Abstract
DNA damage sensing proteins have been shown to localize to the sites of DNA double strand breaks (DSB) within seconds to minutes following ionizing radiation (IR) exposure, resulting in the formation of microscopically visible nuclear domains referred to as radiation-induced foci (RIF). This review characterizes the spatiotemporal properties of RIF at physiological doses, minutes to hours following exposure to ionizing radiation, and it proposes a model describing RIF formation and resolution as a function of radiation quality and chromatin territories. Discussion is limited to RIF formed by three interrelated proteins ATM (Ataxia telangiectasia mutated), 53BP1 (p53 binding protein 1) and γH2AX (phosphorylated variant histone H2AX), with an emphasis on the later. This review discusses the importance of not equating RIF with DSB in all situations and shows how dose and time dependence of RIF frequency is inconsistent with a one to one equivalence. Instead, we propose that RIF mark regions of the chromatin that would serve as scaffolds rigid enough to keep broken DNA from diffusing away, but open enough to allow the repair machinery to access the damage site. We review data indicating clear kinetic and physical differences between RIF emerging from dense and uncondensed regions of the nucleus. We suggest that persistent RIF observed days following exposure to ionizing radiation are nuclear marks of permanent rearrangement of the chromatin architecture. Such chromatin alterations may not always lead to growth arrest as cells have been shown to replicate these in progeny. Thus, heritable persistent RIF spanning over tens of Mbp may reflect persistent changes in the transcriptome of a large progeny of cells. Such model opens the door to a “non-DNA-centric view” of radiation-induced phenotypes.
Keywords: DSB, γH2AX, ATMp, 53BP1, Repair kinetics, Review, Chromatin, Complex damage
1. Introduction
A well accepted paradigm in radiation biology is that ionizing radiation (IR) induced DNA double strand breaks (DSB) are the most deleterious form of DNA damage. It is thought that unrepaired DSB lead to death and misrepaired DSB may lead to viable chromosomal rearrangements. Some of these rearrangements may be instrumental in the development of cancer. DSB happen regularly in cells as consequences of cell exposure to external insults or internal metabolism, such as, oxidative stress or DNA replication errors. Thus cells have evolved efficient and rapid repair responses to maintain the integrity of the genome. Sensor proteins are thought to detect the presence of a DSB, and then recruit transducer proteins which provide the signals to enzymes to repair the break. Depending on the severity of the damage and the cell cycle status of the damaged cell, sensor proteins, also modified by transducers, will induce either cell cycle delay for repair, programmed cell death or senescence.
Sensor proteins have been shown to localize to the sites of DSB within seconds to minutes following IR exposure, resulting in the formation of microscopically visible nuclear domains referred to as radiation-induced foci (RIF). In mammalian cells, Rad51 protein was one of the first proteins identified as forming RIFs in mitotic and meiotic cells [1,2]. Since then, many proteins have been shown to form RIFs and these proteins can be divided into three categories: (1) proteins recruited to damage sites such as 53BP1 [3], MRE11 or NBS1 [4,5]; (2) proteins modified near the damage site, such as the phosphorylation of H2AX (γH2AX [6]); (3) foci resulting from both processes, such as the RIF of phosphorylated (pS1981) ATM (Ataxia Telangiectasia Mutated) [7] and phosphorylated (pT2609, pS2056) DNA-PKcs [8,9]. This review will limit its discussion to three interrelated proteins ATM, 53BP1 and H2AX which form foci minutes following IR. The relationship between RIF and chromatin organization will be discussed with a primary emphasis on γH2AX.
1.1. Spontaneous foci
Many reports have indicated the presence of γH2AX foci in non-irradiated cells. For instance, we showed that about 1.5% of the cells in confluent human fibroblast cultures have 1–4 large γH2AX foci per cell, with an average size of 1.7 μm2 encompassing about 15 Mbp of DNA [10]. Similarly, 6.3% of normal G0 human diploid cells have phosphorylated ATM (ATMp) foci with diameters larger than 1.6 μm (i.e. ~2 μm2) [11]. Other studies have noted large γH2AX foci in senescent human cells and aged mice tissues, and interpreted these foci to be due to unrepairable DSB [12]. Generally, large foci seen spontaneously or in senescent cells have imaging characteristics similar to persistent radiation-induced foci. We will discuss later how these large foci may all reflect a similar chromatin status all having different cellular outcomes.
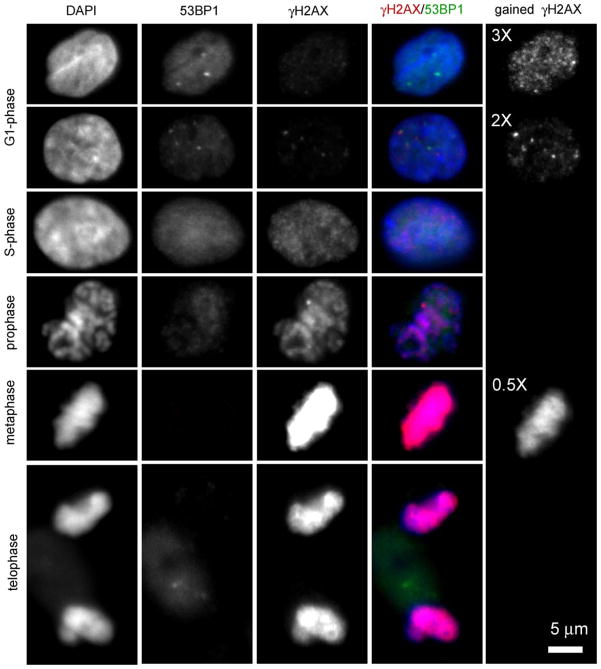
Fig. 1 shows typical images of γH2AX/53BP1 dual staining of cycling non-irradiated normal human mammary epithelial cells (MCF10A). As also observed in fibroblasts [10], a population of growing epithelial cells typically shows a very mixed γH2AX/53BP1 staining pattern. Even though many cells do not appear to contain foci, a significant number of cells show spontaneous foci. Inter-individual differences have been shown to perhaps account for some of this variability as described in a recent paper [13] where 25 untreated normal human fibroblast strains were analyzed in G0/G1 for γH2AX foci. Foci numbers ranged from 0.2 to 2.6 foci/cell on average (with an overall mean ± SD of 1.00 ± 0.57). Another important aspect of γH2AX immunostaining is illustrated in Fig. 1, where the existence of many small and low intensity foci in non-irradiated G1 cells is revealed by digitally enhancing the image. The imaging characteristics of these dim foci have been well described [14,15], and their exhibited pattern is similar to S-phase cells. Although researchers have noted these foci in unexposed cells, similar types of foci have also been detected following IR. The function of these spontaneous or non-DSB related foci is still uncertain [14]. In contrast, 53BP1 shows a uniform staining in the nucleus and exclusion in the nucleolus from G1 to G2, as illustrated in Fig. 1. On the other hand, like γH2AX, 53BP1 also shows occasional spontaneous bright foci. γH2AX foci typically colocalize with 53BP1 foci, but the reverse is not always true as illustrated in the first upper panels of Fig. 1.
Fig. 1.
