Abstract
Background
The chance that a prostate cancer detected by screening is overdiagnosed (ie, it would not have been detected in the absence of screening) can vary widely depending on the patient’s age and tumor characteristics. The purpose of this study is to use age, Gleason score, and prostate-specific antigen (PSA) level to help inform patients with screen-detected prostate cancers about the chances their cancers were overdiagnosed.
Methods
A computer microsimulation model of prostate cancer natural history was used to generate virtual life histories in the presence and absence of PSA screening, including an indicator of whether screen-detected cancers are overdiagnosed. A logistic regression model was fit to nonmetastatic patients diagnosed by screening with PSA less than 10ng/mL, and a nomogram was created to predict the individualized risk of overdiagnosis given age, Gleason score, and PSA at diagnosis.
Results
The calibrated microsimulation model closely reproduces observed incidence trends in the Surveillance, Epidemiology, and End Results registries by age, stage, and Gleason score. The fitted logistic regression predicts risks of overdiagnosis among PSA-detected patients with an area under the curve of 0.75. Chances of overdiagnosis range from 2.9% to 88.1%.
Conclusions
The chances of overdiagnosis vary considerably by age, Gleason score, and PSA at diagnosis. The overdiagnosis nomogram presents tailored estimates of these risks based on patient and tumor information known at diagnosis and can be used to inform decisions about treating PSA-detected prostate cancers.
The recently updated US Preventive Services Task Force recommendations highlight the potential harms associated with prostate-specific antigen (PSA) screening for prostate cancer (1). The harms of greatest concern are overdiagnosis and treatment of overdiagnosed cancers. An overdiagnosed cancer is one that would never have become symptomatic or clinically apparent in the absence of screening. Such a tumor would have posed no risk to the patient and therefore, by definition, does not require treatment. Estimates of overdiagnosis in the US population range from 23% to 42% among all men aged 50 to 84 years at screen detection (2). However, the likelihood that a tumor has been overdiagnosed can vary widely depending on the patient’s age and tumor characteristics (3).
Despite the ubiquity of nomograms in the prostate cancer literature as graphical devices for personalizing the results of predictive models (4), there are no nomograms for predicting the chance that a screen-detected prostate cancer has been overdiagnosed. Although nomograms are available that predict the presence of indolent tumors (5), a nomogram for overdiagnosis may differ considerably from a nomogram for indolent cancer. A tumor is generally classified as indolent based on its biology as expressed by pathologic or clinical characteristics. For example, Kattan et al. (6) define an indolent tumor as pathologically organ confined, 0.5 cc or less in volume, and without poorly differentiated elements. However, whether a tumor is overdiagnosed depends not only on the underlying tumor biology but also on the life expectancy of the patient. In a patient with a very short life expectancy, even a relatively aggressive tumor might be overdiagnosed because death due to other causes may precede progression of the tumor to the point at which it would have become symptomatic.
Why are there dozens of nomograms for different cancer outcomes (including indolent tumors) and no nomograms for overdiagnosed cancer? One reason is that all of the currently existing nomograms pertain to outcomes that are observable in practice. When an outcome is observable in a defined cohort of patients, a standard statistical model such as logistic or Cox regression can be fit to the corresponding data. The nomograms published to date are simply graphical representations of the results of such fitted models. However, whether a tumor has been overdiagnosed is not observable. Once a patient has been diagnosed and treated, we are not privy to the counterfactual course of events that would allow us to know when his disease would have been diagnosed in the absence of screening. Therefore we cannot fit standard regression models to existing clinical cohorts to predict overdiagnosis.
Herein, we present a model of prostate cancer onset, progression, and detection (7,8) that has been calibrated to reflect population patterns of prostate cancer incidence and that includes the counterfactual information necessary to identify whether tumors are overdiagnosed. We extend a version of this model used to project the course of untreated prostate cancer (3) and use it to simulate a population of screen-detected patients for which age, Gleason score, and PSA at diagnosis are known as well as whether the cancer has been overdiagnosed. We fit a logistic regression model to the simulated data and produce the corresponding nomogram for predicting overdiagnosis. The nomogram quantifies contributions of age, Gleason score, and PSA at diagnosis to the predicted likelihood of overdiagnosis and thereby provides a personalized tool for newly diagnosed patients concerned about overdiagnosis and overtreatment.
