Abstract
MADS-domain proteins are important transcription factors involved in many biological processes of plants. In our study, a tomato MADS-box gene, SlFYFL, was isolated. SlFYFL is expressed in all tissues of tomato and significantly higher in mature leave, fruit of different stages, AZ (abscission zone) and sepal. Delayed leaf senescence and fruit ripening, increased storability and longer sepals were observed in 35S:FYFL tomato. The accumulation of carotenoid was reduced, and ethylene content, ethylene biosynthetic and responsive genes were down-regulated in 35S:FYFL fruits. Abscission zone (AZ) did not form normally and abscission zone development related genes were declined in AZs of 35S:FYFL plants. Yeast two-hybrid assay revealed that SlFYFL protein could interact with SlMADS-RIN, SlMADS1 and SlJOINTLESS, respectively. These results suggest that overexpression of SlFYFL regulate fruit ripening and development of AZ via interactions with the ripening and abscission zone-related MADS box proteins.
Ethylene plays important roles in many aspects of plant growth and development, including the processes of leaf senescence, fruit ripening, abscission, other programmed senescence and defense signalling. In many species exogenous ethylene can promote processes that are characteristic of leaf senescence. In these studies, chlorophyll content was used as a marker of leaf senescence1.
Tomato (Solanum lycopersicum) is the primary model for climacteric fruit ripening for a combination of scientific and agricultural reasons. Its fruit plays an important role in the human diet and provides health benefits as a source of vitamins, minerals, and antioxidants (phenolics, folate, lycopene, and β-carotene)2.
Fruit ripening represents a summation of physiological and biochemical processes of fleshy fruits including de-greening and accumulation of colored pigments for attraction, textural changes associated with cell wall metabolism and cell turgor variation leading to softening, and metabolic changes related to flavor and nutrient composition, generally associated with accumulation of sugars, acids and volatiles culminating in a diverse array of tastes and smells varying among species. These changes not only make fruit assisting in seed dispersal, but also provide essential nutrition for human and animal diets3.
Ripening of climacteric fruits is characterized by an autocatalytic increase in respiration and ethylene biosynthesis just prior to the initiation of ripening. Ethylene biosynthesis occurs via a pathway: the first dedicated step is the conversion of s-adenosyl-l-methionine (SAM) to aminocyclopropane-1-carboxylic acid (ACC), by the normally ratelimiting enzyme ACC synthase (ACS)4. ACC is subsequently converted by ACC oxidase (ACO), the so-called ethylene forming enzyme, to ethylene5.
Besides ethylene synthesis, the ability of ethylene perception and response are necessary for fruit ripening. The expression of E4 in fruit is rapidly induced following exogenous ethylene induction6. Meanwhile, the transcripts of E4 in fruit are suppressed through ethylene biosynthesis inhibition7. E8 is a tomato ripening-associated and fruit-specific expression gene8. PG, transcriptionally activated during fruit ripening, is a major cell wall polyuronide degrading enzyme, catalyzes the depolymerization of pectins9. Unraveling the regulation of these gene activities is important to understand the processes of ripening, senescence, abscission, and response to stress10.
To date, a lot of ripening-deficient mutants, such as ripening inhibitor (rin), never ripe (Nr), nonripening (nor) and color nonripening (cnr), have been found and investigated in tomato. They are useful in understanding of the transcriptional control system underlying tomato ripening. The rin mutant displays inhibited fruit ripening and enlarged sepals. This mutant phenotype has been attributed to functions of two MADS-box transcriptional factors, SlMADS-RIN and SlMADS-MC. SlMADS-RIN regulates fruit ripening and SlMADS-MC involves in sepal development and formation of abscission zones11,12. MADS-box proteins have been found playing different and important biological roles in tomato, such as the regulation of inflorescence and fruit ripening13. TDR4 (FUL1)and SLMBP7 (FUL2) both of which have high sequence similarity to Arabidopsis FRUITFULL have been demonstrated to act in fruit ripening via forming MADS-box transcription factor complexes with RIN14,15. The expression of FUL1 is up-regulated during ripening15,16. TM6 transcripts mainly accumulate in the carpel primordial and young fruits in tomato and have been considered to be involved in fruit ripening17,18. RNAi suppression of TAG1 in tomato leads to misshapen fruits and homeotic conversion of stamens into petaloid organs18. The antisense suppression of TAGL1 results in ripening inhibition and pericarp thickness reduction19. Interestingly, all of those MADS-box proteins play as positive regulators of ripening.
Recently, we found that a tomato MADS-box transcription factor, SlMADS1 acted as a negative regulator of fruit ripening and interacted with SlMADS-RIN20. An APETALA2 transcription factor (SlAP2a), belonging to the AP2/ERF (Ethylene Response Element) family and encoding putative transcription factors21, was identified through transcriptional profiling of fruit maturation. RNAi repression of SlAP2a results in fruits that over-produce ethylene, ripen early and modify carotenoid accumulation profiles by altering carotenoid pathway flux22. These suggest that SlMADS1 and SlAP2a function as modulators of ripening and act to balance the activities of positive ripening regulators.
Therefore, there are positive and negative regulation factors to balance fruit ripening process. While only one negative regulator of fruit ripening has been reported in MADS-box family20,23. Although previous researches have done a lot of contributions to tomato fruit ripening, the developmental mechanisms undoubtedly need further study.
Abscission in plants is a crucial process used to shed organs such as leaves, flowers, and fruits when they are senescent, damaged, or mature. Abscission occurs at predetermined positions called abscission zones (AZs), which have several layers of small, densely cytoplasmic cells at the junction of organ and the main body of plant24,25. Control of abscission in fruit and grain crops is a key agricultural concern. For example, during cereal crop domestication, mutants that reduce seed shattering have been preferentially selected, because shattering is a major limiting factor for yield26. Defective AZs in fruit pedicel facilitates large-scale harvesting of tomato by saving time removing the calices, because when the jointless fruit is harvested, the calyx remains attached to the plant, not to the fruit27.
In tomato, JOINTLESS encodes a short vegetative phase group MADS-box transcription factor, and its mutant phenotype is called jointless, which is characterized by defective AZs in fruit pedicels25,27. The mutant lateral suppressor (ls) suppress the development of pedicel AZs28. SLMADS-MC previously identified as a sepal size regulator, physically interacts with JOINTLESS to regulate the development of tomato pedicel AZs12,25. Transcriptome analyses of pedicels at the preabscission stage reveal that MC and JOINTLESS regulate the expression of LeWUS, Bl, GOB and Ls, which are homologs of WUSCHEL, REGULATOR OF AXILLARY MERISTEMS, CUP-SHAPED COTYLEDON, and LATERAL SUPPRESSOR in Arabidopsis, respectively, and these transcription factors play key roles in pedicel AZ development12. In addition, ethylene-induced abscission is correlated with an increase in poly-galacturonase (PG) and endo-β-l, 4-D-glucanase (cellulase) activity in tomato29,30. TAPG1 and cell wall hydrolase Cel2 are also required for abscission at the AZ30. Although the regulation of fruit abscission is essential for agriculture, the developmental mechanisms remain unclear.
Here, we describe a novel tomato MADS-box transcription factor SlFYFL (GenBank No. KF709444), an ortholog of Arabidopsis FYF (FOREVER YOUNG FLOWER), AtAGL42, which acts as a repressor controlling floral organ senescence and abscission31. In this study, 35S:FYFL plants were generated to investigate the exact role of SlFYFL in tomato, and the results revealed that overexpression of SlFYFL can delay fruit ripening, leaf and sepal senescence and the development of fruit pedicel AZs.
