated biphenyls on neuronal cells.241 Dopamine signaling from PC-12 cells was measured. In addition to being able to rapidly quantitate the amount of a drug in tissues after administration, it is critical to understand how drugs are distributed and metabolized in the body. μTAS has potential advantages over conventional instrumentation in terms of examining limited volume samples taken from small animals for pharmacokinetic investigations. A good demonstration of these advantages was reported using a LC-chip-MS/MS device for quantitating fluoxetine and norfluoxetine in rat serum.242
Environmental Health and Safety, Food and Water Analysis
The capability to inexpensively separate and detect a variety of environmental, food and water contaminants is key to providing safe living conditions in society. Several μTAS for performing such analyses have already been discussed including the separation of banned aromatic amines,7 the identification of pathogenic bacteria,116,243 the detection of nutrients in water,139 and the detection of pesticide residues on vegetables244 Another example includes the real time detection of trace explosive vapors at levels of 1 ppb. In this device gas phase analytes were detected on-chip using SERS.245 Other μTAS assays focused on the development of toxicity tests. In one example, the toxicity of quantum dots on 3-D cell culturing models was assessed.244 Toxicity screening against algae with an integrated gradient generator was also reported.246 This device streamlined the screening method through the integration of culturing, addition and dosing of toxin, and detection of cellular endpoints in one assay. In addition to toxicity assays, the general detection of potentially pathogenic organisms was of special note. While some pathogen detection devices were already discussed above, several other devices that focused on foods and fluids analyses have been demonstrated, including the detection of E. coli in drinking water.247 The E. coli in this device were trapped on Ag-Au nanoparticles immobilized on the microchannel surface. The device was able to differentiate between safe and contaminated levels of E. coli in water. Other systems incorporated PCR for the detection of a broader range of pathogens in water,248 or for the detection of C. sakazakkii in milk.249 In an especially creative approach, a novel microfluidic device was used to investigate the potential for immunomodulatory effects of dairy food by creating a miniaturized human gastrointestinal tract on-chip.250
Extreme Conditions
All of the advantages of μTAS can be utilized to create devices capable of operating under extreme environmental conditions. One illustrative example of this is the integration of μTAS with an autonomous underwater vehicle for explosives detection (Figure 10A,B,C).251 High-throughput microfluidic immunosensors were used to detect trinitrotoluene at concentrations of 20-175 ppb. The system proved robust under the conditions tested.
Figure 10.

Extreme Environments. (a) TNT immunoassay used to detect ppb levels of explosives in water. (b) AUV used for analysis of minute levels of explosives in water with microfluidic chip mounted on the front. (c) Chip mounting on AUV. Reprinted with permission from Adams, A. A.; Charles, P. T.; Veitch, S. P.; Hanson, A.; Deschamps, J. R.; Kusterbeck, A. W. Anal Bioanal Chem 2013, 405, 5171-8. Copyright 2013 Springer.
Conclusions and Outlooks
The last 12 months have seen considerable progress toward the goal of truly sample-in/answer-out μTAS. This progress is especially apparent in the areas of nucleic acid, water, and food analysis. Many of these devices are still in the research laboratory and have not been tested under realistic conditions yet, but the rate of movement out of the lab and into the clinic and field should begin to increase in the near future. In addition to the development of true μTAS, the unique capabilities of these platforms in various areas of cellular analysis continue to be apparent. Many sophisticated, multistep analyses examining cell-cell interactions, toxicity, responses to external physical and chemical stimuli, gene expression, and chemotaxis have been developed. The range of applications is likely to broaden significantly over the next few years, and the pace of integration should quicken. On a similar note, expect to see a move toward more sophisticated tissue and organ models that can realistically mimic in vivo conditions for significant amounts of time. As the focus on applications slowly moves from the academic laboratory to the clinic, the migration to substrates amenable high volume manufacturing will increase, although PDMS will probably be the material of choice for most academic labs for the foreseeable future. In addition, because of potential problems with PDMS as a substrate for cell culturing, there should be steady interest in improving fabrication methods for poly(styrene) devices. Finally, attempts to minimize external components especially for pumping and detection to create inexpensive, portable devices for clinical and resource-poor situations will continue.
Acknowledgments
This work was supported by NIH grant R21NS061202. An NSF REU program co-sponsored by DOD ASSURE (CHE-1004991) supported M.P.
Biographies
Samantha A. Stewart-James received her B.S. degree in Chemistry from Minnesota State, University Moorhead in 2012. She is currently a graduate student in the Chemistry Dept. at Kansas State University. Her current research is focused on developing microanalytical tools to study the immune response of Anopheles gambiae mosquitoes to Plasmodium parasites.
Kathleen A. Sellens received her B.S. in Chemistry from McKendree University in 2012. Currently, she is a graduate in the Chemistry Department at the Kansas State University. Her research focuses on isolating Anopheles gambiae immune response protein complexes using capillary immunoaffinity chromatography.
Tom G. Mickleburgh is an undergraduate biochemistry student at Kansas State who will soon be pursuing graduate studies within microfluidics. His research interests involve high throughput microfluidic devices for single cell analysis.
Melissa Pressnall is currently an undergraduate student at Bethany College in Lindsborg, KS. She plans on graduating with a B.A. in Chemistry and Biology in May 2014. She worked in Dr. Culbertson's lab at Kansas State University as an REU student in the summer of 2013. She wants to pursue graduate studies in medicinal or pharmaceutical chemistry.
Christopher T. Culbertson received a Ph.D. in Chemistry from the University of North Carolina at Chapel Hill in 1991 followed by a post-doctoral fellowship at Oak Ridge National Laboratory. He is presently an Associate Professor in the Chemistry Department at Kansas State University.
Footnotes
Author contributions: All authors contributed to the wring of the manuscript and have approved the final version.
Notes: The authors declare no competing financial interests.
