Abstract
Immune recognition and elimination of cancerous cells is the primary goal of cancer immunotherapy. However, obstacles including immune tolerance and tumor-induced immunosuppression often limit beneficial immune responses. Vaccination is one proposed intervention that may help to overcome these issues and is an active area of study in cancer immunotherapy. Immunizing with tumor antigenic peptides is a promising, straight-forward vaccine strategy hypothesized to boost preexisting antitumor immunity. However, tumor antigens are often weak T cell agonists, attributable to several mechanisms, including immune self-tolerance and poor immunogenicity of self-derived tumor peptides. One strategy for overcoming these mechanisms is vaccination with mimotopes, or peptide mimics of tumor antigens, which alter the antigen presentation and/or T cell activation to increase the expansion of tumor-specific T cells. Evaluation of mimotope vaccine strategies has revealed that even subtle alterations in peptide sequence can dramatically alter antigen presentation and T cell receptor recognition. Most of this research has been performed using T cell clones, which may not be accurate representations of the naturally occurring antitumor response. The relationship between clones generated after mimotope vaccination and the polyclonal T cell repertoire is unclear. Our work with mimotopes in a mouse model of colon carcinoma has revealed important insights into these issues. We propose that the identification of mimotopes based on stimulation of the naturally responding T cell repertoire will dramatically improve the efficacy of mimotope vaccination.
Keywords: Mimotope, Affinity, Vaccine, Altered peptide ligands, Modified peptide, Heteroclitic peptide
Introduction
In the early nineteenth century, a New York surgeon named William Coley developed a treatment based on the observation of spontaneous regression of sarcomas in patients with erysipelas, an acute streptococcus bacterial infection [1]. By injecting live or inactivated Streptococcus pyogenes into the tumors, Coley created an inflammation storm that resulted in destruction of tumor cells by the immune system in up to 40 % of his patients. Although few of the mechanisms were understood at the time, Coley’s trials demonstrated the power of activating the immune system to combat cancer cell growth. A half-century later, the cancer immunosurveillance hypothesis was proposed by Burnet and Thomas, which postulated that the immune system monitors and eliminates tumor growth by recognizing the transforming mutations as neo-antigens [2, 3].
Since then, the role of the immune system in cancer surveillance, development, and elimination has been debated [4, 5]. For example, CBA/H nude mice, lacking T cells, were often cited as not having increased susceptibility to spontaneous tumor formation, suggesting the immune system does not monitor tumor growth [6, 7]. However, nude mice are not completely immunodeficient, retaining some αβ T cells and natural killer (NK) cells, which play an important role in eliminating tumor cells [8]. More recent findings that reinvigorated the concept of immunosurveillance include observations that mice deficient in key components of T cell-mediated immunity (RAG−/−, STAT−/−, and IFNγ/IFNγR−/−) are more susceptible to spontaneous, transplantable, and chemically induced tumors [9, 10]. Furthermore, adoptively transferred autologous CD8+ T cells from melanoma patients result in tumor regression, definitively demonstrating that the immune system can be utilized to target and eliminate tumor cells [11].
Evidence that T cells of the immune system can monitor and prevent tumor growth is significant, yet there is also evidence that the immune system is involved in ‘sculpting’ the tumor to avoid further immune detection [12]. Schreiber and colleagues collated evidence that encompasses the interaction between the immune system and cancer into a model called the three E’s of cancer immunoediting: elimination, equilibrium, and escape [12–14]. Most of what is described in the immunosurveillance hypothesis is also included in the elimination phase of immunoediting, with updates incorporating innate immunity and more molecular details. The equilibrium phase is characterized by the genomic instability of the tumor and the selective pressure against the tumor by the immune response. Tumor escape variants occur in several models, in which the immune system is involved in selecting tumor cells that lose expression of antigens or major histocompatibility complexes (MHC) over time [13, 15–17]. Recently, Matsushita et al. described a T cell-dependent process whereby pre-existing tumor cell clones lacking highly antigenic proteins are preferentially selected for survival [18]. In the escape phase of immunoediting, tumors can produce a wide array of immunosuppressive factors and utilize regulatory arms of the immune system to avoid immune destruction [19–21]. Regulatory T cells (Tregs) and myeloid-derived suppressor cells (MDSCs) are two examples of immune cell types that allow tumors to avoid T cell-mediated destruction.
Most of the focus in tumor immunotherapy has been on enhancing antitumor T cell responses, particularly CD8+ cytotoxic T lymphocytes (CTLs). CTLs recognize short peptide sequences (8–10 amino acids) from proteins within the cell, which are presented on the cell surface in the groove of host MHC class I molecules. CD8+ T cell responses are often initiated by antigen-presenting cells (APCs), primarily dendritic cells (DCs), which process and present peptides derived from self-antigens, tumor cells, or virally infected cells [22]. The discovery and characterization of several tumor-derived antigens recognized by T cells have resulted in an increased interest in exploiting the specificity of CTLs to target cancer cells [23]. Current immunotherapies against cancer that harness the specificity and function of CTLs include adoptive cell transfer of tumor-specific T cells, antibody blockade of T cell inhibitory molecules, and both therapeutic and prophylactic vaccination strategies [5]. While this review focuses on the challenges of inducing immune responses toward tumor antigens, particularly through the use of variant peptide vaccines, or mimotope vaccines, it is important to realize that numerous creative strategies are being developed to overcome tumor-mediated immune inhibitory mechanisms and suppression. Since ‘magic bullet’ discoveries in cancer immunotherapy are rare, success in harnessing the immune system to treat cancer will most likely require combinations of current therapies, as well as the optimization of treatment regimens.
Characteristics of an effective CD8+ T cell response
CD8+ T cells express antigen receptors (TCRs) composed of two polypeptides referred to as the α and β chain. Immense variability within receptors of different T cell clones is generated by random rearrangements of gene segments during development pairing Vα and Jα gene segments and Vβ, Dβ, and Jβ gene segments [24]. Additionally, random nucleotide insertions and deletions in the hypervariable regions within the third complementary-determining regions (CDR3) add additional diversity, and typically, this region of the TCR directly contacts the peptide during antigen recognition [25]. Upon antigen encounter within lymphoid organs, CD8+ T cells rapidly proliferate, differentiate into CTL, and migrate to the site of antigen persistence or inflammation to eliminate cells that express the target antigen. Upon clearance, a majority of the activated CTL undergo apoptosis while a small subset are maintained as memory CD8+ T cells that rapidly proliferate and display effector function upon reencountering antigen [26]. The strength of the pMHC–TCR interaction [27], costimulation [28], and cytokine milieu [29] all contribute to the magnitude of expansion and differentiation profile of the responding T cells. Other factors, including CD4+ T cell help, contribute to the differentiation of effective memory cells [30]. As discussed below, the quality of the T cell response is often attributed to the recognition of peptide-MHC (pMHC) and is reflective of the affinity and the kinetics of pMHC–TCR interactions.
The functional avidity of T cells is the sensitivity of a T cell to activation by pMHC molecules [31]. Expansion of high-avidity antigen-specific T cells correlates with enhanced antiviral and antitumor immunity in several mouse and human diseases [32]. Additionally, there is a relative lack of correlation between the frequency of antigen-specific CD8+ T cells and the control of virus or tumor growth, indicating that the quality of the T cell response is a determining factor for antigen clearance [33]. The density and affinity of several cognate receptor–ligand interactions, including pMHC–TCR, the CD8 co-receptor, CD28/CD80/CD86, and ICAM-1/LFA-1, influence functional avidity (reviewed in [34]). T cells with increased functional avidity produce cytokines and kill antigen-expressing targets at lower antigen concentrations than T cells with low functional avidity. T cell responses to HIV and influenza demonstrate that T cells with increased functional avidity also produce multiple cytokines (IFNγ, TNFα, IL-2, and MIP-1β) and possess potent antiviral and antitumor activity [35, 36] (Fig. 1). Mobilization of effector molecules is triggered more quickly in highly sensitive T cells, suggesting these T cells may be desirable to fight pathogens [37]. Consequently, high-avidity T cells, which proliferate extensively, may become exhausted or lose replicative potential if exposure to antigen is too high [32]. Nevertheless, generating high-avidity T cell responses is hypothesized to provide the best opportunity for success in cancer immunotherapy. Unfortunately, several mechanisms that have evolved to protect the host from autoimmunity may also limit high-avidity T cell responses against tumor antigens.
