Abstract
Background
Clinical outcome of patients with triple-negative breast cancer (TNBC) is highly variable. This study aims to identify and validate a prognostic protein signature for TNBC patients to reduce unnecessary adjuvant systemic therapy.
Methods
Frozen primary tumors were collected from 126 lymph node–negative and adjuvant therapy–naive TNBC patients. These samples were used for global proteome profiling in two series: an in-house training (n = 63) and a multicenter test (n = 63) set. Patients who remained free of distant metastasis for a minimum of 5 years after surgery were defined as having good prognosis. Cox regression analysis was performed to develop a prognostic signature, which was independently validated. All statistical tests were two-sided.
Results
An 11-protein signature was developed in the training set (median follow-up for good-prognosis patients = 117 months) and subsequently validated in the test set (median follow-up for good-prognosis patients = 108 months) showing 89.5% sensitivity (95% confidence interval [CI] = 69.2% to 98.1%), 70.5% specificity (95% CI = 61.7% to 74.2%), 56.7% positive predictive value (95% CI = 43.8% to 62.1%), and 93.9% negative predictive value (95% CI = 82.3% to 98.9%) for poor-prognosis patients. The predicted poor-prognosis patients had higher risk to develop distant metastasis than the predicted good-prognosis patients in univariate (hazard ratio [HR] = 13.15; 95% CI = 3.03 to 57.07; P = .001) and multivariable (HR = 12.45; 95% CI = 2.67 to 58.11; P = .001) analysis. Furthermore, the predicted poor-prognosis group had statistically significantly more breast cancer–specific mortality. Using our signature as guidance, more than 60% of patients would have been exempted from unnecessary adjuvant chemotherapy compared with conventional prognostic guidelines.
Conclusions
We report the first validated proteomic signature to assess the natural course of clinical TNBC.
Triple-negative breast cancer (TNBC) is one of the most aggressive breast cancer subtypes. To date, there is no clinically available targeted therapy for patients diagnosed with TNBC. Current guidelines for breast cancer treatment recommend that the majority of lymph node–negative (LNN) breast cancer patients, including TNBC patients, be treated with adjuvant chemotherapy (1) . Eventually approximately 30% of LNN TNBC patients develop distant metastasis (2) and could thus potentially benefit from adjuvant chemotherapy; this indicates that the majority of patients are currently being overtreated. The lack of highly sensitive and specific prognostic markers is a major obstacle to predicting prognosis of TNBC patients. Moreover, there is an urgent need to identify potentially useful targets for therapy. Some commonly applied approaches for biomarker discovery, such as gene expression profiling, have not yet succeeded in identifying highly sensitive and specific markers related to disease progression of TNBC (3). Although a few publications have claimed to identify gene signatures that predict prognosis of TNBC (4–8), the reported signatures have limited clinical value because of their experimental design and lack of sufficient sensitivity and specificity. Therefore, it is desirable to identify a highly sensitive and specific prognostic signature for clinical application.
Quantitative proteome analysis using nanoscale liquid chromatography and tandem mass spectrometry (nLC-MS/MS) may complement these efforts because nLC-MS/MS quantitatively profiles a large portion of the human proteome (9). Some protein markers predicting prognosis (10) and therapy resistance (11) of breast cancer have already been identified using nLC-MS/MS–based comparative proteome profiling. In this study, we identified and independently validated a protein signature to predict 5-year metastasis-free survival of LNN patients who did not receive systemic adjuvant therapy, which would allow for the selection of TNBC patients that can be spared the toxicity of adjuvant chemotherapy.
Methods
Study Design
Patient specimens used to develop and validate a prognostic signature were collected in five European institutes between 1985 and 2005. Triple negativity of tumor tissues was confirmed based on mRNA expression using real-time quantitative polymerase chain reaction with a reported criterion of estrogen receptor (ESR1) less than 0.2, progesterone receptor (PGR) less than 0.1, and human epidermal growth factor receptor (HER2/ERBB2) genes BB2 less than 18 (12,13). With this criterion, we identified 271 and 61 TNBC tumor tissues from local tissue bank and European Organization for Research and Treatment of Cancer (EORTC) collaborators, respectively. Tissues used in this study were restricted to LNN patients who had not received systemic adjuvant therapy. Patients who developed distant metastasis as first event within 5 years after removal of the primary tumor were considered to have poor prognosis, whereas patients who remained free of distant metastasis for 5 years were defined as having good prognosis. Patients who had bilateral breast cancer were excluded. In addition, inclusion for microdissection and mass spectrometry (MS) profiling depended on sufficient invasive tumor area for microdissection and morphological quality. With the above-mentioned criteria, samples were subsequently rejected on the basis of clinical reasons [patients were diagnosed with positive lymph-nodes (92), received adjuvant chemotherapy (3), had insufficient clinical follow-up (19), and developed local relapse before distant metastasis (26)] and technical reasons [insufficient tumor tissues (33), indistinguishable morphology, and insufficient tumor area for microdissection (32)]. As a consequence, 63 and 64 tumors from training and test sets were prepared for nLC-MS/MS profiling, respectively. One sample was not successfully measured because of machinery failure. Clinical characteristics of the included (n = 126) and excluded (n = 206) subjects are summarized in Supplementary Table 1 (available online). nLC-MS/MS data from 63 samples were used for development of the protein signature (training set), and data from the other 63 samples were used for independent validation (test set) (Table 1; Supplementary Table 2, available online). This study was approved by the local institutional medical ethics committee (MEC 02.953), and wherever possible we adhered to the Reporting Recommendations for Tumor Marker Prognostic Studies (14).
Table 1.