Example of typical γH2AX/53BP1 dual staining in cycling normal non-irradiated human mammary epithelial cells (MCF10A). In these images, as previously described [26], γH2AX has been fluorescently labeled in red with mouse monoclonal anti phospho-histone H2AX (Ser139) antibody (1.42 μg/ml; lot #27505; Upstate Cell Signaling Solutions Inc. Charlottesville, VA) and secondary Alexa 594 (at 1:300 from Molecular Probes, Invitrogen, Carlsbad, CA). 53BP1 has been fluorescently labeled in green with rabbit polyclonal anti 53BP1 (5 μg/ml, lot #A300-272A, Bethyl Lab, Montgomery, TX) and secondary Alexa 488 (at 1:300 from Molecular Probes, Invitrogen, Carlsbad, CA). Cells have been counter stained with DAPI which labels nuclear DNA (blue). Each channel represents one center slice of a cell acquired with the same exposure time and digital camera gain. Each row depicts a different phase of MCF10A, going from G1 (top) to mitosis (bottom). G1 cells typically show no γH2AX foci or few bright γH2AX foci. However, if the γH2AX channel gain is increased by a factor 3, the presence of many dim foci is then visible (upper right panel). In contrast, 53BP1 shows a pattern in G1 that typically matches DAPI signal, with some spontaneous foci as well. DAPI and 53BP1 pattern similarity disappears during S-phase, even though 53BP1 signal remains uniform and elevated. γH2AX immunoreactivity is significantly increased during S-phase with a pattern similar to the dim foci revealed by gained enhancement in G1. As cells move to mitosis, γH2AX immunoreactivity further increases as depicted with a fully saturated signal in metaphase that needs to be acquired with half the gain in order to not saturate the image. γH2AX pattern in mitosis matches DAPI, revealing full phosphorylation of H2AX in the condensed chromosomes. In contrast, 53BP1 seems to be progressively excluded from the nucleus during mitosis.
Using the fact that PCNA is bright in S-phase cells [16], we previously showed that S-phase cells typically have a diffuse and high background intensity with discrete punctuate small γH2AX foci [10]. MCF10A nuclei with such imaging characteristics were visually selected and are displayed in Fig. 1. γH2AX foci in these cells may be an indication of stalled or broken replication forks in S-phase [17]. It was in fact hypothesized in a recent review [18] that ATRIP could phosphorylate H2AX at stalled replication forks, since ATRIP recognizes single-stranded regions in the DNA similar to the regions formed during S-phase. Thus ATR mediated γH2AX could lead to foci which do not necessarily mark DSB. Flow cytometry studies confirmed immunofluorescence results by showing that the intensity of γH2AX staining increases as a cell moves through the cell cycle with cells in S/G2/M phases having a much larger fluorescence than the expected ~2-fold increase from G1 levels [17,19]. In contrast, no distinct pattern is typically observed for 53BP1 in S-phase (see Fig. 1 and ref. [20]).
Cells in metaphase contain γH2AX foci that are ATM-dependent and may reflect a conserved mitotic function for this modification [15]. Fig. 1 illustrates the very strong uniform staining of γH2AX in mitosis, which appears maximum in metaphase and reduces in telophase. Similar observations have been previously published in vivo on mice germ cell mitotic chromosomes [21]. In contrast, a complete loss of 53BP1 immunoreactivity is noted in the nucleus during mitosis, suggesting diffusion of 53BP1 foci into the cytoplasm [20].
To conclude this section, cell cycle and inter-individuality are factors affecting the presence of spontaneous foci. The general consensus in the literature links these spontaneous foci to unrepairable DNA damage, transient DSB or genomic instability. However one could challenge this “DNA-centric” view, given that the binding of various repair factors to chromatin has been shown to be sufficient to trigger foci formation in an ATM- and DNAPK-dependent manner in the absence of DNA damage [22].
1.2. RIF: imaging characteristics of early response proteins following low-LET radiation
Numerous studies have detailed the appearance of RIF containing various proteins following exposure to different radiation qualities and quantified the induced foci microscopically by eye or with computational analysis. The first approach limits the analysis to small numbers of cells, and the amount of information one can extract from the images, such as foci shape, size or intensity, and is prone to observer bias. Thus, in Table 1 we only summarize computer analyzed RIF data reported within an hour following exposure to low-LET IR. Note that “normal cells” here means that these cells are not neoplastic and does not necessarily mean they are primary cell lines. One result that can be concluded from Table 1 is that even though DSB are generated immediately upon exposure to radiation, not all RIF appear immediately. RIF frequencies reach a maximum of ~10–40 RIF/nucleus/Gy approximately 15–30 min after exposure to low-LET. Similar delays were shown using biochemical assays such as two-dimensional gel analysis, reaching half-maximal value at 1 min and maximal value at 9–30 min post exposure [6] or reaching maximum intensity at 15–30 min using flow cytometry [23]. In contrast, delays in DSB induction are not observed using pulse field gel electrophoresis—PFGE, the standard method for detection of DSB. PFGE data show initial values of 25–35 DSB/Gy, with breaks immediately decreasing exponentially following IR [24,25]. Illustration of such disparity is shown in Fig. 2A. There are many explanations for the weak RIF detection prior to 30 min which are not mutually exclusive: (1) some DSB are repaired by mechanisms that do not require foci formation; (2) some RIF remain below detectable levels; (3) extra time is required to assemble enough molecules at some sites before they become detectable (i.e. if the site is less accessible).
Table 1.
Characteristics RIF frequencies and sizes for low-LET induced foci.
| Species | Marker | Cell | Dose (Gy) | Tmax (min) | Max (RIF/Gy) | Foci size (μm2) | Ref. | |
|---|---|---|---|---|---|---|---|---|
| “Normal” human | Breast epithelial | γH2AX | HMEC184 | 1 | 30 | 15.9 | 0.35 @ 30 min | [26] (Table I) |
| Skin fibroblast | γH2AX | HCA2 | 0.1–3 | 45–120 | 13 | 0.2–0.35 (5 min–1 h) | [10] (Fig. 3A and B) | |
| Fibroblast | γH2AX | HF19 | 1 | 20–30 | 19.1 | – | [27] (Fig. 3) | |
| Fibroblast | γH2AX | 25 strains | 0.05–0.25 | 10–30 | 14–21 | – | [13] | |
| Breast epithelial | ATMpa | HMEC184 | 1 | 30 | 16.0 | 0.35 @ 30 min | [26] (Table I) | |
| Skin fibroblast | ATMpa | HCA2 | 0.1–3 | 30 | 35 | 0.14 (5 min–2 h) | [10] (Fig. 5A and B) | |
| Diploid fibroblast | ATMpa | HE49 | 0.1–1 | 15 | 36.9 | ~0.28 @ 1 h (Ø ~0.6 μm) | [11] (Fig. 1A and C) | |
| Breast epithelial | 53BP1 | HMEC184 | 1 | 30 | 16.3 | 0.35 @ 30 min | [26] (Table I) | |
| Diploid fibroblast | 53BP1 | HE49 | 0.1–1 | 15 | ~37 | ~0.28 @ 1 h (Ø ~0.6 μm) | [11] (Fig. 4A and C) | |
| Human tumor | Melanoma | γH2AX | HT144 | 2 | 30–60a | 22 | 0.1–0.12 | [23] (Fig. 3) |
| Cervical | γH2AX | SiHa | 2 | 30–60a | 16 | 0.1–0.12 | [23] (Fig. 3) | |
| Osteosarcoma | 53BP1 | U2OS | 0.5–8 | 30 | 23 | – | [3] (Fig. 4B and D) | |
| “Normal” hamster | Ovary | γH2AX | CHO | 0.1–1 | 20 | 20 | – | [28] (Fig. 2) |
| Lung | γH2AX | V79-4 | 1 | 20–30 | 12.2 | 0.2 | [27] (Figs. 1 and 3) | |
| Lung | γH2AX | V79 | 2 | 30a | 25 or 10 | 0.1–0.12 | [23] (Fig. 2) |
ATMp: phosphorylated (pS1981) ATM.
Information provided in text, not shown in figures.
