Abstract
Biomarkers are the only feasible way to detect and monitor presymptomatic Alzheimer's disease (AD). No single biomarker can predict future cognitive decline with an acceptable level of accuracy. In addition to designing powerful multimodal diagnostic platforms, a careful investigation of the major sources of disease heterogeneity and their influence on biomarker changes is needed. Here we investigated the accuracy of a novel multimodal biomarker classifier for differentiating cognitively normal (NC), mild cognitive impairment (MCI) and AD subjects with and without stratification by ApoE4 genotype. 111 NC, 182 MCI and 95 AD ADNI participants provided both structural MRI and CSF data at baseline. We used an automated machine-learning classifier to test the ability of hippocampal volume and CSF Aβ, t-tau and p-tau levels, both separately and in combination, to differentiate NC, MCI and AD subjects, and predict conversion. We hypothesized that the combined hippocampal/CSF biomarker classifier model would achieve the highest accuracy in differentiating between the three diagnostic groups and that ApoE4 genotype will affect both diagnostic accuracy and biomarker selection. The combined hippocampal/CSF classifier performed better than hippocampus-only classifier in differentiating NC from MCI and NC from AD. It also outperformed the CSF-only classifier in differentiating NC vs. AD. Our amyloid marker played a role in discriminating NC from MCI or AD but not for MCI vs. AD. Neurodegenerative markers contributed to accurate discrimination of AD from NC and MCI but not NC from MCI. Classifiers predicting MCI conversion performed well only after ApoE4 stratification. Hippocampal volume and sex achieved AUC = 0.68 for predicting conversion in the ApoE4-positive MCI, while CSF p-tau, education and sex achieved AUC = 0.89 for predicting conversion in ApoE4-negative MCI. These observations support the proposed biomarker trajectory in AD, which postulates that amyloid markers become abnormal early in the disease course while markers of neurodegeneration become abnormal later in the disease course and suggests that ApoE4 could be at least partially responsible for some of the observed disease heterogeneity.
Abbreviations: Aβ, Amyloid beta; Aβ42, Amyloid beta with 42 amino acid residues; AD, Alzheimer's disease; ADNI, Alzheimer's Disease Neuroimaging Initiative; ApoE, apolipoprotein E; AUC, area under the curve; CSF, cerebrospinal fluid; ICBM, International Consortium for Brain Mapping; MCI, mild cognitive impairment; MCIc, MCI converters; MCInc, MCI nonconverters; MMSE, Mini-Mental State Examination; NC, normal control; ROC, receiver operating curve; SVM, support vector machine; t-tau, total tau protein; p-tau, phosphorylated tau protein
Keywords: Alzheimer's disease, Abeta, Tau, Hippocampus atrophy, ADNI, Diagnosis
Highlights
-
•
Multimodal classifiers have better predictive power than unimodal classifier.
-
•
ApoE4 significantly affects diagnostic discriminability in the MCI and dementia stages.
-
•
Our data supports the hypothesized biomarker trajectory in AD.
1. Introduction
Alzheimer's disease (AD), the most common neurodegenerative disorder, is increasingly prevalent among those aged 65 years and over. AD prevalence is projected to triple by the year 2050 (Hebert et al., 2001) making it vital to achieve early and accurate diagnosis and to discover disease-modifying therapies. The only feasible approach for presymptomatic diagnosis to date is through the use of biomarkers.
Hippocampal atrophy is the most established AD structural imaging biomarker. Hippocampal atrophy is seen in normal aging, yet in the latent AD stages hippocampal atrophy becomes greatly accelerated (Apostolova et al., 2006, Apostolova et al., in press, Apostolova et al., 2010a, Jack et al., 1997, Jack et al., 1998, Jack et al., 2000). Hippocampal atrophy shows strong correlation with cognitive decline (de Toledo-Morrell et al., 2000, Fleischman et al., 2005, Mortimer et al., 2004) and with AD pathologic markers such as neuronal and neurofibrillary tangle counts and Braak and Braak pathological stages (Apostolova et al., 2010b, Bobinski et al., 1995, Bobinski et al., 1997, Schonheit et al., 2004, Zarow et al., 2005).
Cerebrospinal fluid (CSF) measures of amyloid beta protein (Aβ) and tau are the most established AD fluid biomarkers. Pathologic Aβ deposition in the brain tissue is thought to occur early in the disease course and is associated with low CSF Aβ42 levels (Blennow and Hampel, 2003). CSF total tau (t-tau) and phosphorylated tau (p-tau) are significantly elevated in subjects with AD (Andreasen et al., 2001, Blennow et al., 1995, Clark et al., 2003, Galasko et al., 1998) and are thought to reflect neurodegeneration of tau-containing neurons. Unlike CSF Aβ42 (Wallin et al., 2006), CSF t-tau and p-tau changes occur later in the disease course and are associated with cognitive decline (Buerger et al., 2002, Buerger et al., 2005, Riemenschneider et al., 2002, Wallin et al., 2006).
Several research groups have independently investigated the individual accuracy of these biomarkers to differentiate cognitively normal elderly, MCI and AD subjects (Andreasen et al., 2001, Brys et al., 2009, de Leon et al., 2006, Frisoni et al., 2009, Galasko et al., 1998, Hampel et al., 2004, Mattsson et al., 2009, Shaw et al., 2009). Yet while biomarker changes are clearly present years before AD is diagnosed, no single biomarker can adequately predict conversion to AD or serve as a diagnostic tool with an acceptable level of accuracy.
Many groups including ours have made important strides towards multimodal biomarker diagnostic discrimination (Cui et al., 2011, Davatzikos et al., 2011, Ewers et al., 2012, Kohannim et al., 2010, Vos et al., 2012, Walhovd et al., 2010, Westman et al., 2012). Here we combined imaging and CSF biomarker data from the Alzheimer's Disease Neuroimaging Initiative-1 (ADNI-1) dataset to compare the performance of unimodal (hippocampal atrophy or CSF biomarkers alone) and multimodal (hippocampal atrophy, CSF variables and ApoE genotype combined) biomarker classifiers for differentiating NC, MCI and AD. We also examined to what extent the amyloid (i.e., CSF Aβ42) and the neurodegenerative biomarkers (CSF tau, CSF p-tau and hippocampal atrophy) play a role for differentiating different disease stages from each other and for predestining conversion to AD. In addition we sought to determine the effect of ApoE4 genotype on diagnostic accuracy and biomarker selection.
2. Materials and methods
2.1. Subjects
Data used preparing this article were obtained from the ADNI database (www.loni.ucla.edu/ADNI). ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations; subjects have been recruited from over 50 sites across the U.S. and Canada. For up-to-date information, please see www.adni-info.org.
ADNI-1 enrolled approximately 400 amnestic MCI, 200 mild AD and 200 NC subjects, aged 55–90. Written informed consent was obtained from all participants. The clinical description of the ADNI-1 cohort was recently published (Petersen et al., 2010). The full list of inclusion/exclusion criteria may be accessed online at http://www.adni-info.org/Scientists/ADNIGrant.aspx.
As all ADNI subjects had serial 1.5T MRI images, their inclusion in our analyses was largely determined by the availability of CSF data. CSF measures were performed in only a subset of the ADNI subjects. 111 NC, 182 MCI and 95 AD ADNI participants had both structural MRI and CSF assessment at baseline. 191 subjects (49%; 27 NC, 99 MCI, 65 AD) were apolipoprotein E4 (ApoE4) carriers and 197 (51%; 84 NC, 83 MCI, 30 AD) were noncarriers. Additionally 21 NC, 14 MCI and 3 AD subjects were ApoE2 carriers.
As one of the main goals of ADNI is to carefully track biomarker changes in NC and MCI predestined to convert to AD, we also analyzed which combination of biomarkers can predict conversion to AD. We used all available MCI subjects who had either converted to AD at any point between baseline and month 36 (MCI converters, N = 80) or remained stable all the way to month 36 (MCI nonconverters, N = 80). 22 MCI subjects dropped out before month 36 without converting to AD and were excluded from our analyses.
2.2. CSF biomarker data
We downloaded the baseline CSF Aβ42, t-tau and the tau phosphorylated at threonine at position 181 (p-tau181) data from the ADNI website (http://www.loni.ucla.edu/ADNI) in October 2008. CSF collection and transportation protocols, and procedural details on CSF Aβ42, t-tau and the p-tau181 measurements are provided in the ADNI procedural manual posted at http://www.adni-info.org and in a recent publication by Shaw et al. (Shaw et al., 2009).
2.3. MRI preprocessing
All subjects were scanned with a standardized high-resolution MRI protocol (http://www.loni.ucla.edu/ADNI/Research/Cores/index.shtml) on scanners developed by one of three manufacturers (General Electric Healthcare, Siemens Medical Solutions and Philips Medical Systems) with a protocol optimized for best contrast to noise in a feasible acquisition time (Jack et al., 2008, Leow et al., 2006). Raw data with an acquisition matrix of 192 × 192 × 166 and voxel size 1.25 × 1.25 × 1.2 mm3 in the x-, y-, and z-dimensions was subjected to in-plane, zero-filled reconstruction (i.e., sinc interpolation) resulting in a 256 × 256 matrix and voxel size of 0.9375 × 0.9375 × 1.2 mm3. Image quality was inspected at the ADNI MRI quality control center at the Mayo Clinic (in Rochester, MN, USA) (Jack et al., 2008). Phantom-based geometric corrections, image non-uniformity and bias field corrections were applied (Gunter et al., 2006, Jack et al., 2008, Jovicich et al., 2006, Sled et al., 1998). Both the uncorrected and corrected image files are freely available to interested researchers at http://www.loni.ucla.edu/ADNI.
