Abstract
Increased male prevalence has been repeatedly reported in several neurodevelopmental disorders (NDs), leading to the concept of a “female protective model.” We investigated the molecular basis of this sex-based difference in liability and demonstrated an excess of deleterious autosomal copy-number variants (CNVs) in females compared to males (odds ratio [OR] = 1.46, p = 8 × 10−10) in a cohort of 15,585 probands ascertained for NDs. In an independent autism spectrum disorder (ASD) cohort of 762 families, we found a 3-fold increase in deleterious autosomal CNVs (p = 7 × 10−4) and an excess of private deleterious single-nucleotide variants (SNVs) in female compared to male probands (OR = 1.34, p = 0.03). We also showed that the deleteriousness of autosomal SNVs was significantly higher in female probands (p = 0.0006). A similar bias was observed in parents of probands ascertained for NDs. Deleterious CNVs (>400 kb) were maternally inherited more often (up to 64%, p = 10−15) than small CNVs < 400 kb (OR = 1.45, p = 0.0003). In the ASD cohort, increased maternal transmission was also observed for deleterious CNVs and SNVs. Although ASD females showed higher mutational burden and lower cognition, the excess mutational burden remained, even after adjustment for those cognitive differences. These results strongly suggest that females have an increased etiological burden unlinked to rare deleterious variants on the X chromosome. Carefully phenotyped and genotyped cohorts will be required for identifying the symptoms, which show gender-specific liability to mutational burden.
Introduction
Gender bias has been repeatedly observed in neurodevelopmental disorders (NDs), including neuropsychiatric disorders. Epidemiologic studies in schools and institutions caring for individuals with intellectual disability (ID) have shown a 30%–50% excess of males over females.1 In autism spectrum disorder (ASD), the male-to-female ratio is 4:1. It increases to 7:1 for high-functioning autism and drops to 2:1 for individuals with moderate to severe ID.2 Several studies have attempted to gather evidence in favor of a female protective effect (or male susceptibility because this is a relative concept). In a general-population dizygotic-twin cohort, a recent study showed that the gender of the proband with autistic traits influenced the level of autistic traits in the twin sibling: higher autistic traits were measured in the sibling when the proband was a female.3 This suggests that there is a greater etiological load in female probands and their relatives. Other studies, however, did not observe such findings.4–6 It has been suggested that sample size and ascertainment methods might be responsible for these discrepancies. Levy et al.7 suggested a trend toward a higher frequency of mostly autosomal de novo copy-number variants (CNVs) in autistic females than in autistic males (11.7% in females versus 7.4% in males, p = 0.16) given that de novo CNVs encompassed more genes in female probands (15.5 in females versus 2.0 in males, p = 0.05). A similar trend was reported for CNVs8 and disrupting de novo single-nucleotide variants (SNVs) (p = 0.07).9
Studies of specific genomic disorders have also reported gender bias, such as the 2-fold increase in the frequency of males carrying a 16p11.2 deletion among individuals ascertained for NDs.10,11 The same bias was observed for individuals who carried the reciprocal duplication and who were referred for NDs, and the opposite bias was seen in nonmedically ascertained carriers (transmitting parents and carriers in the general population).11 This suggests that males are more likely to be referred for genetic testing than females carrying the same autosomal variant. Recently, studies have also reported increased maternal inheritance of deleterious CNVs, but the statistical power was limited or the observation was done in a specific context, such as the inheritance of a secondary CNV was conditioned on the presence of an initial pathogenic CNV.12–14
The factors underlying this excess in males ascertained for NDs remain unknown. X-linked variants are obvious candidates, but several studies have pointed out that “monogenic” X-linked ID is too infrequent (5%–8% of ID in males1,15) to account for the 30% excess of males with ID. The clinical manifestations underlying this excess referral are also undetermined.10
To pursue the investigation of gender differences beyond the aforementioned observations, we systematically explored the distribution of deleterious autosomal variants (CNVs and SNVs) in males and females ascertained for ND. We considered two disease cohorts in the study (Table S1, available online). The first cohort consists of 9,206 male and 6,379 female ND-affected individuals referred to diagnostic labs for CNV testing by array comparative genomic hybridization (CGH); the second is the Simons Simplex Collection (SSC), which was ascertained on the basis of simplex cases of ASD. In the latter, data on two partially overlapping subsets were available: 226 male and 96 female probands (and their relatives) whose exomes had been sequenced and 762 families with available CNV data. Examining the parent-of-origin and sex differences, we observed a systematic excess of deleterious variants in females ascertained for NDs. We also found that mothers not medically ascertained also showed an increased mutational burden in comparison to fathers of probands ascertained for NDs. This supports the “female protective model,” suggesting that the clinical manifestations of NDs require a higher “mutational burden” for females.
Material and Methods
Data sets associated with cases in this study have been previously published.16–18 Raw data for control CNVs were obtained from SNP microarray data in dbGaP. All data were collected and analyzed in accordance with the ethical standards of the local institutional review boards.
Disease Cohorts and CNV Analyses
CNV data from individuals with NDs and ASD were obtained from Signature Genomic Laboratories16 and the SSC. The Signature Genomics data set consists of 15,767 DNA samples from individuals referred to Signature Genomics by multiple clinical genetic centers across the United States and Canada for diagnostic purposes. Information on gender is available for 15,585 cases (9,206 males and 6,379 females). Samples were analyzed across nine custom array CGH platforms, and most were tested on an Agilent array with 97,000 probes. CNV calls were detected and previously deposited into dbVar (accession number nstd54). The reason for referral in this data set was ID in the vast majority of individuals; autism was noted in 1,379 cases, and epilepsy was noted in 1,776 cases. A constellation of congenital malformations, including congenital heart disease (n = 575), was reported. Twelve percent of the cases were not annotated. Details on motives for referral were previously described.16,19
CNVs were detected and validated as previously published.16,20,21 A whole-genome bacterial artificial chromosome (BAC) microarray chip (SignatureChipWG) and an oligo-based chip (SignatureChipOS) (either 105K custom designed by Signature Genomics and manufactured by Agilent Technologies or 135K custom designed by Signature Genomics and manufactured by Roche NimbleGen) were used for CNV detection. Microarray hybridizations were performed as described previously.21–23 CNVs from the Signature Genomics collection were then rigorously assessed for eliminating potential size-estimation errors associated with low probe densities, intensity noise resulting from high-copy duplications, rearrangements associated with immune genes, reference-sample CNVs, and other potential artifacts. We filtered CNVs according to the following criteria: CNV region count < 158 (1% of individuals), CNVs < 1% population frequency, and <10% of the CNV overlapping with an “artifact” list. The list of artifact-prone loci was defined by regions with immune system genes prone to rearrangement, known reference-sample CNVs, very large blocks of highly similar segmental duplications, and artifacts identified as part of a batch effect. The artifact list included the following: chr2: 88,937,989–89,411,302; chr2: 89,589,457–89,897,555; chr2: 196,517,337–196,847,645; chr3: 30,618,438–30,728,248; chr7: 105,609,512–105,811,026; chr14: 21,159,851–22,090,936; chr14: 105,065,301–106,352,275; chr15: 0–20,060,121; chr15: 91,157,836–91,364,629; chr16: 87,299,650–87,418,927; chr22: 20,602,619–20,926,359; and chr22: 20,715,572–21,595,082.