References
- 1.Lopacinska JM, Emneus J, Dufva M. PLoS One. 2013;8:e53107. doi: 10.1371/journal.pone.0053107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chia GLP, Bollgruen P, Egunov AI, Mager D, Malloggi F, Korvink JG, Luchnikov VA. Lab Chip. 2013;13:3827–3831. doi: 10.1039/c3lc50542a. [DOI] [PubMed] [Google Scholar]
- 3.Dandin M, Abshire P, Smela E. J Micromech Microeng. 2012;22:095018/1–095018/9. [Google Scholar]
- 4.Lee HW, Bien DCS, Badaruddin SAM, Teh AS. Microsyst Technol. 2013;19:253–259. [Google Scholar]
- 5.Rojas L, Norarat R, Napari M, Kivisto H, Chienthavorn O, Whitlow HJ. Nucl Instrum Methods Phys Res, Sect B. 2013;306:296–298. [Google Scholar]
- 6.Lounsbury JA, Karlsson A, Miranian DC, Cronk SM, Nelson DA, Li J, Haverstick DM, Kinnon P, Saul DJ, Landers JP. Lab Chip. 2013;13:1384–1393. doi: 10.1039/c3lc41326h. [DOI] [PubMed] [Google Scholar]
- 7.Li R, Wang L, Gao X, Du G, Zhai H, Wang X, Guo G, Pu Q. J Hazard Mater. 2013;248-249:268–275. doi: 10.1016/j.jhazmat.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Rahmanian O, DeVoe DL. Lab Chip. 2013;13:1102–1108. doi: 10.1039/c2lc41057e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson AS, Anderson KB, Halpin ST, Kirkpatrick DC, Spence DM, Martin RS. Analyst. 2013;138:129–136. doi: 10.1039/c2an36168j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson KB, Halpin ST, Johnson AS, Martin RS, Spence DM. Analyst. 2013;138:137–43. doi: 10.1039/c2an36171j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berthier E, Young EWK, Beebe D. Lab Chip. 2012;12:1224–1237. doi: 10.1039/c2lc20982a. [DOI] [PubMed] [Google Scholar]
- 12.Gabriel EFM, Duarte GF, Jr, Garcia PdT, de JDP, Coltro WKT. Electrophoresis. 2012;33:2660–2667. doi: 10.1002/elps.201200009. [DOI] [PubMed] [Google Scholar]
- 13.Burlage K, Gerhardy C, Praefke H, Liauw MA, Schomburg WK. Chem Eng J. 2013;227:111–115. [Google Scholar]
- 14.Mokkapati VRSS, Bethge O, Hainberger R, Brueckl H. IEEE Electron Compon Technol Conf. 2012;62nd:1965–1969. [Google Scholar]
- 15.Pei Y, Wang X, Huang W, Liu P, Zhang L. Cellulose. 2013;20:1897–1909. [Google Scholar]
- 16.Baker BM, Trappmann B, Stapleton SC, Toro E, Chen CS. Lab Chip. 2013;13:3246–3252. doi: 10.1039/c3lc50493j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kemna EWM, Segerink LI, Wolbers F, Vermes I, van d B A. Analyst. 2013;138:4585–4592. doi: 10.1039/c3an00569k. [DOI] [PubMed] [Google Scholar]
- 18.Marshall LA, Wu LL, Babikian S, Bachman M, Santiago JG. Anal Chem. 2012;84:9640–9645. doi: 10.1021/ac302622v. [DOI] [PubMed] [Google Scholar]
- 19.Huang Y, Mason AJ. Lab Chip. 2013;13:3929–3934. doi: 10.1039/c3lc50437a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B, Dong Q, Korman CE, Li Z, Zaghloul ME. Sci Rep. 2013;3:1098, 8. [Google Scholar]
- 21.Hitzbleck M, Lovchik RD, Delamarche E. Adv Mater. 2013;25:2672–2676. doi: 10.1002/adma.201204854. [DOI] [PubMed] [Google Scholar]
- 22.Renault C, Li X, Fosdick SE, Crooks RM. Anal Chem. 2013;85:7976–7979. doi: 10.1021/ac401786h. [DOI] [PubMed] [Google Scholar]
- 23.Xing S, Jiang J, Pan T. Lab Chip. 2013;13:1937–1947. doi: 10.1039/c3lc41255e. [DOI] [PubMed] [Google Scholar]
- 24.Auad P, Ueda E, Levkin PA. ACS Appl Mater Interfaces. 2013;5:8053–7. doi: 10.1021/am402135e. [DOI] [PubMed] [Google Scholar]
- 25.Kim M, Song KH, Doh J. Colloids Surf B Biointerfaces. 2013;112C:134–138. doi: 10.1016/j.colsurfb.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 26.You JB, Min KI, Lee B, Kim DP, Im SG. Lab Chip. 2013;13:1266–1272. doi: 10.1039/c2lc41266g. [DOI] [PubMed] [Google Scholar]
- 27.Li L, Bi X, Yu J, Ren CL, Liu Z. Electrophoresis. 2012;33:2591–2597. doi: 10.1002/elps.201200206. [DOI] [PubMed] [Google Scholar]
- 28.Chen Cf, Gerlach TF. RSC Adv. 2013;3:14066–14072. [Google Scholar]
- 29.Lu JC, Liao WH, Tung YC. J Micromech Microeng. 2012;22:075006/1–075006/8. [Google Scholar]
- 30.Diaz-Gonzalez M, Baldi A. Anal Chem. 2012;84:7838–7844. doi: 10.1021/ac301512f. [DOI] [PubMed] [Google Scholar]
- 31.Douma MD, Brown L, Koerner T, Hugh HJ, Oleschuk RD. Microfluid Nanofluid. 2013;14:133–143. [Google Scholar]
- 32.Jackson JM, Witek MA, Hupert ML, Brady C, Pullagurla S, Kamande J, Aufforth RD, Tignanelli CJ, Torphy RJ, Yeh JJ, Soper SA. Lab Chip. 2013 doi: 10.1039/c3lc50618e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parra-Cabrera C, Sporer C, Rodriguez-Villareal I, Rodriguez-Trujillo R, Homs-Corbera A, Samitier J. Lab Chip. 2012;12:4143–50. doi: 10.1039/c2lc40107j. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, Feng X, Xu F, Hu X, Li P, Liu BF. Anal Methods. 2013;5:4694–4700. [Google Scholar]
- 35.Liu CM, Liang RP, Wang XN, Wang JW, Qiu JD. J Chromatogr A. 2013;1294:145–151. doi: 10.1016/j.chroma.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 36.Tu Q, Wang JC, Liu R, He J, Zhang Y, Shen S, Xu J, Liu J, Yuan MS, Wang J. Colloids Surf, B. 2013;102:361–370. doi: 10.1016/j.colsurfb.2012.08.033. [DOI] [PubMed] [Google Scholar]
- 37.Wu T, Suzuki H, Su Y, Tang Z, Zhang L, Yomo T. Soft Matter. 2013;9:3473–3477. [Google Scholar]
- 38.Kwong P, Gupta M. Anal Chem. 2012;84:10129–10135. doi: 10.1021/ac302861v. [DOI] [PubMed] [Google Scholar]
- 39.Style RW, Che Y, Park SJ, Weon BM, Je JH, Hyland C, German GK, Power MP, Wilen LA, Wettlaufer JS, Dufresne ER. Proc Natl Acad Sci U S A. 2013;110:12541–12544. doi: 10.1073/pnas.1307122110. S12541/1-S12541/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W, Choi DS, Nguyen YH, Chang J, Qin L. Sci Rep. 2013;3:2332. doi: 10.1038/srep02332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sengupta A, Bahr C, Herminghaus S. Soft Matter. 2013;9:7251–7260. [Google Scholar]
- 42.Sochol RD, Li S, Lee LP, Lin L. Lab Chip. 2012;12:4168–4177. doi: 10.1039/c2lc40610a. [DOI] [PubMed] [Google Scholar]
- 43.Bao H, Zhang L, Chen G. J Chromatogr A. 2013;1310:74–81. doi: 10.1016/j.chroma.2013.08.040. [DOI] [PubMed] [Google Scholar]
- 44.Lu YT, Zhao L, Shen Q, Garcia MA, Wu D, Hou S, Song M, Xu X, Ouyang WH, Ouyang WWL, Lichterman J, Luo Z, Xuan X, Huang J, Chung LWK, Rettig M, Tseng HR, Shao C, Posadas EM. Methods. 2013 in press, corrected proof http://dx.doi.org/10.1016/j.ymeth.2013.06.019.