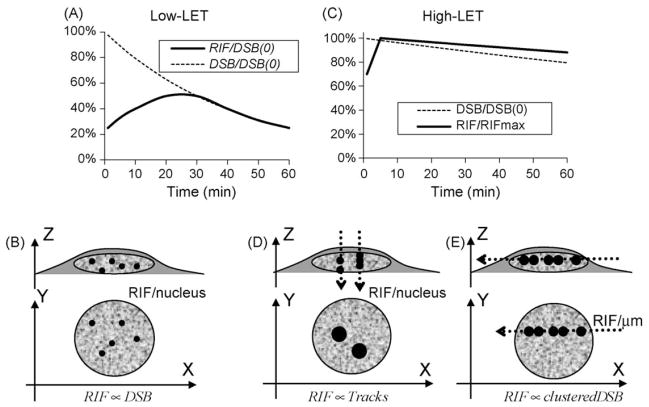
Fig. 2.
Hypothesized foci frequency curves for different radiation qualities and exposure regimens. Upper panels (A and C) depict relative RIF frequencies expected with each radiation quality compared to the expected relative DSB kinetics as measured by PFGE; lower panels (B, D and E) depict geometrical configuration of cells grown as monolayer during irradiation, with dotted lines representing direction of high-LET beam across cells (D and E); the XY plane depicts the way RIF will be visualized microscopically, with representative RIF sizes. (A and B) Schematize the low-LET RIF kinetics, where geometrical configuration has no effect. Both percentages of RIF (solid line) and DSB (dotted line) per nucleus with respect to the initial expected number of DSB (DSB(0)) are graphed with the curves reflecting the lack of foci detection for DSB repaired within the first 30 min. Kinetic curves are based on the assumption of a 30 min half-life for DSB repair after low-LET and show good correlation with DSB kinetic after 30 min (symbolized by RIF ∝ DSB). (C) Schematizes the relative RIF frequency normalized to its maximum value following high-LET exposure. Normalizing to the expected number of DSB is not done here as RIF for high-LET reflects more DSB clustering. High-LET typically induces slower DSB repair and is approximated here with a 2.5 h half-life for a LET ~150 keV/μm [38,39]. In contrast, high-LET RIF have been shown to have an even slower resolution half-life of 5 h [26]. Two possible geometries can be applied for high-LET, with a beam perpendicular to the plate (D), leading to multiple DSB in a single focus per track when visualizing foci or with beam parallel to the plate (E). In the perpendicular configuration, foci frequencies correlate with track traversal (symbolized by RIF ∝ tracks), not DSB. The horizontal configuration (E) leads to visual track with multiple larger foci along it. Such geometry permits evaluation of the number of RIF/μm along the track instead of the classic RIF/nucleus. The slower kinetic for high-LET reflects repair of complex damages as well as clustering of these damages into single foci. One must note here that high energy particles (HZE) are more favorable for such a geometrical configuration, since particles must go through mm to cm of media and plastic. As has been previously described, for lower particle energies, one has to angle slides in such a manner as to allow the beam to hit the bottom of the slide to avoid traversal through large amounts of medium [40,75].
Foci frequencies have been shown to be proportional to the amount of dose delivered to a cell in the low dose range. One study in which γH2AX frequencies were quantified by eye reported a constant value of 35 RIF/Gy 3 min following IR using doses ranging from 1.2 mGy to 2 Gy [29]. There are however many confounding factors affecting an accurate RIF quantification. First, γH2AX occurs as a normal process during the cell cycle as discussed previously. For example, IR induces higher levels of γH2AX in S-phase cells as measured by microscopy [10] but there is less induction per Gy in S-phase cells as compared to G1 cells as have been shown by flow cytometry [17,19]. Second, the sensitivity of the optics used to acquire images and the type of algorithms used to detect a focus or the criteria used to score a RIF by eye can lead to additional discrepancies between labs. Finally, statistical significance is hard to achieve at low doses. For example, in the study previously mentioned [29], ~16–32 RIFs/cell were scored by eye in a total of 400–800 primary human lung MRC-5 fibroblasts exposed to 1.2 mGy, leading to an average of 16/400/1.2e–3 = 33.3 RIF/cell/ Gy. One may question how analysis of such a small number of cells allowed these researchers to detect differences at such low doses. The authors suggest that the very low level of spontaneous foci observed in their cell line allowed them to detect differences at these very low doses (control cells exhibited 0.05 foci/cell, and 0.04 RIF/cell were induced over controls at a 1.2 mGy dose). One recent study circumvented the foci background issue by looking at live cells where one knows exactly how many foci there is before IR allowing identification of true RIF after exposure to IR. In this study [30], these authors also show a linear response following 5 mGy to 1 Gy of low-LET radiation in the human epithelial fibrosarcoma cell line HT1080 stably transfected with 53BP1-YFP 30 min post-IR. Note however that even though this study reported linearity, very different levels of damage were observed depending on the cell line used, (i.e. 16–20 RIF/Gy) for both γH2AX and 53BP1-YFP in HT1080 versus 60 RIF/Gy for a hTERT immortalized normal human bronchial epithelial line (HBECs) that stably express (EGFP)-tagged-53BP1. RIF numbers were determined using the same microscope, the same quantification, and the same optics, again highlighting the importance of cell type in the number of RIF. Such discrepancies clearly weaken the usage of RIF as a pure indicator of DNA DSB, as physics predicts similar numbers of DSB for the same dose and genome size.
In the high dose range, departure from linearity has been observed. For example, we showed γH2AX RIF yields are 30–40% lower between 1 and 3 Gy than below 1 Gy, 1 h post-IR in normal human fibroblasts [10]. Lower RIF yields per Gy at doses larger than 1 Gy may be due in part to a resolution problem, as with higher doses there is more potential for overlapping foci. However we and others have also shown that smaller γH2AX foci are produced following doses greater than 0.5 Gy in the first hour post-irradiation, which should reduce foci overlapping at higher doses [10,23]. In addition, it was recently shown in 18 different normal human fibroblast strains at low doses, that foci were unlikely to overlap, and that RIF yield decreased consistently between 5 and 25 cGy, with averages from 21 to 17 RIF/Gy respectively [13]. These changes are evidently small and difficult to quantify statistically at low dose. However, other methods that do not need to resolve foci such as measuring the total γH2AX intensity per cell or using flow cytometry also suggest saturation at higher doses [23]. In reviewing the data, it is noteworthy that dose response slopes are 2–3 times higher between 0 and 1 Gy than between 1 and 8 Gy for various human and hamster cell lines [23]. In contrast, an other study looking at 10 different cell lines suggested linearity for the total intensity of γH2AX from 1 to 4 Gy, in all lines studied except the human breast line MCF7 and the HT1080 line, which showed lower relative immunofluorescence at 3 and 4 Gy [31]. In this latter study however, it is hard to conclude there was no saturation for the 8 other cell lines as no measurements below 1 Gy were available making it impossible to estimate the initial slope in the low dose range.
Overall, the literature cited here suggests a loss of detection with increasing dose, with clear saturation taking place in general above 1 Gy. As for spontaneous foci, large inter-individual variations were noted in this phenotype as well. Lower yield at higher dose most likely reflects saturation at the kinase level (e.g. lower foci size for higher dose suggests this) and it would be interesting to see if the level of p53 in different cells correlate with sensitivity to saturation, as p53 has been suggested to play a role in modulating the levels of γH2AX in different cells [32].