Methods
Our model was developed by investigators in the prostate working group of the Cancer Intervention and Surveillance Modeling Network (CISNET; http://cisnet.cancer.gov/) with the goal of quantifying the role of PSA screening in explaining prostate cancer mortality declines in the United States (9). The model is a microsimulation model that produces virtual life histories for a hypothetical population representing US men aged 50 to 84 years in calendar years 1975 to 2005. The virtual life histories include both observable and unobservable (latent) events. Examples of observable events are age at diagnosis for a patient diagnosed by PSA screening in 1995, cancer stage at diagnosis, and age at death. Although the latter may be censored, it is still, in principle, observable given sufficient follow-up. Examples of unobservable events are age at onset of disease and the age and stage had the cancer been diagnosed in the absence of PSA screening.
For each virtual life history, the model generates the point of cancer onset and PSA levels before and after onset. PSA growth curves are estimated using serial PSA measurements from the control arm of the Prostate Cancer Prevention Trial (10,11). At the point of onset, the model also generates Gleason score (Gleason ≤6 vs ≥7), with higher Gleason score associated with faster PSA growth. The risk of cancer onset and the risk that a cancer has a higher Gleason score increase with age. The risks of progression from a nonmetastatic (M0) to a metastatic (M1) state and from a latent tumor to clinical diagnosis increase with the PSA level. Other-cause mortality is generated using US life tables. A description of the natural history model is presented in Gulati et al. (7,8) with supplementary details available at http://cisnet.cancer.gov/prostate/profiles.html.
To reproduce prostate cancer incidence trends observed in the Surveillance, Epidemiology, and End Results (SEER) registries for the period 1975 to 2005, we superimpose PSA screening on this natural history model. Because PSA screening dissemination was not tracked in real time, we rely on a reconstruction by Mariotto et al. (12). We assume that the cutoff for a positive test is 4ng/mL, as was standard in the United States in the 1990s, and that biopsy frequencies after a positive test depend on age and PSA, as observed among participants in the Prostate, Lung, Colorectal, and Ovarian cancer screening trial (13). Biopsies after a screening test are assumed to detect an existing, latent tumor with a probability that improves over time as more extensive biopsy schemes became popular, from 80% for sextant biopsies in the early 1990s to 93% for extended core biopsies by 2000 and remaining constant thereafter (14,15).
Model-projected prostate cancer incidence in pre-PSA and PSA-era years are compared with observed SEER incidence for corresponding years using a Poisson likelihood. We then calibrate the model’s risk parameters governing cancer onset, progression, and clinical detection by maximizing this likelihood. The calibration procedure and results have been previously described (7,8). The calibrated model produces virtual life histories that aggregate to produce incidence that approximates observed incidence rates by age, year, stage, and Gleason score at diagnosis. Major racial/ethnic groups are not modeled.
Statistical Analysis
Once the model has been calibrated, we use it to simulate a population of virtual life histories representative of the experience of US men in the period from 1975 to 2005. The simulated data include dates of diagnosis with and without screening and the date of other-cause death. We extract records for nonmetastatic patients diagnosed by screening in the year 2005 with PSA less than 10ng/mL and label each as overdiagnosed (if the model-generated date of other-cause death precedes clinical diagnosis) or not. We focus on nonmetastatic cases with modest PSA values to represent individuals detected early in their natural history by PSA screening. A sample of 10000 such patients is used to fit a logistic regression model with the overdiagnosis indicator as the response variable and age, Gleason score, and PSA level at diagnosis as the predictor variables. Statistical significance of predictor variables is assessed using two-sided Wald tests; a P value of less than .05 is considered statistically significant. A nomogram is then constructed to represent the results of the logistic regression fit. To illustrate the output of the nomogram, we compute the predicted overdiagnosis proportions for specified Gleason scores and PSA levels by age at diagnosis.
We also construct a receiver operating curve with the overdiagnosis indicator as the response variable and the probability of overdiagnosis from the fitted logistic regression model as the evaluated signal. To assess the accuracy of the prediction model, we estimate the area under the receiver operating curve. All analyses were performed using the R (16) statistical computing package rms (17) to fit the logistic regression model and create the nomogram.
Results
The calibrated model reasonably approximates age-, year-, stage-, and grade-specific incidence patterns (7,8). Under the calibrated model, 33% of men have disease onset in their lifetimes and 38% of these would be diagnosed in the absence of screening, with an average interval from onset to diagnosis of 14 years. These results are comparable with those from other prostate cancer models (3).
