Abstract
Toxin-antitoxin (TA) systems are unique modules that effect plasmid stabilization via post-segregational killing of the bacterial host. The genes encoding TA systems also exist on bacterial chromosomes, where they are speculated to be involved in a variety of cellular processes. Interest in TA systems has increased dramatically over the past five years as the ubiquitous nature of TA genes on bacterial genomes has been revealed. The exploitation of TA systems as an antibacterial strategy via artificial activation of the toxin has been proposed and has considerable potential; however, efforts in this area remain in the early stages, and several major questions remain. This review will investigate the tractability of targeting TA systems to kill bacteria, including fundamental requirements for success, recent advances, and challenges associated with artificial toxin activation.
Keywords: addiction modules, mazEF, EDF, antibiotics
Toxin-Antitoxin systems and the induction of bacterial cell death
Type II toxin-antitoxin (TA) systems consist of an antitoxin protein that binds to and inactivates a toxin protein. In relative terms, the toxin is considerably more stable (longer cellular half-life) than the antitoxin, as the less ordered structure of the antitoxin makes it more susceptible to proteolytic degradation. The TA functionality capitalizes on this differential stability between the two proteins (Figure 1). TA systems were discovered in 1983 to confer plasmid stabilization via toxin-induced post-segregational killing (PSK) [1-3]. If the plasmid encoding the TA system is not inherited by a daughter cell, the antitoxin is degraded by cellular proteases and not replenished, liberating the latent toxin to kill the cell and thereby diminishing the population of plasmid-free cells [4]. Many such “addiction” modules stabilize plasmids that carry drug-resistance determinants in important pathogens, notably the axe-txe and ε-ζ TA systems (Box 1) commonly found on vanA plasmids in vancomycin-resistant enterococci (VRE) [5,6].
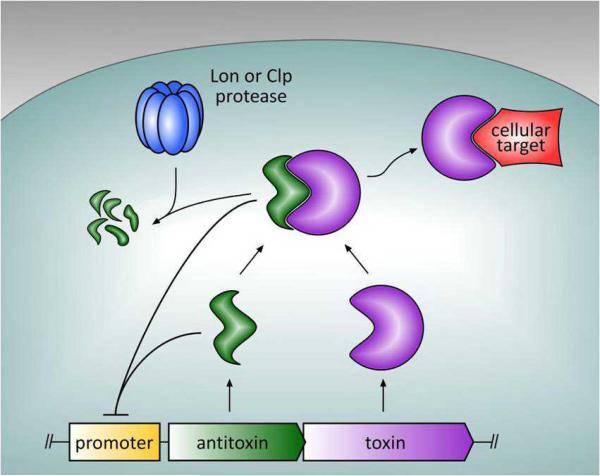
Figure 1.
Proteic Type II toxin-antitoxin (TA) systems encode antitoxin and toxin proteins, with both the antitoxin and the TA complex negatively regulating the TA promoter. The antitoxin is subject to proteolytic degradation by either Lon or Clp proteases, which releases the toxin and allows it to act on its cellular target, resulting in growth inhibition or cell death.
Box 1. Classification of TA systems.
There are three known classes of TA systems; however this review focuses exclusively on the Type II proteic modules. The 10 toxin families within the Type II proteic systems are listed in Table I, organized by cellular target and toxin mode of action (adapted from and reviewed in [8,29,40]).
Although the role of plasmid-encoded TA systems is clear, there is no such consensus for chromosomal TA genes, and in fact there are at least ten proposed biological roles for such systems [7-9]. Once regarded as superfluous genetic material with ambiguous benefit to the bacterial cell [10], chromosomal TA systems have recently been proposed to be involved in numerous cellular pathways including starvation-induced cell stasis [11,12], stress response [13,14], genetic stabilization [15,16], programmed cell death [17,18], biofilm formation [19,20], quorum sensing [9], antiphage protection [21,22], virulence [23], persistence [24,25], and gene regulation [26].
There are many excellent reviews covering the physiological functions of TA systems [7,8,27-30], their role in stress response [31,32], their mechanism of action [33], their structures [34,35], their involvement in multicellular bacterial behavior [36], their induction by antibiotics [37], their applications in biotechnology [38,39], or some combination thereof [40-43]; these aspects of TA systems will not be discussed here. This review will focus on recent experiments aimed at exploitation of TA systems as an antibacterial strategy and challenges therein.