Results
SlFYFL isolation and expression pattern analyse
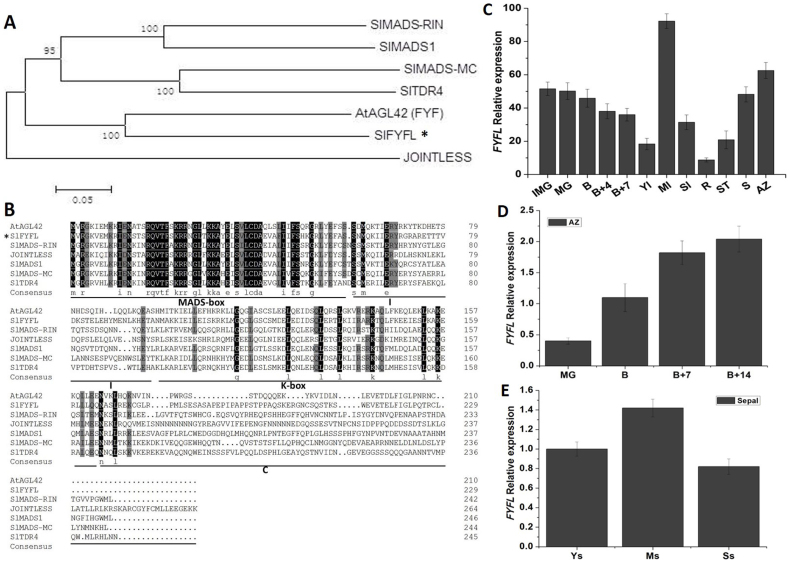
Based on the sequence in GenBank (accession No.KF709444), full-length cDNA of SlFYFL was cloned with specific primers SlFYFL-F and SlFYFL-R (Table 1S) from tomato (Solanum lycopersicon Mill. cv. Ailsa Craig) and sequenced. Phylogenetic and amino acid homology analysis showed that SlFYFL was highly homologous to AtFYF (AtAGL42) (Fig. 1 A) and belonged to a very conservative MADS-box transcription factor family (Fig. 1 B). Quantitative real-time PCR technology was performed to analyze the expression of SlFYFL. The results showed that the expression level of SlFYFL was high in mature leave, AZ, sepal and immature green fruit than other tissues, and its expression decreased slightly after the onset of ripening (Fig. 1 C). During the development of fruit abscission zones in tomato, the SlFYFL expression increased gradually (Fig. 1 D). In flower sepals, the SlFYFL mRNA was highly accumulated in young sepals and increased to the highest level in mature sepals, then decreased slightly in senescence sepals (Fig. 1 E), which expression pattern was similar to that in leaves (Fig. 1 C). These results indicate that SlFYFL may play an important role in fruit ripening and development process of AZ, leaf and sepal.
Figure 1. SlFYFL sequence and expression analysis in AC (WT).
(A) Phylogenetic analysis of FYFL and other MADS-Box proteins was constructed by the neighbor-joining method, bootstrap analysis of 1000 replicates. FYFL is marked with asterisk. Accession numbers for the proteins listed are as follows: SlMADS-MC (NP_001234665), SlTDR4 (FUL1) (NM_001247244), SlMADS-RIN (NM_001247741.1), SlMADS1 (NP_001234380), AtAGL42 (AY141213), SlFYFL(KF709444), JOINTLESS (AAG09811). (B) Multiple sequence alignment of FYFL and other MADS-Box proteins. SlFYFL is marked with asterisk. Identical amino acids are shaded in black, and similar amino acids are shaded in gray. (C) The relative expression patterns of FYFL in AC. IMG, immature green fruit; MG, mature green fruit; B, breaker fruit; B + 4, 4 days after breaker fruit; B + 7, 7 days after breaker fruit; Yl, young leaf; Ml, muture leaf; Sl, senescent leaf; R, root; ST, stem; S, sepal of flower in anthesis; AZ, abscission zone of flower in anthesis. The relative expression of FYFL in different stages AZ (D) and different stages sepal (E) of AC. MG, mature green fruit; B, breaker fruit; B + 4, 4 days after breaker fruit; B + 7, 7 days after breaker fruit; B + 14, 14 days after breaker fruit. Ys, young sepal; Ms, mature sepal; Ss, senescent sepal. The data represent mean from three replicates with three biological repeats. Error bars indicate SE.
35S:FYFL plant delayed leaf and sepal senescence
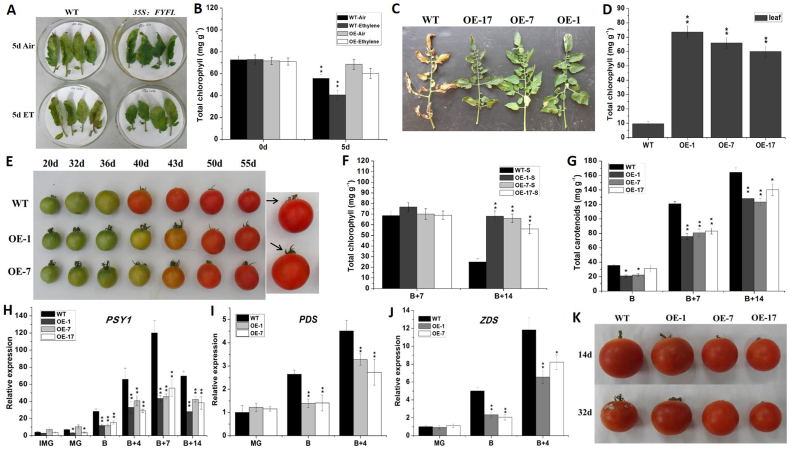
Mature leaves were detached and incubated on wet filter paper under air condition. Five days later, the detached leaves from wild type exhibited senescence, while that from 35S:FYFL plant remained green. The detached leaves from the wild type and transgenic plants both became yellowing after 5 days of ethylene treatment, but wild type leaves exhibited more yellowing than transgenic lines (Fig. 2 A). The chlorophyll contents of wild type leaves were dramatically reduced after 5 days of treatment with air or ethylene, whereas that of transgenic plant had no significant difference (Fig. 2 B). In addition, 120 days after colonization, the wild type leaves next to the first inflorescence were completely yellow, while only edge of transgenic leaves exhibited senescence (Fig. 2 C, D). We also observed that sepal senescence was delayed in transgenic plants. At B + 14 stage, wild-type sepals have become senescent, while transgenic sepals were still green (Fig. 2 E). The chlorophyll content of wild-type sepal decreased dramatically at B + 14 stage, while that in transgenic lines still remained at high level (Fig. 2 F).
Figure 2. Phenotype and physiological indices, and related genes expression of wild-type and 35S:FYFL lines.
(A) Leaf phenotype of wild type and transgenic lines after 5 day treatment of ethylene (ET) and the air (Air). (B) Total chlorophyll content of leaves of wild type and transgenic lines before and after 5 day treatment of ethylene (ET) and the air (Air). Leaf phenotype (C) and total chlorophyll content of leaves (D) of wild type and transgenic lines of 120 days old. The leaves next to the first inflorescence. (E) Fruits phenotype of wild-type and 35S:FYFL lines. 20 d–55 d, statistical time starting from the pollination. 35S:FYFL lines displayed 3–5 days delayed fruit ripening and longer and younger sepals than wild type. (F) Total chlorophyll content of different stages sepal of wild-type and transgenic lines. (G) Total carotenoid content of wild-type and 35S:FYFL lines. (H) to (J) respectively represent expression analysis of PSY1, PDS, ZDS in different stages fruits of wild-type and transgenic lines. (K) Fruits storability phenotype of wild-type and transgenic lines. 14 d and 32 d, post-harvest storage time. WT, wild type; OE-1, OE-7, OE-17, different transgenic lines. IMG, immature green fruit; MG, mature green fruit; B, breaker fruit; B + 4, 4 days after breaker fruit; B + 7, 7 days after breaker fruit; B + 14, 14 days after breaker fruit. The data represent mean from three replicates with three biological repeats. *, indicate P < 0.05, **, indicate P < 0.01 between the wild type and others by t-test. Error bars indicate SE.