- 45.Gurkan UA, Tasoglu S, Akkaynak D, Avci O, Unluisler S, Canikyan S, MacCallum N, Demirci U. Adv Healthcare Mater. 2012;1:661–668. doi: 10.1002/adhm.201200009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu J, Nguyen TH, Pei R, Stojanovic M, Lin Q. Lab Chip. 2012;12:3504–3513. doi: 10.1039/c2lc40411g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hosseini Y, Zellner P, Agah M. J Microelectromech Syst. 2013;22:356–362. [Google Scholar]
- 48.Chen Y, Pei W, Tang R, Chen S, Chen H. Sens Actuators, A. 2013;189:143–150. [Google Scholar]
- 49.Swaminathan S, Harris T, McClellan D, Cui Y. Mater Lett. 2013;106:208–212. [Google Scholar]
- 50.Puttaraksa N, Napari M, Merilainen L, Whitlow HJ, Sajavaara T, Gilbert L. Nucl Instrum Methods Phys Res, Sect B. 2013;306:302–306. [Google Scholar]
- 51.Farshchian B, Park S, Choi J, Amirsadeghi A, Lee J, Park S. Lab Chip. 2012;12:4764–71. doi: 10.1039/c2lc40572e. [DOI] [PubMed] [Google Scholar]
- 52.Rekstyte S, Malinauskas M, Juodkazis S. Opt Express. 2013;21:17028–17041. doi: 10.1364/OE.21.017028. 14 pp. [DOI] [PubMed] [Google Scholar]
- 53.Nie J, Liang Y, Zhang Y, Le S, Li D, Zhang S. Analyst. 2013;138:671–676. doi: 10.1039/c2an36219h. [DOI] [PubMed] [Google Scholar]
- 54.Travagliati M, Girardo S, Pisignano D, Beltram F, Cecchini M. Anal Chem. 2013;85:8080–8084. doi: 10.1021/ac4019796. [DOI] [PubMed] [Google Scholar]
- 55.Abi-Samra K, Kim TH, Park DK, Kim N, Kim J, Kim H, Cho YK, Madou M. Lab Chip. 2013;13:3253–3260. doi: 10.1039/c3lc50472g. [DOI] [PubMed] [Google Scholar]
- 56.Nguyen TV, Duncan PN, Ahrar S, Hui EE. Lab Chip. 2012;12:3991–3994. doi: 10.1039/c2lc40466d. [DOI] [PubMed] [Google Scholar]
- 57.Devaraju NSGK, Unger MA. Lab Chip. 2012;12:4809–4815. doi: 10.1039/c2lc21155f. [DOI] [PubMed] [Google Scholar]
- 58.Yu F, Horowitz MA, Quake SR. Lab Chip. 2013;13:1911–1918. doi: 10.1039/c3lc41394b. [DOI] [PubMed] [Google Scholar]
- 59.Weston MC, Nash CK, Homesley JJ, Fritsch I. Anal Chem. 2012;84:9402–9409. doi: 10.1021/ac302063a. [DOI] [PubMed] [Google Scholar]
- 60.Korten S, Albet-Torres N, Paderi F, ten SL, Diez S, Korten T, te KG, Mansson A. Lab Chip. 2013;13:866–876. doi: 10.1039/c2lc41099k. [DOI] [PubMed] [Google Scholar]
- 61.Jingmin L, Chong L, Zheng X, Kaiping Z, Xue K, Liding W. PLoS One. 2012;7:e50320. doi: 10.1371/journal.pone.0050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arata H, Komatsu H, Hosokawa K, Maeda M. PLoS One. 2012;7:e48329. doi: 10.1371/journal.pone.0048329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim SJ, Paczesny S, Takayama S, Kurabayashi K. Lab Chip. 2013;13:2091–2098. doi: 10.1039/c3lc50187f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Novo P, Volpetti F, Chu V, Conde JP. Lab Chip. 2013;13:641–645. doi: 10.1039/c2lc41083d. [DOI] [PubMed] [Google Scholar]
- 65.Gong MM, MacDonald BD, Nguyen TV, Sinton D. Biomicrofluidics. 2012;6:044102/1–044102/13. [Google Scholar]
- 66.Wang G, Ho HP, Chen Q, Yang AKL, Kwok HC, Wu SY, Kong SK, Kwan YW, Zhang X. Lab Chip. 2013;13:3698–3706. doi: 10.1039/c3lc50545f. [DOI] [PubMed] [Google Scholar]
- 67.Thio THG, Ibrahim F, Al-Faqheri W, Moebius J, Khalid NS, Soin N, Kahar MKBA, Madou M. Lab Chip. 2013;13:3199–3209. doi: 10.1039/c3lc00004d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen LC, Wu CC, Wu RG, Chang HC. Langmuir. 2012;28:11281–5. doi: 10.1021/la302186d. [DOI] [PubMed] [Google Scholar]
- 69.Murray C, McCoul D, Sollier E, Ruggiero T, Niu X, Pei Q, Di CD. Microfluid Nanofluid. 2013;14:345–358. [Google Scholar]
- 70.Wu X, Langan TJ, Durney BC, Holland LA. Electrophoresis. 2012;33:2674–2681. doi: 10.1002/elps.201200173. [DOI] [PubMed] [Google Scholar]
- 71.He Y, Huang BL, Lu DX, Zhao J, Xu BB, Zhang R, Lin XF, Chen QD, Wang J, Zhang YL, Sun HB. Lab Chip. 2012;12:3866–3869. doi: 10.1039/c2lc40401j. [DOI] [PubMed] [Google Scholar]
- 72.Chiu YJ, Cho SH, Mei Z, Lien V, Wu TF, Lo YH. Lab Chip. 2013;13:1803–9. doi: 10.1039/c3lc41202d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Burke KS, Parul D, Reddish MJ, Dyer RB. Lab Chip. 2013;13:2912–21. doi: 10.1039/c3lc50497b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.den Dulk RC, Schmidt KA, Sabatte G, Liebana S, Prins MWJ. Lab Chip. 2013;13:106–118. doi: 10.1039/c2lc40929a. [DOI] [PubMed] [Google Scholar]
- 75.Li X, Zwanenburg P, Liu X. Lab Chip. 2013;13:2609–2614. doi: 10.1039/c3lc00006k. [DOI] [PubMed] [Google Scholar]
- 76.Collino RR, Reilly-Shapiro N, Foresman B, Xu K, Utz M, Landers JP, Begley MR. Lab Chip. 2013;13:3668–3674. doi: 10.1039/c3lc50481f. [DOI] [PubMed] [Google Scholar]
- 77.Chen CY, Chen CY, Lin CY, Hu YT. Lab Chip. 2013;13:2834–2839. doi: 10.1039/c3lc50407g. [DOI] [PubMed] [Google Scholar]
- 78.Jensen EC, Stockton AM, Chiesl TN, Kim J, Bera A, Mathies RA. Lab Chip. 2013;13:288–296. doi: 10.1039/c2lc40861a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ahmed D, Chan CY, Lin SCS, Muddana HS, Nama N, Benkovic SJ, Huang TJ. Lab Chip. 2013;13:328–31. doi: 10.1039/c2lc40923b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Adler M, Erickstad M, Gutierrez E, Groisman A. Lab Chip. 2012;12:4835–4847. doi: 10.1039/c2lc21006a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim C, Kreppenhofer K, Kashef J, Gradl D, Herrmann D, Schneider M, Ahrens R, Guber A, Wedlich D. Lab Chip. 2012;12:5186–5194. doi: 10.1039/c2lc40172j. [DOI] [PubMed] [Google Scholar]