The SSC is a cohort of simplex families with one proband ascertained for moderate to severe autistic symptoms and with a mean full-scale IQ ≈ 80.24 Full details on inclusion and exclusion criteria are available at the Simons Foundation Autism Research Initiative (SFARI) website. CNVs in the SSC were previously studied by Sanders et al.8 We reanalyzed CNVs by using the algorithm described by Itsara et al.22,25 to include CNVs that would be excluded as a result of the stringent size filtering in the Sanders study, and we filtered CNVs by their frequency in both unrelated parents (<1%, corresponding to an occurrence in fewer than 15 of 1,524 unrelated parents) and an independent cohort of controls (<1%, corresponding to an occurrence in fewer than 25 of 2,515 controls) also profiled on Illumina SNP arrays with similar density.16 We filtered CNV calls to exclude those not detectable on all three Illumina 1M platforms. The final data set included all family members of 653 male and 109 female probands.
Parental Origin of CNVs
The International Standards for Cytogenomic Arrays (ISCA) Consortium includes genomic copy-number data from 15,749 individuals referred for clinical chromosomal-microarray testing in the context of development delay (DD), ID, ASD, or multiple congenital anomalies.26 Information on parental transmission was available for a subset of 1,735 CNVs. In the Signature Genomics data set described above, information on parental transmission was available for 1,826 CNVs. In the SSC (n = 762 probands), information on parental transmission was available for all CNVs (n = 11,078).
Control CNV Data
Control specimens included samples from the following previously described adult controls profiled on Illumina and Affymetrix SNP arrays.16,19
Cohort 1
From the Wellcome Trust Case Control Consortium 2 National Blood Services Cohort (WTCCC2 NBS), 1,213 females and 1,302 males of European descent from the UK Blood Service control group (age range of blood donors = 18–69 years) were genotyped on a custom Illumina 1.2M SNP array.20 CNVs were called as described previously.22 In brief, a hidden Markov model based on both allele frequencies and total intensity values (logR) was used for identifying putative alterations (overall precision of 0.892 in identifying large CNVs > 100 kb16). Subsequently, manual inspection of large CNVs (>100 probes and >1 Mb) was performed in conjunction with user-guided merging of nearby calls (<1 Mb between CNVs for arrays with fewer than one million probes and <200 kb between CNVs for arrays with more than one million probes).
Cohort 2
Affymetrix SNP Array 6.0 profiles were obtained from the Atherosclerosis Risk in Communities (ARIC) study control set (dbGaP accession phs000090.v1.p1) and processed with Affymetrix Genotyping Console 4.1 with hg18 (UCSC Genome Browser) chromosome annotations. Samples were filtered with the default contrast quality-control parameters, and segmentation was also performed with default settings. Samples that demonstrated significantly lower-than-expected log ratios in conjunction with high CNV counts, as well as cases with excessive CNV counts (>72 CNVs per case), were removed. After quality-control filtering, the final control set consisted of 4,806 females and 3,927 males.
SNV Data
We used all available raw exome sequencing data from the SCC. This was essential for reprocessing and recalling data sets with the same methods as for limiting technical artifacts. Data originated from two autism exome sequencing studies.17,18 Reads were mapped to a custom GRCh37/hg19 build of the human reference genome with the Burrows-Wheeler Aligner v.0.5.6.25 Read qualities were recalibrated with the Genome Analysis Toolkit (GATK) Table Recalibration 1.0.2905, and Picard-tools 1.14 was used for flagging duplicate reads. Genotypes were generated with the GATK Unified Genotyper27 with FILTER = “HRun >4 || SB >=0.10 || QUAL ≤ 50.0 || QD < 5.0” and the default SnpCluster and low-quality filters. Multisample calling was performed on two sets. The first set consisted of 188 mother-father-proband-sibling quads and 20 mother-father-proband trios18 and the 95 individuals from the Environmental Genome Project Panel 2 (National Institute of Environmental Health Sciences). The second consisted of 31 autism quads and 168 trios.17 Probands and siblings with fewer than 20 million reads and parents with fewer than ten million reads in their exomes were removed. Samples were tested for validity on the basis of the program PRIMUS,28 which tests the identity-by-descent (IBD) region for relatedness and allows for determination of sample membership. If PRIMUS determined that one individual was unrelated, the entire family was removed from further analysis, except when the discordant IBD analysis concerned a sibling. In the latter case, the remaining proband and parents were kept as a trio. Filtering included the removal of the following from analyses: (1) Y chromosomes, (2) sites falling in tandem repeats or segmental duplications, (3) dbSNP132 variants > 1% frequency, (4) sites covered by fewer than ten reads and alleles covered by fewer than six reads, (5) sites where the child was homozygous for the reference allele, (6) trios with children with >25 Mendelian-inheritance errors (successive errors suggesting the presence of an indel were not included in this count), (7) known pseudogenes, and (8) all 14 families of non-European descent. In total, 324 families, including 226 male and 98 female probands, remained. We selected variants present only in a single family (“private variants”) and annotated them with ANNOVAR (last updated February 21, 2013) and RefSeq gene annotation (GRCh37, accessed December 9, 2013).
