Abstract
The successful use of man-made proteins to advance synthetic biology requires both the fabrication of functional artificial proteins in a living environment, and the ability of these proteins to interact productively with other proteins and substrates in that environment. Proteins made by the maquette method integrate sophisticated oxidoreductase function into evolutionarily naive, non-computationally designed protein constructs with sequences that are entirely unrelated to any natural protein. Nevertheless, we show here that we can efficiently interface with the natural cellular machinery that covalently incorporates heme into natural cytochromes c to produce in vivo an artificial c-type cytochrome maquette. Furthermore, this c-type cytochrome maquette is designed with a displaceable histidine heme ligand that opens to allow functional oxygen binding, the primary event in more sophisticated functions ranging from oxygen storage and transport to catalytic hydroxylation. To exploit the range of functions that comes from the freedom to bind a variety of redox cofactors within a single maquette framework, this c-type cytochrome maquette is designed with a second, non-heme C, tetrapyrrole binding site, enabling the construction of an elementary electron transport chain, and when the heme C iron is replaced with zinc to create a Zn porphyrin, a light-activatable artificial redox protein. The work we describe here represents a major advance in de novo protein design, offering a robust platform for new c-type heme based oxidoreductase designs and an equally important proof-of-principle that cofactor-equipped man-made proteins can be expressed in living cells, paving the way for constructing functionally useful man-made proteins in vivo.
Introduction
Man-made proteins have an important role to play in the synthetic biology goal of constructing functional parts and devices for incorporation into either explicitly biological or artificial organisms or systems.1 The program to integrate sophisticated oxidoreductase function into evolutionarily naive, non-computationally designed protein constructs is called the maquette approach.2,3 By beginning with a generic protein sequence designed only to adopt a simple helical bundle fold and iteratively adding engineering elements onto this stripped-down protein chassis, the maquette approach allows the designer to avoid the layers of complexity that hinder the redesign of natural protein scaffolds. Man-made protein maquettes self-assemble in vitro with a wide range of the redox cofactors seen in nature, including hemes, chlorins, metal ions, flavins and quinones.2 However synthetic biology requires that functional artificial proteins and enzymes interact productively with natural proteins and substrates. They must also fully and functionally assemble in vivo, meaning that cofactor binding sites must be occupied by the target cofactors to carry out the desired function.
Here we test the ability of a novel man-made protein to interface with natural proteins in vivo in order to assemble a functional redox protein. Despite the completely unnatural protein sequence, the natural post-translational machinery of E. coli (Fig. 1A)4 successfully inserts heme B and forms two covalent links between the heme vinyls and protein cysteines to create a synthetic heme C cytochrome with exceptional efficiency. This man-made cytochrome c successfully forms a heme oxy-ferrous state with a stability akin to natural oxygen transport proteins containing heme B,5 but with entirely unrelated sequence or structure. As part of a program to design increasingly sophisticated man-made oxidoreductases, this protein is equipped with an intraprotein electron-transfer chain by including a second non-heme C binding site that self-assembles in vitro with heme B. Light activated function is added to this dyad by replacing the Fe of the original heme C with Zn to create a Zn-porphyrin photo-center.
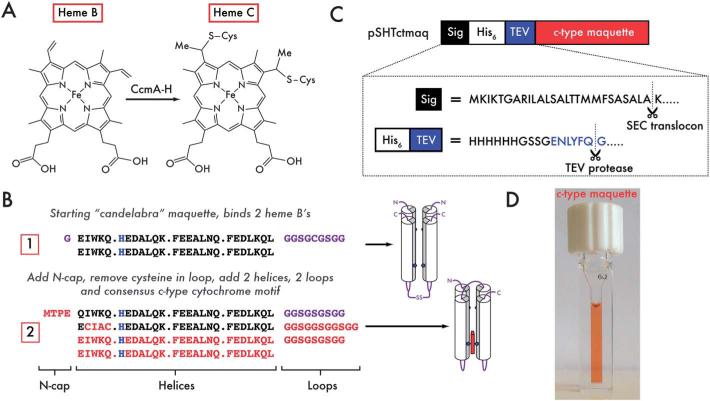
Fig. 1.
Design and expression of a single-chain, artificial c-type cytochrome. (A) Heme B is covalently attached to the substrate protein backbone via thioether linkages between the peripheral vinyl substituents on the porphyrin and the cysteine sidechains within the consensus CXXCH motif. This is achieved in E. coli with the cytochrome c maturation proteins (CcmA-H) located in the periplasmic space. (B) Starting with the designed oxygen transport maquette, 1, 2 loops (comprising glycine and serine) and a 4-residue N-cap (MTPE) are added to allow symmetry breaking across the 4-helix bundle and to add rigidity at the N-terminus. The CXXCH motif is then added into the sequence for covalent heme incorporation by the E. coli cytochrome c maturation apparatus (2). Changes to the amino acid sequence between iterations are highlighted in red, heme ligating histidine residues in blue and loops and N-termini in purple. Cartoon representations of the proteins are indicated to the right of the sequences: heme-ligating histidines are represented by blue pentagons, loops and N-/C-termini by purple lines, heme C positioning by the red rectangular box. (C) The N-terminal signal sequence from the periplasmic E. coli maltose binding protein (MBP, black) is cleaved between A and K on transport across the inner membrane. A hexa-histidine tag (white) facilitates purification on Ni-NTA, and can be subsequently cleaved through the addition of a TEV protease cleavage site (blue – ENLYFQ-G). (D) This expression system yields highly efficient heme incorporation into the expressed 2.