- 82.Casadevall iSX, Turek V, Prodromakis T, Edel JB. Lab Chip. 2012;12:4049–4054. doi: 10.1039/c2lc40746a. [DOI] [PubMed] [Google Scholar]
- 83.Hansson J, Karlsson JM, Haraldsson T, Brismar H, van d W W, Russom A. Lab Chip. 2012;12:4644–4650. doi: 10.1039/c2lc40241f. [DOI] [PubMed] [Google Scholar]
- 84.Collins DJ, Alan T, Helmerson K, Neild A. Lab Chip. 2013;13:3225–3231. doi: 10.1039/c3lc50372k. [DOI] [PubMed] [Google Scholar]
- 85.Zeng Y, Shin M, Wang T. Lab Chip. 2013;13:267–273. doi: 10.1039/c2lc40906b. [DOI] [PubMed] [Google Scholar]
- 86.Sun X, Tang K, Smith RD, Kelly RT. Microfluid Nanofluid. 2013;15:117–126. doi: 10.1007/s10404-012-1133-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zec H, Rane TD, Wang TH. Lab Chip. 2012;12:3055–3062. doi: 10.1039/c2lc40399d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dangla R, Kayi SC, Baroud CN. Proc Natl Acad Sci U S A. 2013;110:853–858. doi: 10.1073/pnas.1209186110. S853/1-S853/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu J, Zhang M, Li X, Wen W. Anal Chem. 2012;84:9689–9693. doi: 10.1021/ac302249h. [DOI] [PubMed] [Google Scholar]
- 90.Boybay MS, Jiao A, Glawdel T, Ren CL. Lab Chip. 2013;13:3840–3846. doi: 10.1039/c3lc50418b. [DOI] [PubMed] [Google Scholar]
- 91.Zhang Y, Ho YP, Chiu YL, Chan HF, Chlebina B, Schuhmann T, You L, Leong KW. Biomaterials. 2013;34:4564–4572. doi: 10.1016/j.biomaterials.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schoeman RM, Kemna EWM, Wolbers F, van d B A. Electrophoresis. 2013 in press, corrected proof. [Google Scholar]
- 93.Wu L, Chen P, Dong Y, Feng X, Liu BF. Biomed Microdevices. 2013;15:553–560. doi: 10.1007/s10544-013-9754-z. [DOI] [PubMed] [Google Scholar]
- 94.Zhou H, Yao S. Lab Chip. 2013;13:962–969. doi: 10.1039/c2lc41060e. [DOI] [PubMed] [Google Scholar]
- 95.Li S, Ding X, Guo F, Chen Y, Lapsley MI, Lin SCS, Wang L, McCoy JP, Cameron CE, Huang TJ. Anal Chem. 2013;85:5468–5474. doi: 10.1021/ac400548d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Park W, Han S, Lee H, Kwon S. Microfluid Nanofluid. 2012;13:511–518. [Google Scholar]
- 97.Casavant BP, Berthier E, Theberge AB, Berthier J, Montanez-Sauri SI, Bischel LL, Brakke K, Hedman CJ, Bushman W, Keller NP, Beebe DJ. Proc Natl Acad Sci U S A. 2013;110:10111–10116. doi: 10.1073/pnas.1302566110. S10111/1-S10111/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Albrecht JC, Kotani A, Lin JS, Soper SA, Barron AE. Electrophoresis. 2013;34:590–597. doi: 10.1002/elps.201200462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Manage DP, Elliott DG, Backhouse CJ. Electrophoresis. 2012;33:3213–3221. doi: 10.1002/elps.201200188. [DOI] [PubMed] [Google Scholar]
- 100.Hurth C, Gu J, Aboud M, Estes MD, Nordquist AR, McCord B, Zenhausern F. Electrophoresis. 2012;33:2604–2611. doi: 10.1002/elps.201200148. [DOI] [PubMed] [Google Scholar]
- 101.Huft J, Haynes CA, Hansen CL. Anal Chem. 2013;85:1797–1802. doi: 10.1021/ac303153a. [DOI] [PubMed] [Google Scholar]
- 102.Sukas S, De MW, Desmet G, Gardeniers HJGE. Anal Chem. 2012;84:9996–10004. doi: 10.1021/ac302450z. [DOI] [PubMed] [Google Scholar]
- 103.Guan Q, Noblitt SD, Henry CS. Electrophoresis. 2012;33:2875–2883. doi: 10.1002/elps.201200255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tentori AM, Hughes AJ, Herr AE. Anal Chem. 2013;85:4538–4545. doi: 10.1021/ac4001767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang S, Njoroge SK, Battle K, Zhang C, Hollins BC, Soper SA, Feng J. Lab Chip. 2012;12:3362–3369. doi: 10.1039/c2lc40132k. [DOI] [PubMed] [Google Scholar]
- 106.Hughes AJ, Herr AE. Proc Natl Acad Sci U S A. 2012;109:21450–5. doi: 10.1073/pnas.1207754110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Regel A, Lunte S. Electrophoresis. 2013;34:2101–2106. doi: 10.1002/elps.201300055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Selimovic A, Martin RS. Electrophoresis. 2013;34:2092–100. doi: 10.1002/elps.201300163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Parisi J, Liu Y, Su L, Lei Y. RSC Adv. 2013;3:1388–1396. [Google Scholar]
- 110.Scott DE, Grigsby RJ, Lunte SM. Chemphyschem. 2013;14:2288–94. doi: 10.1002/cphc.201300449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Delaney JL, Doeven EH, Harsant AJ, Hogan CF. Anal Chim Acta. 2013;790:56–60. doi: 10.1016/j.aca.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 112.Godino N, Gorkin R, Bourke K, Ducree J. Lab Chip. 2012;12:3281–3284. doi: 10.1039/c2lc40223h. [DOI] [PubMed] [Google Scholar]
- 113.Matharu Z, Enomoto J, Revzin A. Anal Chem. 2013;85:932–939. doi: 10.1021/ac3025619. [DOI] [PubMed] [Google Scholar]
- 114.Fernandez-la-Villa A, Sanchez-Barragan D, Pozo-Ayuso DF, Castano-Alvarez M. Electrophoresis. 2012;33:2733–2742. doi: 10.1002/elps.201200236. [DOI] [PubMed] [Google Scholar]
- 115.Gaudry AJ, Breadmore MC, Guijt RM. Electrophoresis. 2013;34:2980–2987. doi: 10.1002/elps.201300256. [DOI] [PubMed] [Google Scholar]
- 116.Lillehoj PB, Kaplan CW, He J, Shi W, Ho CM. J Lab Autom. 2013 doi: 10.1177/2211068213495207. http://jla.sagepub.com/content/early/2013/07/12/2211068213495207. [DOI] [PubMed]
- 117.Andreou C, Hoonejani MR, Barmi MR, Moskovits M, Meinhart CD. ACS Nano. 2013;7:7157–7164. doi: 10.1021/nn402563f. [DOI] [PubMed] [Google Scholar]