2.4. Hippocampal segmentation
The preprocessed baseline 1.5 T 3D T1-weighted scans were downloaded and linearly registered to the International Consortium for Brain Mapping (ICBM-53) brain template (Mazziotta et al., 2001) using the Minctracc algorithm and 9-parameter (9P) transformation (3 translations, 3 rotations, 3 scales) (Collins et al., 1994). The aligned images were resampled in an isotropic space of 220 voxels along each axis (x, y, and z) resulting in a final voxel size of 1 mm3. The hippocampi were segmented with our recently developed and validated automated machine-learning hippocampal segmentation technique (AdaBoost) which uses the adaptive boosting approach originally proposed by Freund and Shapire (1997) as previously described (Apostolova et al., 2010d, Morra et al., 2008a, Morra et al., 2008b, Morra et al., 2009a, Morra et al., 2009b). Hippocampal volumes were extracted.
2.5. Statistical methods
2.5.1. Demographic comparisons
We used one-way Analyses of Variance (ANOVA) with post hoc Bonferroni correction for multiple comparisons to examine diagnostic differences in age, education, MMSE, CSF biomarker levels and hippocampal volume at baseline, and chi-squared test to determine differences in sex distribution between each diagnostic group. For comparison of baseline demographic and biomarker measures between ApoE4-positive and negative subjects and MCI converters (MCIc) and MCI nonconverters (MCInc) we used a two-tailed Student's t-test for continuous variables, and a chi-squared test for categorical variables.
2.5.2. Support vector machines classifier
SVMs are popular machine learning algorithms, formulated to learn patterns in training data and classify new testing data. The mathematical principle by which SVM performs pattern recognition is by finding a multidimensional plane that maximizes the margin between data points in different classes (Vapnik, 1995). SVMs have been particularly successful in biological classification problems, as non-linear kernels can be introduced to the algorithm so that non-planar, multidimensional surfaces can instead be used to classify patterns of data. In our study, we implemented the radial basis function (RBF) kernel and optimized its width or γ parameter (as well as the SVM cost or C parameter) through grid search using the e1071 package (Dimitriadou et al., 2006) in R (http://cran.r-project.org). Often, addition of non-contributory features can reduce classification performance. For this reason, we ranked our features based on the elements of a linear SVM's normal vector (i.e., |wi|; (Guyon et al., 2002)) and iteratively removed those with lower weights to find sets of features that yield maximal classification accuracies.
We trained the SVM algorithm with CSF, ApoE4 and imaging measures for subjects with known diagnoses and used the leave-one-out approach to predict each new subject's diagnostic category. This process was repeated n times and the machine's predictive accuracy was measured by summing up the correct and incorrect classifications. All classifiers included age, sex, and educational level (in years). Next, we obtained receiver operating characteristic (ROC) curves for the predictions to additionally assess the classifier's sensitivity, specificity and area under the curve (AUC) characteristics.
Our classifier results were further subjected to multiple comparisons correction by permutation analyses. We ran 10,000 permutations of the dependent variable (clinical diagnosis) against the sets of individual biomarker characteristics for each individual classifier and defined a final single corrected p-value for each ROC.
3. Results
3.1. Demographic characteristics
The demographic characteristics of the diagnostic groups are shown in Table 1, Table 2, Table 3. There were no significant age or educational differences between the diagnostic groups. The MCI group had significantly more males relative to both the NC and the AD groups (p = 0.023). As expected, AD subjects had the lowest mean MMSE and CSF Aβ42 and NC the highest (p < 0.001). The opposite was seen for CSF t-tau and p-tau (both p < 0.001). The MCI group was intermediate on these variables (Table 1).
Table 1.
Mean demographic and biomarker data.
| Variable at baseline | NC N = 111 | MCI N = 182 | AD N = 95 | One-way ANOVA/chi squared test, p-value |
|---|---|---|---|---|
| Age, years | 75.5 (5.2) | 74.2 (7.4) | 74.6 (7.9) | 0.3 |
| Gender, M:F | 56:55 | 121:61 | 55:40 | 0.023 |
| Education, years | 15.8 (2.8) | 15.8 (3.0) | 15.3 (3.0) | 0.3 |
| MMSE | 29.1 (1.0) | 26.9 (1.8) | 23.6 (1.9) | < 0.001 |
| CSF Aβ42 level, pg/ml | 206 (55) | 163 (55) | 143 (40) | < 0.001 |
| CSF t-tau level, pg/ml | 69 (30) | 103 (61) | 124 (58) | < 0.001 |
| CSF p-tau181 level, pg/ml | 25 (15) | 35 (18) | 43 (20) | < 0.001 |
| Mean hippocampal volume, mm3 | 4100 (586) | 3779 (631) | 3518 (604) | < 0.001 |
| ApoE4 positive subjects | ||||
| Variable at baseline | NC N = 27 | MCI N = 99 | AD N = 65 | One-way ANOVA/chi squared test, p-value |
| Age, years | 75.8 (5.8) | 73.5 (6.6) | 74.0 (7.4) | 0.3 |
| Gender, M:F | 18:9 | 60:39 | 39:26 | 0.8 |
| Education, years | 15.6 (2.8) | 15.7 (2.8) | 14.8 (3.0) | 0.2 |
| MMSE | 28.9 (1.1) | 27.0 (1.8) | 23.6 (1.9) | < 0.001 |
| CSF Aβ42 level, pg/ml | 157(49) | 143 (41) | 131 (27) | 0.012 |
| CSF t-tau level, pg/ml | 80 (40) | 117 (67) | 122 (53) | 0.007 |
| CSF p-tau181 level, pg/ml | 32 (21) | 40 (18) | 43 (19) | 0.05 |
| Mean hippocampal volume, mm3 | 4175 (443) | 3708 (608) | 3476 (608) | < 0.001 |
| ApoE4 negative subjects | ||||
| Variable at baseline | NC N = 84 | MCI N = 83 | AD N = 30 | One-way ANOVA/chi squared test, p-value |
| Age, years | 75.4 (5.0) | 74.9 (8.2) | 75.9 (8.9) | 0.8 |
| Gender, M:F | 38:46 | 61:22 | 14:16 | 0.001 |
| Education, years | 15.8 (2.7) | 16.0 (3.2) | 16.3 (2.8) | 0.7 |
| MMSE | 29.1 (1.0) | 26.8 (1.8) | 23.5 (1.9) | < 0.001 |
| CSF Aβ42 level, pg/ml | 222 (48) | 187 (60) | 168 (52) | < 0.001 |
| CSF t-tau level, pg/ml | 65 (25) | 85 (48) | 127 (69) | < 0.001 |
| CSF p-tau181 level, pg/ml | 22 (11) | 30 (16) | 42 (22) | < 0.001 |
| Mean hippocampal volume, mm3 | 4077 (625) | 3731 (573) | 3610 (564) | < 0.001 |
Bold values indicate significance at p < 0.05.
Table 2.
Demographic and biomarker comparisons by ApoE genotype using a two-tailed t-test for continuous and a chi-squared test for categorical variables (p-values are shown; for mean and SD for each variable, please see Table 1).
| Variable at baseline | MCI ApoE4 + vs ApoE4 - | MCI ApoE4 + vs. ApoE4 - | AD ApoE4 + vs ApoE4 - |
|---|---|---|---|
| Age, years | 0.7 | 0.2 | 0.3 |
| Gender, M:F | 0.053 | 0.07 | 0.5 |
| Education, years | 0.8 | 0.5 | 0.02 |
| MMSE | 0.4 | 0.6 | 0.8 |
| CSF Aβ42 level, pg/ml | < 0.001 | < 0.001 | 0.001 |
| CSF t-tau level, pg/ml | 0.07 | < 0.001 | 0.8 |
| CSF p-tau181 level, pg/ml | 0.024 | < 0.001 | 0.9 |
| Mean hippocampal volume, mm3 | 0.5 | 0.8 | 0.3 |
Bold values indicate significance at p < 0.05.
Table 3.
Baseline demographic and biomarker comparisons of MCI converters vs. nonconverters using a two-tailed t-test for continuous and a chi-squared test for categorical variables.
| Variable at baseline | MCI converters | MCI nonconverters | Two-tailed t-test/chi squared test, p-value |
|---|---|---|---|
| Age, years | 74.8 (7.1) | 73.6 (7.5) | 0.3 |
| Gender, M:F | 48:32 | 55:25 | 0.3 |
| Education, years | 15.5 (3.0) | 16.3 (2.8) | 0.1 |
| ApoE4 positive:negative | 53:27 | 35:45 | 0.004 |
| MMSE | 26.6 (1.8) | 27.3 (1.7) | 0.014 |
| CSF Aβ42 level, pg/ml | 145 (40) | 172 (60) | 0.001 |
| CSF t-tau level, pg/ml | 113 (51) | 88 (47) | 0.002 |
| CSF p-tau181 level, pg/ml | 40 (16) | 31 (17) | 0.001 |
| Mean hippocampal volume, mm3 | 3600 (569) | 3803 (542) | 0.022 |
Bold values indicate significance at p < 0.05.