1.3. DNA damage sensing as a function of radiation quality
High-LET particles deposit energy along their trajectory and therefore present interesting opportunities for studying the spatial organization of RIF. Another distinction between high and low-LET is in the complexity of the generated DSB. For the following discussion we will designate a DSB with one (or more) break(s) within 10 bp as simple DSB, and a DSB with two or more breaks on each strand within 10 bp as a complex DSB. As LET increases, 30% of DSB are simple and 70% are complex [33,34]. In contrast, only 30% of the DSB induced by low-LET are complex. When monolayers of cells are exposed perpendicular to a high-LET particle beam, each impact induces many complex DSB within a very restricted area and the RIF frequencies reflect particle fluence instead of individual DSB [10,27,35]. As illustrated in Fig. 2D, it is difficult to resolve individual foci within the tracks produced when cells are irradiated in this manner due to the much poorer resolution of a microscope along the Z-axis. We have previously shown that RIF formation is faster following exposure to high-LET N ions (132 keV/μm), with a maximum number of foci detected 10–15 min post-IR instead of 30–45 min for low-LET in normal human fibroblasts [10]. In addition, high-LET RIF typically detect 100% of the tracks as shown by us and others [10,35] and their size increases twice as fast as for low-LET, resulting in a three-fold increase during a 2 h period [10]. Since high-LET particles induce more complex DSB, these data suggest that severe lesions seem to induce a faster and more robust RIF formation.
On the other hand, the speed at which RIF are resolved remains unclear at this point as there are contradicting reports. In normal human fibroblasts, we have shown that foci frequency remains high up to 2 h following N ions (150 keV/μm), whereas other investigators have shown in V79 Chinese hamster cells using computer-based analysis that foci loss matches PFGE DSB rates after exposure to alpha particles (3.31 MeV, 120 keV/μm) [27]. However, one could argue that the kinetic response in hamsters is different and perhaps faster. In addition, exposing cells perpendicular to the beam as done in this latter study, complicates the interpretation of the response as the amount of DSB per focus will vary greatly with the shape of the cells, the LET, the atomic number, and the energy of the particle. Differences in foci frequency or kinetics could also be attributed to physical differences in the radiation quality. To circumvent this problem and better resolve damage along high-LET tracks, cells can be irradiated with high-LET particle beams parallel to cell layer [26,36,37]. As illustrated in Fig. 2E, since the XY resolution of a microscope is much higher than the in the Z direction, and cells grown as monolayer are elongated in the XY plane, foci can be differentiated more easily when irradiation is performed this way. Using such a configuration we have been able to show in human epithelial cells exposed to high-LET Fe ions (150 keV/μm) that γH2AX and 53BP1 RIF are maximal as early as 5 min post-IR with maximum (frequencies ~0.7–0.9 RIF/μm along Fe ion tracks) [26]. In addition, we show a half-life of RIF resolution of between 4–6 h, which is much slower than has been reported for PFGE measurements in human glioma cells exposed to 10 Gy of 125 keV/μm N ions with a reported half-life of 2 h [38] or in normal human fibroblast cells in G1 after 80 Gy of 150 keV/μm Fe ions which exhibited a half-life of 3 h [39]. The difference between DSB and γH2AX kinetics is illustrated further in Fig. 2C assuming for illustration purposes a 2.5 h half-life for the theoretical DSB kinetics. Interestingly, studies have shown that increasing LET does not seem to change the number of RIF along a track, as ~0.5–1 γH2AX RIF/μm have been observed following Carbon ions of 200 keV/μm and 0.96 XRCC1 RIF/μm have been noted for Uranium ions at 14,300 keV/μm [40]. Theoretical computations for these high-LET horizontal tracks predicted the number of DSBs/μm to be 1.1, 2.6 and 187 for Fe, C and U respectively IR [26,40]. Thus, as energy deposition along track increases with LET, the number of foci remains the same but more DSBs must be comprised in each one of them, suggesting a mechanism for the high dose saturation previously mentioned for low LET. The slower foci resolution for high-LET may then simply reflect the fact that multiple DSBs are within each focus and thus it takes longer to repair all DSB within one focus.
2. DNA damage response is modulated by chromatin density
Evidence from recent years suggests that chromatin organization mediates the response to DNA damage. The mechanism by which this happens remains unclear, but local chromatin structure appears to play a role. Chromatin decondensation around the DSB is believed to be an important trigger for ATM dimer dissociation and subsequent ATM autophosphorylation and activation [7,41]. Similarly, DSB induce a local higher order chromatin change unmasking methylated lysine 79 on histone H3, which serves as the binding site for 53BP1, a critical DNA repair protein [42]. The phosphorylation of histone H2AX by ATM, ATR and DNA PKcs near the DSB is also a chromatin modification critical to the repair process [6,43]. γH2AX has been proposed to play a major role in chromatin remodeling itself by promoting biochemical interactions between multiple proteins following exposure to radiation [44]. The rapid outward spread of this histone modification from the site of the DSB has recently been suggested to be a key event in homologous recombination during G2 phase [45].
In our recent study on 1 GeV/amu Fe track induced damage [26], we noted that γH2AX, pATM and 53BP1 RIF distribution along a track was not random, and was characterized by a regular spacing of 1.2 μm between consecutive RIF instead of the more likely 0.5 μm spacing predicted by theoretical modeling. Optical properties of the microscope and physical characteristics of Fe ion energy deposition were all taken into account in this theoretical computation. It is noteworthy that the deviation from randomness was significant at the time points measured, 5–30 min post-IR. In addition, simulations also predicted that DSB should be more likely in regions with more DNA (i.e. heterochromatic regions) whereas our experimental results actually showed more RIF in low-DAPI regions (i.e. euchromatin) or at the euchromatin/heterochromatin interfaces. A recent study further confirmed this finding by co-staining with specific markers such as non-histone chromatin protein HP1 and trimethylated-H3K9 [46]. The authors concluded that DSB-inducing agents failed to efficiently generate γH2AX foci in heterochromatin, perhaps due to the epigenetic or packaging properties of the heterochromatin. Similarly, other investigators have shown that detection of γH2AX using CHIP assays was significantly lower on heterochromatic satellite 2 sequences and α-satellite repeats [47].
There are many possible reasons explaining the observed RIF spatial distribution with respect to chromatin density. For example, reactive oxygen species generated by IR may be more efficiently scavenged by the higher concentration of histone in the heterochromatin than in the euchromatin. This would lead to the observed lower number of RIF in the heterochromatin [26,46,47]. Supporting this idea, hypotonic treatments, which lead to the swelling of the nucleus and thus poorer radical scavenging, can induce a 3–5-fold increase in DSB yield [48,49]. Similarly, histone deacetylase (HDAC) inhibitors have gained considerable interest recently in enhancing anti-cancer therapy by increasing the acetylation of core histones, resulting in an open chromatin configuration that is more accessible to DNA-targeting agents. When HDAC inhibitors were used in conjunction with radiation, more γH2AX RIF were induced, they often decayed slower and tumor cells were sensitized to radiation [50–53]. Of course, one could also interpret these results as HDAC participating in the repair directly and not necessarily participating in chromatin relaxation.