Ethylene biosynthetic genes were expressed at low levels in the leaves and sepals of transgenic plant
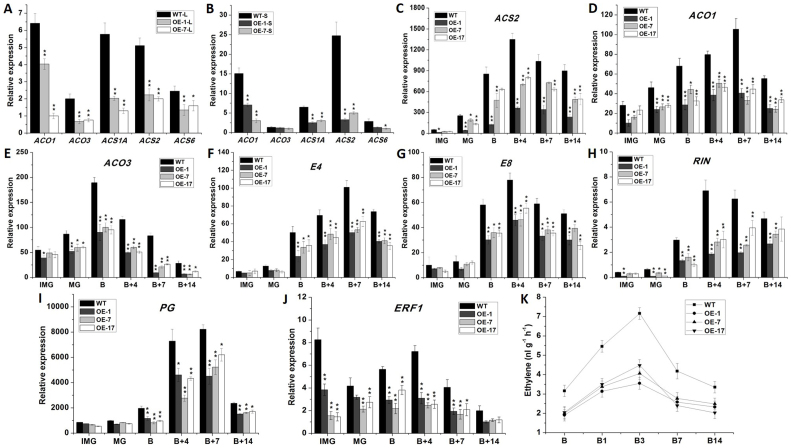
As ethylene plays an important role in senescence, we examined the expression of ethylene biosynthesis genes, such as ACO1, ACO3, ACS1A, ACS2 and ACS6 in leaf and sepal of wild-type and transgenic plants. The results showed that these five ethylene biosynthesis genes were down-regulated significantly in leaf of transgenic plant (Fig. 3 A). In sepal of transgenic plant, except ACO3, expression level of other four genes were all decreased by 50–80% (Fig. 3 B). These results suggest that reduced expression of ethylene biosynthesis genes might affect the ethylene biosynthesis, thus delay the senescence of transgenic leaves and sepals.
Figure 3. Ethylene synthesis genes expression in 120 days old leaves, next to the first inflorescence, and in B + 14 stage sepals of wild-type and transgenic lines, Ethylene synthesis and Fruit ripening-related genes expression, and Ethylene content of different stages fruits of wild-type and transgenic lines.
(A) Ethylene biosynthesis genes expression in 120 days old leaves, next to the first inflorescence of wild-type and transgenic lines. (B) Ethylene biosynthesis genes expression in B + 14 stage sepals of wild-type and transgenic lines. (C) to (J) respectively represent expression analysis of ACS2, ACO1, ACO3, E4, E8, RIN, PG and ERF1 in different stages fruits of wild-type and transgenic lines. (K) Ethylene content in different stages fruits of wild type and transgenic lines. WT, wild type; OE-1, OE-7, OE-17, different transgenic lines. IMG, immature green fruit; MG, mature green fruit; B, breaker fruit; B + 1, 1 days after breaker fruit; B + 3, 3 days after breaker fruit; B + 4, 4 days after breaker fruit; B + 7, 7 days after breaker fruit; B + 14, 14 days after breaker fruit. The data represent mean from three replicates with three biological repeats. *, indicate P < 0.05, **, indicate P < 0.01 between the wild type and others by t-test. Error bars indicate SE.
35S:FYFL fruit ripening was delayed
During the process of fruit development, we measured the time from pollination to ripening. We observed that the color of 35S:FYFL fruits changed later than the wild type (Fig. 2 E), and its ripening time was delayed 3 to 5 days (Table 2S). It was reported that the dramatic change of pigmentation in ripening tomato fruits was caused by accumulation of carotenoids32. Thus, the carotenoids in transgenic and wild type fruits at B (the colour change from green to yellow), B + 7 (7 days after B) and B + 14 (14 days after B) stage were extracted and determined. As shown in Fig. 2 G, the accumulation of carotenoid in transgenic lines was down-regulated by 30–40% than wild type. Real-time PCR analysis results revealed that expression level of PSY1 (Phytone synthease1), PDS (phytone desaturase) and ZDS(ζ-carotene desaturase) were down-regulated by 35–50% in transgenic lines compared with wild type at the stages of fruit ripening (Fig. 2 H, I and G). These results indicate that overexpression of SlFYFL gene affects the fruit ripening of tomato.
Ethylene-related and ripening-related genes were significantly down-regulated in 35S:FYFL fruits
To further characterize the molecular regulation mechanism of SlFYFL in fruit ripening, a set of ethylene-related and ripening-related genes in wide type and transgenic tomato fruits were examined. Three ethylene biosynthesis genes, ACS2, ACO1 and ACO3, and two ripening-related genes, E4 and E8, which responded specifically to ethylene, were down-regulated to different degrees in 35S:FYFL fruits at B, B + 4, B + 7 and B + 14 stages (Fig. 3 C–G). SlMADS-RIN mRNA level was 30–75% lower in 35S:FYFL fruits than that in wild type (Fig. 3 H). Expression of PG was decreased by 20–60% in 35S:FYFL fruits at B, B + 4 and B + 7 stages (Fig. 3 I). These results indicated that overexpression of SlFYFL might inhibit fruit ripening by impacting ethylene biosynthesis or ethylene response. Additionally, ethylene-responsive factor ERF1 associated with defense responses, was also down-regulated in transgenic fruits at all stages (Fig. 3 J), suggesting that SlFYFL might play a role in stress response.
Ethylene production was reduced significantly in 35S:FYFL fruit
To further investigate the relationship between SlFYFL and ethylene, we measured the ethylene production during the fruit ripening process. 35S:FYFL fruit exhibited a climacteric rise with the peak of ethylene production occurring at day 3 after ripening was initiated and declined at day 7 when fruits entered the senescence stage as the wild type did, but 35S:FYFL fruit produced only about half level of the ethylene as wild type did during fruit ripening (Fig. 3 K).
The storability of 35S:FYFL fruits were improved
Fruits of wild-type and transgenic lines were harvested at B + 7 stage and stored under the same conditions. Fourteen days after harvested, wild type tomatoes began to soften, darken, yet transgenic fruits remained hardness and lighter in color. Thirty-two days after harvested, wild-type tomatoes were soft, dehydrated and moldy, while transgenic tomatoes just began to soften (Fig. 2 K).
