Abstract
Prokaryote-specific sugars, including N,N′-diacetylbacillosamine (diNAcBac) and pseudaminic acid, have experienced a renaissance in the past decade because of their discovery in glycans related to microbial pathogenicity. DiNAcBac is found at the reducing end of oligosaccharides of N- and O-linked bacterial protein glycosylation pathways of Gram-negative pathogens, including Campylobacter jejuni and Neisseria gonorrhoeae. Further derivatization of diNAcBac results in the nonulosonic acid known as legionaminic acid, which was first characterized in the O-antigen of the lipopolysaccharide (LPS) in Legionella pneumophila. Pseudaminic acid, an isomer of legionaminic acid, is also important in pathogenic bacteria such as Helicobacter pylori because of its occurrence in O-linked glycosylation of flagellin proteins, which plays an important role in flagellar assembly and motility. Here, we present recent advances in the characterization of the biosynthetic pathways leading to these highly modified sugars and investigation of the roles that each plays in bacterial fitness and pathogenicity.
Glycosylation is an abundant eukaryotic protein modification reaction, which modulates a variety of cellular processes, including protein folding, trafficking, cell–cell interactions, cell signaling, and the host immune response.1−4 It is now recognized that selected bacteria also possess the machinery necessary to glycosylate proteins, and there is evidence that this modification may play an important role in bacterial fitness and pathogenicity.5−8 In some bacterial protein glycosylation pathways, the glycan is first assembled in a stepwise fashion onto a polyprenyl diphosphate-linked carrier on the cytoplasmic face of the inner membrane prior to being translocated to the periplasmic face of the same membrane in readiness for transfer onto an acceptor protein. In this case, attachment of bacterial glycans onto proteins is accomplished by oligosaccharyltransferase-mediated en bloc transfer onto asparagine (N-linked) or serine/threonine (O-linked) residues.9,10 Protein glycosylation can also occur in a stepwise manner with nucleotide-activated sugars by Leloir glycosyltransferases. Highly modified bacterial sugars, including 2,4-diacetamido-2,4,6-trideoxy-d-glucose (N,N′-diacetylbacillosamine or diNAcBac) and 5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-l-manno-nonulosonic acid (pseudaminic acid or Pse), are prevalent features of bacterial glycans, and in some cases, the presence of such sugars has been related to pathogenicity.11,12 For example, diNAcBac is found at the reducing end of glycans in N- and O-linked glycoproteins, and Pse is featured in the O-linked glycans of flagellar proteins of significant human pathogens.
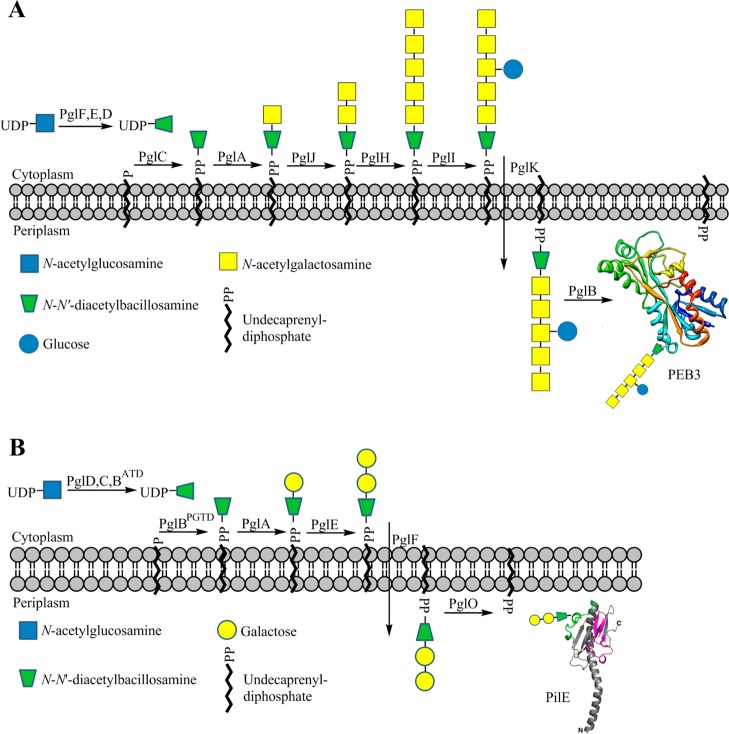
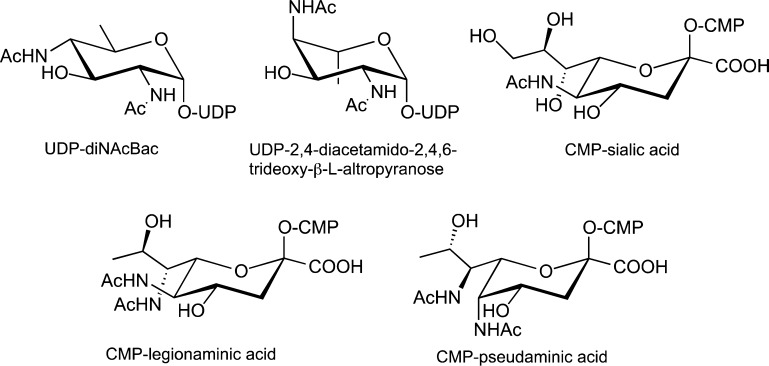
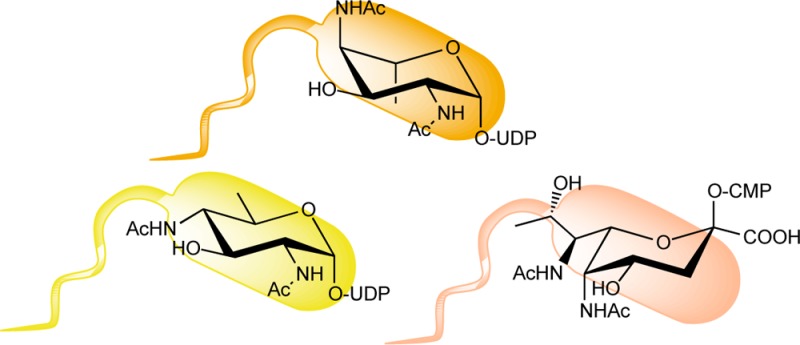
The Campylobacter jejuni N-linked protein glycosylation (pgl) pathway, which affords a heptasaccharide for transfer to proteins, is one of the best characterized bacterial glycosylation pathways to date (Figure 1A). N-Linked glycans resulting from this pathway have been identified on more than 65 C. jejuni proteins.13 An analogous O-linked pathway, found in Neisseria gonorrhoeae, generates a trisaccharide (Figure 1B) that has been identified on 19 glycoproteins, including the pilin protein PilE.14 In each pathway, diNAcBac is first biosynthesized as the corresponding UDP-sugar from UDP-N-acetylglucosamine (UDP-GlcNAc) and then diNAcBac is assembled into a polyprenyl diphosphate-linked glycan for transfer to acceptor proteins. In other pathways, further modification of diNAcBac through a series of enzymes results in legionaminic acid, a nonulosonic acid, which is structurally related to sialic acid (N-acetylneuraminic acid) (Figure 2). Additionally, an analogous pathway, which utilizes 2,4-diacetamido-2,4,6-trideoxy-l-altropyranose, an isomer of diNAcBac, produces pseudaminic acid (Figure 2). In contrast to the N- and O-linked glycosylation pathways that incorporate diNAcBac as part of a preassembled glycan, these elaborated 2,4-diacetamido-2,4,6-trideoxy-α-d-hexose (DATDH) derivatives are transferred directly from the corresponding CMP derivatives to serine and threonine side chains to afford O-linked glycoproteins. Legionaminic and pseudaminic acids are essential for flagellar assembly in Campylobacter spp., Legionella pneumophila, and Helicobacter pylori.15 The biosynthetic pathways responsible for these unique sugars have recently been linked to bacterial pathogenesis16−18 and therefore may present novel targets for the development of antivirulence agents, which could potentially complement current efforts to develop new antibiotics to address the growing challenges with antibiotic resistance.
Figure 1.
(A) N-linked protein glycosylation pathway from C. jejuni showing the heptasaccharide glycan attached to the PEB3 protein. (B) O-linked protein glycosylation pathway from N. gonorrhoeae showing the trisaccharide glycan attached to the PilE protein. Both pathways utilize the unique, bacterial sugar diNAcBac at the reducing end of the glycan. Abbreviations: ATD, acetyltransferase domain; PGTD, phosphoglycosyltransferase domain.
Figure 2.
Structural comparison of modified carbohydrates found in bacteria that are discussed in this review.
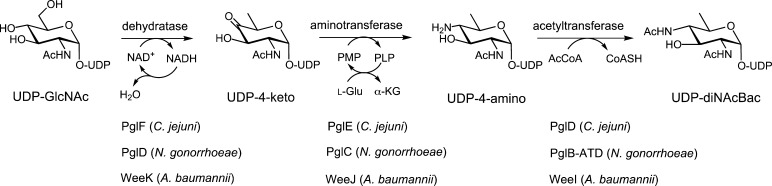
This review focuses on the pathways leading to diNAcBac and pseudaminic acid, which are two particularly important prokaryote-specific sugars. UDP-diNAcBac is biosynthesized from UDP-GlcNAc using a series of three conserved enzymes, including a dehydratase, an aminotransferase, and an acetyltransferase (Figure 3). These three enzymes have been extensively studied in the Gram-negative bacterium C. jejuni.19−21 Subsequent to the work on the C. jejuni N-linked protein glycosylation pathway, diNAcBac was also discovered in glycans, which modify serine and threonine residues (O-linked) in other pathogenic bacteria, including N. gonorrhoeae,22 and UDP-diNAcBac biosynthesis enzymes have also been identified in the AYE strain of Acinetobacter baumannii.23 Importantly, diNAcBac and the related DATDH based on l-altropyranose, which is isomeric at C4 and C5, serve as starting points for the biosynthesis of legionaminic and pseudaminic acid. In this review, the enzymes responsible for the biosynthesis of these unique bacterial carbohydrates will be explored in detail. Our current understanding of the biosynthesis and incorporation of these highly modified sugars into glycoprotein virulence factors provides a strong motivation for future investigations of the molecular basis of their roles in bacterial pathogenicity.
Figure 3.
Biosynthetic pathway for the conversion of UDP-GlcNAc into UDP-diNAcBac in C. jejuni, N. gonorrhoeae, and A. baumannii (AYE strain).