Variants were annotated with Combined Annotation Dependent Depletion (CADD), a method that integrates functional annotations, conservation, and gene-model information into a single metric. For variant inclusion, we required the scaled C-scores obtained from CADD to be greater or equal to 20. These scores are on a PHRED-like scale; a score of 20 indicates that the variant is as damaging as 1% of the single-nucleotide substitutions that can be generated from the human reference genome.29
Gene Lists
In order to develop a list of ND-associated genes (ND genes),13 we searched for all genes that were strongly associated with NDs. We conducted searches in the OMIM database with the following terms: “mental retardation” “intellectual disabilities,” “autism,” “schizophrenia,” “psychosis,” and “epilepsy.” We also included SFARI autism candidate genes with association scores ranging from 1 to 4 (n = 155 genes). In addition, all ND candidate genes in known genomic disorders were included.16 A total list of 1,560 genes was established on the basis of those search terms and criteria. No manual curation was performed. This list therefore included genes that might have been falsely associated in the literature with a particular ND. The sensitivity and specificity of the list reflected the current state of the literature and OMIM. The list was not specific to any particular ND, but it was highly enriched with genes involved in NDs. The brain-expressed genes (BE genes) were previously described;13 in brief, we defined a gene to be brain expressed if its expression ranked in the top ≈5% of all genes. Brain expression was defined as the mean expression across 18 brain regions. Gene-expression data were from the Human U133A/GNF1H Gene Atlas (Gene Expression Omnibus accession number GSE1133), comprising 79 human tissues, including 18 nervous system tissues.30 Expression values were averaged across multiple probes when available.
Statistical Analyses
For the regression analyses, each proband was assigned a mutational burden consisting of the sum of the lengths of CNVs containing ND genes. The association between IQ and the mutational burden was tested by a linear regression, including gender as a covariate. The association between gender and the mutational burden was tested by a logistic regression, including IQ as a covariate. All analyses were performed with standard packages written in R.
Results
Excess of Deleterious Autosomal CNVs in Females Ascertained for Nonspecified NDs
We investigated previously published CNV calls from a group of 9,206 males and 6,379 females referred by physicians for diagnostic purposes.16 Overall, 73% of the cases presented with DD, ID, and/or ASD. Individuals could also show one or several congenital malformations. All individuals were referred for diagnostic arrays. The 44% excess of males in this sample is similar to what has been reported in ID and DD.1
We hypothesized that the “female protective model” is associated with an increase in deleterious CNVs in females ascertained for NDs. Large (>400 kb), rare (prevalence < 1%), and de novo CNVs are criteria significantly associated with ND16 and were therefore used in this study. To further predict deleteriousness, we established a list of ND genes (n = 1,560; see Material and Methods).13 We compared the CNV burden between genders by using the aforementioned variables to stratify the data set (Figure 1). Small (<400 kb) and rare (<1%) CNVs were equally distributed across gender, but those larger than 400 kb or 1 Mb were significantly enriched in females (odds ratio [OR] = 1.18, p = 1 × 10−6 and OR = 1.28, p = 9 × 10−9, respectively). Only taking into account large and very rare CNVs (<1/1,000) that include ND genes further increased this bias (OR = 1.28, p = 3 × 10−6 for CNVs > 400 kb and OR = 1.46, p = 8 × 10−10 for CNVs > 1 Mb). De novo CNVs > 400 kb and > 1 Mb showed a similar high and significant excess in females (OR = 1.39 and 1.46, respectively) (Figure 1). CNV size and frequency and the probability of haploinsufficiency of an ND gene are not independent criteria. However, a logistic regression including all three variables showed that they each explained a significant proportion of the excess mutational burden in females (CNV size, p = 2 × 10−5; presence of ND genes, p = 0.016; CNV frequency, p = 0.05).
Figure 1.
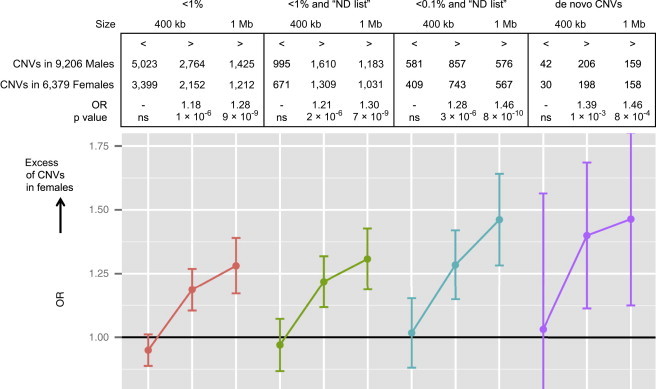
Excess of Autosomal CNVs in Females Ascertained for NDs
Odds ratios (ORs) and associated p values represent the enrichment of CNVs in females compared to males ascertained for NDs. The CNVs are stratified on the basis of criteria previously associated with deleteriousness:16 frequency (<1% and <1/1,000), size (400 kb and 1 Mb as cutoffs), and de novo variants. An additional and previously published filter13 was applied on the basis of the presence of an ND gene (see Material and Methods). ORs and p values were calculated with a two-tailed Fisher’s exact test (ns, not significant). Data on gender were available for 476 de novo CNVs.
Excess Burden of Deleterious Autosomal CNVs in Females Ascertained for ASD
We confirmed this increased burden in a subset of 653 male and 109 female probands from an independent cohort of individuals ascertained for ASD. The SSC is associated with a remarkable excess of males (six males per one female) previously described in high-functioning autism.24 Females showed a 2-fold increase in large CNVs (>400 kb) in comparison to males (OR = 2, p = 0.003), and after exclusion of CNVs without ND genes, the excess further increased to 3-fold (OR = 3, p = 7 × 10−4) (Table 1).
Table 1.
Excess of Autosomal CNVs in SSC Females Ascertained for ASD
|
<400 kb |
>400 kb |
|||
|---|---|---|---|---|
| All | ND Genes | All | ND Genes | |
| CNVs in Males | 4,482a | 326 (47%b) | 108 (16%) | 36 (5%) |
| CNVs in Females | 788a | 53 (48%) | 32 (29%) | 17 (15%) |
| OR (p value)c | NAa | 1 (ns) | 2 (3 × 10−3) | 3 (7 × 10−4) |
CNVs in 653 male and 109 female probands from the SSC.
Fisher’s exact test did not apply because all individuals carried more than one small CNV, but the binomial test showed that the proportion of small CNVs was similar in males and females (p = 0.14).
Frequency of individuals carrying at least one variant (326 variants were present in 308 male probands).
Two-sided Fisher’s exact test. Only rare CNVs present in <1% (<25/2,515) of the general population and <1% (<15/1,520) of the unrelated parents of this subgroup of the SSC were included in the analysis.