SlFYFL affected the development of tomato fruit stalk abscission zone
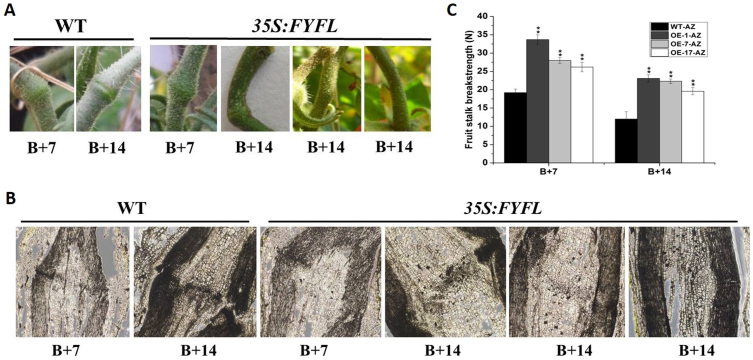
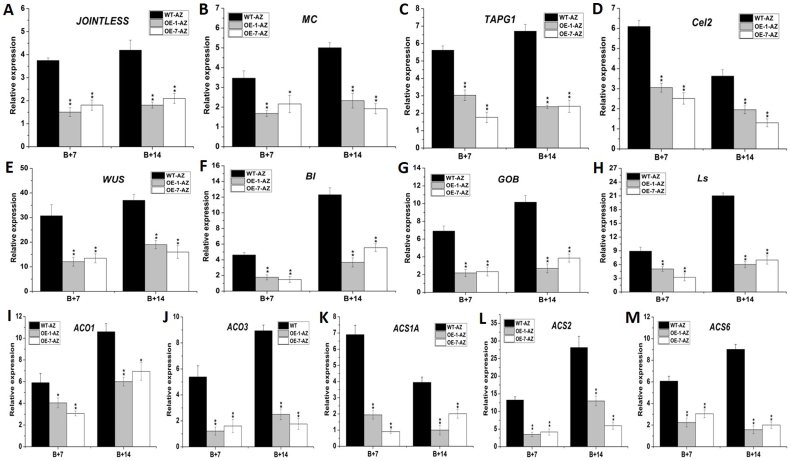
In 35S:FYFL lines, another striking phenotype is that the formation of fruit stalk abscission zone (AZs) is obviously suppressed, even non-AZ-forming is observed in B + 14 fruit stalk of transgenic lines (Fig. 4 A). To clearly observe the development of fruit stalk AZs, microscopy was used to examine the morphological changes in the wild type and 35S:FYFL AZs. At B + 7 stage, AZ cells were observed obviously at the longitudinal section plane of fruit stalk AZs (edge and central) in wild-type plant, while a smooth pedicel without obvious AZ cells was observed at the center of 35S:FYFL plant AZs. At B + 14 stage, the AZ cells of 35S:FYFL plants began to be observed obviously, and even non-AZ-forming was detected in some transgenic lines (Fig. 4 B). To further explore the differences in abscission between the wild type and 35S:FYFL plants, a breakstrength meter (Fig. 2 S) was used to quantitatively measure the force needed to break fruit stalk from the AZs. As shown in Fig. 4 C, the breakstrength of the 35S:FYFL pedicel were significantly larger than wild type at B + 7 and B + 14 stages, and the breakstrength of 35S:FYFL pedicel at B + 14 stage were approximately equal to that of wild type at B + 7 stage (Fig. 4 C), suggesting that the timing of AZ cell separation should be delayed in the 35S:FYFL fruit stalk AZs. In addition, a set of AZ-related genes, such as J, MC, TAPG1, Cel2, WUS, Bl, GOB and Ls, were examined in fruit stalk abscission zone. These genes in transgenic tomato AZs were all significantly down-regulated (Fig. 5 A–H), impling that SlFYFL might inhibit fruit AZs development by impacting cell wall modification and degradation at fruit AZ. The ethylene biosynthesis genes, ACO1, ACO3, ACS1A, ACS2 and ACS6, were also detected in B + 7 and B + 14 AZs of wild-type and transgenic fruits. The results showed that expression of these five ethylene biosynthesis genes were decreased by 30–80% in transgenic AZs (Fig. 5 I–M), indicating that the ethylene biosynthesis was affected, which might be one of the reasons of delaying the formation of AZs in transgenic lines.
Figure 4. Phenotype and breakstrength of AZs of wild-type and 35S:FYFL lines.
(A) AZs phenotype of wild-type and 35S:FYFL lines. (B) AZs microscopic observation phenotype of wild type and 35S:FYFL lines. (C) Breakstrength of different stages fruit stalk AZs of wild-type and 35S:FYFL lines. WT, wild type; OE-1, OE-7, OE-17, different transgenic lines. B + 7, 7 days after breaker fruit; B + 14, 14 days after breaker fruit. The data represent mean from three replicates with three biological repeats. *, indicate P < 0.05, **, indicate P < 0.01 between the wild type and others by t-test. Error bars indicate SE.
Figure 5. Relative expression of abscission zone formation-related genes and ethylene biosynthetic genes of wild-type and 35S:FYFL lines.
(A) to (M) respectively represent expression analysis of JOINTLESS, MC, TAPG1, Cel2, WUS, Bl, GOB, Ls, ACO1, ACO3, ACS1A, ACS2 and ACS6 in different stages AZs of wild type and transgenic lines. WT, wild type; OE-1, OE-7, different transgenic lines. B + 7, 7 days after breaker fruit; B + 14, 14 days after breaker fruit. The data represent mean from three replicates with three biological repeats. *, indicate P < 0.05, **, indicate P < 0.01 between the wild type and others by t-test. Error bars indicate SE.
Yeast two-hybrid assay demonstrated SlFYFL interacted with SlMADS-RIN, SlMADS1 and SlJOINTLESS, respectively
To further explore the relationship between SlFYFL with other MADS-box proteins, two essential ripening-related regulators of SlMADS-RIN and SlMADS1 and AZs development related protein SlJOINTLESS were preferentially selected for yeast two-hybrid assay. The open reading frame of SlFYFL was amplified and cloned into pGBKT7 as the bait. Self-activation of FYFL-pGBKT7 was tested, and the result was minus. The open reading frame of SlMADS-RIN, SlMADS1 and SlJOINTLESS were amplified and cloned into pGADT7 as the prey, respectively. An empty prey and bait vector were used as negative controls with each bait and prey construct, respectively. Fig. 6 showed that the yeast grew on selective media and turned blue on X-α-gal indicator plate, suggesting that there exist interaction between SlFYFL and SlMADS-RIN, SlJOINTLESS and SlMADS1 in vivo respectively.
Figure 6. Yeast Two-hybrid Assay for SlFYFL & SlMADS-RIN, SlFYFL & SlMADS1 and SlFYFL & SlJOINTLESS Proteins.
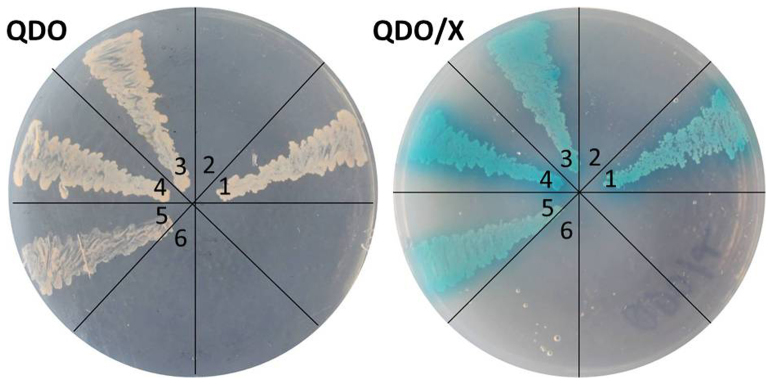
QDO, SD medium without Trp, Leu, His and Ade; QDO/X, QDO medium with X-α-Gal. 1.pGBKT7-53 & pGADT7-T (positive control); 2.pGBKT7-Lam & pGADT7-T (negative control); 3.pGBKT7-FYFL & pGADT7-RIN; 4.pGBKT7-FYFL & pGADT7- MADS1; 5.pGBKT7-FYFL & pGADT7-JOINTLESS; 6.Blank. Empty bait vector, empty prey vector and autoactivation assay were growth of none yeast.