ApoE4-positive NC subjects had significantly lower CSF Aβ42 (p < 0.001) and higher CSF p-tau levels (p = 0.024) relative to ApoE4-negative NC. ApoE4-positive MCI subjects also showed significantly lower CSF Aβ42 (p < 0.001), higher CSF tau and p-tau (both p < 0.001) relative to ApoE4-negative MCI subjects. ApoE4-positive AD subjects were significantly less educated (p = 0.02) and had lower CSF Aβ42 levels (p = 0.001) relative to ApoE4-negative AD subjects (Table 2).
Relative to MCI nonconverters, MCI converters had significantly lower MMSE (p = 0.014), higher proportion of ApoE4 carriers (p = 0.004), lower hippocampal volume (p = 0.022) and CSF Aβ42 (p = 0.001), and higher CSF tau (p = 0.002) and p-tau (p = 0.001, Table 3).
3.2. Classifier results
3.2.1. Cross-sectional classifiers
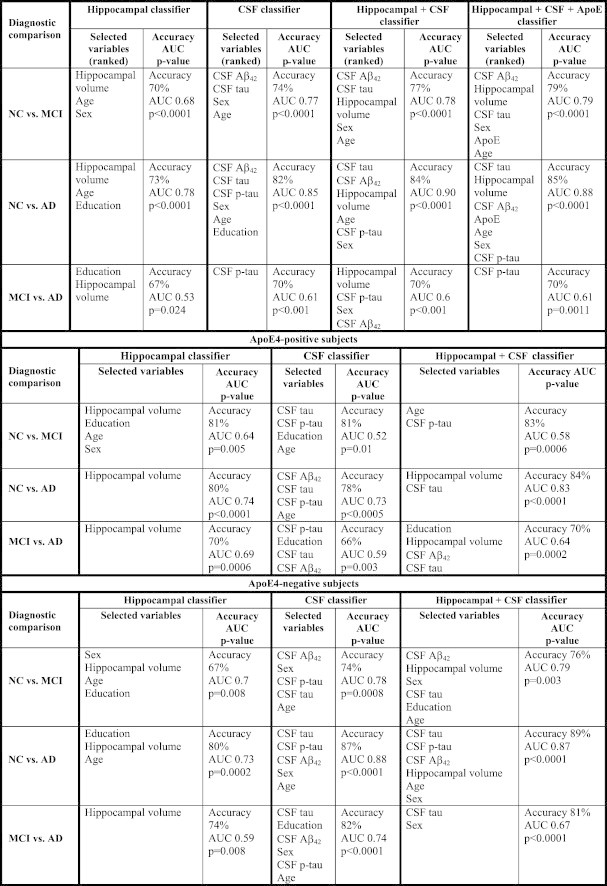
Fig. 1 and Table 4 show the cross-sectional classifier ROCs, classifier performance metrics, ranking of variables selected by each classifier and permutation corrected classifier significance.
Fig. 1.
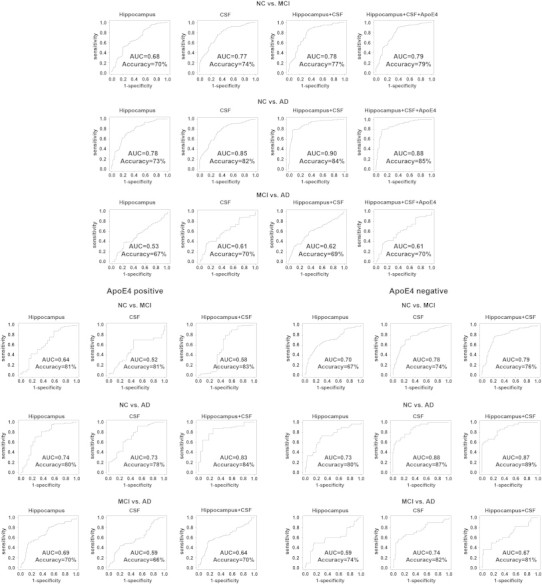
Receiver Operation Characteristic (ROC) for the cross-sectional classifiers.
Table 4.
Classifier performance metrics, ranking of variables selected by each classifier, and permutation corrected classifier significance.
3.2.1.1. NC vs. MCI classifiers (Fig. 1 top portion first row)
The hippocampal NC vs. MCI classifier achieved an AUC of 0.68. The features selected by the classifier included hippocampal volume, age and sex. The permutation corrected classifier significance was pcorrected < 0.0001.
The CSF NC vs. MCI classifier achieved an AUC of 0.77. The features selected by the classifier included CSF Aβ42, CSF tau, sex and age. The permutation corrected classifier significance was pcorrected < 0.0001.
ApoE4 genotype performed as well as hippocampal volume and CSF biomarkers regardless of whether ApoE2 carriers were excluded or not (all subjects AUC = 0.7, pcorrected < 0.0001, without ApoE2 carriers AUC = 0.71, pcorrected < 0.0001). Substituting the binary ApoE4-positive vs. negative predictor variable with a variable reflecting the number of ApoE4 alleles (0, 1 or 2) did not result in overall improvement in classifier performance (AUC = 0.67, pcorrected < 0.0001).
The multimodal NC vs. MCI classifier presented with all three CSF variables, hippocampal volume, age, sex and education achieved an AUC of 0.78. The variables selected by the classifier included CSF Aβ42, CSF tau, hippocampal volume, sex, and age. The permutation corrected classifier significance was pcorrected < 0.0001. The addition of ApoE4 genotype did not seem to affect the overall multimodal classifier results (AUC = 0.79, pcorrected < 0.0001, Table 6).
Table 6.
Statistical comparisons of classifiers (p-values).
| Diagnostic comparison | Classifier Comparison | p-value |
|---|---|---|
| NC vs. MCI | Hippocampal vs. CSF classifier | 0.01 |
| Hippocampus + CSF vs. hippocampal classifier | 0.0044 | |
| Hippocampus + CSF vs. CSF classifier | NS | |
| Hippocampus + CSF + ApoE vs. hippocampus + CSF classifier | NS | |
| NC vs. AD | Hippocampal vs. CSF classifier | 0.04 |
| Hippocampus + CSF vs. hippocampal classifier | 0.0001 | |
| Hippocampus + CSF vs. CSF classifier | 0.03 | |
| Hippocampus + CSF + ApoE vs. hippocampus + CSF | NS | |
| MCI vs. AD | Hippocampal vs. CSF classifier | NS |
| Hippocampus + CSF vs. hippocampal classifier | NS | |
| Hippocampus + CSF vs. CSF classifier | NS | |
| Hippocampus + CSF + ApoE vs. hippocampus + CSF classifier | NS | |
| ApoE4 + NC vs. MCI | Hippocampal vs. CSF classifier | NS |
| Hippocampus + CSF vs. hippocampal classifier | NS | |
| Hippocampus + CSF vs. CSF classifier | NS | |
| Hippocampus + CSF + ApoE vs. hippocampus + CSF classifier | NS | |
| ApoE4 + NC vs. AD | Hippocampal vs. CSF classifier | NS |
| Hippocampus + CSF vs. hippocampal classifier | NS | |
| Hippocampus + CSF vs. CSF classifier | NS | |
| Hippocampus + CSF + ApoE vs. hippocampus + CSF classifier | NS | |
| ApoE4 + MCI vs. AD | Hippocampal vs. CSF classifier | NS |
| Hippocampus + CSF vs. hippocampal classifier | NS | |
| Hippocampus + CSF vs. CSF classifier | NS | |
| Hippocampus + CSF + ApoE vs. hippocampus + CSF classifier | NS | |
| ApoE4 − NC vs. MCI | Hippocampal vs. CSF classifier | NS |
| Hippocampus + CSF vs. hippocampal classifier | 0.012 | |
| Hippocampus + CSF vs. CSF classifier | NS | |
| Hippocampus + CSF + ApoE vs. hippocampus + CSF classifier | NS | |
| ApoE4 − NC vs. AD | Hippocampal vs. CSF classifier | 0.012 |
| Hippocampus + CSF vs. hippocampal classifier | 0.008 | |
| Hippocampus + CSF vs. CSF classifier | NS | |
| Hippocampus + CSF + ApoE vs. hippocampus + CSF classifier | NS | |
| ApoE4 − MCI vs. AD | Hippocampal vs. CSF classifier | NS |
| Hippocampus + CSF vs. hippocampal classifier | NS | |
| Hippocampus + CSF vs. CSF classifier | NS | |
| Hippocampus + CSF + ApoE vs. hippocampus + CSF classifier | NS |
NS—not significant.