Excess Burden of Deleterious Autosomal SNVs in Females Ascertained for ASD
The distribution of SNVs and indels was studied in a subset of 226 males and 98 females from the SSC. Only rare variants were taken into account (Table 2). Variant annotation was performed via the CADD method, which integrates many annotations into a single metric. The resulting C-scores (scaled) were considered most likely deleterious if greater than 20 (see Material and Methods).29 Rare truncating SNVs were in slight excess in females (OR = 1.1, one-sided p = 0.03), and this enrichment was more apparent when only variants truncating ND or BE genes (defined as genes with expression levels ranking in the top 5% of all genes in the brain, see Material and Methods and Krumm et al.13) were considered (OR = 1.34, one-sided p = 0.04). The same trend was observed for rare truncating variants only present in probands. However, in siblings, the distribution of truncating SNVs was balanced across gender (Table 2). There was no excess of missense mutations in female probands, even after filtering for deleterious C-scores intersecting ND or BE genes (Table 2).
Table 2.
Autosomal SNVs in Females Compared to Males Ascertained for ASD
|
Truncating Variants |
Missense Variants C-Score > 20 |
Truncating Variants in ND or BE Genes |
Missense Variants C-Score > 20 in ND or BE Genes |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | ORa | pa | n | ORa | pa | n | ORa | pa | n | ORa | pa | |
| Probands | ||||||||||||
| Male | 1,220 | 1.1 | 0.03 | 17,625 | 1 | ns | 104 | 1.34 | 0.035 | 2,661 | 1 | ns |
| Female | 576 | 1.1 | 0.03 | 7,456 | 1 | ns | 59 | 1.34 | 0.035 | 1,133 | 1 | ns |
| Unaffected Siblings | ||||||||||||
| Male | 387 | 0.86 | ns | 5,110 | 0.98 | ns | 40 | 0.86 | ns | 775 | 0.98 | ns |
| Female | 411 | 0.86 | ns | 5,890 | 0.98 | ns | 40 | 0.86 | ns | 879 | 0.98 | ns |
SNVs were called in 226 male and 98 female European-descent probands ascertained for ASD (SSC) and 70 male and 82 female siblings. There was an increase in deleterious variants in female probands ascertained for ASD. The following abbreviations are used: ND genes, genes associated with neurodevelopmental disorders (see Material and Methods); and BE genes, genes with an expression ranked in the top 5% of all genes expressed in the brain (see Material and Methods).
A one-sided Fisher’s exact test counted the number of nonsynonymous variants with and without the specific mutation type and filters detailed in the header of the table (e.g., truncating variants versus nonsynonymous variants, which are not truncating).
We further explored the involvement of autosomal SNVs and indels in gender bias by comparing their deleteriousness across gender (Figures 2A and 2B). We hypothesized that deleterious variants are only present at the tail of the C-score distribution and performed our analyses in the top 1%, 5%, 10%, 20%, 30%, and 50% of raw C-scores. Females showed significantly higher C-scores than did males in the top percentiles of the distribution, which was driven by variants truncating ND genes (p = 0.0006 from Wilcoxon rank-sum test for the top 1% of C-scores corresponding to deleterious variants in ND genes). Missense mutations showed no increase in the deleteriousness of C-scores in females (Figure 2 and Table S2B).
Figure 2.
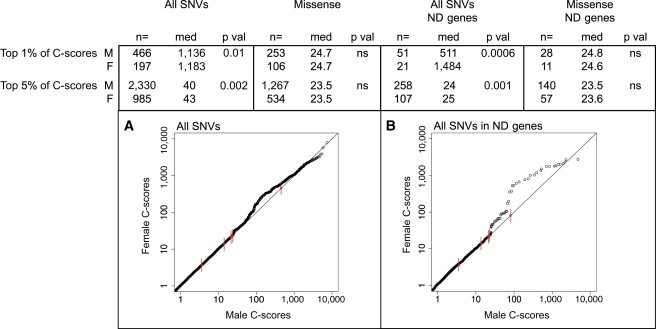
The Deleteriousness of Autosomal SNVs in Males and Females Ascertained for ASD
The deleteriousness of SNVs was significantly higher in females than in males ascertained for ASD. Truncating variants (gain or loss of stop mutations and frameshift mutations) mainly drove this increased burden in females, and the gender bias was most apparent for variants involving ND genes. Missense variants showed no or only a marginal excess of deleteriousness in female probands. Raw C-scores of nonsynonymous variants were compared between males and females. To perform the analysis on the most deleterious variants, we stratified the sample on the basis of the top 1% and 5% of the C-score distribution. The p values were computed by means of a Wilcoxon rank-sum test. Significant p values demonstrate higher C-scores in females than in males. Significant p values with similar medians indicate that the differences lay at the tail of the distribution, as demonstrated by the quantile-quantile (Q-Q) plots. Abbreviations are as follows: M, male; F, female; med, median; p val, p value; and ns, not significant. False-discovery-rate correction was not applied because only one hypothesis was tested in several nested subsets of the same sample.
(A) Q-Q plot comparing the distribution of C-scores for all SNVs in males and females. Red dots with tick marks indicate the top 1%, 5%, 10%, 25%, and 50% for males. The excess of high C-scores (deleterious) in females was only visible in the top 1% and 5% of the C-score distribution. Low C-scores showed equal distribution across gender.
(B) Q-Q plot comparing the distribution of C-scores for all SNVs intersecting ND genes in males and females. Red dots with tick marks indicate the same percentiles as in (A).
Phenotypic Differences Underlying the Gender Bias
This excess of mutations in females might be related to behavioral and/or cognitive phenotypes, which have a gender-specific liability to mutational burden. In the SSC cohort, performance IQ (PIQ) and verbal IQ (VIQ) were lower in females by 8 (p = 3 × 10−7) and 5 (p = 0.02) points, respectively (Table S4). CNV burden (defined as the sum of the lengths of CNVs affecting ND genes) was significantly associated with PIQ (p = 0.001) and, to a lesser extent, with VIQ (p = 0.02), consistent with previous observations.12 Cognitive ability might therefore be an important marker of the mutational burden in ASD females. However, the logistic regression (sex ∼ IQ + CNV burden) showed that CNV burden remained significantly associated with gender (increased in females), even after correction for PIQ or VIQ (p = 0.009 and p = 0.005, respectively) (Table S5). This suggests that traits other than global cognition are associated with increased etiological burden in females. The latter is also supported by the fact that although we observed an increase in truncating SNVs in females, SNV burden was not associated with PIQ or VIQ (Table S6). In the latter analyses, SNV burden was defined as the ratio of truncating SNVs involving ND genes over all nonsynonymous SNVs. Of note, social-responsiveness scores showed no association with CNV or SNV burden. It is unknown whether these results are specific to ASDs or whether they could apply to other NDs given that cognitive and behavior data were not available for the Signature Genomics cohort.