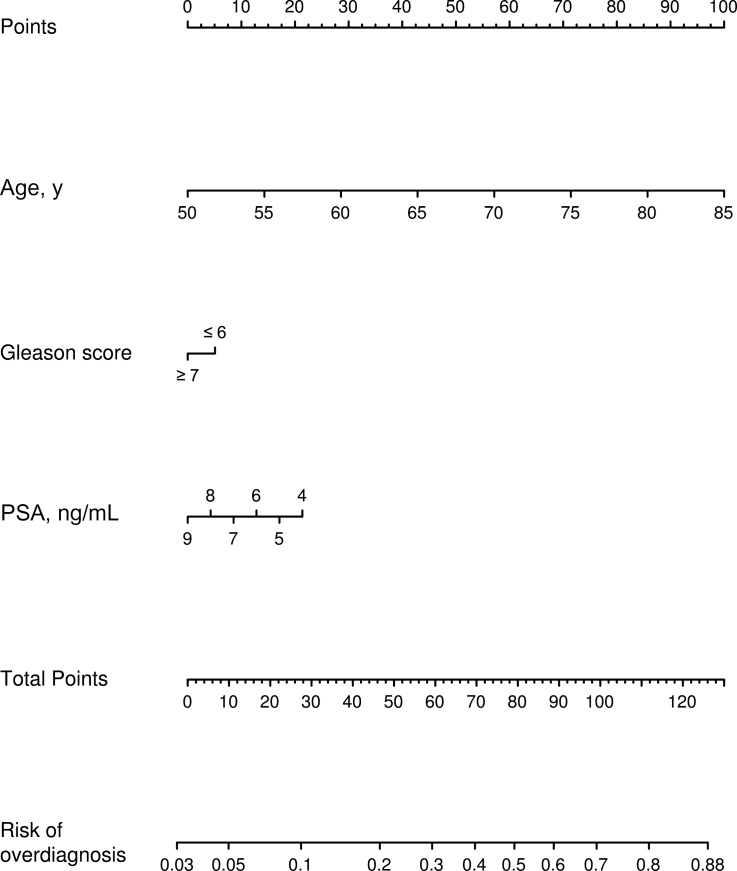
Table 1 summarizes age, Gleason score, and PSA at diagnosis for 10000 nonmetastatic patients diagnosed by PSA screening in the year 2005 simulated by the natural history model. We use these virtual patients to fit a logistic regression model (Table 2). After standard diagnostic assessment of model assumptions, we found that each additional year of age at diagnosis is associated with a 12.9% increase in the odds of overdiagnosis (95% confidence interval [CI] = 12.2% to 13.6%; P < .001), having Gleason score of 7 or greater is associated with a 19.5% decrease in the odds of overdiagnosis relative to Gleason score of 6 or less (95% CI = 11.7% to 26.5%; P < .001), and each additional 1ng/mL of serum PSA up to 10ng/mL is associated with a 16.6% decrease in the odds of overdiagnosis (95% CI = 14.2% to 18.9%; P < .001). Finally, we construct the corresponding nomogram for the fitted logistic regression model (Figure 1). Table 3 presents the corresponding predicted probabilities of overdiagnosis by age for the specified Gleason scores and PSA levels. The predictions are reasonably accurate with an area under the receiver operating curve of 0.75.
Table 1.
Characteristics of 10000 nonmetastatic prostate cancer patients diagnosed by prostate-specific antigen screening simulated by the natural history model*
| Patient characteristic | No. (%) |
|---|---|
| Age, y | |
| 50–54 | 218 (2.2) |
| 55–59 | 1075 (10.8) |
| 60–64 | 1673 (16.7) |
| 65–69 | 1712 (17.1) |
| 70–74 | 2058 (20.6) |
| 75–79 | 1934 (19.3) |
| 80–84 | 1330 (13.3) |
| Gleason score | |
| ≤6 | 6399 (64.0) |
| ≥7 | 3601 (36.0) |
| PSA, ng/mL | |
| 4.0–4.9 | 3964 (39.6) |
| 5.0–5.9 | 2105 (21.1) |
| 6.0–6.9 | 1400 (14.0) |
| 7.0–7.9 | 1125 (11.2) |
| 8.0–8.9 | 812 (8.1) |
| 9.0–9.9 | 594 (5.9) |
* PSA = prostage-specific antigen.
Table 2.
