Abstract
Previous work has demonstrated that northern and southern European ancestries are associated with specific systemic lupus erythematosus (SLE) manifestations. Here, 1855 SLE cases of European descent were genotyped for 4965 single nucleotide polymorphisms and principal components analysis of genotype information was used to define population substructure. The first principal component (PC1) distinguished northern from southern European ancestry, PC2 differentiated eastern from western European ancestry, and PC3 delineated Ashkenazi Jewish ancestry. Compared to northern European ancestry, southern European ancestry was associated with autoantibody production (OR=1.40, 95% CI 1.07-1.83) and renal involvement (OR 1.41, 95% CI 1.06-1.87), and was protective for discoid rash (OR=0.51, 95% CI 0.32-0.82) and photosensitivity (OR=0.74, 95% CI 0.56-0.97). Both serositis (OR=1.46, 95% CI 1.12-1.89) and autoantibody production (OR=1.38, 95% CI 1.06-1.80) were associated with Western compared to Eastern European ancestry. Ashkenazi Jewish ancestry was protective against neurologic manifestations of SLE (OR=0.62, 95% CI 0.40-0.94). Homogeneous clusters of cases defined by multiple PCs demonstrated stronger phenotypic associations. Genetic ancestry may contribute to the development of SLE endophenotypes and should be accounted for in genetic studies of disease characteristics.
Keywords: Systemic lupus erythematosus, epidemiology, population substructure, genetics
Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease that affects multiple organ systems and demonstrates marked clinical heterogeneity. The American College of Rheumatology (ACR) classification criteria for SLE include 11 distinct disease manifestations and individuals with any 4 of these 11 features may be classified as having SLE.1 Some disease manifestations included in the criteria, such as arthritis, photosensitivity, and oral ulcers, are relatively mild, while others, such as seizures, psychosis, renal involvement, and severe hematologic abnormalities, may result in substantial morbidity.
Epidemiologic studies have demonstrated that in the United States, the risk of developing severe SLE manifestations varies by ethnicity, suggesting that genetics may influence disease phenotypes.2-4 Recent studies, particularly in Latino populations, have examined genetic admixture in relation to SLE severity, and have shown a trend toward more severe disease with increasing Amerindian ancestry or decreasing European ancestry.5, 6
Genetic differences within continental populations also provide an opportunity to assess the contribution of genetic ancestry to SLE phenotypes. Within Europe, a genetic gradient spans from northwest to southeast, distinguishing northern European populations, including British, German, Scandinavian, and Eastern European populations, from southern Europeans, including Italian, Greek, and Spanish populations.7-9 We have recently demonstrated that northern and southern European genetic ancestries are associated with specific SLE phenotypes. In particular, northern European ancestry is associated with mucocutaneous manifestations of SLE while southern European ancestry is a risk factor for autoantibody production.10
Genetic diversity among European populations, however, is not limited to north-south differences. Recent studies have used principal components analysis (PCA) of genotyping data to detect and describe multiple axes of genetic variation among individuals of European descent.11, 12 Using PCA, we defined the population genetic substructure for a group of North American SLE cases of European descent. We then examined the relationship between genetic ancestry and specific SLE manifestations to determine whether multiple genetic axes are associated with specific SLE phenotypes.
Results and Discussion
Our study population included 1855 North American SLE cases of self-reported European ancestry. Participants were enrolled in 4 independent case-series (Supplementary Table 1): the University of California, San Francisco (UCSF) Lupus Genetics Project (n=1018), the Autoimmune Biomarkers Collaborative Network (ABCoN, n=338), the University of Pittsburgh Medical Center Lupus Cohort (n=373), and the Multiple Autoimmune Disease Genetics Consortium (MADGC, n=126). In each case series, phenotypic information and diagnosis of SLE were confirmed by chart review. Autoantibody status was determined either by chart review or direct serologic testing. An individual was considered autoantibody positive if presence of the autoantibody was documented at least once either in the medical record or by direct testing. Biological samples and clinical information were collected with written participant consent in accordance with institution-specific protocols and with local Institutional Review Board approval.