Discussion
Overexpression of SlFYFL delays sepal and plant senescence by inhibiting the biosynthesis of ethylene. Senescence is the final stage of the development of organism, which is a process with a series of recessions. Gradual loss of chlorophyll is the most obvious characteristic of plant senescence. Plant hormones are closely associated with senescence and ethylene is a typical senescence promoting hormone33. In our study, wild-type plants and fruit sepals turn yellow earlier than transgenic lines (Fig. 2C, E). Comparing the total chlorophyll content of sepal of wild type and transgenic fruits at B + 14 stage, chlorophyll content in wild-type are significantly reduced, while that in transgenic lines still remain at high level (Fig. 2 F). Expression levels of ethylene biosynthesis genes ACS1A, ACS2, ACS6 and ACO1, are decreased in leaf and sepal of transgenic plants compared with wild-type (Fig. 3 A, B). In addition, detached leaf senescence experiment results demonstrate that senescence of transgenic leaf is considerably delayed after 5 days of treatment with air or ethylene (Fig. 2A, B). These results suggest that overexpression of SlFYFL may decrease the ethylene biosynthesis, thereby delay the senescence of tomato plant. SlFYFL impacts sepal development. To date, five classes of MADS-box genes (A, B, C, D and E) determine the identities of floral organ34,35. In the ABCDE model, sepal structures are specified by genes of A-class. So far, a number of MADS-box genes including SlMADS-MC and TAGL1 were reported to influence sepal development. TAGL1 overexpression induces swelling and ripening of sepals19,36. The mutant rin displays enlarged sepals because of missing of SlMADS-MC11. In addition, SlMADS-MC has been reported to be the homolog of AP1, which is a A-class gene of Arabidopsis11. In our study, the 35S:FYFL lines represented longer sepals (Fig. S3 A–C), and SlMADS-MC was down-regulated in sepals of 35S:FYFL lines (Fig. S3 D). These results indicated that SlFYFL might be a member of A-class gene and regulate the development of sepal. SlFYFL overexpression inhibits ethylene biosynthesis and fruit ripening. In plants, ethylene biosynthesis pathway is well studied. Two modes of ethylene synthesis, system 1 and system 2, have been defined37. System 1 contributes to providing basal ethylene in vegetative tissues and unripe fruits. System 2 produces a large amount of ethylene at the onset of fruit ripening38. Transcriptional regulation of ACS is one of the major control points of ethylene biosynthesis39. SlACS2 is an important factor to transit System 1 to System 240. Antisense suppression of tomato ACS2 prevents ripening in a manner recoverable with exogenous ethylene5,41. SlACS1A and SlACS6 are involved in system 1 and present in tomato fruits before the onset of ripening40. In addition, SlACO1 and SlACO3 have been reported to contribute to triggering fruit ripening5. SlACO3 is induced but transitory at the breaker stage while SlACO1 expression is sustained during ripening39. In this study, we tested the expression of ACS2, ACO1and ACO3 in 35S:FYFL fruits. The results showed that expression levels of these ACC synthase genes in 35S:FYFL lines were 30–60% lower than in wild type (Fig. 3 C, D and E), and half ethylene was produced in the transgenic fruits (Fig. 3 K). These results suggest that SlFYFL overexpression impacts ethylene biosynthesis in fruit. E4 and E8 are well known to be important ethylene response factors impacting fruit ripening42. Our study showed that both of the two genes expressed at lower levels in the transgenic fruits compared with wild type (Fig. 3 F and G). In addition, PSY1, a major regulator of metabolic flux toward downstream carotenoids, induced by ethylene during fruit ripening43, and other two carotenoid biosynthesis enzymes PDS and ZDS, downstream of PSY1, were notably decreased and carotenoid contents were 20–45% lower in transgenic fruits (Fig. 2 G–J). Fig. 2 E also showed the ripening of 35S:FYFL fruits were delayed. These results suggest that overexpression of SlFYFL inhibits tomato fruit ripening. SlFYFL overexpression downregulates the expression of SlMADS-RIN and might affect its activity. Recently, hetero- or homo-dimers or higher-order complexes have been detected in MADS-domain proteins44. SLMADS-RIN is a classical and essential positive regulator of tomato fruit ripening among the MADS-box proteins, and associate with ethylene biosynthesis, ethylene perception and ethylene responsiveness. As previously reported, ACS2 is bound by SlMADS-RIN16,45. ACO1 is indirectly influenced by SlMADS-RIN through a homeobox gene HB116,46. E8 is identified as a novel direct target of SlMADS-RIN, which can be rapidly induced following ethylene induction and during normal fruit ripening16,47. In our study, ACO1, ACS2, E8 and SlMADS-RIN are all down-regulated by 30–80% in 35S:FYFL lines, which suggest that these genes are negatively regulated by SlFYFL (Fig. 3 C, D, G and H). Moreover, the yeast two-hybrid assay indicates that there is an interaction between SlFYFL and SlMADS-RIN (Fig. 6), impling that SlFYFL might bind to SlMADS-RIN and affect its activity. Furthermore, SlMADS1 was reported as a negative regulator of fruit ripening and was interacted with SlMADS-RIN20. In our study, SlFYFL interacts with SlMADS1 in vivo (Fig. 6). Therefore, we suspect that SlFYFL and SlMADS1 may form heterodimers with SlMADS-RIN respectively or together to regulate the expression of ripening related genes, thereby affecting the fruit ripening (Fig. 7). SlFYFL overexpression increases fruits storability. Tomato fruits become senescent and soft quickly after ripening. Therefore, tomato storability is an extremely important quality trait. There are many indicators to measure the storability of tomato fruits, such as fruit color, fruit firmness and pericarp thickness, etc. Ethylene and poly-galacturonic acid enzyme (PG) are two key regulatory factors in the process of tomato fruit ripening48. In our study, ethylene production was reduced and the transcript level of PG was decreased by 30–60% in 35S:FYFL fruits (Fig. 3 I, K), and 35S:FYFL fruits displayed slower rotting and dehydration than wild type (Fig. 2 K). These results indicate that overexpression of SlFYFL improves the storability of tomato fruit, and could be used as a molecular tool to improve fruit storability through modulating the expression of RIN. SlFYFL overexpression inhibits abscission zone development. Abscission is a key agricultural concern and an important trait for tomato to commercial products. The mutant of JOINTLESS has defectiveAZs in fruit pedicels25, which phenotype facilitates large-scale harvesting of tomato fruit by saving time removing the calices27. Another mutant lateral suppressor (ls), encoding a GRAS family transcription factor49, suppress the development of pedicel AZs28. Tomato fruit ripening mutant ripening-inhibitor (rin) develops incomplete pedicel AZs structures that show a knuckle region on the pedicels similar to wild-type plants, but frequently they show insufficient fruit abscission12. The rin mutation is a deletion that affects two tandemly arranged genes, RIN and MC. RIN controls fruit ripening and MC regulates sepal size, inflorescence determinacy and the development of tomato pedicel AZs11,12. In our study, the expression of JOINTLESS, Ls and MC genes in the B + 7 and B + 14 fruit stalk AZs were all down-regulated by 30–82% in transgenic plants (Fig. 5 A, B and H). Transcriptome analyses revealed that LeWUS, Bl and GOB all expressed specifically in pedicel AZs, and these transcription factors might play an important role in abscission processes, such as the regulation of pedicel AZ development, the maintenance of the AZ cells in an undifferentiated state, or the acquisition of competence to respond to abscission signals50,51,52. Mutation in Bl affects the jointless phenotype28, supporting the possibility that Bl acts in AZ development. Thus, the expression of LeWUS, Bl and GOB genes in B + 7 and B + 14 fruit stalk AZs were examined and were all reduced by over 50% in transgenic plants (Fig. 5 E, F and G). Previous studies indicate that the expression of many genes encoding cell wall modification-related proteins, such as Cel2 (endo-1,4-glucanase), polygalacturonase (PG) are drastically up-regulated at the onset of abscission53. Our results showed that Cel2 and TAPG1 in the B + 7 and B + 14 fruit stalk AZs were down-regulated in transgenic plants (Fig. 5 C, D). Meanwhile, ethylene is required to increase the expression of these cell wall modification genes with the onset of abscission54. Expression levels of ethylene biosynthase genes ACO1, ACO3, ACS1A, ACS2 and ASC6 were also reduced in AZs of transgenic plants (Fig. 5 I–M). These results indicate that SlFYFL overexpression may inhibit the expression of AZs related genes and the ethylene biosynthesis, thereby result in a developmental delay in AZs. This was proved in breakstrength test, breakstrength of B + 7 stage AZ in wild-type was approximately equal to that of B + 14 stage AZ in transgenic lines (Fig. 4 C). In addition, yeast two-hybrid assay results intimate that SlFYFL interacts with SlJOINTLESS in vivo (Fig. 6). It was reported that MADS-MC protein interacted physically with JOINTLESS, and regulated fruit abscission55. So we also suspect that FYFL, JOINTLESS and MC might form a complex to control the development of abscission zone (Fig. 7). Moreover, yeast two-hybrid assay results also display that there exist interactions between SlFYFL and SlMADS-RIN and SlMADS1 respectively, impling that SlFYFL might bind to SlMADS-RIN and SlMADS1 to regulate their activity, subsequently inhibit the expression of cell wall modification-related and ethylene biosynthesis genes, ultimately affect the formation of abscission zone.