Direct statistical comparison showed that the CSF NC vs. MCI classifier performed significantly better than the hippocampal NC vs. MCI classifier (p = 0.01). The hippocampal + CSF classifier performed better than the hippocampal-only (p = 0.0044) but not the CSF-only (p = 0.65) classifiers. Adding ApoE4 genotype to the multimodal hippocampal + CSF classifier did not result in a statistically significant difference (p = 0.56).
3.2.1.2. NC vs. AD classifier (Fig. 1 top portion second row)
The hippocampal NC vs. AD classifier achieved an AUC of 0.78. The features selected by the classifier included hippocampal volume, age, and education. The permutation corrected classifier significance was pcorrected < 0.0001.
The CSF NC vs. AD classifier achieved an AUC of 0.85. The features selected by the classifier included CSF Aβ42, CSF tau and p-tau, sex, education and age. The permutation corrected classifier significance was pcorrected < 0.0001.
ApoE4 genotype performed about the same as the hippocampal volume and worse than CSF regardless of whether ApoE2 carriers were excluded or not (all subjects AUC = 0.76, pcorrected < 0.0001, without ApoE2 carriers AUC = 0.74, pcorrected < 0.0001). Substituting the binary ApoE4-positive vs. negative predictor variable with a variable reflecting the number of ApoE4 alleles (0, 1 or 2) did not result in overall improvement in classifier performance (AUC = 0.78, pcorrected < 0.0001).
The multimodal NC vs. AD classifier presented with all CSF variables, hippocampal volume, age, sex and education achieved an AUC of 0.90. The variables selected by the classifier included CSF tau, CSF Aβ42, hippocampal volume, age, CSF p-tau and sex. The permutation corrected classifier significance was pcorrected < 0.0001. The addition of ApoE4 genotype did not affect the overall multimodal classifier results (AUC = 0.88, permutation corrected significance pcorrected < 0.0001, Table 6).
Direct statistical comparison showed that CSF NC vs. AD classifier performed significantly better than the hippocampal NC vs. AD classifier (p = 0.04), and the hippocampal + CSF classifier performed better than the hippocampal-only (p = 0.0001) and the CSF-only (p = 0.03) classifiers. Adding ApoE4 genotype to the multimodal hippocampal + CSF classifier did not result in a statistically significant difference (p = 0.43).
3.2.1.3. MCI vs. AD classifier (Fig. 1 top portion bottom row)
The hippocampal MCI vs. AD classifier achieved an AUC of 0.53. The features selected by the classifier included education and hippocampal volume. The permutation corrected classifier significance was pcorrected = 0.024.
The CSF MCI vs. AD classifier achieved an AUC of 0.61. The single feature selected by the classifier was CSF p-tau. The permutation corrected classifier significance was pcorrected < 0.001.
ApoE4 genotype performed about the same as the hippocampal volume and worse than CSF regardless of whether ApoE2 carriers were excluded or not (all subjects AUC = 0.52, pcorrected < 0.0001, without ApoE2 carriers AUC = 0.51, pcorrected < 0.0001). Substituting the binary ApoE4-positive vs. negative predictor variable with a variable reflecting the number of ApoE4 alleles (0, 1 or 2) did not result in overall improvement in classifier performance (AUC = 0.53, pcorrected < 0.0001).
The multimodal MCI vs. AD classifier presented with all CSF variables, hippocamapal volume age, sex and education achieved an AUC of 0.62. The variables selected by the classifier included hippocampal volume, CSF p-tau, sex and CSF tau. The permutation corrected classifier significance was pcorrected < 0.001. The addition of ApoE4 genotype did not seem to affect the overall multimodal classifier results (AUC = 0.61, permutation corrected significance pcorrected = 0.0011, Table 6).
Direct statistical comparison showed no statistically significant improvement in performance between the various classifiers.
3.2.1.4. ApoE4 stratified analyses (Fig. 1 bottom portion)
Hippocampal volume and CSF biomarkers both separately and in combination performed well in discriminating NC vs. AD regardless of ApoE4 genotype (ApoE4-positive subjects: hippocampal classifier AUC = 0.74 pcorrected < 0.0001, CSF classifier AUC = 0.73 pcorrected = 0.0005 and combined classifier AUC = 0.83 pcorrected < 0.0001; ApoE4-negative subjects: hippocampal classifier AUC = 0.73 pcorrected = 0.0002, CSF classifier AUC = 0.88 pcorrected < 0.0001 and combined classifier AUC = 0.87 pcorrected < 0.0001).
In the NC vs. MCI analyses CSF biomarkers alone and the combination of hippocampus and CSF biomarkers achieved reasonable AUCs in the ApoE4-negative sample only (ApoE4-negative subjects: hippocampal lassifier AUC = 0.70 pcorrected = 0.008, CSF classifier AUC = 0.78 pcorrected = 0.0008 and combined classifier AUC = 0.79 pcorrected = 0.003; ApoE4-positive subjects: hippocampal classifier AUC = 0.64 pcorrected = 0.005, CSF classifier AUC = 0.52 pcorrected = 0.01 and combined classifier AUC = 0.58 pcorrected = 0.0006).
In the MCI vs. AD analyses the hippocampal classifier performed best in ApoE4-positive subjects (AUC = 0.69, pcorrected = 0.0006) and the CSF classifier performed best in the ApoE4-negative subjects (AUC = 0.74, pcorrected < 0.0001).
Among ApoE4-positive subjects classifiers did not prove to be statistically significant from each other. Among ApoE4-negative subjects the hippocampal + CSF classifier was significantly better than the hippocampal-only classifier when discriminating NC vs. MCI (p = 0.012) and NC vs. AD (p = 0.008, Table 6). The CSF-only classifier performed significantly better than the hippocampal-only classifier when discriminating Apoe4-negative NC vs. AD subjects (p = 0.012, Table 6).
3.2.2. Predicting conversion
Fig. 2 and Table 5 show the SVM classifier ROCs, classifier performance metrics, ranking of variables selected by each classifier and permutation corrected classifier significance for the MCI conversion classifier analyses.
Fig. 2.
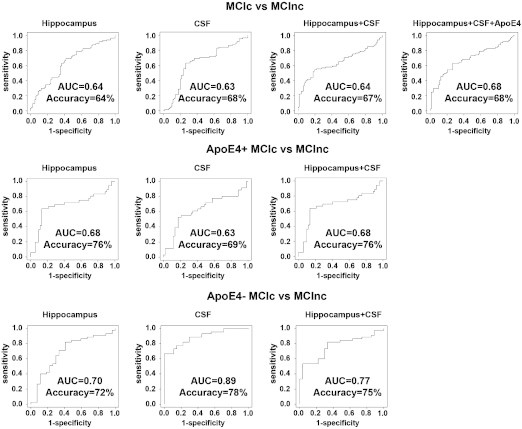
Receiver Operation Characteristic (ROC) for the conversion classifier.
Table 5.
Classifier performance metrics, ranking of variables selected by each classifier and permutation corrected classifier significance in predicting conversion from MCI to AD with and without stratification by ApoE4 genotype.
| Hippocampal classifier |
CSF classifier |
Hippocampal + CSF classifier |
Hippocampal + CSF + ApoE classifier |
|||||
|---|---|---|---|---|---|---|---|---|
| Diagnostic comparison | Selected variables (ranked) | Accuracy AUC p-value | Selected variables (ranked) | Accuracy AUC p-value | Selected variables (ranked) | Accuracy AUC p-value | Selected variables (ranked) | Accuracy AUC p-value |
| MCIc vs MCInc | Hippocampal volume Sex Education |
Accuracy 64% AUC 0.64 p = 0.048 |
CSF Aβ42 | Accuracy 68% AUC 0.63 p = 0.008 |
Hippocampal volume CSF Aβ42 CSF p-tau Education Sex CSF tau |
Accuracy 67% AUC 0.64 p = 0.042 |
ApoE CSF p-tau Hippocampal volume CSF Aβ42 CSF tau Education |
Accuracy 68% AUC 0.68 p = 0.019 |
| ApoE4 stratified MCIc vs. MCInc classifier | ||||||
|---|---|---|---|---|---|---|
| Hippocampal classifier |
CSF classifier |
Hippocampal + CSF classifier |
||||
| Diagnostic comparison | Selected variables | Accuracy AUC p-value | Selected variables | Accuracy AUC p-value | Selected variables | Accuracy AUC p-value |
| ApoE4 + MCIc vs. ApoE4 + MCInc | Hippocampal volume Sex |
Accuracy 76% AUC 0.68 p < 0.0001 |
Age Sex |
Accuracy 69% AUC 0.63 p = 0.002 |
Hippocampal volume Sex |
Accuracy 76% AUC 0.68 p < 0.0001 |
| ApoE4 − MCIc vs. ApoE4 − MCInc | Sex Age |
Accuracy 72% AUC 0.70 p = 0.005 |
CSF p-tau Education Sex |
Accuracy 78% AUC 0.89 p < 0.0001 |
CSF p-tau | Accuracy 75% AUC 0.77 p < 0.001 |
In the classifier model including both ApoE4-positive and negative subjects the hippocampal-only classifier selected hippocampal volume, sex and education and achieved an AUC = 0.64 (pcorrected = 0.048). The CSF-only classifier selected only CSF Aβ42 and achieved an AUC = 0.63 (pcorrected = 0.008). The combined hippocampal-CSF classifier selected hippocampal volume, CSF Aβ42, CSF p-tau, education, sex and CSF tau and achieved an AUC = 0.64 (pcorrected = 0.042). The addition of ApoE4 genotype did not seem to affect the overall multimodal classifier results (AUC = 0.68, pcorrected = 0.019). ApoE4 genotype performed about the same as hippocampal volume and CSF regardless of whether ApoE2 carriers were excluded or not (all subjects AUC = 0.64, pcorrected = 0.002, without ApoE2 carriers AUC = 0.61, pcorrected = 0.004). Substituting the binary ApoE4-positive vs. negative predictor variable with a variable reflecting the number of ApoE4 alleles (0, 1 or 2) did not result in overall improvement in classifier performance (AUC = 0.61, pcorrected = 0.0026).