Exploiting TA systems to kill bacteria: an overview
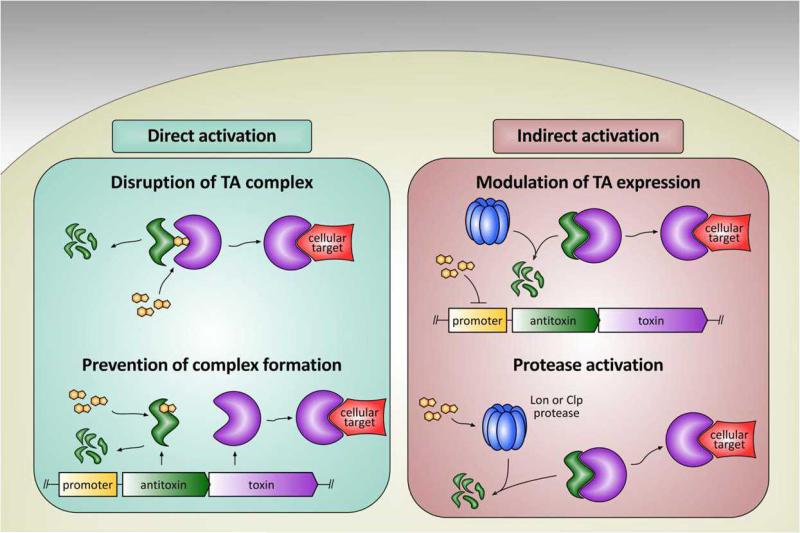
TA genes have no human homologs and appear to be present in the most important bacterial pathogens (as described below); thus, the corresponding protein products could serve as ideal targets for novel antibacterial drugs via one of the mechanisms depicted in Figure 2. In the most straightforward approach, a drug would directly target the TA system proteins and relieve antitoxin inhibition of the toxin. This could be achieved by disruption of the TA complex or prevention of complex formation, as shown in Figure 2. Complete disruption may be required for activation of a toxin such as the ribosome-dependent ribonuclease RelE, as the RelB-RelE complex is likely too large to access the ribosomal A-site [44]. In contrast, activation of a toxin such as MazF, which cleaves free mRNA, may not require full disruption of the complex and instead may be achieved by vacating MazE from the MazF active site or by allosteric activation of MazF in complex with MazE.
Figure 2.
Introduction of a toxin activator (orange fused ring) can activate the TA complex in either a direct or indirect manner. Direct activation (left) can be achieved by either disruption of the TA complex (top) or prevention of complex formation (bottom). Indirect activation (right) may occur either by modulating the expression of the TA complex at the promoter (top) or by activating cellular proteases (bottom) responsible for antitoxin degradation.
Another mode-of-action that can be envisioned is expedited proteolytic degradation of the antitoxin (Figure 2). A molecule that binds promoter DNA and inhibits transcription at the TA locus would prevent replenishment of the antitoxin; there is considerable precedent for such sequence-specific DNA binders [45]. Degradation of the existing antitoxin by Lon or Clp proteases would release the toxin, allowing it to kill the cell. It has also been suggested that toxin activation in response to cellular stress requires increased expression or activity of Lon or Clp, responsible for degrading the antitoxins [11]. Thus, activation of Lon or Clp could serve as an indirect mechanism for toxin activation within the cell, and there is recent precedent for the identification of such compounds in other systems [46,47]. Although this strategy may be generally toxic to the cell, there is evidence that Lon overproduction specifically activated the toxin YoeB from its complex with the YefM antitoxin, resulting in mRNA cleavage and cell lethality [48].
Although the direct and indirect strategies are mechanistically distinct, both artificially activate the toxin from the inert TA complex to kill the bacterial cell. There are two main requirements for successful application of this strategy. The first is an understanding of which TA pairs would serve as ideal targets of an artificial activator. Once specific TA target systems have been identified, molecules must be found that activate the toxin and lead to toxin-mediated cell death.
Which TA systems should be targeted?
Recent genome sequencing and bioinformatic studies have revealed a plethora of TA systems across bacterial species. In 2005, Pandey and Gerdes performed an exhaustive search of 126 sequenced prokaryotic genomes and reported that genes predicted to encode TA systems are highly abundant in free-living bacteria but are absent from the genomes of host-associated bacteria [49]. Shao and co-workers expanded on existing datasets to identify 10,753 putative TA pairs in 1240 sequenced genomes representing 962 bacterial and archaeal strains [50]. More recently, Leplae and co-workers revealed 7034 toxins and 10,829 antitoxins in a search for Type II TA systems in 2181 prokaryotic genomes [51,52]. From this work novel toxins and antitoxins were discovered, some of which were experimentally validated using a host killing and rescue assay in E. coli [51]. In addition to discovering a multitude of TA systems and advancing our understanding of the evolutionary relationships between them, these bioinformatics studies serve as a starting point for more detailed analyses of TA systems within their respective hosts.
Genes for TA systems have been identified in nearly all bacterial pathogens, contributing to their attractiveness as potential antibacterial targets, but which ones will make the best targets? Since many TA systems exist on plasmids or are closely linked with mobile genetic elements, their presence within a given bacterial species is likely to be heterogeneous. Thus, studying TA systems within actual clinical isolates is a necessary and complementary approach to bioinformatics studies. The crucial steps in investigating the tractability of TA systems as antibacterial targets are to determine (i) if TA systems are present in drug-resistant bacterial pathogens, (ii) which TA system are most prevalent, and (iii) whether the TA systems are functional.
In 2007, an examination of TA genes within total genomic DNA from clinical isolates of VRE was reported. Using a PCR-based screen with gene-specific primers for individual TA systems, certain TA genes were found to be widespread across the collection of 75 VRE isolates, namely mazEF (100%), axe-txe (75%), relBE (47%), and ω-ε-ζ (44%) [5]. Many of these TA systems were present on plasmids carrying the vanA gene cassette. Reverse transcription PCR (RT-PCR) analysis showed that the ubiquitous TA system, mazEF, was transcribed in VRE. Furthermore, the mazEF genes, cloned with their native promoter from a VRE isolate, stabilized the unstable enterococcal plasmid pAM401, demonstrating the functionality of this TA system [5]. This epidemiological survey was the first to define which TA systems are most prevalent in clinical isolates of pathogenic bacteria, suggesting these as a viable target for exploitation. Further examination of six axe-txe-positive VRE strains from this study revealed that axe-txe was transcribed in all cases, and physical linkage to the VanA resistance determinant was confirmed by DNA sequencing [53]. Another survey of plasmid DNA isolated from a collection of 93 geographically and epidemiologically diverse Enterococcus faecium strains revealed that 42 (45%) and 18 (19%) harbor genes for axe-txe and ω-ε-ζ, respectively [6]. A smaller study of VRE strains carrying VanB-type vancomycin resistance genes, each from different pulse-field gel electrophoresis (PFGE) types, showed that axe-txe was physically linked to the plasmid encoding vanB in eight of nine strains [54].