Clinical and pathological characteristics of patients and their tumors*
| Variables | Training set | Test set (11-protein signature) | ||
|---|---|---|---|---|
| EMC (n = 63) | Multicenter (n = 63) | EMC (n = 30) | EORTC (n = 33) | |
| Age, y | ||||
| Mean (SD) | 51 (14) | 56 (13) | 54 (13) | 59 (13) |
| ≤40 | 13 (20.6) | 9 (14.3) | 6 (20.0) | 3 (9.1) |
| 41–55 | 25 (39.7) | 23 (36.5) | 12 (40.0) | 11 (33.3) |
| 56–70 | 19 (30.2) | 22 (34.9) | 8 (26.7) | 14 (42.4) |
| >70 | 6 (9.5) | 9 (14.3) | 4 (13.3) | 5 (15.2) |
| Menopausal status | ||||
| Premenopausal | 34 (54.0) | 26 (41.3) | 16 (53.3) | 10 (30.3) |
| Postmenopausal | 29 (46.0) | 37 (58.7) | 14 (46.7) | 23 (69.7) |
| Tumor size, cm | ||||
| Mean (SD)† | 2.9 (1.5) | 2.6 (1.1) | 2.9 (1.2) | 2.3 (0.9) |
| pT1, ≤ 2cm | 22 (34.9) | 22 (34.9) | 6 (20.0) | 16 (48.5) |
| pT2–4, >2cm | 39 (61.9) | 35 (55.6) | 20 (66.7) | 15 (45.5) |
| Unknown | 2 (3.2) | 6 (9.5) | 4 (13.3) | 2 (6.1) |
| Grade | ||||
| Grade 1 | 0 (0.0) | 2 (3.2) | 2 (6.7) | 0 (0.0) |
| Grade 2 | 4 (6.3) | 12 (19.0) | 2 (6.7) | 10 (30.3) |
| Grade 3 | 44 (69.8) | 43 (68.3) | 21 (70.0) | 22 (66.7) |
| Unknown | 15 (23.8) | 6 (9.5) | 5 (16.7) | 1 (3.0) |
| Metastasis within 5 years | ||||
| Yes | 25 (39.7) | 19 (30.2) | 9 (30.0) | 10 (30.3) |
| No | 38 (60.3) | 44 (69.8) | 21 (70.0) | 23 (69.7) |
* Data are No. (%) unless otherwise stated. EMC = Erasmus Medical Center; EORTC = European Organization for Research and Treatment of Cancer; SD = standard deviation.
† Samples with recorded tumor size were used for calculation.
Experimental Procedures
The dedicated procedure of sample preparation was performed (15) (Supplementary Methods, available online). Tissue samples were cut into 8-µm frozen sections and then microdissected to obtain approximately 4000 breast cancer epithelial cells. Proteins were extracted from microdissected cells, and this was followed by denaturation, reduction, and alkylation. Protein samples were digested at 37oC for 4 hours using MS-grade trypsin (Promega, Madison, WI) at a 1:4 (enzyme/protein) ratio and then acidified for further analysis.
Global proteome profiles of the TNBC samples were recorded on an nLC hyphenated LTQ-Orbitrap-XL MS system (ThermoElectron, Bremen, Germany) (Supplementary Methods, available online). Peptide mixtures were separated on the nLC system with a 3-hour binary gradient (mobile phase A: water; mobile phase B: acetonitrile) in a 3-μm C18 silica-packed 50-cm capillary column with 75-μm inner diameter. Mass spectra were acquired over a mass-to-charge ratio (m/z) range of 400 to 1800 at a resolving power of 30000 at 400 m/z. The five most intensive parent ions from the full scan were isolated and fragmented by collisional activated dissociation in the linear ion trap. Dynamic exclusion was used to increase the number of parent ions selected for fragmentation. Recorded raw nLC-MS/MS data have been submitted to ProteomeXchange (accession number: PXD000260).
Statistical Analysis
The recorded MS spectra from the training and test set were separately analyzed in MaxQuant Software (free-ware available from www.maxquant.org, version 1.1.1.36) (16). To identify a prognostic protein signature, Cox regression analysis was performed to associate protein abundance with metastasis-free survival time of patients in the training set. Cox regression coefficients and corresponding P values were determined for all tested proteins. The assumption of proportionality was verified by a test based on the Schoenfeld residuals. Prognostic proteins were selected with a P value cutoff of less than .01. Weighted protein abundance of the prognostic proteins was computed by multiplication of their protein abundance and corresponding Cox regression coefficients. A relapse score was calculated by summation of weighted protein abundance of all protein(s) in a given predictive model, followed by z score transformation of summed score. Multiple predictive models were constructed from the training set by starting with the protein with the lowest P value and stepwise adding one more protein in the next model. Efficiency of these models was then assessed by summation of three parameters: 1) area under the curve (AUC) of receiver operating characteristic (ROC) curve (17), 2) Youden’s index at 100% sensitivity (sensitivity + specificity – 1), and 3) reversed model size (1 − ni/n, where ni is the number of proteins in a defined model, and n is the total number of prognostic proteins). The model with the highest efficiency was considered as the best prognostic signature.
The cutoff of the prognostic protein signature was determined from the ROC curve of the training set to ensure 100% sensitivity and highest specificity. The protein signature was validated in the test set at the determined cutoff. Kaplan–Meier survival curves and log-rank test were performed to evaluate the differences in the time to distant metastasis of predicted good and poor prognosis groups. Univariate and multivariable analyses with Cox proportional hazard model were used to assess the prognostic value of the protein signature with and without consideration of the individual clinical prognostic variables.
Pathway analysis was performed to interpret biological function of signature proteins using gene set enrichment analysis (18). The Supplementary Methods (available online) describe detailed information about MaxQuant settings, statistical software packages, and pathway analysis.