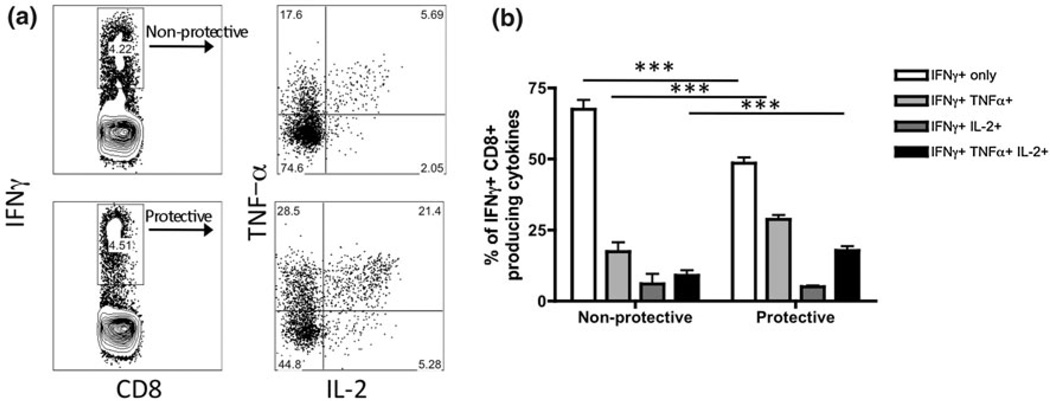
Fig. 1.
Vaccine-elicited T cells that protect mice from tumor challenge produce multiple cytokines. a BALB/c mice were immunized with two different vaccines, which elicit antigen-specific CD8+ T cells that protect mice (protective) from tumor challenge or do not (non-protective). Splenocytes were isolated from mice and stimulated with antigen for 5 h in the presence of monensin. Cells were then stained with antibodies specific for cytokines IFNγ, TNFα, and IL-2. b Combined data plotted from a. *** p < 0.001
Tumor antigens
Early evidence for the existence of tumor antigens was demonstrated bythe immunizationofmice with irradiated tumors, which protected mice from subsequent rechallenge with the same tumor [38]. Since then, numerous T cell-defined tumor antigens have been identified (http://www.cancerimmunity.org/peptidedatabase/Tcellepitopes.htm). Tumor antigens are often categorized as tumor-specific antigens (TSAs) and tumor-associated antigens (TAAs). TSAs typically consist of mutated or virus-derived epitopes and contain unique immunogenic neo-antigens that can be recognized by the immune system. Mutations from cell transformation that affect T cell epitopes, suchas N-RAS and p53, are in this category [39, 40]. Genomic analysis of breast and colorectal tumors revealed that hundreds of mutations occur during the transformation process [41], yet how many of these mutations generate neo-antigens recognized by T cells is unknown. However, epitope algorithm programs have estimated that individual cancers may accumulate on average 7–10 unique HLA-A*0201 neo-antigens that can be recognized by T cells [42]. Unfortunately for cancer immunotherapy, most mutations in cancer are unpredictable and antigenic epitopes need to be identified prior to the treatment.
Conversely, viral antigens exposed during oncogenic virus infection provide unique pathogen-derived targets shared between those infected. Several viral infections linked to human malignancy include human papilloma virus (HPV) [43], Epstein–Barr virus [44], hepatitis B (HBV), and hepatitis C [45, 46]. The recent approval of HPV and HBV vaccines to protect against cervical neo-plasia and hepatocellular carcinoma, respectively, was significant successes for the field [47, 48]. However, many tumors do not express virally derived foreign epitopes, but instead express TAAs, derived from common self-proteins.
Tumor-associated antigens are often shared among patients and different tumor types, making them attractive targets for cancer immunotherapy. Categorized based on their origin or expression pattern, they include differentiation antigens, cancer–testis (CT) antigens, and overex-pressed normal antigens. While differentiation antigens are often restricted to the tissue of origin and normal cells (i.e., Melan-A/MART-1 is expressed in melanoma and normal melanocytes), CT antigens are expressed by multiple tumor types and immunoprivileged sites [49]. Recently, Andersen et al. reported on the antigen recognition pattern of tumor-infiltrating lymphocytes (TILs) for all described tumor-associated antigens in melanoma using a novel technique involving combinatorial coding of peptide multimers. Analyzing several TIL cultures from 19 patients for reactivity against the peptide library of 175 known melanoma-associated epitopes, they reported that responses predominantly targeted differentiation and CT antigens [50]. Thus, differentiation and CT antigens may be ideal targets in cancer immunotherapy, despite some level of risk in generating autoimmune responses. Because these are self-proteins, however, T cells with high avidity for these antigens are often eliminated during selection.
T cell tolerance to tumor antigens
The diverse repertoire of T cells has evolved to combat foreign pathogens and kill unhealthy cells, while limiting reactivity to self-proteins. T cells undergo a rigorous selection process of central tolerance in the thymus, eliminating T cells with high avidity for self-MHC molecules, while allowing those below a certain signal threshold to survive [51]. This selection process protects the host from autoimmunity while the surviving T cells populate the periphery and protect the host from pathogens [52]. However, because many tumor antigens are derived from non-mutated self-proteins (discussed below), central tolerance deletes high-avidity T cells, leaving behind T cells that recognize tumor antigens poorly.
Central tolerance
Several murine tumor models have demonstrated that genetic deletion of genes encoding immunodominant tumor antigens results in the selection of T cells with increased avidity populating the periphery. Our laboratory utilizes the CT26 colon carcinoma model in which the immunodominant MHC class I H-2Ld-restricted epitope, referred to as AH1, is a non-mutated peptide derived from amino acids 423–431 of the gp70 envelope protein of an endogenous ecotropic murine leukemia virus [53]. Mice deficient in gp70 are more resistant to CT26 tumor challenge, and their CD8+ T cells following immunization with the AH1 peptide display increased staining intensity with the AH1-H-2Ld multimers, suggestive of a higher avidity T cell response [54]. Huijbers et al. [55] recently demonstrated similar results using the non-mutated cancer–testis antigen P1A from the mastocytoma tumor P815. Functional CD8+ T cell responses following vaccination with the H-2Ld-restricted P1A35–43 epitope are detectable in DBA/2 mice that express P1A in the thymus, suggesting incomplete central tolerance to this antigen. However, P1A-deficient mice demonstrated enhanced P1A-specific T cell responses and increased tumor protection from P1A-expressing P815 tumor cells. Furthermore, TCR-Vβ usage of P1A-specific T cells was slightly altered, suggesting that ectopic expression of P1A during thymic development skews the peripheral T cell repertoire [55]. These results and others support a role for central tolerance in limiting, not eliminating, T cell responses against self-antigens [56, 57].
Peripheral tolerance
Some T cells specific for self-antigens escape the thymus, despite negative selection. Two mechanisms that explain how T cells avoid negative selection include poor presentation of antigen in the thymus during selection and low affinity of the TCR for pMHC, below the threshold for negative selection [58–60]. Self-specific T cells that escape negative selection and enter the periphery are often held in check by peripheral tolerance mechanisms including low functional avidity for cognate antigen, lack of sufficient dendritic cell maturation or costimulation, deletion, anergy, and suppression by Tregs (reviewed in [61]). Depletion of Tregs protects mice from CT26 tumor challenge and enhances ex vivo IFNγ production from CD8+ T cells in response to the AH1 peptide, demonstrating specific suppression by Tregs of AH1-specific cells [62, 63]. Additionally, as gp70 expression increases with age in BALB/c mice lymphoid tissues, T cell responses following immunization with variant peptides that typically induce immune protection are suppressed [64]. Examination of naïve AH1-specific T cells in these aged mice indicated that these precursor cells are not deleted in the periphery, but instead express high levels of PD-1, indicative of an exhausted phenotype [64]. Recently, antigen expression by lymph node stromal cells has been implicated as a potential source of antigen presentation whereby tolerance against a self-tumor antigen is maintained [65–67]. These results have important implications for future vaccination studies when tumor burden or antigen expression is increased.
Cancer vaccines
Endogenous responses to tumor antigens are detected in both the peripheral blood and tumor of many patients with cancer [37, 68]. Although detectable, endogenous T cells typically fail to control tumor growth, for reasons discussed above. Augmenting T cell responses through vaccination is one strategy to improve anti-tumor immunity. Several vaccine strategies have been studied, including whole-tumor cells irradiated and modified to express immune- modulating cytokines, full-length proteins, autologous DC vaccines pulsed with antigen, and synthetic peptide vaccines (reviewed in [69]). Vaccination as a treatment modality for cancer has resulted in suboptimal success until recently with the first FDA-approved vaccine for hormone-resistant prostate cancer, Sipuleucel-T (Provenge), which extended the life of these terminal patients by 4 months in a Phase III clinical trial [70]. Although a success for cancer immunotherapy, Provenge is a costly and patient-specific autologous cell-based vaccine, causing concern about its practicality as a long-term treatment solution. Alternatively, synthetically derived peptides consisting of known epitopes for CD8+ cytotoxic T cells have been tested in numerous cancer vaccines [49]. While limited by identification of appropriate MHC haplotypes for each epitope, peptide vaccines are less costly, straightforward to produce, safe, and responding T cells can be readily monitored by MHC multimers or stimulation assays [49].
Vaccination with a common Melan-A/MART-1 TAA-peptide in melanoma patients elicits T cells with increased avidity compared with T cells prior to vaccination, but does not result in tumor regression [71]. Several peptide vaccines have been tested in clinical trials, yet clinical outcomes remain poor, with overall objective response rates around 3–4 % (reviewed in [72]). Although vaccination with TAA-peptides can result in the expansion of antigen-specific T cells, the responses are often quantifiably low and of insufficient avidity to mediate tumor destruction, presumably due to poor antigen presentation or tolerance mechanisms [73]. Several TAA-antigens lacking conserved MHC anchor motifs have been described, resulting in a lower affinity of peptide-MHC binding and poor antigen presentation in the thymus [74]. This poor presentation carries over into the periphery, and tumors expressing these antigens may go unrecognized by peripheral T cells. Observations such as these provide the rationale for examining alternative strategies of peptide vaccination, including mimotope vaccines.
