Summary
Poly(N-isopropyl acrylamide) (pNIPAM) undergoes a sharp property change in response to a moderate thermal stimulus at physiological temperatures. In this work, we constructed a radio frequency (RF) plasma reactor for the plasma polymerization of pNIPAM. RF deposition is a method that coats surfaces of any geometry producing surfaces that are sterile and uniform, making this technique useful for forming biocompatible films. The films generated are characterized using X-ray photoelectron spectroscopy (XPS), contact angles, cell culture, and interferometry. We find that a plasma with a decreasing series of power settings (i.e., from 100W to 1W) at a pressure of 140 millitorr yields the most favorable results.
Keywords: biocompatibility, cell culture, contact angle (CA), ESCA/XPS, plasma polymerization
Introduction
Stimulus-responsive polymers (SRPs) are materials that undergo reversible changes in their optical, mechanical, electronic, or other properties in response to environmental cues (e.g., changes in ionic strength,[1] pH,[2] light,[3] electric field,[4] and temperature[5]). Often referred to as “smart” materials, SRPs have found applications in such diverse areas as catalysis,[6] controlled drug release,[7] biosensors,[8] non-fouling surfaces,[9] chemical separations,[10] and tissue engineering.[11] One of the most widely used SRPs is poly(N-isopropyl acrylamide), which is referred to as pNIPAM in this text. In water, pNIPAM has a lower critical solution temperature (LCST) of ~32 °C. Above the LCST, pNIPAM is dehydrated, its solubility decreases dramatically, and the polymer undergoes phase separation.[12] When tethered to a substrate, pNIPAM films undergo a similar change in conformation: above the LCST, the pNIPAM film dehydrates (becomes relatively hydrophobic), and is in a condensed form. Below the LCST, the pNIPAM film rapidly hydrates (becomes relatively hydrophilic), and is in a more expanded conformation.[13]
Since the LCST of pNIPAM (~30–32 °C) is near physiological temperature (37 °C), it has gained interest in the biomedical field. In particular, this change around physiological temperature can be utilized for reversible cell and protein adhesion. There are many applications for reversible cell and protein adhesion which include biofouling (from bacteria[14] and proteins[15]), biological sensors,[16] and tissue engineering.[17–19] For tissue engineering applications, cells are cultured to confluence on a surface coated with pNIPAM. When the temperature is decreased to below the LCST (e.g. room temperature) this causes the cells to release from the surface as an intact cell sheet. Recently, the use of pNIPAM for cell sheet engineering and other biological applications were reviewed by Cooperstein, et al.[20]
Depending on the application, a variety of techniques have been used to deposit pNIPAM onto surfaces. Some of the techniques that have been utilized include: grafting using UV[21,22] or electron beam irradiation,[18,23,24] atom transfer radical polymerization (ATRP),[25–27] solution deposition,[28–30] and vapor-phase plasma polymerization.[17,31] In this work, the technique used to deposit pNIPAM films is plasma polymerization of NIPAM (ppNIPAM). One of the primary advantages of plasma polymerization of NIPAM from the vapor-phase is that surfaces of any chemistry and geometry may be used for deposition. In addition, the technique is sterile, which is important for cell culture applications. Plasma deposition has previously been used to fabricate a number of biocompatible films (e.g., poly(2-hydroxyethyl methacrylate) (pHEMA), acrylic acid (AA), etc.).[32–38]
The construction and characterization of an RF plasma reactor for the deposition of plasma polymerized pNIPAM (ppNIPAM) substrates to be used for tissue engineering and cell culture is presented. After describing the specifications for the reactor and its typical mode of operation, we outline the parameters varied to optimize the films deposited, including monomer use, sample position, flow, pressure, and power.
Experimental Part
Sample Preparation
For all samples used in surface analysis (i.e., by XPS, interferometry, and contact angle measurement), silicon wafers from Siltec Corp. (Salem, OR) were diced into small pieces. 1 cm × 1 cm squares were used for XPS analysis and 0.8 cm×3 cm pieces were used for interferometry and contact angle measurements. The surfaces were cleaned in an ultrasonic cleaner from VWR International (West Chester, PA) twice in each of the following solutions for 5 minutes: dichloromethane, acetone, and methanol from Honeywell Burdick & Jackson (Muskegon, MI). For cell detachment studies, 35 mm Nunc (Rochester, NY) Petri dishes were coated.
X-ray Photoelectron Spectroscopy (XPS)
XPS spectra were acquired using a Surface Science Instruments S-Probe equipped with a monochromatized aluminum Kα X-ray source, an electron flood gun (for charge neutralization), and a hemispherical electron energy analyzer. Three replicates of each sample were analyzed with three survey spectra and one high-resolution spectrum per sample. For compositional analysis, survey scans were acquired at a pass energy of 150eV and high-resolution carbon scans were acquired at a pass energy of 50eV. Data analysis was performed on the Service Physics ESCA 2000A data reduction software. To account for binding energy shifts inherent to insulating samples, the binding energies for the high-resolution C(1s) spectrum were referenced to the C-C/C-H peak at 285eV. High-resolution carbon spectra were peak fitted using the minimum number of peaks possible to obtain random residuals. The peaks were fitted using a 100% Gaussian line shape, with a Shirley function as the background model.
Contact Angles
Contact angles were obtained to determine the surface wettability of ppNIPAM deposited on Si as well as on a Si control chip. The method is quantitative as it yields an angle measurement. These experiments utilized a technique known as captive (or inverted) bubble. An Advanced Goniometer Model #300- UPG equipped with an environmental chamber purchased from ramé-hart instrument co. (Mountain Lakes, NJ) was used. The quartz cell filled with ultra pure water (18 M^) from a Millipore Academic unit (Billerica, MA) is placed in the environmental chamber, which is connected to a temperature controller. The temperature controller was used in order to obtain measurements at room temperature and 37 °C. Measurements were taken using the program DROPimage standard (Mountain Lakes, NJ). Both right and left angles were obtained on three areas of three surfaces.