Results
Protein and vector design
We have previously designed a functional, man-made maquette containing heme B that is capable of reversibly binding molecular oxygen (sequence 1 in Fig. 1B).6 This maquette not only matches the diatomic ligand exchange kinetics and spectroscopy of natural heme containing globins, but also preferentially binds O2 over CO. For a more versatile protein capable of taking advantage of the functionally diverse option of placing a range of different cofactors at two distinct sites, we broke the original dimeric symmetry and united the helices with a long, simple connecting loop composed of just glycine and serine residues. A short, stabilizing N-cap sequence was added to the N-terminus of the protein to increase thermal stability by restricting protein motion (sequence referred herein as 1.5).7
We wished to include a site amenable to covalent heme C attachment for the dual purpose of establishing the interaction of this man-made protein with evolved natural redox proteins in vivo, and secondly to gain a securely attached cofactor that could be exploited under harsh in vitro conditions. The majority of natural c-type cytochrome sequences contain a consensus CX1X2CH motif necessary for in vivo heme incorporation.4 We surveyed the non-redundant PDB for structures with c-type hemes attached to helices (150 structures, ESI, Table S1†) and noted the prevalence of small (A/G) residues at X2 and a general preference for hydrophobic residues at X1 (ESI, Fig. S1†). We selected CIACH as the c-type incorporation motif (sequence 2 in Fig. 1B) to reflect a balance between maintaining the helicity and structure of the protein, and satisfying the very broad substrate specificity of the promiscuous E. coli c-type heme maturation system (Ccm).4 Furthermore, this selection is consistent with a previous analysis of helical porphyrin-binding sites in heme-containing proteins, where the idealized sequence for the most commonly observed histidine rotamer in helical c-type heme sites was identified as CX1ACH.8 To eliminate interference with the added N-cap sequence on helix 1, helix 2 was chosen as the site for the consensus motif, with the distal ligand provided by the histidine on helix 4. The histidines on helices 1 and 3 form an auxiliary binding site for tetrapyrroles such as heme B (Fig. 1B).
In addition to the consensus motif, c-type cytochromes must be exported to the periplasm for covalent heme incorporation by the Ccm machinery.4 We cloned the sequence into the periplasmic expression vector pMal-p4x (NEB, USA), replacing the maltose binding protein (MBP) N-terminal fusion partner with a tobacco etch virus protease (TEV) cleavable hexahistidine tag, leaving the N-terminal signal peptide attached via the cleavable purification tag in the vector pSHTctmaq (Fig. 1C). The signal peptide facilitates periplasmic export through the Sec translocon9,10 and is designed for in vivo cleavage prior to c-type heme incorporation.
In vivo cofactor incorporation
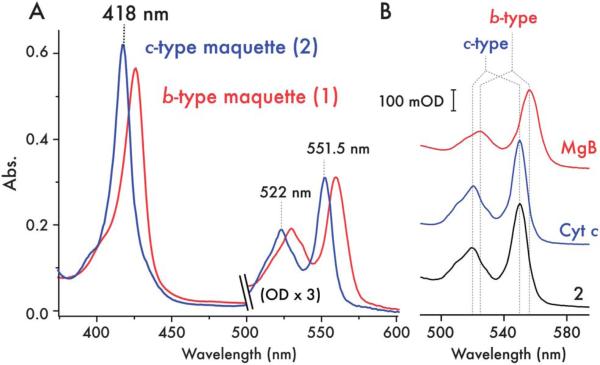
To ensure efficient heme incorporation when expressing c-type cytochromes, the E. coli Ccm machinery must be co-expressed.4,10 This can be achieved at low efficiency through anaerobic growth under nitrate, though the highest levels of heme C incorporation in 2 were observed after aerobic expression in E. coli BL21(DE3) co-transformed with the pEC86 vector harboring ccmA-H.11 In the absence of the pEC86 plasmid, the low levels of Ccm proteins expressed under aerobic conditions lead to very poor heme incorporation (<6%), producing large amounts of apoprotein (ESI, Fig. S2†) and reflecting the low levels of CcmE constitutively expressed under aerobic or micro-aerobic conditions. To further probe the mechanism of heme incorporation, pSHTctmaq was co-transformed with pEC864 – a variant of pEC86 harbouring the catalytically inactive H130A mutant of CcmE12 – into E. coli strain EC65 (ΔccmE).13 The absence of any catalytically active CcmE under these conditions results in a small quantity of b-type heme binding (but no c-type heme binding) to 2, highlighting the necessity of CcmE for efficient c-type heme incorporation into the maquette. Since heme C can be covalently incorporated into some natural c-type cytochromes under reducing conditions in vitro,14,15 we attempted to do so with apo-2 and both hemin and zinc protoporphyrin IX. We were unable to detect appreciable levels of covalent heme incorporation in vitro with either porphyrin, even after a week of incubation, further indicating the necessity of the CcmE protein for efficient maturation. With concomitant pEC86 expression of CcmA-H, the highest levels of expression and heme incorporation (where ~90% of proteins contain heme C) occur during 15 hours aerobic expression in Luria-Bertani broth at 37 °C, the red color of the cell paste serving as a useful indicator of expression levels. Following cell lysis by sonication and Ni-NTA affinity chromatography, the His-tag was cleaved by TEV protease. To remove residual apo-protein and a small quantity of proteolysed protein, the crude protein was further purified by reverse-phase HPLC. MALDI-TOF mass spectrometry confirmed covalent incorporation of heme C (calculated mass = 15 758 Da; observed mass = 15 780 Da, 2 + Na+, ESI, Fig. S3†). Treatment of 2 with acidified 2-butanone16 does not lead to heme partitioning into the organic solvent (ESI, Fig. S4†), a characteristic that differs from proteins containing heme B but one that is shared with covalently bound heme C containing proteins. The UV/vis spectrum of the ferrous protein 2 (Fig. 2A) exhibits the expected blue shift in the Soret/alpha/beta absorption bands compared to ferrous protein 1, consistent with a modification of the vinyl groups on the conjugated porphyrin.17,18 There is a greater degree of spectral shift in comparison with the single thioether-linked heme in the synthetic de novo cytochrome c reported by Isogai17 but a shift similar to that observed by Barker in converting cytochrome b562 to a c-type cytochrome.18 Further confirming the nature of the heme species, the reduced pyridine hemochrome spectrum is identical to that of horse heart cytochrome c and those reported for natural c-type cytochromes19 (Fig. 2B), and signifi-cantly shifted with respect to b-type heme containing proteins and the single thioether-linked heme in the synthetic de novo cytochrome c of Isogai.17 Ferric and ferrous UV/visible (Fig. 2A) and EPR spectra (ESI, Fig. S5†) of 2 are indicative of 6-coordinate bis-histidine ligated heme C.20,21
Fig. 2.
