Abstract
The human genome project has stimulated development of impressive repositories of biological knowledge at the genomic level and new knowledge bases are rapidly being developed in a ‘bottom-up’ fashion. In contrast, higher-level phenomics knowledge bases are underdeveloped, particularly with respect to the complex neuropsychiatric syndrome, symptom, cognitive, and neural systems phenotypes widely acknowledged as critical to advance molecular psychiatry research. This gap limits informatics strategies that could improve both the mining and representation of relevant knowledge, and help prioritize phenotypes for new research. Most existing structured knowledge bases also engage a limited set of contributors, and thus fail to leverage recent developments in social collaborative knowledge-building. We developed a collaborative annotation database to enable representation and sharing of empirical information about phenotypes important to neuropsychiatric research (www.Phenowiki.org). As a proof of concept, we focused on findings relevant to ‘cognitive control’, a neurocognitive construct considered important to multiple neuropsychiatric syndromes. Currently this knowledge base tabulates empirical findings about heritabilities and measurement properties of specific cognitive task and rating scale indicators (n = 449 observations). It is hoped that this new open resource can serve as a starting point that enables broadly collaborative knowledge-building, and help investigators select and prioritize endophenotypes for translational research.
Keywords: phenotype, cognition, heritability, genetics, cognitive control, informatics
Introduction
In the post-genomic era, one of the biggest challenges faced by interdisciplinary biomedical researchers is the lack of tools to manage the complexity of knowledge rapidly being accumulated across widely disparate methods, models and data types. While genetics and genomics knowledge bases have developed rapidly,1–3 higher-level phenomics knowledge bases (that is, comprehensive repositories for phenotype data that can be used on a genome-wide scale) are only now emerging4 (for example, Mouse Phenome Database (www.jax.org/phenome) and Australian Phenomics Centre (www.apf.edu.au)). These informatics resources can advance molecular psychiatry research by helping researchers better define phenotype constructs, select specific phenotypic measures and ultimately develop multilevel models that specify both phene–phene and gene–phene associations.
Given that many important concepts about syndromes, symptoms and cognitive phenotypes remain insufficiently defined and often controversial, refining phenotype constructs is critical in molecular psychiatry research. Multiple inadequacies in the categorical taxonomy of psychiatric syndromes are reflected by the high degree of diagnostic instability over time and difficulties assigning individual cases to unique diagnostic categories (yielding frequent comorbidities that may be real, or may be spurious due to invalid definitions; for example, attention deficits in bipolar disorder). Not surprisingly, both existing treatments and genetic findings generally lack specificity to the syndromal categories. These difficulties have fostered interest in dimensional models of psychopathology,5–7 including increased research on allied phenotypes or endophenotypes— phenotypes presumably intermediate between the syndromal and genetic levels.8–10 These endophenotypes are usually quantitative traits that index genetic susceptibility to the illness (phenotype) in question.11
Many endophenotypes are dimensional constructs that particularly benefit from modeling of their interrelations because the true structures of the traits remain theoretical. For example, ‘working memory’ is not a unitary concept, rather it is a latent construct estimated in a particular study by one or more cognitive tests (observable indicators). The reliability and validity of this construct can be determined with psychometric methods, which enable inferences about true scores of the latent construct.12 A phenomics knowledge base that represents the putative latent traits (endophenotype constructs) and their indicators (test scores and their properties) could therefore provide an empirical basis for evaluating the relative validity of putative endophenotype constructs, and the relative utility of different tests that can be used to measure these constructs.
A phenomics knowledge base could also help researchers select phenotypes for empirical research. Current approaches to phenotype selection for molecular psychiatry research may be well informed given an individual researcher’s vast knowledge, but this knowledge is seldom operationalized formally and objectively.13 For example, most researchers acknowledge that heritability is an important criterion for selecting measures in genetic research, but the justification for selecting a specific phenotype seldom hinges exclusively on heritability, and other factors may be weighed in selecting a specific test for a specific study. Among these, good measurement properties such as reliability are important, because these pose upper limits to validity. Furthermore, when large-scale genetics studies are conducted across many sites, often including multiple countries, the impact of cultural bias on measures may be a factor that must be considered in test selection.14 Psychiatry researchers also are increasingly interested in other forms of validity, including knowledge about neural systems associated with specific endophenotypes. Efforts such as the CNTRICS initiative (R13MH078710) are now aiming to garner expert consensus about the phenotypes most valuable for schizophrenia treatment research, explicitly recognizing validation with respect to neural systems as an important criterion for test selection. A knowledge base capable of summarizing the results of empirical reports could supplement consensus opinions, and create a dynamic resource to advance evidence-based phenotype selection for psychiatry research.
As resources have emerged to accelerate discovery in biomedicine, there has been a parallel but largely independent surge in development of collaborative Web-based tools for knowledge sharing. Wikipedia is perhaps the best-known example, illustrating dramatically the potential for large numbers of users to share knowledge with minimal constraints on input sources. This reflects the ‘bazaar’ approach to knowledge-base development, and can be contrasted to the ‘cathedral’ approach, in which a more rigidly controlled designer group establishes a structure, to which it is hoped users will ultimately visit and contribute. Much of the growth in ontology development has relied on the latter, more structured approach.15 The structured approaches not only have advantages in codifying rules for knowledge sharing, but also the limitation that users may disagree with the structure proposed, or have knowledge to share that deviates from this structure. The ideal informatics tools will ultimately incorporate features of each of these approaches as appropriate to the knowledge domain(s) being represented, and the preferences of the knowledge-base users.