Once stratified by ApoE4 genotype the best results for predicting conversion to AD were achieved by the hippocampus-only classifier in ApoE4-positive (predictors: hippocampal volume and sex; AUC = 0.68, pcorrected < 0.0001) and CSF-only classifier for ApoE4-negative subjects (predictors: CSF p-tau, education and sex; AUC = 0.89, pcorrected < 0.0001).
4. Discussion
All classifier models performed very well in discriminating NC from AD and moderately well in discriminating NC from MCI. The MCI vs. AD ascertainment, as expected, proved to be more challenging for unimodal and multimodal classifiers alike presumably due to the fact that the MCI biomarker pattern is rather similar to the one seen in AD. The multimodal biomarker classifier approach had better diagnostic and predictive power than any unimodal classifier.
Several important observations can be made from the discriminative classifier performance. CSF Aβ42 played a significant role in discriminating NC from MCI and AD but was not selected by the MCI vs. AD classifiers while CSF p-tau contributed to accurate discrimination of AD from both NC and MCI, yet played no role in differentiating NC from MCI. These observations are in agreement with the proposed biomarker trajectory in AD where amyloid markers become abnormal early in the disease course and neurodegenerative markers (here CSF p-tau) become abnormal later in the disease course. Interestingly hippocampal atrophy and CSF t-tau seemed quite ubiquitously used by most classifiers including the NC vs. MCI classifiers, suggesting that these neurodegenerative biomarkers are becoming abnormal somewhere between the CSF Aβ42 and the CSF p-tau changes.
CSF Aβ42 proved to be useful for differentiating NC from MCI only among ApoE4-negative but not ApoE4-positive subjects. This is likely due to the fact that many ApoE4-positive cognitively normal elderly already have significant brain amyloidosis and low CSF Aβ42 rendering amyloid biomarkers insensitive for differentiating NC and MCI. At the same time neurodegenerative biomarkers (CSF tau, CSF p-tau and/or hippocampal atrophy) were readily chosen in both ApoE4-positive and ApoE4-negative NC vs. MCI classifiers establishing their discriminative role for patients with either genotype.
Both amyloid and neurodegenerative biomarkers were readily chosen by the conversion classifiers. Similar to the observations of others (Cui et al., 2011, Davatzikos et al., 2011, Ewers et al., 2012, Vos et al., 2012, Westman et al., 2012) the classifier accuracies in the full sample were marginal at best. However once split by ApoE4 genotype we observed that hippocampal volume and sex were helpful for predicting conversion to AD among ApoE4-positive MCI, while CSF p-tau was helpful for the prediction of conversion to AD among ApoE4-negative MCI subjects. Both classifier algorithms chose only markers of neurodegeneration as one might expect to be the case in the symptomatic MCI stage. These conclusions are well supported by data from a recent paper by Jack et al. showing that among amyloid positive MCI subjects hippocampal atrophy, and not amyloid burden, predicted shorter time to progression to dementia (Jack et al., 2010) because amyloid load plateaus (Jack et al., 2013) while hippocampal volume does not (Jack et al., 2010).
The CSF outperformed the hippocampal metrics in discriminating NC from MCI and AD. Low CSF Aβ42 is tightly linked to the presence of amyloid pathology in the brain while hippocampal atrophy is criticized for being a nonspecific measure observed in many disease states and in normal aging (Apostolova and Thompson, 2008). Techniques capable of detecting hippocampal atrophy in selected subfields are being developed (Apostolova et al., 2010a, Apostolova et al., 2010c, Csernansky et al., 2000, Mueller and Weiner, 2009) and some have even demonstrated ability to detect hippocampal atrophy in the presymptomatic disease stages (Apostolova et al., 2010c). Alternatively more sophisticated MRI measures such as for instance the Structural Abnormality Index (STAND), which captures hippocampal and cortical atrophy, can also improve the ability of MRI for predicting future decline (Vemuri et al., 2009a, Vemuri et al., 2009b).
In contrast to other groups that have published multimodal diagnostic classification papers using ADNI data (Cui et al., 2011, Davatzikos et al., 2011, Ewers et al., 2012, Kohannim et al., 2010, Vos et al., 2012, Walhovd et al., 2010, Westman et al., 2012) we also investigated classifier performance after ApoE4-stratification (i.e., investigated the classifier performance separately in carriers and noncarriers). This led to some interesting observations in respect to the modulatory effect of ApoE4 genotype on biomarker trajectory in AD. It is fascinating that support vector machine classifiers can help uncover interesting observations in respect to AD pathophysiology. This is a novel way of utilization of a statistical methodology thought by many to be only capable of diagnostic discrimination and outcome prediction.
Although multicollinearity can be a problem for multiple linear regression, it is standard for SVMs to be provided with thousands of correlated predictors, and still perform very well. Only certain kinds of machine learning methods, such as naive Bayes methods, assume that the inputs of the classifier are statistically independent. Even when inputs considered to be an n-dimensional vector by SVM, are highly correlated, the SVM will find the optimal hyperplane for separating the samples and making decisions to categorize or classify future data.
Correlation among inputs is less of an issue when making predictions. Redundancy among predictors is common and does not undermine the predictive accuracy of the classifier. However correlated inputs do tend to complicate the interpretation of which predictor variables are driving the effects. This is more of an issue if the classifier is considered as a “descriptive” model — telling us the relative importance of variables for the prediction, but it is less of an issue for predictive accuracy as many of the redundant predictors can be used.
When one fits an SVM with multiple predictors, if the predictors are correlated, SVM might pick some predictors on one occasion and may do equally well on another occasion using other predictors. SVM will always pick the one that minimizes the error. Sometimes the accuracies of certain feature combinations are very close, yet SVM will pick the one with the lowest error. It does not mean that the other dropped variables might not be predictive, only that they are sufficiently redundant with the ones that were used that they were not chosen.
This study has several strengths and limitations. The strengths of ADNI lie in its large size, its detailed cognitive assessment protocol and careful diagnostic ascertainment, as well as in the implementation of unified MRI and CSF collection and processing strategies across multiple sites and the meticulous data quality control. Yet ADNI was designed to inform decisions about future disease modifying clinical trials and as such it employs the rigorous inclusion and exclusion criteria typical of clinical trials. As such, the ADNI cohort is not a complete representation of the elderly population and its findings should be generalized with caution; for example, the classification accuracies may be poorer in populations with a great mix of conditions and co-morbidities. Another relative weakness of our study is the etiologic/pathologic uncertainty in the MCI stage as at least 30% of amnestic MCI subjects have been found to harbor non-AD pathology (Jicha et al., 2006). Even so, etiologic heterogeneity should not be expected to invalidate the biomarker-to-biomarker correlations across the pooled sample.
Another important limitation of our study is the classifiers' exclusive reliance on biomarkers for diagnostic categorization. By doing so we inadvertently compromise our ability to accurately classify AD vs. MCI subjects as the diagnostic distinction between these categories is determined by the presence or absence of functional decline which cannot be ascertained from biomarker data. Thus it is not surprising that our biomarker-based classifier models failed to discriminate between MCI and AD. However we must acknowledge that providing our classifiers with functional or cognitive variables would have introduced a circularity argument, as cognitive variables were used for diagnostic ascertainment of ADNI subjects.
One must also take under consideration that CSF tau and p-tau changes do not accurately reflect (i.e., lag behind) the severity and extent of tau pathology in the AD brain. CSF biomarkers are just peripheral surrogates of actual tau pathology. Several tau-imaging ligands are currently under development. Such technology might provide a much more accurate metric of tau pathology and contribute meaningfully to an improved understanding and staging of the early and presymptomatic stages of AD. Last but not least, our classifier results will benefit from independent validation in a different cohort with similar longitudinal follow-up and biomarker data availability.
Acknowledgments
Data used in preparing this article were obtained from the Alzheimer's Disease Neuroimaging Initiative database (www.loni.ucla.edu/ADNI). Many ADNI investigators have therefore contributed to the design and implementation of ADNI or provided data but did not participate in the analysis or writing of this report. A complete list of ADNI investigators is available at http://adni.loni.ucla.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514.
We thank the members of the ADNI Imaging Core for their contributions to the image pre-processing, the members of the ADNI Biomarker Core for the CSF biomarker analyses and the investigators at the University of Pittsburgh for the PIB SUVR analyses.