Maternal Transmission of Autosomal Variants Involved in NDs
We investigated whether the “female protective model” is associated with a higher rate of deleterious variants in females across different ascertainment methods. Data on inheritance of autosomal CNVs were available for two groups of individuals ascertained for NDs: 1,826 CNVs from the ISCA Consortium and 1,735 CNVs from Signature Genomics. These CNVs were initially selected by cytogeneticists because they were suspected to be important causal factors in the proband’s neurodevelopmental disorder. Subsequently, parental testing was performed on those 3,561 CNVs highly enriched with deleterious variants. Both diagnostic cohorts showed a significant excess of maternally inherited CNVs (p = 1 × 10−8 and p = 3 × 10−7 for ISCA and Signature Genomics, respectively). Overall, there was a 57% rate of maternal inheritance for these autosomal CNVs (p = 2 × 10−14). This excess was mostly driven by deleterious CNVs larger than 400 kb (Figure 3A).
Figure 3.
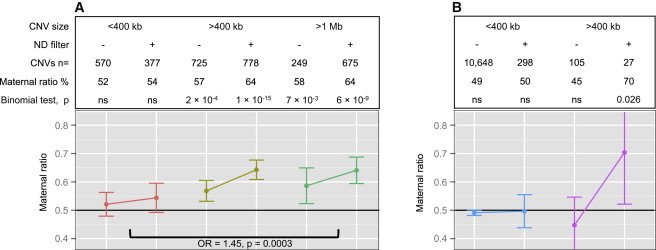
Excess of Maternally Inherited Deleterious Autosomal CNVs
(A) Data on inheritance (maternal, paternal, or de novo) were available for 1,826 and 1,735 CNVs from Signature Genomics and the ISCA, respectively. These CNVs were selected by cytogeneticists, and inheritance was tested on the basis of the likelihood of their association with the proband’s neurodevelopmental phenotype. Maternal ratio in percentage, associated 95% confidence interval, and p values represent the enrichment of maternally versus paternally inherited CNVs. The p values were computed with a binomial test, and the null hypothesis was a balanced 50/50 inheritance. The CNVs were stratified on the basis of size (400 kb and 1 Mb as cutoffs). An additional and previously published filter13 was applied on the basis of the presence of an ND gene (see Material and Methods). Compared to small CNVs, large CNVs showed increased maternal inheritance.
(B) Data on inheritance were available for all CNVs identified in 762 SSC probands ascertained for ASD. The ratio of maternally inherited CNVs is represented with the 95% confidence interval and associated p value. The CNVs were stratified on the basis of size and the disruption of an ND gene. Large CNVs disrupting ND genes were preferentially maternally inherited.
ns, not significant.
Parents are commonly investigated for the presence of inherited deleterious variants identified in their children (probands). Parental testing can, however, be performed sequentially (mother first and then father or vice versa). To account for this possible bias resulting in overestimation or underestimation of maternal transmission, we compared inheritance between different CNV sizes. Maternal inheritance was significantly higher for deleterious CNVs (>400 kb or >1 Mb) than for CNVs < 400 kb (OR = 1.36, p = 2 × 10−4 and OR = 1.45, p = 3 × 10−4, respectively). The same analysis performed on CNVs for which both parents were tested showed the same increase for large CNVs > 1 Mb compared to CNVs < 400 kb (OR = 1.43, p = 0.03).
In the SSC, data on transmission are available for all CNVs regardless of their pathogenicity. In this cohort, specifically ascertained for simplex cases, only a few large CNVs (>400 kb) containing ND genes were transmitted (n = 27). Nevertheless, we observed 70% maternal transmission for CNVs > 400 kb (one-sided p = 0.026) (Figure 3B), confirming the excess of maternal inheritance for deleterious CNVs in the two previous cohorts. Small CNVs (<400 kb, n = 10,648) and large CNVs (>400 kb, n = 105) not containing ND genes showed balanced inheritance (Figure 3B).
SNV inheritance was obtained in a subset of 324 probands and 152 unaffected siblings. Nonsynonymous SNVs with a high C-score (>20) and truncating SNVs overall showed a balanced inheritance (50% and 51% of maternal inheritance, respectively). Those same variants intersecting genes important for neurodevelopment (ND genes) or brain function (BE genes) showed an excess of maternal inheritance (59%, p = 0.017). Proband-specific SNVs (absent in siblings) yielded the same results with a lower significance as a result of a smaller sample size (59% maternal inheritance, p = 0.03) (Table S3A). In the same sample, parental origin was balanced for SNVs identified in siblings regardless of the filtering criteria (Table S3A). Using C-scores (CADD variant-annotation method29), we showed that SNVs inherited from the mother were significantly more deleterious than those inherited from the father. This effect was mostly driven by variants truncating ND genes, and missense mutations only showed a marginal increase in deleteriousness (Table S3B).
Small Contribution of the X Chromosome
The X chromosome was not taken into account in this analysis because of gender-specific deleterious effects of X-linked variants. Comparing the frequency of X-linked variants in both sexes is therefore not straightforward. Presumably, some small but significant proportion of the males with NDs harbor X-linked deleterious variants, adding to the genetic burden in males given that the phenotypic consequences would, on average, be less severe in females carrying the same variant. To estimate how the X-linked CNVs might affect the results of this study, we performed the same analyses presented in Figure 1 but added the additional X-linked CNVs to the previously calculated autosomal burden in males only. In this very conservative approach, we considered the X-linked mutational burden to be null in females, which is incorrect because females have an approximately 2-fold increase in X-linked variants (Table S7), and some of those have phenotypic consequences, including significant ID. In these analyses, we excluded aneuploidies (XXX and XXY) because they are equally distributed across genders (Table S7) and are associated with approximately the same neurodevelopmental effect in both sexes. For large CNVs (>400 kb and >1 Mb), the initial ORs of 1.18 and 1.28, respectively (Figure 1), were recalculated at OR = 1.1 (p = 0.003) and OR = 1.24 (p = 4.5 × 10−7), respectively. For very rare (<0.1%) large CNVs encompassing ND genes, the initial ORs of 1.28 and 1.46 (for >400 kb and >1 Mb CNVs, respectively) were recalculated at OR = 1.2 (p = 0.001) and OR = 1.39 (p = 6 × 10−8), respectively (Table S7). This suggests that rare deleterious variants on the X chromosome account for only a small proportion of the bias observed on the autosomes. A similar reanalysis was performed on X-linked SNV data in the SSC. We identified two rare SNVs truncating ND and BE genes (L1CAM and MAOB) in the female group and one splice-site mutation (in FMR1) in the male group. The initial gender bias in Table 2 therefore remains unchanged (same OR and p values).