Cell Culture
For cell detachment studies, bovine aortic endothelial cells (BAECs) from Genlantis were cultured in Dulbecco's Modified Eagle's Medium (DMEM) from Cellgro. The DMEM was supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, 4.5 g/L glucose, 0.1 mM MEM nonessential amino acids, and 1 mM MEM sodium pyruvate. Cells were incubated at 37 °C in a humid atmosphere with 5% CO2. When confluent, the cells were washed with HyClone Dulbecco’s phosphate buffered saline (DPBS) and lifted from culture flasks with 0.25% trypsin-EDTA from Gibco. Cells were then seeded onto three 35mm Petri dishes coated with ppNIPAM and onto three wells of a six well plate (the three remaining wells were used as blank controls). After the cells became confluent, the media was replaced with 4 °C serum free media and placed on a rotator platform for 2 hours. These conditions have been demonstrated to yield cell detachment from pNIPAM in previous work.[29] Three pictures of each surface were obtained, using a camera attached to a light microscope (Nikon F100, Melville, NY), before the media was replaced and after the two hours. The number of cells in each picture were counted to determine the percentage of cells that detached.
Film Thickness
Film thickness was determined using a NanoSpec Model 210 from Nanometrics Incorporated (Milpitas, CA). The instrument uses optical interferometry to determine the film thickness by measuring the light that is reflected off a surface. It is a non-contact and non-destructive technique to measure film thickness. It has been reported that the refractive index of pNIPAM is similar to polystyrene,[39] so a value of 1.59[40] was used for all measurements. For films that were 500Å or less, program 7: thin oxide on silicon, was used. Reflectance data are taken at 520nm and then compared to the reflectance of bare Si. Program 1 was used for surfaces over 500Å. This program scans using light from 480–800 nm and an average thickness is calculated. Three measurements were taken on three surfaces.
Reactor Design
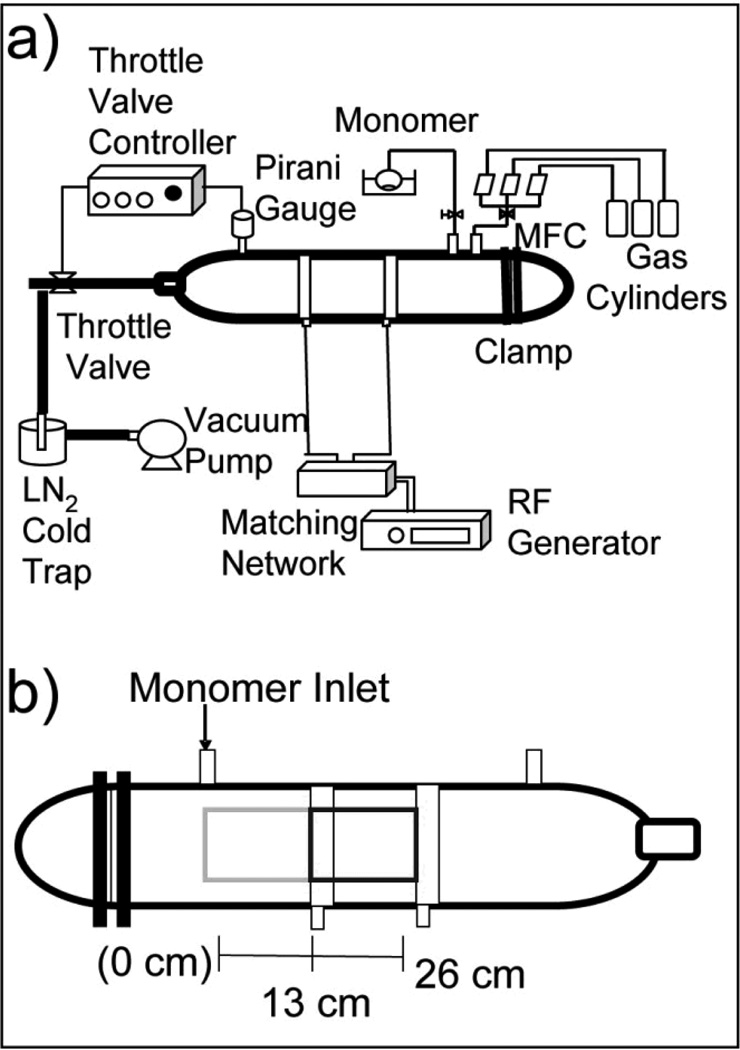
The reactor chamber is 30” long with a 4 ½” diameter. It is made of glass, fabricated to our design specifications by Scientific Glass (Albuquerque, NM). Connected to the chamber is a gas feed line that is connected to an MKS Instruments, Inc. (Andover, MA) and a Unit Instruments Inc. (Yorba Linda, CA) mass flow controller. These mass flow controllers are connected to argon (>99%) from Argyle Welding Supply Co. Inc. (Albuquerque, NM), methane (99.97%) from Matheson Tri-Gas (Parsippany, NJ), and oxygen (99.0%) from Argyle Welding Supply Co. Inc. gas tanks. To control the oxygen and argon flow, a Vacuum General Inc. (San Diego, CA) DynaMass controller is used. To control the methane flow, an MKS Instruments, Inc. type 260 controller is used. In order to read and maintain pressure, a Kurt J Lesker (Clairton, PA) 945 Pirani pressure sensor and Series 345 Pirani gauge is connected to a MKS Instruments, Inc. Type 252A exhaust valve controller, which controls a MKS control valve. (See Figure 1) The monomer flow is controlled via a Swagelok (Solon, OH) needle valve. At the end of the reactor, there is a Nor-Cal Products, Inc. (Yreka, CA) liquid nitrogen cold trap, which is used to condense any organics before they reach the pump. The whole system is evacuated to a pressure of 10−3 Torr using an Alcatel Vacuum Products, Inc. (Hingham, MA) model 2033 mechanical pump.