An additional study investigated the prevalence of TA systems in methicillin-resistant Staphylococcus aureus (MRSA) and P. aeruginosa. This survey demonstrated the ubiquity of mazEFSa in 78 MRSA clinical isolates, and higBAPa and relBEPa in 42 P. aeruginosa clinical isolates [55]. It was also shown that these TA systems are transcribed by their respective hosts, suggesting that they are functional units. Importantly, the PCR-based screen revealed that the parDEPa TA system was present in only 30% of the clinical isolates. Inspection of the three sequenced genomes of P. aeruginosa clinical isolates shows that parDEpa is present in PAO1 and PA7, but not PA14. Furthermore, genotyping of P. aeruginosa isolates using multi-locus variable number tandem repeat analysis (MLVA) revealed that the presence of parDEpa did not correlate with genome relatedness. Thus, the inconsistent presence of parDEPa suggests that activation of ParDEPa would not be a good candidate for a TA-based therapeutic strategy versus P. aeruginosa. A similar study revealed the conservation of relBE2Spn in 70 clinical isolates and 30 sequenced strains of Streptococcus pneumoniae [56].
A combination of factors will determine whether a given TA system is a good antibacterial target. Prevalence and functionality within clinical isolates are absolutely required. The aforementioned epidemiological studies showed that the genes for TA systems were present and transcribed; however, Western blot analysis using antibodies raised to the specific TA systems would lend further support for these protein targets. Additional points to consider revolve around the activity of the toxin itself. The toxin mode of action could influence both the toxicity of the toxin and the propensity for resistance to toxin-activating molecules to arise, leading to reduced efficacy of the strategy. For example, resistance to the toxin CcdB is conferred by a single point mutation within its target, DNA gyrase [57], suggesting limitations with CcdB activation strategy. However, it is more difficult to envision how resistance would arise to a toxin like MazF, which cleaves single-stranded mRNA [58]. Mutational inactivation of the toxin could occur, but presumably the cell would incur a fitness cost associated with such a mutation. While more data must be collected, based on prevalence, functionality, and mode-of-action, MazEF, RelBE, and HigBA appear to be reasonable targets for artificial toxin activation that could lead to bacterial cell death [5,55].
Efforts towards discovering toxin activators
Extracellular death factor
E. coli mazEF is one of the best characterized TA systems and has been implicated in cell stress responses and programmed cell death. A variety of stressors cause MazF-induced cell death, including short-term exposure to antibiotics that target transcription or translation [17], DNA damage due to thymine starvation [59], overproduction of ppGpp [60], and exposure to DNA damaging agents such as mitomycin C or nalidixic acid [13].
The Engelberg-Kulka group has published a series of papers in which they claim to have identified an endogenous peptidic activator of the MazF [9,61-63]. If confirmed, this would be a significant discovery and would lend considerable support to the notion that TA systems are exploitable antibacterial targets. However, as described below, this work is controversial and awaits independent validation. These studies began with the observation that the ability of mazEF to mediate cell death in response to stress was dependent on population size. Brief treatment of cells with rifampicin triggered mazEF-mediated cell death at densities of 3 × 108 or 3×107 cells/mL, but not when the same culture was diluted to 3 × 105 or 3 × 104 cells/mL [9]. Furthermore, transfer of supernatant from a dense culture to a dilute culture followed by a short treatment with rifampicin, chloramphenicol, or trimethoprim resulted in mazEF-dependent cell death. These results suggest that mazEF-dependent cell death requires an “extracellular death factor” (EDF). Subsequent isolation experiments identified EDF as a linear pentapeptide quorum-sensing molecule with the sequence NNWNN. Synthetic NNWNN also induced mazEF-mediated cell death in response to antibiotic stress [9].
The effect of EDF on MazF activity in vitro was assessed using a continuous fluorometric assay for MazF, and in this experiment EDF significantly enhanced the endoribonucleotlytic activity of MazF in both a concentration- and sequence-dependent manner [62]. The derivatives NNGNN and NWN gave no enhancement of MazF activity, whereas other residue substitutions, additions, and deletions were well-tolerated. Furthermore, when increasing concentrations of EDF were mixed with MazE and then added to MazF and fluorogenic substrate, the ability of MazE to inhibit MazF activity was diminished, indicating that EDF prevented the inhibition of MazF by MazE in vitro. A structural model suggests that EDF directly competes with the MazE71-75 sequence IDWGE by placing the EDF Trp3 in the hydrophobic MazF pocket that is typically occupied by MazE Trp73.
Given the experimental results, the authors proposed that the endoribonuclease MazF serves as a cytoplasmic sensor of EDF [9,61,62] and that EDF is required for mazEF-dependent cell death. A possible model for this phenomenon is shown in Figure 3. It is important to note that several laboratories have tried to replicate critical elements of the EDF experiments, but have been unsuccessful [8,29,52]. As such, it is too early to classify EDF as a bona fide TA system activator; a rigorous validation of the NNWNN peptide in various bacterial strains by multiple research groups will be required for the EDF phenomena to be widely accepted.