N,N′-Diacetylbacillosamine
Discovery and Characterization
The serendipitous discovery of bacillosamine occurred in 1957 when Nathan Sharon was exploring polypeptide biosynthesis in Bacillus licheniformis, a Gram-positive bacterium usually found in soil.24 In the course of these studies, an unknown amino sugar component of a B. licheniformis polysaccharide was detected by paper chromatography. Elemental and chemical analysis of this sugar revealed the presence of two nitrogen atoms at positions C2 and C4, with the latter site acetylated. The structure of this carbohydrate was assigned as 4-acetamido-2-amino-2,4,6-trideoxyhexose (4-N-acetylbacillosamine) based upon these initial experiments.25,26 Final confirmation of the structure, including the unambiguous stereochemical assignment, occurred 10 years later through a 12-step chemical synthesis from d-glucosamine.27 More recently, undecaprenyl diphosphate-linked di-N-acetylbacillosamine (diNAcBac)28 and diNAcBac-containing disaccharides29 have also been prepared by chemical synthesis. Since its discovery, bacillosamine and the corresponding N-acetylated derivatives have been found in the glycoconjugates of a variety of pathogenic bacteria. For example, it is found as the reducing-end sugar in N-linked (C. jejuni) and O-linked glycoproteins (Neisseria spp.). Additionally, bacillosamine has been identified in the O-antigen of Pseudomonas reactans(30) and Vibrio cholerae,31 the core region of the lipopolysaccharide (LPS) in Francisella novicida,32 and the capsular polysaccharide (CPS) from Alteromonas sp. CMM155.33 The fundamental question of why bacteria utilize bacillosamine is currently unanswered and remains an important area of research, although some hypotheses suggest that this sugar is not recognized by mammalian hosts and therefore may serve as a decoy to host immune systems and glycan-degrading enzymes.34
Biosynthesis in Bacterial Pathogens
Although the biosynthetic route to bacillosamine was first suggested by Sharon in 1964,35 it took more than 40 years to verify the initial proposal. Following genome sequencing of C. jejuni,36 an operon that was distinct from the lipopolysaccharide cluster was identified, which included several genes that were significantly homologous to previously characterized protein glycosylation genes.37 Selected proteins encoded by the pgl (protein glycosylation) locus were ultimately biochemically characterized and found to be responsible for the biosynthesis of UDP-diNAcBac from UDP-GlcNAc.
Biochemical analysis of Cj1120c, later renamed PglF, resulted in the identification of the first enzyme in the pgl pathway, a membrane-bound NAD+-dependent dehydratase.19 PglF catalyzes a transient NAD+-dependent C4 oxidation of UDP-GlcNAc, which promotes elimination of water across carbons C5 and C6 of the pyran ring. Reduction of the resultant α,β-unsaturated system by hydride addition at C6 produces the UDP-4-keto sugar and regenerates NAD+ (Figure 3). Therefore, PglF catalyzes a net overall dehydration reaction. One- and two-dimensional NMR experiments confirmed the stereochemistry of this product to be UDP-2-acetamido-4-keto-2,4,6-trideoxy-α-d-glucose.19 Unlike the pseudaminic acid dehydratase (Cj1293/PseB) also found in C. jejuni (see below), PglF does not catalyze C5 epimerization. Kinetic characterization of PglF resulted in a kcat/Km of 17 M–1 s–1 for UDP-GlcNAc, making this enzyme the least catalytically efficient on the diNAcBac biosynthesis pathway and thus making this the rate-limiting step.20 Further characterization of PglF homologs in N. gonorrhoeae (PglD) and A. baumannii (WeeK) has resulted in similar kinetic parameters, lending support to the proposal that the dehydratase enzymes may play the role of “gatekeeper” in these pathways.22,23 The diNAcBac dehydratase enzymes have yet to be structurally characterized, in part because of the challenges associated with membrane protein crystallization.
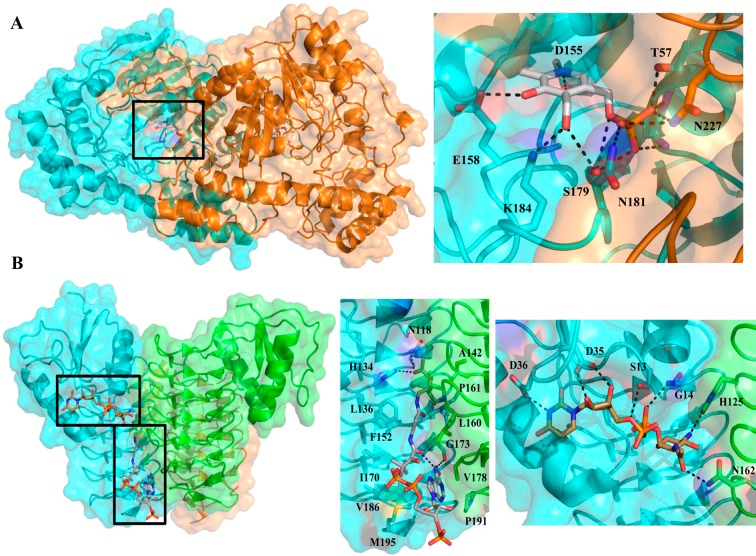
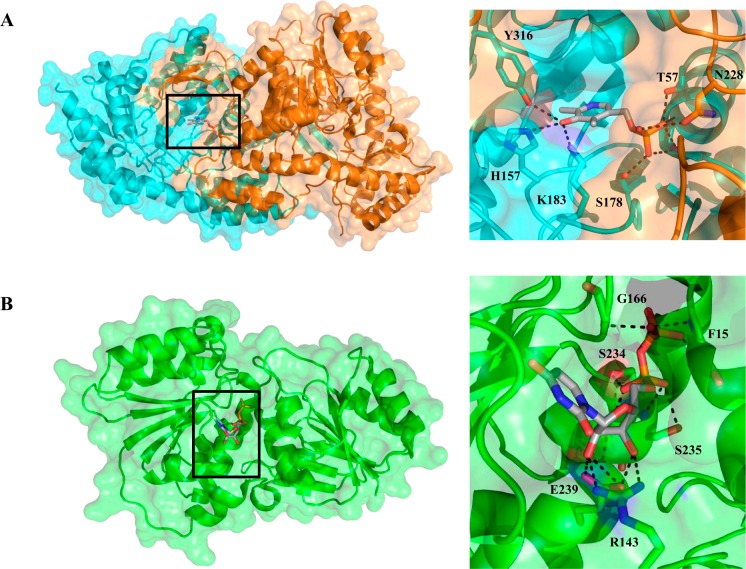
The gene adjacent to pglF in the pgl locus, encoding Cj1121c/PglE, was defined as a pyridoxal 5′-phosphate (PLP)-dependent aminotransferase that catalyzes the transfer of the amino group from l-glutamate to the C4 position of the UDP-4-keto sugar. The transformation occurs in two distinct steps that cycle between the PLP and pyridoxamine 5′-phosphate (PMP) forms of the coenzyme.19,21 Catalysis is initiated by the formation of an imine involving the UDP-4-keto sugar and PMP. Following isomerization to the external aldimine, the UDP-4-amino sugar product is released via transimination of the catalytic lysine residue in the active site. The internal aldimine resulting from this reaction then reacts with l-glutamate to afford α-ketoglutarate, regenerating PMP in the process. Although the preferred amino group donor was determined to be glutamic acid, PglE has also been shown to exhibit moderate activity with methionine, glutamine, alanine, and cysteine.21 The UDP-4-amino sugar product of this reaction was confirmed as UDP-2-acetamido-4-amino-2,4,6-trideoxy-α-d-glucose based upon NMR experiments, including nuclear Overhauser effect (NOE) studies and J coupling constant analysis.21 Kinetically, PglE is a more efficient enzyme than PglF when comparing UDP-sugar substrates (kcat/Km = 6600 M–1 s–1).23 The l-glutamic acid cosubstrate is weakly bound (Km = 11 mM), which is offset by the naturally high intracellular concentrations of the amino acid.38 Studies with the aminotransferase homologs in N. gonorrhoeae (PglC) and A. baumannii (WeeJ) again reveal a low binding affinity for l-glutamic acid, and with respect to UDP-4-keto sugar turnover, both of these enzymes were catalytically less active than PglE. Bacterial sugar aminotransferases such as PglE form homodimers in solution following previous observations with PseC (H. pylori) and WbpE (Pseudomonas aeruginosa).39,40 Furthermore, the crystal structure of PglE supported solution state studies revealing that the enzyme exists as a dimer in the asymmetric unit41 (Figure 4A). The two active sites are located on opposite faces of the dimer interface and are separated by ∼30 Å. At the bottom of each binding pocket resides the PLP coenzyme necessary for the transamination reaction. Structures of the apo and PLP-bound forms of PglE have been determined, while attempts to crystallize the protein in the presence of the UDP-sugar substrate or product have not yet proven to be successful, although it is presumed that substrate binding generally resembles that described in previous studies of related aminotransferases.39 However, because these proteins seem to be highly specific for their cognate UDP-sugars, intriguing questions about the molecular basis for the observed selectivity remain.
Figure 4.
(A) C. jejuni PglE aminotransferase crystal structure (PDB entry 1O61) bound to PLP depicted in ribbon and space-filling format (left). The major interactions between PLP and the PglE active site are depicted with dashed lines (right). The dimer is the biologically active unit, and each protomer has been individually colored for the sake of clarity. (B) Composite C. jejuni PglD acetyltransferase crystal structure constructed from the UDP-4-amino-bound (PDB entry 3BSS) (brown) and AcCoA-bound (PDB entry 3BSY) (gray) structures. For the sake of clarity, the two additional binding pocket substrates have been removed and the protomers individually colored. The biologically active unit is a trimer illustrated in ribbon and space-filling format (left). Major interactions of the PglD active site with AcCoA (center) and UDP-4-amino (right) are depicted with dashed lines.
The final step of UDP-diNAcBac biosynthesis relies on the acetyltransferase PglD (Cj1123c), which acetylates the UDP-4-amino sugar in an acetyl coenzyme A (AcCoA)-dependent reaction. Catalysis involves an active site histidine that acts as a general base to abstract a proton from the UDP-sugar C4 amine, thereby promoting nucleophilic attack on the thioester of AcCoA. Utilizing a combination of radiolabel transfer with [3H]AcCoA, ESI-MS, and NMR, the product was unequivocally shown to be UDP-diNAcBac (UDP-2,4-diacetamido-2,4,6-trideoxy-α-d-glucose).20 The PglD acetyltransferase exhibited the greatest catalytic efficiency among the pathway enzymes for both the UDP-sugar (kcat/Km = 4.0 × 107 M–1 s–1) and AcCoA (kcat/Km = 5.5 × 107 M–1 s–1) substrates.23 Similar catalytic efficiencies were also observed for the N. gonorrhoeae (PglB) and A. baumannii (WeeI) diNAcBac acetyltransferases.42 It is proposed that this enzyme efficiency may create a pathway flux where rapid consumption of the UDP-4-amino sugar drives the rate-limiting step of UDP-4-keto formation by PglF. Interestingly, PglD shows a relaxed substrate specificity based upon its ability to also acetylate UDP-4-amino-4,6-dideoxy-β-l-AltNAc, an intermediate along the pseudaminic pathway,43 which may allow for cross-talk between the two pathways. However, PglD does appear to be specific for sugar nucleotide substrates, as it does not catalyze acetylation of aminoglycoside substrates.43 PglD forms a homotrimer in solution based upon sedimentation velocity analytical ultracentrifugation (AUC) experiments.44 Structural analysis by X-ray crystallography further supports the homotrimeric structure with the C-terminal left-handed β-helix domain of adjacent protomers forming the AcCoA binding site, whereas the N-terminal domain of each monomer contains a β–α–β–α–β Rossmann fold motif to accommodate the UDP-4-amino substrate (Figure 4B).44,45
Connection to Pathogenicity
There is currently considerable interest in identifying and targeting bacterial pathways that are associated with virulence and pathogenicity because of the increasingly frequent occurrence of resistance to currently available bacteriocidal antibiotics.46 This approach is an attractive option because strategies that target virulence, rather than survival, would potentially circumvent the selective pressure to develop resistance associated with many common antibiotics. With respect to virulence, the C. jejuni N-linked protein glycosylation pathway is of significant interest. This bacterial pathogen is a leading cause of gastroenteritis and is also an associated antecedent infection in the development of Guillain-Barré syndrome.47−49 The pgl pathway produces a heptasaccharide containing diNAcBac, which modifies a variety of proteins associated with virulence.50 Therefore, the enzymes responsible for the biosynthesis of diNAcBac may represent appealing antivirulence targets because they are specific only to prokaryotes. It is also noteworthy that C. jejuni has already exhibited increased resistance toward front-line antibiotics, including the macrolides and fluoroquinolones,51 which inhibit bacterial protein synthesis and topoisomerase II, respectively.