We report here initial attempts to develop a collaborative phenotype annotation database, which we refer to as the Phenowiki. This initial application focuses on a specific cognitive phenotype, namely the concept of cognitive control, which has emerged in recent literature as important to multiple psychiatric syndromes. Although specific definitions remain a source of debate, there is consensus that cognitive control may be defined as the set of processes that underlie the ability to initiate, flexibly shape and constrain thoughts and actions in accord with goals. This ability may arise from ‘top-down’ control as proposed by Duncan and Owen (2000), or from a mechanism involved in maintaining an internal representation of a particular stimulus (Miller and Cohen, 2001). Both models are intimately associated with models of prefrontal cortex functioning, making cognitive control a target phenotype for many neuropsychiatric diseases that are putatively associated with frontal lobe dysfunction. Since these theories were put forth, the use of the term cognitive control has increased dramatically in the PubMed literature (see Figure 2), yet a unifying definition of this term remains elusive (see also http://en.wikipedia.org/wiki/Cognitive_control for further description of background and recent research on this concept). We aimed to develop a collaborative knowledge base to summarize important characteristics (that is, heritability, psychometric properties) of specific test indicators used to measure ‘cognitive control’ and its subcomponent constructs.
Figure 2.

‘Velocity’ of cognitive control concept and its subcomponents. Figure displays the publication frequency (number of unique PMID citations) for each term by year. X axis shows the years; y axis shows how many articles for each concept in each year expressed as a percentage of total number of articles over the 10-year span for that concept to normalize for differences in overall literature size.
Materials and methods
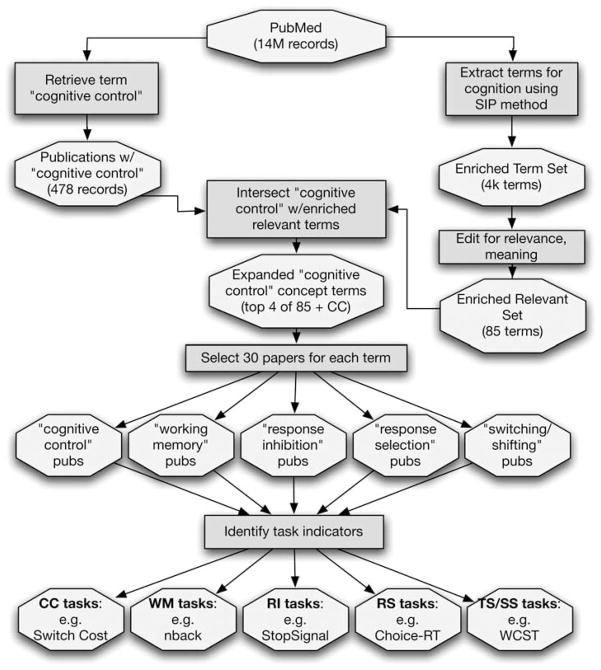
To initialize the collaborative phenotype annotation knowledge base, we developed a database of heritability and psychometric properties of cognitive measures, including those associated with the construct cognitive control. This required (1) defining the scope of cognitive control, including the selection of specific cognitive tests (referred to here as indicators) used to measure this construct; and (2) selecting the specific properties (annotation fields) that would be most useful for phenotype prioritization and selection (Figure 1).
Figure 1.
Methods showing ‘procedures’ (boxes) and ‘products’ (octagons) used to empirically define ‘cognitive control’ via the most strongly associated literature terms and specific task indicators. See ‘Materials and methods’ for details.
Defining the cognitive control construct and its indicators
We used two parallel procedures for defining the cognitive control construct, as illustrated in Figure 2. Although an initial literature search in PubMed using the term ‘cognitive control’ retrieved 478 unique papers, not all of the papers retrieved used ‘cognitive control’ in the sense intended (that is, some papers referred more generally to the cognitive control of behavior rather than using the compound term to refer to a specific construct). In addition, as we were aware of relevant literature that was not retrieved by this search, it was necessary—as a secondary step—to expand and refine the search scope. To do so, we used literature-mining methods to develop a list of potential cognitive concepts associated with cognitive control. First, we employed the statistically improbable phrase method (developed by amazon.com) to extract terms from PubMed (through 2006) that occur more frequently in cognitive neuroscience journals than in the rest of the PubMed database. This led to an enriched set of 6500 cognitive neuroscience terms. From these 6500 terms we eliminated terms that were (1) irrelevant and/or ambiguous (for example, terms like ‘stimuli’ that are overly general); (2) were part of compound phrases, if the additional terms did not further specify distinct constructs (for example, we dropped ‘attention getting’, but kept ‘sustained attention’ and ‘divided attention’; and compound words that delineated unique constructs, such as ‘action observation’ vs ‘action selection’ were retained); or (3) were semantic or syntactic variations of the same word (for example, ‘choice’ vs ‘choose’). This yielded 83 terms representing cognitive constructs (lists available in Supplementary Data online). Next, to find the concepts most closely associated with cognitive control, we examined the co-occurrence of these 83 terms with the term ‘cognitive control’ in the PubMed database using the PubGraph system, which automatically computes and plots measures of association among search terms (see www.pubgraph.org).