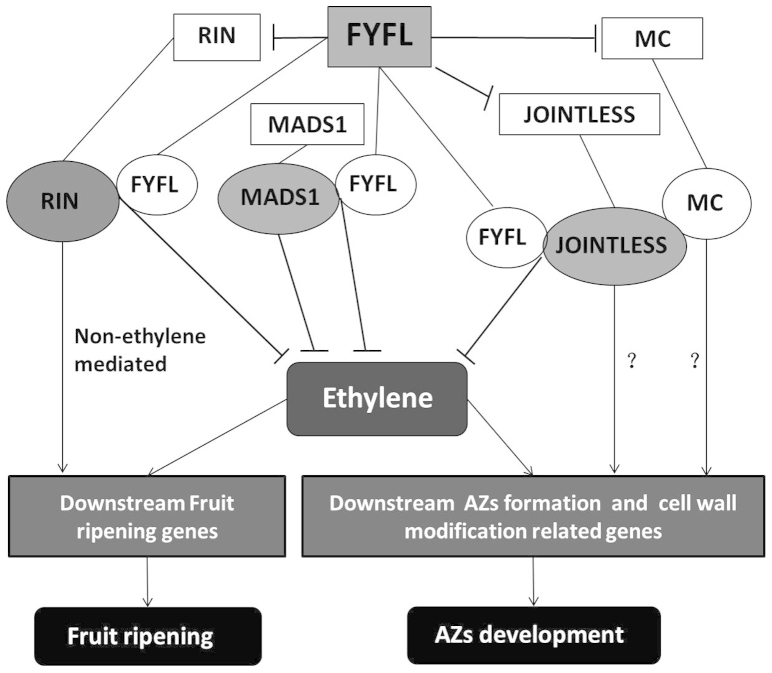
Figure 7. Patterns exist in tomato fruit ripening and abscission zone formation network.
Connection between different regulatory factors based on expression data analysis and yeast two-hybrid test in this study. The overexpression of FYFL inhibits the expression of ripening-related regulatory factor RIN and abscission zone regulatory factors JOINTLESS and MC. Previous studies have shown SlMADS1 was an ethylene biosynthesis inhibitory factor, thereby inhibiting fruit ripening; RIN positively regulated fruit riping both by ethylene and non-ethylene mediated pathways. There were interactions in FYFL & MADS1 and FYFL & RIN. FYFL may regulate the activity of MADS1 and RIN, then control the ethylene and downstream genes, and achieve the purpose of regulating ripening. JOINTLESS and MC were needed in formation of abscission zone of tomato. There was interaction between FYFL & JOINTLESS, moreover, it was reported that there was interaction between JOINTLESS and MC. FYFL may regulate the activity of JOINTLESS and MC, then control the ethylene and downstream genes, subsequently regulate the development of abscission zone. Whether JOINTLESS and MC can directly control the downstream genes or not, it is still not clear.
Methods
Plant materials and growth conditions
In this study, Solanum lycopersicon Mill. cv. Ailsa Craig (AC), a near-isogenic tomato line, was used as the wild type. The plants were planted in greenhouse and managed routinely. Transgenic cultures grew under standard greenhouse conditions. The ripening stages of tomato fruits were divided according to days after pollination (dpa) and fruit colour. In wild type, IMG (Immature green) fruits were defined as 20 dpa. MG (Mature green) fruits were defined as 32 dpa and were characterized as being green and shiny with no obvious colour change. B (Breaker) fruits were defined as the colour change from green to yellow. After breaker the fruit stages were divided B + 4 (4 days after B), B + 7 (7 days after B), B + 14, and so on. All plant samples were immediately frozen with liquid nitrogen and stored at –80°C until further use.
Isolation of SlFYFL and sequence analysis
Total RNA of tomato was extracted using TRIzol (Invitrogen, USA) according to the manufacturer's instructions. Then 1 μg total RNA was used to synthesis first strand cDNA through reverse transcription polymerase chain reaction (M-MLV reverse transcriptase, Takara) with Oligo d(T)18 primer. 1–2 μl cDNA was used to clone the full length of SlFYFL gene with primers SlFYFL-F and SlFYFL-R (Table 1S) through high fidelity PCR (Prime STARTM HS DNA polymerase, Takara). The amplified products were tailed by using DNA -Tailing kit (Takara) and linked with pMD18-T vector (Takara). Positive clones were picked out via Escherichia coli JM109 transformation and confirmed by sequencing (BGI, China). Multiple sequence alignments were performed by DNAMAN version 5.2.2. The phylogenic tree was calculated by MEGA (Molecular Evolutionary Genetics Analysis) version 3.156.
Construction of 35S:FYFL vector and plant transformation
Above-mentioned FYFL-pMD18-T vector was used as the template and was amplified with primers SlFYFL (F + X) and SlFYFL (R + S) which have been tailed with Xba I and SacI restriction site at the 5′ end respectively. Then the amplified products were digested with XbaI and SacI respectively, and linked into the plant binary vector pBI121 with SacI and XbaI restriction sites. The transgene was under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter. The generated binary vectors were transferred into Agrobacterium LBA4404 and Agrobacterium-mediated transformation was performed following the protocols described by Chen et al57. The transgenic plants were detected with primers NPTII-F (5′ GAC AAT CGG CTG CTC TGA 3′) and NPTII-R (5′ AAC TCC AGC ATG AGA TCC 3′). The positive transgenic plants were selected and used for subsequent experiments.
Quantitative real-time PCR analysis
RNA extraction and cDNA synthesis were performed as the above described. The synthesized cDNAs were diluted 1 times in RNase/DNase-free water. Quantitative real-time PCR analysis was carried out using the CFX96™ Real-Time System (C1000™ Thermal Cycler, Bio-Rad). All reactions were performed using the SYBR® Premix Go Taq II kit (Promega, China) in a 10 μl total sample volume (5.0 μl 2 × SYBR Premix Go Taq, 0.5 μl primers, 1.0 μl cDNA, 3.5 μl ddH2O). To remove the effect of genomic DNA and the template from environment, NTC (no template control) and NRT (no reverse transcription control) were performed. Three replications for each sample were used and standard curves were run simultaneously. Melt curve analysis of qPCR samples revealed that there was only one product for each gene primer reaction. The PCR products were sequenced to confirm the specific amplification. SlCAC58 gene was used as internal standard in tomato tissues. The primers SlFYFL(RT)-F and SlFYFL(RT)-R (Table S1) were used to determine the expression level of SlFYFL in wild type and transgenic lines.