Spectroscopic characterization of the c-type cytochrome maquette. (A) UV/visible spectra of ferrous 1 with bound heme B (red) and purified 2 with covalently incorporated heme C (blue) reveals the expected blue-shift in the absorption spectrum of the c-type heme. (B) UV/visible spectra of the pyridine hemochrome (α/β region) preparations of horse heart myoglobin (red, MgB), bovine cytochrome c (blue, Cyt c) and 2 (black). Stereotypical peak positions for b-type heme and c-type heme are indicated by dashed lines, highlighting the spectroscopic fingerprint for c-type heme in 2.
Heme C containing maquette binds O2
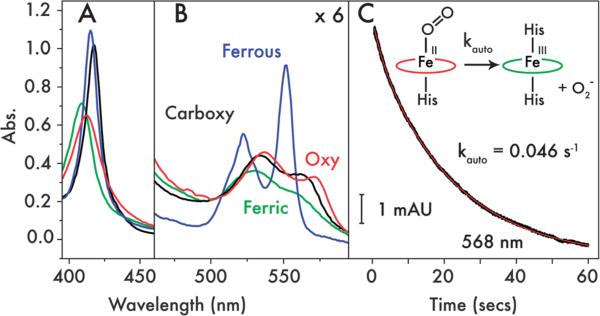
Despite the modifications to the original design (1), 2 retains the ability to undergo ligand exchange between the histidine sidechain occupying the 6th coordination site on heme C and either CO or O2 (Fig. 3A and B). The spectral changes observed on binding CO and O2 are similar to those observed6 for the corresponding heme B-bound species of 1 and the oxyferrous absorbance spectrum is consistent with the oxyferrous spectra of the few natural and engineered c-type cytochromes that can undergo reversible oxygen binding (ESI, Table S2†).22–24 The lifetime of the oxyferrous species of 2 is similar to those observed6 for 1 and 1.5 (1, t1/2 = 9.4 seconds; 1.5, t1/2 = 10 seconds; ESI, Table S3†), though autoxidation at 15 °C occurs less rapidly (t1/2 = 15 seconds), indicating a slight stabilization of the oxyferrous species on switching between hemes B and C. This rate is significantly more rapid than the autoxidation rates of the natural globins25,26 and the natural and engineered c-type cytochromes,27,28 most likely due to the relatively low reduction potential of the hemes in 2 and the other oxygen-binding maquettes,6 but it is within the range of autoxidation rates exhibited by the oxygen binding and activating cytochromes P450 (ESI, Table S3†).29–31
Fig. 3.
Exogenous ligand binding by 2. UV/visible spectra in the (A) Soret (χ), (B) and Q-band (α/β) regions of ferrous (blue), ferric (green), oxyferrous (red) and carbonmonoxyferrous (black) 2. (C) Oxyferrous decay kinetics measured at 568 nm indicating the rate of autoxidation from oxyferrous to ferric 2 & superoxide (O2−). Raw data are displayed in black with a single exponential fit in red.