- 118.Lu X, Samuelson DR, Xu Y, Zhang H, Wang S, Rasco BA, Xu J, Konkel ME. Anal Chem. 2013;85:2320–2327. doi: 10.1021/ac303279u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li B, Zhang W, Chen L, Lin B. Electrophoresis. 2013;34:2162–8. doi: 10.1002/elps.201300138. [DOI] [PubMed] [Google Scholar]
- 120.Benz C, Retzbach H, Nagl S, Belder D. Lab Chip. 2013;13:2808–2814. doi: 10.1039/c3lc00057e. [DOI] [PubMed] [Google Scholar]
- 121.Cao L, Cheng L, Zhang Z, Wang Y, Zhang X, Chen H, Liu B, Zhang S, Kong J. Lab Chip. 2012;12:4864–4869. doi: 10.1039/c2lc40564d. [DOI] [PubMed] [Google Scholar]
- 122.Gitlin L, Hoera C, Meier RJ, Nagl S, Belder D. Lab Chip. 2013;13:4134–4141. doi: 10.1039/c3lc50387a. [DOI] [PubMed] [Google Scholar]
- 123.Salleh MHM, Glidle A, Sorel M, Reboud J, Cooper JM. Chem Commun. 2013;49:3095–3097. doi: 10.1039/c3cc38228a. [DOI] [PubMed] [Google Scholar]
- 124.Teste B, Ali-Cherif A, Viovy JL, Malaquin L. Lab Chip. 2013;13:2344–2349. doi: 10.1039/c3lc50353d. [DOI] [PubMed] [Google Scholar]
- 125.Khan W, Park SY. Lab Chip. 2012;12:4553–4559. doi: 10.1039/c2lc40710h. [DOI] [PubMed] [Google Scholar]
- 126.Kuster SK, Fagerer SR, Verboket PE, Eyer K, Jefimovs K, Zenobi R, Dittrich PS. Anal Chem. 2013;85:1285–1289. doi: 10.1021/ac3033189. [DOI] [PubMed] [Google Scholar]
- 127.Rob T, Gill PK, Golemi-Kotra D, Wilson DJ. Lab Chip. 2013;13:2528–2532. doi: 10.1039/c3lc00007a. [DOI] [PubMed] [Google Scholar]
- 128.SadAbadi H, Badilescu S, Packirisamy M, Wuthrich R. Biosens Bioelectron. 2013;44:77–84. doi: 10.1016/j.bios.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 129.Zhang Y, Tang Y, Hsieh YH, Hsu CY, Xi J, Lin KJ, Jiang X. Lab Chip. 2012;12:3012–3015. doi: 10.1039/c2lc40590c. [DOI] [PubMed] [Google Scholar]
- 130.Lee SH, Lindquist NC, Wittenberg NJ, Jordan LR, Oh SH. Lab Chip. 2012;12:3882–3890. doi: 10.1039/c2lc40455a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stehle R, Goerigk G, Wallacher D, Ballauff M, Seiffert S. Lab Chip. 2013;13:1529–1537. doi: 10.1039/c3lc41291a. [DOI] [PubMed] [Google Scholar]
- 132.Sai KK, Navin CV, Biswas S, Singh V, Ham K, Bovenkamp GL, Theegala CS, Miller JT, Spivey JJ, Kumar CSSR. J Am Chem Soc. 2013;135:5450–5456. doi: 10.1021/ja400434c. [DOI] [PubMed] [Google Scholar]
- 133.Fan F, Gu WH, Wang XH, Chang SJ. Appl Phys Lett. 2013;102:121113/1–121113/4. [Google Scholar]
- 134.Waldbaur A, Kittelmann J, Radtke CP, Hubbuch J, Rapp BE. Lab Chip. 2013;13:2337–2343. doi: 10.1039/c3lc00042g. [DOI] [PubMed] [Google Scholar]
- 135.Scott A, Au AK, Vinckenbosch E, Folch A. Lab Chip. 2013;13:2036–2039. doi: 10.1039/c3lc50201e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Martinez-Rivas A, Mazenq L, Jalabert L, Dollat X, Vieu C, Severac C. Microelectron Eng. 2013;110:461–464. [Google Scholar]
- 137.Sabourin D, Skafte-Pedersen P, Soee MJ, Hemmingsen M, Alberti M, Coman V, Petersen J, Emneus J, Jorg PK, Snakenborg D, Joergensen F, Clausen C, Holmstroem K, Dufva M. J Lab Autom. 2013;18:212–228. doi: 10.1177/2211068212461445. [DOI] [PubMed] [Google Scholar]
- 138.Duford DA, Xi Y, Salin ED. Anal Chem. 2013;85:7834–7841. doi: 10.1021/ac401416w. [DOI] [PubMed] [Google Scholar]
- 139.Hwang H, Kim Y, Cho J, Lee Jy, Choi MS, Cho YK. Anal Chem. 2013;85:2954–2960. doi: 10.1021/ac3036734. [DOI] [PubMed] [Google Scholar]
- 140.Sasso LA, Johnston IH, Zheng M, Gupte RK, Uendar A, Zahn JD. Microfluid Nanofluid. 2012;13:603–612. doi: 10.1007/s10404-012-0980-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lee M, Lee K, Kim KH, Oh KW, Choo J. Lab Chip. 2012;12:3720–7. doi: 10.1039/c2lc40353f. [DOI] [PubMed] [Google Scholar]
- 142.Gan M, Tang Y, Shu Y, Wu H, Chen L. Small. 2012;8:863–7. doi: 10.1002/smll.201102322. [DOI] [PubMed] [Google Scholar]
- 143.Berthier E, Guckenberger DJ, Cavnar P, Huttenlocher A, Keller NP, Beebe DJ. Lab Chip. 2013;13:424–31. doi: 10.1039/c2lc41019b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Petralia S, Verardo R, Klaric E, Cavallaro S, Alessi E, Schneider C. Sens Actuators, B. 2013;187:99–105. [Google Scholar]
- 145.Tantra R, van HH. Lab Chip. 2013;13:2199–2201. doi: 10.1039/c3lc50246e. [DOI] [PubMed] [Google Scholar]
- 146.Myers FB, Henrikson RH, Bone J, Lee LP. PLoS One. 2013;8:e70266. doi: 10.1371/journal.pone.0070266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gao Y, Lam AWY, Chan WCW. ACS Appl Mater Interfaces. 2013;5:2853–2860. doi: 10.1021/am302633h. [DOI] [PubMed] [Google Scholar]
- 148.Song Y, Zhang Y, Bernard PE, Reuben JM, Ueno NT, Arlinghaus RB, Zu Y, Qin L. Nat Commun. 2012;3:2292/1–2292/9. doi: 10.1038/ncomms2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Banerjee A, Liu Y, Heikenfeld J, Papautsky I. Lab Chip. 2012;12:5138–5141. doi: 10.1039/c2lc40723j. [DOI] [PubMed] [Google Scholar]
- 150.Hadwen B, Broder GR, Morganti D, Jacobs A, Brown C, Hector JR, Kubota Y, Morgan H. Lab Chip. 2012;12:3305–3313. doi: 10.1039/c2lc40273d. [DOI] [PubMed] [Google Scholar]
- 151.Kirby AE, Wheeler AR. Anal Chem. 2013;85:6178–6184. doi: 10.1021/ac401150q. [DOI] [PubMed] [Google Scholar]
- 152.Dryden MDM, Rackus DDG, Shamsi MH, Wheeler AR. Anal Chem. 2013;85:8809–8816. doi: 10.1021/ac402003v. [DOI] [PubMed] [Google Scholar]