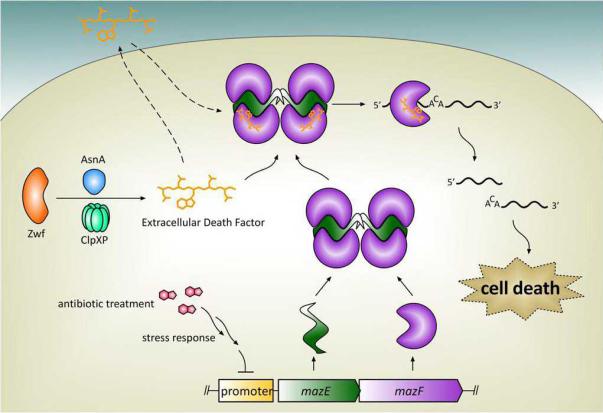
Figure 3.
A possible model for mazEF-dependent cell death induced by extracellular death factor (EDF). EDF production requires primarily the protein Zwf and the protease ClpXP. Preliminary investigation suggests that EDF freely diffuses in and out of the cell. Treatment with an antibiotic triggers the stress response, which inhibits transcription of mazEF. EDF binds to and enhances MazF ribounuclease activity in vitro, although it is not known whether EDF binds to free MazF or to the MazEF complex (as it is depicted here). MazF cleaves single-stranded mRNA in a sequence-specific manner, leading to cell death. It should be noted that there is controversy about EDF, and the role of EDF has not been independently confirmed by multiple laboratories.
Inhibitors of the PemI-PemK/MoxX-MoxT interaction
PemK, of the B. anthracis PemIK TA module, cleaves single-stranded RNAs and is inhibited by the binding of antitoxin PemI [64]. Analysis of PemI deletion variants indicated that the C-terminus is required to bind to PemK. Based on this information, six hepta- and octa-peptides, representing fragments of the antitoxin located in a predicted helical region within the TA binding interface, were analyzed for their ability to inhibit the PemI-PemK interaction [64]. ELISA results revealed that each designed peptide was capable of preventing the PemI-PemK interaction to a certain extent, whereas nonspecific 15- and 9-residue peptides based on the N-terminus of PemI did not affect the PemI-PemK interaction [64].
The authors then examined the effect of the peptides on PemK ribonuclease activity using a fluorogenic chimeric DNA-RNA substrate or a fluorogenic rC substrate [64]. The two peptides that prevented the PemI-PemK interaction to the greatest extent, LLFQHLTE (35% prevention) and RRGYIEMG (30% prevention), inhibited the PemK ribonuclease activity in vitro, while the remaining four peptides did not inhibit PemK ribonuclease activity. This result is not surprising, as one might expect a peptide fragment of the antitoxin to reduce the activity of the toxin. Recently, this B. anthracis TA system was re-named MoxXT [65]. A rationally designed octapeptide, SKIGAWAS, which has potential to form an α-helix and is predicted to occupy the binding interface between MoxX and MoxT, was shown to prevent the MoxXT complex formation in vitro by 42%. However, this peptide also partially inhibited the ribonuclease activity of MoxT [65]. These experiments are encouraging in that they demonstrate that TA interactions can be prevented by peptides; the next step is to design and identify peptides that prevent the protein-protein interaction without inhibiting the toxin enzymatic activity.
Disruptors of ε-ζ
A recent publication disclosed the results from a screen of peptides for disruption of the Streptococcus pyogenes plasmid-derived TA system called ε-ζ [66]. This screen utilized Luc-ε and ζ-GFP fusion proteins in a bioluminescence resonance energy transfer (BRET) assay. An extensive collection of various peptide libraries, including over 4.95 × 107 6-residue peptides, 2.74 × 104 14-residue β-sheet peptides, and 2.74 × 104 17-residue α-helix peptides were evaluated for their ability to disrupt the interaction between Luc-ε and ζK46A-GFP in a cell-free extract. Peptides were tested at both 0.6 μM and 7 μM. Hits were selected based on their ability to decrease the BRET signal relative to untreated controls. No hits were observed at either concentration with the 6- and 14-residue peptide libraries; however, two wells containing members of the 17-residue library decreased the BRET signal [66]. These peptide mixtures were not tested for their ability to activate or inhibit the ζ toxin. When the number of peptides from the sub-libraries that contained the positives tested was reduced, the decrease in BRET signal was lost [66]. Thus, the disruption of the Luc-ε–ζK46A-GFP complex was possibly due to more than one peptide with weak activity. While this could mean that the ε-ζ interaction can be disrupted, more investigation is required to confirm this finding and to determine whether the peptide(s) binds to the antitoxin or to the toxin and if toxin activity is affected in the process.
Challenges for the discovery of toxin activators
Artificial activation of TA systems is a potentially powerful antibacterial strategy. However, the three examples discussed above are the state-of-the art for TA activation with a drug-like compound, thus as of yet there is no molecule convincingly capable of modulating the TA interaction. Such compounds are needed to fully explore the potential of TA disruption and toxin activation as an antibacterial strategy. We have identified five key questions regarding the discovery of an artificial toxin activator.