Furthermore, the expression levels of fruit ripening and ethylene biosynthesis pathway genes E4, E8, PG, PSY1, PDS, ZDS, RIN, ACO1, ACO3, ACS2 and ERF1 were determined in fruits. Above-mentioned abscission zone related genes, JOINTLESS, MC, WUS, GOB, Ls, Bl, Cel2, TAPG1 were detected in abscission zone. Ethylene biosynthesis genes ACO1, ACO3, ACS1A, ACS2 and ACS6 were detected in AZ, leaf and sepal. Primers were shown in Table 1S.
Measurement of carotenoid contents
1.0 g sample was cut from pericarp in a 5 mm wide strip around the equator of B, B + 7 and B + 14 of wild type and 35S:FYFL lines, respectively. Then 10 ml of 60:40 (v/v) hexane-acetone was added respectively and total carotenoids fruits were extracted. The extract was centrifuged at 4000 g for 5 min and the absorbance of supernatant was measured at 450 nm. Carotenoid content was calculated with the following equations: total carotenoid mg ml−1 = 4*(OD450)*10 ml/1 g43,59. Three independent experiments were performed for each sample.
Ethylene measurements
Fruits at B, B + 1, B + 3, B + 7 and B + 14 stages were harvested and placed in open 100 mL jars for 2 h to minimize the effect of wound ethylene caused by picking. Jars were then sealed and incubated at room temperature for 2 h, and 0.5 mL of headspace gas was injected into a Focus GS gas chromatograph (Thermo-Electron) equipped with a flame ionization detector. Samples were compared with reagent grade ethylene standards of known concentration and normalized for fruit weight.
Observation of abscission zone in fruit pedicels
In our experiments, we found that formation of fruit stalk abscission zone in transgenic plants were different from wild type (Fig. 4 A). Therefore, we carried out observation and analysis. The optical microscope (Olympus-BMF) was used to observe fresh abscission zone in fruit pedicel materials at B + 7 and B + 14 stages of transgenic plants and wild type. Repeat twice for each observation.
Breakstrength analysis of abscission zone in fruit pedicels
Every single fruit stalk of wild type and transgenic lines was clipped with a small clamp which was attached to a breakstrength meter. Then the breakstrength of fruit stalk abscission zone was measured using the BOSE ELECTROFORCE 3300 breakstrength meter. The measured value represents the force necessary to break the fruit stalk from the abscission zone. Fruit stalk materials were taken from B + 7 and B + 14 stages fruits, and three measurements were performed at each stage for each plant.
Postharvest storage test
Fruits of wild type and transgenic lines were harvested at B + 7 stage, and placed on filter paper in standard greenhouse conditions. Phenotype was observed once every two days.
Yeast two-hybrid assay
Yeast two-hybrid was performed using the MATCHMAKER TM GAL4 Two-Hybrid System III according to the manufacturer's protocol (Clontech). The open reading frame of SlFYFL was amplified by PCR with the primer pairs SlFYFL(Y)-F and SlFYFL(Y)-R (Table 1S). The PCR products were digested using EcoRI and Pst I and cloned into the EcoR I and Pst I sites of the pGBKT7 bait vector to obtain the vector FYFL-pGBKT7. Then FYFL-pGBKT7 vector was transferred into Y2HGold. The Y2HGold with bait was plated on SD medium lacking Trp (SDO) and SD medium lacking Trp, His, Ade (TDO) to test self-activation of FYFL-pGBKT7. In parallel, the open reading frame of SlRIN, SlJOINTLESS and SlMADS1 were also amplified by primers (SlRIN(Y)-F, SlRIN(Y)-R), (SlJ(Y)-F, SlJ(Y)-R) and (SlMADS1 (Y)-F, SlMADS1 (Y)-R) (Table 1S). The products were cloned into the pGADT7 vector, and introduced into Y187. Subsequently, Y2HGold with bait and Y187 with prey were cultured together in 2 × YPDA medium for 24 h. After that these cultures were cultured on SD medium lacking Trp, Leu (DDO) to select for diploids containing prey and bait vectors. After 2 to 5 days, fresh diploid cells were plated on SD medium lacking Trp, Leu, and His, Ade, with X-α-Gal (QDO/X) to judge whether SlFYFL can interact with SlRIN, SlJOINTLESS and SlMADS1 or not, respectively. Plates were incubated for 3 to7 days at 30°C. An empty prey and bait vector were used as negative controls with each bait and prey construct, respectively. Meanwhile, positive controls were cultured. The assays were repeated at least three times with fresh transformants.
Detached leaf senescence experiment
Mature leaves were cut from 2 month 35S:FYFL and wild-type plants and placed on three layers of wet filter paper in 14 cm Petri dishes, then wrapped in aluminium foil, incubated in the dark at 25°C for 5 d. Some dishes were placed in a glass desiccator with 20 mlL−1 ethylene. The others were incubated under air condition.
Extraction and quantitation of leaf and sepal chlorophyll
Weighted 1 g fresh sepals of B + 7 and B + 14 fruits of wild-type and transgenic lines, pounded to pieces with liquid nitrogen, extracted with 10 ml mixed solution acetone and ethanol (2:1, V/V) for 48 h in dark, centrifuged 5000 rpm for 10 min at 4°C. The absorbance of the supernatant was measured at 645 and 663 nm in a PerkinElmer Lambda 900 UV/VIS/NIR spectrophotometer using above-mentioned mixed solution as a blank. Total chlorophyll content were calculated using the formulas according to the method of Arnon60: Chl (mg/g) = 20.29A645 + 8.02A663. The chlorophyll of each sample was extracted and measured in triplicate. Chlorophyll contents of mature leaves, treated by ethylene/air were measured using the same method.
Statistics of sepal length of flower
In our study, we found that transgenic sepals were longer than the wild type. We measured the length of sepals at the fully open flower stage, at least 10 flowers per plant were measured (Fig. 3S C).
Author Contributions
G.C. and Z.H. designed and managed the research work and improved the manuscript. Q.X., Z.Z., T.D., Z.Z. and B.C. performed the experiments. Q.X. and T.D. wrote the manuscript.
Supplementary Material
Supplementary Info File #1
Acknowledgments
This work was supported by National Natural Science Foundation of China (nos. 31100089, 31171968) and Natural Science Foundation of Chongqing of China (No. cstc, 2011BB1068).
References
- Abeles F. B., Morgan P. W. & Saltveit Jr M. E. The role of Ethylene in agriculture. 264–294 (Academic press, 1992).