Results
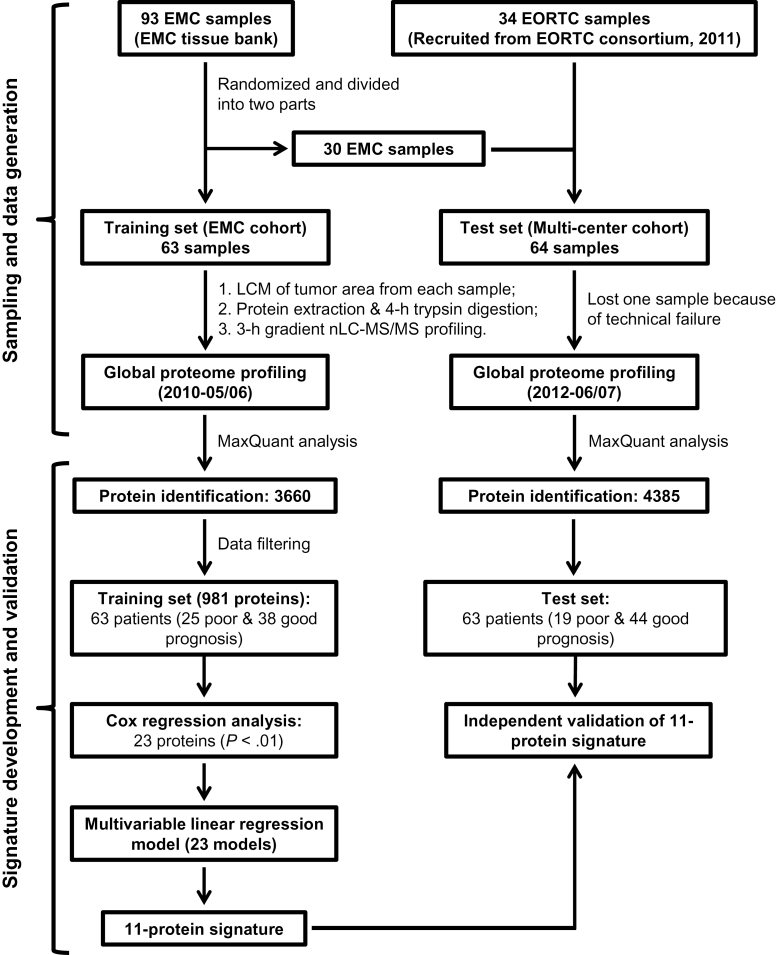
The training and test samples were profiled by nLC-MS/MS in 2010 and 2012, respectively (Figure 1). The baseline clinical features of patients were similar between the Erasmus University Medical Center (EMC) training and multicenter test set, although patients were slightly older and more patients were classified with grade 2 tumors in the test set (Table 1). The median follow-ups for the good-prognosis patients in the training and test set were 117 (range = 61–257) and 108 (range = 61–234) months, respectively. Patients were recruited in five EORTC institutes between 1985 and 2005. Clinical data used for data analysis were updated until October 2012.
Figure 1.
Flow chart of experimental design for the development and validation of an 11-protein signature. EMC = Erasmus University Medical Center; EORTC = European Organization for Research and Treatment of Cancer; LCM = laser capture microdissection; nLC-MS/MS = nanoscale liquid chromatography and tandem mass spectrometry.
Development of a Proteomic Prognostic Signature for TNBC Patients
Proteome profiles for training and test samples were independently generated by nLC-MS/MS (see Supplementary Methods, available online). A stringent filtering procedure resulted in a total of 981 proteins for development of a prognostic signature (Figure 1; Supplementary Figure 1, available online). Good reproducibility and quantitative precision were observed from quantified proteins in replicate samples by performing the same filtering criteria (see Supplementary Methods, available online).
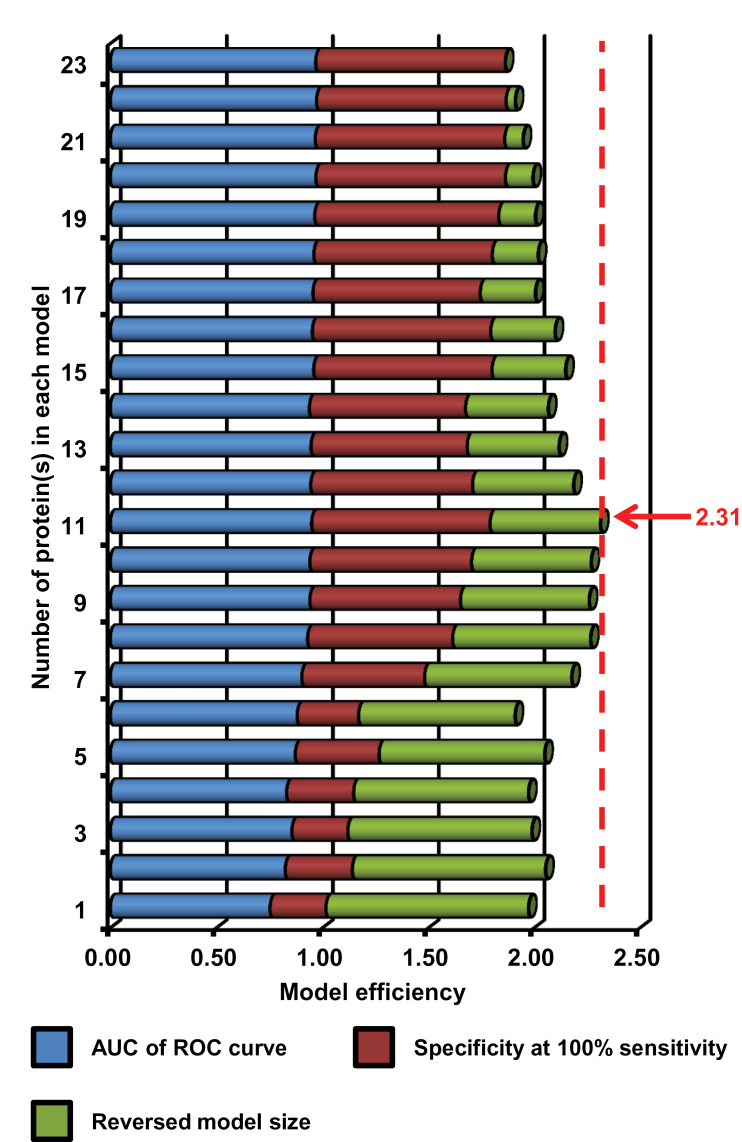
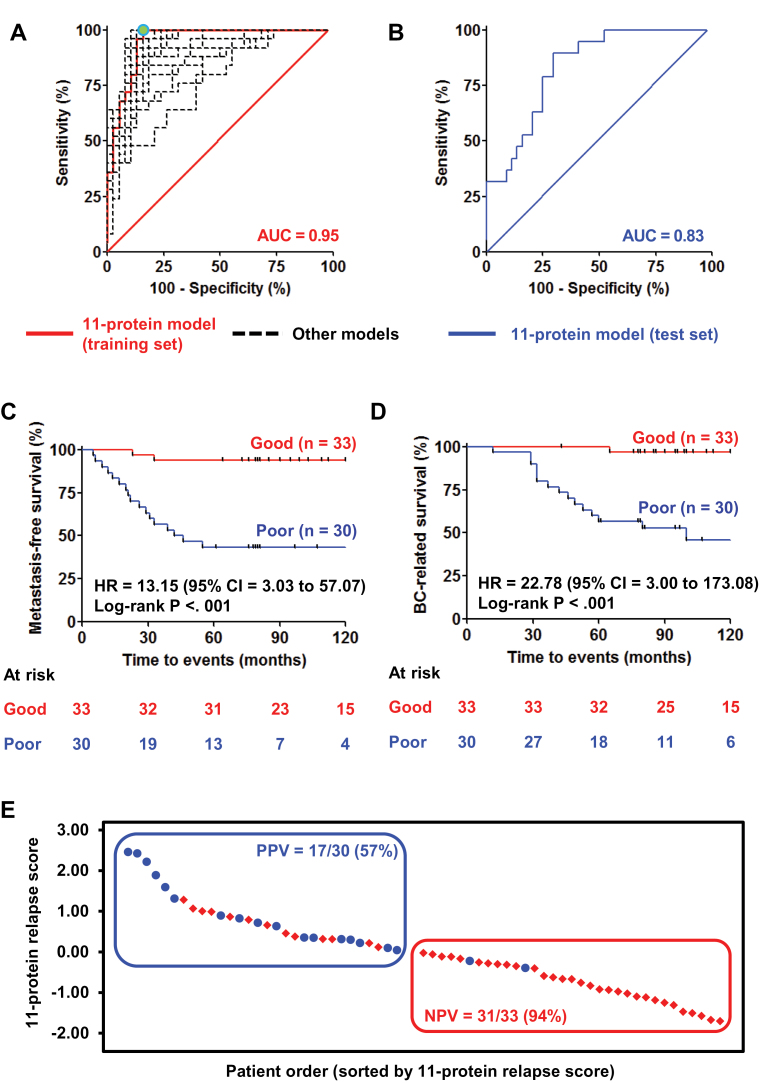
The 981 proteins were used to develop a prognostic signature in the training set. Using univariate Cox analysis, a panel of 23 proteins was statistically significantly associated with metastasis-free survival of patients (P < .01) (Figure 1; Supplementary Figure 1, available online). Twenty-three prognostic models were derived from these 23 proteins using a multivariable linear regression model, from which an 11-protein model performed fairly comparably with the models with 15 or more proteins based on an AUC of ROC curve and specificity at 100% sensitivity to predict poor-prognosis patients. By further considering model size, we calculated efficiency for all 23 models, and the 11-protein model showed the highest model efficiency at 2.31 (Figure 2). Detailed information for these 11 proteins is listed in Table 2 and Supplementary Table 3 (available online). Of these 11 proteins, 10 proteins were upregulated (CMPK1, AIFM1, FTH1, EML4, GANAB, CTNNA1, AP1G1, STX12, AP1M1, and CAPZB), whereas one protein was downregulated (MTHFD1) in good-prognosis patients in the training set. The ROC curve derived from the 11-protein signature showed good sensitivity and specificity with AUC of 0.95 (Figure 3A). A cutoff was selected with a relapse score of zero to identify good-prognosis patients (negative score) at which maximal specificity was reached at 100% sensitivity (Figure 3A, green dot).