Discussion
We investigated molecular characteristics associated with the increased male-to-female ratio in individuals referred for NDs. These results make a strong case for an “increased etiological burden” in females with NDs. Our findings show that females systematically carry more neurodevelopmentally deleterious variants than do males. This is true whether individuals (1) are ascertained for NDs or (2) are parents of a proband referred for those symptoms. These findings are robust and were replicated in several CNV data sets. Remarkably, SNV data also showed an excess burden despite smaller sample sizes and a greater difficulty in distinguishing neutral from deleterious SNVs. Combined, these data bring convincing evidence supporting the “female protective model” in NDs.
An increased prevalence in males has been observed across different NDs (ASD, ID, attention deficit hyperactivity disorder,31 etc.), and multiple comorbidities are common in these individuals.32 It is therefore challenging to identify those symptoms, which show a gender-specific liability to mutation and might subsequently be driving more males into the clinic. We explored clinical traits potentially associated with this increase in CNV and SNV burden in females ascertained for ASD in the SSC. As previously observed,24,33 ascertainment for ASD is associated with lower IQs in females. This difference is more pronounced for PIQ than for VIQ, which also confirms previous observations24,33 (Table S5). Regression analyses showed that PIQ (and to a lesser extent, VIQ) is associated with CNV burden (and SNV burden in males only) (Tables S4 and S5). PIQ could thus be considered a clinical marker of this increased mutational burden in females. However, the increased CNV and SNV burden in females with ND remains after correction for IQ, suggesting that other phenotypes are associated with this excess burden (Tables S4 and S5). An interpretation of this combined increased mutational burden and lower PIQ in females is that lower cognitive abilities are necessary to push females over the ASD diagnostic or referral threshold. The ascertainment of females with lower IQs results in this excess of mutational burden because IQ is associated with deleterious CNVs.12 It is unknown whether these observations for IQ can be generalized across the different cohorts of NDs given that such clinical data are not available for the Signature Genomics cohort.
We are not implying that this gender bias in mutational burden can account completely for the overall dramatic excess of males in high-functioning ASD. Instead, this study suggests that the male brain requires milder alterations to exhibit ASD. The latter might be the basis for what has been described as the “extreme male brain hypothesis,” in which ASD is an extreme expression of the psychological and physiological attributes of the male brain.34 In this hypothesis, female brains would require larger mutational burden to reach the ASD diagnostic threshold.
Phenotype underlying gender bias has recently been explored in a group of 16p11.2 deletion carriers ascertained for NDs; in this group, male carriers significantly outnumbered their female counterparts by 2-fold (112 males and 56 females).10 Although carriers were fully assessed, the study failed to show that female carriers were differentially affected cognitively and/or behaviorally.10 Our analyses of a series of behavioral phenotypes from the SSC did not reveal additional traits with a significant difference across gender (Table S8).
In addition to investigating biological theories, including differences in genetic liability for NDs,35,36 researchers have investigated the “social bias” hypothesis related to gender stereotypes in the diagnosis of ND or ASD,33 e.g., for equal severity of autistic traits, boys were more likely to receive an ASD diagnosis than girls in the ALSPAC cohort.37 Although our study did not investigate this hypothesis, the excess of maternally inherited CNVs and SNVs speaks against it. This inheritance bias is again in favor of sex-differential liability to mutation, resulting in lower adaptive skills in males and thus leading to lower parenting and household-management skills. The excess rate of maternal inheritance was reproducible across the different cohorts (ISCA and Signature Genomics) and reasons for ascertainment (ND and autism), and the increase in maternal inheritance between large and small CNVs (OR ≈ 1.4) was similar to the gender bias observed in probands ascertained for NDs. NDs and parenting skills might thus represent two opposite ascertainment criteria (which enrich for and against ND symptoms, respectively), resulting in an equally increased burden in females. This also highlights the important contribution of inherited autosomal variants in ND, even in the case of the SSC, which has actively ascertained against multiplex families.
Throughout this study, we applied a “candidate-gene filter” (ND genes) based on the hypothesis that mutations disrupting neurodevelopmental processes are underlying this gender bias. This list was designed to exclude a majority of genes unlikely to be involved in neurodevelopment, but its sensitivity and specificity were far from 100%. The results obtained with this filter repeatedly showed that it was a relevant tool for enriching for deleterious variants,13 but in most cases, signal was obtained without application of this candidate list.
Variants truncating ND genes show a consistent pattern of gender bias. Missense variants are weakly associated with this phenomenon and show marginal differences in deleteriousness across gender (Table S2). This difference between truncating and missense mutations might be due to a specific relationship between the sex-differential liability and the categories of mutation, but it might also simply reflect the difficulty in discriminating deleterious from benign missense mutations. This general issue of discriminating deleterious from benign variants also applies to small CNVs (<400 kb) that did not show any gender bias in this analysis. Half of the CNVs in this group were <126 kb, and the vast majority of these were most likely benign. We further investigated the upper range (300 kb < CNV < 400 kb) in this subgroup and found no evidence of gender-related bias (OR = 1), despite the use of additional filters (list of ND genes) to enrich for pathogenic alleles. Unfortunately, the resolution of SNP and clinical microarrays is currently insufficient to map the breakpoints with this level of precision. The reanalysis of a recently published data set of CNVs inferred from exome sequencing13 showed that private (found in one family only) small CNVs truncating ND genes are indeed associated with gender bias (67% of maternal inheritance, p = 0.007).
X-linked variants have been obvious candidates for explaining the gender bias observed in NDs. We note that CNVs and SNVs on the X chromosome were relatively rare in our sample, which is consistent with previous observations,38,39 and our analysis demonstrated that rare deleterious X-linked variants do not account for the increase in autosomal mutational burden in females. We were not able to explore common X-linked variants that might interact with deleterious autosomal mutations.