Fitted logistic regression model of overdiagnosis for nonmetastatic prostate cancer patients diagnosed by prostate-specific antigen (PSA) screening given age, Gleason score, and PSA at diagnosis*
| Patient characteristic | Odds ratio (95% CI) | P |
|---|---|---|
| Age, y | 1.129 (1.122 to 1.136) | <.001 |
| Gleason score | ||
| ≤6 | 1.000 (Referent) | — |
| ≥7 | 0.805 (0.735 to 0.883) | <.001 |
| PSA, ng/mL | 0.834 (0.811 to 0.858) | <.001 |
* Presented are changes to the odds of overdiagnosis for a 1-year increase in age between 50 and 84 years, for a 1ng/mL increase in PSA level between 4 and 10ng/mL, and for Gleason score of 7 or greater relative to 6 or less. Two-sided P values are based on Wald tests. CI = confidence interval.
Figure 1.
Nomogram for predicting overdiagnosis in prostate-specific antigen (PSA)–detected prostate cancer patients. The nomogram is based on a logistic regression model fit to a virtual population of nonmetastatic prostate cancer patients detected by PSA screening in the year 2005. Virtual prostate cancer patients are generated with a competing risks microsimulation model that predicts cancer detection in the presence and absence of PSA screening so that overdiagnosis status is known. The area under the receiver operating curve for the nomogram is 0.75.
Table 3.
Risks (%) of overdiagnosis among nonmetastatic prostate cancer patients diagnosed by prostate-specific antigen screening predicted by the fitted logistic regression model*
| Gleason score | Age, y | PSA, ng/mL | |||||
|---|---|---|---|---|---|---|---|
| 4.0–4.9 | 5.0–5.9 | 6.0–6.9 | 7.0–7.9 | 8.0–8.9 | 9.0–9.9 | ||
| ≤6 | 50–54 | 11.6 | 9.9 | 8.4 | 7.1 | 6.0 | 5.0 |
| 55–59 | 19.5 | 16.8 | 14.4 | 12.3 | 10.5 | 8.9 | |
| 60–64 | 30.7 | 27.0 | 23.6 | 20.5 | 17.7 | 15.2 | |
| 65–69 | 44.9 | 40.4 | 36.1 | 32.1 | 28.2 | 24.7 | |
| 70–74 | 59.9 | 55.5 | 50.9 | 46.4 | 41.9 | 37.6 | |
| 75–79 | 73.3 | 69.6 | 65.6 | 61.4 | 57.0 | 52.5 | |
| 80–84 | 83.4 | 80.8 | 77.8 | 74.5 | 70.9 | 67.0 | |
| ≥7 | 50–54 | 9.6 | 8.1 | 6.9 | 5.8 | 4.9 | 4.1 |
| 55–59 | 16.3 | 14.0 | 11.9 | 10.1 | 8.6 | 7.3 | |
| 60–64 | 26.3 | 22.9 | 19.9 | 17.2 | 14.7 | 12.6 | |
| 65–69 | 39.6 | 35.3 | 31.3 | 27.5 | 24.1 | 20.9 | |
| 70–74 | 54.6 | 50.1 | 45.5 | 41.1 | 36.8 | 32.7 | |
| 75–79 | 68.8 | 64.8 | 60.6 | 56.1 | 51.6 | 47.1 | |
| 80–84 | 80.2 | 77.2 | 73.8 | 70.1 | 66.2 | 62.0 | |
* PSA = prostate-specific antigen.
The nomogram demonstrates the relative importance of age, Gleason score, and PSA in predicting overdiagnosis. Clearly, age is the most important single predictor of overdiagnosis. Among men with Gleason score of 6 or less and slightly elevated PSA levels (4.0–4.9ng/mL), for example, the risk of overdiagnosis increases from 11.6% for ages 50 to 54 years to 59.9% for ages 70 to 74 years and 83.4% for ages 80 to 84 years, a range that may have important clinical implications for decisions about pursuing aggressive treatment. Lower levels of PSA confer a substantially greater risk of overdiagnosis relative to higher levels; the predicted risks of overdiagnosis for PSA just greater than 4ng/mL can be twice as high as those for PSA near 10ng/mL. For a given age and PSA level, the odds of overdiagnosis for men with Gleason score of 7 or greater are only moderately lower relative to men with Gleason score of 6 or less; however, in a model that includes virtual patients with PSA greater than or equal to 10ng/mL and that does not condition on the PSA level, the odds for men with Gleason score of 7 or greater become 31.8% lower (95% CI = 25.7% to 37.5%; P < .001). This difference is because high-grade cancers are associated with higher PSA levels at diagnosis.