The observed RIF spatial distribution may also reveal important distinctions in the way DSB are detected in different chromatin regions. One could hypothesize that only a complex DSB in the heterochromatin leads to a RIF and its formation would require the DSB to first move close enough to the euchromatin. In contrast, any DSB generated in the euchromatin would lead to a rapid induction and resolution of RIF. This would explain why more RIF are generally observed in the euchromatin. Thus, when inducing only simple DSBs one would expect to see fast foci induction. Accordingly, Soutoglou and Misteli observed a fast kinetic of foci formation where breaks were induced by I-SceI endonuclease [54]. In contrast, foci induction would be much slower in the heterochromatin due to the time it takes to move DSB to the interface, and RIF resolution would be much slower due to the complexity of these damages. Such a concept is illustrated in Fig. 3 and indicates the contribution of each type of DSB and chromatin territory in the observed RIF kinetic. In support of this model, recent studies suggest that heterochromatic RIF resolve slowly and that their resolution is ATM-dependent. In addition, observations [55,56] and image quantifications [26] reveal that γH2AX RIF appear preferentially at the periphery of heterochromatic domains rather than within these domains. Physics tells us that preferential location of damage at these interfaces cannot be due to specific deposition of radiation within the nucleus [26], and instead suggests that damages in the heterochromatin may need to be moved towards the euchromatin to be detected and processed. Although the hypothesis of foci movement to the periphery of the heterochomatic domain is highly speculative, and we cannot exclude that other mechanisms prevent the formation of foci in heterochromatin, a relocalization of heterochromatic regions to the periphery of the domain has been previously described for heterochromatin during replication [57]. Moreover, movement of large segments of DNA is not a new concept: it has been shown that some genes become transcriptionally active only upon relocating into open regions of the nucleus [58]. In fact, whole parts of chromosomes have been reported to move over a 1–5 μm distance within a few minutes post transcription activation in mammalian cells [59]. Such movements are hypothetically illustrated in Fig. 3B with arrows indicating the DSB movement from the original position within the heterochromatin (DAPI bright region in the image) to the chromatin interface where detection could take place.
Fig. 3.
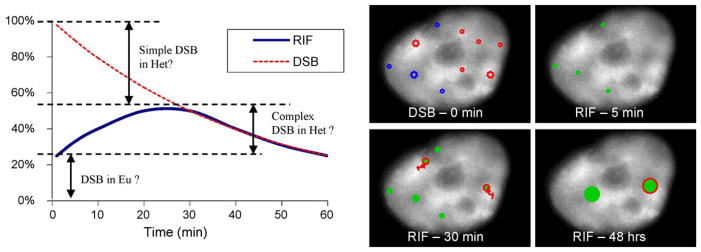
Hypothetical contribution of simple and complex DSB for the classic low-LET RIF kinetic. The left panel repeats the low-LET kinetic curves shown in Fig. 2A with an interpretation of the different types of DSB contributing to the RIF kinetics. The majority of DSB are immediately detected by RIF in the euchromatin (abbreviated Eu) whereas only complex DSB in heterochromatin (abbreviated Het) are detected by RIF and their detection is delayed due to the time it takes to move a DSB to the interface next to euchromatin DNA. The right panel illustrates the kinetic by showing a human cell stained for DAPI with hypothetical regions of DNA damage following IR. Simple DSB are noted as circles and complex DSBs as larger stars. At 0 min, initial damages are shown with blue DSB in low DAPI regions (euchromatin) and red DSB in bright DAPI regions (heterochromatin). At 5 min, only DSB in euchromatin have led to RIF (green full circles), where as complex DSBs in the heterochromatin need to move towards DAPI dim regions as noted by red arrows before being detected at 30 min (shown as green full circles with red edges to note their origin from the heterochromatin). Permanent DNA or chromatin changes are marked by larger RIF sizes at 48 h and are more likely to occur from complex DSB as depicted here.
If the relocalization of heterochromatic DSBs to the periphery of the compact domain turns out to be true, it would be extremely important to establish which mechanism is responsible for this movement. Is it possible, for example, that some sensors of the damage are able to detect it and actively promote its movement at the periphery of the heterochromatic region, despite the compaction of heterochromatin? Would that imply that the involved repair pathways are different in the euchromatin and the heterochromatin? It is also possible that heterochromatin relaxation, occurring in the presence of DSBs, allows the increasing mobility of heterochromatic DNA thus facilitating the stochastic movement of DSBs to a more peripheric area, where they are ultimately detected. Some data suggest that at least some of the DSB sensors are indeed able to access heterochromatin for detecting the lesions. For example, in human cells, the induction of DSBs with laser directed toward the heterochromatic domain results in local HP1b phosphorylation by the casein kinase within 5 min, suggesting that this kinase can access the compact heterochromatic domain [60]. Similarly, the evidence that ATM-phosphorylates Kap1 six min post-IR, which is important for promoting heterochromatin relaxation, implies that the original break in heterochromatin is able to trigger local ATM activity before the relaxation occurs [55,56].
It is interesting to note that recent reports show that HP1, which is known to play a role in stabilizing heterochromatin compaction, is also recruited to laser-induced DSBs. This might suggest the importance of rendering the DSB site less ‘movable’ for facilitating accurate repair [61,62]. In this view, the more rigid structure of heterochromatin might intrinsically protect it from inaccurate repair by stably tethering DNA ends allowing them to stay in place. Such an idea has been supported by reports showing that chromosomal aberrations occur preferentially in the euchromatin and not in the heterochromatin [63]. Bailey and Bedford recently reviewed this topic and summarized a number of studies where radiation-induced translocations were less frequent in the condensed inactive X-chromosome than in its active counterpart [64].
3. Persistent RIF: unrepaired DSB, mis-repaired DNA and/or permanent chromatin modification?
DSB repair measured by electrophoresis or neutral filter-elution [24] has a rapid component (5–30 min half-life), and a slower component likely related to the resolution of complex DSB (4–10 h half-life). Taking into account that ~30% of low-LET radiation DSB are complex DSB [33,34], one would therefore predict that out of the 25 DSB/Gy generated by low-LET [25] about 17 DSB/Gy would be repaired by fast kinetics, and 8 DSB/Gy by a slower kinetics. Thus at 24 h post-IR, we would expect to have at most 1 DSB/Gy still undergoing repair. Although this single or small number of RIF would hard to accurately measure because of spontaneous foci within a cell, several studies have observed persistent foci days following low-LET exposures. For example, normal human fibroblasts irradiated with low-LET showed persistent foci for 5 days following 4 Gy of X-rays even though by that time all DSB should be fully resolved [65]. These persistent γH2AX RIF were large and co-localized with ATMp as well as with p53 phosphorylated at serine 15, suggesting ongoing processing. Rather than concluding that RIF represented un-repaired DSB, these authors concluded that persistent foci were revealing a chromatin alteration, which resulted in the induction of a senescence-like growth arrest following IR. Senescence-like growth arrest is a p53-dependent irreversible G1 arrest thought to suppress radiation-induced telomere dysfunction following genomic instability in fibroblasts. This same group also showed that persistent RIF detected on complete intact metaphase chromosomes 96 h following low-LET exposure, suggesting again that these foci may indicate an aberrant chromatin structure due to illegitimate rejoining [66]. More recently they extended their work to show that large foci have a role in triggering G1 arrest: the larger the foci, the brighter the p53 phosphorylation, the more likely cells would arrest [11].
Other studies looking at earlier time points (2–12 h) also showed that γH2AX RIF could still be observed even at time points when other methods of DSB quantification, such as quantification of chromosomal breaks in Giemsa stained metaphase spreads or PFGE analysis, suggest that repair is complete [21,67]. Even though one might consider such results as a proof of the much greater sensitivity of RIF for detecting DSB, this may also indicate that persistent RIF may not necessarily mark DSB. Similarly, mitotic nuclei exposed to IR have a slower rate of γH2AX foci loss than DSB loss as measured by PFGE [28], likely due to the fact the heterochromatic RIF take longer to resolve [55]. Therefore large persistent foci may be the result of damages occurring in denser regions of the chromatin leading to permanent structural changes.