The analyses reported in this manuscript were funded by the Easton Consortium for Alzheimer's Drug Discovery and Biomarker Development, NIA R01 AG040770, NIA P50 AG16570. Algorithm development was also supported, in part, by NIMH R01 MH097268 and NIA R01 AG040060 (to P.T.). O.K. was supported, in part, by a UCLA Dissertation Year Fellowship, and by the UCLA Medical Scientist Training Program.
Disclosure Statement
Dr. Apostolova serves on the Speakers Bureau for Lilly Pharmaceuticals. She receives research funding from the National Institutes of Health and the Easton Consortium for Alzheimer's Drug Development and Biomarker Discovery. Dr. Apostolova receives research funding from the NIH. Dr. Apostolova had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Dr. Apostolova was responsible for design and conduct of the study; collection, management, analysis, and interpretation of the data, and preparation, review and approval of the manuscript.
Ms. Hwang has no disclosures. Ms. Hwang was responsible for data management, analysis, and interpretation of the data, preparation, review and approval of the manuscript.
Mr. Kohannim has no disclosures. Mr. Kohannim was responsible for management, analysis, and interpretation of the data, review and approval of the manuscript.
Mr. Avila has no disclosures. Mr. Avila was responsible for analysis and interpretation of the data, review and approval of the manuscript.
Dr. Jack serves as a consultant for Janssen, Bristol-Meyer-Squibb, General Electric, Siemens, and Johnson and Johnson and is involved in clinical trials sponsored by Allon and Baxter, Inc. He receives research funding from the National Institutes of Health and the Alexander Family Alzheimer's Disease Research Professorship of the Mayo Foundation. Dr. Jack receives research funding from the NIH. Dr. Jack was responsible for the interpretation of the data, review and approval of the manuscript.
Dr. Shaw receives research funding from the NIH and MJFox Foundation for Parkinson's Research; has served on a technical advisory board for Innogenetics; serves as a consultant for Janssen; advisory board, Saladax, and is involved in clinical trials sponsored by Eisai, Inc. and Baxter, Inc. Dr. Shaw receives research funding from the NIH. Dr. Shaw was responsible for the interpretation of the data, review and approval of the manuscript.
Dr. Trojanowski may accrue revenue in the future on patents submitted by the University of Pennsylvania wherein he is co-Inventor and he received revenue from the sale of Avid to Eli Lily as co-inventor on imaging related patents submitted by the University of Pennsylvania. He receives research support from the NIH, Bristol Myer Squib, Astra Zeneca and several non-profits. Dr. Trojanowski receives research funding from the NIH. Dr. Trojanowski was responsible for the interpretation of the data, review and approval of the manuscript.
Dr. Weiner has served in the past three years on the Scientific Advisory Boards of Lilly, Biogen Idec, Pfizer, Araclon and Institut Catala de Neurociencies Aplicades, Gulf War Veterans Illnesses Advisory Committee, VACO and BOLT Inter-national. He was engaged in consulting activities for Astra Zeneca, Araclon, Medivation/Pfizer, Ipsen, TauRx Therapeutics LTD, Bayer Healthcare, Biogen Idec, Exonhit Therapeutics, Servier, Synarc, Pfizer, Janssen, Harvard University and KLJ Associates. He has received travel funding from NeuroVigil, Inc., CHRU-Hopital Roger Salengro, Siemens, AstraZeneca, Geneva University Hospitals, Lilly, University of California, San Diego — ADNI, Paris University, Institut Catala de Neurociencies Aplicades, University of New Mexico School of Medicine, Ipsen, CTAD (Clinical Trials on Alzheimer's Disease), Pfizer, ADPD meeting, Paul Sabatier University, Novartis, Tohoku University, and Fundacio ACE and Travel eDreams, Inc. He serves on the Editorial Board for Alzheimer's and Dementia and MRI. He has received honoraria from NeuroVigil, Inc., Insitut Catala de Neurociencies Aplicades, PMDA/Japanese Ministry of Health, Labour, and Welfare, Tohoku University and the Alzheimer's Drug Discovery Foundation. He has received research support from Merch and Avid. He owns stock options in Synarc and Elan. Dr. Weiner receives research funding from the NIH. Dr. Weiner was responsible for the interpretation of the data, review and approval of the manuscript.
Dr. Thompson receives research funding from the NIH. Dr. Thompson was responsible for the interpretation of the data, review and approval of the manuscript. Mr. Avila has no disclosures. Mr. Avila was responsible for analysis and interpretation of the data, review and approval of the manuscript.
References
- Andreasen N., Minthon L., Davidsson P., Vanmechelen E., Vanderstichele H., Winblad B., Blennow K. Evaluation of CSF-tau and CSF-Abeta42 as diagnostic markers for Alzheimer disease in clinical practice. Arch. Neurol. 2001;58:373–379. doi: 10.1001/archneur.58.3.373. [DOI] [PubMed] [Google Scholar]
- Apostolova L.G., Thompson P.M. Mapping progressive brain structural changes in early Alzheimer's disease and mild cognitive impairment. Neuropsychologia. 2008;46:1597–1612. doi: 10.1016/j.neuropsychologia.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova L.G., Dutton R.A., Dinov I.D., Hayashi K.M., Toga A.W., Cummings J.L., Thompson P.M. Conversion of mild cognitive impairment to Alzheimer disease predicted by hippocampal atrophy maps. Arch. Neurol. 2006;63:693–699. doi: 10.1001/archneur.63.5.693. [DOI] [PubMed] [Google Scholar]
- Apostolova L.G., Mosconi L., Thompson P.M., Green A.E., Mistur R., Tsui W.H., de Leon M.J. Subregional hippocampal atrophy predicts future decline to Alzheimer's dementia in cognitively normal subjects. Neurobiol. Aging. 2014 doi: 10.1016/j.neurobiolaging.2008.08.008. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova L.G., Thompson P.M., Green A.E., Hwang K.S., Zoumalan C., Jack C.R., Jr., Harvey D.J., Petersen R.C., Thal L.J., Aisen P.S., Toga A.W., Cummings J.L., Decarli C.S. 3D comparison of low, intermediate, and advanced hippocampal atrophy in MCI. Hum. Brain Mapp. 2010;31:786–797. doi: 10.1002/hbm.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova L.G., Hwang K.S., Andrawis J.P., Green A.E., Babakchanian S., Morra J.H., Cummings J.L., Toga A.W., Trojanowski J.Q., Shaw L.M., Jack C.R., Jr., Petersen R.C., Aisen P.S., Jagust W.J., Koeppe R.A., Mathis C.A., Weiner M.W., Thompson P.M. 3D PIB and CSF biomarker associations with hippocampal atrophy in ADNI subjects. Neurobiol. Aging. 2010;31:1284–1303. doi: 10.1016/j.neurobiolaging.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova L.G., Mosconi L., Thompson P.M., Green A.E., Hwang K.S., Ramirez A., Mistur R., Tsui W.H., de Leon M.J. Subregional hippocampal atrophy predicts Alzheimer's dementia in the cognitively normal. Neurobiol. Aging. 2010;31:1077–1088. doi: 10.1016/j.neurobiolaging.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova L.G., Morra J.H., Green A.E., Hwang K.S., Avedissian C., Woo E., Cummings J.L., Toga A.W., Jack C.R., Weiner M.W., Thompson P.M. Automated 3D mapping of baseline and 12-month associations between three verbal memory measures and hippocampal atrophy in 490 ADNI subjects. Neuroimage. 2010;51:488–499. doi: 10.1016/j.neuroimage.2009.12.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blennow K., Hampel H. CSF markers for incipient Alzheimer's disease. Lancet Neurol. 2003;2:605–613. doi: 10.1016/s1474-4422(03)00530-1. [DOI] [PubMed] [Google Scholar]
- Blennow K., Wallin A., Agren H., Spenger C., Siegfried J., Vanmechelen E. Tau protein in cerebrospinal fluid: a biochemical marker for axonal degeneration in Alzheimer disease? Mol. Chem. Neuropathol. 1995;26:231–245. doi: 10.1007/BF02815140. [DOI] [PubMed] [Google Scholar]
- Bobinski M., Wegiel J., Wisniewski H.M., Tarnawski M., Reisberg B., Mlodzik B., de Leon M.J., Miller D.C. Atrophy of hippocampal formation subdivisions correlates with stage and duration of Alzheimer disease. Dementia. 1995;6:205–210. doi: 10.1159/000106948. [DOI] [PubMed] [Google Scholar]