Figure 1.
(A) Schematic of the plasma reactor that was constructed and characterized for ppNIPAM deposition. (B) Schematic of the reactor chamber, illustrating sample locations with respect to the monomer inlet.
To spark a plasma in the chamber, two 2.5 cm copper electrodes are connected to a Dressler (Stolberg, Germany) matching network and a Cesar radio frequency (RF) power generator from Advanced Energy (Fort Collins, CO). The electrode farthest from the monomer inlet is the grounded electrode.
Reactor Operation (ppNIPAM Deposition)
Before deposition, the chamber was cleaned using a 40W oxygen plasma etch for 30 minutes. The oxygen flow was adjusted to maintain a pressure of 8–9×10−2 Torr. The surfaces were then loaded into the reactor at either the monomer inlet (i.e., “upstream”) or 13 cm (i.e., “midstream”) from the inlet. Samples were allowed to degas for 30 minutes, during which time two grams of the monomer, N-isopropyl acrylamide (99%), purchased from Acros Organics (Geel, Belgium), were placed in the hot water bath at 75 °C. After the samples are degassed, the autotransformer is turned on to allow the heating tape to heat to the proper temperature (~82 °C). The next step is an Ar etch, which is done with a gas flow of 10 standard cubic centimeters per minute (sccm) for 2 min and a power of 40W. The pressure of the Ar etch is 3.9×10−1 Torr. The Ar etch is followed by a methane adhesion-promoting layer, which is done using a flow of 7.4 sccm for 5 min and a power of 80W. For this step, the pressure is maintained at 1.4×10−1 Torr.
The next step is the ppNIPAM deposition. First the gas line is closed off, and the needle valve on the monomer line is opened. The number of turns of the needle valve was an experimental variable. The pressure is set to the desired value (another experimental variable). After two minutes, the RF generator was turned on to the first desired power setting (either 80W or 100W) for 5 min, then to 10W for 5 min, then to 5W for 5 min, and then to 1W for 10 min. After the 1W setting, the monomer is left in the hot water bath for 5 min, then put into a room temperature water bath for 2 min, which is followed with an ice bath for 3 min. This series of steps quenches the monomer flow inside the reactor, and condenses the NIPAM in the flask back into a powder.
After the samples are removed from the reactor, they are rinsed with cold DI water in order to remove any uncrosslinked NIPAM off of the surface, and dried with nitrogen. The surfaces are placed in a Petri dish, backfilled with nitrogen, and sealed with Parafilm. The samples are then stored in a desiccator until surface analysis or cell culture are performed.
Results and Discussion
While optimizing the plasma reactor for use to deposit ppNIPAM films, a number of experimental parameters were varied. Table 1 summarizes the different experimental conditions varied during the ppNIPAM deposition. The first parameter tested was upstream vs. midstream, using a pressure of 140 mTorr, 1st use of the monomer, the needle valve was opened 1 turn, and the initial power was 80W. When a setting was determined to work the best, that setting was used for the rest of the experiments (i.e., upstream was determined to work better than midstream, so upstream was used thereafter).
Table 1.
List of the experimental parameters varied to determine optimal conditions for ppNIPAM deposition.
| Monomer Use | 1st | 2nd |
|---|---|---|
| Position in Reactor | 0 cm “Upstream” | 13 cm “Midstream” |
| NIPAM Pressure | 140 mTorr | 250 mTorr |
| # NIPAM Needle Valve Turns | 1 turn | 5 turns |
| Initial Power | 80W | 100W |
Parameter 1: Upstream vs Midstream
The first comparison that was made was between surfaces that were coated “upstream” vs “midstream.” (See Figure 1) The results were also compared to data obtained for plasma films using a reactor that was previously demonstrated to successfully deposit ppNIPAM films at the University of Washington (UW).[41]
Surface chemistry by XPS
XPS analysis was conducted to compare the surface composition and carbon binding environments of the different surfaces. X-ray photoelectron spectroscopy (XPS) is a quantitative technique that allows for the determination of elemental composition, bonding environments, and relative thickness of a surface. From the binding energy and intensity of the photoelectron peak (based on the number of photoelectrons detected) the identity of the elements in the surface, the chemical state of each element, and the quantity of each element with an error of less than 10% may be determined.[42–44]
The elemental compositions are listed in Table 2, and the carbon binding environments are listed in Table 3. As evidenced by Table 2 and 3, XPS is capable of distinguishing between the elemental composition of ppNIPAM films and the silicon controls upon which ppNIPAM is deposited. In comparison to the silicon controls, the composition of the ppNIPAM film generated in the upstream position closely resembles that of the monomer. No Si is detected, indicating that the film is thicker than the sampling depth of XPS (~50–100 Angstrom). However, XPS is not capable of distinguishing between the two sets of surfaces created in our reactor or the UW samples used for comparison. The compositions of these samples generated in our lab compared to those generated previously at UW are within the 3–4% experimental error of the instrument.[42,43]
Table 2.
Elemental composition of a clean Si chip, ppNIPAM deposited upstream, ppNIPAM deposited midstream, and ppNIPAM deposited at the University of Washington[41] determined by XPS analysis. All standard deviations are less that 3% unless noted by an asterisk (C standard deviation for the control is 4.4% and the Si standard deviation for the control is 3.4%).
| C% | N% | O% | Si% | |
|---|---|---|---|---|
| Silicon Control | 35.2* | 0 | 30.1 | 33.7* |
| Upstream | 78.8 | 10.0 | 11.3 | 0 |
| Midstream | 79.4 | 11.6 | 8.8 | 0 |
| UW | 78.8 | 7.2 | 14.0 | 0 |
| Theoretical | 75.0 | 12.5 | 12.5 | 0 |
Table 3.
