ABSTRACT
Outer membrane vesicles (OMV) are spherical membranous structures released from the outer membrane (OM) of Gram-negative bacteria. OMV have been proposed to play several different roles during both pathogenesis and symbiosis. Despite the fact that OMV were described several decades ago, their biogenesis is a poorly characterized process. Whether OMV are produced by an active mechanism or by passive disintegration of the OM is a still matter of controversy. Bacteroides fragilis and Bacteroides thetaiotaomicron are important members of the human microbiota. In this work, we determined and compared the protein compositions of OM and OMV from B. fragilis and B. thetaiotaomicron. SDS-PAGE analysis of both fractions revealed dramatically different protein profiles. Proteomic analysis of OM and OMV in B. fragilis identified more than 40 proteins found exclusively in OMV and more than 30 proteins detectable only in the OM. The OMV-specific proteome showed a high prevalence of glycosidases and proteases, some of which were shown to be active in vitro. Similar results were obtained for B. thetaiotaomicron. Most of the OMV-exclusive proteins were acidic. Based on these results, we propose that these species possess machinery devoted to selectively pack acidic proteins into the OMV. These OMV equipped with hydrolytic enzymes could help in securing nutrients for the benefit of the whole bacterial community present in the microbiota, uncovering a novel function for bacterial OMV.
IMPORTANCE
The members of genus Bacteroides are key players in the symbiosis between the human host and the gut microbiota. It is known for its ability to degrade a wide variety of glycans that are not substrates for human glycosidases. The cleaved glycans can be utilized by Bacteroides and other microbiota members, resulting in the production of short-chain fatty acids that are beneficial for the host. Although members of the genus Bacteroides are known to secrete different hydrolases, their secretion pathways remain uncharacterized. In this article, we show that B. fragilis and B. thetaiotaomicron preferentially pack a large number of hydrolases in outer membrane vesicles (OMV). Most of these hydrolases are acidic and were detected exclusively in OMV. This suggests the presence of a molecular mechanism in Bacteroides responsible for the selection of OMV proteins based on their charge. We propose that OMV contribute to the establishment and balance of the gut microbiota.
INTRODUCTION
The human intestine is colonized by a dense population of bacteria commonly known as the microbiota. The microbiota is beneficial to the host in many ways. These benefits include resistance to invasive pathogens, stimulation of cell turnover, and aiding in the digestion of complex nutrients to provide the host with extra energy and essential vitamins (1). The majority of simple sugars and oligosaccharides ingested by the human host are absorbed through the small intestine (2). Other sugars reach the colon intact, where they serve as the substrates for the microbiota (3, 4). Besides, the host itself produces fermentable products, including glycoproteins and polysaccharides that are consumed by the gut microbiota (4–8). Members of the genus Bacteroides represent the most abundant polysaccharide utilizers in the colon (8, 9). Ten to 20% of the commensal population in the colon consists of representatives of the genus Bacteroides (10). Among these is B. fragilis, an opportunistic pathogen and the most commonly isolated anaerobe from human infections such as intraabdominal and brain abscesses (11–14). Under normal symbiotic conditions, B. fragilis is beneficial to the host and contributes to the modulation of the host immune response through polysaccharide A (PSA) (15), delivered to the host cells via outer membrane vesicles (OMV) (16). Recently, OMV from Bacteroides fragilis and other Bacteroides members were found to participate in the establishment of a cooperative ecosystem in the gut. In the model proposed by Rakoff-Nahoum et al., some species secrete OMV, which are able to break down polysaccharides for the benefit of the other species present in the community (17).
OMV are small blebs originating from the outer membrane (OM) of Gram-negative bacteria. OMV contain lipopolysaccharide (LPS), outer membrane proteins, phospholipids, and periplasmic components (18). The presence of toxins in OMV from different bacteria has led to suggestions that vesicles are long distance delivery tools (18–24). Interestingly, ClyA toxin of enterohemorrhagic Escherichia coli was found to be active only in vesicles, where the proper redox potential supports its oligomerization (25). Additional roles in stress response and quorum sensing have been attributed to OMV (26, 27). Despite the different roles suggested for vesicles, the absence of a clear model for OMV biogenesis has led to disagreement on whether vesiculation is a directed process or just the result of passive membrane disintegration. If vesicles are formed by the latter mechanism, OM and OMV should have the same protein composition. However, previous reports have shown the enrichment of toxins in vesicles from different pathogens, supporting the notion of cargo selection in vesicles (18, 25, 28, 29). A proteomic analysis of OM and OMV in the oral pathogen Porphyromonas gingivalis showed that some OM proteins were enriched in OMV, whereas other OM proteins were excluded from vesicles, and that mutations in the LPS resulted in aberrant cargo selection, providing evidence of the presence of selection machinery that is capable of packing specific cargo into OMV (28).
In this work, we carried out a proteomic analysis of both the OM and OMV of B. fragilis. We found multiple OM proteins that were absent in OMV and an exceptionally large number of proteins exclusively present in vesicles. Many of the OMV proteins were acidic and annotated as hydrolases, mainly proteases and glycosidases, some of which were shown to be active in vitro. Similar results were obtained when the content of the OMV from Bacteroides thetaiotaomicron was analyzed. These results strongly advocate for the existence of yet uncharacterized machinery dedicated to OMV production in Bacteroides.
RESULTS
B. fragilis produces spherical, uniformly sized OMV.
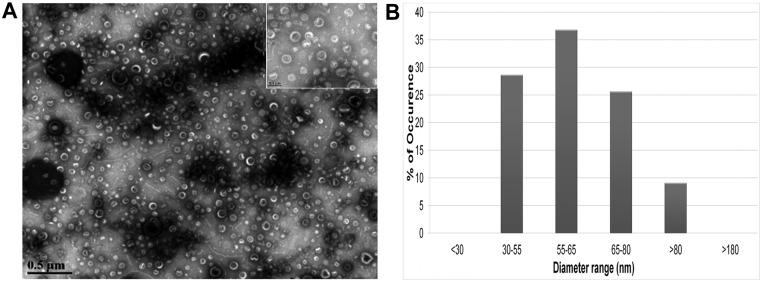
Previous reports showed that, similarly to other Gram-negative bacteria, B. fragilis produces OMV (30). Accordingly, we wanted to ensure that our method of vesicle purification resulted in a cell-free OMV preparation. B. fragilis vesicles were harvested from cell-free supernatant of an overnight culture. The OMV were resuspended in sterile water and then examined using transmission electron microscopy (TEM). As shown in Fig. 1, B. fragilis vesicles were spherical and uniform in size, with diameters ranging from 30 to 80 nm. This range is narrower than what was reported previously for some other Gram-negative bacteria (31). No B. fragilis cells were observed in our OMV preparations.
FIG 1 .
B. fragilis produces outer membrane vesicles (OMV). (A) Transmission electron microscopy of B. fragilis OMV at two different magnifications: ×28,000 and ×140,000 (inset). Scale bars represent 500 nm and 100 nm, respectively. (B) Quantification of the size range of B. fragilis OMV as observed under the electron microscope.
OMV show a different protein profile from OM in B. fragilis.