First, will the strength of the TA interaction preclude disruption with a peptide or small molecule? Most toxin-antitoxin pairs have strong affinities, mediated by extensive electrostatic and hydrophobic interactions [67], resulting in KD values on the order of 1 nM [68,69]. In most cases, the antitoxin wraps around the toxin to form the inactive complex. In contrast, interactions between other protein-protein pairs that have been successfully inhibited are characterized by long, shallow pockets that are accessible to small molecules [70,71]. Perhaps not surprisingly, in all the three examples presented above, peptides were evaluated as toxin-antitoxin disruptors; a small molecule TA modulator has yet to be discovered.
Confounding this issue is the relatively limited information on the specifics governing the TA interaction. Crystal structures have been solved for some TA systems including S. pyogenes ε2-ζ2 [72], and E. coli MazEF [67], and there is a solution structure of E. coli RelBE [73]. However, although there is considerable homology between toxins of the same family and even of different families, the sequence and secondary structure of antitoxins are much more divergent; thus, more structural data is needed. Additionally, minimal data is available regarding the amino acid residues that define the ‘hotspots’ between toxin and antitoxin. Defining these hotspots and minimal TA binding regions through mutational analysis will facilitate the design of molecules specifically targeting these interactions
Second, can the limitations in current assays for toxin activity be overcome? The process of searching for toxin activators is hindered by significant limitations in current in vitro enzymatic assays for toxin activity. Efficient screening of potential activators requires a robust assay, such as the continuous fluorometric assay developed to monitor MazF ribonuclease activity [74]. This assay, utilized in two separate examples in this review, allows for high-throughput analysis of molecules that modulate the enzymatic activity of toxins that cleave free mRNA, such as MazF and PemK. In contrast, toxins such as the ribosome-dependent ribonuclease RelE must be evaluated by reconstituting actively translating ribosomes, which requires a fairly complicated in vitro assay not easily amenable to high-throughput screening. The development of high-throughput methods of assessing ribosome-dependent ribonuclease activity would facilitate the discovery of activators of RelE and HigB.
Additionally, in vitro assays lack key components that exist within the cell, namely the proteases that naturally relieve antitoxin inhibition of the toxin. Thus, current in vitro assays are unable to discover compounds that work through an indirect mechanism. For example, a molecule could increase the proteolytic susceptibility of the antitoxin or modulate TA expression. Discovery of a molecule that acts by one of these mechanisms would best be achieved using cell-based assays.
Third, should the toxin or the antitoxin serve as the target? The examples presented in this review do not give a unified answer. On one hand, EDF (suggested to bind MazF by mimicking a region of the MazE antitoxin) is claimed to both enhance MazF activity and prevent MazE inhibition by directly binding to MazF [62]. On the other hand, peptide fragments of the antitoxin MoxX bind to and inhibit the ribonuclease activity of the toxin MoxT [64,65]. Different antitoxin fragments can have varying effects on the toxin; obviously, if binders to the toxin are developed, they should activate, rather than inhibit. A compound that binds the antitoxin and modulates its interaction with the toxin is most desirable; however, the intrinsically disordered structure of the antitoxin makes it a challenging target for in vitro screens. Nevertheless, a promising new class of molecules that specifically target intrinsically disordered peptides is being developed [75].
On a related note, is full TA disruption is necessary, or is activation of the toxin sufficient for an antibacterial effect? EDF is purported to prevent TA complex formation, but was not shown to activate MazF from the pre-formed MazE-MazF complex [62]. Similarly, the peptide fragments of MoxX prevented complex formation between MoxX and MoxT but were not shown to disrupt the interaction between MoxX and MoxT in a pre-formed complex [64,65]. Is this significant in the cell where the antitoxin is subject to metabolic turnover?
Fourth, will a toxin-activating compound kill bacteria as a single-entity agent? Toxin-mediated cell death is typically studied in response to some outside stimulus, such as amino acid starvation or treatment with an antibiotic. Despite the ability of EDF to enhance MazF activity in vitro, it is unable to induce MazF-dependent cell death on its own, and requires prior activation of the mazEF module via an antibiotic such as rifampicin or chloramphenicol [9,61].
Finally, how does the location of the TA genes (plasmid or chromosomal) influence the TA-targeting strategy? It seems clear that artificial activation of TA proteins encoded on plasmids would kill the cell in a manner analogous to antitoxin degradation, toxin activation, and cell death in post-segregational killing though TA plasmid stabilization mechanisms. When the TA genes are chromosomally-encoded, however, the copy number of resulting TA proteins may not be high enough to induce death after toxin activation. Furthermore, chromosomal TA systems have been implicated in the formation of persister cells (Box 2), which some believe may contribute to chronic infection. There is concern that artificial activation of chromosomally-encoded toxins could potentially induce persister cell formation, an obviously undesirable effect.
Concluding remarks
TA systems present exciting opportunities for the development of novel antibacterial agents. The first requirement, demonstration of the ubiquity and functionality of TA systems in clinical isolates, has been satisfied for key pathogens, including VRE, MRSA and P. aeruginosa and S. pneumoniae. Additional epidemiological surveys and biochemical analyses of toxins will add to the catalog of potential TA systems to target. Although potentially significant, the Extracellular Death Factor story needs further clarification and independent validation. Preliminary work on the disruption of the ε-ζ complex and prevention of the MoxXT complex suggests that peptides can indeed be used to modulate the TA system interaction. Development of a toxin activator and extension of the TA activation strategy to in vivo studies are required to fully assess the potential of this strategy.