Previous studies have examined the importance of global N-linked protein glycosylation in C. jejuni by disrupting the genes responsible for diNAcBac biosynthesis (pglF, pglE, and pglD).52 Utilizing high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) and whole-cell lysate reactivity to the GalNAc-specific SBA lectin, it was demonstrated that the loss of function in these genes resulted in the weakened ability to produce the heptasaccharide-modified proteins in C. jejuni. Additionally, the ΔpglD and ΔpglE C. jejuni strains were examined for their ability to colonize 1-day-old chicks.52 In both cases, strongly attenuated bacterial colonization was detected following inoculation because of the inactivation of the glycosylation pathway, thereby validating these two gene products as targets in pathogenicity. Transposon mutagenesis of C. jejuni verified these results by identifying pglF and pglE as essential genes for colonization of the chick gastrointestinal tract.53 In a related study, the C. jejuni pglE mutant impaired the invasion of intestinal epithelial cells and colonization of intestinal tracts in mice.54
Confirmation of the relationship between pathogenicity and diNAcBac biosynthesis has sharpened the focus on identifying the individual glycoproteins responsible for cell invasion and colonization. The glycoprotein VirB10, a structural component of the type IV secretion system (TFSS), was previously identified in C. jejuni.55 Disruption of the pglE gene resulted in the conclusion that the pgl system glycosylates VirB10 at two sites, N32 and N97. Deletion of the N97 glycosylation site produced a 10-fold decrease in natural competency that could be rescued by complementing with wild-type VirB10. This was the first example of N-linked glycosylation being directly attributed to the stability and function of a known virulence factor. Recently, 16 N-linked glycoproteins were identified and found to be associated with C. jejuni outer membrane vesicles (OMVs), including the known antigenic PEB3 adhesin.56 Pathogens employ OMVs to deliver bacterial proteins into host cells, making this an important finding in the relationship between immunogenic glycoproteins located in the periplasm and pathogenicity.
In parallel with studies exploring the connection between N-linked protein glycosylation and bacterial pathogenicity in C. jejuni, efforts have also focused on a comparable role for prokaryotic O-linked glycosylation. Specifically, the association between O-linked protein glycosylation and pathogenicity has been examined in N. gonorrhoeae. Studies have identified the PilE protein in type IV pilin to be glycosylated with a trisaccharide, including diNAcBac, at a single site (Ser-63).57 Further experiments with the PilE glycoprotein in Neisseria spp. have shown that it is both immunogenic and antigenic.58 Mass spectrometry analysis following two-dimensional gel electrophoresis and immunoblotting identified additional periplasmic glycoproteins that are implicated in protein folding, solute uptake, and respiration.10 Strains of N. gonorrhoeae deficient in the ability to biosynthesize diNAcBac, through disruption of the NAD+-dependent dehydratase gene (pglD), exhibited decreased adherence and invasion of primary human cervical epithelial (pex) cells.59 Similar to N. gonorrhoeae, Neisseria meningitidis encodes a homologous O-linked protein glycosylation pathway that results in modification of PilE with the same trisaccharide.60−62 Recent studies have indicated that this pilin-linked glycan is essential for the adherence of N. meningitidis to human bronchial epithelial cells.63 Further work in an in vivo system is necessary to identify a link between pathogenicity and glycoproteins biosynthesized from this pathway, but this is an exciting and active area of research.
Derivatives of N,N′-Diacetylbacillosamine
Legionaminic Acid
Sialic acid (Figure 2) is a nine-carbon α-keto sugar that is most commonly found incorporated into mammalian cell surface glycoproteins, which are responsible for intercellular interactions.64 Prokaryotes are now also recognized to display sialic acid and related nonulosonic acid derivatives on their outer cell surfaces. For example, selected bacteria produce legionaminic acid (5,7-diacetamido-3,5,7,9-tetradeoxy-d-glycero-d-galacto-nonulosonic acid), which shares the stereochemistry of sialic acid as established by total synthesis.65 It has been hypothesized that bacterial pathogens utilize legionaminic acid as a molecular mimic of sialic acid, which is prominently presented on mammalian cells and is an important factor in immune system regulation and adhesion.65 Legionaminic acid was first identified in a repeating homopolymer in the O-polysaccharide of LPS in L. pneumophila, which is the causative agent of Legionnaires’ disease.66 Recently, legionaminic acid has been found in the glycoconjugates of a variety of other pathogenic bacteria, where it is associated, for example, with the glycoprotein flagella of Campylobacter coli(67) and the O-antigen of A. baumannii,68,69Cronobacter turicensis,70 and Escherichia coli.71 In fact, more than 20% of the 1000 microbial genomes examined to date appear to contain the biosynthetic genes for the legionaminic acid biosynthesis pathway, making this sugar far more widespread than originally believed.72 Work on the O-antigen of Vibrio fischeri has illustrated the importance of legionaminic acid in colonization of the natural host of this bacterium.73 The disruption of this O-antigen through a gene knockout of waaL, the ligase responsible for the assembly of the O-antigen onto the LPS, resulted in a motility defect. Further studies indicate that this O-antigen null strain has a significantly weakened ability to colonize its natural host organism and cannot compete with wild-type V. fischeri in co-colonization assays.73 Although legionaminic acid is found in a number of known bacterial glycoconjugate virulence factors, the relationship between this sugar and host cell interactions remains poorly understood. Further investigation is therefore warranted to determine whether disruption of the biosynthesis of legionaminic acid has an effect on bacterial pathogenicity.
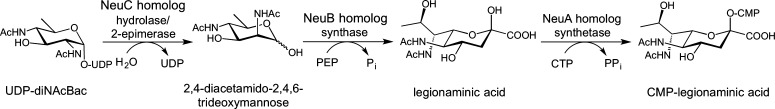
The CMP-activated form of legionaminic acid was originally shown to be biosynthesized in L. pneumophila from UDP-diNAcBac by a series of three enzymes (Figure 5).74,75 These enzymes are necessary for the assembly of functional flagella and show homology to the bacterial sialic acid biosynthetic pathway enzymes NeuC, NeuB, and NeuA. Although it has been hypothesized that sialic acid biosynthesis first emerged in vertebrates and was laterally transferred to bacteria, the legionaminic acid biosynthesis pathway is believed to have evolved convergently from an ancestral nonulosonic acid biosynthesis pathway.72 Additionally, on the basis of a phylogenetic study, it is proposed that sialic acid biosynthesis in pathogenic bacteria may actually be the result of adaptation of an ancient bacterial nonulosonic acid biosynthetic pathway rather than horizontal gene transfer from a eukaryotic host organism.72 The first committed enzyme in the legionaminic acid pathway in L. pneumophila is a NeuC homolog, which affords 2,4-diacetamido-2,4,6-trideoxymannose by an apparent nucleotide hydrolysis and C2 epimerization. In mechanistic studies, a two-dimensional heteronuclear NMR experiment (HMQC) was utilized to establish that the α-anomer is initially formed with retention of stereochemistry at C1, and it was concluded that the mechanism for the NeuC homolog proceeds through anti elimination of UDP followed by syn hydration of the glycal double bond in a fashion similar to that of the homologous enzyme in the bacterial sialic acid biosynthetic pathway.74,76,77 The kinetic parameters for this reaction (kcat/Km = 1.6 × 106 M–1 s–1) suggest that UDP-diNAcBac is the physiological substrate. The inability of the NeuC homolog to turn over UDP-GlcNAc, the natural substrate for the sialic acid pathway, further supports these findings. Following this, the NeuB homolog then utilizes phosphoenolpyruvate (PEP) to condense a three-carbon unit onto 2,4-diacetamido-2,4,6-trideoxymannose to yield N,N′-diacetyllegionaminic acid. This reaction presumably proceeds through an oxocarbenium intermediate following attack on the open chain aldehyde form of 2,4-diacetamido-2,4,6-trideoxymannose by C3 of PEP. Addition of water to this intermediate results in the displacement of phosphate and the formation of the α-keto acid. In these studies, the NeuB homolog showed a rather low level of activity that may be in part due to the assay conditions that were employed.74 The NeuB homolog exhibited no activity with ManNAc, confirming that it is unlikely to be involved in a sialic acid biosynthetic pathway. The final enzymatic step in the process, accomplished by the NeuA homolog, activates the α-keto sugar with CTP to yield CMP-N,N′-diacetyllegionaminic acid. Because of the small amounts of substrate obtained from the previous step, mass spectrometry was utilized to monitor generation of the CMP-activated sugar product.74
Figure 5.
Steps in the conversion of UDP-diNAcBac to CMP-legionaminic acid in L. pneumophila.
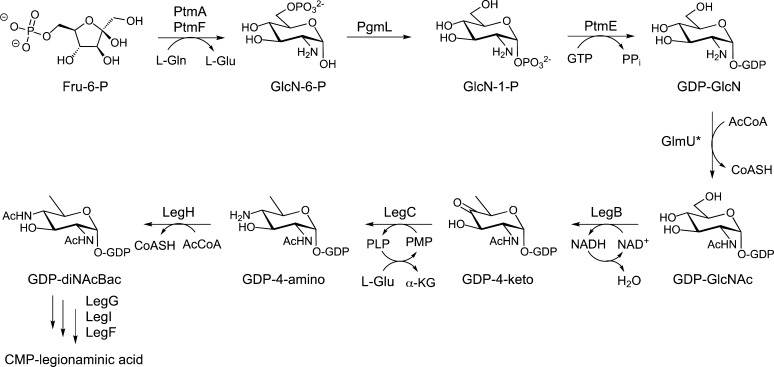
The CMP-legionaminic acid biosynthesis pathway in C. coli VC167 has also been investigated utilizing a targeted metabolomics approach with mass spectrometry and NMR-based approaches.67 Interestingly, in this case, the pathway is found to proceed through alternate intermediates compared to those observed in L. pneumophila. In particular, it was observed that inactivation of the UDP-diNAcBac pathway aminotransferase enzyme PglE in C. coli did not affect the production of CMP-legionaminic acid, and it was apparent in these studies that the biosynthesis of legionaminic acid in C. coli did not proceed through the intermediacy of UDP-diNAcBac. Further work expanded the study of the Campylobacter pathways to C. jejuni where it was demonstrated that the biosynthesis of CMP-legionaminic acid involved GDP rather than UDP-activated sugar intermediates.78 Ultimately, the biosynthesis of CMP-legionaminic acid was shown to occur through a series of enzyme-catalyzed steps that involved conversion of fructose 6-phosphate to GDP-glucosamine (GDP-GlcN) with the nucleotidyltransferase PtmE (Figure 6). For the following step, the N-acetyltransferase responsible for acetylating the C2 amine of GDP-GlcN has not yet been identified; therefore, the C. jejuni GlmU was utilized in the study,78 and efforts to identify the actual enzyme in the pathway are still ongoing. Natively, GlmU is the bifunctional enzyme that first acetylates GlcN-1-P and then catalyzes uridinylation of the sugar phosphate, resulting in the formation of UDP-GlcNAc in C. jejuni.79 The acetylated GDP-GlcNAc in the C. jejuni pathway is then converted to GDP-diNAcBac by a dehydratase (LegB), an aminotransferase (LegC), and an acetyltransferase (LegH) (Figure 6) in a fashion analogous to that of the pgl pathway enzymes that biosynthesize UDP-diNAcBac in C. jejuni. The final three enzymes in the CMP-legionaminic acid pathway in Campylobacter, designated LegG, LegI, and LegF, are homologous to the corresponding enzymes in L. pneumophila (Figure 5). LegG catalyzes the conversion of GDP-diNAcBac to 2,4-diacetamido-2,4,6-trideoxymannose, which is the same as the product from the S. pneumophila NeuC reaction. LegG does not efficiently use the UDP-sugar precursor from the UDP-diNAcBac pathway, further reinforcing the specificity for a GDP-activated sugar. Condensation of PEP with 2,4-diacetamido-2,4,6-trideoxymannose proceeds through the synthase LegI, and activation of this sugar with CTP is accomplished by LegF to yield CMP-legionaminic acid as the final activated sugar for glycoconjugate biosynthesis. It is intriguing that C. jejuni utilizes GDP-linked sugars for the biosynthesis of CMP-legionaminic acid and UDP-linked sugars for the biosynthesis of CMP-pseudaminic acid (used in flagellar O-linked glycosylation) and UDP-diNAcBac in N-linked protein glycosylation. This apparent dichotomy suggests that the alternative GDP-linked sugar pathway to CMP-legionaminic acid may provide some advantage such as a means to control flux through the different biosynthetic pathways.