Mimotopes: what are they?
Although touted for specificity, T cell receptors associate with different but related MHC molecules or peptide antigens [75, 76]. If each T cell in a repertoire had only one cognate antigen, or if the repertoire was non-cross-reactive, T cell responses would be less efficient and the repertoire would be impossibly large. It is estimated that for an effective immune response, at least one T cell in a few thousand must respond to a foreign epitope [77]. Additionally, a recent report suggested that a single TCR can recognize more than a million different peptides [78]. T cell cross-reactivity can be exploited to generate desired immune responses using mimotopes. Mimotopes, also referred to as analog peptides, variant peptides, heteroclitic peptides, or altered peptide ligands, are mimics of peptide epitopes, which can augment or antagonize T cell responses (Table 1). Mimotopes contain amino acid substitutions in the peptide sequence that can improve peptide-binding affinity for the MHC [79, 80] and/or alter the interaction of the pMHC–TCR complex [73, 81, 82]. Although not yet perfected, the intention is that vaccination with mimotopes with strategic amino acid substitutions will result in predictable T cell responses toward a desired antigenic specificity. For instance, in experimental autoimmune encephalomyelitis, a mouse model for multiple sclerosis, mimotopes may tolerize, antagonize, or delete autoreactive T cells, thereby inhibiting disease progression [83, 84]. Conversely, in cancer immunotherapy, the goal of mimotopes is to enhance tumor-specific T cell expansion and functional recognition of tumor cells.
Table 1.
HLA-A*0201-restricted tumor antigens and corresponding mimotopes that have been identified in humans
| Antigen | Tumor Type | Wild Type | Mimotope | Reference |
|---|---|---|---|---|
| Her2/neu (654–662) (GP2) | Breast | IISAVVGIL | ILSAVVGIL | [94] |
| IISAVVGIV | ||||
| IISALVGIL | ||||
| gp100209–217 | Melanoma | ITDQVPFSV | IMDQVPFSV | [79] |
| NY-ESO-1157–165 | Melanoma | SLLMWITQC | SLLMWITQL | [96] |
| Breast | SLLMWITQV | |||
| Several tumors | ||||
| Melan-A/MART-126–35 | Melanoma | EAAGIGILTV | ELAGIGILTV | [117] |
| TERT572 | Breast | RLFFYRKSV | YLFFYRKSV | [118] |
| NSCLC | ||||
| Prostate | ||||
| Wilms Tumor1(WT1)126–134 | Leukemia | RMFPNAPYL | YMFPNAPYL | [97] |
| Other solid tumors | ||||
| Her2/neu369–377 | Breast | KIFGSLAFL | KVFGSLAFV | [119] |
| KLFGSLAFV | ||||
| PSA178–187 | Prostate | VISNDVCAQV | VLSNDVCAQV | [120] |
| gp100154–162 | Melanoma | KTWGQYWQV | KLWGQYWQV | [79] |
| gp100280–288 | Melanoma | YLEPGPVA | YLEPGPVV | [79] |
| Survivin | Melanoma | LTLGEFLKL | LMLGEFLKL | [121] |
| Breast | ||||
| Myeloma |
Mechanisms that limit autoreactive T cell responses, as discussed previously, often prevent sufficient T cell responses to self-TAA. Relative to vaccination with native tumor antigens, mimotopes are often more immunogenic, which increases the expansion of antigen-specific T cells. Thorough examinations of the responses to the gp1002M mimotope in humans, the AH1 mimotopes in mice, and others have been reported. As discussed below, mimotopes may also alter the TCR binding surface, which can affect the quality and repertoire of the responding T cells. The goal of stimulating T cells with mimotopes is to generate increased expansion of T cells with high avidity for tumor antigens. However, the mimotope may also stimulate T cells that do not recognize the native tumor antigen or T cells that interfere with the intended response, which are necessary considerations for rational improvements of mimotope vaccines.
Effect of mimotopes on T cell function
Evidence in several experimental systems suggests that mimotopes elicit stronger T cell responses than the native antigen, yet tumor-specific responses are still suboptimal [72, 85, 86]. One of the major concerns with mimotope vaccines is ensuring that T cells responding to the mimotope will efficiently recognize and respond to the naturally processed and presented native tumor antigen [87]. The gp100209–217 antigen is a well-characterized melanoma antigen, which has a fast dissociation rate from HLA-A*0201 [60]. However, substituting the threonine at P2 with methionine (gp1002M) significantly improves peptide-MHC binding and increases antigen presentation [79]. Several anchor-modified mimotopes similar to the gp1002M mimotope have been tested in humans (Table 2), many of which elicit more tumor-specific T cells following immunization than the wild-type antigen. Shortly following the initial identification of gp1002M, Clay et al. [88] demonstrated that only 25 % of T cell clones expanded from PBMCs of patients vaccinated with the gp1002M peptide cross-react with the natural peptide, and even fewer recognize and kill HLA-A*0201+ gp100+ melanoma cells. Stuge et al. also examined gp1002M-specific clones from vaccinated patients and showed their sensitivity to the native tumor antigen is 2–3 orders of magnitude lower than to the gp1002M peptide [89]. These results are puzzling considering structural analysis reveals no significant differences between wild-type and gp1002M peptide bound to HLA-A*0201, despite the mimotope peptide having a ninefold increase in affinity [80]. These results could reflect artifacts of T cell clones generated in culture, with high-affinity gp1002M-specific T cells that do not efficiently recognize the native gp100 peptide being preferentially selected. Another possible explanation is that the nature of the pMHC–TCR interaction is altered upon interaction with TCR and structural differences in the unbound pMHC are not apparent. There is precedence for TCR recognition of pMHC altering the conformation of the peptide in the binding groove [90, 91]. Contrary to these previous reports, Borbulevych et al. [80] demonstrated a strong correlation in the frequency of T cells producing IFNγ upon stimulation with either peptide from PBMCs isolated from melanoma patients immunized with the gp1002M peptide, suggesting a high level of cross-reactivity. However, not addressed in these results is the level of IFNγ or other cytokines produced by responding T cells, as this may be more reflective of the quality of the response toward each antigen. It is possible that the 2M–elicited T cells, albeit cross-reactive, have lower affinity for the native gp100 peptide that precludes efficient recognition and elimination of tumor cells.
Table 2.
Repertoire analysis examining differences between the tumor-specific T cells responding to mimotope and wild-type peptides
| Wild type | Mimotope | Analysis | Results | Reference |
|---|---|---|---|---|
| MART26–35 endogenous | MART26–35 27L vaccine | Vβ analysis of T cell clones |
Different Vβ Higher FA from endogenous |
[89] |
| MART26–35 vaccine | MART26–35 27L vaccine | Sequence of T cell clones | Different in Jα and CDR3α | [100] |
| MART26–35 in vitro expansion |
MART26–35 27L in vitro expansion |
Vβ, Jβ analysis CDR3 sequence |
Distinct non-overlapping repertoires |
[74] |
| CEA-CAP1 in vitro expansion |
CEA-CAP16D in vitro expansion |
Vβ, CDR3 sequence | Distinct repertoires Different FA |
[122] |
| NY-ESO-1157–165 endogenous |
NY-ESO-1157–165 vaccine | Vβ analysis of clones | Different Vβ families | [123] |
FA functional avidity
Other mimotopes for different TAAs have also resulted in T cells with poor recognition of the native antigen. CAP-16D is a mimotope for CAP-1(605–613), derived from the carcinoembryonic antigen (CEA), which increases TCR interactions with pMHC [81]. Examination of healthy individuals and colorectal carcinoma patients showed that CAP-16D-specific T cells have low avidity for the native antigen and the T cells fail to kill tumor cells in vitro [92]. More recently, Speiser et al. [93] conducted a direct comparison between an unmodified Melan-A/ MART-1(26–35) peptide and the mimotope peptide Melan-A/MART-127L. Interestingly, they observed that vaccination with the mimotope elicits nearly twice as many Melan-A/MART-1-specific T cells as vaccination with the native peptide. However, upon further investigation, T cells expanded by the native peptide have increased functional avidity for the tumor antigen, high levels of T cell activation, and enhanced killing of tumor cells [93]. These results suggest that mimotopes may select different T cells than those expanded by the native peptide, and vaccination of cancer patients with mimotopes may result in expansion of low-avidity tumor-specific T cells that do not adequately kill tumor cells.