Physical chemistry of mixed heme C/B maquettes
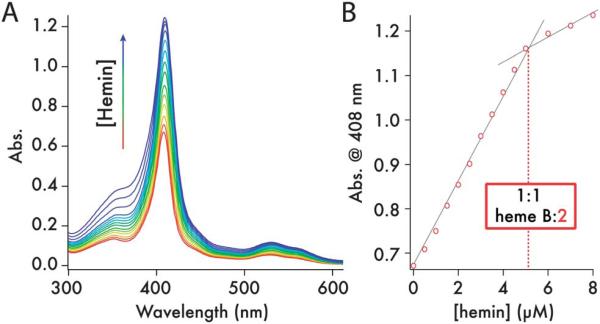
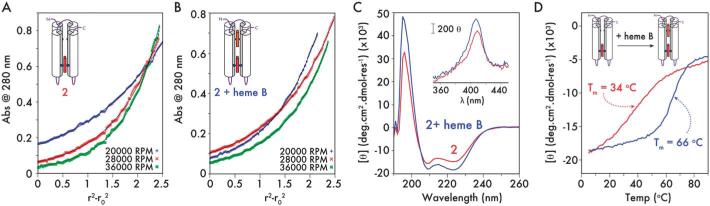
Titration of one equivalent of heme B into 2 creates a mixed heme C/B maquette (Fig. 4). Analytical ultracentrifugation of the C and C/B maquette showed that both sedimented as monomeric 4-helix bundles with the data fitting to species with molecular weights of 18 340 Da and 17 210 Da (Fig. 5A and B – residuals plotted in ESI, Fig. S6†), corresponding to 1.16× and 1.05× the monomer mass respectively (15 758 and 16 374 Da). Circular dichroism spectroscopy indicated both to be highly α-helical with an increase in helicity on heme B ligation (Fig. 5C). Visible CD spectra indicated that the hemes reside in chiral environments in both C and C/B forms. However, ligation of heme B in the auxilliary site confers significantly more thermal stability than does heme C to the heme-free maquette (Fig. 5D; second derivatives of the CD melt data are plotted in ESI, Fig. S7†). Using thermal denaturation to probe the protein stability (monitored at 222 nm in the circular dichroism spectrum), the heme C form melting transition at 34 °C indicates the heme C does not confer significant stability compared to the apo-protein lacking the CXXCH motif (1.5, Tm 42 °C, ESI, Fig. S8†). Given the thermal stability generally imparted on maquettes by the ligation of heme B,2 this is a surprising observation and may reflect a non-idealized and strained binding geometry between heme C and the proximal histidine-containing helix. We were unable to determine the Tm of apo-2 due to the formation of oligomers in the absence of heme C. Binding heme B to the empty site of the heme C form leads to a dramatic 32 °C increase in Tm and a significantly more cooperative melting transition. More support for the stabilizing effect of heme B, as opposed to heme C, is found in the broad 1D 1H NMR spectrum of 2, an indicator of conformational averaging of the protein scaffold and thus a protein with a mobile, liquid interior (ESI, Fig. S9†), compared to the distinct sharpening of the NMR signals on adding heme B.
Fig. 4.
2 binds 1 equivalent of heme B per 4-helix bundle. (A) UV/visible spectra from the titration of 0.5 μM hemin aliquots (0.5 mM hemin in DMSO) into 5 μM 2 (1 cm pathlength cuvette, 25 °C, 20 mM phosphate, 100 mM KCl, pH 7.5). (B) Heme binding isotherm indicating a stoichiometry of 1 heme B per molecule of 2 (dashed line).
Fig. 5.
Structural characterization of 2. (A and B) Analytical ultracentrifugation of 15 μM 2 (A) and 15 μM 2 + heme B (B) in redox buffer (20 mM phosphate, 100 mM KCl, pH 7.5). Data were fitted to models of 1.16× the calculated MW for 2 and 1.05× the calculated MW for 2 + heme B. Residuals for the fits are plotted in ESI Appendix, Fig. S6.† (C) Far-UV circular dichroism spectra of 2 (red) and 2 + heme B (blue) at 5 μM in redox buffer (20 mM phosphate, 100 mM KCl, pH 7.5). Inset, Near-UV/visible CD of 2 (red) and 2 + heme B (blue). (Conditions as for far-UV CD measurements.) (D) Temperature dependence of the CD signal @ 222 nm for 2 (red) and 2 + heme B (blue), highlighting the increase in thermal stability (ΔTm = +32 °C) on occupying the second heme binding site (conditions as for far-UV CD measurements).
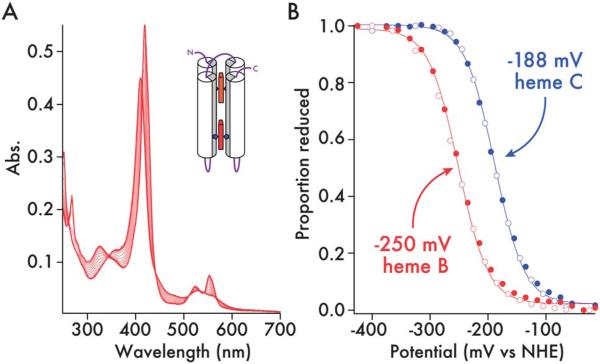
The additions of a connecting inter-helix loop and thioether linkages to the heme macrocycle raise the heme C redox midpoint in 2 by 54 mV compared to the B heme 1 (–252 to –198 mV vs. NHE, ESI, Fig. S10†), possibly accounting for the increase in the lifetime of the oxyferrous state. The C and B heme redox potentials in the mixed C/B maquette are –188 mV and –250 mV respectively (Fig. 6). The greater than 60 mV split in the redox potentials means that electron transfer from B to C will be more than 10 times more favorable than the reverse reaction, providing direction to the nascent electron transfer chain.32
Fig. 6.
Redox potentials of c-type and b-type heme in 2 + heme B. (A) UV/visible spectra of the potentiometric titration of 2 + heme B (61 μM, 50 mM phosphate, 500 mM KCl, 10% glycerol, pH 7.5). (B) Determination of redox potentials of c- and b-type hemes in 2 + heme B. Reductive (closed circles) and oxidative (open circles) data are fitted with 1-electron Nernst curves and quoted vs. the Nernst hydrogen electrode (NHE). The proportion of reduced heme (y-axis) was determined for each sample at wavelengths where the contribution to the absorbance change was exclusive to the type of heme (c-type – 418 nm; b-type – 432 nm).