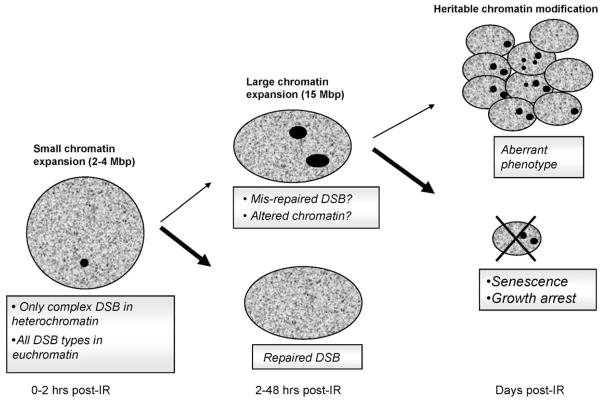
Fig. 4 summarizes the possible fate of cells within the first 48 h post-IR. As previously discussed, the most likely outcome for a fibroblast with persistent foci would be growth arrest. Growth arrest might also be the most likely outcome when RIF are marking sites of permanent DNA damage. On the other hand, when DNA has been fully repaired but the repair process has led to permanent changes in the chromatin structure, leading to persistent RIF, there seems to be no reason for the cell to stop dividing. In fact, RIF have been shown to be replicated in daughter cells [68] thus conserving the chromatin architecture. For example, it was shown in CHO cells stably expressing GFP-histone H2B, that GFP photobleaching patterns could be replicated in daughter cells, suggesting histones were equally segregated at the same nuclear positions in each daughter cell during mitosis [16]. In this study, the authors concluded that duplication of chromatin pattern might be an epigenetic mechanism to maintain cell differentiation. Therefore, if we assume large persistent RIF are marking altered organization of the chromatin, these marks may also relate in some instances to a persistent altered epigenetic programs leading to heritable altered phenotypes.
Fig. 4.
RIF formation/resolution and cell fate. Boxed legends indicate what type of damages foci mark. Bold text indicates corresponding chromatin status for each of these foci types. Small arrows in the flow chart indicate lower probability of events to take place based on discussion in the text. For example, cells with persistent RIF related to unrepaired DNA will most likely be eliminated (large arrow, cross). On the other hand, when a RIF marks chromatin changes where DNA damage was repaired successfully, there should be no obstacles for a cell to resume division (small arrow) allowing replication of its aberrant chromatin.
In summary, euchromatin damages are repaired faster but may lead to more chromosomal rearrangements, whereas, damages in the heterochromatin may be more accurately repaired but may lead to irreversible chromatin structural changes. We hypothesize that the more condensed the chromatin is, the more reorganization it will undergo during repair, and the more likely permanent changes of chromatin structure will be observed. Indeed, the slower RIF kinetics in mitotic DNA or heterochromatic regions previously discussed suggest that chromatin modifications have difficulty forming in dense regions of the DNA. In addition, RIF in mammalian cells have been estimated by gel electrophoresis to cover 1–4 Mbp of DNA at early times post exposure [6], with maximum sizes of 15–30 Mbp being reached at 2 h post-IR [10,18,69,70]. We may then wonder what is more deleterious: the loss of a few kbp of DNA, or epigenetic alterations over tens of Mbp? In the former case, mis-repaired DNA rarely leads to deleterious effects (e.g. ~10−5 to 10−6 mutations/cell/Gy for HPRT locus [71]), whereas changes of chromatin architecture over Mbp will definitely have an impact on the transcriptome of a cell.
Thinking of chromatin as a target of ionizing radiation and permanent chromatin alterations as a mark detectable by persistent RIF opens the door to an unexplored mechanism for radiation-induced phenotypes. For example, epigenetic changes marked by persistent RIF could be another factor influencing radiation-induced genomic instability. As reviewed by various investigators [72,73], a mis-repaired DSB is typically considered to be an important potential inducer of genomic instability. However a large study on a panel of NCI-60 tumor cell lines correlated the frequency of chromosomal rearrangements with γH2AX foci frequency and concluded that chromatin instability might be partially responsible for the high foci frequencies in tumor lines [74]. In addition, the relationship between mutations (i.e. a measure of mis-repaired DSB) and genomic instability is difficult to reconcile, given that there is a large discrepancy between the very small rate of DNA mutation induced by low-LET and the high yield of radiation-induced genomically unstable cells (10%) [72]. If DNA mutations were the cause of genomic instability, one should observe much lower frequencies of genomic instability. On the other hand, with a reported persistent RIF frequency between 30 and 40%, 24–120 h following low-LET exposure in normal fibroblasts [65,66], persistent changes in chromatin marked by RIF match more closely the rates of genomic instability. This is an interesting speculation that should be further investigated.
4. Conclusion
The assumption that RIF only reflect the presence of a DSB has caused a number of misconceptions in the field of radiation biology, as scientists often refer to them as a DSB when in fact they are only marks of chromatin modifications. It is our hope that we have provided evidence to indicate that damages other than DSB, such as architectural changes in the chromatin can result in RIF. This manuscript emphasized the importance of not equating RIF with DSB in all situations and showed how dose and time dependence of RIF frequency is inconsistent with a one to one equivalence.
As summarized in Fig. 4, we tried to reconcile data from the literature by adding chromatin status as a main factor in the foci response. Briefly, upon irradiation, DSB are generated and cause immediate chromatin decondensation in euchromatin and rapid formation of RIF. In heterochromatin, the packaging of DNA moderates this response and only complex breaks elicit RIF which have slower formation and resolution. In addition, for doses larger than 1 Gy, or after exposure to high-LET, RIF most likely reflect clusters of multiple DSBs and RIF remain longer in the nucleus. If repair has failed or has led to chromatin alterations that cannot be restored, the mechanical forces signaling DNA sensing proteins remain active leading to persistent RIF. Persistent RIF or large foci seen spontaneously in non-irradiated cell lines may reflect regions where chromatin architecture is damaged or is undergoing remodeling. In fibroblasts, such alterations have been linked to permanent growth arrest. On the other hand, one could hypothesize that if DNA has been repaired but chromatin organization could not be restored, a cell would resume its cell cycle allowing replication of RIF. Therefore, heritable persistent RIF spanning over tens of Mbp may affect the transcriptome of a large progeny of cells leading to the emergence of new and stable phenotypes. Such a model opens the door to a “non-DNA-centric view” of radiation-induced phenotypes.
Acknowledgments
We apologize to all authors whose publications have not been cited due to space limitation and conciseness of the manuscript. We would like to thank Dr. G. Karpen, Dr. P. Olive, Dr. B. Rydberg and Dr. T. Groesser for their useful comments on the manuscript and their engaging scientific discussions. We also thank the National Aeronautics and Space Administration, which sponsors our research on RIF under grant no. T6275W, NASA Specialized Center for Research in Radiation Health Effects.