Effects of amino acid substitutions on antigen structure
One explanation for why mimotopes may elicit T cells with low avidity for the native peptide is that alterations in the native peptide may affect how the peptide binds to the MHC or TCR, each of which could impact TCR interactions. Several mimotopes to date have been compared structurally to the native antigens, some revealing obvious differences that may account for differences in T cell function. Significant structural deviations of key TCR contact residues were revealed between the native HER2/ neu antigens (GP2) and the anchor-modified peptide antigen [94]. Additionally, co-crystallization of an NY-ESO-1157–165-specific TCR bound to HLA-A*0201 wild-type (C9) antigen or to mimotopes (V9 or L9) revealed changes in the surface presented for TCR recognition as an indirect result of position 9 modifications [95, 96]. More recently, a peptide mimotope has been described for the HLA-A*0201-restricted Wilm’s tumor antigen (WT1126–134), which substitutes arginine in position 1 of the peptide with a tyrosine, a technique predicted to improve peptide-binding affinity and pMHC stability [97]. However, recent structural analysis has indicated that while this alteration does not affect the conformation of the peptide in the binding groove, it does alter the positions of charged side chains and significantly reduces the peptide/MHC electrostatic surface potential, which may severely affect TCR recognition [98].
The gp1002M and Melan-A/MART-127L mimotopes have both been analyzed structurally, revealing no significant conformational changes from their native peptide counterparts [80, 99]. Interestingly, Cole et al. [74] demonstrated that the Melan-A/MART-1-specific TCR, MEL5, bound the 27L anchor-modified pMHC with lower affinity than the native pMHC, suggesting that this anchor modification can affect TCR interactions. These results highlight the need for careful consideration in the identification of mimotopes, even anchor-modified mimotopes, as any change in the amino acid sequence of peptide antigens is likely to affect some aspect of TCR–pMHC recognition. As a result, the repertoire of T cells may be altered and T cells responding with high affinity to these substitutions may not be functionally reactive toward the tumor antigen.
Effect of mimotope vaccination on T cell repertoire
Enhancing the affinity for MHC by altering peptide anchor residues was hypothesized to be the safest method of preserving crucial pMHC–TCR contacts, eliciting a repertoire of T cells specific for native antigen [87]. However, a comparison of the T cells responding to wild-type and mimotope peptides showed that, in fact, they have distinct TCR sequences, which result in different antitumor responses (Table 2). Recent sequencing analysis of T cell clones generated from patients vaccinated with either Melan-A/MART-126–35 or the mimotope 27L differ slightly in Jα and CDR3α regions [100]. TCR-Vβ-chain usage is broader after vaccination with the wild-type peptide and shared a ‘public’ CDR3β motif between patients. However, the motif is not among the dominant clones identified in each patient, and T cells expressing the motif do not have increased functional avidity for native antigen. The authors concluded that the repertoires responding to mimotope and native antigen are subtly different, but overlapping and the observed functional differences are attributed to subtle structural changes in the TCR [100]. Additionally, PBMCs isolated from melanoma patients expanded with either the native Melan-A/MART-1 peptide or the 27L mimotope result in distinct T cell repertoires [74], supporting previous data that immunization with these peptides results in T cells with distinct functional properties toward the native antigen [93]. Much of the repertoire analysis is generated from T cell clones following in vitro stimulation and culture, which may skew the T cell population and may not be a true representation of endogenous responses [101]. Future experiments aimed at analyzing the TAA-specific repertoire directly ex vivo in immunized patients may provide more insight into differences between mimotope and native antigen vaccines.
Targeting the ‘optimal’ T cells with mimotope vaccination
How can we predict the most beneficial mimotopes that will activate T cells with high functional avidity for tumor antigens? Of the available antigen-specific T cells, which ones will mediate the most effective antitumor immune response? Answers to these questions are becoming more essential for mimotope identification as the realization that anchor modifications can significantly alter TCR interactions. Shifting attention away from the pMHC, it may be beneficial to evaluate the T cells responding naturally to TAA and identify mimotopes that enhance the expansion and activation of these T cells. Using the CT26 cancer model to study the T cell responses in mice provides the opportunity to test many hypotheses about how to make an effective mimotope vaccine. We have generated a panel of mimotopes for AH1, the immunodominant antigen expressed by CT26, which provide a range of protection from challenge with tumor cells (Table 3). Importantly, immunization with the native AH1 peptide results in weak expansion of AH1-specific T cells that do not protect mice from tumor growth [73, 102, 103]. These mimotope peptides were identified from an ‘alanine-scan’ of the AH1 peptide [102], a combinatorial peptide library [73], and genetically encoded libraries [104]. We selected our mimotopes based on improved binding to and activation of the CT clone, an AH1-specific T cell clone that was expanded in culture using CT26.B7 to stimulate growth [102]. The TCR sequences in 100 % (6/6) of these clones were identical, and adoptive transfer of this T cell clone in mice eliminated 3-day established tumors.
Table 3.
Mimotopes for the immunodominant tumor antigen in CT26, AH1, identified using CT clone
| Peptide/ Mimotope |
Amino acids | % of mimotope cells that cross-react with AH1 |
% of AH1-specific T cells with TCR that express the CDR3β motif |
% of tumor-free survival 60 days after challenge |
Reference |
|---|---|---|---|---|---|
| AH1 | SPSYVYHQF | 100 | 60 | 0 | [53] |
| A5 | SPSYAYHQF | 60 | 45 | 90–100 | [102] |
| F1A5 | FPSYAYHQF | 60 | 75 | 90–100 | [104] |
| 39 | MNKYAYHML | 30 | 40 | 50–60 | [73] |
| 15 | MPKYAYHML | 15 | 43 | 10–20 | [73] |
| WMF | SPTYAYWMF | 10 | 10 | 0–10 | [104] |
Amino acid substitutions in AH1, particularly substitution of the position 5 valine for alanine, dramatically improve binding between the pMHC and the CT-TCR. More importantly, vaccination with the A5 mimotope dramatically increased the expansion of T cells that recognize the native AH1 peptide and increased survival in mice following CT26 challenge [102]. The mechanism as to how the alanine substitution in the A5 peptide improves T cell responses is being addressed structurally. Preliminary structural analysis of the AH1 and A5 peptides bound to H-2Ld reveals subtle changes between the two peptides and indicates that position 5 may be a secondary MHC contact residue (unpublished results). However, we have observed no differences in the binding affinity of each peptide with H-2Ld [103]. The structure of the CT-TCR binding to the A5 or AH1-H-2Ld complex may help elucidate important structural differences occurring during antigen recognition. However, this will only provide a mechanistic explanation as it pertains to a single AH1-specific TCR, while the response following immunization in vivo is polyclonal.
Each of the mimotopes tested in our CT26 model were identified using the AH1-specific CT clone, yet they varied significantly in protection of mice from tumor challenge. Interestingly, the CT clone has a monomeric affinity for AH1 of 5.8 µM, surprisingly high for a self-antigen [102]. Similarly, two well-studied TCRs specific for foreign antigens, the 2C TCR and OT-I TCR, have affinities of 3.9 µM and 5.9 µM for pMHC, respectively [105]. Several mimotopes that stimulated IFNγ production from the CT clone were identified with varying affinities for the CT-TCR, each with increased affinity compared to the AH1 peptide. Protective responses to mimotopes with the highest affinity (KD 1.1–2.0 µM), intermediate affinity (KD 2.5–3.6 µM), and lowest affinity (KD 3.2–4.6 µM) for CT-TCR were compared and the results revealed that interactions of intermediate affinity for this single T cell clone resulted in enhanced T cell function and protective antitumor immunity in vivo [73]. Other reports have since supported these findings, suggesting that high-affinity interactions may not stimulate the most productive T cell responses, or may even lead to a more anergic phenotype when examined in vivo [106]. Importantly, the CT clone identified several high-affinity mimotopes that were non-protective when tested in vivo.
Further analysis indicated that mimotopes that protect mice from tumor growth elicit an increased proportion of mimotope-specific cells that functionally recognize the native tumor antigen [103]. Further characterization of these responses revealed that the repertoire of cross-reactive T cells elicited by protective mimotopes is similar in sequence to those that respond to native AH1 vaccination [107]. Utilizing high-throughput sequencing, we analyzed the TCR CDR3β regions of Vβ8+ AH1-multimer+ T cells following immunization. The limited population of AH1-specific cells elicited by the native AH1 peptide revealed a dominant CDR3β motif in these cells. Additionally, protective mimotopes had increased expansion of AH1-specific T cells expressing the same motif and the frequency of motif-expressing cells correlated positively with tumor-free survival [107]. Importantly, this motif is also enriched in AH1-specific T cells infiltrating the tumors of unvaccinated mice, and these T cells produce more IFNγ upon stimulation with protective mimotopes (unpublished data). These results suggest that effective mimotopes increase the expansion of endogenously responding T cells and the identification of mimotopes that preferentially target these cells may contribute to the best immunotherapies.
One interesting observation from the deep sequencing results from AH1-specific T cells is that the CT clone used to identify each of the mimotopes in our system was only identified one time throughout thousands of sequences (unpublished data). Additionally, the CT-TCR does not express the dominant Vβ8.3 CDR3-motif and pairs with Vα4.11, while each of the motif-expressing clones expanded from our protective mimotope vaccines expressed Vα6 ([107] and unpublished data). These results raise doubts about how representative the CT clone is during the anti-tumor immune response and whether this T cell clone is optimal for identifying mimotope peptides. Alternatively, using endogenously responding antitumor T cells, rather than T cell clones, may improve the identification and efficacy of mimotopes for vaccines. With improved sequencing technologies, it may be possible to gain a better understanding of the CDR3-regions that are preferentially selected during endogenous T cell responses against tumors. It will be interesting to understand how these T cells relate to clones generated in culture and whether targeting mimotope vaccines to these cells will improve antitumor responses.