All participants had been previously genotyped as part of one of two parent studies. A subset of 1450 participants was genotyped for a genome-wide association study of SLE susceptibility using an Illumina HumanHap550 BeadChip as previously described.13 The majority of these participants was also included in our previous study of European population substructure and SLE phenotypes.10 A set of controls (n=2398) included in the genome wide association study for SLE susceptibility was also included in these analyses for improved PCA differentiation and interpretation (Figure 1). An additional 405 participants not included in our previous study were genotyped for 12,000 single nucleotide polymorphisms (SNPs) using an Illumina custom iSELECT Infinium II array.
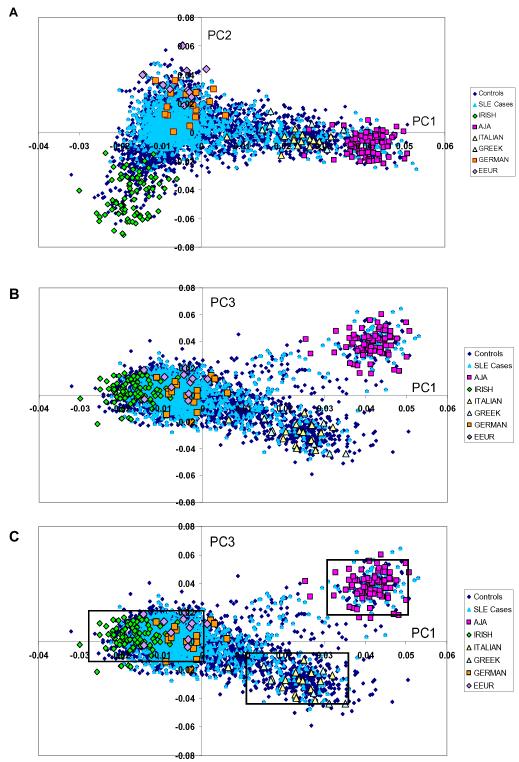
Figure 1.
Principal components analysis (PCA) shows European population substructure.
(a) Panel 1a plots principal component (PC) 2 against PC1. (b) Panel 1b plots PC3 against PC1. (c) Panel 1c identifies homogenous clusters (northern European, southern European, Ashkenazi Jewish) defined by PC1 and PC3. Points on the graphs represent individual study participants; individuals are color coded according to SLE status and grandparental country of origin, AJA=Ashkenazi Jewish ancestry, EEUR=Eastern European.
Population genetic substructure was determined using PCA of 4965 single nucleotide polymorphisms (SNPs) in EIGENSTRAT (Cambridge, MA). Of these SNPs, 2617 were informative for European population substructure and the remainder were informative for continental ancestry (187 SNPs) or East Asian substructure (2161 SNPs). These SNPs are distributed throughout the autosomal genome and exclude regions of extended linkage disequilibrium (inversion regions and the major histocompatibility complex region). Additionally, pairs of SNPs with r2 >0.5 in European population groups, SNPs not in Hardy-Weinberg equilibrium, and SNPs missing in >10% of participants were excluded.11
To improve PC differentiation and interpretation, we also included a set of 2398 controls genotyped for the same set of SNPs. Controls were adults of self-reported European ancestry enrolled in the New York Cancer Project who did not have SLE.19 Information on grandparental country of origin was collected for controls, and controls with all 4 grandparents from a single European country or region or who shared a single ethnic identity were used for PC interpretation.
Prior to PCA, we screened the study population for non-European ancestry using a set of 187 ancestry informative markers included in the SNP panel. We used STRUCTURE, a program that applies a model based, non-hierarchical clustering method for individual ancestry estimation.20 Participants with >10% non-European ancestry were excluded prior to PCA to determine European substructure. Participants were also excluded for outlying PC values (>6 standard deviations from the mean).