Figure 6.
Legionaminic acid biosynthetic pathway in C. jejuni. The initial steps in the pathway afford the GDP-GlcNAc intermediate utilized in the formation of GDP-diNAcBac. The bifunctional GlmU* was used as the acetyltransferase for the formation of GDP-GlcNAc in these studies;78 however, the enzyme responsible for this reaction in vivo has not been characterized. The final three steps in the conversion of GDP-diNAcBac into CMP-legionaminic acid are conducted by C. jejuni homologs of LegG, LegI, and LegF.
Pseudaminic Acid
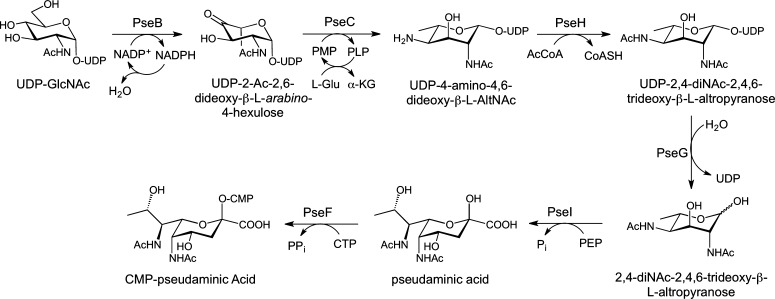
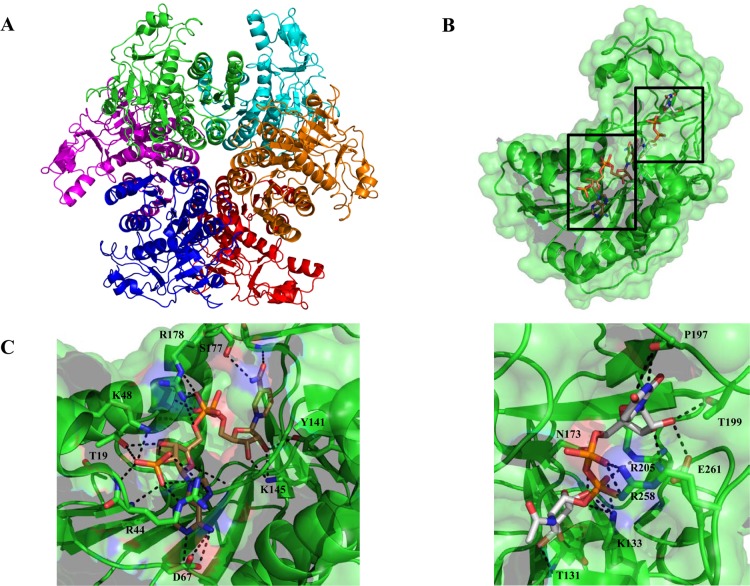
Similar to legionaminic acid, pseudaminic acid is a nine-carbon sialic acid analog that is found on flagellin proteins in C. jejuni and H. pylori (Figure 2).15,80 These glycoproteins are essential for the proper assembly of functional flagella, and consequently bacterial motility, making the pseudaminic acid biosynthesis pathway a potentially important virulence target.81,82 Pseudaminic acid is an isomer of legionaminic acid that is biosynthesized from UDP-GlcNAc through the intermediacy of UDP-2,4-diacetamido-2,4,6-trideoxy-β-l-altropyranose, which is a UDP-DATDH that has stereochemistry at C4 and C5 inverted relative to that of UDP-diNAcBac (Figure 7). The first enzyme in the pathway, PseB, is a member of the short chain dehydrogenase/reductase (SDR) superfamily and lacks the N-terminal transmembrane domain observed in the corresponding enzyme from the C. jejuni pathway, PglF.83 PseB catalyzes both redox-dependent dehydratase and C5 epimerase activities15,19 to generate UDP-2-acetamido-2,6-dideoxy-β-l-arabino-4-hexulose through the action of the catalytic triad of Ser (Thr), Tyr, and Lys in the active site.84,85 It is interesting to note that PseB utilizes NADP+ to oxidize UDP-GlcNAc at C4 rather than NAD+, which is the cofactor in the PglF dehydratase reaction on the diNAcBac pathway.19,86,87 PseB binds UDP-GlcNAc with a much greater affinity (140-fold lower Km) with respect to the diNAcBac biosynthesis enzyme, PglF, resulting in a higher catalytic efficiency (31-fold).88 Upon accumulation of the 4-keto sugar, PseB can also catalyze an additional C5 epimerization to generate UDP-2-acetamido-4-keto-2,4,6-trideoxy-α-d-glucose, the UDP-4-keto sugar utilized in the diNAcBac pathway.89 This side reaction allows for cross talk between the two pathways and potentially establishes another level of control in the production of these sugars for the assembly of bacterial glycoconjugates, with respect to their involvement in pathogenicity. Electron microscopy and X-ray structural analysis of PseB from H. pylori revealed a hexameric assembly in the form of a compact doughnut-like structure (Figure 8).86 Each protomer is bilobal containing an N-terminal domain that binds NADP+ and a smaller C-terminal lobe responsible for binding UDP-GlcNAc. Site-directed mutagenesis of the active site lysine (K133M and K133E) in PseB resulted in the attenuation of enzyme activity, confirming the involvement of this residue in catalysis. Previous crystallographic studies of the active site tyrosine90 in a homologous SDR also confirmed the importance of this residue in catalysis. Further mutagenesis studies with the PseB homolog in C. jejuni were confirmatory in nature, and Y135F and K127A mutations resulted in severe attenuation of enzyme activity.89 The proposed PseB reaction mechanism can be dissected into three distinct steps starting with oxidation at the C4 position of UDP-GlcNAc through a catalytic base (Y141) and reduction of NADP+. This is followed by abstraction of the C5 proton (most likely by K133 acting as a general base), which results in the elimination of water and formation of an enone intermediate. Finally, the transfer of a hydride from NADPH to C6 and protonation of C5 on the opposite face from the dehydration step result in the final UDP-4-keto sugar with inversion of stereochemistry at C5 and regeneration of NADP+. The keto sugar generated from PseB is utilized by the aminotransferase PseC in a PLP-dependent reaction to form the 4-amino sugar product.19,39,91 The diNAcBac C. jejuni aminotransferase (PglE) produces an isomer of this sugar that varies in the stereochemistry at positions C4 and C5. Interestingly, PglE is a more efficient enzyme with respect to PseC, showing a 44-fold increase in kcat/Km. The aminotransferases from the pseudaminic and diNAcBac pathways display no cross talk with their respective substrates, demonstrating the stereospecificity of each enzyme. PseC forms a homodimer in the crystal structure with each protomer containing two α/β/α domains (Figure 9A).39 This enzyme has been crystallized in the apo form, as an internal aldimine with PLP, and as an external aldimine between the UDP-sugar product and PLP.39 Interestingly, no significant conformational changes are apparent among these PseC structures. The UDP-4-keto sugar substrate binds to both protomers in the homodimer; however, few direct interactions are observed between the sugar moiety and PseC, resulting in the conclusion that flexibility at this position may be important for catalysis. The PseC structure with an external aldimine between PLP and the UDP-sugar product does not yet reveal how these aminotransferases differentiate between UDP-4-keto substrates that vary in stereochemistry only at position C5. However, a comparison between the external aldimine structures of PseC and DesI from Streptomyces venezuelae led the authors to conclude that axial versus equatorial addition of the amino group is the result of the different conformations in which the substrates bind.92 A structure of the C. jejuni PglE bound to either the substrate or product UDP-sugar could address the question of how these enzymes accomplish the transformations with complete substrate specificity.
Figure 7.
H. pylori CMP-pseudaminic acid biosynthesis pathway that proceeds through the intermediacy of UDP-2,4-diNAc-2,4,6-trideoxy-β-l-altropyranose.
Figure 8.
(A) Ribbon representation of the H. pylori dehydratase PseB biological hexamer unit with each protomer individually colored for the sake of clarity. (B) Ribbon and space-filling representation of the monomer bound to UDP-GlcNAc (gray) and NADP+ (brown) (PDB entry 2GN6). (C) Major PseB active site interactions depicted as dashed lines with NADP+ (left) and UDP-GFlcNAc (right).
Figure 9.
(A) H. pylori aminotransferase PseC crystal structure (PDB entry 2FN6) represented in ribbon and space-filling format (left) with bound PLP (gray). The major interactions between PLP and the PseC active site are depicted with dashed lines (right). The biological unit is a dimer with each subunit individually colored for the sake of clarity. (B) C. jejuni UDP-sugar hydrolase PseG (PDB entry 3HBN) in ribbon and space-filling format (left). The biological unit is a monomer consisting of two domains that each include a Rossmann fold. PseG binds to UDP (gray) between these two subunits. Major interactions of the PseG active site with UDP are depicted as dashed lines (right).
The acetyltransferase PseH then acetylates the 4-amino sugar in an AcCoA-dependent manner forming UDP-2,4-diacetamido-2,4,6-trideoxy-β-l-altropyranose.93 Although the PseH crystal structure remains undetermined, sequence analysis reveals that this enzyme belongs to the GCN5-related N-acetyltransferase (GNAT) superfamily and bears no structural resemblance to their left-handed β-helix counterparts in UDP-diNAcBac biosynthesis.94 Few bacterial acetyltransferases that adopt a GNAT fold have been characterized; however, WecD from E. coli is one example in which an apo structure and an AcCoA-bound structure have been determined.95 It is interesting that the Pse and diNAcBac pathway N-acetyltransferases are completely divergent based upon their sequence and respective folds, and it is likely that these biosynthetic pathways evolved independently.