Box 2. Persister cell induction as a caveat for TA-targeting antibacterials.
Persister cells are defined as a small fraction of a bacterial population that tolerate antibiotics not by mutation or acquisition of resistance determinants but by entering a state of dormancy [30]. Further culturing of these dormant cells restores normal growth, and subsequent application of antibiotic selects for a new sub-population of persister cells [76]. One model proposes that persisters arise when a small fraction of cells in a mid-exponential phase experience stochastic changes in gene expression, producing individual dormant cells that are recalcitrant to subsequent antibiotic treatment [77,78]. This phenotypic switch has been shown to be induced by activation of chromosomally-encoded toxin genes [25]. Additionally, upregulation of the transcripts for TA genes have been observed in persister cells [77], and the occurrence of persister cells progressively diminished with successive deletion of the ten ribonuclease-encoding toxin genes in E. coli [25]. A separate model proposes that the SOS response induces persister cell formation via the Type I TA system tisAB/istR. Treatment with fluoroquinolones induces the SOS response, causing transcription of the LexA-controlled tisB, which encodes a membrane acting toxin [79]. TisB decreases the proton motive force, which leads to decreased ATP levels and a state of dormancy [80]. Although additional work is required to describe the exact mechanism of persister cell formation and resuscitation, it will be important to determine whether artificial toxin activators induce persister cell formation as this may contribute to chronic infections [30].
Table I.
The seven toxin families
| Activity-based toxin family | Activity of toxin | Cellular process inhibited |
|---|---|---|
| CcdB, ParE | DNA-gyrase complex poison | Replication |
| MazF, HicA | Ribosome-independent mRNA interferases | Translation |
| RelE, HigB | Ribosome-dependent mRNA interferases | Translation |
| ζ | Phospohorylates UDP-Glc-NAc* | Peptidoglycan synthesis |
| HipA | Phosphorylates EF-Tu† | Translation |
| Doc | Binds 30S ribosomal subunit | Translation |
| VapC | Cleaves tRNAfMet | Translation |
UDP-Glc-NAC: Uridine diphosphate-N-acetylglucosamine
EF-Tu: Elongation factor
Acknowledgements
We thank the National Institutes of Health (R01GM068385) for support of our research. JJW was partially supported by NIH Cell and Molecular Biology Training grant T32 GM007283.
References
- 1.Gerdes K, et al. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci U S A. 1986;83(10):3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogura T, Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci U S A. 1983;80(15):4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsuchimoto S, et al. Two genes, pemK and pemI, responsible for stable maintenance of resistance plasmid R100. J Bacteriol. 1988;170(4):1461–1466. doi: 10.1128/jb.170.4.1461-1466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherny I, et al. Structural and thermodynamic characterization of the Escherichia coli RelBE toxin-antitoxin system: indication for a functional role of differential stability. Biochemistry. 2007;46(43):12152–12163. doi: 10.1021/bi701037e. [DOI] [PubMed] [Google Scholar]
- 5.Moritz EM, Hergenrother PJ. Toxin-antitoxin systems are ubiquitous and plasmid-encoded in vancomycin-resistant enterococci. Proc Natl Acad Sci U S A. 2007;104(1):311–316. doi: 10.1073/pnas.0601168104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosvoll TC, et al. PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501- and pHTbeta-related replicons associated with glycopeptide resistance and stabilizing toxin-antitoxin systems. FEMS Immunol Med Microbiol. 2009;58(2):254–268. doi: 10.1111/j.1574-695X.2009.00633.x. [DOI] [PubMed] [Google Scholar]
- 7.Magnuson RD. Hypothetical functions of toxin-antitoxin systems. J Bacteriol. 2007;189(17):6089–6092. doi: 10.1128/JB.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Melderen L. Toxin-antitoxin systems: why so many, what for? Curr Opin Microbiol. 2010;13(6):781–785. doi: 10.1016/j.mib.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Kolodkin-Gal I, et al. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science. 2007;318(5850):652–655. doi: 10.1126/science.1147248. [DOI] [PubMed] [Google Scholar]
- 10.Tsilibaris V, et al. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J Bacteriol. 2007;189(17):6101–6108. doi: 10.1128/JB.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen SK, et al. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci U S A. 2001;98(25):14328–14333. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pedersen K, et al. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol. 2002;45(2):501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- 13.Hazan R, et al. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J Bacteriol. 2004;186(11):3663–3669. doi: 10.1128/JB.186.11.3663-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donegan NP, Cheung AL. Regulation of the mazEF toxin-antitoxin module in Staphylococcus aureus and its impact on sigB expression. J Bacteriol. 2009;191(8):2795–2805. doi: 10.1128/JB.01713-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan J, et al. The three Vibrio cholerae chromosome II-encoded ParE toxins degrade chromosome I following loss of chromosome II. J Bacteriol. 2011;193(3):611–619. doi: 10.1128/JB.01185-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wozniak RA, Waldor MK. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 2009;5(3):e1000439. doi: 10.1371/journal.pgen.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sat B, et al. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J Bacteriol. 2001;183(6):2041–2045. doi: 10.1128/JB.183.6.2041-2045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nariya H, Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132(1):55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y, et al. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J Bacteriol. 2009;191(4):1258–1267. doi: 10.1128/JB.01465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat Chem Biol. 2011;7(6):359–366. doi: 10.1038/nchembio.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazan R, Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol Genet Genomics. 2004;272(2):227–234. doi: 10.1007/s00438-004-1048-y. [DOI] [PubMed] [Google Scholar]