- 153.Pyne DG, Salman WM, Abdelgawad M, Sun Y. Appl Phys Lett. 2013;103:024103/1–024103/4. [Google Scholar]
- 154.Ng AHC, Choi K, Luoma RP, Robinson JM, Wheeler AR. Anal Chem. 2012;84:8805–8812. doi: 10.1021/ac3020627. [DOI] [PubMed] [Google Scholar]
- 155.Fobel R, Fobel C, Wheeler AR. Appl Phys Lett. 2013;102:193513/1–193513/5. [Google Scholar]
- 156.Sjostrom SL, Joensson HN, Svahn HA. Lab Chip. 2013;13:1754–1761. doi: 10.1039/c3lc41398e. [DOI] [PubMed] [Google Scholar]
- 157.Chang C, Sustarich J, Bharadwaj R, Chandrasekaran A, Adams PD, Singh AK. Lab Chip. 2013;13:1817–1822. doi: 10.1039/c3lc41418c. [DOI] [PubMed] [Google Scholar]
- 158.Chen CH, Miller MA, Sarkar A, Beste MT, Isaacson KB, Lauffenburger DA, Griffith LG, Han J. J Am Chem Soc. 2013;135:1645–1648. doi: 10.1021/ja307866z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rane TD, Zec HC, Puleo C, Lee AP, Wang TH. Lab Chip. 2012;12:3341–3347. doi: 10.1039/c2lc40537g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rahman MT, Barikbin Z, Badruddoza AZM, Doyle PS, Khan SA. Langmuir. 2013;29:9535–9543. doi: 10.1021/la401613w. [DOI] [PubMed] [Google Scholar]
- 161.Fodera V, Pagliara S, Otto O, Keyser UF, Donald AM. J Phys Chem Lett. 2012;3:2803–2807. [Google Scholar]
- 162.Zhao M, Schiro PG, Kuo JS, Koehler KM, Sabath DE, Popov V, Feng Q, Chiu DT. Anal Chem. 2013;85:2465–2471. doi: 10.1021/ac400193b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ju Y, Song J, Geng Z, Zhang H, Wang W, Xie L, Yao W, Li Z. Lab Chip. 2012;12:4355–4362. doi: 10.1039/c2lc40332c. [DOI] [PubMed] [Google Scholar]
- 164.Liu Y, Barua D, Liu P, Wilson BS, Oliver JM, Hlavacek WS, Singh AK. PLoS One. 2013;8:e60159. doi: 10.1371/journal.pone.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Huang KW, Wu YC, Lee JA, Chiou PY. Lab Chip. 2013;13:3721–3727. doi: 10.1039/c3lc50607j. [DOI] [PubMed] [Google Scholar]
- 166.Vahey MD, Quiros PL, Svensson JP, Samson LD, Voldman J. Lab Chip. 2013;13:2754–2763. doi: 10.1039/c3lc50162k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.El DB, Utharala R, Balyasnikova IV, Griffiths AD, Merten CA. Proc Natl Acad Sci U S A. 2012;109:11570–11575. doi: 10.1073/pnas.1204514109. S11570/1-S11570/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhou J, Giridhar PV, Kasper S, Papautsky I. Lab Chip. 2013;13:1919–1929. doi: 10.1039/c3lc50101a. [DOI] [PubMed] [Google Scholar]
- 169.Lee MG, Shin JH, Bae CY, Choi S, Park JK. Anal Chem. 2013;85:6213–6218. doi: 10.1021/ac4006149. [DOI] [PubMed] [Google Scholar]
- 170.Hur SC, Brinckerhoff TZ, Walthers CM, Dunn JCY, Di CD. PLoS One. 2012;7:e46550. doi: 10.1371/journal.pone.0046550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wu L, Guan G, Hou HW, Bhagat AAS, Han J. Anal Chem. 2012;84:9324–9331. doi: 10.1021/ac302085y. [DOI] [PubMed] [Google Scholar]
- 172.Zhang W, Kai K, Choi DS, Iwamoto T, Hguyen YH, Wong H, Landis MD, Ueno NT, Chang J, Qin L. Proc Natl Acad Sci U S A. 2012;109:18707–18712. doi: 10.1073/pnas.1209893109. S18707/1-S18707/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhu T, Cheng R, Lee SA, Rajaraman E, Eiteman MA, Querec TD, Unger ER, Mao L. Microfluid Nanofluid. 2012;13:645–654. doi: 10.1007/s10404-012-1004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nam J, Lim H, Kim C, Kang JY, Shin S. Biomicrofluidics. 2012;6:4718719, 11. doi: 10.1063/1.4718719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yang AHJ, Soh HT. Anal Chem. 2012;84:10756–10762. doi: 10.1021/ac3026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bose S, Singh R, Hanewich-Hollatz M, Shen C, Lee CH, Dorfman DM, Karp JM, Karnik R. Sci Rep. 2013;3:2329. doi: 10.1038/srep02329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cao Z, Chen F, Bao N, He H, Xu P, Jana S, Jung S, Lian H, Lu C. Lab Chip. 2013;13:171–178. doi: 10.1039/c2lc40950j. [DOI] [PubMed] [Google Scholar]
- 178.Hou S, Zhao L, Shen Q, Yu J, Ng C, Kong X, Wu D, Song M, Shi X, Xu X, OuYang WH, He R, Zhao XZ, Lee T, Brunicardi FC, Garcia MA, Ribas A, Lo RS, Tseng HR. Angew Chem, Int Ed. 2013;52:3379–3383. doi: 10.1002/anie.201208452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Casavant BP, Mosher R, Warrick JW, Maccoux LJ, Berry SMF, Becker JT, Chen V, Lang JM, McNeel DG, Beebe DJ. Methods. 2013 doi: 10.1016/j.ymeth.2013.05.027. in press, corrected proof http://jla.sagepub.com/content/early/2013/07/12/2211068213495207. [DOI] [PMC free article] [PubMed]
- 180.Wu CH, Huang YY, Chen P, Hoshino K, Liu H, Frenkel EP, Zhang JXJ, Sokolov KV. ACS Nano. 2013;7:8816–8823. doi: 10.1021/nn403281e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kim S, Han SI, Park MJ, Jeon CW, Joo YD, Choi IH, Han KH. Anal Chem. 2013;85:2779–86. doi: 10.1021/ac303284u. [DOI] [PubMed] [Google Scholar]
- 182.Zhao W, Cui CH, Bose S, Guo D, Shen C, Wong WP, Halvorsen K, Farokhzad OC, Teo GSL, Phillips JA, Dorfman DM, Karnik R, Karp JM. Proc Natl Acad Sci U S A. 2012;109:19626–31. doi: 10.1073/pnas.1211234109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sheng W, Chen T, Tan W, Fan ZH. ACS Nano. 2013;7:7067–7076. doi: 10.1021/nn4023747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Helleso OG, Lovhaugen P, Subramanian AZ, Wilkinson JS, Ahluwalia BS. Lab Chip. 2012;12:3436–3440. doi: 10.1039/c2lc40375g. [DOI] [PubMed] [Google Scholar]