Clinical and demographic characteristics of the 1631 SLE cases included in the final analysis (see Supplementary Table 1 for exclusions), including the prevalence of SLE manifestations that comprise the 11 ACR classification criteria, are detailed in Supplementary Table 2. As expected for SLE, most participants were female (93.1%), with a mean age at disease onset of 34.9 years (standard deviation 13.2). The median disease duration was 7 years (interquartile range 3-14). The most common SLE manifestations were anti-nuclear antibody production (97.0%), arthritis (75.2%), and photosensitivity (72.3%). Less common disease manifestations included discoid rash (8.4%) and fulfillment of the neurologic disorder criterion (9.6%).
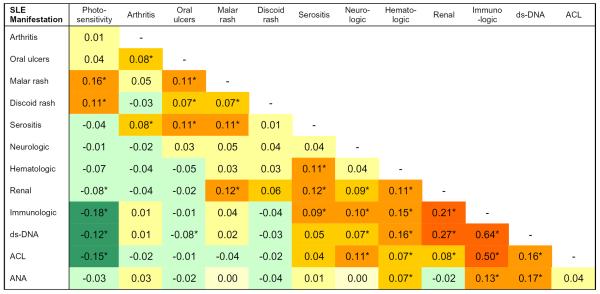
Several SLE manifestations tended to occur together and pairwise correlation coefficients for diseases manifestations are shown in Figure 2. Correlation between disease manifestations was assessed using the Spearman rank-order correlation coefficient. The Spearman rank-order correlation coefficient was chosen because it makes no assumptions about the nature of the correlation between two variables and can be applied to non-normally distributed variables. We found that the immunologic disorder and renal disorder criteria were correlated; each was also correlated with the neurologic disorder and hematologic disorder criteria (p ≤ 0.0008). The cutaneous manifestations—photosensitivity, malar rash, and discoid rash—were correlated with one another (p ≤ 0.007) while photosensitivity was inversely correlated with fulfillment of the renal and immunologic disorder criteria (p ≤ 0.0008).
Figure 2.
Correlation coefficients for SLE manifestations. We used the Spearman rank correlation coefficient, which does not require a normal distribution of values for a variable, to assess the correlation between pairs of SLE manifestations included in the American College of Rheumatology classification criteria for SLE and also for specific autoantibodies. Shading reflects the strength and direction of the correlation between SLE phenotypes.
*p<0.01; ds-DNA=anti-double stranded DNA antibody, ACL=anti-cardiolipin antibody, ANA=anti-nuclear antibody
We applied PCA to a set of 4965 SNPs common to both genotyping platforms for ancestry determination. The first three PCs explained 54% of the variance among the first 10 PCs and had the following eigenvalues and variance: 19.4 (33.3%), 6.7 (11.8%), and 5.1 (9%). The eigenvalues for PC4-PC10 showed a plateau, suggesting that the first three PCs account for the population substructure in this analysis.
We used grandparental country of origin of controls to interpret PCs (Figure 1). Positive values for PC1 corresponded to southern European ancestry while negative values for PC1 corresponded to northern European ancestry. PC2 defined an east-west gradient, distinguishing Irish from German and eastern European ancestry. PC3 distinguished individuals of Ashkenazi Jewish ancestry from other European ancestries. The large variance in PC1 is consistent with previous studies indicating that the north-south geographic axis corresponds to the largest component of population substructure within Europe.8, 9, 11, 14
Next, we examined the relationship between genetic ancestry, represented by the first three PCs, and each of the 11 disease manifestations included in the ACR classification criteria for SLE. We first assessed the bivariate association between each PC, as a continuous variable, and the disease manifestations included in the ACR classification criteria for SLE. For these analyses, we used the Wilcoxon rank-sum test because the PCs, particularly PC1, were not normally distributed. PCs that were associated with a disease manifestation in bivariate analysis (p<0.1) were further evaluated using logistic regression and are included in Table 1.