Carbon binding environments of a Si chip, ppNIPAM deposited upstream, ppNIPAM deposited midstream, and ppNIPAM deposited at the University of Washington[41] as determined by peak fitting the high resolution C1s XPS spectra. The standard deviation is less than 2%.
| C-H/C-C | C-N/C-O | N-C=0 | |
|---|---|---|---|
| (285 eV) | (+1.5 eV) | (+3 eV) | |
| Silicon Control | 92.9 | 7.1 | 0 |
| Upstream | 66.5 | 21.5 | 12.0 |
| Midstream | 68.2 | 23.3 | 8.5 |
| UW | 71.5 | 17.8 | 10.8 |
| Theoretical | 66.7 | 16.7 | 16.7 |
The same trend is observed in the carbon species present on the samples: the species present on ppNIPAM films and the silicon control are easily distinguished (see Table 3). The silicon control has two carbon binding environments present. The first is carbon bound to hydrogen or carbon (at 285 eV), and the second is carbon bound to either nitrogen or oxygen (shifted 1.5 eV). On the control, there are C-H/C-C bonds C-N/C-O bonds. On the ppNIPAM surfaces, there are three carbon environments present: two of which are the same as the control (C-C/C-H and C-O/C-N) and the third is N-C=O (amide). However, the two films created by our reactor and the data obtained at UW for comparison, cannot be distinguished by high-resolution carbon XPS. When the 3–4% error of the instrumentation is taken into consideration,[42,43] there is little to no variance in the data. Taking into account the 3–4% error, as well as the standard deviation, using different XPS instruments, different reactors, and different deposition parameters, XPS is not capable of distinguishing between these films. Therefore, XPS was not used for later comparisons of film optimization parameters. Instead, subsequent analysis relied on thickness, contact angles, and cell detachment.
Film thickness by Interferometry
From the XPS results, it is clear that film thickness for all parameters is greater than 100 Å. To obtain higher resolution data for film thickness, thickness measurements were obtained via interferometry. Previously, it was reported that the thickness of a pNIPAM layer has an effect on cell growth.[39] It was shown that BAECs grew on surfaces that were 15.5 nm thick, but not on surfaces that were 29.3 nm thick.
The thickness values obtained for ppNIPAM surfaces generated using the reactor at UNM are presented in Table 4. Sample position seemed to have a large impact on the thickness and thermoresponsive behavior of the film. Surfaces deposited downstream were twice as thick as those deposited upstream (40.0nm for upstream and 88.3nm for downstream).
Table 4.
Thickness measurements obtained via interferometry for ppNIPAM samples deposited using various operating conditions. A refractive index of 1.59 was used for all surfaces.
| Thickness | |
|---|---|
| (nm) | |
| Upstream | 40.0±13 |
| Midstream | 88.3±24 |
| 1st Use | 60.8±9 |
| 2nd Use | 36.1±23 |
| 140 mTorr | 52.3±11 |
| 250 mTorr | 31.6±5 |
| 1 Turn | 38.8±6 |
| 5 Turns | 89.6±16 |
| 80W | 36.4±12 |
| 100W | 49.2±14.4 |
Thermoresponse by Contact Angle Goniometry
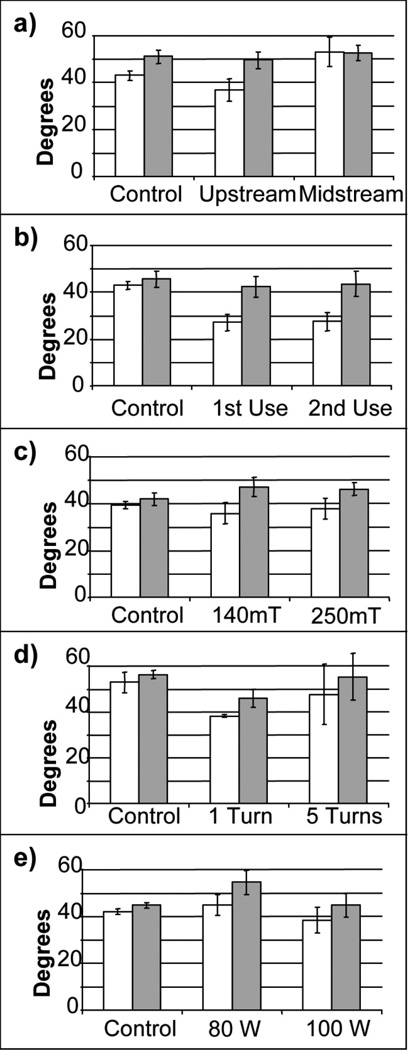
The next technique used to characterize ppNIPAM films generated from the reactor was contact angles obtained above and below the LCST of pNIPAM (~32 °C). This technique will determine if the different operating conditions have an effect on the thermoresponse of the surface. Figure 2 and Table 5 show the contact angle data obtained at both room temperature and 37 °C for all of the parameters tested. Contact angles varied from about 27°–60°. It has been reported that the deposition parameters have an impact on the contact angle.[45] The contact angle values that have been reported for ppNIPAM range from about 5°–65°.[15,45,46] Films deposited upstream retained thermoresponse (36.9°±4.7 at room temperature and 49.5°±3.4 at 37 °C, a 4.5° change at the bounds of the standard deviation), while films deposited midstream did not retain thermoresponse (53.1°±6.1 at room temperature and 52.6°±3.3 at 37 °C).
Figure 2.
Contact angle measurements using the inverted bubble technique at both room temperature (white) and 37 °C (black) for the parameters varied: (a) upstream vs. midstream, (b) 1st use vs. 2nd use of the monomer, (c) 140mT vs. 250 mT, (d) 1 vs. 5 turns of the monomer needle valve, and (e) 80W vs. 100W for the initial power. The control is a Si chip.