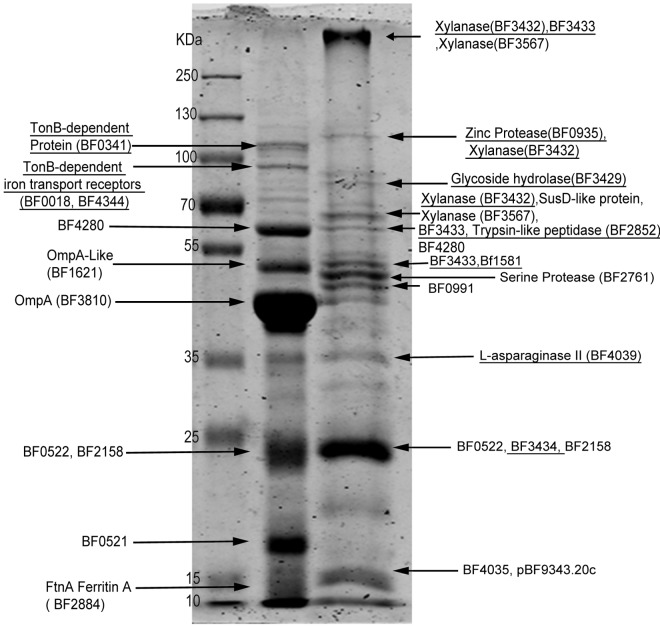
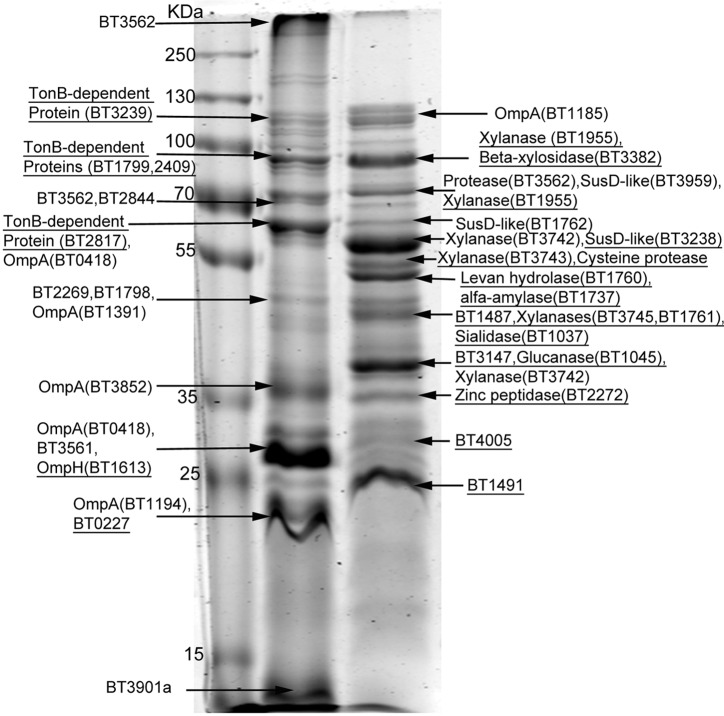
Recent work has shown that OMV from P. gingivalis, Serratia marcescens, and Neisseria meningitidis carry, to a variable extent, different proteins compared to the OM from which they originated (28, 29, 32). We tested whether OMV cargo selection occurs in B. fragilis, which is an important member of the gut microbiota and genetically close to P. gingivalis (10, 33). To study protein sorting in B. fragilis OMV, both OM and OMV proteins were separated by SDS-PAGE followed by Coomassie staining. B. fragilis OMV exhibited a very different protein profile from the OM of the same cells (Fig. 2). Bands were excised from gel, tryptically digested, and analyzed by mass spectrometry (MS) for protein identification. A total of 115 proteins were detected in OMV, while 102 proteins were found in the OM of B. fragilis. Only predicted outer membrane and periplasmic proteins were found in OMV, with no inner membrane or cytosolic proteins detected.
FIG 2 .
OMV show different protein profile from that of OM. Ten micrograms of purified OM and OMV proteins were run on 10% SDS-PAGE followed by Coomassie staining. Protein bands were excised from the gel and digested with trypsin. The resulting peptides were enriched using ZipTip C18 columns then analyzed via liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) followed by protein identification with Mascot search engine using the NCBInr database. The arrows point to the most abundant proteins in each band. Underlined proteins were detected only in the corresponding compartment.
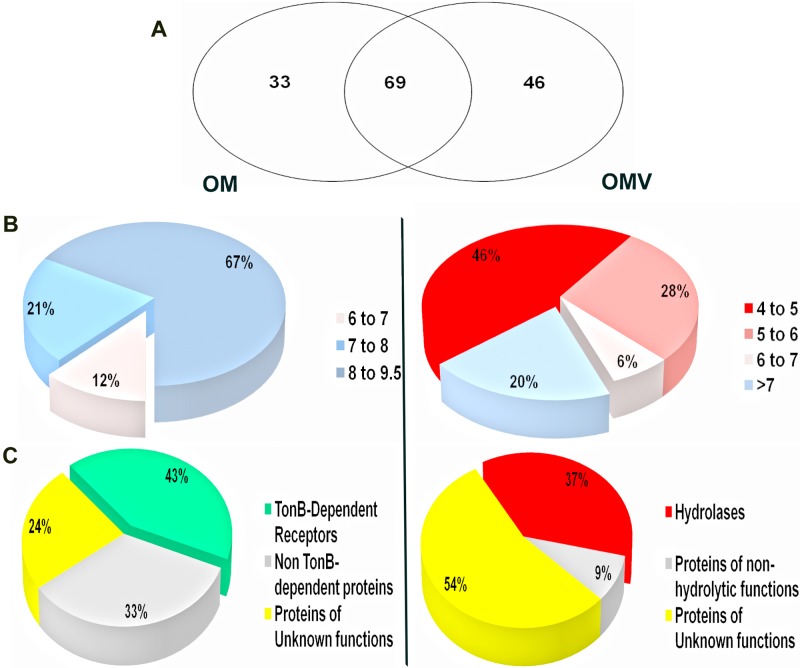
Sixty-nine proteins were common between OMV and OM (Fig. 3A; see Table S2 in the supplemental material). Among the common proteins were several proteins annotated as “SusD-like.” SusD proteins belong to the “starch utilization system” (Sus) family, composed of a very large number of glycan binding proteins. Sus proteins can bind to starch and its derivatives, like maltooligosaccharides, amylose, amylopectin, and pullulan (34–36). In Bacteroides thetaiotaomicron, an important human gut symbiont that is genetically related to B. fragilis, the Sus proteins account for about 18% of the genome (35). Some of the Sus components were predicted to be surface-exposed lipoproteins (37).
FIG 3 .
B. fragilis OMV and OM display different distributions of proteins based on pIs and function. (A) Schematic representation of protein compartmentalization in both OMV and OM. Proteomic analysis identified 69 proteins common to OM and OMV, and 46 proteins only detected in OMV. (B) Pie charts showing the distributions of OM and OMV exclusive proteins according to their pI. OMV displayed preferential sorting toward acidic proteins, while alkaline proteins resided in the membrane. (C) Pie charts showing the functional distributions of OM and OMV exclusive proteins. OMV were enriched in hydrolases compared to OM, which displayed a large percentage of TonB-dependent receptors.
Forty-six proteins were found exclusively in OMV (Table 1 and Fig. 3A; see Table S1 in the supplemental material). Although enrichment of a few proteins in OMV has been previously described, the dramatic difference in the compositions of OM and OMV observed in B. fragilis has not been previously reported. For those proteins that are not annotated in GenBank, we relied on BLASTP (38), PFAM (39), and HHPRED algorithms (40) for homology-based annotation of the identified proteins. Interestingly, we found about one-quarter of OMV proteins to be homologous to hydrolases (Fig. 3C). These included 11 glycosidases, 11 peptidases, one phosphatase, and one lipase. Most of these glycosidases (8/11) and peptidases (7/11) were not detected in OM. (Table 1). Such abundance of hydrolytic enzymes was only previously reported in OMV of Myxococcus xanthus (41, 42). On the other hand, MS analysis of OM proteins revealed 33 unique proteins that were not detected in vesicles (Table 2). Most proteins in the OM fraction are involved in ligand binding and transport, albeit some putative hydrolases were also detected but at lower abundance compared to OMV. The majority of the OM proteins were TonB-dependent receptors (TBDTs). TBDTs are bacterial outer membrane proteins that bind and transport different substrates, like siderophores, carbohydrates, and vitamin B12, using energy provided by the inner membrane proton motive force (43). Other proteins involved in heme binding and transport, like HmuY, were also found in the OM (Fig. 3C; see Table S2 in the supplemental material).
TABLE 1 .