The fourth step of the pathway relies on UDP hydrolysis by PseG, resulting in 2,4-diacetamido-2,4,6-trideoxy-l-altropyranose. Mechanistic studies of this enzyme established that the hydrolysis of UDP-sugar proceeded in a concerted fashion with attack by a water molecule at C1 and cleavage of the C–O anomeric bond.96 Additionally, apo and UDP-bound PseG crystal structures allowed for the identification of His-17 as the general base utilized for activating the nucleophilic water molecule.97 The C. jejuni UDP-sugar hydrolase PseG is a monomer in solution and in the crystal structure, with the enzyme exhibiting a GT-B fold that comprises two separate Rossmann domains with UDP binding between these domains (Figure 9B). Although the UDP-sugar-bound PseG structure could not be obtained, molecular dynamics studies suggested that the sugar conformation adopts a 5S1 twist-boat conformation in the enzyme active site.97
The PseI synthase catalyzes the condensation of PEP with 2,4-diacetamido-2,4,6-trideoxy-l-altrose, generating pseudaminic acid in a fashion similar to that of C. jejuni LegI (the NeuB homolog) in the legionaminic acid pathway.98 Analysis of this enzyme revealed the requirement of a divalent metal ion for catalysis and that the formation of pseudaminic acid proceeds through a tetrahedral intermediate after attack of C3 from PEP on the open chain aldehyde sugar. Following collapse of this intermediate, inorganic phosphate is released followed by cyclization to the pyranose form of pseudaminic acid. Although there is currently no known crystal structure of PseI, the structure of the homologous enzyme from the sialic acid biosynthetic pathway in N. meningitidis (NeuB) has been determined.99,100 This enzyme forms a homodimer with each protomer active site consisting of an N-terminal TIM barrel domain that binds to the sugar substrate in the open chain form.
The final step in the pathway, which forms CMP-activated pseudaminic acid, is catalyzed by PseF.93 This reaction is dependent upon alkaline pH and Mg2+. In connection with their studies on pseudaminic acid biosynthesis, Schoenhofen, Logan, and co-workers also demonstrated that CMP-Pse can be produced by combining all of the pathway enzymes in a “one-pot” biotransformation. However, it was noted that the final CMP-Pse yield in this process was severely reduced in the presence of all of the CMP-Pse enzymes when compared to the yields of individual reactions in the pathway. Further studies revealed that CMP-Pse acid was a potent inhibitor of the first enzyme (PseB) in the pathway with a Ki(app) of 18.7 μM, allowing for negative feedback inhibition of the biosynthesis of the final CMP-Pse product.89 A metabolomics approach, utilizing gene knockouts and detection of nucleotide intermediates by capillary electrophoresis–electrospray mass spectrometry, confirmed the direct involvement of the Pse proteins in pseudaminic acid biosynthesis.101 Recently, pseudaminic acid has been chemically synthesized from GlcNAc, which will allow for more extensive studies for improving our understanding of the relationship between this sugar and bacterial O-linked glycosylation and pathogenicity.102 Although some of the pathway enzymes described above have been examined in an in vitro biochemical setting, a comprehensive kinetic analysis is still lacking. It is likely that such studies would aid in the understanding of the interplay between substrates and enzymes and will be important for developing these enzymes as potential antivirulence targets.
The two best studied bacterial pathogens in terms of flagellin glycosylation with pseudaminic acid are C. jejuni and H. pylori; however, other species, including Clostridium botulinum and Aeromonas caviae, are also currently under investigation.103−109 The C. jejuni flagellin protein, FlaA, is glycosylated at 19 serine/threonine residues, with eight of these sites making a major contribution to motility and autoagglutination of the bacteria.80,110 Disruption of the pseudaminic acid biosynthesis genes pseB and pseC resulted in nonglycosylated flagellin, confirming their essentiality in the bacterial flagellum assembly. Studies with pseF, pseG, and pseH mutants were also conducted in C. jejuni to verify their role in pseudaminic acid production,82 and in each case, disruption of these genes resulted in nonmotile phenotypes because of the lack of flagella filaments and hook structures. These mutated strains also exhibited a reduction in adherence to and invasion of intestinal epithelial cell layers. Recent studies have demonstrated that flagellin proteins can undergo spontaneous antigenic variation through O-dimethylglyceric acid derivatives of pseudaminic acid.111,112 Although the homopolymeric tract-containing Cj1295 gene is responsible for this modification, the question of why this occurs remains unanswered. This type of structural diversity, which is observed in variant pilin glycoproteins in N. gonorrhoeae,113,114 may be important for evading the host immune response during colonization. H. pylori has also been extensively studied, resulting in the identification of the FlaA and FlaB flagellin proteins that are modified with pseudaminic acid.115,116 It was previously found that flagellar motility is a requirement for colonization in both in vitro and in vivo model systems.117,118 Insertional mutagenesis led to the discovery of three genes (HP0326A, HP0326B, and HP0178) that are directly involved in the biosynthesis of pseudaminic acid.81 Disruption of each gene resulted in nonmotile phenotypes, a decreased level of flagellin glycoprotein, and accumulation of UDP-sugar nucleotide precursors. H. pylori is also able to regulate pathogen motility through deglycosylation of pseudaminic acid on FlaA through the HP0518 protein.119 The HP0518 knockout mutant exhibited hypermotility and a superior ability to colonize C57BL/6 mice in vivo, confirming the role that H. pylori flagella play in pathogen–host interactions. A comprehensive effort has led to the identification of two flagellin proteins (FlaA and FlaB) that play a role in pathogenesis; however, the glycome of H. pylori is still poorly understood. Recently, glycan metabolic labeling coupled with mass spectrometry analysis has resulted in the identification of 125 O-linked glycoproteins, many of which are linked to pathogenesis.120 Work still remains on the characterization of these putative glycoproteins and the role glycosylation plays in H. pylori pathogenesis.
Conclusions
The ever-increasing resistance toward present-day antibiotics has resulted in the search for novel agents to address this challenge. Bacterial protein glycosylation, using highly modified carbohydrates, as it relates to pathogenicity may represent an important area of future focus because of the absence of these unique building blocks in eukaryotes and the decrease in pathogenicity when their biosynthesis is disrupted. It has been fewer than 15 years since the discovery that N-linked protein glycosylation, once thought to be exclusive to eukaryotes, also occurs in bacteria. The biochemical characterization of the C. jejuni pgl N-linked pathway has dramatically improved our understanding of bacterial glycosylation at the molecular level. Furthermore, the biosynthetic protein glycosylation machinery is also present as an O-linked system for example in Neisseria spp. Although these pathways glycosylate diverse proteins with different glycans, the one remaining constant is the reducing-end sugar diNAcBac. This highly modified bacterial sugar is biosynthesized by a series of three enzymes that are conserved in these pathogens. The presence of diNAcBac has been shown to be of utmost importance to the formation of the fully assembled glycan as disruption of the genes leading to its biosynthesis in the native hosts results in the significant attenuation of protein glycosylation. Importantly, some of the protein targets of this pathway are known virulence factors that play a key role in pathogenicity. For example, disrupting glycosylation of VirB10 from the type IV secretion system in C. jejuni and PilE from type IV pilin in Neisseria spp. leads to decreases in competency and cell adherence, respectively. What is still unknown is why bacteria utilize diNAcBac in N- and O-linked glycosylation as well as in the O-antigen and core region of LPS. In contrast, eukaryotes feature GlcNAc as the reducing-end sugar in N-linked glycosylation. One possibility is that diNAcBac acts as a decoy to avoid detection and hydrolysis in eukaryotic hosts. However, this is just one possibility, and clearly, further investigation into understanding the precise role of diNAcBac is warranted.
The importance of carbohydrates such as diNAcBac and related stereoisomers in bacterial pathogenicity has been further underscored by the characterization of O-linked legionaminic and pseudaminic acid biosynthetic pathways. These nine-carbon α-keto sugars are molecular mimics of sialic acid, a carbohydrate found predominately on the exterior of mammalian cells that is essential for cell–cell communication and adhesion. CMP-legionaminic acid is produced from nucleotide-activated (UDP or GDP) diNAcBac by the actions of three enzymes that hydrolyze the nucleotide diphosphate, invert the stereochemistry at the C2 position, and condense a three-carbon unit from PEP onto the sugar. The final enzymatic step activates legionaminic acid through CTP, forming the CMP-sugar. Although experimental evidence has pointed to the presence of legionaminic acid in flagella and the O-antigen of LPS, in vivo validation of the essentiality of the sugar is still lacking. Future studies are needed to elucidate the role that legionaminic acid plays in the assembly of these virulence factors and in bacterial pathogenicity. Pseudaminic acid is produced by a series of enzymes, which are homologous to those in the legionaminic acid pathway and utilize the DATDH isomer 2,4-diacetamido-2,4,6-trideoxy-l-altropyranose as an intermediate. Pseudaminic acid is found on multiple sites in flagellar proteins, and disruption of the genes responsible for the biosynthesis of this sugar has adverse effects on motility, adherence, and invasion. Further investigation into the glycome of these pathogenic bacteria in the context of pseudaminic acid glycosylation is still necessary.
Although each of the diNAcBac-related pathways utilizes similar starting substrates, the question of how pathogenic bacteria elicit control over these systems and under what circumstances remains. Similarly, there appears to be a high level of cross talk between some of the homologous enzymes that act on different substrates responsible for pathogenicity. Is this an evolutionary remnant that is controlled by segregation of pathways in the host organisms, or is this an additional mechanism whereby bacteria can elicit a level of control based upon selective pressures in the environment? In conclusion, great strides have been made in understanding the biosynthesis of unique, bacterial sugars in the context of pathogenicity. A tremendous amount of work still remains to validate the pathway enzymes of bacterial sugars as true antivirulence targets; however, we are ever closer to understanding the biological context of protein glycosylation in relationship to pathogenicity.
Acknowledgments
We thank Dr. Angelyn Larkin, Dr. Meredith Hartley, and Austin Travis for critical reading of the manuscript.
Glossary
Abbreviations
- AcCoA
acetyl-coenzyme A
- AUC
analytical ultracentrifugation
- CMP
cytidine monophosphate
- CTP
cytidine triphosphate
- DATDH
2,4-diacetamido-2,4,6-trideoxy-α-d-hexose
- DiNAcBac
N,N′-diacetylbacillosamine
- ESI-MS
electrospray ionization mass spectrometry
- Gal
galactose
- GalNAc
N-acetylgalactosamine
- GDP
guanosine diphosphate
- Glc
glucose
- GlcNAc
N-acetylglucosamine
- legionaminic acid
5,7-diacetamido-3,5,7,9-tetradeoxy-d-glycero-d-galacto-nonulosonic acid
- GNAT
GCN5-related N-acetyltransferase
- LPS
lipopolysaccharide
- N-linked
asparagine-linked
- NAD+
nicotinamide adenine dinucleotide
- NADP+
nicotinamide adenine dinucleotide phosphate
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- O-linked
serine- or threonine-linked
- OMV
outer membrane vesicle
- PDB
Protein Data Bank
- PEP
phosphoenolpyruvate
- Pgl
protein glycosylation
- PglB-ATD
acetyltransferase domain of PglB
- PglB-PGTD
phosphoglycosyltransferase domain of PglB
- Pse or pseudaminic acid
5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-l-manno-nonulosonic acid
- PLP
pyridoxal 5′-phosphate
- PMP
pyridoxamine 5′-phosphate
- SDR
short chain dehydrogenase/reductase
- sialic acid
N-acetylneuraminic acid
- TFSS
type IV secretion system
- UDP
uridine diphosphate
- UDP-4-amino
UDP-2-acetamido-4-amino-2,4,6-trideoxy-α-d-glucose
- UDP-4-keto
UDP-2-acetamido-4-keto-2,4,6-trideoxy-α-d-glucose
- UDP-diNAcBac
UDP-N,N′-diacetylbacillosamine or UDP-2,4-diacetamido-2,4,6-trideoxy-α-d-glucose
- Und-P
undecaprenyl phosphate
- Und-PP
undecaprenyl diphosphate.