Future of mimotope identification and vaccination
We propose that mimotopes selected to stimulate the endogenous T cells will be more productive and limit the less-cross-reactive T cells. High-throughput sequencing may facilitate the identification of the naturally responding TAA-specific T cells, as demonstrated in the CT26 system [107]. We identified a dominant Vβ CDR3 motif expressed by AH1-specific T cells that correlated with tumor protection, which led us to generate a ‘representative’ TCR to screen baculovirus-encoding pMHC libraries [108]. Preliminary results show that this TCR does not bind the non-protective mimotopes identified using the CT clone ([107] and unpublished results). Additionally, immunizing mice with mimotopes identified by the representative TCR elicits more AH1-specific T cells, compared to mimotopes identified within the same library by the non-representative AH1-specific CT-TCR (unpublished results). Interestingly, preliminary results suggest that the affinity of the TCR for AH1–H–2Ld may be lower than that of the CT-TCR. Examination of two different Melan-A/MART-1-specific TCRs with different affinities for mimotope versus native peptide suggests that TCRs with high affinity may be preferred targets for vaccination strategies [74]. However, the CT-TCR that we originally identified had high affinity for the tumor antigen and it was not observed in the endogenous response, potentially explaining why several mimotopes identified by this TCR were not effective. While we agree that targeting high-affinity T cells during vaccination is logical, our results stress the importance of targeting representative tumor-specific T cells with mimotope vaccination.
Based on preliminary work in mouse models, it is possible to establish a system to screen large pMHC libraries using naturally responding polyclonal T cells. TIL may also be screened and sorted using MHC multimers for common immunodominant antigenic epitopes (i.e., MAGE, NY-ESO1, gp100, etc.). High-avidity T cells may be sorted based on their intensity of multimer staining, a common indicator of higher avidity T cells [109]. The TAA-specific T cells may be used to screened for mimotopes using several epitope identification techniques including combinatorial peptide libraries [110], bacterial/yeast display [111], or baculovirus-encoding libraries [108]. We are currently using baculovirus as a display platform and have observed that baculovirus virions expressing appropriate pMHC will bind T cells via pMHC–TCR interactions (Fig. 2). Mimotope candidates can be enriched and verified using stimulation assays (proliferation, cytokines) and T cells purified from tumors (Fig. 3). Thus, TIL can be used to identify the most suitable mimotopes.
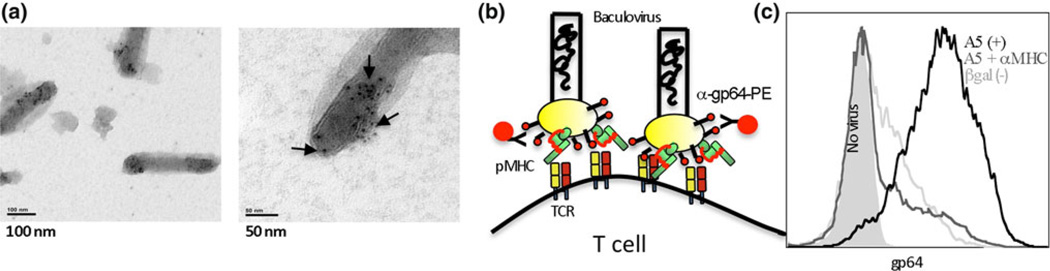
Fig. 2.
Baculovirus-encoding peptide-MHC display libraries are well-characterized systems for screening and identification of new epitopes or mimotopes. Baculovirus (BV) are large, enveloped, double-stranded DNA viruses that have been utilized extensively for expression and production of recombinant proteins. Chimeric proteins encoding the extracellular domain of mammalian MHC, linked to various peptides, and the transmembrane domain of viral glycoprotein gp64 are expressed on the surface of infected insect cells. During the infection cycle, BV ‘bud’ from the cell membrane of infected insect cells taking the membrane-bound recombinant protein with them, resulting in expression on the virion surface [124]. a BV-expressing pMHC complexes were visualized using electron microscopy and immunogold-conjugated antibody specific for MHC. Our work supports previous data demonstrating that ligands on BV bind to cell-bound cognate receptors [125]. b Briefly, rBV-expressing pMHC is incubated with antigen-specific T cells, the excess is washed away, and bound virus is detected using a monoclonal antibody specific for the gp64 viral protein. c We observe that BV-expressing peptides specific for their cognate T cell receptor (A5) bind in a pMHC–TCR complex, while non-specific pMHC (βgal) do not bind as efficiently. Furthermore, blocking with an MHC-specific antibody reduces the A5–H–2Ld interaction. Using this technology, tumor-specific T cells may be used to screen large peptide-MHC libraries expressed by BV. Bound viruses are recovered by trypsinizing the T cell surface proteins and used to infect more insect cells, enriching for peptides that bind specifically to T cells. Individual peptides can be cloned by limiting dilution and screened for antigen-specific function by stimulating T cells to proliferate, produce cytokine, or kill target cells
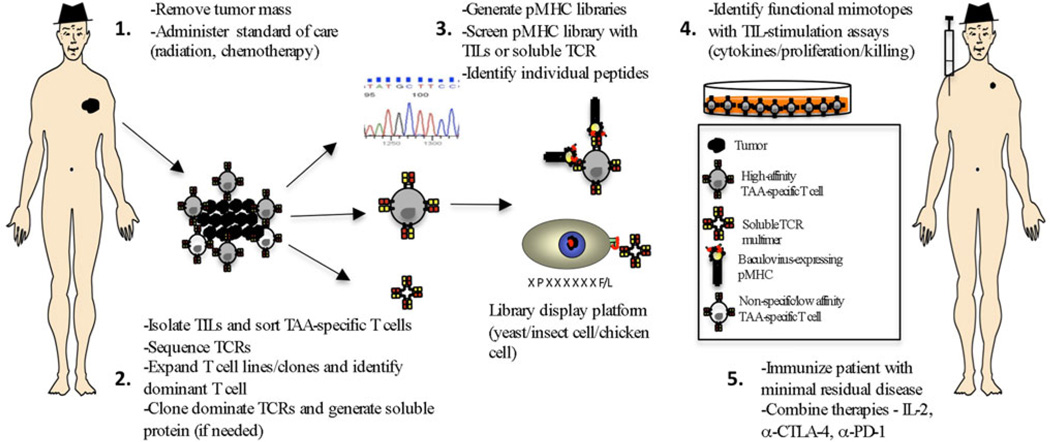
Fig. 3.
Proposed schematic representation using TIL to screen baculovirus-encoding pMHC libraries for mimotopes
Improving the quality of T cell responses elicited by mimotope vaccines will improve antitumor immunity. Optimizing mimotope vaccine regimens by incorporating native antigen as boosts can enhance the quality and quantity of tumor-specific T cells over either vaccine alone (Buhrman et al., submitted, [112]). This strategy selects for high-avidity TAA-specific T cells elicited by the mimotope and may also limit the expansion of mimotope-specific T cells that do not cross-react with the tumor antigen. Peptide vaccines have become more complex, involving combinations of multiple peptides targeting different antigens, which may limit tumor variant outgrowth and escape [113]. Additionally, long-peptide vaccines (~20 amino acids) are a promising strategy because they require processing prior to MHC class I presentation, which may mimic tumor antigen presentation more accurately than short peptides that load directly on APCs without prior processing [114]. Recently, vaccination with a long peptide containing the Melan-A/MART-1 anchor modification 27L resulted in sustained cross-presentation by monocyte-derived DCs and elicited long-lived tumor-reactive T cells [115]. Long peptides without the anchor modification primed T cells poorly, while the short peptide containing the 27L substitution elicited short-lived T cells [115]. Long-peptide vaccines and multi-peptide vaccines may also contain CD4+ T cell epitopes, which provide CD4 help to CD8+ T cells, boosting the response and improving memory [5]. However, some reports have recently suggested that incorporation of CD4+ epitopes in both long-peptide and multi-epitope vaccines results in decreased CD8+ T cell responses [116].
Immunization in combination with other therapies or antibodies targeting PD-1/PDL-1 or CTLA-4 may further improve T cell responses elicited by mimotopes [5]. Lastly, vaccination with mimotopes may be more effective during stages of reduced tumor burden, following resection, for example, or upon clinically diagnosed minimal residual disease. In this way mimotopes will be better suited to activate TAA-specific T cells in the absence of the immunosuppressive tumor microenvironment and may be an important contributory factor in eliminating residual tumor cells. Combining current treatment modalities with mimotope vaccines that stimulate the naturally responding tumor-specific T cells may significantly improve antitumor immunity.
Acknowledgments
This work was supported by ACS RSG-08-184-01-L1B and CA109560. Jonathan D. Buhrman was supported by the Cancer Research Institute Pre-doctoral Emphasis Pathway in Tumor Immunology Fellowship.