Conversion to a photoactive maquette
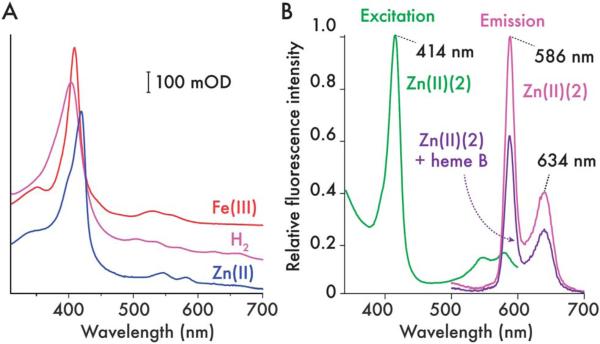
By covalently appending the cofactor to the protein backbone, diverse functionality can be incorporated into the heme C site of the maquette whilst maintaining a robust scaffold with the capacity for a secondary or complementary function at the auxiliary tetrapyrrole binding site. Since c-type cytochromes undergo facile demetallation in the presence of hydrofluoric acid and remetallation by zinc to produce light sensitive porphyrins, the heme C in 2 can be modified to create a maquette capable of simple light-activated electron/energy transfer. Using a protocol first devised by Vanderkooi,33 lyophilized heme C maquette is dissolved in 70% hydrogen fluoride in pyridine, the almost instantaneous color change to purple indicates the formation of the metal-free form. Following removal of the excess hydrogen fluoride and pyridine, reloading with zinc by stirring with an excess of Zn2+ at pH 2 produces Zn cytochrome c form (Zn(II) (2)), which retains the auxiliary tetrapyrrole binding site. The UV/visible spectra and fluorescence excitation and emission spectra of metal-free and Zn porphyrin forms (Fig. 7) are consistent with those obtained for the metal free and zinc horse heart cytochrome c.33 Adding 1 equivalent of heme B resulting in the mixed ZnC/B form (ESI, Fig. S11†). This binding of heme B to the auxiliary site quenched the fluorescence of the covalently appended zinc porphyrin by approximately 40%, indicating the presence of an efficient energy transfer mechanism between chromophores with overlapping emission/absorbance spectra, a feature common to natural light harvesting complexes.34
Fig. 7.
Generation of a covalently-appended, light-sensitive cofactor in 2. (A) UV/visible spectra of ferric (Fe(III) (2), red), demetallated (H2(2), pink) and zinc(II) (Zn(II) (2), blue) 2 (5 μM protein, 20 mM phosphate, 100 mM KCl, pH 7.5). Iron is removed from the porphyrin with HF–pyridine solution, then after purification of demetallated 2 (H2(2)), Zn2+ can be inserted to create a covalently bound, photosensitive cofactor. (B) Fluorescence excitation (green) and emission (magenta) spectra of Zn(II) (2) (900 nM, 20 mM CHES, 150 mM KCl. pH 9.0). Emission spectra were recorded with an excitation wavelength of 420 nm in both heme-free and heme-bound Zn(II) (2) and normalized to the Zn(II) (2) maximum at 586 nm.
Discussion
We have shown that it is plausible to practice synthetic biology with completely artificial redox proteins that plug in to natural biochemical pathways and are functionalized in vivo. Furthermore, with maquettes we can potentially customize inter-protein interactions by adjusting the patterning of the maquette external charges to be complementary with localized regions of natural protein surface charge, independent of the incorporation of hemes in the interior. We have also described a tractable procedure whereby a designer 4-helix bundle capable of non-covalently binding heme cofactors can be converted into a functional artificial c-type cytochrome. The design criteria are simple: take an existing heme B binding protein maquette and convert to a single chain 4-helix bundle to break the bundle symmetry and satisfy the required heme C ligation, in this case 6-coordinate bis-histidine ligation; incorporate the consensus CX1X2CH motif derived from natural c-type cytochrome sequences and co-express in the E. coli periplasm with the natural cytochrome c maturation genes. Efficient translocation to the periplasm is achieved by appending the natural N-terminal signal sequence from MBP, demonstrating the compatibility of artificial protein sequences with complex cellular machinery, resulting in a very high percentage of the translocated protein. Efficient heme incorporation into the maquette is only achieved in the E. coli periplasm in the presence of active, co-expressed heme lyase, CcmE (ESI, Fig. S2†).4
We caution that working with in vivo Ccm maturation proteins may require that our artificial proteins designs are not overly robust and thermally stable. Apo-horse heart cytochrome c is largely unfolded under physiological conditions,35 only attaining the native state on heme B binding or heme C incorporation.36 A similar situation is likely to exist for the double cysteine mutant of cytochrome b562 engineered for c-type heme incorporation (cytochrome cb562): wild type apocytochrome b562 exists in a molten globule-like state,37 while the mature cytochrome cb562 adopts a native state almost identical to that of the b-type heme containing version.38,39 Such behavior is indicative of a preference of the maturation system for protein substrates with heme binding sites that exhibit disordered or at least molten globular characteristics when unoccupied, thus decreasing the kinetic barrier of chaperone-presented heme binding to the protein (via the CcmE heme lyase in the E. coli), the first step in covalent heme incorporation.4 Since our c-type cytochrome maquette exhibits a relatively low Tm, we surmise that the high efficiency of heme incorporation can be attributed to the dynamic, molten state of the maquette.