The authors declare no competing financial interest.
This work was supported by National Institutes of Health (NIH) Grant GM097241 (B.I.) and NIH Biotechnology Training Program T32-GM08334 (M.J.M.).
Funding Statement
National Institutes of Health, United States
References
- Varki A. (1993) Biological roles of oligosaccharides: All of the theories are correct. Glycobiology 3, 97–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra N.; Sinha S.; Ramya T. N.; Surolia A. (2006) N-Linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem. Sci. 31, 156–163. [DOI] [PubMed] [Google Scholar]
- Rudd P. M.; Elliott T.; Cresswell P.; Wilson I. A.; Dwek R. A. (2001) Glycosylation and the immune system. Science 291, 2370–2376. [DOI] [PubMed] [Google Scholar]
- Larkin A.; Imperiali B. (2011) The expanding horizons of asparagine-linked glycosylation. Biochemistry 50, 4411–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H.; Szymanski C. M. (2013) Bacterial protein N-glycosylation: New perspectives and applications. J. Biol. Chem. 288, 6912–6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemka A.; Nothaft H.; Zheng J.; Szymanski C. M. (2013) N-glycosylation of Campylobacter jejuni surface proteins promotes bacterial fitness. Infect. Immun. 81, 1674–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemka A.; Clyne M.; Shanahan F.; Tomkins T.; Corcionivoschi N.; Bourke B. (2011) Probiotic colonization of the adherent mucus layer of HT29MTXE12 cells attenuates Campylobacter jejuni virulence properties. Infect. Immun. 78, 2812–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deun K.; Pasmans F.; Ducatelle R.; Flahou B.; Vissenberg K.; Martel A.; Van den Broeck W.; Van Immerseel F.; Haesebrouck F. (2008) Colonization strategy of Campylobacter jejuni results in persistent infection of the chicken gut. Vet. Microbiol. 130, 285–297. [DOI] [PubMed] [Google Scholar]
- Young N. M.; Brisson J. R.; Kelly J.; Watson D. C.; Tessier L.; Lanthier P. H.; Jarrell H. C.; Cadotte N.; St. Michael F.; Aberg E.; Szymanski C. M. (2002) Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 8, 42530–42539. [DOI] [PubMed] [Google Scholar]
- Vik A.; Aas F. E.; Anonsen J. H.; Biosborough S.; Schneider A.; Edde-Jacobsen W.; Koomey M. (2009) Broad-spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U.S.A. 106, 4447–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. A.; Riley L. W.; Benz I. (2003) Sweet new world: Glycoproteins in bacterial pathogens. Trends Microbiol. 11, 554–561. [DOI] [PubMed] [Google Scholar]
- Upreti R. K.; Kumar M.; Shankar V. (2003) Bacterial glycoproteins: Functions, biosynthesis and applications. Proteomics 3, 363–379. [DOI] [PubMed] [Google Scholar]
- Scott N. E.; Parker B. L.; Connolly A. M.; Paulech J.; Edwards A. V.; Crossett B.; Falconer L.; Kolarich D.; Djordjevic S. P.; Højrup P.; Packer N. H.; Larsen M. R.; Cordwell S. J. (2011) Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 10, M000031-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonsen J. H.; Vik A.; Egge-Jacobsen W.; Koomey M. (2012) An extended spectrum of target proteins and modification sites in the general O-linked protein glycosylation system in Neisseria gonorrhoeae. J. Proteome Res. 11, 5781–5793. [DOI] [PubMed] [Google Scholar]
- Goon S.; Kelly J. F.; Logan S. M.; Ewing C. P.; Guerry P. (2003) Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 50, 659–671. [DOI] [PubMed] [Google Scholar]
- Szymanski C. M.; Yao R.; Ewing C. P.; Trust T. J.; Guerry P. (1999) Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32, 1022–1030. [DOI] [PubMed] [Google Scholar]
- Szymanski C. M.; Burr D. H.; Guerry P. (2002) Campylobacter protein glycosylation affects host cell interactions. Infect. Immun. 70, 2242–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L.; Pique M. E.; Tainer J. A. (2004) Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378. [DOI] [PubMed] [Google Scholar]
- Schoenhofen I. C.; McNally D. J.; Vinogradov E.; Whitfield D.; Young N. M.; Dick S.; Wakarchuk W. W.; Brisson J. R.; Logan S. M. (2005) Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter. J. Biol. Chem. 281, 723–732. [DOI] [PubMed] [Google Scholar]
- Olivier N. B.; Chen M. M.; Behr J. R.; Imperiali B. (2006) In vitro biosynthesis of UDP-N,N′-diacetylbacillosamine by enzymes of the Campylobacter jejuni general protein glycosylation system. Biochemistry 45, 13659–13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykumar S.; Merkx-Jacques A.; Ratnayake D. B.; Gryski I.; Obhi R. K.; Houle S.; Dozois C. M.; Creuzenet C. (2006) Cj1121c, a novel UDP-4-keto-6-deoxy-GlcNAc C-4 aminotransferase essential for protein glycosylation and virulence in Campylobacter jejuni. J. Biol. Chem. 281, 27733–27743. [DOI] [PubMed] [Google Scholar]
- Hartley M. D.; Morrison M. J.; Aas F. E.; Borud B.; Koomey M.; Imperiali B. (2011) Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for biosynthesis of protein glycans containing N,N′-diacetylbacillosamine. Biochemistry 50, 4936–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison M. J.; Imperiali B. (2013) Biosynthesis of UDP-N,N′-diacetylbacillosamine in Acinetobacter baumannii: Biochemical characterization and correlation to existing pathways. Arch. Biochem. Biophys. 536, 72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon N. (2007) Celebrating the golden anniversary of the discovery of bacillosamine, the diamino sugar of a Bacillus. Glycobiology 17, 1150–1155. [DOI] [PubMed] [Google Scholar]
- Sharon N.; Jeanloz R. W. (1959) The isolation of an aminohexose from Bacillus subtilis. Biochim. Biophys. Acta 31, 277–278. [DOI] [PubMed] [Google Scholar]
- Sharon N.; Jeanloz R. W. (1960) The diaminohexose component of a polysaccharide isolated from Bacillus subtilis. J. Biol. Chem. 235, 1–5. [PubMed] [Google Scholar]
- Liav A.; Hildesheim J.; Zehavi U.; Sharon N. (1974) Synthesis of 2-acetamido-2,6-dideoxy-d-glucose (N-acetyl-d-quinovosamine), 2-acetamido-2,6-dideoxy-d-galactose (N-acetyl-d-fucosamine), and 2,4-diacetamido-2,4,6-trideoxy-d-glucose from 2-acetamido-2-deoxy-d-glucose. Carbohydr. Res. 33, 217–227. [Google Scholar]
- Weerapana E.; Glover K. J.; Chen M. M.; Imperiali B. (2005) Investigating bacterial N-linked glycosylation: Synthesis and glycosyl acceptor activity of the undecaprenyl pyrophosphate-linked bacillosamine. J. Am. Chem. Soc. 127, 13766–13767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedini E.; Esposito D.; Parrilli M. (2006) A versatile strategy for the synthesis of N-acetyl-bacillosamine-containing disaccharide building blocks related to bacterial O-antigens. Synlett 6, 825–830. [Google Scholar]
- Molinaro A.; Evidente A.; Iacobellis N. S.; Lanzetta R.; Cantore P. L.; Mancino A.; Parrilli M. (2002) O-Specific chain structure from the lipopolysaccharide fraction of Pseudomonas reactans: A pathogen of the cultivated mushrooms. Carbohydr. Res. 337, 467–471. [DOI] [PubMed] [Google Scholar]
- Chowdhury T. A.; Jansson P. E.; Lindberg B.; Lindberg J.; Gustafsson B.; Holme T. (1991) Structural studies of the Vibrio cholerae O:3 O-antigen poly-saccharide. Carbohydr. Res. 215, 303–314. [DOI] [PubMed] [Google Scholar]
- Vinogradov E.; Perry M. B. (2004) Characterisation of the core part of the lipopolysaccharide O-antigen of Francisella novicida (U112). Carbohydr. Res. 339, 1643–1648. [DOI] [PubMed] [Google Scholar]
- Zubkov V. A.; Nazarenko E. L.; Gorshkova R. P.; Ivanova E. P.; Shashkov A. S.; Knirel Y. A.; Paramonov N. A.; Ovodov Y. S. (1995) Structure of the capsular polysaccharide from Alteromonas sp. CMM 155. Carbohydr. Res. 275, 147–154. [DOI] [PubMed] [Google Scholar]
- Moran A. P.; Prendergast M. M. (2001) Molecular mimicry in Campylobacter jejuni and Helicobacter pylori lipopolysaccharides: Contribution of gastrointestinal infections to autoimmunity. J. Autoimmun. 16, 241–256. [DOI] [PubMed] [Google Scholar]
- Sharon N.; Shiff I.; Zehavi U. (1964) The isolation of d-fucosamine (2-amino-2,6-dideoxy-d-galactose) from polysaccharides of Bacillus spp. Biochem. J. 93, 210–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J.; Wren B. W.; Mungall K.; Ketley J. M.; Churcher C.; Basham D.; Chillingworth T.; Davies R. M.; Feltwell T.; Holroyd S.; Jagels K.; Karlyshev A. V.; Moule S.; Pallen M. J.; Penn C. W.; Quail M. A.; Rajandream M. A.; Rutherford K. M.; van Vliet C. W.; Whitehead S.; Barrell B. G. (2000) The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668. [DOI] [PubMed] [Google Scholar]
- Szymanski C. M.; Yao R.; Ewing C. P.; Turst T. J.; Guerry P. (1999) Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32, 1022–1030. [DOI] [PubMed] [Google Scholar]
- Bennett B. D.; Kimball E. H.; Gao M.; Osterhout R.; Van Dien S. J.; Rabinowitz J. D. (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenhofen I. C.; Lunin V. V.; Julien J. P.; Li Y.; Ajamian E.; Matte A.; Cygler M.; Brisson J. R.; Aubry A.; Logan S. M.; Bhatia S.; Wakarchuk W. W.; Young N. M. (2006) Structural and functional characterization of PseC, an aminotransferase involved in the biosynthesis of pseudaminic acid, an essential flagellar modification in Helicobacter pylori. J. Biol. Chem. 281, 8907–8916. [DOI] [PubMed] [Google Scholar]
- Larkin A.; Olivier N. B.; Imperiali B. (2010) Structural analysis of WbpE from Pseudomonas aeruginosa PAO1: A nucleotide sugar aminotransferase involved in O-antigen assembly. Biochemistry 49, 7227–7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger J.; Sauder J. M.; Adams J. M.; Antonysamy S.; Bain K.; Bergseid M. G.; Buchanan S. G.; Buchanan M. D.; Batiyenko Y.; Christopher J. A.; Emtage S.; Eroshkina A.; Feil I.; Furlong E. B.; Gajiwala K. S.; Gao X.; He D.; Hendle J.; Huber A.; Hoda K.; Kearins P.; Kissinger C.; Laubert B.; Lewis H. A.; Lin J.; Loomis K.; Lorimer D.; Louie G.; Maletic M.; Marsh C. D.; Miller I.; Molinari J.; Muller-Dieckmann H. J.; Newman J. M.; Noland B. W.; Pagarigan B.; Park F.; Peat T. S.; Post K. W.; Radojicic S.; Ramos A.; Romero R.; Rutter M. E.; Sanderson W. E.; Schwinn K. D.; Tresser J.; Winhoven J.; Wright T. A.; Wu L.; Xu J.; Harris T. J. R. (2005) Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins 60, 787–796. [DOI] [PubMed] [Google Scholar]