Biography
![]()
Jill E. Slansky Jonathan D. Buhrman
Contributor Information
Jonathan D. Buhrman, Integrated Department of Immunology, University of Colorado, School of Medicine, National Jewish Health, 1400 Jackson Street, Room K511, Denver, CO 80206, USA
Jill E. Slansky, Integrated Department of Immunology, University of Colorado, School of Medicine, National Jewish Health, 1400 Jackson Street, Room K511, Denver, CO 80206, USA, Jill.Slansky@ucdenver.edu
References
- 1.Coley WB., II Contribution to the knowledge of Sarcoma. Ann Surg. 1891;14:199–220. doi: 10.1097/00000658-189112000-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burnet M. Cancer: a biological approach. III. Viruses associated with neoplastic conditions IV Practical applications. Br Med J. 1957;1:841–847. doi: 10.1136/bmj.1.5023.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas L. In Cellular and humoral aspects of the hypersensitive states. In: Lawrence eH., editor. Discussion. New York: Hoeber-Harper; 1959. [Google Scholar]
- 4.Quezada SA, et al. Shifting the equilibrium in cancer immunoediting: from tumor tolerance to eradication. Immunol Rev. 2011;241:104–118. doi: 10.1111/j.1600-065X.2011.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stutman O. Chemical carcinogenesis in nude mice: comparison between nude mice from homozygous matings and heterozygous matings and effect of age and carcinogen dose. J Natl Cancer Inst. 1979;62:353–358. [PubMed] [Google Scholar]
- 7.Stutman O. Tumor development after 3-methylcholanthrene in immunologically deficient athymic-nude mice. Science. 1974;183:534–536. doi: 10.1126/science.183.4124.534. [DOI] [PubMed] [Google Scholar]
- 8.Maleckar JR, Sherman LA. The composition of the T cell receptor repertoire in nude mice. J Immunol. 1987;138:3873–3876. [PubMed] [Google Scholar]
- 9.Street SE, et al. Suppression of lymphoma and epithelial malignancies effected by interferon gamma. J Exp Med. 2002;196:129–134. doi: 10.1084/jem.20020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shankaran V, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 11.Dudley ME, et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;298:850–854. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koebel CM, et al. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- 13.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 14.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 15.Maeurer MJ, et al. Tumor escape from immune recognition: lethal recurrent melanoma in a patient associated with down-regulation of the peptide transporter protein TAP-1 and loss of expression of the immunodominant MART-1/Melan-A antigen. J Clin Invest. 1996;98:1633–1641. doi: 10.1172/JCI118958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeurer MJ, et al. Tumor escape from immune recognition: loss of HLA-A2 melanoma cell surface expression is associated with a complex rearrangement of the short arm of chromosome 6. Clin Cancer Res. 1996;2:641–652. [PubMed] [Google Scholar]
- 17.Zhou G, et al. Reciprocal changes in tumor antigenicity and antigen-specific T cell function during tumor progression. J Exp Med. 2004;200:1581–1592. doi: 10.1084/jem.20041240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsushita H, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarnicki AG, et al. Suppression of antitumor immunity by IL-10 and TGF-beta-producing T cells infiltrating the growing tumor: influence of tumor environment on the induction of CD4+ and CD8+ regulatory T cells. J Immunol. 2006;177:896–904. doi: 10.4049/jimmunol.177.2.896. [DOI] [PubMed] [Google Scholar]
- 20.Lizee G, et al. Immunosuppression in melanoma immunotherapy: potential opportunities for intervention. Clin Cancer Res. 2006;12:2359s–65s. doi: 10.1158/1078-0432.CCR-05-2537. [DOI] [PubMed] [Google Scholar]
- 21.Gross S, et al. Immunosuppressive mechanisms in cancer: consequences for the development of therapeutic vaccines. Vaccine. 2009;27:3398–3400. doi: 10.1016/j.vaccine.2009.01.070. [DOI] [PubMed] [Google Scholar]
- 22.Zhang N, Bevan MJ. CD8(+) T cells: foot soldiers of the immune system. Immunity. 2011;35:161–168. doi: 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buonaguro L, et al. Translating tumor antigens into cancer vaccines. Clin Vaccine Immunol. 2010;18:23–34. doi: 10.1128/CVI.00286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson RK, et al. Structure, organization and polymorphism of murine and human T-cell receptor alpha and beta chain gene families. Immunol Rev. 1988;101:149–172. doi: 10.1111/j.1600-065x.1988.tb00736.x. [DOI] [PubMed] [Google Scholar]
- 25.Krogsgaard M, Davis MM. How T cells ‘see’ antigen. Nat Immunol. 2005;6:239–245. doi: 10.1038/ni1173. [DOI] [PubMed] [Google Scholar]
- 26.Lefrancois L, Obar JJ. Once a killer, always a killer: from cytotoxic T cell to memory cell. Immunol Rev. 2010;235:206–218. doi: 10.1111/j.0105-2896.2010.00895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenschow DJ, Walunas TL, Bluestone JA. CD28/B7 system of T cell costimulation. Annu Rev Immunol. 1996;14:233–58. doi: 10.1146/annurev.immunol.14.1.233. [DOI] [PubMed] [Google Scholar]
- 29.Curtsinger JM, et al. Type I IFNs provide a third signal to CD8 T cells to stimulate clonal expansion and differentiation. J Immunol. 2005;174:4465–4469. doi: 10.4049/jimmunol.174.8.4465. [DOI] [PubMed] [Google Scholar]
- 30.Sun JC, Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science. 2003;300:339–342. doi: 10.1126/science.1083317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slifka MK, Whitton JL. Functional avidity maturation of CD8(+) T cells without selection of higher affinity TCR. Nat Immunol. 2001;2:711–717. doi: 10.1038/90650. [DOI] [PubMed] [Google Scholar]
- 32.Appay V, Douek DC, Price DA. CD8+ T cell efficacy in vaccination and disease. Nat Med. 2008;14:623–628. doi: 10.1038/nm.f.1774. [DOI] [PubMed] [Google Scholar]
- 33.Dunbar PR, et al. A shift in the phenotype of melan-A-specific CTL identifies melanoma patients with an active tumor-specific immune response. J Immunol. 2000;165:6644–6652. doi: 10.4049/jimmunol.165.11.6644. [DOI] [PubMed] [Google Scholar]
- 34.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.La Gruta NL, Doherty PC, Turner SJ. A correlation between function and selected measures of T cell avidity in influenza virus-specific CD8+ T cell responses. Eur J Immunol. 2006;36:2951–299. doi: 10.1002/eji.200636390. [DOI] [PubMed] [Google Scholar]
- 36.Almeida JR, et al. Antigen sensitivity is a major determinant of CD8 + T-cell polyfunctionality and HIV-suppressive activity. Blood. 2009;113:6351–6360. doi: 10.1182/blood-2009-02-206557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee PP, et al. Characterization of circulating T cells specific for tumor-associated antigens in melanoma patients. Nat Med. 1999;5:677–685. doi: 10.1038/9525. [DOI] [PubMed] [Google Scholar]
- 38.Prehn RT. Main JM Immunity to methylcholanthrene-induced sarcomas. J Natl Cancer Inst. 1957;18:769–778. [PubMed] [Google Scholar]
- 39.Linard B, et al. A ras-mutated peptide targeted by CTL infiltrating a human melanoma lesion. J Immunol. 2002;168:4802–4808. doi: 10.4049/jimmunol.168.9.4802. [DOI] [PubMed] [Google Scholar]
- 40.Ito D, et al. Immunological characterization of missense mutations occurring within cytotoxic T cell-defined p53 epitopes in HLA-A*0201 + squamous cell carcinomas of the head and neck. Int J Cancer. 2007;120:2618–2624. doi: 10.1002/ijc.22584. [DOI] [PubMed] [Google Scholar]
- 41.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 42.Segal NH, et al. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68:889–892. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 43.Beaudenon S, et al. A novel type of human papillomavirus associated with genital neoplasias. Nature. 1986;321:246–249. doi: 10.1038/321246a0. [DOI] [PubMed] [Google Scholar]
- 44.List AF, Greco FA, Vogler LB. Lymphoproliferative diseases in immunocompromised hosts: the role of Epstein-Barr virus. J Clin Oncol. 1987;5:1673–1689. doi: 10.1200/JCO.1987.5.10.1673. [DOI] [PubMed] [Google Scholar]
- 45.Tsukuma H, et al. Risk factors for hepatocellular carcinoma among patients with chronic liver disease. N Engl J Med. 1993;328:1797–1801. doi: 10.1056/NEJM199306243282501. [DOI] [PubMed] [Google Scholar]
- 46.Beasley RP, et al. Hepatocellular carcinoma and hepatitis Bvirus. A prospective study of 22 707 men in Taiwan. Lancet. 1981;2:1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 47.Koutsky LA, et al. A controlled trial of a human papillomavirus type 16 vaccine. N Engl J Med. 2002;347:1645–1651. doi: 10.1056/NEJMoa020586. [DOI] [PubMed] [Google Scholar]
- 48.Chang MH, et al. Universal hepatitis Bvaccination in Taiwan and the incidence of hepatocellular carcinoma in children. Taiwan Childhood Hepatoma Study Group. N Engl J Med. 1997;336:1855–1859. doi: 10.1056/NEJM199706263362602. [DOI] [PubMed] [Google Scholar]