Abbreviations
- RIF
radiation-induced foci
- DSB
double strand break
- IR
ionizing radiation
- Post-IR
following exposure to ionizing radiation
- ATM
Ataxia telangiectasia mutated
- ATMp
ATM phosphorylated at serine 1981
- γ-H2AX
histone H2AX phosphorylated at serine 139
- 53BP1
p53 binding protein 1
- PFGE
pulse field gel electrophoresis
- LET
linear energy transfer (typical unit: keV/μm)
- HZE
ions with high energy and high atomic number
References
- 1.Haaf T, Golub EI, Reddy G, Radding CM, Ward DC. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci USA. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashley T, Plug AW, Xu J, Solari AJ, Reddy G, Golub EI, Ward DC. Dynamic changes in Rad51 distribution on chromatin during meiosis in male and female vertebrates. Chromosoma. 1995;104:19–28. doi: 10.1007/BF00352222. [DOI] [PubMed] [Google Scholar]
- 3.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 Binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maser RS, Monsen KJ, Nelms BE, Petrini JH. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- 6.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 7.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 8.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, Chen DJ. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, Botvinick E, Qin J, Chen DJ. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem. 2005;280:14709–14715. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- 10.Costes SV, Boissiere A, Ravani S, Romano R, Parvin B, Barcellos-Hoff MH. Imaging features that discriminate between foci induced by high- and low-LET radiation in human fibroblasts. Radiat Res. 2006;165:505–515. doi: 10.1667/RR3538.1. [DOI] [PubMed] [Google Scholar]
- 11.Yamauchi M, Oka Y, Yamamoto M, Niimura K, Uchida M, Kodama S, Watanabe M, Sekine I, Yamashita S, Suzuki K. Growth of persistent foci of DNA damage checkpoint factors is essential for amplification of G1 checkpoint signalling. DNA Rep (Amst) 2008;7:405–417. doi: 10.1016/j.dnarep.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Sedelnikova OA, Horikawa I, Zimonjic DB, Popescu NC, Bonner WM, Barrett JC. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat Cell Biol. 2004;6:168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- 13.Wilson PF, Nham PB, Urbin SS, Hinz JM, Jones IM, Thompson LH. Inter-individual variation in DNA double-strand break repair in human fibroblasts before and after exposure to low doses of ionizing radiation. Mutat Res. 2010;683:91–97. doi: 10.1016/j.mrfmmm.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 14.Han J, Hendzel MJ, Allalunis-Turner J. Quantitative analysis reveals asynchronous and more than DSB-associated histone H2AX phosphorylation after exposure to ionizing radiation. Radiat Res. 2006;165:283–292. doi: 10.1667/rr3516.1. [DOI] [PubMed] [Google Scholar]
- 15.McManus KJ, Hendzel MJ. ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol Biol Cell. 2005;16:5013–5025. doi: 10.1091/mbc.E05-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Essers J, van Cappellen WA, Theil AF, van Drunen E, Jaspers NG, Hoeij-makers JH, Wyman C, Vermeulen W, Kanaar R. Dynamics of relative chromosome position during the cell cycle. Mol Biol Cell. 2005;16:769–775. doi: 10.1091/mbc.E04-10-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacPhail SH, Banath JP, Yu Y, Chu E, Olive PL. Cell cycle-dependent expression of phosphorylated histone H2AX: reduced expression in unirradiated but not X-irradiated G1-phase cells. Radiat Res. 2003;159:759–767. doi: 10.1667/rr3003. [DOI] [PubMed] [Google Scholar]
- 18.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whalen MK, Gurai SK, Zahed-Kargaran H, Pluth JM. Specific ATM-mediated phosphorylation dependent on radiation quality. Radiat Res. 2008;170:353–364. doi: 10.1667/RR1354.1. [DOI] [PubMed] [Google Scholar]
- 20.Anderson L, Henderson C, Adachi Y. Phosphorylation and Rapid Relocalization of 53BP1 to Nuclear foci upon DNA Damage. Mol Cell Biol. 2001;21:1719–1729. doi: 10.1128/MCB.21.5.1719-1729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forand A, Dutrillaux B, Bernardino-Sgherri J. Gamma-H2AX expression pattern in non-irradiated neonatal mouse germ cells and after low-dose gamma-radiation: relationships between chromatid breaks and DNA double-strand breaks. Biol Reprod. 2004;71:643–649. doi: 10.1095/biolreprod.104.027466. [DOI] [PubMed] [Google Scholar]
- 22.Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacPhail SH, Banath JP, Yu TY, Chu EH, Lambur H, Olive PL. Expression of phosphorylated histone H2AX in cultured cell lines following exposure to X-rays. Int J Radiat Biol. 2003;79:351–358. doi: 10.1080/0955300032000093128. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Zeng ZC, Bui TA, Sonoda E, Takata M, Takeda S, Iliakis G. Efficient rejoining of radiation-induced DNA double-strand breaks in vertebrate cells deficient in genes of the RAD52 epistasis group. Oncogene. 2001;20:2212–2224. doi: 10.1038/sj.onc.1204350. [DOI] [PubMed] [Google Scholar]
- 25.Stenerlow B, Karlsson KH, Cooper B, Rydberg B. Measurement of prompt DNA double-strand breaks in mammalian cells without including heat-labile sites: results for cells deficient in nonhomologous end joining. Radiat Res. 2003;159:502–510. doi: 10.1667/0033-7587(2003)159[0502:mopdds]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 26.Costes SV, Ponomarev A, Chen JL, Nguyen D, Cucinotta FA, Barcellos-Hoff MH. Image-based modeling reveals dynamic redistribution of DNA damage into nuclear sub-domains. PLoS Comput Biol. 2007;3:e155. doi: 10.1371/journal.pcbi.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leatherbarrow EL, Harper JV, Cucinotta FA, O’Neill P. Induction and quantification of gamma-H2AX foci following low and high LET-irradiation. Int J Radiat Biol. 2006;82:111–118. doi: 10.1080/09553000600599783. [DOI] [PubMed] [Google Scholar]
- 28.Kato TA, Okayasu R, Bedford JS. Comparison of the induction and disappearance of DNA double strand breaks and gamma-H2AX foci after irradiation of chromosomes in G1-phase or in condensed metaphase cells. Mutat Res. 2008;639:108–112. doi: 10.1016/j.mrfmmm.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low X-ray doses. Proc Natl Acad Sci USA. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asaithamby A, Chen DJ. Cellular responses to DNA double-strand breaks after low-dose gamma-irradiation. Nucleic Acids Res. 2009;37:3912–3923. doi: 10.1093/nar/gkp237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahrhofer H, Burger S, Oppitz U, Flentje M, Djuzenova CS. Radiation induced DNA damage and damage repair in human tumor and fibroblast cell lines assessed by histone H2AX phosphorylation. Int J Radiat Oncol Biol Phys. 2006;64:573–580. doi: 10.1016/j.ijrobp.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka T, Kurose A, Huang X, Traganos F, Dai W, Darzynkiewicz Z. Extent of constitutive histone H2AX phosphorylation on Ser-139 varies in cells with different TP53 status. Cell Prolif. 2006;39:313–323. doi: 10.1111/j.1365-2184.2006.00387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikjoo H, O’Neill P, Goodhead DT, Terrissol M. Computational modelling of low-energy electron-induced DNA damage by early physical and chemical events. Int J Radiat Biol. 1997;71:467–483. doi: 10.1080/095530097143798. [DOI] [PubMed] [Google Scholar]
- 34.Nikjoo H, O’Neill P, Wilson WE, Goodhead DT. Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat Res. 2001;156:577–583. doi: 10.1667/0033-7587(2001)156[0577:cafdts]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 35.Karlsson KH, Stenerlow B. Focus formation of DNA repair proteins in normal and repair-deficient cells irradiated with high-LET ions. Radiat Res. 2004;161:517–527. doi: 10.1667/rr3171. [DOI] [PubMed] [Google Scholar]