Table 5.
Thickness measurements obtained via interferometry for ppNIPAM samples deposited using various operating conditions. A refractive index of 1.59 was used for all surfaces.
| 25 °C | 37 °C | |
|---|---|---|
| (nm) | (nm) | |
| Upstream | 36.9 °±4.7 | 49.5 °±3.4 |
| Midstream | 53.1 °±6.1 | 52.6 °±3.3 |
| 1st Use | 27.1 °±3.4 | 42.4 °±4.3 |
| 2nd Use | 27.5 °±3.8 | 43.5 °±5.3 |
| 140 mTorr | 35.8 °±4.6 | 47.1 °±3.9 |
| 250 mTorr | 37.9 °±4.4 | 46.1 °±2.8 |
| 1 Turn | 38.2 °±0.5 | 46.0 °±3.8 |
| 5 Turns | 47.5 °±13.1 | 55.2 °±10.1 |
| 80W | 44.9 °±4.4 | 54.6 °±5.1 |
| 100W | 38.5 °±5.4 | 44.8 °±5.1 |
Reversible cell adhesion
Previously, it was demonstrated that mammalian cells will adhere and grow to confluence on pNIPAM at 37 °C as well as detach when the media is replaced with 4 °C serum free media.[29] This behavior has previously been exploited for a variety of tissue engineering applications, which require intact cell sheets be harvested. To test the suitability of these thermoresponsive films for cell sheet engineering, cell detachment studies were conducted on the surfaces using Bovine Aortic Endothelial Cells (BAEC) grown to confluence to determine if cells would grow at 37 °C and then detach when the temperature is decreased below the LCST.
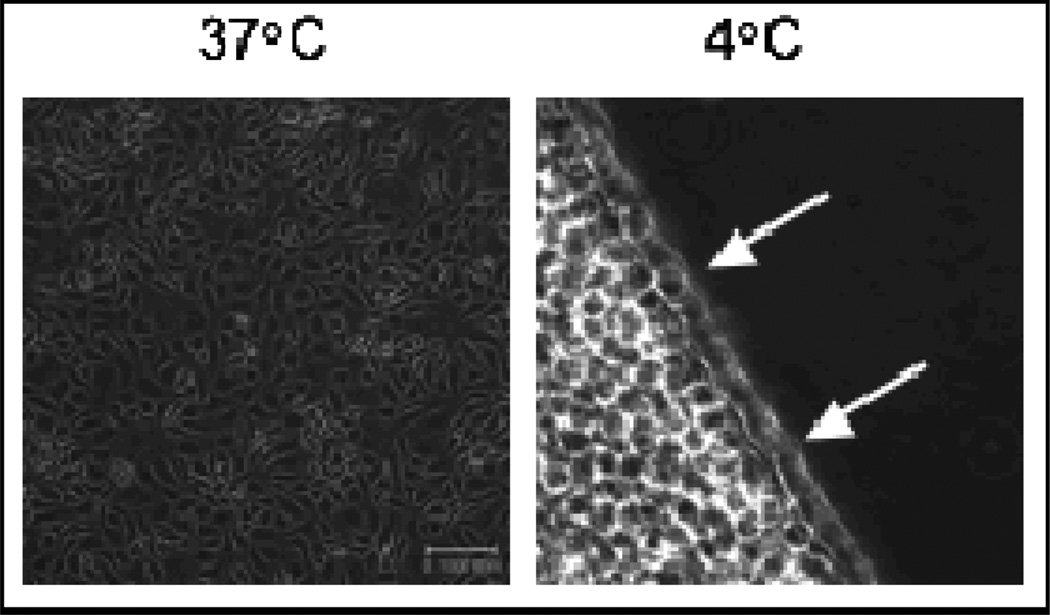
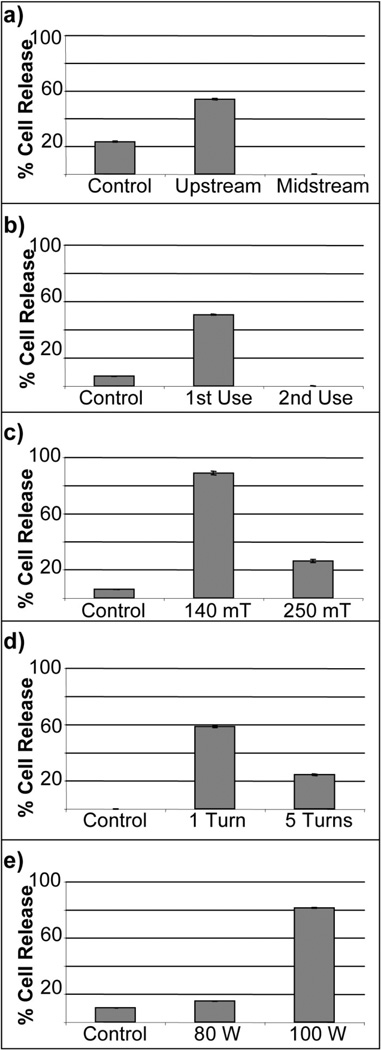
Figure 3 shows cells releasing from a ppNIPAM surface fabricated using this reactor. After two hours, approximately half of the cells remain on the surfaces. Figure 4 shows the amount of cell release from ppNIPAM using the different deposition conditions. Cell response on these surfaces showed a little over half of the cells were detached when the samples were coated upstream, while no cells detached from surfaces coated in the midstream position. It is interesting to note that this does correspond to the contact angle measurements (see Figure 2 b). The upstream surfaces retained the thermoresponsive property of pNIPAM, while the midstream surfaces did not demonstrate a statistically relevant change.
Figure 3.
Bright field mincroscopy images of BAECs grown to confluence (left) at 37 °C and releasing from a ppNIPAM surface (right) at 4 °C. Arrows indicate cell release. (Scale bar= 100µm)
Figure 4.