Figure 2.
Selection of the best predictive signature from 23 prognostic models developed in the training set. The different models were created from 23 prognostic proteins (Cox regression analysis: P < .01), starting with the protein with the lowest P value and gradually adding one more protein at a time, thereby constructing 23 different prognostic models. Model efficiency was considered by three aspects: 1) area under the curve (AUC) of the receiver operating characteristic (ROC) curve, 2) specificity at 100% sensitivity, and 3) reversed model size (1 − 1/n, where n is number of used protein[s] for the model). The model with the highest model efficiency was considered as the best model, resulting in selection of the 11-protein signature (model efficiency = 2.31) for validation.
Table 2.
Eleven-signature proteins and their prognostic information in training set
| Protein ID | Gene name | Protein description | Subcellular localization* | Cox coefficient (Hazard ratio)† | P (FDR)‡ |
|---|---|---|---|---|---|
| P30085 | CMPK1 | UMP-CMP kinase | Nucleus/cytoplasm | −0.587 (0.556) | .00 (0.123) |
| O95831 | AIFM1 | Apoptosis-inducing factor 1, mitochondrial | Mitochondrion/nucleus/ cytoplasm | −0.860 (0.423) | .001 (0.199) |
| P02794 | FTH1 | Ferritin heavy chain | Cytoplasm/intracellular matrix | −0.400 (0.670) | .001 (0.199) |
| P11586 | MTHFD1 | C-1-tetrahydrofolate synthase, cytoplasmic | Cytoplasm | 1.178 (3.247) | .001 (0.199) |
| Q9HC35 | EML4 | Echinoderm microtubule- associated protein- like 4 | Cytoplasm/ cytoskeleton/ microtubule | −0.576 (0.562) | .001 (0.267) |
| Q14697 | GANAB | Neutral alpha- glucosidase AB | Endoplasmic reticulum/ Golgi apparatus/ melanosome | −1.050 (0.350) | .002 (0.312) |
| P35221 | CTNNA1 | Catenin alpha-1 | Cytoplasm/ cytoskeleton/ cell junction/Cell membrane | −1.115 (0.328) | .002 (0.312) |
| O43747 | AP1G1 | AP-1 complex subunit gamma-1 | Golgi apparatus/ cytoplasmic vesicle/ clathrin-coated vesicle membrane | −0.997 (0.369) | .003 (0.332) |
| Q86Y82 | STX12 | Syntaxin-12 | Endosome membrane/ Golgi apparatus membrane | −0.675 (0.509) | .003 (0.332) |
| Q9BXS5 | AP1M1 | AP-1 complex subunit mu-1 | Golgi apparatus/ cytoplasmic vesicle/ clathrin-coated vesicle membrane | −0.846 (0.429) | .004 (0.398) |
| P47756 | CAPZB | F-actin-capping protein subunit beta | Cytoplasm/cytoskeleton | −0.916 (0.400) | .005 (0.398) |
* Adapted from UniProt Knowledgebase.