- 49.Speiser DE, Romero P. Molecularly defined vaccines for cancer immunotherapy, and protective T cell immunity. Semin Immunol. 2010;22:144–154. doi: 10.1016/j.smim.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 50.Sick Andersen R, et al. Dissection of T cell antigen specificity in human melanoma. Cancer Res. 2012;72:1642–1650. doi: 10.1158/0008-5472.CAN-11-2614. [DOI] [PubMed] [Google Scholar]
- 51.Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. Nat Rev Immunol. 2005;5:772–782. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- 52.Kappler JW, Roehm N, Marrack P. T cell tolerance by clonal elimination in the thymus. Cell. 1987;49:273–280. doi: 10.1016/0092-8674(87)90568-x. [DOI] [PubMed] [Google Scholar]
- 53.Huang AY, et al. The immunodominant major histocompatibility complex class I-restricted antigen of a murine colon tumor derives from an endogenous retroviral gene product. Proc Natl Acad Sci USA. 1996;93:9730–9735. doi: 10.1073/pnas.93.18.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McWilliams JA, et al. Age-dependent tolerance to an endogenous tumor-associated antigen. Vaccine. 2008;26:1863–1873. doi: 10.1016/j.vaccine.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huijbers IJ, et al. Minimal tolerance to a tumor antigen encoded by a cancer-germline gene. J Immunol. 2012;188:111–121. doi: 10.4049/jimmunol.1002612. [DOI] [PubMed] [Google Scholar]
- 56.de Visser KE, et al. Low-avidity self-specific T cells display a pronounced expansion defect that can be overcome by altered peptide ligands. J Immunol. 2001;167:3818–3828. doi: 10.4049/jimmunol.167.7.3818. [DOI] [PubMed] [Google Scholar]
- 57.Zehn D, Bevan MJ. T cells with low avidity for a tissue-restricted antigen routinely evade central and peripheral tolerance and cause autoimmunity. Immunity. 2006;25:261–170. doi: 10.1016/j.immuni.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mamula MJ. The inability to process a self-peptide allows autoreactive T cells to escape tolerance. J Exp Med. 1993;177:567–571. doi: 10.1084/jem.177.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colella TA, et al. Self-tolerance to the murine homologue of a tyrosinase-derived melanoma antigen: implications for tumor immunotherapy. J Exp Med. 2000;191:1221–1232. doi: 10.1084/jem.191.7.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu Z, et al. Poor immunogenicity of a self/tumor antigen derives from peptide-MHC-I instability and is independent of tolerance. J Clin Invest. 2004;114:551–559. doi: 10.1172/JCI21695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Redmond WL, Sherman LA. Peripheral tolerance of CD8 T lymphocytes. Immunity. 2005;22:275–284. doi: 10.1016/j.immuni.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 62.Casares N, et al. Immunization with a tumor-associated CTL epitope plus a tumor-related or unrelated Th1 helper peptide elicits protective CTL immunity. Eur J Immunol. 2001;31:1780–1789. doi: 10.1002/1521-4141(200106)31:6<1780::aid-immu1780>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 63.Casares N, et al. CD4 +/CD25 + regulatory cells inhibit activation of tumor-primed CD4 + T cells with IFN-gamma-dependent antiangiogenic activity, as well as long-lasting tumor immunity elicited by peptide vaccination. J Immunol. 2003;171:5931–5939. doi: 10.4049/jimmunol.171.11.5931. [DOI] [PubMed] [Google Scholar]
- 64.Kemmler CB, et al. Elevated tumor-associated antigen expression suppresses variant Peptide vaccine responses. J Immunol. 2011;187:4431–4439. doi: 10.4049/jimmunol.1101555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fletcher AL, Malhotra D, Turley SJ. Lymph node stroma broaden the peripheral tolerance paradigm. Trends Immunol. 2010;32:12–18. doi: 10.1016/j.it.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cohen JN, et al. Lymph node-resident lymphatic endothelial cells mediate peripheral tolerance via Aire-independent direct antigen presentation. J Exp Med. 2010;207:681–688. doi: 10.1084/jem.20092465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lund AW, et al. VEGF-C promotes immune tolerance in B16 melanomas and cross-presentation of tumor antigens by lymph node lymphatics. Cell Rep. 2012;1:191–199. doi: 10.1016/j.celrep.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 68.Kawakami Y, et al. Identification of a human melanoma antigen recognized by tumor-infiltrating lymphocytes associated with in vivo tumor rejection. Proc Natl Acad Sci USA. 1994;91:6458–6462. doi: 10.1073/pnas.91.14.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dougan M, Dranoff G. Immune therapy for cancer. Annu Rev Immunol. 2009;27:83–117. doi: 10.1146/annurev.immunol.021908.132544. [DOI] [PubMed] [Google Scholar]
- 70.Kantoff PW, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010;363:411–422. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 71.Dutoit V, et al. Dissecting TCR-MHC/peptide complex interactions with A2/peptide multimers incorporating tumor antigen peptide variants: crucial role of interaction kinetics on functional outcomes. Eur J Immunol. 2002;32:3285–393. doi: 10.1002/1521-4141(200211)32:11<3285::AID-IMMU3285>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 72.Klebanoff CA, et al. Therapeutic cancer vaccines: are we there yet? Immunol Rev. 2011;239:27–44. doi: 10.1111/j.1600-065X.2010.00979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McMahan RH, et al. Relating TCR-peptide-MHC affinity to immunogenicity for the design of tumor vaccines. J Clin Invest. 2006;116:2543–2551. doi: 10.1172/JCI26936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cole DK, et al. Modification of MHC anchor residues generates heteroclitic peptides that alter TCR binding and T cell recognition. J Immunol. 2010;185:2600–2610. doi: 10.4049/jimmunol.1000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kersh GJ, Allen PM. Structural basis for T cell recognition of altered peptide ligands: a single T cell receptor can productively recognize a large continuum of related ligands. J Exp Med. 1996;184:1259–1268. doi: 10.1084/jem.184.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sloan-Lancaster J, Allen PM. Altered peptide ligand-induced partial T cell activation: molecular mechanisms and role in T cell biology. Annu Rev Immunol. 1996;14:1–27. doi: 10.1146/annurev.immunol.14.1.1. [DOI] [PubMed] [Google Scholar]
- 77.Mason D. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol Today. 1998;19:395–404. doi: 10.1016/s0167-5699(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 78.Wooldridge L, et al. A single autoimmune T cell receptor recognizes more than a million different peptides. J Biol Chem. 2011;287:1168–1177. doi: 10.1074/jbc.M111.289488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parkhurst MR, et al. Improved induction of melanoma-reactive CTL with peptides from the melanoma antigen gp100 modified at HLA-A*0201-binding residues. J Immunol. 1996;157:2539–2548. [PubMed] [Google Scholar]
- 80.Borbulevych OY, et al. Increased immunogenicity of an anchor-modified tumor-associated antigen is due to the enhanced stability of the peptide/MHC complex: implications for vaccine design. J Immunol. 2005;174:4812–4820. doi: 10.4049/jimmunol.174.8.4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zaremba S, et al. Identification of an enhancer agonist cytotoxic T lymphocyte peptide from human carcinoembryonic antigen. Cancer Res. 1997;57:4570–4577. [PubMed] [Google Scholar]
- 82.Salazar E, et al. Agonist peptide from a cytotoxic t-lymphocyte epitope of human carcinoembryonic antigen stimulates production of tc1-type cytokines and increases tyrosine phosphorylation more efficiently than cognate peptide. Int J Cancer. 2000;85:829–838. doi: 10.1002/(sici)1097-0215(20000315)85:6<829::aid-ijc16>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 83.Wasserman HA, et al. MHC variant peptide-mediated anergy of encephalitogenic T cells requires SHP-1. J Immunol. 2008;181:6843–6849. doi: 10.4049/jimmunol.181.10.6843. [DOI] [PubMed] [Google Scholar]
- 84.Katsara M, et al. The good, the bad and the ugly: how altered peptide ligands modulate immunity. Exp Opin Biol Ther. 2008;8:1873–1884. doi: 10.1517/14712590802494501. [DOI] [PubMed] [Google Scholar]
- 85.Tang Y, et al. An altered peptide ligand for naive cytotoxic T lymphocyte epitope of TRP-2(180–188) enhanced immunogenicity. Cancer Immunol Immunother. 2007;56:319–329. doi: 10.1007/s00262-006-0195-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mimura K, et al. Substitution analog peptide derived from HER-2 can efficiently induce HER-2-specific, HLA-A24 restricted CTLs. Cancer Immunol Immunother. 2006;55:1358–1366. doi: 10.1007/s00262-006-0123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iero M, et al. Modified peptides in anti-cancer vaccines: are we eventually improving anti-tumour immunity? Cancer Immunol Immunother. 2009;58:1159–1167. doi: 10.1007/s00262-008-0610-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Clay TM, et al. Changes in the fine specificity of gp100 (209–217)-reactive T cells in patients following vaccination with a peptide modified at an HLA-A2.1 anchor residue. J Immunol. 1999;162:1749–1755. [PubMed] [Google Scholar]