Putative hydrolases identified by MS/MS exclusively in OMV of B. fragilis
| No. | Function | Transmembrane domain or lipoprotein signal | pI |
|---|---|---|---|
| BF3432 | Xylanase | Yes | Acidic |
| BF3429 | Glycosyl hydrolase | No | Acidic |
| BF3562 | Xylanase | Yes | Acidic |
| BF0339 | β-Glucosidase | Yes | Acidic |
| BF3793 | β-Galactosidase | Yes | Acidic |
| BF0810 | α-l-Fucosidase | Yes | Alkaline |
| BF4060 | β-Glucanase | Yes | Acidic |
| BF3083 | α-l-Fucosidase | No | Alkaline |
| BF0935 | Zinc protease | Yes | Acidic |
| BF2852 | Trypsin-like peptidase | No | Acidic |
| BF2757 | Clostripain-related peptidase | No | Acidic |
| BF2157 | Peptidase | Yes | Acidic |
| BF3010 | Trypsin-like peptidase | Yes | Acidic |
| BF1408 | Dipeptidase | Yes | Acidic |
| BF4039 | l-Asparaginase II | Yes | Alkaline |
TABLE 2 .
Putative hydrolases identified by MS/MS exclusively in OMV of B. thetaiotaomicron
| No. | Function | Transmembrane domain or lipoprotein signal | pI |
|---|---|---|---|
| BT_1955 | Xylanase | Yes | Acidic |
| BT_3382 | β-Xylosidase | Yes | Acidic |
| BT_3727 | Glycosyl hydrolase | Yes | Acidic |
| BT_0278 | Glucanase | Yes | Acidic |
| BT_3859 | Hydrolase | Yes | Acidic |
| BT_3742 | Xylanase | Yes | Acidic |
| BT_3743 | Xylanase | Yes | Acidic |
| BT_3960 | Cysteine proteinases | Yes | Acidic |
| BT_1760 | Levan hydrolase | Yes | Acidic |
| BT_1737 | α-Amylase | Yes | Alkaline |
| BT_3987 | Endo-β-N-acetylglucosaminidase | Yes | Acidic |
| BT_3523 | Glucanases | Yes | Acidic |
| BT_3381 | β-Xylosidase | Yes | Acidic |
| BT_1037 | α-Sialidase | Yes | Acidic |
| BT_3745 | Xylanase | Yes | Acidic |
| BT_1761 | Xylanase | Yes | Acidic |
| BT_3985 | Chitinase | Yes | Acidic |
| BT_1038 | Chitinase | Yes | Acidic |
| BT_1045 | Glucanases | Yes | Acidic |
| BT_3861 | Glucanases | Yes | Acidic |
| BT_2239 | Protease | Yes | Acidic |
| BT_3577 | Serine protease | No | Acidic |
| BT_2272 | Zinc peptidase | No | Acidic |
| BT_3522 | Glucanases | Yes | Acidic |
| BT_3237 | Zinc peptidase | Yes | Acidic |
To purify the OM and separate it from the inner membrane, we used a Triton X-100-based method (28). To exclude the possibility that the differences in the protein contents were due to the treatment with this detergent, we repeated our analysis using total membrane preparations, which includes inner and outer membrane proteins. After the lipids were removed, the total membrane proteins were digested with trypsin. The resulting peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify the proteins. None of the 46 proteins unique to OMV was found in the total membrane, indicating that the presence of proteins only in OMV is not an artifact of the preparation (data not shown). Taken together, these results indicate that mechanisms for sorting and exclusion of proteins into OMV must exist in B. fragilis.
OMV exclusive proteins are mostly acidic, and OM unique proteins are mostly basic.
Another interesting trend discovered in this analysis is related to the isolectric points (pIs) of OM and OMV proteins. According to their calculated pIs, about 80% of OMV-specific proteins are acidic (Table 1; Fig. 3B). In contrast with vesicular proteins, only 2 of the 33 OM unique proteins were found to have acidic isoelectric points, while the majority were found to be alkaline. As shown in Table 1 here and Table S1 in the supplemental material, the majority of the OMV proteins were predicted to have either a lipoprotein signal or transmembrane domains. This suggests that these proteins are genuine membrane proteins, which must be targeted to the OM before their inclusion in OMV, and not secreted proteins interacting with OMV after OMV are formed.
OMV show proteolytic activity.
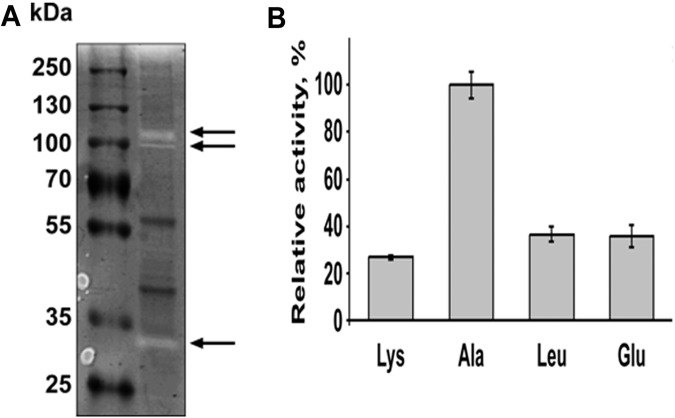
As previously mentioned, we identified 11 proteins in OMV that were annotated as proteases. Therefore, we tested if they display activity via a zymogram analysis. OMV were harvested from B. fragilis and run on an SDS-PAGE gel containing gelatin as a protease substrate. Following renaturation and incubation at 37°C, the gel was stained with Coomassie to detect proteolytic activity. As shown in Fig. 4A, several clear bands were visualized after staining the gel, indicating that gelatin was digested at these sites. This allowed us to conclude that at least some of the proteases detected in the vesicles are active. In addition, this experiment was repeated with heat-inactivated OMV, and no hydrolase activity was observed (data not shown). To confirm the protease activity of vesicles, we performed in vitro protease assays using amino acids (lysine, alanine, leucine, and glutamate) linked to p-nitroanilide. p-Nitroanilide is a chromogenic compound that will show maximum absorbance at λ = 405 nm when the substrate is cleaved. All 4 substrates were cleaved by the OMV, showing higher activity when using p-nitroanilide alanine as the substrate (Fig. 4B).
FIG 4 .
OMV display protease activity. (A) Ten micrograms of OMV proteins was loaded on 10% Tris-glycine gel with 0.1% gelatin as the substrate. Following separation at denaturing conditions, proteins were renatured and then incubated at 37°C for 2 days to allow substrate cleavage. Colloidal Coomassie brilliant blue G-250 was used to stain the gel. Clear bands marked by the arrows indicate the digestion of gelatin by proteases. (B) Peptidase activity of B. fragilis vesicles was tested using different p-nitroanilide-linked amino acids. Ten micrograms of purified OMV proteins was incubated with different substrates for 1 h at 37°C. Activities were determined by measuring the absorbance of the released p-nitroanilide at 405 nm. All activities are represented relative to the alanine-peptidase activity. All experiments were done in triplicates.
Sugar-hydrolyzing activity of OMV.