† Cox coefficients were calculated by natural logarithm (ln) of hazard ratios.
‡ P values were computed by Cox regression analysis, and the corresponding false discovery rate (FDR) of the proteins is reported in parentheses.
Figure 3.
Development and multicenter validation of the 11-protein signature. A) Receiver operating characteristic (ROC) curve of the 11-protein signature in the training set (red solid line: area under the curve [AUC]). A cutoff was chosen to ensure maximal sensitivity to identify the good-prognosis patients with the highest possible specificity in the training set (green dot). Other models developed from 23 prognostic proteins were also plotted as ROC curves (black dashed lines). B) ROC curve of the 11-protein signature in the test set. Kaplan–Meier analysis shows that the 11-protein signature is strongly predictive of metastasis-free survival (C) and breast cancer (BC)–related survival (D) CI = confidence interval; HR = hazard ratio. E) Waterfall plot stratifies two groups of triple-negative breast cancer patients with different predicted prognosis in the test set. NPV = negative predictive value; PPV = positive predictive value.
All statistical tests were two-sided.
Validation of the 11-Protein Signature in a Multicenter Cohort of TNBC Patients
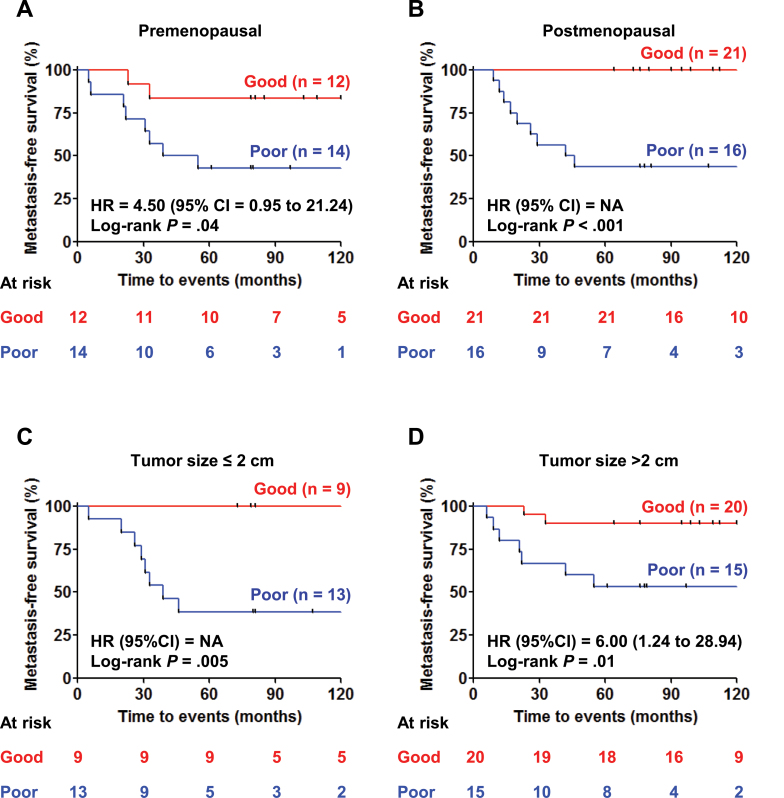
The 11-protein signature was validated on the test cohort, resulting in an ROC curve with AUC of 0.83 (Figure 3B). Using the predetermined cutoff (relapse score = 0), patients predicted as having a poor prognosis (positive score) had much worse 5-year metastasis-free survival (log-rank P < .001) (Figure 3C) and breast cancer–related survival (log-rank P < .001) (Figure 3D) than predicted good-prognosis patients. In the predicted good-prognosis group, 31 of 33 patients did not develop distant metastasis (negative predictive value = 93.9%; 95% confidence interval [CI] = 82.3% to 98.9%) (Figure 3, C and E). In contrast, 17 of 30 patients developed distant metastasis in the predicted poor-prognosis group (positive predictive value =56.7%; 95% CI = 43.8% to 62.1%) (Figure 3, C and E). Overall, our signature results in a sensitivity of 89.5% (95% CI = 69.2% to 98.1%) and specificity of 70.5% (95% CI = 61.7% to 74.2%) to predict poor-prognosis patients in the test set. In univariate Cox analyses, the poor-prognosis patients also have statistically significantly higher risk of developing distant metastases (hazard ratio [HR] = 13.15; 95% CI = 3.03 to 57.07; P = .001) (Figure 3C; Table 3) and breast cancer–related death (HR = 22.78; 95% CI = 3.00 to 173.08; P = .003) (Figure 3D; Table 3) than those predicted to have good prognosis. The 11-protein signature was also prognostic in subgroups of patients with different menopausal status (Figure 4, A and B) and tumor size (Figure 4, C and D). Furthermore, the 11-protein signature was a strong independent prognostic factor to predict risk of distant metastasis (HR = 12.45; 95% CI = 2.67 to 58.11; P = .001) and of breast cancer–related death (HR = 36.08; 95% CI = 4.00 to 325.67; P = .001) in multivariable Cox regression model after correction for the contribution of traditional prognostic factors (Table 3). No other prognostic factors, such as age, menopausal status, tumor size, or tumor grade of the patients, were statistically significantly associated with metastasis-free survival in univariate and multivariable analyses (Table 3).
Table 3.