Table 1. Association between Genetic Ancestry and SLE Manifestations.
| Principal Component 1 | ||||
|---|---|---|---|---|
|
| ||||
| Characteristic1 | Crude OR2 (95% CI) |
p-value | Adjusted OR3 (95%CI) |
p-value |
| Discoid rash | 0.57 (0.37-0.90) | 0.016 | 0.51 (0.32-0.82) | 0.005 |
| Photosensitivity | 0.68 (0.52-0.89) | 0.004 | 0.74 (0.56-0.97) | 0.032 |
| Oral ulcers | 1.29 (1.00-1.64) | 0.046 | 1.15 (0.89-1.49) | 0.300 |
| Neurologic disorder | 1.42 (0.94-2.15) | 0.093 | 1.34 (0.88-2.04) | 0.179 |
| Immunologic disorder | 1.48 (1.14-1.92) | 0.003 | 1.40 (1.07-1.83) | 0.014 |
| Anti-Smith | 1.20 (0.82-1.76) | 0.345 | 1.19 (0.80-1.75) | 0.393 |
| Anti-cardiolipin | 1.52 (1.17-1.98) | 0.002 | 1.40 (1.06-1.84) | 0.017 |
| Anti-dsDNA | 1.37 (1.07-1.75) | 0.013 | 1.42 (1.10-1.83) | 0.006 |
| Renal disorder | 1.54 (1.17-2.03) | 0.002 | 1.41 (1.06-1.87) | 0.019 |
| Principal Component 2 | ||||
|---|---|---|---|---|
|
| ||||
| Characteristic1 | Crude OR2 (95%CI) |
p-value | Adjusted OR3 (95%CI) |
p-value |
| Malar rash | 1.28 (1.00-1.63) | 0.047 | 1.28 (1.00-1.64) | 0.049 |
| Immunologic disorder | 1.27 (0.98-1.64) | 0.066 | 1.38 (1.06-1.80) | 0.018 |
| Anti-Smith | 1.00 (0.69-1.45) | 0.996 | 1.03 (0.71-1.49) | 0.883 |
| Anti-dsDNA | 1.07 (0.84-1.36) | 0.594 | 1.07 (0.83-1.37) | 0.612 |
| Anti-cardiolipin | 1.16 (0.89-1.50) | 0.271 | 1.22 (0.93-1.60) | 0.154 |
| Serositis | 1.35 (1.06-1.71) | 0.017 | 1.46 (1.12-1.89) | 0.005 |
| Pleuritis | 1.29 (0.99-1.69) | 0.061 | 1.33 (1.01-1.75) | 0.046 |
| Pericarditis | 1.69 (1.17-2.44) | 0.005 | 1.75 (1.20-2.56) | 0.004 |
| Principal Component 3 | ||||
|---|---|---|---|---|
|
| ||||
| Characteristic1 | Crude OR2 (95%CI) |
p-value | Adjusted OR3 (95%CI) |
p-value |
| Neurologic disorder | 0.60 (0.40-0.91) | 0.018 | 0.62 (0.40-0.94) | 0.024 |
| Psychosis | 0.49 (0.20-1.24) | 0.132 | 0.50 (0.20-1.26) | 0.142 |
| Seizure | 0.77 (0.48-1.24) | 0.287 | 0.77 (0.48-1.25) | 0.298 |
| Malar rash | 0.80 (0.63-1.72) | 0.075 | 0.79 (0.61-1.01) | 0.056 |
The American College of Rheumatology (ACR) criteria included in this table are disease manifestations that were first shown to be associated with a PC in bivariate analysis (p≤0.1) using the Wilcoxon rank-sum test. For composite criteria that may be fulfilled in several ways, such as the immunologic, hematologic, and neurologic disorder criteria, we examined the specific disease manifestations that comprise those criteria if the criteria were associated with the PCs after adjustment for covariates (p≤0.1).