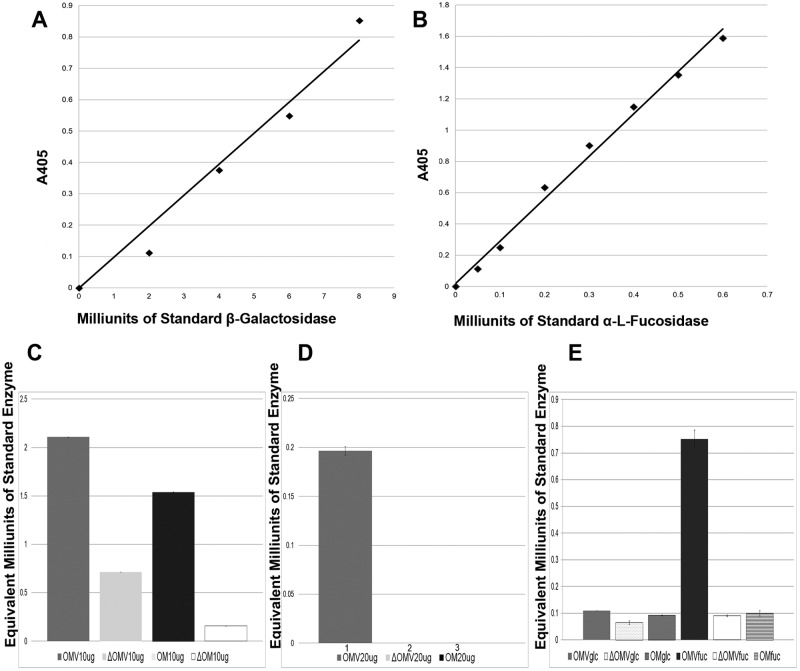
The proteomic analysis of OMV revealed the presence of several proteins annotated as sugar hydrolases (see Table 1; also see Table S2 in the supplemental material). These included α-1,2-mannosidase (BF3236), xylanase (BF3567), and a peptide-N-glycosidase (BF0811). The previous proteins were detected in both the OM and OMV fractions. However, OMV possessed several unique putative sugar hydrolases that were not detected in the OM (Table 1). These hydrolases were two xylanases (BF3432 and BF3562), two α-l-fucosidases (BF0810 and BF3083), a putative glycoside hydrolase (BF3429), β-glucosidase (BF0339), β-galactosidase (BF3793), and β-glucanase (BF4060). Since these proteins are annotated based on in silico analysis, we decided to test the activities of some of them. We compared the β-galactosidase and the α-l-fucosidase activities in both OMV and O.M. For these experiments, we employed specific sugars linked to a chromogenic group that is released upon enzymatic cleavage, allowing absorbance to be measured at λ = 405 nm. Enzymatic activities of different membrane compartments were determined by comparison against standard curves produced using commercial enzymes incubated with the same chromogenic substrates (for details, see Materials and Methods). Both OMV and OM displayed comparable β-galactosidase activities, which were severely reduced by heat treatment (Fig. 5C). Interestingly, α-l-fucosidase activity was detected exclusively in OMV, with both the OM and heat-inactivated OMV showing no fucosidase activity (Fig. 5D). It has been previously established that α-l-fucosidase activity in Bacteroides can be induced in the presence of fucosylated human milk oligosaccharides (44). We hypothesized that B. fragilis can sense fucose monomers, inducing secretion of fucosidases in OMV. To test this hypothesis, B. fragilis was grown on defined media using either glucose or fucose as the carbon source. OM and OMV were collected under both conditions, and α-l-fucosidase activity was determined. In the presence of fucose, OMV fucosidase activity was induced about 7-fold compared to OMV collected from B. fragilis grown on glucose (Fig. 5E). This suggests the hydrolase content of the OMV is regulated to optimize the breakdown of the nutrients.
FIG 5 .
B. fragilis vesicles possess sugar-hydrolyzing activity. OMV were tested for the presence of β-galactosidase and α-l-fucosidase activities. (A) A standard curve for β-galactosidase activity was developed using commercial β-galactosidase from E. coli and 4-nitrophenyl-β-d-galactopyranoside as a substrate. (B) A standard curve for α-l-fucosidase activity was developed using commercial α-l-fucosidase from Thermotoga maritima and 2-chloro-4-nitrophenyl-α-l-fucopyranoside as a substrate (upper panel). (C) Ten micrograms of purified OMV and OM proteins were incubated with 4-nitrophenyl β-d-galactopyranoside for 1 h at 37°C. Reactions halted by the addition of 50 µl 1 M Na2CO3 followed by reading the absorbance at λ = 405. β-Galactosidase activities of OMV and OM were calculated from the standard curve using A405 readings and then plotted (lower panel). (D) α-l-Fucosidase activities of OM and OMV were measured by incubation with 2-chloro-4-nitrophenyl-α-l-fucopyranoside for 2.5 h at 37°C. A405 readings of OM and OMV were used to calculate α-l-fucosidase activities from the standard curve. (E) Fucosidase activity of cells grown in defined media with glucose or fucose as the carbon source. Heat-killed controls are marked by open triangles. All experiments were done in triplicate.
Acidic hydrolases are also packed in Bacteroides thetaiotaomicron OMV.
Since B. fragilis OMV were enriched in acidic proteins, including hydrolases, we investigated whether this phenomenon is specific to B. fragilis or a common feature among the genus Bacteroides. We extended our proteomic analysis to OMV and OM of B. thetaiotaomicron, which counts for 12% of all Bacteroidetes in the adult colon (45). As shown in Fig. 6, the B. thetaiotaomicron OMV protein profile was different from that of OM. The MS-based protein identification revealed the packing of 84 proteins in B. thetaiotaomicron OMV that were not detectable in OM. Eighty-two percent of these 84 OMV proteins are acidic. Additionally, 25 of the OMV unique proteins in B. thetaiotaomicron were predicted to be hydrolases. With the exception of one protein, all of the OMV unique hydrolases have acidic isoelectric points (Table 2).
FIG 6 .
B. thetaiotaomicron OMV display different protein content from OM. Ten micrograms of purified OM and OMV proteins was run on 10% SDS-PAGE. Coomassie-stained bands were excised from the gel and tryptically digested. The resulting peptides were analyzed via liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) followed by protein identification with the Mascot search engine using the NCBInr database. The arrows point to the most abundant proteins in each band. The underlined proteins were detected only in the corresponding compartment.
B. fragilis selectively packs nonnative acidic hydrolases into OMV.
BACOVA_04502 is an inulinase that was shown to be secreted in OMV of Bacteroides ovatus (17). We tested if this enzyme is also recognized and selectively packed in OMV produced by B. fragilis. We expressed a His-tagged version of this inulinase in B. fragilis and determined the presence of the protein in whole cells, OM, and OMV. As controls, we employed His-tagged versions of two proteins identified in our proteomic analysis, BF1581 and BF0018. BF1581 is an acidic protein detected only in OMV, while BF0018 is an alkaline TonB-dependent protein that was found only in OM of B. fragilis. OM and OMV were collected from the B. fragilis strains, each expressing one of the three proteins. Samples were normalized according to total protein content and separated on SDS-PAGE followed by Western blotting. As shown in Fig. 7, BF1581 only appeared in OMV, while BF0018 was detected only in OM, confirming the MS results. The B. ovatus acidic inulinase was greatly enriched in the OMV of B. fragilis (Fig. 7C). This suggests that the B. fragilis machinery is capable of recognizing and sorting nonnative acidic hydrolases into the OMV.
FIG 7 .
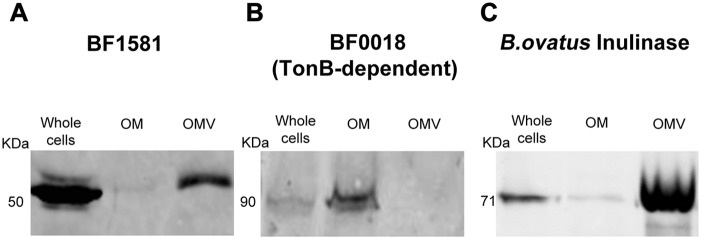
B. fragilis packs B. ovatus inulinase into its OMV. Western blots showing the distribution of BF1581 (A), BF0018 (B), and B. ovatus acidic inulinase BACOVA_04502 (C) in OM and OMV of B. fragilis. Ten micrograms of OM and OMV proteins of each strain were run on 10% SDS-PAGE followed by Western blotting.
Lipid A compositions are similar in OMV and OM.