- 89.Stuge TB, et al. Diversity and recognition efficiency of T cell responses to cancer. PLoS Med. 2004;1:e28. doi: 10.1371/journal.pmed.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tynan FE, et al. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat Immunol. 2007;8:268–276. doi: 10.1038/ni1432. [DOI] [PubMed] [Google Scholar]
- 91.Tynan FE, et al. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat Immunol. 2005;6:1114–1122. doi: 10.1038/ni1257. [DOI] [PubMed] [Google Scholar]
- 92.Iero M, et al. Low TCR avidity and lack of tumor cell recognition in CD8(+) T cells primed with the CEA-analogue CAP1–6D peptide. Cancer Immunol Immunother. 2007;56:1979–1991. doi: 10.1007/s00262-007-0342-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Speiser DE, et al. Unmodified self antigen triggers human CD8 T cells with stronger tumor reactivity than altered antigen. Proc Natl Acad Sci USA. 2008;105:3849–3854. doi: 10.1073/pnas.0800080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sharma AK, et al. Class I major histocompatibility complex anchor substitutions alter the conformation of T cell receptor contacts. J Biol Chem. 2001;276:21443–21449. doi: 10.1074/jbc.M010791200. [DOI] [PubMed] [Google Scholar]
- 95.Chen JL, et al. Ca2 + release from the endoplasmic reticulum of NY-ESO-1-specific T cells is modulated by the affinity of TCR and by the use of the CD8 coreceptor. J Immunol. 2010;184:1829–1839. doi: 10.4049/jimmunol.0902103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen JL, et al. Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J Exp Med. 2005;201:1243–1255. doi: 10.1084/jem.20042323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pinilla-Ibarz J, et al. Improved human T-cell responses against synthetic HLA-0201 analog peptides derived from the WT1 oncoprotein. Leukemia. 2006;20:2025–2033. doi: 10.1038/sj.leu.2404380. [DOI] [PubMed] [Google Scholar]
- 98.Borbulevych OY, Do P, Baker BM. Structures of native and affinity-enhanced WT1 epitopes bound to HLA-A*0201: implications for WT1-based cancer therapeutics. Mol Immunol. 2010;47:2519–2524. doi: 10.1016/j.molimm.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Borbulevych OY, et al. Structures of MART-126/27–35 Peptide/ HLA-A2 complexes reveal a remarkable disconnect between antigen structural homology and T cell recognition. J Mol Biol. 2007;372:1123–1136. doi: 10.1016/j.jmb.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wieckowski S, et al. Fine structural variations of alphabet-aTCRs selected by vaccination with natural versus altered self-antigen in melanoma patients. J Immunol. 2009;183:5397–5406. doi: 10.4049/jimmunol.0901460. [DOI] [PubMed] [Google Scholar]
- 101.Dietrich PY, et al. TCR analysis reveals significant repertoire selection during in vitro lymphocyte culture. Int Immunol. 1997;9:1073–1083. doi: 10.1093/intimm/9.8.1073. [DOI] [PubMed] [Google Scholar]
- 102.Slansky JE, et al. Enhanced antigen-specific antitumor immunity with altered peptide ligands that stabilize the MHC-peptide-TCR complex. Immunity. 2000;13:529–538. doi: 10.1016/s1074-7613(00)00052-2. [DOI] [PubMed] [Google Scholar]
- 103.Jordan KR, et al. Peptide vaccines prevent tumor growth by activating T cells that respond to native tumor antigens. Proc Natl Acad Sci USA. 2010;107:4652–4657. doi: 10.1073/pnas.0914879107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jordan KR, et al. Baculovirus-infected insect cells expressing peptide-MHC complexes elicit protective antitumor immunity. J Immunol. 2008;180:188–197. doi: 10.4049/jimmunol.180.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stone JD, Chervin AS, Kranz DM. T-cell receptor binding affinities and kinetics: impact on T-cell activity and specificity. Immunology. 2009;126:165–176. doi: 10.1111/j.1365-2567.2008.03015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Corse E, et al. Attenuated T cell responses to a high-potency ligand in vivo. PLoS Biol. 2010;8(9):e1000481. doi: 10.1371/journal.pbio.1000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jordan KR, et al. TCR hypervariable regions expressed by T cells that respond to effective tumor vaccines. Cancer Immunol Immunother. 2012 doi: 10.1007/s00262-012-1217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Crawford F, et al. Use of baculovirus MHC/peptide display libraries to characterize T-cell receptor ligands. Immunol Rev. 2006;210:156–170. doi: 10.1111/j.0105-2896.2006.00365.x. [DOI] [PubMed] [Google Scholar]
- 109.Crawford F, et al. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity. 1998;8:675–682. doi: 10.1016/s1074-7613(00)80572-5. [DOI] [PubMed] [Google Scholar]
- 110.Pinilla C, et al. Combinatorial peptide libraries as an alternative approach to the identification of ligands for tumor-reactive cytolytic T lymphocytes. Cancer Res. 2001;61:5153–5160. [PubMed] [Google Scholar]
- 111.Adams JJ, et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity. 2011;35:681–693. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vetsika EK, et al. Sequential administration of the native TERT572 cryptic peptide enhances the immune response initiated by its optimized variant TERT(572Y) in cancer patients. J Immunother. 2011;34:641–650. doi: 10.1097/CJI.0b013e31823284a6. [DOI] [PubMed] [Google Scholar]
- 113.Rosenberg SA, et al. Altered CD8(+) T-cell responses when immunizing with multiepitope peptide vaccines. J Immunother. 2006;29:224–231. doi: 10.1097/01.cji.0000190399.98802.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Slingluff CL, Jr, Engelhard VH, Ferrone S. Peptide and dendritic cell vaccines. Clin Cancer Res. 2006;12:2342s–2345s. doi: 10.1158/1078-0432.CCR-05-2541. [DOI] [PubMed] [Google Scholar]
- 115.Chauvin JM, et al. HLA anchor optimization of the melan-A-HLA-A2 epitope within a long peptide is required for efficient cross-priming of human tumor-reactive T cells. J Immunol. 2012;188:2102–2110. doi: 10.4049/jimmunol.1101807. [DOI] [PubMed] [Google Scholar]
- 116.Slingluff CL, Jr, et al. Randomized multicenter trial of the effects of melanoma-associated helper peptides and cyclophosphamide on the immunogenicity of a multipeptide melanoma vaccine. J Clin Oncol. 2011;29:2924–2932. doi: 10.1200/JCO.2010.33.8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Valmori D, et al. Enhanced generation of specific tumor-reactive CTL in vitro by selected Melan-A/MART-1 immunodominant peptide analogues. J Immunol. 1998;160:1750–1758. [PubMed] [Google Scholar]
- 118.Gross DA, et al. High vaccination efficiency of low-affinity epitopes in antitumor immunotherapy. J Clin Invest. 2004;113:425–433. doi: 10.1172/JCI19418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Keogh E, et al. Identification of new epitopes from four different tumor-associated antigens: recognition of naturally processed epitopes correlates with HLA-A*0201-binding affinity. J Immunol. 2001;167:787–796. doi: 10.4049/jimmunol.167.2.787. [DOI] [PubMed] [Google Scholar]
- 120.Terasawa H, et al. Identification and characterization of a human agonist cytotoxic T-lymphocyte epitope of human prostate-specific antigen. Clin Cancer Res. 2002;8:41–53. [PubMed] [Google Scholar]
- 121.Andersen MH, et al. Identification of a cytotoxic T lymphocyte response to the apoptosis inhibitor protein survivin in cancer patients. Cancer Res. 2001;61:869–872. [PubMed] [Google Scholar]
- 122.Hou Y, Kavanagh B, Fong L. Distinct CD8 + T cell repertoires primed with agonist and native peptides derived from a tumor-associated antigen. J Immunol. 2008;180:1526–1534. doi: 10.4049/jimmunol.180.3.1526. [DOI] [PubMed] [Google Scholar]
- 123.Le Gal FA, et al. Distinct structural TCR repertoires in naturally occurring versus vaccine-induced CD8 + T-cell responses to the tumor-specific antigen NY-ESO-1. J Immunother. 2005;28:252–257. doi: 10.1097/01.cji.0000161398.34701.26. [DOI] [PubMed] [Google Scholar]
- 124.Grabherr R, et al. Developments in the use of baculoviruses for the surface display of complex eukaryotic proteins. Trends Biotechnol. 2001;19:231–236. doi: 10.1016/s0167-7799(01)01610-9. [DOI] [PubMed] [Google Scholar]
- 125.Sakihama T, et al. A simple detection method for low-affinity membrane protein interactions by baculoviral display. PLoS One. 2008;3:e4024. doi: 10.1371/journal.pone.0004024. [DOI] [PMC free article] [PubMed] [Google Scholar]