Percent of BAECs detached from ppNIPAM at different reactor operating conditions: (a) sample position, (b) use of the monomer, (c) pressure, (d) monomer flow, and (e) initial power settings.
From this information, it is clear that the samples positioned upstream yielded better films than those positioned midstream, or between the electrodes. This may be due to the fact that the plasma is mostly observed as occurring between the electrodes, and is less intense from the grounded electrode (upstream). This will cause more fragmentation of the monomer between the electrodes, which corresponds well with the lack of thermoresponse observed on the midstream surfaces (see Figure 2 a).
Parameter 2: 1st vs 2nd use of the monomer
Film thickness by Interferometry
The thickness for samples deposited with the 1st use of the monomer were approximately 1.5 times thicker than surfaces deposited with the 2nd use of the monomer (60.8 nm for the 1st use and 36.1 nm for the 2nd use). It was also observed that the surfaces deposited midstream are approximately twice as thick as surfaces deposited upstream (88.3 nm for midstream and 40.0 nm for upstream), which may affect the thermoresponse and the cell attachment/detachment.[39]
Thermoresponse by Contact Angle Goniometry
When comparing the monomer use, it is clear that ppNIPAM films created using both the 1st and 2nd use retain their thermoresponse. The 1st use had an angle change from 27.1°±3.4 at room temperature to 42.4°±4.3 at 37 °C, which gives a change of about 7.6°. The 2nd use had an angle change from 27.5°±3.8 at room temperature to 43.5°±5.3 at 37 °C, which gives a change of ~6.9°. This is larger than the change reported for previous ppNIPAM films (~4.5° change).[15]
Reversible cell adhesion
Approximately half of the cells detached after two hours using the first use, while no cells detached with the second use. When the cell culture results are compared with contact angle data for identical surfaces, it is found that both sets of surfaces retained thermoresponse (see Figure 2 b). This result explains why the cells detach with the first use surfaces.
However, the fact that the second use surfaces did not have any cell release cannot be explained from a lack of thermoresponse. In general, better results were found using the first use of the monomer compared to the second use. This may be due to the fact that, with the 1st use, more of the monomer reaches the gas phase. After it is condensed, following the 1st use, it may need to be heated to a higher temperature to reach a gas phase. This is consistent with the observation of a majority of the monomer in liquid phase during deposition. It is also consistent with the thickness data (see Table 4), the first use does obtain a thicker surface than the second use. It may also be that the monomer may start to polymerize in the flask due to the heating before and during each use.
Parameter 3: Pressure settings
Film thickness by Interferometry
Surfaces deposited using a pressure of 140 mTorr were approximately 1.5 times thicker than those deposited using 250 mTorr (52.3 nm for 140 mTorr and 31.6 nm for 250 mTorr), which is similar to the difference seen in the 1st vs 2nd use of the monomer.
Thermoresponse by Contact Angle Goniometry
Unlike with the monomer use, there was no significant thermoresponse observed at both of the pressures that were varied. For 140 mTorr, 35.8°±4.6 at room temperature and 47.1°±3.9 at 37 °C, a 2.8° change. For 250 mTorr, 37.9°±4.4 at room temperature and 46.1°±2.8 at 37 °C, a 1° change.
Reversible cell adhesion
Variations in the deposition pressure showed significant cell release, ~90%, from the surfaces deposited at 140 mTorr. Some cell release was observed on surfaces deposited at 250 mTorr, but only ~25%. It is interesting to note that no significant thermoresponse was observed on either set of surfaces (see Figure 2 c). Thus, a pressure of 140 mTorr yielded better results than those obtained with 250 mTorr. This may result from a decrease in the mean free path, and an increase in inelastic losses. Thus, fewer molecules will have the energy to reach a plasma state and become polymerized. This corresponds to the thickness (see Table 4). The surfaces coated at 140 mTorr are thinner than those deposited at 250 mTorr.
Parameter 4: PNIPAM flow rate
Film thickness by Interferometry
In this home built RF reactor, the pNIPAM flow rate was controlled using a needle valve. The flow was varied from low (1 turn) to high (5 turns). When the number of needle valve turns was varied, it was observed that surfaces deposited using 5 turns yielded surfaces approximately twice as thick as surfaces deposited using 1 turn (89.6 nm for 5 turns and 38.8 nm for 1 turn).
Thermoresponse by Contact Angle Goniometry
There was slight thermoresponse on surfaces deposited with one turn (38.2°±0.5 at room temperature and 46.0°±3.8 at 37 °C, a 3.5° change). No thermoresponse was observed on surfaces that were deposited using five turns (47.5°±13.1 at room temperature and 55.2°±10.1 at 37 °C).
Reversible cell adhesion
Cell release was observed on surfaces that were deposited using one and five needle valve turns. However, there was more significant release with one turn as oppose to five turns (60% versus 25%). This again corresponds to the contact angle data (see Figure 2 d). The one turn surfaces were thermoresponsive, while the five turn surfaces were not. For the optimal number of turns of the needle valve, one turn of the worked better than five turns. One potential reason for this may be that opening the needle valve to the completely open state (i.e., 5 turns) allows the majority of the monomer vapor phase to be delivered into the reactor as a bolus.
Parameter 5: Initial power setting
Film thickness by Interferometry
The relative amount of crosslinking in a film is controlled via the power setting of the RF generator: a higher wattage will increase crosslinking. The surfaces deposited at both 80W and 100W had approximately the same thickness (36.4 nm for 80W and 49.2 nm for 100W).
Thermoresponse by Contact Angle Goniometry
There was no significant change in the angles with respect to temperature for either setting (44.9°±4.4 at room temperature and 54.6°±5.1 at 37 °C for 80W and 38.5°±5.4 at room temperature and 44.8°±5.1 at 37 °C for 100W).