The prokaryotic E. coli Ccm proteins are capable of incorporating heme C into a wider variety of c-type cytochromes compared to the eukaryotic system, which display marked specificity for eukaryotic cytochromes c and c1 (the only known eukaryotic c-type cytochromes).4 A recent study by Kranz40 has demonstrated that the S. cerevisiae maturation apparatus can process prokaryotic homologues of eukaryotic cytochrome c, provided that a sequence recognition motif41 is included preceding the CXXCH motif at the N-terminus of the apoprotein substrate. Furthermore, the complex of holocytochrome c synthase (HCCS), a eukaryotic cytochrome c maturation protein responsible for terminal heme transfer and attachment, and apocytochrome c is primarily mediated by interactions with heme at the protein–protein interface.40 Since the heme ligands of the nascent heme C binding site are provided by a histidine from the CXXCH motif of the apocytochrome c substrate and a histidine from HCCS, it would appear that effective interfacing with HCCS and thus maturation with this system would require an unfolded apocytochrome substrate. We therefore postulate that the inclusion of the additional recognition sequence to our c-type maquette coupled with the partially unfolded nature of the apo-maquette under physiological conditions may enable efficient heme incorporation through this maturation system, thereby facilitating the in vivo assembly of artificial c-type cytochromes in eukaryotic organisms.
There is only one known example of a natural, oxygen binding c-type cytochrome that forms the oxyferrous state in the wild-type protein: Sphaeroides Heme Protein (SHP), a class 1 cytochrome c from Rhodobacter sphaeroides and related proteobacteria.24,28 SHP adopts the fold of a typical cytochrome c, but with either the unusual His/Asp or the more common bis-His ligation scheme. Both ligation schemes free up the sixth heme coordination site for diatomic ligand binding when reduced, favoring mono-His ligation in the ferrous state.42 Two examples also exist of oxyferrous states in engineered variants of cytochrome c (horse heart cytochrome c, S. cerevisiae iso-1-cytochrome c)22,23,27 where the distal heme-ligating methionine ligands are replaced with alanine, freeing the sixth coordination site for ligand binding. The oxyferrous state is exceptionally long-lived in the engineered iso-1-cytochrome c,27 exhibiting an autoxidation rate that is significantly lower than that of myoglobin.26 Taken with the oxygen-binding capabilities of 2, we therefore conclude that the covalent linkages of heme C do not interfere with or limit the oxygen binding capabilities of c-type cytochromes, and that the lack of characterized, natural oxygen-binding c-type cytochromes is not due to an intrinsic issue with the biophysical properties of heme C per se. We also believe our heme C maquettes have the capacity for further incorporation of functional elements that may support the activation of bound molecular oxygen for catalytic hydroxylation. Indeed our cytochrome c maquette displays an oxyferrous autoxidation rate that falls well within the range of the oxygen binding and activating cytochromes P450,29–31 thus potentially providing a robust scaffold for studying oxygen activation in the context of an artificial protein.
In engineering artificial redox proteins, there is a great advantage to covalently securing the tetrapyrrole redox cofactor. The covalent modification confers on the protein an effectively infinite affinity for heme. Heme C redox and biophysical properties can thus be modulated by selecting alternative ligation schemes such as 6-coordinate His/Met and 5-coordinate mono-His, facilitating site differentiation in a maquette over hundreds of millivolts scale and the formation of 5-coordinate catalytic heme sites for small molecule binding and subsequent redox catalysis. Such modulation of cofactor reduction potential is a key factor in the functional engineering of natural heme-containing oxidoreductases, largely dictating the enzymatic function of the cofactor,2 or, when incorporated into a chain of redox cofactors, the driving force for and direction of electron transfer.32
Furthermore, the covalent incorporation of heme should enable dense packing of hemes within an artificial cytochrome. Natural b-type cytochromes rarely contain more than 2 hemes per polypeptide chain or discrete functional unit, while there are many examples of c-type cytochromes with between 4 and 34 hemes per polypeptide chain.43 These multiheme c-type cytochromes often arrange their heme cofactors in close proximity to support very rapid electron transfer rates across long distances, functioning very much like a protein-insulated wire. The protein maquettes reported here might be redesigned for multi-electron reduction and oxidation of small molecules or expressible, self-assembling molecular wires for extracellular metal reduction.
Since maquettes have a demonstrated ability to accommodate a wide range of cofactors in a relatively malleable interior, further development of these artificial multi-cofactor oxidoreductases can exploit a rich combinatorial diversity of cofactor moderated functions within a single protein,44 often with only minor modification of the protein framework, ranging from reversible oxygen binding (an obligate intermediate in cytochrome P450 and heme dioxygenase catalysis3) to light-activated electron transfer.