The four terms that co-occurred most frequently with the term ‘cognitive control’ (as reflected by the highest Jaccard coefficients (The Jaccard coefficient is a measure of association defined as the size of the intersection divided by the union of sample sets; in this instance the intersection is the number of publications containing both search terms, and the union is the sum of all publications containing either term in their title or abstract.)) were ‘working memory’, ‘task switching/set shifting’ (because searches revealed that the terms ‘task switching’ and ‘set shifting’ were associated with a highly redundant set of test indicators (both terms were associated with Wisconsin Card Sorting Test (WCST); CANTAB intradimensional/ extra-dimensional shift test; Trail making (part B) test; and letter/number, color/shape, or tone/digit switching tests), we collapsed these two terms into a single concept using the logical OR function (‘task switching’ OR ‘set shifting’)), ‘response selection’, and ‘response inhibition’. The term frequencies and co-occurrence statistics are shown in Table 1. These five terms (that is, ‘cognitive control’ plus the four most highly associated terms) were used to operationally define the scope of the cognitive control construct.
Table 1.
Co-occurrence statistics
| Terms | Co-occurrence statistics
|
||||
|---|---|---|---|---|---|
| Cognitive control | Response inhibition | Response selection | Task switching OR set shifting | Working memory | |
| Cognitive control | 478 | ||||
| Response inhibition | 23 | ||||
|
1151 e(−3.9) |
696 | ||||
| Response selection | 10 | 10 | |||
|
1054 e(−4.6) |
1272 e(−4.8) |
586 | |||
| Task switching OR set shifting | 42 | 24 | 23 | ||
|
1053 e(−3.2) |
1289 e(−4.0) |
1180 e(−3.9) |
617 | ||
| Working memory | 54 | 71 | 41 | 128 | |
|
7313 e(−4.9) |
7514 e(−4.6) |
7434 e(−5.2) |
7378 e(−4.0) |
6889 | |
Numbers are the number of papers (unique PMID identifiers) found in the PubMed database. Diagonals show size of the paper in PubMed. Conjunctions (in italics) represent the union of both concepts (i.e., OR), while intersection (in bold) shows papers that include both concepts (i.e., AND). The Jaccard coefficient (in parentheses) measures similarity between two datasets by dividing the union (OR) from the intersection (AND).
To identify the specific behavioral tasks used to measure cognitive control, we randomly selected 150 publications, including 30 for each of our five terms, published in the last decade (1997–2006). To ensure that these publications would represent changing term usage over this period, we selected papers in direct proportion to the total number of publications for each term in each year (Figure 2 plots the publication frequency for each term by year). For each of the 150 papers, the specific behavioral measure used to measure the construct of interest was determined by review of the full-text manuscript by the first author (FWS). Papers that did not report using a behavioral test to measure the construct were excluded. This exclusion was most significant for the ‘response selection’ concept, which was frequently tested using electrophysiological measures (event-related potentials; for example references 16–18). Most papers clearly identified a specific cognitive test as putatively measuring a specific cognitive construct of interest. A noteworthy exception was the term ‘cognitive control’ itself. Only three of thirty papers selected to contain this term indicated that ‘cognitive control’ was measured using a specific test,19–21 and in two of these three cases the tests were already identified as relevant to one of the other component constructs (one set-shifting/task-switching and one response inhibition paper). In most cases, the measurement of ‘cognitive control’ was indirect, that is, the construct of cognitive control was first operationally defined via another concept (for example, ‘response inhibition’), and then this was measured (for example, with the stop-signal test). We therefore relied only on the indirect references to cognitive control via its objectively identified component concepts.
For each test, we determined the specific variables that were used to measure the construct of interest, since some tests generate many variables (indicators), only some of which are relevant to the construct (for example, for the Go-NoGo test, response inhibition is indicated by reduced accuracy on NoGo trials, or increased response time on those trials, while the variable Go reaction time may also be reported, but is not considered a measure of ‘response inhibition’). Table 2 lists the behavioral tasks most frequently used to measure each concept. The complete tabulation of 150 papers, indexed by cognitive concept, and listing the specific tasks and task indicator(s) extracted from each, is available in Supplementary Data.
Table 2.
Most commonly used cognitive control tasks
| Concept and no. of papers reporting each measure
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Working memory | No. of papers | Response selection | No. of papers | Response inhibition | No. of papers | Task switching/ set shifting | No. of papers | |
| Tasks | N-back | 7 | Choice RT | 6 | Stop-signal | 10 | WCST | 9 |
| Digit Span | 4 | Changing response mapping tasks | 3 | Go-NoGo | 7 (3) | Switch between 2 Stimuli | 7 (2) | |
| Sternberg (set size) tasks | 3 | The Simon task | 2 (1) | Stroop | 6 (3) | ID/ED (CANTAB) | 6 | |
| Delayed Match2Sample | 3 | Anti-saccade | 2 | Trails | 5 | |||
| Box&Token task (Spatial) | 2 | |||||||
| Total no. of papers/ concept | 19 | 11 (1) | 25 (6) | 27 (2) | ||||
Abbreviations: CANTAB, Cambridge Neuropsychological Test Automated Battery; ED, extra-dimensional; ID, intradimensional; RT, reaction time; WCST, Wisconsin Card Sorting Test.