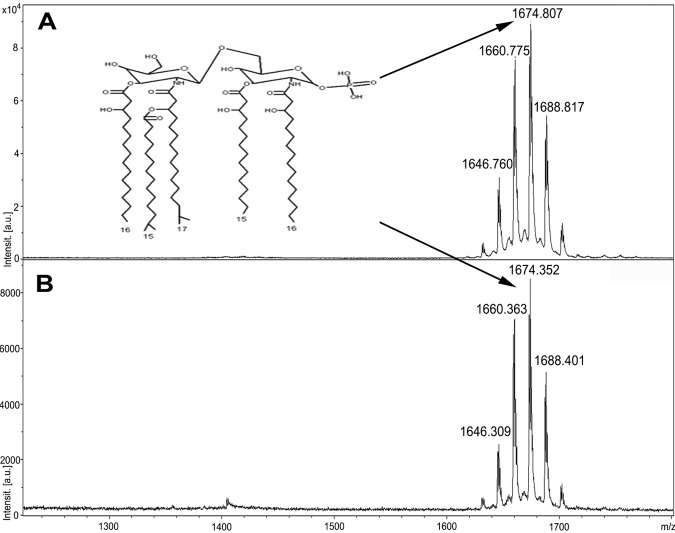
In previous work, we showed that deacylated lipid A accumulated in OMV of the dental pathogen P. gingivalis (28). Since B. fragilis and P. gingivalis are evolutionarily related, we investigated if the same phenomenon occurs in B. fragilis by comparing the lipid A compositions in OM and OMV using MS (33). We used the Trizol method to purify LPS from Bacteroides cells and OMV. Lipid A was then obtained through mild acid hydrolysis of LPS (46, 47). As shown in Fig. 8, matrix-assisted laser desorption ionization–time of flight MS (MALDI-TOF MS) analysis of the purified lipid A showed no difference between OM and OMV. Both spectra displayed a cluster of peaks (m/z 1,632, 1,646, 1,660, 1,674, 1,688, and 1,702) representing the penta-acylated lipid A of B. fragilis. The 14-atomic-mass-unit (amu) differences are caused by fatty acid length variations known to occur in B. fragilis (46).
FIG 8 .
OMV and OM share the same lipid A species. Lipid A was extracted from both whole cells and vesicles of B. fragilis by mild acid hydrolysis of LPS in 1% SDS–10 mM sodium acetate (pH 4.5). Lipid A was dissolved in water and mixed with 2′,4′,6′-trihydroxyacetophenone monohydrate matrix (THAP) in 1:1 ratio followed by MALDI-TOF MS analysis in negative linear mode. MS analysis of lipid A from cells (A) and vesicles (B) showed the same cluster of peaks representing the penta-acylated monophosphorylated lipid A. The 14-amu differences are caused by fatty acid length variation.
DISCUSSION
Membrane vesicles in eukaryotic cells are well known for their role in storage, trafficking, and digestion of cellular components according to their location and function (48). Vesiculation appears to be ubiquitous among Gram-negative bacteria, which suggests OMV play an important physiological role (42, 49–51). Different functions have been assigned to vesicles, including toxin delivery, interbacterial communication, biofilm formation, horizontal gene transfer, and disposal of misfolded proteins (19, 52–56). In this article, we characterized the protein contents of OMV from B. fragilis and B. thetaiotaomicron. One of the most striking characteristics of these vesicles is the abundance of acidic hydrolytic enzymes, mainly glycosidases and proteases. Previous reports showed the presence of hydrolase activity in the vesicles of Bacteroides, albeit none of these studies included a total proteome investigation of OMV (30, 57). It is well-recognized that glycan metabolism plays an important role in the establishment, composition, and balance of the gut microbiota (58). Species from the genus Bacteroides carry multiple systems to bind and degrade polysaccharides known as “Sus-like proteins.” The SusCDEFG proteins form a surface-localized complex, where SusG is an α-amylase, SusC is a TonB-dependent porin, and SusDEF are glycan-binding proteins (59). Glycan metabolism is very important for these bacteria, to the point that Sus-like proteins can constitute up to 20% of their genome. The presence of active glycosidases in OMV, some of them selectively packed, suggests that B. fragilis secretes hydrolytic OMV to maximize the chances to degrade polysaccharides and other glycoconjugates. While the Sus-like proteins would be carrying a “selfish” activity, because the polysaccharides are degraded at the bacterial surface and the sugars are immediately taken up, the hydrolases in OMV may carry a “social” function, as the resulting oligo- and monosaccharides would be available for other bacteria to utilize. Recently it has been described that members of the genus Bacteroides participate in a complex polysaccharide utilization network based on the release and use of OMV, which act as “public goods” (17). The authors showed that OMV produced from a microorganism can support the growth of another bacterium that is unable to degrade a given polysaccharide. Thus, OMV produce nutrients that can be utilized by other members of the microbiota. However, the sugars liberated by OMV may also be exploited by pathogenic bacteria. For instance, Campylobacter jejuni does not produce fucosidases, but it can utilize fucose liberated by other bacteria in the gut (60). Finally, it has been recently demonstrated that Salmonella enterica serovar Typhimurium and Clostridium difficile are also capable of utilizing sugars, such as fucose and sialic acid liberated by B. thetaiotaomicron, to successfully colonize the gut (61). Our results suggest that OMV produced by Bacteroides play an important role in sugar metabolism in the gut. Several sugar hydrolases are only induced in the presence of the specific polysaccharide (5). We showed the levels of fucosidase activity packed in OMV are increased in the presence of fucose, suggesting that the content of OMV is also dependent on the nutrients.
Fucose is of utmost importance for Bacteroides because it is incorporated in both its capsular polysaccharides and glycoproteins. The Bacteroides mutant lacking fucose in its proteins or capsule was found to be less fit in competitive colonization experiments with wild-type Bacteroides (62). We detected two fucosidases (BF0810 and BF3083) exclusively in Bacteroides vesicles. BF0810 is in the same operon with BF0811, which is annotated as a peptide-N-glycosidase and was found in OMV as well. This suggests that the two hydrolases might be functionally related. BF0811 might cleave the human glycans from glycoproteins, allowing BF0810 to cleave the fucose. Similar to B. fragilis, B. thetaiotaomicron OMV displayed enrichment in acidic hydrolases. Among these is the levan hydrolase BT_1760, which was shown to be involved in supporting the growth of other Bacteroides species unable to degrade levan (17). In an interesting analogy, OMV produced by the bacterial predator Myxococcus xanthus were shown to contain a high number of digestive hydrolases. These OMV played a role in predation and possessed lytic activity against E. coli (41, 42). Another example of hydrolase-containing OMV is the cellulolytic rumen bacterium Bacteroides succinogenes, which was shown to have 50% of its cellulose-digesting enzymes associating with vesicles (63). Additionally, these hydrolases might also target human molecules, such as mucus glycans, allowing Bacteroides to modulate host pathways. P. gingivalis OMV are rich in proteases that can degrade some of the host molecules, leading to impairment of normal cellular function (64).