Reversible cell adhesion
There was very little cell release from surfaces deposited using an initial power of 80W, about 15%, while there was significant cell release from surfaces deposited using an initial power of 100W, about 80%. Neither set of surfaces showed thermoresponse with contact angles (see Figure 2 e). As wattage applied to the reactor during the initial stage is high (i.e., 80W), the resulting film will be highly crosslinked and fragmented (as evidenced by the lack of thermoresponse). If the remaining monomer in the flask does not escape into the vapor phase (as is the case with 2nd use), there may be insufficient monomer to achieve a less crosslinked film in the remaining steps. The results obtained indicate that an initial power of 100W results in better ppNIPAM films an initial power of 80W. This result may be due to the crosslinking and fragmentation of the monomer. The 1st step is foundation for the rest of the film. Therefore, the higher power (i.e., 100W) will result in a foundation that is more fragmented and crosslinked.
Conclusion
In this work, the pertinent characteristics (monomer use, sample position, operating pressure, needle valve turns, and initial power) of ppNIPAM films generated by a home-built RF reactor were characterized by XPS, contact angles, cell detachment, and interferometry. Our data were also compared to data obtained from a similar reactor operated at the University of Washington. Our results indicate that for this reactor, fresh monomer (i.e., the “1st use”) should be used, and the reactor evacuated to 140 mTorr. NIPAM is added into the chamber by turning the needle valve 1 turn to ensure addition of the polymer is relatively constant over time, as opposed to a bolus. The initial power setting of the RF generator should be 100W, with the samples upstream (i.e., 0 cm from the monomer inlet). These settings resulted in the most cell detachment, which was the primary goal for these biocompatible films.
For ppNIPAM surfaces generated from this reactor, it appears that cell adhesion and growth did not depend on the surface thickness being in the range of 15.5–29.3 nm, as has been predicted in previous studies. Instead, cells grew on all of the surfaces, which ranged in thickness from 31.6 nm to 88.3 nm. However, we did find an optimal thickness for cell release: from 38.8 nm to 60.8 nm. This discrepancy is optimal thickness is most likely a factor of the method of film deposition, which would change the overall characteristics of a film. However, as previously suggested by Okano, these thermoresponsive films are more effective if they are relatively thin. To achieve this parameter with our reactor, these films must be formed using a less intense plasma with a shorter mean free path to create thinner films with a stable base of highly fragmented and crosslinked polymer.
It is interesting to note that for samples obtained from this reactor, thermoresponse as indicated by contact angle measurements did not always correspond to the relative degree of cell detachment observed; in some cases, cell detachment was observed, yet there was not significant thermoresponse observed via contact angle measurements. This result is significant because contact angles are often used as a relatively fast and inexpensive technique to determining the thermoresponse, and therefore the suitability of a film for mammalian cell release. Due to this discrepancy, it is necessary to use a confirmatory technique (such as atomic force microscopy) to study the swelling behavior of our ppNIPAM surfaces.
Acknowledgements
The authors thank Professors Elizabeth Hedberg-Dirk and Gabriel López for helpful discussions and expertise, as well as Steven Candelaria and Laura Pawlikowski. XPS spectra were obtained by Lara Gamble at the National ESCA and Surface Analysis Center for Biomedical Problems (NESAC/BIO), which is supported by NIBIB grant EB-002027.
This work was supported by NSF-Partnerships for Research and Education in Materials (PREM) program grant # DMR-0611616, Sandia-University Research Program grant # 739577, as well as funding from 3M Corporation and the UNM Center for Biomedical Engineering. JAR was supported by an NSF Graduate Research Fellowship.
Reference
- 1.Duracher D, Sauzedde F, Elaissari A, Perrin A, Pichot C. Colloid. Polym. Sci. 1998;276:219–231. [Google Scholar]
- 2.Moselhy J, Wu XY, Nicholov R, Kodaria K. J. Biomater. Sci. Polym. Ed. 2000;11:123–147. doi: 10.1163/156856200743616. [DOI] [PubMed] [Google Scholar]
- 3.Sershen SR, Westcott SL, Halas NJ, West JL. J. Biomed. Mater. Res. 2000;51:293–298. doi: 10.1002/1097-4636(20000905)51:3<293::aid-jbm1>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka T, Nishio I, Sun ST, Uenonishio S. Science. 1982;218:467–469. doi: 10.1126/science.218.4571.467. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Tanaka T. J. Chem. Phys. 1990;92:1365–1371. [Google Scholar]