Materials and methods
Molecular biology, expression and purification
Synthetic genes were obtained from DNA2.0 (USA), and expressed from a modified variant of pMal-p4x (pSHT) and the proteins were purified using a combination of standard nickel-afinity chromatography and high performance liquid chromatography (as described in the ESI†). The preparation of metal-free and Zn-substituted 2 is described in the ESI.†
Spectroscopy and biophysical characterization
Detailed procedures for maquette characterization by UV/visible, fluorescence, EPR, NMR and CD spectroscopies, stopped-flow spectrophotometry, AUC, MALDI-TOF mass spectrometry and spectroelectrochemistry are contained within the Materials and methods section of the ESI.†
Supplementary Material
Acknowledgements
This work was supported at the University of Bristol by the BBSRC (grant no.: BBI014063/1), the Royal Society, through a University Research Fellowship to JLRA. Research on the development of genes, molecular biology supplies for cloning and mutagenesis, protein and heme C expression and purification from E. coli, and the work on the biophysical and spectroscopic characterization of the heme C maquette as an oxygen transporter was supported at the University of Pennsylvania by the US National Institutes of Health, General Medical Institutes [RO1 GM 41048]. Research on converting the C-heme maquette into a light-active metal free, Zn-porphyrin maquettes and their fluorescence characterization was supported by the US Department of Energy Office of Basic Energy Sciences, Energy Frontier Research Center (PARC) (DE-PSE02-08ER15944 to P.L.D. and C.C.M.). Heme B binding to Zn-porphyrin maquette experiments and its fluorescence characterization is supported by the US Department of Energy, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering [DE-FG02-05ER46223 to P.L.D.]. The Authors wish to thank Prof. Dek Woolfson (Bristol) for kindly providing access to his equipment, Prof. Stuart Ferguson (Oxford) for his kind gift of the pEC864 vector and E. coli strain EC65, and Prof. Graeme Reid (Edinburgh) for his kind gift of the pEC86 vector.
Footnotes
Electronic supplementary information (ESI) available: Experimental methods, supplementary Tables S1–3 and supplementary Fig. S1–11. See DOI: 10.1039/c3sc52019f
Notes and references
- 1.Channon K, Bromley EH, Woolfson DN. Curr. Opin. Struct. Biol. 2008;18:491–498. doi: 10.1016/j.sbi.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Lichtenstein BR, Farid TA, Kodali G, Solomon LA, Anderson JLR, Sheehan MM, Ennist NM, Fry BA, Chobot SE, Bialas C, Mancini JA, Armstrong CT, Zhao Z, Esipova TV, Snell D, Vinogradov SA, Discher BM, Moser CC, Dutton PL. Biochem. Soc. Trans. 2012;40:561–566. doi: 10.1042/BST20120067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong CT, Watkins DW, Anderson JLR. Dalton Trans. 2013;42:3136–3150. doi: 10.1039/c2dt32010j. [DOI] [PubMed] [Google Scholar]
- 4.Stevens JM, Daltrop O, Allen JWA, Ferguson SJ. Acc. Chem. Res. 2004;37:999–1007. doi: 10.1021/ar030266l. [DOI] [PubMed] [Google Scholar]
- 5.Kakar S, Hoffman FG, Storz JF, Fabian M, Hargrove MS. Biophys. Chem. 2010;152:1–14. doi: 10.1016/j.bpc.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koder RL, Anderson JLR, Solomon LA, Reddy KS, Moser CC, Dutton PL. Nature. 2009;458:305–309. doi: 10.1038/nature07841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakrabartty A, Doig AJ, Baldwin RJ. Proc. Natl. Acad. Sci. U. S. A. 1993;90:11332–11336. doi: 10.1073/pnas.90.23.11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Negron C, Fufezan C, Koder RL. Proteins. 2009;74:400–416. doi: 10.1002/prot.22143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.di Guan C, Li P, Riggs PD, Inouye H. Gene. 1988;67:21–30. doi: 10.1016/0378-1119(88)90004-2. [DOI] [PubMed] [Google Scholar]
- 10.Braun M, Thony-Meyer L. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12830–12835. doi: 10.1073/pnas.0402435101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arslan E, Schulz H, Zuffery R, Kunzler P, Thony-Meyer L. Biochem. Biophys. Res. Commun. 1998;251:744–747. doi: 10.1006/bbrc.1998.9549. [DOI] [PubMed] [Google Scholar]
- 12.Mavridou DAI, Stevens JM, Monkemeyer L, Daltrop O, di Gleria K, Kessler BM, Ferguson SJ, Allen JWA. J. Biol. Chem. 2011;287:2342–2352. doi: 10.1074/jbc.M111.313692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulz H, Thony-Meyer L. J. Bacteriol. 2000;182:6831–6833. doi: 10.1128/jb.182.23.6831-6833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daltrop O, Allen JW, Willis AC, Ferguson SJ. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7872–7876. doi: 10.1073/pnas.132259099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daltrop O, Ferguson SJ. J. Biol. Chem. 2004;279:45347–45353. doi: 10.1074/jbc.M408637200. [DOI] [PubMed] [Google Scholar]