Measures most frequently used in the literature to assess each of the four subcomponents of cognitive control. Tasks are ranked by frequency of occurrence in each 30-paper subset. Tasks used in the 30-paper subset for ‘cognitive control’ itself were allocated to the relevant subcomponent are shown in parentheses. Only tasks used in more than one paper are listed.
Annotation fields and content retrieval for phenotype selection database
Having defined the specific terms and test indicators used to define the concept of ‘cognitive control’, we created a database summarizing key aspects of studies utilizing these cognitive tasks. Our initial goal was to provide annotation specifically about heritability statistics, key features of experimental design, sample sizes, task versions and psychometric properties of the measures (for example, test–retest reliability statistics).
Following initial entries into this database using literature already identified, we used additional searches to retrieve further relevant literature. Along with the relevant cognitive terms (cognition, cognitive control, working memory, response inhibition, task switching, set shifting, response selection), we added conjunctions (AND) for heritability (heritab*, and inherit*) as well as ‘twin’ and ‘family’. From this literature, we also reviewed reference sections and keywords to further expand searches. Papers were included if they reported heritability estimates, or if they reported intraclass correlation coefficients (ICC) for monozygotic and dizygotic twins from which we could calculate heritability estimates. This yielded 41 papers reporting heritability estimates, of which 26 papers used one of the tasks we had previously identified as within the domain of cognitive control. These 26 papers revealed 30 independent results relevant to the ‘cognitive control’ concept, from which heritability estimates could be identified. Details regarding the methods and results of these papers are now available online for both review, and user contributions (www.Phenowiki.org). In addition to heritability estimates, the database also contains fields for intraclass correlations in the relevant study groups, heritability modeling software and type of model used, test–retest and coefficient α statistics (if reported), sample characteristics (for example, mean age, sample size(s)) and other task-specific variables (for example, task version and contrast used to determine heritability). The database also links directly with the PubMed entry, which allows search by author, journal, year and keyword (see Table 3 for full list of fields).
Table 3.
Phenowiki fields
| Field | Brief field description | Examples |
|---|---|---|
| Paper ID | ||
| *PMID | Hypertext link of the ID no. to national PubMed database | 17555989 |
| First author | First author’s last name | Bilder |
| Year | Year published | 2004 |
| Task ID | ||
| Cognitive domain | Latent construct defined in the manuscript | Working memory |
| Task | Name of behavioral task employed | N-back |
| Task version | If not a novel design, provide reference paper | Cohen 1997 |
| Sample characteristics (1→n)a | ||
| *Sample group | Who was run in the study (clinical/age/etc.) | SCZ or children or healthy |
| *Study design | (case–control, family, healthy, meta-analysis, twin) | Case–Control |
| *Female (%) | Percentage of females in the sample | 50 |
| *Mean age | Average age of participants in the sample | 50 |
| *Sample size | Number of participants in this sample | 50 |
| Metric characteristics (1→n)a | ||
| *Condition/contrast | Condition or comparison used to generate the stat value | 3-Back accuracy |
| *Cognitive metric | Statistical test parameter | F |
| Group 1 | First comparison group | Patients |
| Group 2 (if applicable) | Second comparison group | Controls |
| *Metric statistical value | Value of metric obtained from testing | 7.52 |
| 95% CI Min | Lower boundary of 95% confidence interval | 3.5 |
| 95% CI Max | Upper boundary of 95% confidence interval | 7.2 |
| Group 1 Mean | Average value for group 1 | 0.48 |
| Group 1 SD | Standard Deviation for group 1 | 0.25 |
| Group 2 Mean | Average value for group 2 | 0.78 |
| Group 2 SD | Standard Deviation for group 2 | 0.15 |
| P-value significance | Two-tailed significance value | 0.001 |
| Genetic validity (1→n)a | ||
| MZ ICC | Intraclass correlation coefficient for monozygotic twins | 0.47 |
| DZ ICC | Intraclass correlation coefficient for dizygotic twins | 0.2 |
| Heredity modeling package | N/A, Falconer’s h2, LISREL, MX, SOLAR | MX |
| Best fitting heritability model | Description of best fitting model | ACE |
| Heritability statistical value | Value for heritability statistic | 0.55 |
| Psychometric validity (1→n)a | ||
| Test–retest metric | Measure used to establish test–retest reliability | r |
| Test–retest statistic | Test–retest reliability value | 0.85 |
| Internal consistency | Coefficient α | 0.9 |
| Neural systems validity (1→n)a | ||
| Brain region of interest | Region tested | Inferior frontal gyrus |
| Spatial extent (voxels) | Spatial extent of that region, if defined functionally | 50 |
| Spatial smoothing kernel (mm) | Smoothing kernel used during pre-processing | 8 |
| X coordinate | X coordinate in standard space of region | −45 |
| Y coordinate | Y coordinate in standard space of region | 14 |
| Z coordinate | Z coordinate in standard space of region | 24 |
| Group registration atlas | (N/A, MNI, Talairach, none) | Talairach |
| Free text/notes annotation author | Additional notes, comments or description | |
| *Author | Link to individual who uploaded the information | Fws |
Table 3 shows each field in our online database, and provides a short description and an example for each. All fields are either numeric or available in dropdown menus generated from look-up tables; ‘Other’ is an option in these dropdown menus, which if selected triggers a dialog box asking user to enter a short value and value label; these additions are automatically sent to system administrator and Phenowiki project team for periodic review. Items marked with a * are required to start a record.