Univariate and multivariable analyses* of the 11-protein signature for its prognostic value (n = 63)
| Test set (multicenter cohort) | Metastasis-free survival | Breast cancer–related survival | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariable analysis | Univariate analysis | Multivariable analysis | |||||
| Variables | Hazard ratio (95% CI) | P | Hazard ratio (95% CI) | P | Hazard ratio (95% CI) | P | Hazard ratio (95% CI) | P |
| Age, y | 0.99 (0.95 to 1.02) | .42 | 1.02 (0.96 to 1.07) | .59 | 0.99 (0.95 to 1.03) | .68 | 1.02 (0.96 to 1.08) | .61 |
| Menopausal status, post- vs premenopausal | 0.60 (0.24 to 1.48) | .27 | 0.41 (0.08 to 1.96) | .26 | 0.71 (0.27 to 1.89) | .49 | 0.50 (0.09 to 2.66) | .42 |
| Tumor size, >2cm & unknown vs ≤2 cm | 0.74 (0.30 to 1.83) | .51 | 0.70 (0.28 to 1.75) | .45 | 0.94 (0.34 to 2.60) | .91 | 0.93 (0.34 to 2.58) | .89 |
| Tumor grade† | ||||||||
| Grade 3 vs 1 and 2 | 2.79 (0.64 to 12.22) | .17 | 3.22 (0.71 to 14.52) | .13 | 2.09 (0.47 to 9.33) | .34 | 2.31 (0.50 to 10.75) | .29 |
| Grade unknown vs 1 and 2 | 2.61 (0.37 to 18.55) | .34 | 2.70 (0.37 to 19.80) | .33 | 2.67 (0.37 to 19.05) | .33 | 2.88 (0.39 to 21.48) | .30 |
| 11-protein signature, predicted poor vs good | 13.15 (3.03 to 57.07) | .001 | 12.45 (2.67 to 58.11) | .001 | 22.78 (3.00 to 173.08) | .003 | 36.08 (4.00 to 325.67) | .001 |
* Univariate and multivariable analyses were performed based on Cox regression model. All statistical tests were two-sided. CI = confidence interval
† Tumor grade classification according to Bloom and Richardson scoring system (34).
Figure 4.
Kaplan–Meier analysis of metastasis-free survival in subgroups of triple-negative breast cancer patients in the test set. A) Premenopausal patients. B) Postmenopausal patients. Hazard ratio (HR) and 95% confidence interval (CI) could not be computed because of the absence of metastatic events in one of the tested groups. NA = not applicable. C) Patients with tumor size of 2cm or less. Hazard ratio and 95% confidence interval could not be computed because of the absence of metastatic events in one of the tested groups. D) Patients with tumor size greater than 2cm. All statistical tests were two-sided.
Currently, there is no specific clinical guideline of recommend treatment for TNBC patients. Two clinical consensus criteria, St. Gallen (19) and National Institutes of Health (NIH) (20), are often applied to guide treatment of breast cancer cases. In our test set, 91% (n = 40 of 44) and 95% (n = 38 of 40) of the good-prognosis patients would be classified as high risk and therefore would be guided to receive apparently unnecessary adjuvant chemotherapy using St. Gallen and NIH criteria, respectively (Table 4). On the other hand, only 30% (n = 13 of 44) of the good-prognosis patients would be classified for adjuvant chemotherapy using the 11-protein signature (Table 4). Therefore, more than 60% of patients in the test set would have been exempted from unnecessary adjuvant chemotherapy using the 11-protein signature as guidance, compared with St. Gallen and NIH criteria.
Table 4.
Comparison of the 11-protein signature with currently applied clinical consensus criteria on treatment of breast cancer
| Method | Patients guided to receive adjuvant chemotherapy in the test set | |
|---|---|---|
| Poor prognosis | Good prognosis | |
| St. Gallen* | 18/18 (100%)‡ | 40/44 (91%) |
| NIH† | 14/17 (82%)‡ | 38/40 (95%)‡ |
| 11-protein signature | 17/19 (89%) | 13/44 (30%)§ |
* St. Gallen consensus criteria: tumor ≥2cm, ESR1 negative, grade 2–3, patient aged <35 years (one of these criteria).
† National Institutes of Health (NIH) guideline: tumor >1cm.
‡ Patients with missing clinical information were excluded from these analyses.
§ A statistically significant improvement.
Pathway Analysis of Prognostic Signature
The function of the 11 signature proteins related to progression of TNBC was interpreted in silico by gene set enrichment analysis. To overcome low identification rate of our proteomic data (see Supplementary Methods, available online), we used gene expression data from 63 in-house available TNBC samples, of which 47 samples were also included in our global proteome profiling. An overall statistically significant correlation between transcriptome and proteome in the 47 samples indicated the validity of using gene expression to interpret the molecular functions related to TNBC progression and expression of signature proteins (see Supplementary Data, available online). In total, ten proteins were matched with their coding genes. Biological pathways related to good prognosis of the TNBC patients were mainly associated with immune response (eg, modulation of cytokines, antigen processing and presentation, and activation of T cells, B cells, and natural killer cells) and cell death (eg, ceramide signaling, tumor necrosis factor–mediated apoptosis, FAS-FAS ligand–mediated apoptosis, and caspase cascade). Nine of 10 genes were associated with good prognosis of TNBC patients (CMPK1, AIFM1, FTH1, EML4, GANAB, CTNNA1, AP1G1, STX12, and CAPZB), of which FTH1, GANAB, and STX12 were associated with 58, 18, and three enriched pathways related to immune response or cell death with recommended false discovery rate less than 0.25 (Supplementary Table 4, available online). On the other hand, cell metabolism (eg, metabolism of nucleotides and noncoding RNA) and transport of macromolecules (eg, transport of mature transcripts and ribonucleoproteins and export of proteins) were key pathways related to poor prognosis of TNBC patients. MTHFD1 was the only protein upregulated in TNBC patients with poor prognosis and was associated with two metabolic pathways (metabolism of nucleotides and noncoding RNA) with false discovery rate less than 0.25 (Supplementary Table 4, available online).