For calculation of odds ratios, we used logistic regression. PCs were divided into tertiles and we compared the highest and lowest tertiles. The reference group for PC1 is the lowest tertile (northern European ancestry). For PC2, the reference group is the highest tertile (eastern European ancestry). For PC3 the reference group is the lowest tertile (European, non-Ashkenazi Jewish ancestry).
For multivariable analyses, we again used logistic regression. PCs were divided into tertiles in the same way as described when calculating crude odds ratios. Models were adjusted for sex, disease duration, and case collection. Analyses were performed using Stata 10.0 (StataCorp, College Station, TX)
For logistic regression analyses, we first divided each PC into tertiles. Division of the PCs into quantiles was done so that we could compare high and low values within a PC and thus improve the interpretability of our results. Tertiles were specifically chosen because using three groups maintains quantile size while still preserving some specificity within quantiles. We then used logistic regression to assess the association between PC tertile and disease characteristics, comparing the highest and lowest tertiles within each PC (Table 1). After adjustment for disease duration, sex, and case collection, we found that southern European ancestry, represented by the highest tertile of PC1, was protective for photosensitivity (ORhigh-low 0.74, 95% CI 0.56-0.97) and discoid rash (OR high-low 0.51, 95% CI 0.32-0.82) and was associated with fulfillment of the renal (OR high-low 1.41, 95% CI 1.06-1.87) and immunologic (OR high-low=1.40, 95% CI 1.07-1.83) disorder criteria when compared to northern European ancestry.
Because several autoantibodies are included in the immunologic disorder criterion, we further investigated the relationship between genetic ancestry and production of specific autoantibodies. We found that after adjustment for disease duration, sex, and case collection, southern European ancestry was associated with anti-double stranded DNA (dsDNA, OR high-low=1.42, 95% CI 1.10-1.83) and anti-cardiolipin (OR high-low =1.40, 95% CI 1.06-1.84) antibody production (Table 1).
PC2 and PC3 had fewer phenotypic associations (Table 1). Western European ancestry, represented by the lowest tertile of PC2, was a risk factor for serositis (OR low-high=1.46, 95% CI 1.12-1.89) and fulfillment of the immunologic disorder criterion (OR low-high=1.38, 95% CI 1.06-1.80) when compared to eastern European ancestry (highest tertile of PC2). Membership in the highest tertile of PC3, which included individuals of Ashkenazi Jewish ancestry, was protective against fulfillment of the neurologic disorder criterion compared to other European ancestries (OR high-low=0.62, 95% CI 0.40-0.94).
In a subgroup analysis, we examined whether differences in socioeconomic status among participants could explain our results. Data on educational attainment, a stable measure of socioeconomic status, were available for some participants from the UCSF and ABCoN case collections (n=799). We repeated the analyses presented in Table 1 controlling for disease duration, case collection, sex, and also for educational attainment. We found that the magnitude and direction of the odds ratios presented in Table 1 were largely unchanged, though many results did not reach statistical significance because of the sharply reduced sample size and power.
Differing environmental exposures, such as sunlight, could also potentially explain our results. Data on place of birth, which may capture some information about environmental exposures, were available for some participants from the UCSF collection (n=762). We repeated the analyses presented in Table 1, controlling for disease duration, sex, case collection, and place of birth. Again, we found that the magnitude and direction of our results presented in Table 1 were largely unchanged, though because of reduced sample size, many results did not reach statistical significance. Place of birth was not an independent predictor of any disease manifestation except for malar rash.
Lastly, we observed that when graphed, PC1 and PC3 together defined three distinct clusters of subjects (Figure 1c): one near controls who reported Ashkenazi Jewish ancestry, a second near controls who reported southern European ancestry (Greek and Italian) and a third large cluster near those who reported northern European ancestry (German, Irish, eastern European). We then investigated whether these homogeneous clusters of subjects, defined by two PCs rather than a single PC, demonstrated particular phenotypic associations.