Separate descriptions for each effect (1 through n) in the study for applicable items in the category if necessary. Additional items will be required depending on the study design (e.g., ICC values for heritability studies or brain region for imaging study). Example values are not from a single source, as one paper would be unlikely to contain data enabling completion of all fields. More information is available online at (www.phenowiki.org).
Results
As outlined above, we identified 26 publications reporting 30 heritability statistics relevant to our definition of cognitive control. For some measures there was good consistency across studies in the use of a specific indicator (for example, for Digit Span Backwards, only one indicator was used: correctly recalled digits). In contrast, there was considerable variation in the specific indicator used for other tests (for example, for Go-NoGo performance, each of three studies examined different indicators, and often used different versions of the test as well). This lack of consistency greatly increases the difficulty of pooling data and interpreting results across studies; as the field moves increasingly toward large-scale phenomics research, standardization of tests and specific indicators used across studies will clearly be a high priority (Table 4).
Table 4.
Heritability statistics for ‘cognitive control’ phenotypes
| Concept/task | Mean h2 | h2 range | Total sample size | Indicator(s) | Total no. of results | No. of publications |
|---|---|---|---|---|---|---|
| Working memory | ||||||
| N-back | 0.42 | — | 220 | 2-Back accuracy | 1 | 1 |
| Digit Span backwards | 0.45 | 0.19–0.52 | 1836 | Correctly recalled digits | 6 | 5 |
| Delayed match2sample | 0.48 | — | 780 | Median accuracy | 1 | 1 |
| Sternberg | 0.45 | 0.45–0.47 | 596 | Accuracy, number of correct trials | 2 | 2 |
| Response inhibition | ||||||
| Stop-signal | (2) | — | 220 | Stop-signal RT | 1 | 1 |
| Go-NoGo | 0.41 | 0.31–0.93 | 1844 | False alarms, no. of correct Trials | 3 | 3 |
| Stroop | 0.51 | 0.49–0.58 | 1116 | Incong-cong scores; no. of correct incong trials | 3 | 3 |
| Anti-saccade | (2) | — | 220 | Percent correct | 1 | 1 |
| Task switch/set shift | ||||||
| WCST | 0.441 | 520 | Perseverative errors | 2 | 2 | |
| ID/ED (CANTAB battery) | — | — | — | — | — | — |
| Trails-B | 0.461 | 0.41–0.50 | 944 | Completion time | 4 | 4 |
| Switch between 2 stimuli | 0.64 | 0.58–0.70 | 440 | Switch–unswitch RT | 2 | 1 |
| Response selection | ||||||
| Choice RT | 0.60 | 0.52–0.70 | 2766 | 2-, 4- or 8-choice RT | 4 | 2 |
| Simon task | — | — | — | — | — | — |
| Resp. map. tasks | — | — | — | — | — | — |
| Cognitive control | 0.50 | 30 | 26 | |||
Abbreviations: CANTAB, Cambridge Neuropsychological Test Automated Battery; ED, extra-dimensional; ID, intradimensional; RT, reaction time; WCST, Wisconsin Card Sorting Test.
Heritability statistics are averaged across studies, weighted by sample sizes. For a description of each task see online supplemental information.
Also has relative risk scores 2.0 for WCST; 4.0 for Trails-B and
measures showing a nonadditive genetic influence.
Inspection of the heritability statistics indicated that ‘cognitive control’ had a mean h2 of 0.50 across all indicators. Most components of cognitive control had reasonable consistency of h2 values across indicators although some studies reported potential outlying values, with a minimum h2 of 0.19 and a maximum of 0.93. Some indicators have been used more than others, despite relatively low heritability statistics (for example, the three most published measures, Digit Span, WCST and Go-NoGo all have h2 ≤ 0.41).
Our data also reveal that for certain tests with multiple indicators (for example, WCST), heritability estimates vary widely across indicators. In our table we present only heritability for one of the primary dependent measures, perseverative errors; however, in the study of Anokhin 2003, the authors examine multiple indicators. WCST ‘failure to maintain set’ had an estimated heritability of only 0.04, while ‘perseverative responses’ had h2 of 0.46, suggesting that perseverative responses may be a more useful cognitive trait to examine in genetic studies. Similarly, heritability estimates for the choice reaction time (RT) task reveal an association between h2 and task difficulty/complexity, with higher h2 for the more complex eight-choice condition, suggesting that heritability may increase with increasing task difficulty and/or increases in true score variance that may characterize this condition; however, further work is needed to clarify this relationship.
Discussion
We developed a collaborative annotation database to catalog empirical observations relevant to cognitive endophenotype selection for molecular psychiatry research. As a proof of concept, we populated our database with empirical findings documenting the heritability and psychometric properties for key indicators of the construct ‘cognitive control’, which is currently considered an important endophenotype across multiple syndromes, including schizophrenia, bipolar disorder and attention deficit/hyperactivity disorder. We believe this database, while currently in a simple form, can serve as the seed for collaborative knowledge-building that can be of high value to molecular psychiatry researchers by codifying the empirical bases for phenotype selection.
In the course of assembling this demonstration project, we used several methods to help define more clearly the scope of relevant literature and the concepts themselves. For example, several literature-mining approaches were used to better specify the construct of ‘cognitive control’ in terms of four major components (working memory, response inhibition, set shifting/task switching and response selection) and the specific tasks with their measures used as indicators of these constructs (Figure 3). While individual investigators committed to studying the cognitive control construct may disagree about the true ontology and component structure of this construct, the approach we used has virtues of clear operationalization, objectivity, and derives directly from term usage statistics in existing literature. The Phenowiki knowledge base itself does not specify or otherwise constrain an individual investigator’s concept selections, rather it provides the basis for assembly of whatever model the investigator wishes to interrogate.