Discussion
Although other studies have quantified the likelihood of overdiagnosis associated with PSA screening for the US population as a whole, the results presented here are likely to be considerably more useful for screen-detected patients trying to understand the severity of their newly diagnosed prostate cancer. Depending on a man’s age, Gleason score, and PSA level, the likelihood that his tumor has been overdiagnosed ranges from 2.9% to 88.1%. Determining where in this range a patient lies is clearly vital for his ability to make informed decisions about whether to consider immediate curative treatment for his disease.
Our overdiagnosis nomogram is clearly different from nomograms for indolent tumors because it includes age, which turns out to be the most critical predictor of overdiagnosis. Nomograms for indolent and overdiagnosed tumors will be similar for younger men. However, in older men, many nonindolent tumors will be overdiagnosed because of shorter remaining life expectancy. Thus, in an older population we would expect a considerably higher predicted probability of overdiagnosis than of an indolent tumor. Even for younger men, our results are not directly comparable with those of the Kattan et al. nomogram for indolent tumors (5) because their prediction model excluded patients with features that the investigators felt would disqualify them from having an indolent cancer, such as primary or secondary Gleason pattern 4 or 5, greater than 50% positive cores, and PSA greater than 20ng/mL.
When simulating virtual disease histories, we superimpose population screening and biopsy patterns on the underlying disease progression process. Given that the standard for biopsy referral in the United States from 1990 to 2005 was a PSA level greater than 4ng/mL, men in the simulation are only eligible for biopsy (and diagnosis) after screening if their PSA exceeds this threshold. Therefore, all screen-detected patients in the simulation and in the regression model behind the nomogram have a PSA level of at least 4ng/mL. Information on biopsy patterns in this population is available and is used to generate biopsies and screen detections in the model. In principle, the model could accommodate a lower PSA threshold for biopsy referral; however, the prevalence of biopsy among men with lower PSA levels during the 1990s was not well documented, and without this information, the model cannot be properly calibrated.
Our nomogram includes three predictors—namely, age, Gleason score, and PSA at diagnosis. Other clinico-pathologic features of the cancer, such as volume of tumor on biopsy or the percentage of biopsy cores positive for cancer, have been shown to be predictive of pathologic stage and disease prognosis and may impact the likelihood of overdiagnosis, but they are not included in SEER, and a model including those features could not easily be calibrated to SEER data. Similarly, clinical stage is likely associated with overdiagnosis; future versions of the model will include this predictor, but the current version of the model is calibrated to SEER data and therefore only distinguishes nonmetastatic (M0) and metastatic (M1) patients. The recognition that overdiagnosis is ultimately a function of remaining life expectancy implies that care should be taken to accurately assess the risk of other-cause death in these patients. It is possible that screen-detected prostate cancer patients may be at a lower risk of other-cause death than the general population (18). If this is the case, then we may be slightly overestimating the frequency of overdiagnosis. In addition, comorbidity status will affect the likelihood that a screen-detected patient has been overdiagnosed. Ultimately it will be of value to incorporate comorbidity into overdiagnosis predictions. A future extension may also incorporate race. Although we accounted for population-based frequencies of biopsy after an abnormal PSA test given a man’s age and PSA level, we did not explicitly model confirmation PSA or rectal exams that may also be associated with the decision to biopsy. Finally, a potential limitation of our results is that they are based on a single model. Previous comparisons with other prostate cancer models show that estimates of overdiagnosis can be sensitive to assumptions about natural history (3), although variability in estimates across models is often smaller than variability in estimates due to age or PSA at diagnosis.
In summary, the results of this study extend our understanding of the range of risks of overdiagnosis in US men detected by PSA screening and how they depend on patient and tumor characteristics. It is hoped that the resulting nomogram, tailored to individual patient characteristics known at diagnosis, will provide useful information for patients and their physicians seeking to weigh the likely harms and benefits of the treatment options available for contemporary screen-detected prostate cancers. Recently publicized results from the long-awaited Prostate Intervention Versus Observation Trial have indicated that low-risk patients are unlikely to benefit from immediate surgery (19). This information, coupled with the relatively high frequencies of overdiagnosis projected by the model for these patients, should provide a compelling reason to carefully consider the appropriateness of active surveillance for low-risk disease, particularly in older men.
Funding
This work was supported by the National Cancer Institute of the National Institutes of Health and the Centers for Disease Control and Prevention (grant U01 CA157224).