An overrepresentation of females who carry a specific deleterious CNV has been previously observed in general population cohorts (e.g., 16p11.2 duplication11). We investigated whether this same observation could be replicated and generalized in an aggregate analysis of all deleterious autosomal CNVs in general population cohorts. In 1,213 females and 1,302 males from the WTCCC2 NBS, large CNVs (>400 kb) encompassing ND genes and deletions from the latter group were overrepresented in females (p = 0.007 and p = 0.03, respectively) (Figure S1). As expected, this observation relied on a small number of large deleterious CNVs (n = 24) (Figure S1). CNVs < 400 kb were equally distributed across gender. In an independent general-population cohort (ARIC) of 4,806 females and 3,927 males, this finding was not replicated, suggesting that larger cohorts of unaffected individuals will be required for determining whether increased prevalence of female carriers of large CNVs is a general property of the human population. In fact, a general-population cohort of 150,000 individuals would be required for studying a sample of moderately deleterious variants (e.g., enrichment of 10-fold in individuals with NDs) similar in size to what is available in the Signature Genomics cohort.
This study is a strong case in favor of an increased mutational burden in females ascertained across different NDs. This effect was observed for CNVs and SNVs disrupting genes involved in neurodevelopment. In ASD populations, PIQ is a good clinical marker of this increased burden, but additional clinical symptoms are also implicated in this phenomenon. Inheritance analyses also demonstrated an overrepresentation of ND susceptibility alleles in mothers as compared to fathers, suggesting that mating, parenting, and/or household-management skills show a gender-specific mutation liability similar to what is observed for symptoms driving the gender bias in NDs.
Acknowledgments
This work was supported by the Fond de Relève Académique, Université de Lausanne (S.J.), Swiss National Science Foundation (PP00P3_144902), Simons Foundation Autism Research Initiative (137578 and 191889 to E.E.E.), and National Institutes of Health MH101221 (E.E.E.). E.E.E. is an investigator of the Howard Hughes Medical Institute, is on the scientific advisory board (SAB) of SynapDx Corp., and was an SAB member of Pacific Biosciences Inc. (2009–2013) and DNAnexus Inc. (2011–2013). J.R. is an employee of Signature Genomic Laboratories, a subsidiary of PerkinElmer Inc. S.J. has acted as a consultant for Novartis Pharma AG and has received grants for the clinical investigation of mavoglurant. We thank all families at the participating Simons Simplex Collection (SSC) sites and the principal investigators (A. Beaudet, R.B., J. Constantino, E. Cook, E. Fombonne, D. Geschwind, E. Hanson, D. Grice, A. Klin, R. Kochel, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, K. Pelphrey, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C. Walsh, Z. Warren, and E. Wijsman). We acknowledge M. State and the SSC Genetics Consortium for providing Illumina genotyping and T. Lehner and the Autism Sequencing Consortium for data exchange among the participating groups. This study used data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to data generation is available at http://www.wtccc.org.uk/. We are grateful for manuscript preparation from T. Brown and helpful discussion from M. Kircher and Eichler lab members.
Contributor Information
Sébastien Jacquemont, Email: sebastien.jacquemont@chuv.ch.
Evan E. Eichler, Email: eee@gs.washington.edu.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Combined Annotation Dependent Depletion (CADD), http://cadd.gs.washington.edu
National Institute of Environmental Health Sciences (NIEHS) Environmental Genome Project (EGP), http://evs.gs.washington.edu/niehsExome/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
SFARI (Simons Foundation Autism Research Initiative), www.sfari.org
References
- 1.Stevenson R.E., Schwartz C.E., Schroer R.J. Oxford University Press; New York: 2000. X-linked mental retardation. [Google Scholar]
- 2.Fombonne E. Epidemiology of pervasive developmental disorders. Pediatr. Res. 2009;65:591–598. doi: 10.1203/PDR.0b013e31819e7203. [DOI] [PubMed] [Google Scholar]
- 3.Robinson E.B., Lichtenstein P., Anckarsäter H., Happé F., Ronald A. Examining and interpreting the female protective effect against autistic behavior. Proc. Natl. Acad. Sci. USA. 2013;110:5258–5262. doi: 10.1073/pnas.1211070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goin-Kochel R.P., Abbacchi A., Constantino J.N., Autism Genetic Resource Exchange Consortium Lack of evidence for increased genetic loading for autism among families of affected females: a replication from family history data in two large samples. Autism. 2007;11:279–286. doi: 10.1177/1362361307076857. [DOI] [PubMed] [Google Scholar]
- 5.Ozonoff S., Young G.S., Carter A., Messinger D., Yirmiya N., Zwaigenbaum L., Bryson S., Carver L.J., Constantino J.N., Dobkins K. Recurrence risk for autism spectrum disorders: a Baby Siblings Research Consortium study. Pediatrics. 2011;128:e488–e495. doi: 10.1542/peds.2010-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zwaigenbaum L., Bryson S.E., Szatmari P., Brian J., Smith I.M., Roberts W., Vaillancourt T., Roncadin C. Sex differences in children with autism spectrum disorder identified within a high-risk infant cohort. J. Autism Dev. Disord. 2012;42:2585–2596. doi: 10.1007/s10803-012-1515-y. [DOI] [PubMed] [Google Scholar]