The lack of an accepted mechanism for OMV biogenesis has generated the following controversy: Are OMV the result of a directed and still poorly understood active cellular process, or are OMV formed by passive membrane disintegration or membrane lysis? Several of our results favor the first hypothesis for OMV biogenesis in Bacteroidetes. First, the simple comparison of the OMV and OM proteins (Fig. 2 and 6) shows dramatically different profiles. Second, the MS analysis identified more than 40 proteins in the OMV that were not present at detectable amounts at the OM. Third, a significant percentage of the proteins that are preferentially packed into OMV are mostly hydrolytic enzymes. Random clustering of such functionally related enzymes is unlikely. Fourth, proteins directed to the OMV are predominantly acidic. Previous reports showed that OMV and OM vary in terms of lipid and protein compositions. As evidence for lipid variation, different LPS were detected in membrane and vesicles of the same bacteria. For example, vesicles from Pseudomonas aeruginosa were found enriched in the negatively charged LPS species, which was suggested to play a role in OMV biogenesis through charge repulsion (31). In Serratia marcescens, unique proteins were detected in vesicles but not in the outer membrane. Among the proteins detected in Serratia vesicles are lipases, phospholipases, and chitinases, which play a role in virulence (29). N. meningitidis provides another example of specific proteins’ enrichment into OMV. Lappann et al. compared the protein contents of both OM and OMV of N. meningitidis quantitatively. Some proteins were found to be enriched in OMV compared to OM and vice versa. Both preparations contained cytoplasmic proteins (32). In addition, we previously showed that vesicles of P. gingivalis varied from the OM in their protein composition. Multiple proteins in P. gingivalis OM were excluded from the vesicles. Concurrently, a few other proteins were enriched in vesicles compared to the membrane, suggesting the presence of a protein sorting mechanism. In an interesting homology to the role of galectins in the eukaryotic system, LPS was suggested to participate in the cargo selection process occurring during vesiculation of P. gingivalis (28). More proteomic studies on OMV of various organisms showed that they were enriched in different virulence factors (51). However, the dramatic differences between OM and OMV protein compositions determined in B. fragilis and B. thetaiotaomicron are unprecedented. Furthermore, for the first time, a common signature, acidity, was found for the proteins preferentially sorted into the OMV. The observation that an acidic inulinase from B. ovatus can be selectively packed in OMV by B. fragilis indicates that whatever mechanism is employed is likely common within the genus Bacteroides. Another component that is variable between OMV and OM are the capsular polysaccharides. In B. fragilis, important members of the human gut microbiota, polysaccharides A and B (PSA and PSB, respectively) were detected in vesicles. PSA is known to have an immunomodulatory role in the human gut, where it indirectly activates regulatory T cells (Tregs) to secrete interleukin-10. Interestingly, it was shown that B. fragilis uses its OMV to deliver PSA to dendritic cells which subsequently leads to Treg activation (16). Unlike the protein differences that we found between OMV and OM in B. fragilis, no variation in lipid A between the two compartments was found. Previously, we showed that the lipid A components of LPS were different between vesicles and membrane of P. gingivalis. The latter observation implied the presence of a lipid selection mechanism in vesicles additional to the protein selection. Although P. gingivalis is genetically close to B. fragilis, they seem to behave differently when packing lipids into their vesicles (33). This indicates that the vesiculation process might be variable among Gram-negative bacteria even in closely related species.
Rakoff-Nahoum et al. demonstrated that OMV produced by members of the microbiome can break down complex polysaccharides, supporting the growth of other bacterial species unable to degrade those polysaccharides. Diverse species appear to have coevolved to be responsible for the degradation of specific polysaccharides and now may rely on each other’s OMV for efficient utilization of the polysaccharides present in the gut. Our work shows that sugar hydrolases and proteases are preferentially packed into OMV from these organisms, in which there appears to be a pI-dependent mechanism. Our results support the model that OMV produced by all these bacteria contribute to the syntrophy required for the establishment and maintenance of the human microbiota.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacteroides fragilis NCTC 9343 and Bacteroides thetaiotaomicron VPI-5482 were grown either on blood agar plates containing 5% defibrinated horse blood or brain heart infusion broth supplemented with hemin (5 µg/ml) and menadione (1 µg/ml) in an anaerobic atmosphere of 90% N2, 5% H2, and 5% CO2. When needed, B. fragilis was grown on basal medium supplemented with 0.5% glucose or fucose.
OMV purification.
Two hundred fifty milliliters of 24-h cultures of B. fragilis was centrifuged at 10,000 rpm at 4°C. In order to remove residual cells, the supernatant was filtered using a 0.45-µm-pore membrane followed by a 0.2-µm-pore polyvinylidene difluoride (PVDF) membrane (Millex GV; Millipore). The filtrate was subjected to ultracentrifugation at 100,000 × g for 2 h (Optima L-90K ultracentrifuge; Beckman Coulter). The supernatant was discarded, the pellet was washed with sterile PBS, and the ultracentrifugation step was repeated. The vesicle pellet was resuspended in distilled water or 50 mM Tris-HCl buffer (pH 6.5) and quantified using the 2D-quant kit (GE Healthcare Life Sciences).
Membrane purification.
Cells of overnight cultures were harvested by centrifugation at 10,000 rpm for 10 min at 4°C. The pellets were gently resuspended in a mixture of 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 50 mM MgCl2 containing complete EDTA-free protease inhibitor mixture (Roche Applied Science) followed by sonication. Total membranes were collected by ultracentrifugation at 100,000 × g for 1 h at 4°C. The OM was isolated by differential extraction with the same buffer and 1.5% (vol/vol) Triton X-100 and incubated at 4°C overnight. The OM fractions were recovered by centrifugation at 100,000 × g for 1 h at 4° C.
Transmission electron microscopy.
Three microliters of the OMV preparations was adsorbed onto carbon-coated copper grids (3 min). Liquid excess was discarded, and the samples were negatively stained with 2% (wt/vol) uranyl acetate for 3 min and evaluated in a Morgagni (FEI) transmission electron microscope. OMV sizes were determined using the images obtained.
Mass spectrometry analysis of the OM and OMV.
Purified OM and OMV were run on 10% SDS-PAGE and stained with Coomassie brilliant blue (28). Protein bands were excised from gels. The excised protein bands were in-gel digested using sequencing grade-modified trypsin (Promega). Peptide fragments were eluted from the gel piece, desalted using ZipTip C18 columns (Millipore) according to the supplier protocol, and dissolved in 0.1% formic acid. For in-solution digestion of membranes, lipids were removed from lyophilized pellets using trifluoroethanol. In-solution trypsin digestion was then carried out as before. A hybrid quadrupole orthogonal acceleration time-of-flight (Q-TOF) mass spectrometer, Premier (Waters), equipped with a nanoACQUITY Ultraperformance liquid chromatography system (Waters) was used for MS/MS analyses of the peptides, and the resulting mass spectra were used for the identification of the proteins by the Mascot search engine using the NCBI nr database.
OMV zymography.
OMV were solubilized in urea buffer, and the total protein content was quantified using the 2D-quant kit (GE Healthcare Life Sciences). Zymography of OMV (10 µg) was performed using Novex zymogram gel (10% Tris-glycine gel with 0.1% gelatin as the substrate) according to the manufacturer’s protocol (Life Technologies). Following separation under denaturing conditions, the proteins were renatured to allow substrate cleavage. Colloidal Coomassie brilliant blue G-250 staining of gel was used to visualize areas where the substrate (gelatin) was digested by proteases.
Detection of OMV peptidase activity.
The peptidase enzyme activity was determined by incubation of 10 µg of the quantified OMV proteins in a 50 mM Tris-HCl buffer (pH 6.5), with 200 µl of 50 mM l-lysine-p-nitroanilide dihydrobromide, l-alanine-p-nitroanilide hydrochloride, l-leucine-p-nitroanilide, or pyroglutamic acid-p-nitroanilide (Sigma-Aldrich), for 1 h at 37°C. Reactions were stopped by the addition of 100 µl of 1 M Na2CO3. The release of the p-nitroanilide group was then detected spectrophotometrically by measuring the absorbance at 405 nm. All experiments were done in triplicates.
Detection of OMV glycoside hydrolase activity.
Bacteroides vesicles were tested for the presence of β-galactosidases using 4-nitrophenyl-β-d-galactopyranoside as a substrate. Purified OMV and OM were normalized according to protein content and incubated with the substrate separately for 1 h at 37°C. Fifty microliters 1 M Na2CO3 was added to stop the reaction. α-l-Fucosidase activity was measured by incubating 2-chloro-4-nitrophenyl-α-l-fucopyranoside with different membrane fractions for 2.5 h at 37°C. In both assays, the absorbances of the hydrolyzed products were measured spectrophotometrically at 405 nm. Standard curves were generated using the same substrates and commercial enzymes (Escherichia coli β-galactosidase from Sigma-Aldrich and Thermotoga maritima α-l-fucosidase from Megazyme). Sugar-hydrolyzing activities of OMV and OM were calculated using the corresponding A405 readings and standard curves. All experiments were done in triplicates.
Protein localization assays in OM and OMV.