- 185.Zand K, Pham T, Davila A, Jr, Wallace DC, Burke PJ. Anal Chem. 2013;85:6018–25. doi: 10.1021/ac4010088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Su X, Theberge AB, January CT, Beebe DJ. Anal Chem. 2013;85:1562–1570. doi: 10.1021/ac3027228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Donato SS, Chu V, Prazeres DMF, Conde JP. Electrophoresis. 2013;34:575–582. doi: 10.1002/elps.201200292. [DOI] [PubMed] [Google Scholar]
- 188.Fritzsch FSO, Rosenthal K, Kampert A, Howitz S, Dusny C, Blank LM, Schmid A. Lab Chip. 2013;13:397–408. doi: 10.1039/c2lc41092c. [DOI] [PubMed] [Google Scholar]
- 189.Vasdekis AE. RSC Adv. 2013:3, 6343–6346. [Google Scholar]
- 190.Gundabala VR, Martinez-Escobar S, Marquez SM, Marquez M, Fernandez-Nieves A. J Phys D: Appl Phys. 2013;46:114006, 4. [Google Scholar]
- 191.Eyer K, Stratz S, Kuhn P, Kuster SK, Dittrich PS. Anal Chem. 2013;85:3280–3287. doi: 10.1021/ac303628j. [DOI] [PubMed] [Google Scholar]
- 192.Deng H, Wang C, Su M, Fang Y. Anal Chem. 2012;84:8232–8239. doi: 10.1021/ac301495n. [DOI] [PubMed] [Google Scholar]
- 193.Jin BJ, Ko EA, Namkung W, Verkman AS. Lab Chip. 2013;13:3862–3867. doi: 10.1039/c3lc50821h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hamon M, Jambovane S, Bradley L, Khademhosseini A, Hong JW. Anal Chem. 2013;85:5249–54. doi: 10.1021/ac400743w. [DOI] [PubMed] [Google Scholar]
- 195.Jastrzebska E, Flis S, Rakowska A, Chudy M, Jastrzebski Z, Dybko A, Brzozka Z. Microchim Acta. 2013;180:895–901. doi: 10.1007/s00604-013-1009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Peng CC, Liao WH, Chen YH, Wu CY, Tung YC. Lab Chip. 2013;13:3239–3245. doi: 10.1039/c3lc50388g. [DOI] [PubMed] [Google Scholar]
- 197.Huang NT, Chen W, Oh BR, Cornell TT, Shanley TP, Fu J, Kurabayashi K. Lab Chip. 2012;12:4093–4101. doi: 10.1039/c2lc40619e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Xu T, Yue W, Li CW, Yao X, Yang M. Lab Chip. 2013;13:1060–1069. doi: 10.1039/c3lc40880a. [DOI] [PubMed] [Google Scholar]
- 199.Uhlendorf J, Miermont A, Delaveau T, Charvin G, Fages F, Bottani S, Batt G, Hersen P. Proc Natl Acad Sci U S A. 2012;109:14271–14276. doi: 10.1073/pnas.1206810109. S14271/1-S14271/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ullman G, Wallden M, Marklund EG, Mahmutovic A, Razinkov I, Elf J. Philos Trans R Soc, B. 2013;368:20120025/1–20120025/8. doi: 10.1098/rstb.2012.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Xie Z, Zhang Y, Zou K, Brandman O, Luo C, Ouyang Q, Li H. Aging Cell. 2012;11:599–606. doi: 10.1111/j.1474-9726.2012.00821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ricicova M, Hamidi M, Quiring A, Niemisto A, Emberly E, Hansen CL. Proc Natl Acad Sci U S A. 2013;110:11403–11408. doi: 10.1073/pnas.1215850110. S11403/1-S11403/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Denervaud N, Becker J, Delgado-Gonzalo R, Damay P, Rajkumar AS, Unser M, Shore D, Naef F, Maerkl SJ. Proc Natl Acad Sci U S A. 2013;110:15842–15847. doi: 10.1073/pnas.1308265110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lee H, Crane MM, Zhang Y, Lu H. Integr Biol. 2013;5:372–380. doi: 10.1039/c2ib20078c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Cimetta E, Sirabella D, Yeager K, Davidson K, Simon J, Moon RT, Vunjak-Novakovic G. Lab Chip. 2013;13:355–64. doi: 10.1039/c2lc40836h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ciftlik AT, Lehr HA, Gijs MAM. Proc Natl Acad Sci U S A. 2013;110:5363–5368. doi: 10.1073/pnas.1211273110. S5363/1-S5363/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Song JW, Bazou D, Munn LL. Integr Biol. 2012;4:857–862. doi: 10.1039/c2ib20061a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Liu A, Liu W, Wang Y, Wang JC, Tu Q, Liu R, Xu J, Shen S, Wang J. Microfluid Nanofluid. 2013;14:515–526. [Google Scholar]
- 209.Dinh ND, Chiang YY, Hardelauf H, Baumann J, Jackson E, Waide S, Sisnaiske J, Frimat JP, van TC, Janasek D, Peyrin JM, West J. Lab Chip. 2013;13:1402–1412. doi: 10.1039/c3lc41224e. [DOI] [PubMed] [Google Scholar]
- 210.Mao S, Zhang J, Li H, Lin JM. Anal Chem. 2013;85:868–76. doi: 10.1021/ac303164b. [DOI] [PubMed] [Google Scholar]
- 211.Markov DA, Lu JQ, Samson PC, Wikswo JP, McCawley LJ. Lab Chip. 2012;12:4560–4568. doi: 10.1039/c2lc40304h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Prabhakarpandian B, Shen MC, Nichols JB, Mills IR, Sidoryk-Wegrzynowicz M, Aschner M, Pant K. Lab Chip. 2013;13:1093–1101. doi: 10.1039/c2lc41208j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ren L, Liu W, Wang Y, Wang JC, Tu Q, Xu J, Liu R, Shen SF, Wang J. Anal Chem. 2012;85:235–244. doi: 10.1021/ac3025812. [DOI] [PubMed] [Google Scholar]
- 214.Li L, Lieleg O, Jang S, Ribbeck K, Han J. Lab Chip. 2012;12:4071–4079. doi: 10.1039/C2LC40161D. [DOI] [PubMed] [Google Scholar]
- 215.Baudoin M, Song Y, Manneville P, Baroud CN. Proc Natl Acad Sci U S A. 2013;110:859–64. doi: 10.1073/pnas.1211706110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Shin YS, Choi TS, Kim H, Beauchamp JL, Heath JR, Kim HI. Lab Chip. 2012;12:5243–5248. doi: 10.1039/c2lc40940b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Graff JR, Forschner-Dancause SR, Menden-Deuer S, Long RA, Rowley DC. Front Aquat Microbiol. 2013;4:8. doi: 10.3389/fmicb.2013.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yang J, Chen Z, Ching P, Shi Q, Li X. Lab Chip. 2013;13:3373–3382. doi: 10.1039/c3lc50264c. [DOI] [PubMed] [Google Scholar]