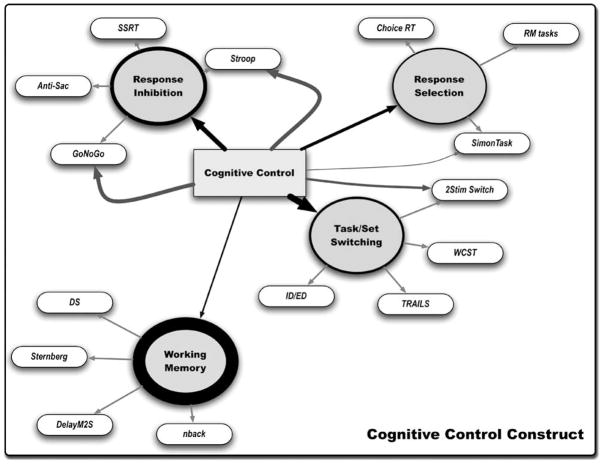
Figure 3.
Components of the construct ‘cognitive control’. Figure displays a graphical representation of the construct ‘cognitive control’ as defined by the literature and expert review of behavioral tasks. Circles represent concepts most closely associated in the literature with the term cognitive control. Thickness of the circles represents the size of each literature. Arrows show where the term cognitive control was linked directly with a behavioral task, without first being related to one of the four concepts. Thickness of these lines represents number of occurrences. Bubbles depict cognitive tasks associated with each cognitive concept as determined by expert review of the literature. Distances between bubble and concepts, as well as between concepts, represent the strength of association (that is, number of co-occurrences).
Our proof-of-concept example, focused on examining the heritability of ‘cognitive control’, revealed generally high consistency and moderate heritability for many widely used indicators of this construct. The compiled data reveal that some indicators with lower heritability also have lower test–retest reliability or internal consistency as indicated by coefficient α. Unfortunately, details about psychometric properties of the indicators are often not reported, limiting the ability to determine the extent to which differences among observed heritability statistics may be explained by psychometric characteristics alone. For example, we found a wide range of heritability scores for WCST task indicators,22 which may be due to the poor psychometric properties of indicators like ‘the failure to maintain set’ variable (h2 = 0.04), which in most samples shows strong positive skew. Similarly, heritability estimates for the choice RT task reveal an association between h2 and task difficulty/complexity, with higher h2 for the more complex eight-choice condition.23 As this is the only study to consider the heritability of complex choice RT conditions, the generalizability of this finding is not clear. Nevertheless, these observations highlight the importance of taking into account the psychometric properties of the various tests, which influence the error term in structural equation models estimating heritability. Since reliability poses an upper limit to all forms of validity including heritability, it is clear that such measures are important and inclusion of such data may represent a valuable publication standard for heritability and other validity studies. When available, however, these psychometric data offer one objective means for instrument selection; or in cases where reliability is poor but the investigator’s desire to measure the specific construct is important, these data highlight the urgency to modify tasks to enhance their psychometric properties.
The findings also illustrate how existing heritability data may reflect some ‘instrumentation inertia.’ In other words, there are more data regarding heritability of older ‘classical’ neuropsychological tests (such as Digit Span and the Trail Making Test), relative to newer measures developed to assess the functioning of more specific neural systems (for example, N-back). These findings suggest that literature-based association measures may benefit from considering carefully the time course of both concept definitions and their relations with other measures, since concepts may change over time, and both the terms used to refer to time-invariant concepts and their relations to other measures also change over time. ‘Velocity’ measures (increased frequency of occurrence or association over time) may help characterize those concepts that are emerging as promising but may so far lack a large literature precisely because of their novelty. For example, literature-based approaches showed how the concept of ‘cognitive control’ is increasing rapidly in usage, while use of the component terms is continuing, but at a steady pace (Figure 2). Such methods may overcome the instrumentation inertia that leads to continued use and reification of previously validated measurement tools despite investigators’ acknowledgment that the measures may be suboptimal, and also highlight those measurement domains and specific instruments that may benefit from psychometric refinement or development of alternate methods.
This initial proof-of-concept example offers a template that can be expanded readily to include other data types and relations. This paper describes only one component focused on psychometric test findings relevant to cognitive control and their heritability. Findings can be added to help determine the validity of the underlying constructs, by representing the associations among different putative indicators of the same construct. For example, it is possible to incorporate measures of association between the different test variables presumably measuring cognitive control. If the literature association statistics supporting the validity of this construct are valid, then the different indicators of this construct should also covary in empirical studies. While final tests of construct validity would demand direct examination of covariance among all putative indicators in the same sample, knowledge about the pair-wise associations between sets of test variables obtained across different studies could help refine the design of the most appropriate and efficient experiments. We note further that heritability is only one form of ‘biological validity’ (presumably due to shared genetic effects). We already have implemented additional database components for collaborative annotation to assess other forms of validity. For example, we have tabulated effects relating cognitive test findings to diagnostic group membership (for example, ‘What is the standardized effect size on the N-back test distinguishing people with the diagnosis of schizophrenia from healthy comparison groups?’), to neuroimaging findings (for example, ‘What are the coordinates and peak intensities of activations elicited in functional MRI by contrasts between experimental conditions on the N-back test?’) and to polymorphisms in certain candidate genes (for example, ‘What is the standardized effect size distinguishing individuals with different allelic variations of the Val158-Met polymorphism?’). By populating the Phenowiki with data describing these kinds of relations, it is possible to assemble a multilevel model describing complex scientific hypothesis (for example, ‘How strongly associated is a polymorphism in the Val158-Met gene to altered prefrontal activation, impaired N-back test performance and the diagnosis of schizophrenia?’).