For this analysis, we identified all cases with PC1 and PC3 values within two standard deviations of the mean PC1 and PC3 values for controls of self-reported Italian and Greek ancestry (n=66). We repeated this procedure to identify cases near controls of Ashkenazi Jewish ancestry (n=68) and cases near controls of northern European ancestry (n=1188). We then compared both the southern European (Italian/Greek) ancestry cluster and the Ashkenazi Jewish ancestry cluster to the northern European (German, Irish, eastern European) cluster to examine whether these more homogeneous clusters of participants have specific phenotypic associations.
Compared to cases in the northern European ancestry cluster, cases in the Ashkenazi Jewish ancestry cluster were more likely to meet the immunologic disorder criterion (OR=1.98, 95% CI 1.09-3.60, p=0.025) after adjustment for disease duration, sex, and case collection. Those in the Ashkenazi Jewish ancestry cluster also had a higher odds of anti-dsDNA antibody production (OR= 1.91, 95% CI 1.13-3.22, p=0.016) and anti-cardiolipin antibody production (OR=1.80, 95% CI 1.05-3.10, p=0.033) compared to cases in the northern European cluster after adjustment for disease duration, sex, and case collection.
Cases in the southern European ancestry cluster also had an increased odds of meeting the immunologic disorder criterion (OR=1.26, 95% CI 0.70-2.27, p=0.5), anti-dsDNA antibody production (1.47, 95% CI 0.87-2.49, p=0.2) and anti-cardiolipin antibody production (OR=1.41, 95% CI 0.82-2.44, p=0.2) compared to cases of northern European ancestry after adjustment for disease duration, sex, and cohort. These associations, though, did not reach statistical significance.
While the Ashkenzi Jewish ancestry cluster had larger, statistically significant odds ratios for autoantibody production and the southern European ancestry cluster had smaller, non statistically significant associations, Wald tests directly comparing the odds ratios for each phenotype (immunologic disorder criterion, anti-dsDNA antibody production, anti-cardiolipin production) across the southern European and Ashkenazi Jewish subgroups were not statistically significant (p>0.2).
Using a non-parametric method for ancestry estimation, an expanded SNP set, and an extended sample, we have identified multiple axes of genetic variation among a population of individuals of European descent. We have also demonstrated that southern European ancestry is a risk factor for renal disease and autoantibody production and is protective against photosensitivity and discoid rash. Renal disease and autoantibody production are correlated, as are the cutaneous manifestations of SLE, and our findings suggest that a common genetic background may contribute to the development of a particular constellation of SLE manifestations. These findings are consistent with our previous work, which showed that southern European ancestry is associated with autoantibody production and northern European ancestry with mucocutaneous manifestations of SLE.10 The alternative analytic technique and extended sample used here provide further evidence for a more severe SLE phenotype among southern Europeans.
Epidemiologic studies have suggested that SLE may be more prevalent in southern European countries such as Spain and Italy than in northern European countries.15 Intriguingly, the PD1.3A allele of PDCD1, a variant associated with renal manifestations of SLE, is more common among southern Europeans than northern Europeans.16, 17 By contrast, the R620W polymorphism of PTPN22, which has been associated both with the development of SLE and with other autoimmune diseases that demonstrate autoantibody production, is more common among northern Europeans.18 These contrasting examples illustrate the complex relationships between genes, populations, and development of disease endophenotypes. Nonetheless, geographic variation in alleles associated with SLE and SLE phenotypes supports a plausible role for genetic ancestry in contributing to the development of specific SLE endophenotypes.
In addition to differentiating northern and southern European ancestry, PCA also distinguished eastern and western European ancestry and separated individuals of Ashkenazi Jewish ancestry from other European ancestries, a finding consistent with previous studies of European population substructure.11 These additional axes of genetic variation captured less of the variance in the dataset and had fewer independent associations with SLE endophenotypes. Nonetheless, we did identify some phenotypic associations with the second and third PC, suggesting that other dimensions of genetic variation beyond north-south differences may play a role in the development of specific SLE manifestations.