- 219.Wong BG, Paz A, Corrado MA, Ramos BR, Cinquin A, Cinquin O, Hui EE. Integr Biol. 2013;5:976–982. doi: 10.1039/c3ib20291g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Reboud J, Bourquin Y, Wilson R, Pall GS, Jiwaji M, Pitt AR, Graham A, Waters AR, Cooper JM. Proc Natl Acad Sci U S A. 2012;109:15162–15167. doi: 10.1073/pnas.1206055109. S15162/1-S15162/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lu Y, Gao J, Zhang DD, Gau V, Liao JC, Wong PK. Anal Chem. 2013;85:3971–3976. doi: 10.1021/ac4004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zhang R, Gong HQ, Zeng XD, Sze CC. Anal Bioanal Chem. 2013;405:4277–4282. doi: 10.1007/s00216-013-6826-0. [DOI] [PubMed] [Google Scholar]
- 223.Sanati NA, Naghavi M, Packirisamy M, Bhat R, Geitmann A. Proc Natl Acad Sci U S A. 2013;110:8093–8. doi: 10.1073/pnas.1221677110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Perozziello G, Simone G, Malara N, La RR, Tallerico R, Catalano R, Pardeo F, Candeloro P, Cuda G, Carbone E, Di FE. Electrophoresis. 2013;34:1845–1851. doi: 10.1002/elps.201300106. [DOI] [PubMed] [Google Scholar]
- 225.Kim E, Schueller O, Sweetnam PM. Lab Chip. 2012;12:2255–64. doi: 10.1039/c2lc21078a. [DOI] [PubMed] [Google Scholar]
- 226.Funamoto K, Zervantonakis IK, Liu Y, Ochs CJ, Kim C, Kamm RD. Lab Chip. 2012;12:4855–4863. doi: 10.1039/c2lc40306d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zhu Q, Gao Y, Yu B, Ren H, Qiu L, Han S, Jin W, Jin Q, Mu Y. Lab Chip. 2012;12:4755–4763. doi: 10.1039/c2lc40774d. [DOI] [PubMed] [Google Scholar]
- 228.Zhang R, Gong HQ, Zeng X, Lou C, Sze C. Anal Chem. 2013;85:1484–1491. doi: 10.1021/ac3026509. [DOI] [PubMed] [Google Scholar]
- 229.Lounsbury JA, Landers JP. J Forensic Sci. 2013;58:866–874. doi: 10.1111/1556-4029.12162. [DOI] [PubMed] [Google Scholar]
- 230.Martinez-Quijada J, Caverhill-Godkewitsch S, Reynolds M, Gutierrez-Rivera L, Johnstone RW, Elliott DG, Sameoto D, Backhouse CJ. Sens Actuators, A. 2013;193:170–181. [Google Scholar]
- 231.Ohlander A, Zilio C, Hammerle T, Zelenin S, Klink G, Chiari M, Bock K, Russom A. Lab Chip. 2013;13:2075–2082. doi: 10.1039/c3lc50171j. [DOI] [PubMed] [Google Scholar]
- 232.Penchovsky R. Lab Chip. 2013;13:2370–2380. doi: 10.1039/c3lc50208b. [DOI] [PubMed] [Google Scholar]
- 233.Chiou CH, Shin DJ, Zhang Y, Wang TH. Biosens Bioelectron. 2013;50:91–9. doi: 10.1016/j.bios.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Zhu J, Palla M, Ronca S, Wapner R, Ju J, Lin Q. Sens Actuators, A. 2013;195:175–182. doi: 10.1016/j.sna.2012.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Eastburn DJ, Sciambi A, Abate AR. PLoS One. 2013;8:e62961. doi: 10.1371/journal.pone.0062961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Benitez JJ, Topolancik J, Tian HC, Wallin CB, Latulippe DR, Szeto K, Murphy PJ, Cipriany BR, Levy SL, Soloway PD, Craighead HG. Lab Chip. 2012;12:4848–4854. doi: 10.1039/c2lc40955k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Li Y, Feng X, Du W, Li Y, Liu BF. Anal Chem. 2013;85:4066–73. doi: 10.1021/ac4000893. [DOI] [PubMed] [Google Scholar]
- 238.Schumm JW, Gutierrez-Mateo C, Tan E, Selden R. J Forensic Sci. 2013;58:1584–1592. doi: 10.1111/1556-4029.12214. [DOI] [PubMed] [Google Scholar]
- 239.Scanlon TC, Dostal SM, Griswold KE. Biotechnol Bioeng. 2013 doi: 10.1002/bit.25019. early view. [DOI] [PubMed] [Google Scholar]
- 240.Jakiela S, Kaminski TS, Cybulski O, Weibel DB, Garstecki P. Angew Chem Int Ed Engl. 2013;52:8908–11. doi: 10.1002/anie.201301524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Park S, Choi JW, Kim YK. J Biomed Nanotechnol. 2013;9:880–5. doi: 10.1166/jbn.2013.1497. [DOI] [PubMed] [Google Scholar]
- 242.Houbart V, Servais AC, Charlier TD, Pawluski JL, Abts F, Fillet M. Electrophoresis. 2012;33:3370–3379. doi: 10.1002/elps.201200168. [DOI] [PubMed] [Google Scholar]
- 243.Shu B, Zhang C, Xing D. Microfluid Nanofluid. 2013;15:161–172. [Google Scholar]
- 244.Wu J, Chen Q, Liu W, Zhang Y, Lin JM. Lab Chip. 2012;12:3474–3480. doi: 10.1039/c2lc40502d. [DOI] [PubMed] [Google Scholar]
- 245.Piorek BD, Lee SJ, Moskovits M, Meinhart CD. Anal Chem. 2012;84:9700–9705. doi: 10.1021/ac302497y. [DOI] [PubMed] [Google Scholar]
- 246.Zheng G, Wang Y, Wang Z, Zhong W, Wang H, Li Y. Mar Pollut Bull. 2013;72:231–243. doi: 10.1016/j.marpolbul.2013.03.035. [DOI] [PubMed] [Google Scholar]
- 247.Kim WJ, Kim S, Kim AR, Yoo DJ. Ind Eng Chem Res. 2013;52:7282–7288. [Google Scholar]
- 248.Ramadan Q, Gijs MAM. Microfluid Nanofluid. 2012;13:529–542. [Google Scholar]
- 249.El-Sharoud WM, Darwish MS, Batt CA. Int Dairy J. 2013;33:67–74. [Google Scholar]
- 250.Vergeres G, Bogicevic B, Buri C, Carrara S, Chollet M, Corbino-Giunta L, Egger L, Gille D, Kopf-Bolanz K, Laederach K, Portmann R, Ramadan Q, Ramsden J, Schwander F, Silacci P, Walther B, Gijs M. Br J Nutr. 2012;108:762–8. doi: 10.1017/S0007114512002693. [DOI] [PubMed] [Google Scholar]
- 251.Adams AA, Charles PT, Veitch SP, Hanson A, Deschamps JR, Kusterbeck AW. Anal Bioanal Chem. 2013;405:5171–8. doi: 10.1007/s00216-013-6853-x. [DOI] [PubMed] [Google Scholar]