These extensions comprise initial steps toward the development of a ‘phenomics atlas’ (Figure 4), enabling investigators to test competing models to determine ‘goodness of fit’ with the underlying evidence, and conduct exploratory analyses to identify previously unsuspected links that would improve their original hypotheses. Similar methods could enable automated discovery of the strongest evidence chains linking diverse levels of biological knowledge; for example, investigators might enter starting nodes reflecting selected phenotype concepts, and the system could then be used to identify the most relevant phenotype indicators, along with catalogs of possibly relevant genes, proteins or pharmacological agents, by linking to other repositories of biological knowledge (for example, other components of the National Library of Medicine’s existing systems, such as gene, protein and nucleotide databases, PubMed and Online Mendelian Inheritance in Man).
Figure 4.
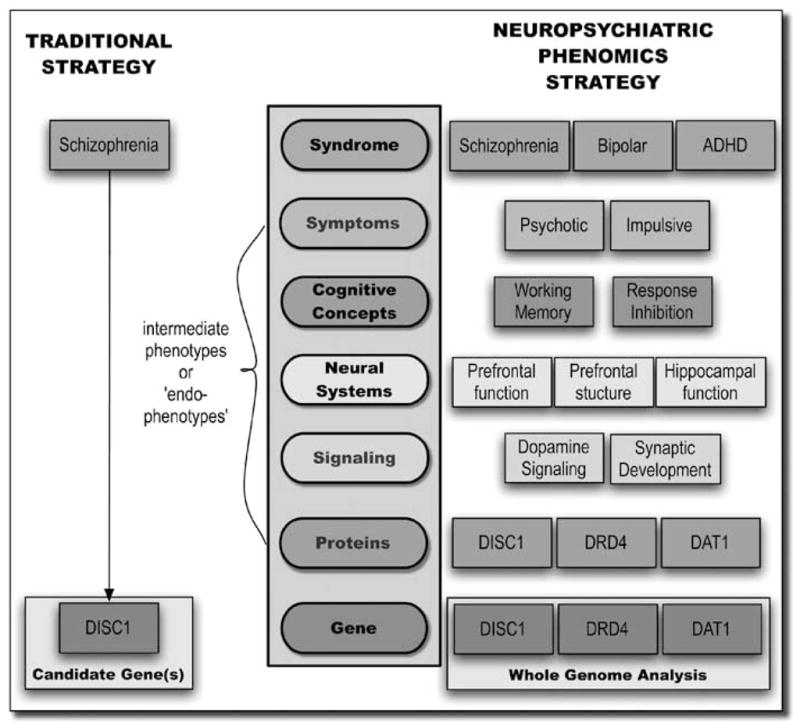
The neuropsychiatric phenomics approach contrasts with the traditional approach to psychiatric genetics studies, in which gene–syndrome relationships are assessed directly. The neuropsychiatric phenomics strategy involves analysis of multiple levels of intermediate traits, across multiple syndromal categories.
The example we describe here is currently open but structured. Future extensions are planned to enable both user specification of additional structured tables and free text annotation, ‘voting’ to develop and represent consensus opinions of the field, and other features including free text and image annotations that have been developed in other Mediawiki projects like Wikipedia. Like any collaborative knowledge base, the Phenowiki will ultimately depend on the ‘wisdom of crowds’ to develop in these and other directions, and manifest its full potential. A challenge to any of these methods is the extent to which the ‘Field of Dreams’ model will be successful (‘if we build it, will anyone come?’). While the barriers to participation are low, the incentives to participation remain in question. Our discussions with potential collaborators have been promising, and we believe the strongest incentives come from those who have vested interests in the definitions of the constructs, the best measurement methods to examine these constructs and the pressing questions about the validity of their own hypotheses. We hope that the investigator community, including prominently the readership of Molecular Psychiatry, will be eager to contribute and advance the collation of empirical knowledge that helps bridge the currently wide gap between genome and syndrome.
Supplementary Material
Acknowledgments
We thank Russ Poldrack, Tyrone Cannon and Nikki Kittur for comments and guidance in the development of this article. FW Sabb is currently supported by a T32 (MH014584; Nuechterlein, PI). This project was supported by the Consortium for Neuropsychiatric Phenomics (under P20 RR020750; Bilder, PI).