- 36.Jakob B, Scholz M, Taucher-Scholz G. Biological imaging of heavy charged-particle tracks. Radiat Res. 2003;159:676–684. doi: 10.1667/0033-7587(2003)159[0676:biohct]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 37.Desai N, Davis E, O’Neill P, Durante M, Cucinotta FA, Wu H. Immunofluores-cence detection of clustered gamma-H2AX foci induced by HZE-particle radiation. Radiat Res. 2005;164:518–522. doi: 10.1667/rr3431.1. [DOI] [PubMed] [Google Scholar]
- 38.Stenerlow B, Blomquist E, Grusell E, Hartman T, Carlsson J. Rejoining of DNA double-strand breaks induced by accelerated nitrogen ions. Int J Radiat Biol. 1996;70:413–420. doi: 10.1080/095530096144888. [DOI] [PubMed] [Google Scholar]
- 39.Lobrich M, Cooper PK, Rydberg B. Joining of correct and incorrect DNA ends at double-strand breaks produced by high-linear energy transfer radiation in human fibroblasts. Radiat Res. 1998;150:619–626. [PubMed] [Google Scholar]
- 40.Jakob B, Splinter J, Taucher-Scholz G. Positional stability of damaged chromatin domains along radiation tracks in mammalian cells. Radiat Res. 2009;171:405–418. doi: 10.1667/RR1520.1. [DOI] [PubMed] [Google Scholar]
- 41.Zgheib O, Huyen Y, DiTullio RA, Jr, Snyder A, Venere M, Stavridi ES, Halazonetis TD. ATM signaling and 53BP1. Radiother Oncol. 2005;76:119–122. doi: 10.1016/j.radonc.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 42.Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 43.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 44.Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- 45.Lowndes NF, Toh GW. DNA repair: the importance of phosphorylating histone H2AX. Curr Biol. 2005;15:R99–R102. doi: 10.1016/j.cub.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 46.Cowell IG, Sunter NJ, Singh PB, Austin CA, Durkacz BW, Tilby MJ. gamma-H2AX foci form preferentially in euchromatin after ionising-radiation. PLoS ONE. 2007;2:e1057. doi: 10.1371/journal.pone.0001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karagiannis TC, Harikrishnan KN, El-Osta A. Disparity of histone deacetylase inhibition on repair of radiation-induced DNA damage on euchromatin and constitutive heterochromatin compartments. Oncogene. 2007;26:3963–3971. doi: 10.1038/sj.onc.1210174. [DOI] [PubMed] [Google Scholar]
- 48.Warters RL, Lyons BW. Variation in radiation-induced formation of DNA double-strand breaks as a function of chromatin structure. Radiat Res. 1992;130:309–318. [PubMed] [Google Scholar]
- 49.Nygren J, Ljungman M, Ahnstrom G. Chromatin structure and radiation-induced DNA strand breaks in human cells: soluble scavengers and DNA-bound proteins offer a better protection against single- than double-strand breaks. Int J Radiat Biol. 1995;68:11–18. doi: 10.1080/09553009514550861. [DOI] [PubMed] [Google Scholar]
- 50.Munshi A, Tanaka T, Hobbs ML, Tucker SL, Richon VM, Meyn RE. Vorinostat, a histone deacetylase inhibitor, enhances the response of human tumor cells to ionizing radiation through prolongation of gamma-H2AX foci. Mol Cancer Ther. 2006;5:1967–1974. doi: 10.1158/1535-7163.MCT-06-0022. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Adachi M, Zou H, Hareyama M, Imai K, Shinomura Y. Histone deacetylase inhibitors enhance phosphorylation of histone H2AX after ionizing radiation. Int J Radiat Oncol Biol Phys. 2006;65:859–866. doi: 10.1016/j.ijrobp.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 52.Camphausen K, Burgan W, Cerra M, Oswald KA, Trepel JB, Lee MJ, Tofilon PJ. Enhanced radiation-induced cell killing and prolongation of gammaH2AX foci expression by the histone deacetylase inhibitor MS-275. Cancer Res. 2004;64:316–321. doi: 10.1158/0008-5472.can-03-2630. [DOI] [PubMed] [Google Scholar]
- 53.Banuelos CA, Banath JP, MacPhail SH, Zhao J, Reitsema T, Olive PL. Radio-sensitization by the histone deacetylase inhibitor PCI-24781. Clin Cancer Res. 2007;13:6816–6826. doi: 10.1158/1078-0432.CCR-07-1126. [DOI] [PubMed] [Google Scholar]
- 54.Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with hetero-chromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 56.Goodarzi AA, Noon AT, Jeggo PA. The impact of heterochromatin on DSB repair. Biochem Soc Trans. 2009;37:569–576. doi: 10.1042/BST0370569. [DOI] [PubMed] [Google Scholar]
- 57.Quivy JP, Gerard A, Cook AJ, Roche D, Almouzni G. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat Struct Mol Biol. 2008;15:972–979. doi: 10.1038/nsmb.1470. [DOI] [PubMed] [Google Scholar]
- 58.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 59.Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 60.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 61.Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, Warmerdam DO, Lindh M, Brink MC, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–586. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zarebski M, Wiernasz E, Dobrucki JW. Recruitment of heterochromatin protein 1 to DNA repair sites. Cytometry A. 2009;75:619–625. doi: 10.1002/cyto.a.20734. [DOI] [PubMed] [Google Scholar]
- 63.Obe G, Pfeiffer P, Savage JR, Johannes C, Goedecke W, Jeppesen P, Natarajan AT, Martinez-Lopez W, Folle GA, Drets ME. Chromosomal aberrations: formation, identification and distribution. Mutat Res. 2002;504:17–36. doi: 10.1016/s0027-5107(02)00076-3. [DOI] [PubMed] [Google Scholar]
- 64.Bailey SM, Bedford JS. Studies on chromosome aberration induction: what can they tell us about DNA repair? DNA Repair (Amst) 2006;5:1171–1181. doi: 10.1016/j.dnarep.2006.05.033. [DOI] [PubMed] [Google Scholar]
- 65.Suzuki M, Suzuki K, Kodama S, Watanabe M. Interstitial chromatin alteration causes persistent p53 activation involved in the radiation-induced senescence-like growth arrest. Biochem Biophys Res Commun. 2006;340:145–150. doi: 10.1016/j.bbrc.2005.11.167. [DOI] [PubMed] [Google Scholar]
- 66.Suzuki M, Suzuki K, Kodama S, Watanabe M. Phosphorylated histone H2AX foci persist on rejoined mitotic chromosomes in normal human diploid cells exposed to ionizing radiation. Radiat Res. 2006;165:269–276. doi: 10.1667/rr3508.1. [DOI] [PubMed] [Google Scholar]
- 67.Markova E, Schultz N, Belyaev IY. Kinetics and dose-response of residual 53BP1/ gamma-H2AX foci: co-localization, relationship with DSB repair and clonogenic survival. Int J Radiat Biol. 2007;83:319–329. doi: 10.1080/09553000601170469. [DOI] [PubMed] [Google Scholar]
- 68.Klokov D, MacPhail SM, Banath JP, Byrne JP, Olive PL. Phosphorylated histone H2AX in relation to cell survival in tumor cells and xenografts exposed to single and fractionated doses of X-rays. Radiother Oncol. 2006;80:223–229. doi: 10.1016/j.radonc.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 69.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pilch DR, Sedelnikova OA, Redon C, Celeste A, Nussenzweig A, Bonner WM. Characteristics of gamma-H2AX foci at DNA double-strand breaks sites. Biochem Cell Biol. 2003;81:123–129. doi: 10.1139/o03-042. [DOI] [PubMed] [Google Scholar]
- 71.Costes S, Sachs R, Hlatky L, Vannais D, Waldren C, Fouladi B. Large-mutation spectra induced at hemizygous loci by low-LET radiation: evidence for intrachro-mosomal proximity effects. Radiat Res. 2001;156:545–557. doi: 10.1667/0033-7587(2001)156[0545:lmsiah]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 72.Little JB. Radiation carcinogenesis. Carcinogenesis. 2000;21:397–404. doi: 10.1093/carcin/21.3.397. [DOI] [PubMed] [Google Scholar]
- 73.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 74.Yu T, MacPhail SH, Banath JP, Klokov D, Olive PL. Endogenous expression of phosphorylated histone H2AX in tumors in relation to DNA double-strand breaks and genomic instability. DNA Rep (Amst) 2006;5:935–946. doi: 10.1016/j.dnarep.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 75.Jakob B, Splinter J, Durante M, Taucher-Scholz G. Live cell microscopy analysis of radiation-induced DNA double-strand break motion. Proc Natl Acad Sci USA. 2009;106:3172–3177. doi: 10.1073/pnas.0810987106. [DOI] [PMC free article] [PubMed] [Google Scholar]