- Bobinski M., Wegiel J., Tarnawski M., Bobinski M., Reisberg B., de Leon M.J., Miller D.C., Wisniewski H.M. Relationships between regional neuronal loss and neurofibrillary changes in the hippocampal formation and duration and severity of Alzheimer disease. J. Neuropathol. Exp. Neurol. 1997;56:414–420. doi: 10.1097/00005072-199704000-00010. [DOI] [PubMed] [Google Scholar]
- Brys M., Pirraglia E., Rich K., Rolstad S., Mosconi L., Switalski R., Glodzik-Sobanska L., De Santi S., Zinkowski R., Mehta P., Pratico D., Saint Louis L.A., Wallin A., Blennow K., de Leon M.J. Prediction and longitudinal study of CSF biomarkers in mild cognitive impairment. Neurobiol. Aging. 2009;30:682–690. doi: 10.1016/j.neurobiolaging.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerger K., Teipel S.J., Zinkowski R., Blennow K., Arai H., Engel R., Hofmann-Kiefer K., McCulloch C., Ptok U., Heun R., Andreasen N., DeBernardis J., Kerkman D., Moeller H., Davies P., Hampel H. CSF tau protein phosphorylated at threonine 231 correlates with cognitive decline in MCI subjects. Neurology. 2002;59:627–629. doi: 10.1212/wnl.59.4.627. [DOI] [PubMed] [Google Scholar]
- Buerger K., Ewers M., Andreasen N., Zinkowski R., Ishiguro K., Vanmechelen E., Teipel S.J., Graz C., Blennow K., Hampel H. Phosphorylated tau predicts rate of cognitive decline in MCI subjects: a comparative CSF study. Neurology. 2005;65:1502–1503. doi: 10.1212/01.wnl.0000183284.92920.f2. [DOI] [PubMed] [Google Scholar]
- Clark C.M., Xie S., Chittams J., Ewbank D., Peskind E., Galasko D., Morris J.C., McKeel D.W., Jr., Farlow M., Weitlauf S.L., Quinn J., Kaye J., Knopman D., Arai H., Doody R.S., DeCarli C., Leight S., Lee V.M., Trojanowski J.Q. Cerebrospinal fluid tau and beta-amyloid: how well do these biomarkers reflect autopsy-confirmed dementia diagnoses? Arch. Neurol. 2003;60:1696–1702. doi: 10.1001/archneur.60.12.1696. [DOI] [PubMed] [Google Scholar]
- Collins D.L., Neelin P., Peters T.M., Evans A.C. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J. Comput. Assist. Tomogr. 1994;18:192–205. [PubMed] [Google Scholar]
- Csernansky J.G., Wang L., Joshi S., Miller J.P., Gado M., Kido D., McKeel D., Morris J.C., Miller M.I. Early DAT is distinguished from aging by high-dimensional mapping of the hippocampus. Dementia of the Alzheimer type. Neurology. 2000;55:1636–1643. doi: 10.1212/wnl.55.11.1636. [DOI] [PubMed] [Google Scholar]
- Cui Y., Liu B., Luo S., Zhen X., Fan M., Liu T., Zhu W., Park M., Jiang T., Jin J.S. Identification of conversion from mild cognitive impairment to Alzheimer's disease using multivariate predictors. PloS One. 2011;6:e21896. doi: 10.1371/journal.pone.0021896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C., Bhatt P., Shaw L.M., Batmanghelich K.N., Trojanowski J.Q. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol. Aging. 2011;32:e2319–e2327. doi: 10.1016/j.neurobiolaging.2010.05.023. (2322) [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leon M.J., DeSanti S., Zinkowski R., Mehta P.D., Pratico D., Segal S., Rusinek H., Li J., Tsui W., Saint Louis L.A., Clark C.M., Tarshish C., Li Y., Lair L., Javier E., Rich K., Lesbre P., Mosconi L., Reisberg B., Sadowski M., DeBernadis J.F., Kerkman D.J., Hampel H., Wahlund L.O., Davies P. Longitudinal CSF and MRI biomarkers improve the diagnosis of mild cognitive impairment. Neurobiol. Aging. 2006;27:394–401. doi: 10.1016/j.neurobiolaging.2005.07.003. [DOI] [PubMed] [Google Scholar]
- de Toledo-Morrell L., Dickerson B., Sullivan M.P., Spanovic C., Wilson R., Bennett D.A. Hemispheric differences in hippocampal volume predict verbal and spatial memory performance in patients with Alzheimer's disease. Hippocampus. 2000;10:136–142. doi: 10.1002/(SICI)1098-1063(2000)10:2<136::AID-HIPO2>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Dimitriadou E., Hornik K., Leisch F., Meyer D., Weingessel A. TU Wien; 2006. e1071: Misc Functions of the Department of Statistics; p. e1071. (Available from http://cran.R-project.org) [Google Scholar]
- Ewers M., Walsh C., Trojanowski J.Q., Shaw L.M., Petersen R.C., Jack C.R., Jr., Feldman H.H., Bokde A.L., Alexander G.E., Scheltens P., Vellas B., Dubois B., Weiner M., Hampel H. Prediction of conversion from mild cognitive impairment to Alzheimer's disease dementia based upon biomarkers and neuropsychological test performance. Neurobiol. Aging. 2012;33:1203–1214. doi: 10.1016/j.neurobiolaging.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischman D.A., Wilson R.S., Gabrieli J.D., Schneider J.A., Bienias J.L., Bennett D.A. Implicit memory and Alzheimer's disease neuropathology. Brain. 2005;128:2006–2015. doi: 10.1093/brain/awh559. [DOI] [PubMed] [Google Scholar]
- Freund Y., Shapire R. A decision-theoretic generalization of online learning and an application to boosting. J. Comp. Syst. Sci. 1997;55:119–139. [Google Scholar]
- Frisoni G.B., Prestia A., Zanetti O., Galluzzi S., Romano M., Cotelli M., Gennarelli M., Binetti G., Bocchio L., Paghera B., Amicucci G., Bonetti M., Benussi L., Ghidoni R., Geroldi C. Markers of Alzheimer's disease in a population attending a memory clinic. Alzheimers Dement. 2009;5:307–317. doi: 10.1016/j.jalz.2009.04.1235. [DOI] [PubMed] [Google Scholar]
- Galasko D., Chang L., Motter R., Clark C.M., Kaye J., Knopman D., Thomas R., Kholodenko D., Schenk D., Lieberburg I., Miller B., Green R., Basherad R., Kertiles L., Boss M.A., Seubert P. High cerebrospinal fluid tau and low amyloid beta42 levels in the clinical diagnosis of Alzheimer disease and relation to apolipoprotein E genotype. Arch. Neurol. 1998;55:937–945. doi: 10.1001/archneur.55.7.937. [DOI] [PubMed] [Google Scholar]
- Gunter J., Bernstein M., Borowski B., Felmlee J., Blezek D., Mallozzi R. ISMRM 14th Scientific Meeting and Exhibition. 2006. Validation testing of the MRI calibration phantom for the Alzheimer's Disease Neuroimaging Initiative Study. [Google Scholar]
- Guyon I., Waeston J., Barnhill S., Vapnik V. Cancer classification using support vector machines. Mach. Learn. 2002:389–422. [Google Scholar]
- Hampel H., Teipel S.J., Fuchsberger T., Andreasen N., Wiltfang J., Otto M., Shen Y., Dodel R., Du Y., Farlow M., Moller H.J., Blennow K., Buerger K. Value of CSF beta-amyloid1-42 and tau as predictors of Alzheimer's disease in patients with mild cognitive impairment. Mol. Psychiatry. 2004;9:705–710. doi: 10.1038/sj.mp.4001473. [DOI] [PubMed] [Google Scholar]
- Hebert L.E., Beckett L.A., Scherr P.A., Evans D.A. Annual incidence of Alzheimer disease in the United States projected to the years 2000 through 2050. Alzheimer Dis. Assoc. Disord. 2001;15:169–173. doi: 10.1097/00002093-200110000-00002. [DOI] [PubMed] [Google Scholar]
- Jack C.R., Jr., Petersen R.C., Xu Y.C., Waring S.C., O'Brien P.C., Tangalos E.G., Smith G.E., Ivnik R.J., Kokmen E. Medial temporal atrophy on MRI in normal aging and very mild Alzheimer's disease. Neurology. 1997;49:786–794. doi: 10.1212/wnl.49.3.786. (see comment) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Petersen R.C., Xu Y., O'Brien P.C., Smith G.E., Ivnik R.J., Tangalos E.G., Kokmen E. Rate of medial temporal lobe atrophy in typical aging and Alzheimer's disease. Neurology. 1998;51:993–999. doi: 10.1212/wnl.51.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Petersen R.C., Xu Y., O'Brien P.C., Smith G.E., Ivnik R.J., Boeve B.F., Tangalos E.G., Kokmen E. Rates of hippocampal atrophy correlate with change in clinical status in aging and AD. Neurology. 2000;55:484–489. doi: 10.1212/wnl.55.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Bernstein M.A., Fox N.C., Thompson P., Alexander G., Harvey D., Borowski B., Britson P.J., L.W. J, Ward C., Dale A.M., Felmlee J.P., Gunter J.L., Hill D.L., Killiany R., Schuff N., Fox-Bosetti S., Lin C., Studholme C., DeCarli C.S., Krueger G., Ward H.A., Metzger G.J., Scott K.T., Mallozzi R., Blezek D., Levy J., Debbins J.P., Fleisher A.S., Albert M., Green R., Bartzokis G., Glover G., Mugler J., Weiner M.W. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J. Magn. Reson. Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Wiste H.J., Vemuri P., Weigand S.D., Senjem M.L., Zeng G., Bernstein M.A., Gunter J.L., Pankratz V.S., Aisen P.S., Weiner M.W., Petersen R.C., Shaw L.M., Trojanowski J.Q., Knopman D.S. Brain beta-amyloid measures and magnetic resonance imaging atrophy both predict time-to-progression from mild cognitive impairment to Alzheimer's disease. Brain. 2010;133:3336–3348. doi: 10.1093/brain/awq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Wiste H.J., Lesnick T.G., Weigand S.D., Knopman D.S., Vemuri P., Pankratz V.S., Senjem M.L., Gunter J.L., Mielke M.M., Lowe V.J., Boeve B.F., Petersen R.C. Brain beta-amyloid load approaches a plateau. Neurology. 2013;80:890–896. doi: 10.1212/WNL.0b013e3182840bbe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jicha G.A., Parisi J.E., Dickson D.W., Johnson K., Cha R., Ivnik R.J., Tangalos E.G., Boeve B.F., Knopman D.S., Braak H., Petersen R.C. Neuropathologic outcome of mild cognitive impairment following progression to clinical dementia. Arch. Neurol. 2006;63:674–681. doi: 10.1001/archneur.63.5.674. [DOI] [PubMed] [Google Scholar]