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
References
- 1.Goh CS, Gianoulis TA, Liu Y, Li J, Paccanaro A, Lussier YA, et al. Integration of curated databases to identify genotype–phenotype associations. BMC Genomics. 2006;7:257. doi: 10.1186/1471-2164-7-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korbel JO, Doerks T, Jensen LJ, Perez-Iratxeta C, Kaczanowski S, Hooper SD, et al. Systematic association of genes to phenotypes by genome and literature mining. PLoS Biol. 2005;3:e134. doi: 10.1371/journal.pbio.0030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lussier YA, Liu Y. Computational approaches to phenotyping: high-throughput phenomics. Proc Am Thorac Soc. 2007;4:18–25. doi: 10.1513/pats.200607-142JG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Driel MA, Bruggeman J, Vriend G, Brunner HG, Leunissen JA. A text-mining analysis of the human phenome. Eur J Hum Genet. 2006;14:535–542. doi: 10.1038/sj.ejhg.5201585. [DOI] [PubMed] [Google Scholar]
- 5.Craddock N, O’Donovan MC, Owen MJ. Symptom dimensions and the Kraepelinian dichotomy. Br J Psychiatry. 2007;190:361. doi: 10.1192/bjp.190.4.361. author reply 361–362. [DOI] [PubMed] [Google Scholar]
- 6.Craddock N, Owen MJ. Rethinking psychosis: the disadvantages of a dichotomous classification now outweigh the advantages. World Psychiatry. 2007;6:20–27. [PMC free article] [PubMed] [Google Scholar]
- 7.Haslam N, Kim H. Categories and continua: a review of taxometric research. Genet Soc Gen Psychol Monogr. 2002;128:271–320. [PubMed] [Google Scholar]
- 8.Bearden CE, Freimer NB. Endophenotypes for psychiatric disorders: ready for primetime? Trends Genet. 2006;22:306–313. doi: 10.1016/j.tig.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Freimer N, Sabatti C. The human phenome project. Nat Genet. 2003;34:15–21. doi: 10.1038/ng0503-15. [DOI] [PubMed] [Google Scholar]
- 10.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 11.Almasy L, Blangero J. Endophenotypes as quantitative risk factors for psychiatric disease: rationale and study design. Am J Med Genet. 2001;105:42–44. [PubMed] [Google Scholar]
- 12.Nunnally JC. Introduction to Statistics for Psychology and Education. McGraw-Hill; New York: 1975. [Google Scholar]
- 13.Flint J, Munafo MR. The endophenotype concept in psychiatric genetics. Psychol Med. 2007;37:163–180. doi: 10.1017/S0033291706008750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glahn DC, Almasy L, Blangero J, Burk GM, Estrada J, Peralta JM, et al. Adjudicating neurocognitive endophenotypes for schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2007;144:242–249. doi: 10.1002/ajmg.b.30446. [DOI] [PubMed] [Google Scholar]
- 15.Noy NF, Crubezy M, Fergerson RW, Knublauch H, Tu SW, Vendetti J, et al. Protege-2000: an open-source ontology-development and knowledge-acquisition environment. AMIA Annu Symp Proc. 2003:953. [PMC free article] [PubMed] [Google Scholar]
- 16.Hill H, Ott F, Herbert C, Weisbrod M. Response execution in lexical decision tasks obscures sex-specific lateralization effects in language processing: evidence from event-related potential measures during word reading. Cereb Cortex. 2006;16:978–989. doi: 10.1093/cercor/bhj040. [DOI] [PubMed] [Google Scholar]
- 17.Lobmann R, Smid HG, Pottag G, Wagner K, Heinze HJ, Lehnert H. Impairment and recovery of elementary cognitive function induced by hypoglycemia in type-1 diabetic patients and healthy controls. J Clin Endocrinol Metab. 2000;85:2758–2766. doi: 10.1210/jcem.85.8.6737. [DOI] [PubMed] [Google Scholar]
- 18.Smid HG, Trumper BG, Pottag G, Wagner K, Lobmann R, Scheich H, et al. Differentiation of hypoglycaemia induced cognitive impairments. An electrophysiological approach. Brain. 1997;120(Part 6):1041–1056. doi: 10.1093/brain/120.6.1041. [DOI] [PubMed] [Google Scholar]
- 19.Grachev ID, Kumar R, Ramachandran TS, Szeverenyi NM. Cognitive interference is associated with neuronal marker N-acetyl aspartate in the anterior cingulate cortex: an in vivo (1)H-MRS study of the Stroop Color-Word task. Mol Psychiatry. 2001;6:496, 529–539. doi: 10.1038/sj.mp.4000940. [DOI] [PubMed] [Google Scholar]
- 20.Habel U, Klein M, Shah NJ, Toni I, Zilles K, Falkai P, et al. Genetic load on amygdala hypofunction during sadness in nonaffected brothers of schizophrenia patients. Am J Psychiatry. 2004;161:1806–1813. doi: 10.1176/ajp.161.10.1806. [DOI] [PubMed] [Google Scholar]
- 21.Kray J, Lindenberger U. Adult age differences in task switching. Psychol Aging. 2000;15:126–147. doi: 10.1037//0882-7974.15.1.126. [DOI] [PubMed] [Google Scholar]
- 22.Anokhin AP, Heath AC, Myers E. Genetics, prefrontal cortex, and cognitive control: a twin study of event-related brain potentials in a response inhibition task. Neurosci Lett. 2004;368:314–318. doi: 10.1016/j.neulet.2004.07.036. [DOI] [PubMed] [Google Scholar]
- 23.Luciano M, Posthuma D, Wright MJ, de Geus EJ, Smith GA, Geffen GM, et al. Perceptual speed does not cause intelligence, and intelligence does not cause perceptual speed. Biol Psychol. 2005;70:1–8. doi: 10.1016/j.biopsycho.2004.11.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.