- 16.Teale FW. Biochim. Biophys. Acta. 1959;35:543. doi: 10.1016/0006-3002(59)90407-x. [DOI] [PubMed] [Google Scholar]
- 17.Ishida M, Dohmae N, Shiro Y, Oku T, Iizuka T, Isogai Y. Biochemistry. 2004;43:9823–9833. doi: 10.1021/bi049546e. [DOI] [PubMed] [Google Scholar]
- 18.Barker PD, Nerou EP, Freund SMV, Fearnley IM. Biochemistry. 1995;34:15191–15203. doi: 10.1021/bi00046a027. [DOI] [PubMed] [Google Scholar]
- 19.Berry EA, Trumpower BL. Anal. Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 20.Yamanaka T. The Biochemistry of Bacterial Cytochromes. Japan Scientific Societies Press and Springer-Verlag; 1992. [Google Scholar]
- 21.Iakovleva O, Reiner M, Rau H, Haehnel W, Parak F. Phys. Chem. Chem. Phys. 2002;4:655–660. [Google Scholar]
- 22.Lu Y, Casimiro DR, Bren KL, Richards JH, Gray HB. Proc. Natl. Acad. Sci. U. S. A. 1993;90:11456–11459. doi: 10.1073/pnas.90.24.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallace CJA, Clark-Lewis I. J. Biol. Chem. 1992;267:3852–3861. [PubMed] [Google Scholar]
- 24.Meyer TE, Cusanovich MA. Biochim. Biophys. Acta. 1985;80:308–319. doi: 10.1016/0005-2728(85)90263-4. [DOI] [PubMed] [Google Scholar]
- 25.Dewilde S, Kiger L, Burmester T, Hankeln T, Baudin-Creuza V, Aerts T, Marden MC, Caubergs R, Moens L. J. Biol. Chem. 2001;276:38949–38955. doi: 10.1074/jbc.M106438200. [DOI] [PubMed] [Google Scholar]
- 26.Shibata T, Matsumoto D, Nishimura R, Tai H, Matsuoka A, Nagao S, Matsuo T, Hirota S, Imai K, Neya S, Suzuki A, Yamamoto Y. Inorg. Chem. 2012;51:11955–11960. doi: 10.1021/ic301848t. [DOI] [PubMed] [Google Scholar]
- 27.Bren K, Gray HB. J. Inorg. Biochem. 1993;51:111. [Google Scholar]
- 28.Klarskov K, Van Driessche G, Backers K, Dumortier C, Meyer TE, Tollin G, Cusanovich MA, Van Beeumen JJ. Biochemistry. 1998;37:5995–6002. doi: 10.1021/bi972498w. [DOI] [PubMed] [Google Scholar]
- 29.Eisenstein L, Debey P, Douzou D. Biochem. Biophys. Res. Commun. 1977;77:1377–1383. doi: 10.1016/s0006-291x(77)80131-9. [DOI] [PubMed] [Google Scholar]
- 30.Denisov IG, Grinkova YV, McLean MA, Sligar SG. J. Biol. Chem. 2007;282:26865–26873. doi: 10.1074/jbc.M704747200. [DOI] [PubMed] [Google Scholar]
- 31.Sevrioukova IF, Peterson JA. Arch. Biochem. Biophys. 1995;317:397–404. doi: 10.1006/abbi.1995.1180. [DOI] [PubMed] [Google Scholar]
- 32.Page CC, Moser CC, Chen X, Dutton PL. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 33.Vanderkooi JM, Landesberg R, Hayden GW, Owen CS. Eur. J. Biochem. 1977;81:339–347. doi: 10.1111/j.1432-1033.1977.tb11957.x. [DOI] [PubMed] [Google Scholar]
- 34.Sener M, Strumpfer J, Hsin J, Chandler D, Scheuring S, Hunter CN, Schulten K. ChemPhysChem. 2011;12:518–531. doi: 10.1002/cphc.201000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elove GA, Bhuyan AK, Roder H. Biochemistry. 1994;33:6925–6935. doi: 10.1021/bi00188a023. [DOI] [PubMed] [Google Scholar]
- 36.Dumont ME, Corin AF, Campbell GA. Biochemistry. 1994;33:7368–7378. doi: 10.1021/bi00189a043. [DOI] [PubMed] [Google Scholar]
- 37.Feng Y, Sligar SG, Wand AJ. Nat. Struct. Biol. 1994;1:30–35. doi: 10.1038/nsb0194-30. [DOI] [PubMed] [Google Scholar]
- 38.Arnesano F, Banci L, Bertini I, Ciofi-Baffoni S, Woodyear T. de L., Johnson CM, Barker PD. Biochemistry. 2000;39:1499–1514. doi: 10.1021/bi991831o. [DOI] [PubMed] [Google Scholar]
- 39.Faraone-Menella J, Tezcan FA, Gray HB, Winkler JR. Biochemistry. 2006;45:10504–10511. doi: 10.1021/bi060242x. [DOI] [PubMed] [Google Scholar]
- 40.San Francisco B, Bretsynder EC, Kranz RG. Proc. Natl. Acad. Sci. U. S. A. 2012;110:E788–E797. doi: 10.1073/pnas.1213897109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asher WB, Bren KL. Chem. Commun. 2012;48:8344–8346. doi: 10.1039/c2cc31112g. [DOI] [PubMed] [Google Scholar]
- 42.Quintas PO, Catarino T, Todorovic S, Turner DL. Biochemistry. 2011;50:5624–5632. doi: 10.1021/bi200480a. [DOI] [PubMed] [Google Scholar]
- 43.Mowat CG, Chapman SK. Dalton Trans. 2005;7:3381–3389. doi: 10.1039/b505184c. [DOI] [PubMed] [Google Scholar]
- 44.Farid TA, Kodali G, Solomon LA, Lichtenstein BR, Sheehan MM, Fry BA, Bialas C, Ennist NM, Siedlecki JA, Zhao Z, Stetz MA, Valentine KG, Anderson JLR, Wand AJ, Discher BM, Moser CC, Dutton PL. Nat. Chem. Biol. 2013 doi: 10.1038/nchembio.1362. DOI: 10.1038/NCHEMBIO.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.