- Jovicich J., Czanner S., Greve D., Haley E., van der Kouwe A., Gollub R., Kennedy D., Schmitt F., Brown G., Macfall J., Fischl B., Dale A. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Kohannim O., Hua X., Hibar D.P., Lee S., Chou Y.Y., Toga A.W., Jack C.R., Jr., Weiner M.W., Thompson P.M. Boosting power for clinical trials using classifiers based on multiple biomarkers. Neurobiol. Aging. 2010;31:1429–1442. doi: 10.1016/j.neurobiolaging.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leow A.D., Klunder A.D., Jack C.R., Jr., Toga A.W., Dale A.M., Bernstein M.A., Britson P.J., Gunter J.L., Ward C.P., Whitwell J.L., Borowski B.J., Fleisher A.S., Fox N.C., Harvey D., Kornak J., Schuff N., Studholme C., Alexander G.E., Weiner M.W., Thompson P.M. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage. 2006;31:627–640. doi: 10.1016/j.neuroimage.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson N., Zetterberg H., Hansson O., Andreasen N., Parnetti L., Jonsson M., Herukka S.K., van der Flier W.M., Blankenstein M.A., Ewers M., Rich K., Kaiser E., Verbeek M., Tsolaki M., Mulugeta E., Rosen E., Aarsland D., Visser P.J., Schroder J., Marcusson J., de Leon M., Hampel H., Scheltens P., Pirttila T., Wallin A., Jonhagen M.E., Minthon L., Winblad B., Blennow K. CSF biomarkers and incipient Alzheimer disease in patients with mild cognitive impairment. JAMA. 2009;302:385–393. doi: 10.1001/jama.2009.1064. [DOI] [PubMed] [Google Scholar]
- Mazziotta J., Toga A., Evans A., Fox P., Lancaster J., Zilles K., Woods R., Paus T., Simpson G., Pike B., Holmes C., Collins L., Thompson P., MacDonald D., Iacoboni M., Schormann T., Amunts K., Palomero-Gallagher N., Geyer S., Parsons L., Narr K., Kabani N., Le Goualher G., Boomsma D., Cannon T., Kawashima R., Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra J.H., Tu Z., Apostolova L.G., Green A.E., Avedissian C., Madsen S.K., Parikshak N., Hua X., Toga A.W., Jack C.R., Jr., Weiner M.W., Thompson P.M. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer's disease mild cognitive impairment, and elderly controls. Neuroimage. 2008;43:59–68. doi: 10.1016/j.neuroimage.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra J.H., Tu Z., Apostolova L.G., Green A.E., Toga A.W., Thompson P.M. Automatic subcortical segmentation using a contextual model. Med. Image Comput. Comput. Assist. Interv. 2008;11:194–201. doi: 10.1007/978-3-540-85988-8_24. [DOI] [PubMed] [Google Scholar]
- Morra J.H., Tu Z., Apostolova L.G., Green A.E., Avedissian C., Madsen S.K., Parikshak N., Toga A.W., Jack C.R., Jr., Schuff N., Weiner M.W., Thompson P.M. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Neuroimage. 2009;45:S3–15. doi: 10.1016/j.neuroimage.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra J.H., Tu Z., Apostolova L.G., Green A., Toga A.W., Thompson P.M. Comparison of adaboost and support vector machines for detecting Alzheimer's disease through automated hippocampal segmentation. IEEE Trans. Med. Imaging. 2009;29:30–43. doi: 10.1109/TMI.2009.2021941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer J.A., Gosche K.M., Riley K.P., Markesbery W.R., Snowdon D.A. Delayed recall, hippocampal volume and Alzheimer neuropathology: findings from the Nun Study. Neurology. 2004;62:428–432. doi: 10.1212/01.wnl.0000106463.66966.65. [DOI] [PubMed] [Google Scholar]
- Mueller S.G., Weiner M.W. Selective effect of age, Apo e4, and Alzheimer's disease on hippocampal subfields. Hippocampus. 2009;19:558–564. doi: 10.1002/hipo.20614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen R.C., Aisen P.S., Beckett L.A., Donohue M.C., Gamst A.C., Harvey D.J., Jack C.R., Jr., Jagust W.J., Shaw L.M., Toga A.W., Trojanowski J.Q., Weiner M.W. Alzheimer's Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74:201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemenschneider M., Lautenschlager N., Wagenpfeil S., Diehl J., Drzezga A., Kurz A. Cerebrospinal fluid tau and beta-amyloid 42 proteins identify Alzheimer disease in subjects with mild cognitive impairment. Arch. Neurol. 2002;59:1729–1734. doi: 10.1001/archneur.59.11.1729. [DOI] [PubMed] [Google Scholar]
- Schonheit B., Zarski R., Ohm T.G. Spatial and temporal relationships between plaques and tangles in Alzheimer-pathology. Neurobiol. Aging. 2004;25:697–711. doi: 10.1016/j.neurobiolaging.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Shaw L.M., Vanderstichele H., Knapik-Czajka M., Clark C.M., Aisen P.S., Petersen R.C., Blennow K., Soares H., Simon A., Lewczuk P., Dean R., Siemers E., Potter W., Lee V.M., Trojanowski J.Q. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann. Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled J.G., Zijdenbos A.P., Evans A.C. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Vapnik V. Springer; New York: 1995. The Nature of Statistical Learning Theory. [Google Scholar]
- Vemuri P., Wiste H.J., Weigand S.D., Shaw L.M., Trojanowski J.Q., Weiner M.W., Knopman D.S., Petersen R.C., Jack C.R., Jr. MRI and CSF biomarkers in normal, MCI, and AD subjects: predicting future clinical change. Neurology. 2009;73:294–301. doi: 10.1212/WNL.0b013e3181af79fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri P., Wiste H.J., Weigand S.D., Shaw L.M., Trojanowski J.Q., Weiner M.W., Knopman D.S., Petersen R.C., Jack C.R., Jr. MRI and CSF biomarkers in normal, MCI, and AD subjects: diagnostic discrimination and cognitive correlations. Neurology. 2009;73:287–293. doi: 10.1212/WNL.0b013e3181af79e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos S., van Rossum I., Burns L., Knol D., Scheltens P., Soininen H., Wahlund L.O., Hampel H., Tsolaki M., Minthon L., Handels R., L'Italien G., van der Flier W., Aalten P., Teunissen C., Barkhof F., Blennow K., Wolz R., Rueckert D., Verhey F., Visser P.J. Test sequence of CSF and MRI biomarkers for prediction of AD in subjects with MCI. Neurobiol. Aging. 2012;33:2272–2281. doi: 10.1016/j.neurobiolaging.2011.12.017. [DOI] [PubMed] [Google Scholar]
- Walhovd K.B., Fjell A.M., Brewer J., McEvoy L.K., Fennema-Notestine C., Hagler D.J., Jr., Jennings R.G., Karow D., Dale A.M. Combining MR imaging, positron-emission tomography, and CSF biomarkers in the diagnosis and prognosis of Alzheimer disease. AJNR Am. J. Neuroradiol. 2010;31:347–354. doi: 10.3174/ajnr.A1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin A.K., Blennow K., Andreasen N., Minthon L. CSF biomarkers for Alzheimer's disease: levels of beta-amyloid, tau, phosphorylated tau relate to clinical symptoms and survival. Dement. Geriatr. Cogn. Disord. 2006;21:131–138. doi: 10.1159/000090631. [DOI] [PubMed] [Google Scholar]
- Westman E., Muehlboeck J.S., Simmons A. Combining MRI and CSF measures for classification of Alzheimer's disease and prediction of mild cognitive impairment conversion. Neuroimage. 2012;62:229–238. doi: 10.1016/j.neuroimage.2012.04.056. [DOI] [PubMed] [Google Scholar]
- Zarow C., Vinters H.V., Ellis W.G., Weiner M.W., Mungas D., White L., Chui H.C. Correlates of hippocampal neuron number in Alzheimer's disease and ischemic vascular dementia. Ann. Neurol. 2005;57:896–903. doi: 10.1002/ana.20503. [DOI] [PMC free article] [PubMed] [Google Scholar]