Discussion
Prognosis of LNN TNBC patients is extremely heterogeneous and often not associated with conventional prognostic parameters (patient age, tumor size, and tumor grade) (21), concordant with this study. Approximately 30% of TNBC patients eventually experience distant relapse (2). Therefore, a substantial proportion of patients are being overtreated with systemic adjuvant therapy. In this study, we identified and independently validated a clinically relevant prognostic 11-protein signature for TNBC, which could be used to reduce the number of patients that would receive unnecessary adjuvant chemotherapy.
To translate the 11-protein signature into a clinically useful assay, two future steps are essential. First, a quantitative, targeted, and ready-to-use assay needs to be developed for absolute quantification of the signature proteins. The most promising candidate for such an assay would be a mass spectrometry–based selected reaction monitoring assay (22). Second, such a targeted assay can then be incorporated into future clinical trials and subsequently used for clinical decision making for treatment options. Furthermore, some of the signature proteins may serve as potential targets for novel therapies.
Some signature proteins have been reported to be prognostic in breast cancer or other cancers. The prognostic role of these proteins is summarized in three categories. First, expression of some proteins is directly linked to disease progression. For instance, decreased levels of FTH1 protein have been associated with lymph node metastasis in colorectal cancer (23). Similarly, abnormal expression of CTNNA1 protein has been related with poor patient survival in invasive breast cancer (24). Also, association has been observed between increased expression of GANAB protein and favorable prognosis in head and neck cancer (25). Second, genetic polymorphisms of some proteins showed important prognostic values. Single nucleotide polymorphisms of CMPK1 have been associated with prognosis of non–small cell lung cancer (26) and pancreatic cancer (27) patients treated with gemcitabine-based chemotherapeutics. A correlation has also been reported between a 1958AA single nucleotide polymorphism of MTHFD1 gene and poor clinical outcome of premenopausal breast cancer patients (28). Moreover, genetic mutations of certain protein-coding genes may also be prognostic. EML-ALK fusion gene has been suggested to be associated with relatively favorable prognosis of non–small cell lung cancer patients (29). We speculate that there may be a correlation between expression of these proteins and their genetic variance, which needs to be further investigated.
Pathway analysis showed that immune response, cell death, cell metabolism, and transport of macromolecules are the major underlying molecular mechanisms of the signature proteins related to prognosis of TNBC patients. These findings are in concordance with previous observations. Lehmann and colleagues identified an immunomodulatory subtype that was associated with favorable clinical outcome (30). Rody and colleagues identified a signature of high B-cell and low IL-8 metagenes that predicted good prognosis of TNBC patients (5), and Yau and coworkers reported a 14-gene signature linked to immune/inflammatory cytokine regulation in which the majority of genes were associated with good prognosis of TNBC patients (8). In our prognostic signature, three proteins (FTH1, GANAB, and STX12) are associated with immunomodulation and cell death–associated pathways. FTH1 is an iron storage protein and indirectly regulates the ratio of CD4+ T cells and CD8+ T cells by altering iron distribution (31). Increased CD8+ T cells may help to eliminate tumor cells (32), which would be favorable for survival of TNBC patients. Molecular functions of GANAB and STX12 related to immune response and cell death have not been well studied. Therefore, further studies are required to understand the functions of these proteins in disease progression. On the other hand, it is known that the luminal androgen receptor subtype of TNBC, with aberrant alteration of cell metabolism, is related to adverse clinical outcome of TNBC patients (30). In our signature, MTHFD1 is associated with poor prognosis and is involved in folate metabolism. It has been suggested that increased concentration of folate resulted in a dose-dependent downregulation of tumor suppressor genes in breast cancer cell lines due to increased DNA methylation of their promoter region (33), which may indicate the importance of MTHFD1 in TNBC progression.
Our study has some limitations. First, microdissection-based sample preparation is tedious and may introduce additional biases between diagnostic individuals and laboratories. Second, the nLC-MS/MS–based platform is difficult to apply in routine clinical practice. Therefore, our future work will focus on developing simple sampling strategies and high-throughput selected reaction monitoring assay to reliably measure the 11-protein signature.
In summary, we have developed an 11-protein signature to predict TNBC patients who develop a distant metastasis with high sensitivity, specificity, positive predictive value, and negative predictive value (82.3% to 98.9%). Therefore, our signature could aid in clinical practice to avoid unnecessary treatment with adjuvant chemotherapy of LNN TNBC patients. Future prospective clinical trials are needed to further consolidate the validity of the 11-protein signature.
Funding
This study was financially supported by the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research (The Hague, The Netherlands), Center for Translational Molecular Medicine (Eindhoven, The Netherlands) Breast CARE project 030-104, and Dutch Cancer Society, (Amsterdam, The Netherlands) project number EMCR 2009–4319.
The authors are solely responsible for the study design, data generation, analysis and interpretation of the data, writing the manuscript, and decision to submit the manuscript for publication.
The authors gratefully acknowledge Dr Lennard J.M. Dekker and Lona Zeneyedpour for technical assistance on nLC-MS/MS measurements and Dr Elizabeth A. McClellan and Dr Andrew Stubbs for technical support on bioinformatics analysis.