- Morrison M. J.; Imperiali B. (2013) Biochemical analysis and structure determination of bacterial acetyltransferases responsible for the biosynthesis of UDP-N,N′-diacetylbacillosamine. J. Biol. Chem. 288, 32248–32260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demendi M.; Creuzenet C. (2009) Cj1123c (PglD), a multifaceted acetyltransferase from Campylobacter jejuni. Biochem. Cell Biol. 87, 469–483. [DOI] [PubMed] [Google Scholar]
- Olivier N. B.; Imperiali B. (2008) Crystal structure and catalytic mechanism of PglD from Campylobacter jejuni. J. Biol. Chem. 283, 27937–27946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan E. S.; Ruane K. M.; Sulea T.; Watson D. C.; Proteau A.; Leclerc S.; Cygler M.; Matte A.; Young N. M. (2008) Structure and active site residues of PglD, an N-acetyltransferase from the bacillosamine synthetic pathway required for N-glycan synthesis in Campylobacter jejuni. Biochemistry 47, 1827–1836. [DOI] [PubMed] [Google Scholar]
- Escaich S. (2008) Antivirulence as a new antibacterial approach for chemotherapy. Curr. Opin. Chem. Biol. 12, 400–408. [DOI] [PubMed] [Google Scholar]
- Hughes R. (2004) Campylobacter jejuni in Guillain–Barré syndrome. Lancet Neurol. 3, 644. [DOI] [PubMed] [Google Scholar]
- Komagamine T.; Yuki N. (2006) Ganglioside mimicry as a cause of Guillain–Barré syndrome. CNS Neurol. Disord.: Drug Targets 5, 391–400. [DOI] [PubMed] [Google Scholar]
- Yu R. K.; Usuki S.; Ariga T. (2006) Ganglioside molecular mimicry and its pathological roles in Guillain–Barré syndrome and related diseases. Infect. Immun. 74, 6517–6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H.; Szymanski C. M. (2010) Protein glycosylation in bacteria: Sweeter than ever. Nat. Rev. Microbiol. 8, 765–778. [DOI] [PubMed] [Google Scholar]
- Luangtongkum T.; Jeon B.; Han J.; Plummer P.; Logue C. M.; Zhang Q. (2009) Antibiotic resistance in Campylobacter: Emergence, transmission and persistence. Future Microbiol. 4, 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J.; Jarrell H.; Millar L.; Tessier L.; Fiori L. M.; Lau P. C.; Allan B.; Szymanski C. M. (2006) Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. J. Bacteriol. 188, 2427–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrixson D. R.; DiRita V. J. (2004) Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52, 471–484. [DOI] [PubMed] [Google Scholar]
- Szymanski C. M.; Burr D. H.; Guerry P. (2001) Campylobacter protein glycosylation affects host-cell interactions. Infect. Immun. 70, 2242–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J. C.; Szymanski C. M.; Guerry P. (2004) N-Linked protein glycosylation is required for full competence in Campylobacter jejuni 81-176. J. Bacteriol. 186, 6508–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmi A.; Watson E.; Sandu P.; Gundogdu O.; Mills D. C.; Inglis N. F.; Manson E.; Imrie L.; Bajaj-Elliott M.; Wren B. W.; Smith D. G. E.; Dorrell N. (2012) Campylobacter jejuni outer membrane vesicles play an important role in bacterial interactions with human intestinal epithelial cells. Infect. Immun. 80, 4089–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aas F. E.; Egge-Jacobsen W.; Winther-Larsen H. C.; Lovold C.; Hitchen P. G.; Dell A.; Koomey M. (2006) Neisseria gonorrhoeae Type IV pili undergo multisite, hierarchical modifications with phosphoethanolamine and phosphocholine requiring an enzyme structurally related to lipopolysaccharide phosphoethanolamine transferases. J. Biol. Chem. 281, 27712–27723. [DOI] [PubMed] [Google Scholar]
- Borud B.; Aas F. E.; Vik A.; Winther-Larsen H. C.; Egge-Jacobson W.; Koomey M. (2010) Genetic, structural, and antigenic analyses of glycan diversity in the O-linked protein glycosylation systems of human Neisseria species. J. Bacteriol. 192, 2816–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings M. P.; Jen F. E.-C.; Roddam L. F.; Apicella M. A.; Edwards J. L. (2011) Neisseria gonorrhoeae pilin glycan contributes to CR3 activation during challenge of primary cervical epithelial cells. Cell. Microbiol. 13, 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimson E.; Virji M.; Makepeace K.; Dell A.; Morris H. R.; Payne G.; Saunders J. R.; Jennings M. P.; Barker S.; Panico M.; Blench I.; Moxon E. R. (1995) Meningococcal pilin: A glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol. Microbiol. 17, 1201–1214. [DOI] [PubMed] [Google Scholar]
- Jennings M. P.; Virji M.; Evans D.; Foster V.; Srikhanta Y. N.; Steeghs L.; van der Ley P.; Moxon E. R. (1998) Identification of a novel gene involved in pilin glycosylation in Neisseria meningitidis. Mol. Microbiol. 29, 975–984. [DOI] [PubMed] [Google Scholar]
- Power P. M.; Roddam L. F.; Rutter K.; Fitzpatrick S. Z.; Srikhanta Y. N.; Jennings M. P. (2003) Genetic characterization of pilin glycosylation and phase variation in Neisseria meningitidis. Mol. Microbiol. 49, 833–847. [DOI] [PubMed] [Google Scholar]
- Jen F. E.-C.; Warren M. J.; Schulz B. L.; Power P. M.; Swords W. E.; Weiser J. N.; Apicella M. A.; Edwards J. L.; Jennings M. P. (2013) Dual pili post-translational modifications synergize to mediate meningococcal adherence to platelet activating factor receptor on human airway cells. PLoS Pathog. 9, e1003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer R. (2009) Sialic acids as regulators of molecular and cellular interactions. Curr. Opin. Struct. Biol. 19, 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov Y. E.; Shashkov A. S.; Knirel Y. A.; Zahringer U. (2001) Synthesis and identification in bacterial lipopolysaccharides of 5,7-diacetamido-3,5,7,9-tetradeoxy-d-glycero-d-galacto- and -d-glycero-d-talo-non-2-ulosonic acids. Carbohydr. Res. 331, 233–237. [DOI] [PubMed] [Google Scholar]
- Knirel Y. A.; Rietschel E. T.; Marre R.; Zahringer U. (1994) The structure of the O-specific chain of Legionella pneumophila serogroup 1 lipopolysaccharide. Eur. J. Biochem. 221, 239–245. [DOI] [PubMed] [Google Scholar]
- McNally D. J.; Aubry A. J.; Hui J. P.; Khieu N. H.; Whitfield D.; Ewing C. P.; Guerry P.; Brisson J. R.; Logan S. M.; Soo E. C. (2007) Targeted metabolomics analysis of Campylobacter coli VC167 reveals legionaminic acid derivatives as novel flagellar glycans. J. Biol. Chem. 282, 14463–14475. [DOI] [PubMed] [Google Scholar]
- Haseley S. R.; Wilkinson S. G. (1997) Structural studies of the putative O-specific polysaccharide of Acinetobacter baumannii O24 containing 5,7-diamino-3,5,7,9-tetradeoxy-l-glycero-d-galactononulosonic acid. Eur. J. Biochem. 250, 617–623. [DOI] [PubMed] [Google Scholar]
- Hu D.; Liu B.; Dijkshoorn L.; Wang L.; Reeves P. R. (2013) Diversity in the major polysaccharide antigen of Acinetobacter baumannii assessed by DNA sequencing, and development of a molecular serotyping scheme. PLoS One 8, e70329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean L. L.; Vinogradov E.; Pagotto F.; Perry M. B. (2011) Characterization of the lipopolysaccharide O-antigen of Cronobacter turicensis HPB3287 as a polysaccharide containing a 5,7-diacetamido-3,5,7,9-tetradeoxy-d-glycero-d-galacto-non-2-ulosonic acid (legionaminic acid) residue. Carbohydr. Res. 346, 2589–2594. [DOI] [PubMed] [Google Scholar]
- Li X.; Perepelov A. V.; Wang Q.; Senchenkova S. N.; Liu B.; Shevelev S. D.; Guo X.; Shashkov A. S.; Chen W.; Wang L.; Knirel Y. A. (2010) Structural and genetic characterization of the O-antigen of Escherichia coli O161 containing a derivative of a higher acidic diamino sugar, legionaminic acid. Carbohydr. Res. 345, 1581–1587. [DOI] [PubMed] [Google Scholar]
- Lewis A. L.; Desa N.; Hansen E. E.; Knirel Y. A.; Gordon J. I.; Gagneux P.; Nizet V.; Varki A. (2009) Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc. Natl. Acad. Sci. U.S.A. 106, 13552–13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post D. M. B.; Yu L.; Krasity B. C.; Choudhury B.; Mandel M. J.; Brennan C. A.; Ruby E. G.; McFall-Ngai M. J.; Gibson B. W.; Apicella M. A. (2012) O-antigen and core carbohydrate of Vibrio fischeri lipopolysaccharide: Composition and analysis of their role in Euprymna scolopes light organ colonization. J. Biol. Chem. 287, 8515–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaze P. A.; Watson D. C.; Young N. M.; Tanner M. E. (2008) Biosynthesis of CMP-N,N′-diacetyllegionaminic acid from UDP-N,N′-diacetylbacillosamine in Legionella pneumophila. Biochemistry 47, 3272–3282. [DOI] [PubMed] [Google Scholar]
- Lüneberg E.; Zetzmann N.; Alber D.; Knirel Y. A.; Kooistra O.; Zähringer U.; Frosch M. (2000) Cloning and functional characterization of a 30 kb gene locus required for lipopolysaccharide biosynthesis in Legionella pneumophila. Int. J. Med. Microbiol. 290, 37–39. [DOI] [PubMed] [Google Scholar]
- Murkin A. S.; Chou W. K.; Wakarchuk W. W.; Tanner M. E. (2004) Identification and mechanism of a bacterial hydrolyzing UDP-N-acetylglucosamine 2-epimerase. Biochemistry 43, 14290–14298. [DOI] [PubMed] [Google Scholar]
- Chou W. K.; Hinderlich S.; Reutter W.; Tanner M. E. (2003) Sialic acid biosynthesis: Stereochemistry and mechanism of the reaction catalyzed by the mammalian UDP-N-acetylglucosamine 2-epimerase. J. Am. Chem. Soc. 125, 2455–2461. [DOI] [PubMed] [Google Scholar]
- Schoenhofen I. C.; Vinogradov E.; Whitfield D. M.; Brisson J. R.; Logan S. M. (2009) The CMP-legionaminic acid pathway in Campylobacter: Biosynthesis involving novel GDP-linked precursors. Glycobiology 19, 715–725. [DOI] [PubMed] [Google Scholar]
- Mengin-Lecreulx D.; van Heijenoort J. (1994) Copurification of glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase activities of Escherichia coli: Characterization of the glmU gene product as a bifunctional enzyme catalyzing two subsequent steps in the pathway for UDP-N-acetylglucosamine synthesis. J. Bacteriol. 176, 5788–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault P.; Logan S. M.; Kelly J. F.; Brisson J. R.; Ewing C. P.; Trust T. J.; Guerry P. (2001) Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276, 34862–34870. [DOI] [PubMed] [Google Scholar]