BF1581 and BF0018 of B. fragilis NCTC 9343 were PCR amplified using primers listed in Table S5 in the supplemental material. The BF1581 PCR product was digested with BamHI and SacI and then inserted into pFD340 using the same cut sites. BF0018 PCR product was digested with BamHI and HindIII and cloned into pEXT20 and then subcloned into BamHI and SmaI sites of pFD340. pLEC280 expressing His-tagged BACOVA_04502 from B. ovatus was a generous gift from L. E. Comstock. The recombinant vectors were transferred to B. fragilis via E. coli harboring pRK231. OM and OMV were harvested from the recombinant strains and then normalized according to their total protein content using the 2D-quant kit (GE Healthcare Life Sciences). Both compartments in each strain were run on SDS-PAGE followed by Western blotting into nitrocellulose membranes. Localization of proteins was detected using anti-His rabbit antibody (primary antibody) followed by Alex Fluor 680-labeled anti-rabbit goat antibody (secondary antibody). Images were taken using the LI-COR Odyssey Imaging system.
MALDI MS analysis of lipid A.
Lipid A from vesicles and cells was prepared in duplicates using 10 mg of sample for each preparation according to the procedure of Yi and Hackett (47). The purified lipid A was resuspended in 6 µl of methanol-dichloromethane (1:1). One microliter of the mixture was loaded on the MALDI plate followed by addition of 0.5 µl of 2′,4′,6′-trihydroxyacetophenone monohydrate (THAP) as the matrix. MALDI MS was then performed on a Bruker Daltonics (Bremen, Germany) UltrafleXtreme MALDI-TOF/TOF mass spectrometer in the linear negative mode.
SUPPLEMENTAL MATERIAL
Proteins identified by MS/MS exclusively in OMV of B. fragilis
Proteins identified by MS/MS in OM of B. fragilis
Proteins identified by MS/MS exclusively in OMV of B. thetaiotaomicron
Proteins identified by MS/MS in OM of B. thetaiotaomicron
Primers used in cloning BF1581 and BF0018. Restriction sites are in bold. Histidine tags and stop codons are underlined.
ACKNOWLEDGMENTS
We thank Laurie Comstock for providing us with the B. fragilis and B. thetaiotaomicron strains and plasmids, as well as Nicolas Vozza, Maria Florencia Haurat, and Brent Weber for critical reading of the manuscript. Furthermore, we thank Arlene Oatway (Biological Sciences Microscopy Facility, University of Alberta) for assistance with transmission electron microscopy imaging and Bela Reiz, Jing Zheng, and Xiaoxia Ye (Department of Chemistry, University of Alberta) for assistance with mass spectrometry. Moreover, we thank Rajinder Dubb for helping with the cloning.
This work was supported by grants from the Alberta Glycomics Centre and the Natural Sciences and Engineering Research Council of Canada (NSERC) to M.F.F. M.F.F. is a CIHR new investigator.
The authors declare they have no financial or nonfinancial competing interests.
Footnotes
Citation Elhenawy W, Debelyy MO, Feldman MF. 2014. Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio 5(2):e00909-14. doi:10.1128/mBio.00909-14.
REFERENCES
- 1. Boleij A, Tjalsma H. 2012. Gut bacteria in health and disease: a survey on the interface between intestinal microbiology and colorectal cancer. Biol. Rev. Camb. Philos. Soc. 87:701–730. 10.1111/j.1469-185X.2012.00218.x [DOI] [PubMed] [Google Scholar]
- 2. Bond JH, Currier BE, Buchwald H, Levitt MD. 1980. Colonic conservation of malabsorbed carbohydrate. Gastroenterology 78:444–447 [PubMed] [Google Scholar]
- 3. Cummings JH, Gibson GR, Macfarlane GT. 1989. Quantitative estimates of fermentation in the hind gut of man. Acta Vet. Scand. Suppl. 86:76–82 [PubMed] [Google Scholar]
- 4. Gibson GR, Roberfroid MB. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401–1412 [DOI] [PubMed] [Google Scholar]
- 5. Macfarlane GT, Gibson GR. 1991. Formation of glycoprotein degrading enzymes by Bacteroides fragilis. FEMS Microbiol. Lett. 61:289–293 [DOI] [PubMed] [Google Scholar]
- 6. Roberton AM, Stanley RA. 1982. In vitro utilization of mucin by Bacteroides fragilis. Appl. Environ. Microbiol. 43:325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Salyers AA, O’Brien M, Kotarski SF. 1982. Utilization of chondroitin sulfate by Bacteroides thetaiotaomicron growing in carbohydrate-limited continuous culture. J. Bacteriol. 150:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Salyers AA, West SE, Vercellotti JR, Wilkins TD. 1977. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl. Environ. Microbiol. 34:529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Macy JM, Probst I. 1979. The biology of gastrointestinal Bacteroides. Annu. Rev. Microbiol. 33:561–594. 10.1146/annurev.mi.33.100179.003021 [DOI] [PubMed] [Google Scholar]
- 10. Fletcher CM, Coyne MJ, Villa OF, Chatzidaki-Livanis M, Comstock LE. 2009. A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell 137:321–331. 10.1016/j.cell.2009.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brook I. 1989. Aerobic and anaerobic microbiology of intra-abdominal abscesses in children. South. Med. J. 82:1479–1482. 10.1097/00007611-198912000-00006 [DOI] [PubMed] [Google Scholar]
- 12. Duerden BI. 1980. The identification of gram-negative anaerobic bacilli isolated from clinical infections. J. Hyg. (Lond) 84:301–313. 10.1017/S0022172400026802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sherwood JE, Fraser S, Citron DM, Wexler H, Blakely G, Jobling K, Patrick S. 2011. Multi-drug resistant Bacteroides fragilis recovered from blood and severe leg wounds caused by an improvised explosive device (IED) in Afghanistan. Anaerobe 17:152–155. 10.1016/j.anaerobe.2011.02.007 [DOI] [PubMed] [Google Scholar]
- 14. Wexler HM. 2007. Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20:593–621. 10.1128/CMR.00008-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mazmanian SK, Round JL, Kasper DL. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453:620–625. 10.1038/nature07008 [DOI] [PubMed] [Google Scholar]
- 16. Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. 2012. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12:509–520. 10.1016/j.chom.2012.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rakoff-Nahoum S, Coyne MJ, Comstock LE. 2014. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr. Biol. 24:40–49. 10.1016/j.cub.2013.10.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ellis TN, Kuehn MJ. 2010. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 74:81–94. 10.1128/MMBR.00031-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O’Toole GA, Stanton BA. 2009. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 5:e1000382. 10.1371/journal.ppat.1000382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chitcholtan K, Hampton MB, Keenan JI. 2008. Outer membrane vesicles enhance the carcinogenic potential of Helicobacter pylori. Carcinogenesis 29:2400–2405. 10.1093/carcin/bgn218 [DOI] [PubMed] [Google Scholar]
- 21. Jin JS, Kwon SO, Moon DC, Gurung M, Lee JH, Kim SI, Lee JC. 2011. Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles. PLoS One 6:e17027. 10.1371/journal.pone.0017027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kadurugamuwa JL, Beveridge TJ. 1997. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J. Antimicrob. Chemother. 40:615–621. 10.1093/jac/40.5.615 [DOI] [PubMed] [Google Scholar]
- 23. Kato S, Kowashi Y, Demuth DR. 2002. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb. Pathog. 32:1–13. 10.1006/mpat.2001.0474 [DOI] [PubMed] [Google Scholar]
- 24. Kolling GL, Matthews KR. 1999. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:1843–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wai SN, Lindmark B, Söderblom T, Takade A, Westermark M, Oscarsson J, Jass J, Richter-Dahlfors A, Mizunoe Y, Uhlin BE. 2003. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115:25–35. 10.1016/S0092-8674(03)00754-2 [DOI] [PubMed] [Google Scholar]
- 26. Mashburn LM, Whiteley M. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437:422–425. 10.1038/nature03925 [DOI] [PubMed] [Google Scholar]