- 7.Levy D., Ronemus M., Yamrom B., Lee Y.-H., Leotta A., Kendall J., Marks S., Lakshmi B., Pai D., Ye K. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Sanders S.J., Ercan-Sencicek A.G., Hus V., Luo R., Murtha M.T., Moreno-De-Luca D., Chu S.H., Moreau M.P., Gupta A.R., Thomson S.A. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iossifov I., Ronemus M., Levy D., Wang Z., Hakker I., Rosenbaum J., Yamrom B., Lee Y.-H., Narzisi G., Leotta A. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zufferey F., Sherr E.H., Beckmann N.D., Hanson E., Maillard A.M., Hippolyte L., Macé A., Ferrari C., Kutalik Z., Andrieux J., Simons VIP Consortium. 16p11.2 European Consortium A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J. Med. Genet. 2012;49:660–668. doi: 10.1136/jmedgenet-2012-101203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacquemont S., Reymond A., Zufferey F., Harewood L., Walters R.G., Kutalik Z., Martinet D., Shen Y., Valsesia A., Beckmann N.D. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girirajan S., Rosenfeld J.A., Coe B.P., Parikh S., Friedman N., Goldstein A., Filipink R.A., McConnell J.S., Angle B., Meschino W.S. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N. Engl. J. Med. 2012;367:1321–1331. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krumm N., O’Roak B.J., Karakoc E., Mohajeri K., Nelson B., Vives L., Jacquemont S., Munson J., Bernier R., Eichler E.E. Transmission disequilibrium of small CNVs in simplex autism. Am. J. Hum. Genet. 2013;93:595–606. doi: 10.1016/j.ajhg.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vulto-van Silfhout A.T., Hehir-Kwa J.Y., van Bon B.W.M., Schuurs-Hoeijmakers J.H.M., Meader S., Hellebrekers C.J.M., Thoonen I.J.M., de Brouwer A.P.M., Brunner H.G., Webber C. Clinical significance of de novo and inherited copy-number variation. Hum. Mutat. 2013;34:1679–1687. doi: 10.1002/humu.22442. [DOI] [PubMed] [Google Scholar]
- 15.Mandel J.-L., Chelly J. Monogenic X-linked mental retardation: is it as frequent as currently estimated? The paradox of the ARX (Aristaless X) mutations. Eur. J. Hum. Genet. 2004;12:689–693. doi: 10.1038/sj.ejhg.5201247. [DOI] [PubMed] [Google Scholar]
- 16.Cooper G.M., Coe B.P., Girirajan S., Rosenfeld J.A., Vu T.H., Baker C., Williams C., Stalker H., Hamid R., Hannig V. A copy number variation morbidity map of developmental delay. Nat. Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders S.J., Murtha M.T., Gupta A.R., Murdoch J.D., Raubeson M.J., Willsey A.J., Ercan-Sencicek A.G., DiLullo N.M., Parikshak N.N., Stein J.L. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenfeld J.A., Coe B.P., Eichler E.E., Cuckle H., Shaffer L.G. Estimates of penetrance for recurrent pathogenic copy-number variations. Genet. Med. 2013;15:478–481. doi: 10.1038/gim.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Craddock N., Hurles M.E., Cardin N., Pearson R.D., Plagnol V., Robson S., Vukcevic D., Barnes C., Conrad D.F., Giannoulatou E., Wellcome Trust Case Control Consortium Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duker A.L., Ballif B.C., Bawle E.V., Person R.E., Mahadevan S., Alliman S., Thompson R., Traylor R., Bejjani B.A., Shaffer L.G. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur. J. Hum. Genet. 2010;18:1196–1201. doi: 10.1038/ejhg.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itsara A., Cooper G.M., Baker C., Girirajan S., Li J., Absher D., Krauss R.M., Myers R.M., Ridker P.M., Chasman D.I. Population analysis of large copy number variants and hotspots of human genetic disease. Am. J. Hum. Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ballif B.C., Theisen A., McDonald-McGinn D.M., Zackai E.H., Hersh J.H., Bejjani B.A., Shaffer L.G. Identification of a previously unrecognized microdeletion syndrome of 16q11.2q12.2. Clin. Genet. 2008;74:469–475. doi: 10.1111/j.1399-0004.2008.01094.x. [DOI] [PubMed] [Google Scholar]
- 24.Fischbach G.D., Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaminsky E.B., Kaul V., Paschall J., Church D.M., Bunke B., Kunig D., Moreno-De-Luca D., Moreno-De-Luca A., Mulle J.G., Warren S.T. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet. Med. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staples J., Nickerson D.A., Below J.E. Utilizing graph theory to select the largest set of unrelated individuals for genetic analysis. Genet. Epidemiol. 2013;37:136–141. doi: 10.1002/gepi.21684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014 doi: 10.1038/ng.2892. Published online February 2, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abikoff H.B., Jensen P.S., Arnold L.L.E., Hoza B., Hechtman L., Pollack S., Martin D., Alvir J., March J.S., Hinshaw S. Observed classroom behavior of children with ADHD: relationship to gender and comorbidity. J. Abnorm. Child Psychol. 2002;30:349–359. doi: 10.1023/a:1015713807297. [DOI] [PubMed] [Google Scholar]
- 32.Moreno-De-Luca A., Myers S.M., Challman T.D., Moreno-De-Luca D., Evans D.W., Ledbetter D.H. Developmental brain dysfunction: revival and expansion of old concepts based on new genetic evidence. Lancet Neurol. 2013;12:406–414. doi: 10.1016/S1474-4422(13)70011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dworzynski K., Ronald A., Bolton P., Happé F. How different are girls and boys above and below the diagnostic threshold for autism spectrum disorders? J. Am. Acad. Child Adolesc. Psychiatry. 2012;51:788–797. doi: 10.1016/j.jaac.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 34.Baron-Cohen S. The extreme male brain theory of autism. Trends Cogn. Sci. 2002;6:248–254. doi: 10.1016/s1364-6613(02)01904-6. [DOI] [PubMed] [Google Scholar]
- 35.Szatmari P., Liu X.-Q., Goldberg J., Zwaigenbaum L., Paterson A.D., Woodbury-Smith M., Georgiades S., Duku E., Thompson A. Sex differences in repetitive stereotyped behaviors in autism: implications for genetic liability. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2012;159B:5–12. doi: 10.1002/ajmg.b.31238. [DOI] [PubMed] [Google Scholar]
- 36.Baron-Cohen S., Lombardo M.V., Auyeung B., Ashwin E., Chakrabarti B., Knickmeyer R. Why are autism spectrum conditions more prevalent in males? PLoS Biol. 2011;9:e1001081. doi: 10.1371/journal.pbio.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell G., Steer C., Golding J. Social and demographic factors that influence the diagnosis of autistic spectrum disorders. Soc. Psychiatry Psychiatr. Epidemiol. 2011;46:1283–1293. doi: 10.1007/s00127-010-0294-z. [DOI] [PubMed] [Google Scholar]
- 38.Stankiewicz P., Thiele H., Schlicker M., Cseke-Friedrich A., Bartel-Friedrich S., Yatsenko S.A., Lupski J.R., Hansmann I. Duplication of Xq26.2-q27.1, including SOX3, in a mother and daughter with short stature and dyslalia. Am. J. Med. Genet. A. 2005;138:11–17. doi: 10.1002/ajmg.a.30910. [DOI] [PubMed] [Google Scholar]
- 39.Møller R.S., Jensen L.R., Maas S.M., Filmus J., Capurro M., Hansen C., Marcelis C.L.M., Ravn K., Andrieux J., Mathieu M. X-linked congenital ptosis and associated intellectual disability, short stature, microcephaly, cleft palate, digital and genital abnormalities define novel Xq25q26 duplication syndrome. Hum. Genet. 2013 doi: 10.1007/s00439-013-1403-3. Published online December 11, 2013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