References
- 1. Harris L, Fritsche H, Mennel R, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25(33):5287–5312 [DOI] [PubMed] [Google Scholar]
- 2. Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13(15 Pt 1):4429–4434 [DOI] [PubMed] [Google Scholar]
- 3. Weigelt B, Pusztai L, Ashworth A, Reis-Filho JS. Challenges translating breast cancer gene signatures into the clinic. Nat Rev Clin Oncol. 2012;9(1):58–64 [DOI] [PubMed] [Google Scholar]
- 4. Cheang MCU, Voduc D, Bajdik C, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14(5):1368–1376 [DOI] [PubMed] [Google Scholar]
- 5. Rody A, Karn T, Liedtke C, et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011;13(5):R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hallett RM, Dvorkin-Gheva A, Bane A, Hassell JA. A gene signature for predicting outcome in patients with basal-like breast cancer. Sci Rep. 2012;2:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuo W-H, Chang Y-Y, Lai L-C, et al. Molecular characteristics and metastasis predictor genes of triple-negative breast cancer: a clinical study of triple-negative breast carcinomas. PloS One. 2012;7(9):e45831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yau C, Esserman L, Moore DH, Waldman F, Sninsky J, Benz CC. A multigene predictor of metastatic outcome in early stage hormone receptor-negative and triple-negative breast cancer. Breast Cancer Res. 2010;12(5):R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nagaraj N, Wisniewski JR, Geiger T, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Geiger T, Madden SF, Gallagher WM, Cox J, Mann M. Proteomic portrait of human breast cancer progression identifies novel prognostic markers. Cancer Res. 2012;72(9):2428–2439 [DOI] [PubMed] [Google Scholar]
- 11. Umar A, Kang H, Timmermans AM, et al. Identification of a putative protein profile associated with tamoxifen therapy resistance in breast cancer. Mol Cell Proteomics. 2009;8(6):1278–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sieuwerts AM, Usher PA, Meijer-van Gelder ME, et al. Concentrations of TIMP1 mRNA splice variants and TIMP-1 protein are differentially associated with prognosis in primary breast cancer. Clin Chem. 2007;53(7):1280–1288 [DOI] [PubMed] [Google Scholar]
- 13. Van Agthoven T, Sieuwerts AM, Meijer-van Gelder ME, et al. Relevance of breast cancer antiestrogen resistance genes in human breast cancer progression and tamoxifen resistance. J Clin Oncol. 2009;27(4):542–549 [DOI] [PubMed] [Google Scholar]
- 14. McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst. 2005;97(16):1180–1184 [DOI] [PubMed] [Google Scholar]
- 15. Liu NQ, Braakman RBH, Stingl C, et al. Proteomics pipeline for biomarker discovery of laser capture microdissected breast cancer tissue. J Mammary Gland Biol Neoplasia. 2012;17(2):155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372 [DOI] [PubMed] [Google Scholar]
- 17. Mossman D, Somoza E. ROC curves, test accuracy, and the description of diagnostic tests. J Neuropsychiatry Clin Neurosci. 1991;3(3):330–333 [DOI] [PubMed] [Google Scholar]
- 18. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goldhirsch A, Wood WC, Gelber RD, Coates AS, Thürlimann B, Senn H-J. Meeting highlights: updated international expert consensus on the primary therapy of early breast cancer. J Clin Oncol. 2003;21(17):3357–3365 [DOI] [PubMed] [Google Scholar]
- 20. Eifel P, Axelson JA, Costa J, et al. National Institutes of Health Consensus Development Conference Statement: adjuvant therapy for breast cancer, November 1–3, 2000. J Natl Cancer Inst. 2001;93(13):979–989 [DOI] [PubMed] [Google Scholar]
- 21. Rakha EA, El-Sayed ME, Green AR, Lee AHS, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109(1):25–32 [DOI] [PubMed] [Google Scholar]
- 22. Liu NQ, Dekker LJM, Stingl C, et al. Quantitative proteomic analysis of microdissected breast cancer tissues: comparison of label-free and SILAC based quantification with shotgun, directed and targeted MS approaches [published online ahead of print August 20, 2013]. J Proteome Res. 2013;12(10):4627–4641 [DOI] [PubMed] [Google Scholar]
- 23. Ma Y, Zhao M, Zhong J, et al. Proteomic profiling of proteins associated with lymph node metastasis in colorectal cancer. J Cell Biochem. 2010;110(6):1512–1519 [DOI] [PubMed] [Google Scholar]
- 24. Nakopoulou L, Gakiopoulou-Givalou H, Karayiannakis AJ, et al. Abnormal alpha-catenin expression in invasive breast cancer correlates with poor patient survival. Histopathology. 2002;40(6):536–546 [DOI] [PubMed] [Google Scholar]
- 25. Chiu C-C, Lin C-Y, Lee L-Y, et al. Molecular chaperones as a common set of proteins that regulate the invasion phenotype of head and neck cancer. Clin Cancer Res. 2011;17(14):4629–4641 [DOI] [PubMed] [Google Scholar]
- 26. Ryu J-S, Shin E-S, Nam H-S, et al. Differential effect of polymorphisms of CMPK1 and RRM1 on survival in advanced non-small cell lung cancer patients treated with gemcitabine or taxane/cisplatinum. J Thorac Oncol. 2011;6(8):1320–1329 [DOI] [PubMed] [Google Scholar]
- 27. Woo HI, Kim K-K, Choi H, et al. Effect of genetic polymorphisms on therapeutic response and clinical outcomes in pancreatic cancer patients treated with gemcitabine. Pharmacogenomics. 2012;13(9):1023–1035 [DOI] [PubMed] [Google Scholar]
- 28. Babyshkina N, Malinovskaya E, Nazarenko M, et al. The effect of folate-related SNPs on clinicopathological features, response to neoadjuvant treatment and survival in pre- and postmenopausal breast cancer patients. Gene. 2013;518(2):397–404 [DOI] [PubMed] [Google Scholar]
- 29. Takahashi T, Sonobe M, Kobayashi M, et al. Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol. 2010;17(3):889–897 [DOI] [PubMed] [Google Scholar]
- 30. Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Knovich MA, Storey JA, Coffman LG, Torti SV, Torti FM. Ferritin for the clinician. Blood Rev. 2009;23(3):95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Halpern M, Zahalka MA, Traub L, Moroz C. Antibodies to placental immunoregulatory ferritin with transfer of polyclonal lymphocytes arrest MCF-7 human breast cancer growth in a nude mouse model. Neoplasia. 2007;9(6):487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lubecka-Pietruszewska K, Kaufman-Szymczyk A, Stefanska B, Fabianowska-Majewska K. Folic acid enforces DNA methylation-mediated transcriptional silencing of PTEN, APC and RARbeta2 tumour suppressor genes in breast cancer. Biochem Biophys Res Commun. 2013;430(2):623–628 [DOI] [PubMed] [Google Scholar]
- 34. Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403–410 [DOI] [PubMed] [Google Scholar]