Importantly, these additional PCs also provided information for defining more homogeneous clusters of participants among northern and southern Europeans. We used information from PC3 to delineate two distinct clusters of participants among those with high PC1 values: those of Ashkenazi Jewish ancestry and those of southern European ancestry. These two groups shared similar PC1 values and thus demonstrated some genetic similarity. Still, when compared to the same reference group, northern Europeans, these two groups had varying risks of developing specific SLE manifestations. While small sample sizes within each subgroup limited our ability to demonstrate that these subgroups definitively differed from one another with respect to phenotypic associations, the varying magnitudes of associations by subgroup suggest that even relatively fine differences in genetic ancestry may confer differential risk for developing specific SLE manifestations.
This study has several limitations. First, participants’ clinical data were collected retrospectively, which may result in some misclassification of outcomes due to underreporting or failure to capture disease manifestations that evolve over time. Any misclassification, however, should not differ with respect to genetic ancestry and should bias results toward the null. Second, we examined the association between a number of SLE phenotypes and each of the PCs. This approach led to a large number of hypothesis tests and consequently, an increased likelihood of committing a type I error. However, a traditional Bonferroni correction is overly conservative, since, as we have shown, lupus phenotypes are not independent. We have presented unadjusted p-values to allow readers to assess statistical significance. Lastly, unmeasured confounders, such as cultural differences or differing environmental exposures among populations may have contributed to these results. Data on place of residence and socioeconomic status were not available for the entire study population, but our subgroup analyses demonstrate that controlling for birthplace and educational attainment did not substantively alter our results.
In summary, our study has provided further evidence for a more severe SLE phenotype among individuals of southern European ancestry and suggests that even relatively small differences in population genetics may contribute to disease endophenotypes. Future studies that investigate specific genes that contribute to these phenotypic differences will likely provide invaluable insights into the clinical variability of SLE.
Supplementary Material
Acknowledgements
This work has been supported by NIH/NIAMS grants P60 AR053308, K24 AR02175, R01 AR052300, and a Kirkland Scholar Award to Dr. Criswell, NIH/NIAMS and NIH/NIAID grants R01 AI68759 and N01 AR1-2256 to Dr. Gregersen, NIH/NIAMS grants R01 AR46588-05 and K24 AR002213-05 to Dr. Manzi, and NIH/NIAMS grant R01 AR050267 to Dr. Seldin.
These studies were performed in part in the University of California, San Francisco, General Clinical Research Center (Moffitt Hospital), the University of Pittsburgh General Clinical Research Center, and the Johns Hopkins University General Clinical Research Center with funds provided by the U.S. Public Health Service National Center for Research Resources, M01 RR-00079, M01 RR-000056, and M01 RR-00052. This publication was also supported by NIH/NCRR/OD UCSF-CTSI Grant Number TL1 RR024129. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Dr. Behrens and Mr. Ortmann are employees of Genentech, Inc.
Footnotes
Disclosures/Conflict of Interest: Dr. Behrens and Mr. Ortmann are employees of Genentech, Inc.
Supplementary information is available at the Genes and Immunity website (http://www.nature.com/gene/index.html).