- 22.Fineran PC, et al. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci U S A. 2009;106(3):894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown JS, et al. A locus contained within a variable region of pneumococcal pathogenicity island 1 contributes to virulence in mice. Infect Immun. 2004;72(3):1587–1593. doi: 10.1128/IAI.72.3.1587-1593.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y, Wood TK. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem Biophys Res Commun. 2010;391(1):209–213. doi: 10.1016/j.bbrc.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maisonneuve E, et al. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci U S A. 2011;108(32):13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Amitai S, et al. Escherichia coli MazF leads to the simultaneous selective synthesis of both "death proteins" and "survival proteins". PLoS Genet. 2009;5(3):e1000390. doi: 10.1371/journal.pgen.1000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamaguchi Y, Inouye M. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat Rev Microbiol. 2011;9(11):779–790. doi: 10.1038/nrmicro2651. [DOI] [PubMed] [Google Scholar]
- 28.Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301(5639):1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 29.Hayes F, Van Melderen L. Toxins-antitoxins: diversity, evolution and function. Crit Rev Biochem Mol Biol. 2011;46(5):386–408. doi: 10.3109/10409238.2011.600437. [DOI] [PubMed] [Google Scholar]
- 30.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 31.Gerdes K, et al. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3(5):371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Wood TK. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol. 2011;77(16):5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diaz-Orejas R, et al. Bacterial toxin-antitoxin systems targeting translation. Journal of Applied Biomedicine. 2010;8(4):179–188. [Google Scholar]
- 34.Kamphuis MB, et al. Structure and function of bacterial kid-kis and related toxin-antitoxin systems. Protein Pept Lett. 2007;14(2):113–124. doi: 10.2174/092986607779816096. [DOI] [PubMed] [Google Scholar]
- 35.Blower TR, et al. Balancing at survival's edge: the structure and adaptive benefits of prokaryotic toxin-antitoxin partners. Curr Opin Struct Biol. 2011;21(1):109–118. doi: 10.1016/j.sbi.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 36.Engelberg-Kulka H, et al. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2(10):e135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engelberg-Kulka H, et al. Bacterial programmed cell death systems as targets for antibiotics. Trends Microbiol. 2004;12(2):66–71. doi: 10.1016/j.tim.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 38.Stieber D, et al. The art of selective killing: plasmid toxin/antitoxin systems and their technological applications. Biotechniques. 2008;45(3):344–346. doi: 10.2144/000112955. [DOI] [PubMed] [Google Scholar]
- 39.Bukowski M, et al. Prokaryotic toxin-antitoxin systems--the role in bacterial physiology and application in molecular biology. Acta Biochim Pol. 2011;58(1):1–9. [PubMed] [Google Scholar]
- 40.Mutschler H, Meinhart A. epsilon/zeta systems: their role in resistance, virulence, and their potential for antibiotic development. J Mol Med (Berl) 2011 doi: 10.1007/s00109-011-0797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeNap JC, Hergenrother PJ. Bacterial death comes full circle: targeting plasmid replication in drug-resistant bacteria. Org Biomol Chem. 2005;3(6):959–966. doi: 10.1039/b500182j. [DOI] [PubMed] [Google Scholar]
- 42.Engelberg-Kulka H, et al. mazEF: a chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J Cell Sci. 2005;118:4327–4332. doi: 10.1242/jcs.02619. Pt 19. [DOI] [PubMed] [Google Scholar]
- 43.Williams JJ, Hergenrother PJ. Exposing plasmids as the Achilles' heel of drug-resistant bacteria. Curr Opin Chem Biol. 2008;12(4):389–399. doi: 10.1016/j.cbpa.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takagi H, et al. Crystal structure of archaeal toxin-antitoxin RelE-RelB complex with implications for toxin activity and antitoxin effects. Nat Struct Mol Biol. 2005;12(4):327–331. doi: 10.1038/nsmb911. [DOI] [PubMed] [Google Scholar]
- 45.Raskatov JA, et al. Modulation of NF-kappaB-dependent gene transcription using programmable DNA minor groove binders. Proc Natl Acad Sci U S A. 2012;109(4):1023–1028. doi: 10.1073/pnas.1118506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sass P, et al. Antibiotic acyldepsipeptides activate ClpP peptidase to degrade the cell division protein FtsZ. Proc Natl Acad Sci U S A. 2011;108(42):17474–17479. doi: 10.1073/pnas.1110385108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leung E, et al. Activators of cylindrical proteases as antimicrobials: identification and development of small molecule activators of ClpP protease. Chem Biol. 2011;18(9):1167–1178. doi: 10.1016/j.chembiol.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 48.Christensen SK, et al. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol Microbiol. 2004;51(6):1705–1717. doi: 10.1046/j.1365-2958.2003.03941.x. [DOI] [PubMed] [Google Scholar]