The study sponsor had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute, the National Institute of Health, or the Centers for Disease Control and Prevention.
We are grateful to Dr Alex Tsodikov for updating our previously developed model of PSA screening patterns in the United States to reflect contemporary practice.
References
- 1. Chou R, Croswell JM, Dana T, et al. Screening for prostate cancer: a review of the evidence for the U.S. Preventive Services Task Force. Ann Intern Med. 2011;155(11):762–771 [DOI] [PubMed] [Google Scholar]
- 2. Draisma G, Etzioni R, Tsodikov A, et al. Lead time and overdiagnosis in prostate-specific antigen screening: importance of methods and context. J Natl Cancer Inst. 2009;101(6):374–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gulati R, Wever EM, Tsodikov A, et al. What if i don’t treat my PSA-detected prostate cancer? Answers from three natural history models. Cancer Epidemiol Biomarkers Prev. 2011;20(5):740–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kattan MW. Do we need more nomograms for predicting outcomes in patients with prostate cancer? Nat Clin Pract Urol. 2008;5(7):366–367 [DOI] [PubMed] [Google Scholar]
- 5. Kattan MW, Eastham JA, Wheeler TM, et al. Counseling men with prostate cancer: a nomogram for predicting the presence of small, moderately differentiated, confined tumors. J Urol. 2003;170(5):1792–1797 [DOI] [PubMed] [Google Scholar]
- 6. Partin AW, Kattan MW, Subong EN, et al. Combination of prostate-specific antigen, clinical stage, and Gleason score to predict pathological stage of localized prostate cancer. A multi-institutional update. JAMA. 1997;277(18):1445–1451 [PubMed] [Google Scholar]
- 7. Gulati R, Inoue L, Katcher J, et al. Calibrating disease progression models using population data: a critical precursor to policy development in cancer control. Biostatistics. 2010;11(4):707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gulati R, Gore JL, Etzioni R. Comparative effectiveness of alternative prostate-specific antigen-based prostate cancer screening strategies: model estimates of potential benefits and harms. Ann Intern Med. 2013;158(3):145–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Etzioni R, Tsodikov A, Mariotto A, et al. Quantifying the role of PSA screening in the US prostate cancer mortality decline. Cancer Causes Control. 2008;19(2):175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thompson IM, Goodman PJ, Tangen CM, et al. The influence of finasteride on the development of prostate cancer. New Engl J Med. 2003;349(3):215–224 [DOI] [PubMed] [Google Scholar]
- 11. Etzioni RD, Howlader N, Shaw PA, et al. Long-term effects of finasteride on prostate specific antigen levels: results from the prostate cancer prevention trial. J Urol. 2005;174(3):877–881 [DOI] [PubMed] [Google Scholar]
- 12. Mariotto A, Etzioni R, Krapcho M, et al. Reconstructing prostate-specific antigen (PSA) testing patterns among black and white men in the US from Medicare claims and the National Health Interview Survey. Cancer. 2007;109(9):1877–1886 [DOI] [PubMed] [Google Scholar]
- 13. Pinsky PF, Andriole GL, Kramer BS, et al. Prostate biopsy following a positive screen in the Prostate, Lung, Colorectal and Ovarian cancer screening trial. J Urol. 2005;173(3):746–750; discussion 750–751. [DOI] [PubMed] [Google Scholar]
- 14. Babaian RJ, Toi A, Kamoi K, et al. A comparative analysis of sextant and an extended 11-core multisite directed biopsy strategy. J Urol. 2000;163(1):152–157 [PubMed] [Google Scholar]
- 15. Presti JCJ, Chang JJ, Bhargava V, et al. The optimal systematic prostate biopsy scheme should include 8 rather than 6 biopsies: results of a prospective clinical trial. J Urol. 2000;163(1):163–166; discussion 166–167. [PubMed] [Google Scholar]
- 16. R Development Core Team R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013
- 17. Harrell FJ. rms: regression modeling strategies. R package version 3.6-3. 2013. http://cran.r-project.org/package=rms Accessed July 1, 2013.
- 18. Cho H, Mariotto A, Mann BS, et al. Assessing non-cancer-related health status of US cancer patients: other-cause survival and comorbidity prevalence. Am J Epidemiol. 2013;178(3)339–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilt T, Brawer MK, Jones K, et al. Radical prostatectomy versus observation for localized prostate cancer. New Engl J Med. 2012;367(3):203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]