References
- 1.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis and rheumatism. 1982;25(11):1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 2.Siegel M, Reilly EB, Lee SL, Fuerst HT, Seelenfreund M. Epidemiology of Systemic Lupus Erythematosus: Time Trend and Racial Differences. American journal of public health and the nation’s health. 1964;54:33–43. doi: 10.2105/ajph.54.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alarcon GS, McGwin G, Jr., Petri M, Reveille JD, Ramsey-Goldman R, Kimberly RP. Baseline characteristics of a multiethnic lupus cohort: PROFILE. Lupus. 2002;11(2):95–101. doi: 10.1191/0961203302lu155oa. [DOI] [PubMed] [Google Scholar]
- 4.Alarcon GS, Friedman AW, Straaton KV, Moulds JM, Lisse J, Bastian HM, et al. Systemic lupus erythematosus in three ethnic groups: III. A comparison of characteristics early in the natural history of the LUMINA cohort. LUpus in MInority populations: NAture vs. Nurture. Lupus. 1999;8(3):197–209. doi: 10.1191/096120399678847704. [DOI] [PubMed] [Google Scholar]
- 5.Seldin MF, Qi L, Scherbarth HR, Tian C, Ransom M, Silva G, et al. Amerindian ancestry in Argentina is associated with increased risk for systemic lupus erythematosus. Genes and immunity. 2008;9(4):389–93. doi: 10.1038/gene.2008.25. [DOI] [PubMed] [Google Scholar]
- 6.Alarcon GS, Bastian HM, Beasley TM, Roseman JM, Tan FK, Fessler BJ, et al. Systemic lupus erythematosus in a multi-ethnic cohort (LUMINA) XXXII: [corrected] contributions of admixture and socioeconomic status to renal involvement. Lupus. 2006;15(1):26–31. doi: 10.1191/0961203306lu2260oa. [DOI] [PubMed] [Google Scholar]
- 7.Cavalli-Sforza LL, Menozzi P, Piazza A. Demic expansions and human evolution. Science (New York, N.Y. 1993;259(5095):639–46. doi: 10.1126/science.8430313. [DOI] [PubMed] [Google Scholar]
- 8.Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, et al. Genes mirror geography within Europe. Nature. 2008;456(7218):98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seldin MF, Shigeta R, Villoslada P, Selmi C, Tuomilehto J, Silva G, et al. European population substructure: clustering of northern and southern populations. PLoS genetics. 2006;2(9):e143. doi: 10.1371/journal.pgen.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung SA, Tian C, Taylor KE, Lee AT, Ortmann WA, Hom G, et al. European population substructure is associated with mucocutaneous manifestations and autoantibody production in systemic lupus erythematosus. Arthritis and rheumatism. 2009;60(8):2448–56. doi: 10.1002/art.24707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian C, Plenge RM, Ransom M, Lee A, Villoslada P, Selmi C, et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS genetics. 2008;4(1):e4. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 13.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. The New England journal of medicine. 2008;358(9):900–9. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 14.Lao O, Lu TT, Nothnagel M, Junge O, Freitag-Wolf S, Caliebe A, et al. Correlation between genetic and geographic structure in Europe. Curr Biol. 2008;18(16):1241–8. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 15.Danchenko N, Satia JA, Anthony MS. Epidemiology of systemic lupus erythematosus: a comparison of worldwide disease burden. Lupus. 2006;15(5):308–18. doi: 10.1191/0961203306lu2305xx. [DOI] [PubMed] [Google Scholar]
- 16.Ferreiros-Vidal I, D’Alfonso S, Papasteriades C, Skopouli FN, Marchini M, Scorza R, et al. Bias in association studies of systemic lupus erythematosus susceptibility due to geographical variation in the frequency of a programmed cell death 1 polymorphism across Europe. Genes and immunity. 2007;8(2):138–46. doi: 10.1038/sj.gene.6364370. [DOI] [PubMed] [Google Scholar]
- 17.Lee YH, Woo JH, Choi SJ, Ji JD, Song GG. Association of programmed cell death 1 polymorphisms and systemic lupus erythematosus: a meta-analysis. Lupus. 2009;18(1):9–15. doi: 10.1177/0961203308093923. [DOI] [PubMed] [Google Scholar]
- 18.Gregersen PK, Lee HS, Batliwalla F, Begovich AB. PTPN22: setting thresholds for autoimmunity. Seminars in immunology. 2006;18(4):214–23. doi: 10.1016/j.smim.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell MK, Gregersen PK, Johnson S, Parsons R, Vlahov D. The New York Cancer Project: rationale, organization, design, and baseline characteristics. J Urban Health. 2004;81(2):301–10. doi: 10.1093/jurban/jth116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Human mutation. 2009;30(1):69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.