- 27. McBroom AJ, Kuehn MJ. 2007. Release of outer membrane vesicles by gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63:545–558. 10.1111/j.1365-2958.2006.05522.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haurat MF, Aduse-Opoku J, Rangarajan M, Dorobantu L, Gray MR, Curtis MA, Feldman MF. 2011. Selective sorting of cargo proteins into bacterial membrane vesicles. J. Biol. Chem. 286:1269–1276. 10.1074/jbc.M110.185744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McMahon KJ, Castelli ME, García Vescovi E, Feldman MF. 2012. Biogenesis of outer membrane vesicles in Serratia marcescens is thermoregulated and can be induced by activation of the Rcs phosphorelay system. J. Bacteriol. 194:3241–3249. 10.1128/JB.00016-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patrick S, McKenna JP, O’Hagan S, Dermott E. 1996. A comparison of the haemagglutinating and enzymic activities of Bacteroides fragilis whole cells and outer membrane vesicles. Microb. Pathog. 20:191–202. 10.1006/mpat.1996.0018 [DOI] [PubMed] [Google Scholar]
- 31. Nguyen TT, Saxena A, Beveridge TJ. 2003. Effect of surface lipopolysaccharide on the nature of membrane vesicles liberated from the gram-negative bacterium Pseudomonas aeruginosa. J. Electron Microsc. (Tokyo) 52:465–469. 10.1093/jmicro/52.5.465 [DOI] [PubMed] [Google Scholar]
- 32. Lappann M, Otto A, Becher D, Vogel U. 2013. Comparative proteome analysis of spontaneous outer membrane vesicles and purified outer membranes of Neisseria meningitidis. J. Bacteriol. 195:4425–4435. 10.1128/JB.00625-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, Daugherty SC, Dodson RJ, Durkin AS, Gwinn M, Haft DH, Kolonay JF, Nelson WC, Mason T, Tallon L, Gray J, Granger D, Tettelin H, Dong H, Galvin JL, Duncan MJ, Dewhirst FE, Fraser CM. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591–5601. 10.1128/JB.185.18.5591-5601.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cho KH, Salyers AA. 2001. Biochemical analysis of interactions between outer membrane proteins that contribute to starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 183:7224–7230. 10.1128/JB.183.24.7224-7230.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koropatkin NM, Smith TJ. 2010. SusG: a unique cell-membrane-associated alpha-amylase from a prominent human gut symbiont targets complex starch molecules. Structure 18:200–215. 10.1016/j.str.2009.12.010 [DOI] [PubMed] [Google Scholar]
- 36. Shipman JA, Berleman JE, Salyers AA. 2000. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J. Bacteriol. 182:5365–5372. 10.1128/JB.182.19.5365-5372.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martens EC, Chiang HC, Gordon JI. 2008. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4:447–457. 10.1016/j.chom.2008.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138–D141. 10.1093/nar/gkh121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248. 10.1093/nar/gki162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kahnt J, Aguiluz K, Koch J, Treuner-Lange A, Konovalova A, Huntley S, Hoppert M, Søgaard-Andersen L, Hedderich R. 2010. Profiling the outer membrane proteome during growth and development of the social bacterium Myxococcus xanthus by selective biotinylation and analyses of outer membrane vesicles. J. Proteome Res. 9:5197–5208. 10.1021/pr1004983 [DOI] [PubMed] [Google Scholar]
- 42. Whitworth DE. 2011. Myxobacterial vesicles death at a distance? Adv. Appl. Microbiol. 75:1–31. 10.1016/B978-0-12-387046-9.00001-3 [DOI] [PubMed] [Google Scholar]
- 43. Noinaj N, Guillier M, Barnard TJ, Buchanan SK. 2010. TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64:43–60. 10.1146/annurev.micro.112408.134247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu ZT, Chen C, Newburg DS. 2013. Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology 23:1281–1292. 10.1093/glycob/cwt065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. 10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Berezow AB, Ernst RK, Coats SR, Braham PH, Karimi-Naser LM, Darveau RP. 2009. The structurally similar, penta-acylated lipopolysaccharides of Porphyromonas gingivalis and Bacteroides elicit strikingly different innate immune responses. Microb. Pathog. 47:68–77. 10.1016/j.micpath.2009.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yi EC, Hackett M. 2000. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. Analyst 125:651–656. 10.1039/b000368i [DOI] [PubMed] [Google Scholar]
- 48. Nieuwland R, Sturk A. 2010. Why do cells release vesicles? Thromb. Res. 125(Suppl 1):S49–S51. 10.1016/j.thromres.2010.01.037 [DOI] [PubMed] [Google Scholar]
- 49. Kulp A, Kuehn MJ. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64:163–184. 10.1146/annurev.micro.091208.073413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mashburn-Warren LM, Whiteley M. 2006. Special delivery: vesicle trafficking in prokaryotes. Mol. Microbiol. 61:839–846. 10.1111/j.1365-2958.2006.05272.x [DOI] [PubMed] [Google Scholar]
- 51. Schwechheimer C, Sullivan CJ, Kuehn MJ. 2013. Envelope control of outer membrane vesicle production in gram-negative bacteria. Biochemistry 52:3031–3040. 10.1021/bi400164t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dutta S, Iida K, Takade A, Meno Y, Nair GB, Yoshida S. 2004. Release of Shiga toxin by membrane vesicles in Shigella dysenteriae serotype 1 strains and in vitro effects of antimicrobials on toxin production and release. Microbiol. Immunol. 48:965–969. 10.1111/j.1348-0421.2004.tb03626.x [DOI] [PubMed] [Google Scholar]
- 53. Horstman AL, Kuehn MJ. 2000. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 275:12489–12496. 10.1074/jbc.275.17.12489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Horstman AL, Kuehn MJ. 2002. Bacterial surface association of heat-labile enterotoxin through lipopolysaccharide after secretion via the general secretory pathway. J. Biol. Chem. 277:32538–32545. 10.1074/jbc.M203740200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kadurugamuwa JL, Beveridge TJ. 1995. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 177:3998–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Keenan JI, Davis KA, Beaugie CR, McGovern JJ, Moran AP. 2008. Alterations in Helicobacter pylori outer membrane and outer membrane vesicle-associated lipopolysaccharides under iron-limiting growth conditions. Innate Immun. 14:279–290. 10.1177/1753425908096857 [DOI] [PubMed] [Google Scholar]
- 57. Forsberg CW, Beveridge TJ, Hellstrom A. 1981. Cellulase and xylanase release from Bacteroides succinogenes and its importance in the rumen environment. Appl. Environ. Microbiol. 42:886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 10:323–335. 10.1038/nrmicro2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J. Biol. Chem. 284:24673–24677. 10.1074/jbc.R109.022848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stahl M, Friis LM, Nothaft H, Liu X, Li J, Szymanski CM, Stintzi A. 2011. L-fucose utilization provides Campylobacter jejuni with a competitive advantage. Proc. Natl. Acad. Sci. U. S. A. 108:7194–7199. 10.1073/pnas.1014125108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502:96–99. 10.1038/502S96a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Coyne MJ, Reinap B, Lee MM, Comstock LE. 2005. Human symbionts use a host-like pathway for surface fucosylation. Science 307:1778–1781. 10.1126/science.1106469 [DOI] [PubMed] [Google Scholar]
- 63. Groleau D, Forsberg CW. 1981. Cellulolytic activity of the rumen bacterium Bacteroides succinogenes. Can. J. Microbiol. 27:517–530. 10.1139/m81-077 [DOI] [PubMed] [Google Scholar]
- 64. Furuta N, Takeuchi H, Amano A. 2009. Entry of Porphyromonas gingivalis outer membrane vesicles into epithelial cells causes cellular functional impairment. Infect. Immun. 77:4761–4770. 10.1128/IAI.00841-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteins identified by MS/MS exclusively in OMV of B. fragilis
Proteins identified by MS/MS in OM of B. fragilis
Proteins identified by MS/MS exclusively in OMV of B. thetaiotaomicron
Proteins identified by MS/MS in OM of B. thetaiotaomicron
Primers used in cloning BF1581 and BF0018. Restriction sites are in bold. Histidine tags and stop codons are underlined.