- Fraser P. D., Enfissi E. & Bramley P. M. Genetic engineering of carotenoid formation in tomato fruit and the potential application of systems and synthetic biology approaches. Arch. Biochem. Biophys. 483, 196–204 (2009). [DOI] [PubMed] [Google Scholar]
- Klee H. J. & Giovannoni J. J. Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45, 41–59 (2011). [DOI] [PubMed] [Google Scholar]
- Adams D. & Yang S. Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. P. Natl. Acad. Sci. USA 76, 170–174 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander L. & Grierson D. Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J. Exp. Bot. 53, 2039–2055 (2002). [DOI] [PubMed] [Google Scholar]
- Xu R., Goldman S., Coupe S. & Deikman J. Ethylene control of E4 transcription during tomato fruit ripening involves two cooperativecis elements. Plant Mol. Biol. 31, 1117–1127 (1996). [DOI] [PubMed] [Google Scholar]
- Lincoln J. E. & Fischer R. L. Regulation of gene expression by ethylene in wild-type and rin tomato (Lycopersicon esculentum) fruit. Plant Physiol. 88, 370–374 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deikman J. & Fischer R. L. Interaction of a DNA binding factor with the 5′-flanking region of an ethylene-responsive fruit ripening gene from tomato. EMBO J. 7, 3315 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery J., Pollard V., Deikman J. & Fischer R. L. Positive and negative regulatory regions control the spatial distribution of polygalacturonase transcription in tomato fruit pericarp. Plant Cell 5, 1049–1062 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R. et al. Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23, 923–941 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J. et al. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Sci. 296, 343–346 (2002). [DOI] [PubMed] [Google Scholar]
- Nakano T. et al. MACROCALYX and JOINTLESS interact in the transcriptional regulation of tomato fruit abscission zone development. Plant Physiol. 158, 439–450 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman L. C. et al. Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Mol. Biol. Evol. 23, 2245–2258 (2006). [DOI] [PubMed] [Google Scholar]
- Shima Y. et al. Tomato FRUITFULL homologues act in fruit ripening via forming MADS-box transcription factor complexes with RIN. Plant Mol. Biol. 1–12 (2013). [DOI] [PubMed] [Google Scholar]
- Bemer M. et al. The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant cell 24, 4437–4451 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C., Vrebalov J., Tafelmeyer P. & Giovannoni J. J. The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 157, 1568–1579 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busi M. V. et al. MADS-box genes expressed during tomato seed and fruit development. Plant Mol. Biol. 52, 801–815 (2003). [DOI] [PubMed] [Google Scholar]
- Pnueli L., Hareven D., Rounsley S. D., Yanofsky M. F. & Lifschitz E. Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell 6, 163–173 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J. et al. Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21, 3041–3062 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T. et al. A tomato MADS-box transcription factor, SlMADS1, acts as a negative regulator of fruit ripening. Plant Physiol. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigyo M., Hasebe M. & Ito M. Molecular evolution of the AP2 subfamily. Gene 366, 256–265 (2006). [DOI] [PubMed] [Google Scholar]
- Chung M. Y. et al. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 64, 936–947 (2010). [DOI] [PubMed] [Google Scholar]
- Ferrandiz C., Liljegren S. J. & Yanofsky M. F. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289, 436–438 (2000). [DOI] [PubMed] [Google Scholar]
- Brown K. M. Ethylene and abscission. Physiol. Plantarum 100, 567–576 (1997). [Google Scholar]
- Mao L. et al. JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 406, 910–913 (2000). [DOI] [PubMed] [Google Scholar]
- Li C., Zhou A. & Sang T. Rice domestication by reducing shattering. Science 311, 1936–1939 (2006). [DOI] [PubMed] [Google Scholar]
- Zahara M. B. & Scheuerman R. W. Hand-harvesting jointless vs. jointed-stem tomatoes. Calif. Agr. 42, 14–14 (1988). [Google Scholar]
- Roberts J. A., Elliott K. A. & Gonzalez-Carranza Z. H. Abscission, dehiscence, and other cell separation processes. Annu. Rev. Plant Biol. 53, 131–158 (2002). [DOI] [PubMed] [Google Scholar]
- Roberts J. A., Schindler C. B. & Tucker G. A. Ethylene-promoted tomato flower abscission and the possible involvement of an inhibitor. Planta 160, 159–163 (1984). [DOI] [PubMed] [Google Scholar]
- Brummell D. A., Hall B. D. & Bennett A. B. Antisense suppression of tomato endo-1, 4-β-glucanase Cel2 mRNA accumulation increases the force required to break fruit abscission zones but does not affect fruit softening. Plant Mol. Biol. 40, 615–622 (1999). [DOI] [PubMed] [Google Scholar]
- Chen M. K. et al. The MADS box gene, FOREVER YOUNG FLOWER, acts as a repressor controlling floral organ senescence and abscission in Arabidopsis. Plant J. 68, 168–185 (2011). [DOI] [PubMed] [Google Scholar]
- Fraser P. D., Truesdale M. R., Bird C. R., Schuch W. & Bramley P. M. Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression). Plant Physiol. 105, 405–413 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić V. & Bleecker A. B. Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 8, 595–602 (1995). [Google Scholar]
- Coen E. S. & Meyerowitz E. M. The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37 (1991). [DOI] [PubMed] [Google Scholar]
- Theiβen G. & Saedler H. Floral quartets. Nature 409, 469–471 (2001). [DOI] [PubMed] [Google Scholar]
- Itkin M. et al. TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 60, 1081–1095 (2009). [DOI] [PubMed] [Google Scholar]
- McMurchie E., McGlasson W. & Eaks I. Treatment of fruit with propylene gives information about the biogenesis of ethylene. Nature 237, 235–236 (1972). [DOI] [PubMed] [Google Scholar]
- Yang S. & Oetiker J. The role of ethylene in fruit ripening. Postharvest Physiol. Fru. 398, 167–178 (1994). [Google Scholar]
- Barry C. S. et al. Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J. 9, 525–535 (1996). [DOI] [PubMed] [Google Scholar]
- Barry C. S., Llop-Tous M. I. & Grierson D. The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 123, 979–986 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeller P. W., Lu M., Taylor L. P., Pike D. A. & Theologis A. Reversible inhibition of tomato fruit senescence by antisense RNA. Science 254, 437–439 (1991). [DOI] [PubMed] [Google Scholar]
- Lincoln J. E. & Fischer R. L. Diverse mechanisms for the regulation of ethylene-inducible gene expression. Mol. Gen. Genet. 212, 71–75 (1988). [DOI] [PubMed] [Google Scholar]
- Fray R. G. & Grierson D. Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol. Biol. 22, 589–602 (1993). [DOI] [PubMed] [Google Scholar]
- Favaro R. et al. Ovule-specific MADS-box proteins have conserved protein-protein interactions in monocot and dicot plants. Mol. Genet. Genomics 268, 152–159 (2002). [DOI] [PubMed] [Google Scholar]
- Ito Y. et al. DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 55, 212–223 (2008). [DOI] [PubMed] [Google Scholar]
- Lin Z. et al. A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 55, 301–310 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Wang Y., Cao B., Wang W. & Tian S. Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J. (2012). [DOI] [PubMed] [Google Scholar]
- Crookes P. R. & Grierson D. Ultrastructure of tomato fruit ripening and the role of polygalacturonase isoenzymes in cell wall degradation. Plant Physiol. 72, 1088–1093 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K., Schmitt T., Rossberg M., Schmitz G. & Theres K. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. P. Natl. Acad. Sci. USA 96, 290–295 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T. et al. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Gene. Dev. 17, 1175–1187 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T., Abbott J., Moritz T. & Doerner P. Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18, 598–611 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyhle F., Sarkar A. K., Tucker E. J. & Laux T. <i> WUSCHEL</i> regulates cell differentiation during anther development. Dev. Biol. 302, 154–159 (2007). [DOI] [PubMed] [Google Scholar]
- Kalaitzis P., Koehler S. M. & Tucker M. L. Cloning of a tomato polygalacturonase expressed in abscission. Plant Mol. Biol. 28, 647–656 (1995). [DOI] [PubMed] [Google Scholar]
- Meir S. et al. Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol. 154, 1929–1956 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leseberg C. H. et al. Interaction study of MADS-domain proteins in tomato. J. Exp. Bot. 59, 2253–2265 (2008). [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K. & Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5, 150–163 (2004). [DOI] [PubMed] [Google Scholar]
- Chen G. et al. Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol. 136, 2641–2651 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expósito-Rodríguez M., Borges A. A., Borges-Pérez A. & Pérez J. A. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 8, 131 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forth D. & Pyke K. A. The suffulta mutation in tomato reveals a novel method of plastid replication during fruit ripening. J. Exp. Bot. 57, 1971–1979 (2006). [DOI] [PubMed] [Google Scholar]
- Arnon D. I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1 (1949). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Info File #1