- Schirm M.; Soo E. C.; Aubry A. J.; Austin J.; Thibault P.; Logan S. M. (2003) Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 48, 1579–1592. [DOI] [PubMed] [Google Scholar]
- Guerry P.; Ewing C. P.; Schirm M.; Lorenzo M.; Kelly J. F.; Pattarini D.; Majam G.; Thibault P.; Logan S. M. (2006) Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 60, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallberg Y.; Oppermann U.; Persson B. (2010) Classification of the short-chain dehydrogenase/reductase superfamily using hidden Markov models. FEBS J. 277, 2375–2386. [DOI] [PubMed] [Google Scholar]
- Oppermann U.; Filling C.; Hult M.; Shafqat N.; Wu X.; Lindh M.; Shafqat J.; Nordling E.; Kallberg Y.; Persson B.; Jornvall H. (2003) Short-chain dehydrogenases/reductases (SDR): The 2002 update. Chem.-Biol. Interact. 143–144, 247–253. [DOI] [PubMed] [Google Scholar]
- Filling C.; Berndt K. D.; Benach J.; Knapp S.; Prozorovski T.; Nordling E.; Ladenstein R.; Jornvall H.; Oppermann U. (2002) Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 277, 25677–25684. [DOI] [PubMed] [Google Scholar]
- Ishiyama N.; Creuzenet C.; Miller W. L.; Demendi M.; Anderson E. M.; Harauz G.; Lam J. S.; Berghuis A. M. (2006) Structural studies of FlaA1 from Helicobacter pylori reveal the mechanism for inverting 4,6-dehydratase activity. J. Biol. Chem. 281, 24489–24495. [DOI] [PubMed] [Google Scholar]
- Morrison J. P.; Schoenhofen I. C.; Tanner M. E. (2008) Mechanistic studies on PseB of pseudaminic acid biosynthesis: A UDP-N-acetylglucosamine 5-inverting 4,6-dehydratase. Bioorg. Chem. 36, 312–320. [DOI] [PubMed] [Google Scholar]
- Creuzenet C. (2004) Characterization of CJ1293, a new UDP-GlcNAc C6 dehydratase from Campylobacter jejuni. FEBS Lett. 559, 136–140. [DOI] [PubMed] [Google Scholar]
- McNally D. J.; Schoenhofen I. C.; Houliston R. S.; Khieu N. H.; Whitfield D. M.; Logan S. M.; Jarrell H. C.; Brisson J. R. (2008) CMP-pseudaminic acid is a natural potent inhibitor of PseB, the first enzyme of the pseudaminic acid pathway in Campylobacter jejuni and Helicobacter pylori. ChemMedChem 3, 55–59. [DOI] [PubMed] [Google Scholar]
- Thoden J. B.; Wohlers T. M.; Fridovich-Keil J. L.; Holden H. M. (2000) Crystallographic evidence for Tyr157 functioning as the active site base in human UDP-galactose 4-epimerase. Biochemistry 39, 5691–5701. [DOI] [PubMed] [Google Scholar]
- Obhi R. K.; Creuzenet C. (2005) Biochemical characterization of the Campylobacter jejuni Cj1294, a novel UDP-4-keto-6-deoxy-GlcNAc aminotransferase that generates UDP-4-amino-4,6-dideoxy-GalNAc. J. Biol. Chem. 280, 20902–20908. [DOI] [PubMed] [Google Scholar]
- Burgie E. S.; Holden H. M. (2007) Molecular architecture of DesI: A key enzyme in the biosynthesis of desosamine. Biochemistry 46, 8999–9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenhofen I. C.; McNally D. J.; Brisson J. R.; Logan S. M. (2006) Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: Synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology 16, 8C–14C. [DOI] [PubMed] [Google Scholar]
- Matte A., Schoenhofen I. C., Sulea T., Cygler M., and Young N. M. (2012) Synthesis of 4-acetamidohexoses in bacteria: Structural insights from the bacillosamine and nonulosonic acid pathways. In Bacterial Glycomics: Current Research, Technology, and Applications (Reid C. W., Reid A. N., and Twine S. M., Eds.) 1st ed., pp 193–212, Caister Academic Press, Norfolk, U.K. [Google Scholar]
- Hung M. N.; Rangarajan E.; Munger C.; Nadeau G.; Sulea T.; Matte A. (2006) Crystal structure of TDP-fucosamine acetyltransferase (WecD) from Escherichia coli, an enzyme required for enterobacterial common antigen synthesis. J. Bacteriol. 188, 5606–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F.; Tanner M. E. (2006) PseG of pseudaminic acid biosynthesis: A UDP-sugar hydrolase as a masked glycosyltransferase. J. Biol. Chem. 281, 20902–20909. [DOI] [PubMed] [Google Scholar]
- Rangarajan E. S.; Proteau A.; Cui Q.; Logan S. M.; Potetinova Z.; Whitfield D.; Purisima E. O.; Cygler M.; Matte A.; Sulea T.; Schoenhofen I. C. (2009) Structural and functional analysis of Campylobacter jejuni PseG: A UDP-sugar hydrolase from the pseudaminic acid biosynthetic pathway. J. Biol. Chem. 284, 20989–21000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou W. K.; Dick S.; Wakarchuk W. W.; Tanner M. E. (2005) Identification and characterization of NeuB3 from Campylobacter jejuni as a pseudaminic acid synthase. J. Biol. Chem. 280, 35922–35928. [DOI] [PubMed] [Google Scholar]
- Gunawan J.; Simard D.; Gilbert M.; Lovering A. L.; Wakarchuk W. W.; Tanner M. E.; Strynadka N. (2005) Structural and mechanistic analysis of sialic acid synthase NeuB from Neisseria meningitidis in complex with Mn2+, phosphoenolpyruvate, and N-acetylmannosaminitol. J. Biol. Chem. 280, 3555–3563. [DOI] [PubMed] [Google Scholar]
- Liu F.; Lee H. J.; Strynadka N.; Tanner M. E. (2009) Inhibition of Neisseria meningitidis sialic acid synthase by a tetrahedral intermediate analogue. Biochemistry 48, 9194–9201. [DOI] [PubMed] [Google Scholar]
- McNally D. J.; Hui J. P. M.; Aubry A. J.; Mui K. K. K.; Guerry P.; Brisson J. R.; Logan S. M.; Soo E. C. (2006) Functional characterization of the flagellar glycosylation locus in Campylobacter jejuni 81-176 using a focused metabolomics approach. J. Biol. Chem. 281, 18489–18498. [DOI] [PubMed] [Google Scholar]
- Lee Y. J.; Kubota A.; Ishiwata A.; Ito Y. (2011) Synthesis of pseudaminic acid, a unique nonulopyranoside derived from pathogenic bacteria through 6-deoxy-AltdiNAc. Tetrahedron Lett. 52, 418–421. [Google Scholar]
- Carter A. T.; Paul C. J.; Mason D. R.; Twine S. M.; Alston M.; Logan S. M.; Austin J. W.; Peck M. W. (2009) Independent evolution of neurotoxin and flagellar genetic loci in proteolytic Clostridium botulinum. BMC Genomics 10, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. L.; Day-Williams M. J.; Tomas J. M.; Stafford G. P.; Shaw J. G. (2012) Identification of a putative glycosyltransferase responsible for the transfer of pseudaminic acid onto the polar flagellin of Aeromonas caviae Sch3N. MicrobiologyOpen 1, 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush N. O.; Deshpande N. P.; Wilkins M. R.; Raftery M. J.; Janitz K.; Mitchell H. (2011) Comparative analyses of Campylobacter concisus strains reveal the genome of the reference strain BAA-1457 is not representative of the species. Gut Pathog. 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandiba L.; Eichler J. (2013) Analysis of putative nonulosonic acid biosynthesis pathways in Archaea reveals a complex evolutionary history. FEMS Microbiol. Lett. 345, 110–120. [DOI] [PubMed] [Google Scholar]
- Vinogradov E.; Frimmelova M.; Toman R. (2013) Chemical structure of the carbohydrate backbone of the lipopolysaccharide from Piscirickettsia salmonis. Carbohydr. Res. 378, 108–113. [DOI] [PubMed] [Google Scholar]
- Logan S. M.; Kelly J. F.; Thibault P.; Ewing C. P.; Guerry P. (2002) Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol. Microbiol. 46, 587–597. [DOI] [PubMed] [Google Scholar]
- Josenhans C.; Ferrero R.; Labigne A.; Suerbaum S. (1999) Cloning and allelic exchange mutagenesis of two flagellin genes of Helicobacter felis. Mol. Microbiol. 33, 350–362. [DOI] [PubMed] [Google Scholar]
- Ewing C. P.; Ekaterina A.; Guerry P. (2009) Functional characterization of flagellin glycosylation in Campylobacter jejuni 81-176. J. Bacteriol. 191, 7086–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchen P.; Brzostek J.; Panico M.; Butler J. A.; Morris H. R.; Dell A.; Linton D. (2010) Modification of the Campylobacter jejuni flagellin glycan by the product of the Cj1295 homopolymeric-tract-containing gene. Microbiology 156, 1953–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampronio C. G.; Blackwell G.; Penn C. W.; Cooper H. J. (2011) Novel glycosylation sites localized in Campylobacter jejuni flagellin FlaA by liquid chromatography electron capture dissociation tandem mass spectrometry. J. Proteome Res. 10, 1238–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen C.; Koomey M.; Borud B. (2012) Hypomorphic glycosyltransferase alleles and recoding at contingency loci influence glycan microheterogeneity in the protein glycosylation system of Neisseria species. J. Bacteriol. 194, 5034–5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borud B.; Aas F. E.; Vik A.; Winther-Larsen H. C.; Egge-Jacobsen W.; Koomey M. (2010) Genetic, structural, and antigenic analyses of glycan diversity in the O-linked protein glycosylation systems of human Neisseria species. J. Bacteriol. 192, 2816–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrzynska M.; Betts J. D.; Austin J. W.; Trust T. J. (1991) Identification, characterization and spatial localization of two flagellin species in Helicobacter pylori flagella. J. Bacteriol. 173, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerbaum S.; Josenhans C.; Labigne A. (1993) Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J. Bacteriol. 175, 3278–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton K. A.; Suerbaum S.; Josenhans C.; Krakowka S. (1996) Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64, 2445–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottemann K. M.; Lowenthal A. C. (2002) Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect. Immun. 70, 1984–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura H.; Churin Y.; Bauer B.; Boettcher J. P.; Bartfield S.; Hashii N.; Kawasaki N.; Mollenkopf H. J.; Jungblut P. R.; Brinkmann V.; Meyer T. F. (2010) Helicobacter pylori HP0518 affects flagellin glycosylation to alter bacterial motility. Mol. Microbiol. 78, 1130–1144. [DOI] [PubMed] [Google Scholar]
- Champasa K.; Longwell S. A.; Eldridge A. M.; Stemmler E. A.; Dube D. H. (2013) Targeted identification of glycosylated proteins in the gastric pathogen Helicobacter pylori (Hp). Mol. Cell. Proteomics 12, 2568–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]