- 49.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33(3):966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao Y, et al. TADB: a web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011;39:D606–611. doi: 10.1093/nar/gkq908. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leplae R, et al. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011;39(13):5513–5525. doi: 10.1093/nar/gkr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guglielmini J, Van Melderen L. Bacterial toxin-antitoxin systems: Translation inhibitors everywhere. Mobile Genetic Elements. 2011;1(4):1–8. doi: 10.4161/mge.18477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Halvorsen EM, et al. Txe, an endoribonuclease of the enterococcal Axe-Txe toxin-antitoxin system, cleaves mRNA and inhibits protein synthesis. Microbiology. 2011;157:387–397. doi: 10.1099/mic.0.045492-0. Pt 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bjorkeng E, et al. Clustering of polyclonal VanB-type vancomycin-resistant Enterococcus faecium in a low-endemic area was associated with CC17-genogroup strains harbouring transferable vanB2-Tn5382 and pRUM-like repA containing plasmids with axe-txe plasmid addiction systems. APMIS. 2011;119(4-5):247–258. doi: 10.1111/j.1600-0463.2011.02724.x. [DOI] [PubMed] [Google Scholar]
- 55.Williams JJ, et al. Toxin-antitoxin (TA) systems are prevalent and transcribed in clinical isolates of Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett. 2011;322(1):41–50. doi: 10.1111/j.1574-6968.2011.02330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nieto C, et al. The relBE2Spn toxin-antitoxin system of Streptococcus pneumoniae: role in antibiotic tolerance and functional conservation in clinical isolates. PLoS One. 2010;5(6):e11289. doi: 10.1371/journal.pone.0011289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernard P, Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol. 1992;226(3):735–745. doi: 10.1016/0022-2836(92)90629-x. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, et al. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell. 2003;12(4):913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 59.Sat B, et al. The Escherichia coli mazEF suicide module mediates thymineless death. J Bacteriol. 2003;185(6):1803–1807. doi: 10.1128/JB.185.6.1803-1807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aizenman E, et al. An Escherichia coli chromosomal "addiction module" regulated by guanosine [corrected] 3',5'-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci U S A. 1996;93(12):6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kolodkin-Gal I, Engelberg-Kulka H. The extracellular death factor: physiological and genetic factors influencing its production and response in Escherichia coli. J Bacteriol. 2008;190(9):3169–3175. doi: 10.1128/JB.01918-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belitsky M, et al. The Escherichia coli extracellular death factor EDF induces the endoribonucleolytic activities of the toxins MazF and ChpBK. Mol Cell. 2011;41(6):625–635. doi: 10.1016/j.molcel.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 63.Kolodkin-Gal I, et al. The communication factor EDF and the toxin-antitoxin module mazEF determine the mode of action of antibiotics. PLoS Biol. 2008;6(12):e319. doi: 10.1371/journal.pbio.0060319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agarwal S, et al. PemK toxin of Bacillus anthracis is a ribonuclease: an insight into its active site, structure, and function. J Biol Chem. 2010;285(10):7254–7270. doi: 10.1074/jbc.M109.073387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chopra N, et al. Modeling of the structure and interactions of the B. anthracis antitoxin, MoxX: deletion mutant studies highlight its modular structure and repressor function. J Comput Aided Mol Des. 2011;25(3):275–291. doi: 10.1007/s10822-011-9419-z. [DOI] [PubMed] [Google Scholar]
- 66.Lioy VS, et al. A toxin-antitoxin module as a target for antimicrobial development. Plasmid. 2009 doi: 10.1016/j.plasmid.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 67.Kamada K, et al. Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol Cell. 2003;11(4):875–884. doi: 10.1016/s1097-2765(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 68.Overgaard M, et al. RelB and RelE of Escherichia coli form a tight complex that represses transcription via the ribbon-helix-helix motif in RelB. J Mol Biol. 2009;394(2):183–196. doi: 10.1016/j.jmb.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li GY, et al. Characterization of dual substrate binding sites in the homodimeric structure of Escherichia coli mRNA interferase MazF. J Mol Biol. 2006;357(1):139–150. doi: 10.1016/j.jmb.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 70.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450(7172):1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 71.Heeres JT, et al. Identifying modulators of protein-protein interactions using photonic crystal biosensors. J Am Chem Soc. 2009;131(51):18202–18203. doi: 10.1021/ja907066r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meinhart A, et al. Crystal structure of the plasmid maintenance system epsilon/zeta: functional mechanism of toxin zeta and inactivation by epsilon 2 zeta 2 complex formation. Proc Natl Acad Sci U S A. 2003;100(4):1661–1666. doi: 10.1073/pnas.0434325100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li GY, et al. Inhibitory mechanism of Escherichia coli RelE-RelB toxin-antitoxin module involves a helix displacement near an mRNA interferase active site. J Biol Chem. 2009;284(21):14628–14636. doi: 10.1074/jbc.M809656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang NR, Hergenrother PJ. A continuous fluorometric assay for the assessment of MazF ribonuclease activity. Anal Biochem. 2007;371(2):173–183. doi: 10.1016/j.ab.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Metallo SJ. Intrinsically disordered proteins are potential drug targets. Curr Opin Chem Biol. 2010;14(4):481–488. doi: 10.1016/j.cbpa.2010.06.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keren I, et al. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230(1):13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 77.Shah D, et al. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Balaban NQ, et al. Bacterial persistence as a phenotypic switch. Science. 2004;305(5690):1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 79.Dorr T, et al. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8(2):e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Unoson C, Wagner EG. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol Microbiol. 2008;70(1):258–270. doi: 10.1111/j.1365-2958.2008.06416.x. [DOI] [PubMed] [Google Scholar]