- 6.Heskins M, Guillet JE. J. Macromol. Sci. Part A. 1968;2:1441–1455. [Google Scholar]
- 7.Lopez VC, Raghavan SL, Snowden MJ. J. React. Func. Polym. 2004;58:175–185. [Google Scholar]
- 8.Gerlach G, Guenther M, Suchaneck G, Sorber J, Arndt KF, Richter A. Macromol. Symp. 2004;210:403–410. [Google Scholar]
- 9.Ista LK, Perez-Luna VH, Lopez GP. Appl. Environ. Microbiol. 1999;65:1603–1609. doi: 10.1128/aem.65.4.1603-1609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasgoz H, Ozgumus S, Orbay M. Polymer. 2003;44:1785–1793. [Google Scholar]
- 11.Lee KY, Mooney DJ. J. Chem. Rev. 2001;101:1869–1879. doi: 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- 12.Chiantore O, Guaita M, Trossarelli L. Macromole. Chem. Phys. 1979;180:969–973. [Google Scholar]
- 13.Takezawa T, Mori Y, Yoshizato K. Bio-tech. 1990;8:854–856. doi: 10.1038/nbt0990-854. [DOI] [PubMed] [Google Scholar]
- 14.Ista LK, Lopez GP. J. Ind. Microbiol. Biotechnol. 1998;20:121–125. [Google Scholar]
- 15.Cheng XH, Canavan HE, Stein MJ, Hull JR, Kweskin SJ, Wagner MS, Somorjai GA, Castner DG, Ratner BD. Langmuir. 2005;21:7833–7841. doi: 10.1021/la050417o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okajima S, Sakai Y, Yamaguchi T. Langmuir. 2005;21:4043–4049. doi: 10.1021/la046994e. [DOI] [PubMed] [Google Scholar]
- 17.Canavan HE, Cheng XH, Graham DJ, Ratner BD, Castner DG. PPP. 2006;3:516–523. [Google Scholar]
- 18.Okano T, Yamada N, Sakai H, Sakurai Y. J. Biomed. Mater. Res. 1993;27:1243–1251. doi: 10.1002/jbm.820271005. [DOI] [PubMed] [Google Scholar]
- 19.Ohya S, Nakayama Y, Matsuda T. Biomacromolecules. 2001;2:856–863. doi: 10.1021/bm010040a. [DOI] [PubMed] [Google Scholar]
- 20.Cooperstein MA, Canavan HE. Langmuir. 2009 [Google Scholar]
- 21.Luo QZ, Mutlu S, Gianchandani YB, Svec F, Frechet JMJ. Electrophoresis. 2003;24:3694–3702. doi: 10.1002/elps.200305577. [DOI] [PubMed] [Google Scholar]
- 22.Yang B, Yang W. J. Membr. Sci. 2003;218:247–255. [Google Scholar]
- 23.Shiroyanagi Y, Yamato M, Yamazaki Y, Toma H, Okano T. Tissue Eng. 2003;9:1005–1012. doi: 10.1089/107632703322495646. [DOI] [PubMed] [Google Scholar]
- 24.Okano T, Yamada N, Okuhara M, Sakai H, Sakurai Y. Biomat. 1995;16:297–303. doi: 10.1016/0142-9612(95)93257-e. [DOI] [PubMed] [Google Scholar]
- 25.Frimpong RA, Hilt JZ. 2008;19 doi: 10.1088/0957-4484/19/17/175101. [DOI] [PubMed] [Google Scholar]
- 26.Jones DM, Smith JR, Huck WTS, Alexander C. Adv. Mater. 2002;14:1130–1134. [Google Scholar]
- 27.Mizutani A, Kikuchi A, Yamato M, Kanazawa H, Okano T. Biomater. 2008;29:2073–2081. doi: 10.1016/j.biomaterials.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 28.Endoh KI, Ueno K, Takezawa T, Yamazaki M, Mori Y, Satoh T. J. Toxicol. Sci. 1993;18:381. [Google Scholar]
- 29.Reed JA, Lucero AE, Cooperstein MA, Canavan HE. J. App. Biomat. Biomech. 2008;6:81–88. [PMC free article] [PubMed] [Google Scholar]
- 30.Takezawa T, Yamazaki M, Mori Y, Yonaha T, Yoshizato K. J. Cell. Sci. 1992;101:495–501. doi: 10.1242/jcs.101.3.495. [DOI] [PubMed] [Google Scholar]
- 31.Pan YV, Wesley RA, Luginbuhl R, Denton DD, Ratner BD. Biomacromolecules. 2001;2:32–36. doi: 10.1021/bm0000642. [DOI] [PubMed] [Google Scholar]
- 32.Lopez GP, Ratner BD, Tidwell CD, Haycox CL, Rapoza RJ, Horbett TA. J. Biomed. Mat. Res. 1992;26:413–439. doi: 10.1002/jbm.820260402. [DOI] [PubMed] [Google Scholar]
- 33.Lopez GP, Ratner BD, Rapoza RJ, Horbett TA. 1993;26:3247–3253. [Google Scholar]
- 34.Godek ML, Malkov GS, Fisher ER, Grainger DW. 2006;3:485–497. doi: 10.1002/ppap.200600007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Detomaso L, Gristina R, d'Agostino R, Senesi GS, Favia P. 2005;200:1022–1025. doi: 10.1016/j.biomaterials.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 36.Colley HE, Mishra G, Scutt AM, McArthur SL. 2009;6:831–839. [Google Scholar]
- 37.Wickson BM, Brash JL. Colloids Surf. A. 1999;156:201–213. [Google Scholar]
- 38.Siow KS, Britcher L, Kumar S, Griesser HJ. PPP. 2006;3:392–418. [Google Scholar]
- 39.Akiyama Y, Kikuchi A, Yamato M, Okano T. Langmuir. 2004;20:5506–5511. doi: 10.1021/la036139f. [DOI] [PubMed] [Google Scholar]
- 40.Wei Y, Yang DC, Tang LG, Hutchins MGK. J. Mater. Res. 1993;8:1143–1152. [Google Scholar]
- 41.Canavan HE, Cheng XH, Graham DJ, Ratner BD, Castner DG. Langmuir. 2005;21:1949–1955. doi: 10.1021/la048546c. [DOI] [PubMed] [Google Scholar]
- 42.Escamilla R, Huerta L. Supercond. Sci. Technol. 2006;19:623–628. [Google Scholar]
- 43.Hesse R, Chasse T, Streubel P, Szargan R. Surf. Interface Anal. 2004;36:1373–1383. [Google Scholar]
- 44.Ratner BD, Castner DG, Vickerman JC, editors. Chichester: John Wiley and Sons; 1997. pp. 43–98. [Google Scholar]
- 45.Tamirisa PA, Koskinen J, Hess DW. Thin Solid Films. 2006;515:2618–2624. [Google Scholar]
- 46.Teare DOH, Barwick DC, Schofield WCE, Garrod RP, Beeby A, Badyal JPS. J. Phys. Chem. B. 2005;109:22407–22412. doi: 10.1021/jp052869f. [DOI] [PubMed] [Google Scholar]