Abstract
Plants have evolved a number of monitoring systems to sense their surroundings and to coordinate their growth and development accordingly. Vernalization is one example, in which flowering is promoted after plants have been exposed to a long-term cold temperature (i.e. winter). Vernalization results in the repression of floral repressor genes that inhibit the floral transition in many plant species. Here, we describe recent advances in our understanding of the vernalization-mediated promotion of flowering in Arabidopsis and other flowering plants. In Arabidopsis, the vernalization response includes the recruitment of chromatin-modifying complexes to floral repressors and thus results in the enrichment of repressive histone marks that ensure the stable repression of floral repressor genes. Changes in histone modifications at floral repressor loci are stably maintained after cold exposure, establishing the competence to flower the following spring. We also discuss similarities and differences in regulatory circuits in vernalization responses among Arabidopsis and other plants.
INTRODUCTION
The floral transition is a critical developmental change in plant life cycle. Plants ensure their reproductive success, in part, by flowering under favorable conditions. Plants undergo the floral transition only during certain seasons of the year through multiple regulatory networks that interpret environmental signals, such as day lengths and temperature fluctuations (Figure 1). In the model plant Arabidopsis thaliana, four major flowering pathways have been defined by classical genetic analyses: the photoperiod pathway, the autonomous pathway, the gibberellin (GA) pathway, and the vernalization pathway (Figure 1 ). In particular, many plant species in temperate climates need to be exposed to a certain period of winter cold to initiate the floral transition in following spring (Chouard, 1960; Lang, 1965; Bernier et al., 1981). This requirement for exposure to long-term cold for spring flowering is known as vernalization (Lang, 1965; Henderson and Dean, 2004; Sung and Amasino, 2006; Kim et al., 2009). The requirement for vernalization serves in part to prevent flowering in the fall season prior to winter, but permits flowering the following spring.
Figure 1.
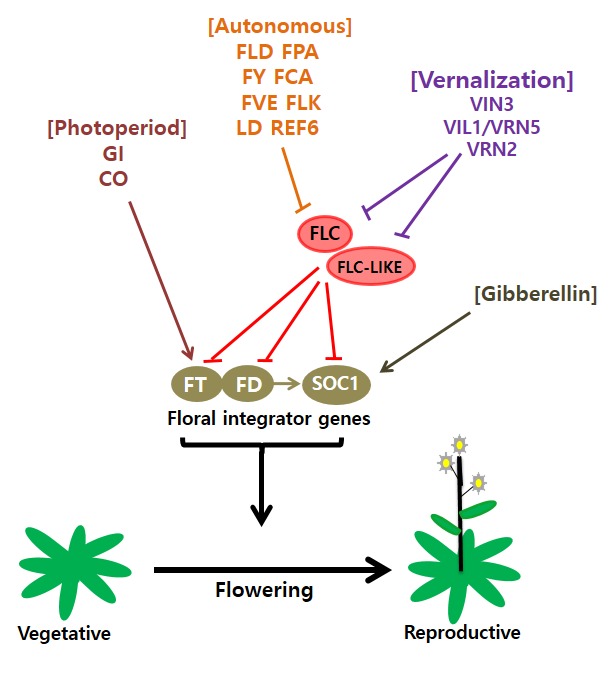
Overview on flowering pathways in Arabidopsis.
FLC and other FLC-related proteins repress floral integrator genes, including FT, FD and SOC1, in Arabidopsis. Upon the activation of floral integrators, the floral transition ensues. FT is induced by the photoperiod pathway through the activation of CO. FT protein is a mobile flowering signal that physically interacts with FD protein at meristem to activate SOC1 and other floral activators. Therefore, FLC and CO antagonistically determine proper timing of flowering in Arabidopsis. Two genetically independent pathways, vernalization and autonomous pathways, repress the transcription of FLC. The autonomous pathway is required for repression of FLC regardless of environment stimuli. The vernalization pathway triggers stable repression of FLC. Gibberellin, a phytohormone, independently promotes flowering through the activation of SOC1 and other floral activator genes.
Plants integrate multiple internal and external flowering cues through gene expression changes. In Arabidopsis, a series of changes in the levels of floral gene expression in environmental and internal flowering pathways converge to regulate floral integrator genes, including FLOWERING LOCUS T (FT:At1 g65480), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1:At2g45660), and AGAMOUS-LIKE 24 (AGL24:At4g24540) (Kim et al., 2009). To ensure optimal flowering time, plants use regulatory circuitries to control expression of floral integrator genes. Major components of these regulatory circuits are two major flowering time genes, CONSTANS (CO: At5g15840) and FLOWERING LOCUS C (FLC: At5g10140) (Figure 1 ). CO acts as a floral activator, whereas FLC acts as a floral repressor. CO is induced by inductive photoperiod (i.e. long days) and activates expressions of downstream floral integrators, thus promoting the floral transition. On the other hand, FLC acts to inhibit the floral transition through suppression of downstream floral integrators (Searle et al., 2006; Kim et al., 2009). Both the autonomous pathway and the vernalization pathway repress the floral repressor FLC. When FLC expression is high, flowering is inhibited even under inductive long days. Therefore, high levels of FLC are responsible for the vernalization requirement in Arabidopsis (Sheldon et al., 1999; Michaels and Amasino, 1999; Johanson et al., 2000). Over the past few decades, extensive molecular genetic studies have elucidated the molecular mechanisms governing the floral transition by both endogenous and environmental cues in Arabidopsis. Here, we describe our current understanding of vernalization-mediated flowering- time regulation.
VERNALIZATION AS A RESPONSE TO COLD TEMPERATURES
Cold temperature initiates a series of physiological and molecular responses in plants. Notably, a number of genes are rapidly induced by above-freezing cold temperatures to trigger cold acclimation in plants (Thomashow, 2001 ; Chinnusamy et al., 2007). Vernalization is similar to cold acclimation in that both responses are triggered by similar above-freezing temperatures (Lang, 1965; Bond et al., 2011; Zografos and Sung, 2012). However, there are two major differences. Cold acclimation occurs in a wide variety of plant tissues, including mature leaves. On the other hand, vernalization is effective either at the shoot apical meristem (SAM) or young leaves, indicating that rapidly dividing cells are responsive to vernalizing cold (Wellensiek, 1964; Lang, 1965; Zeevaart, 1976). In addition, vernalization requires longer period of cold exposure than cold acclimation. For example, cold acclimation can be achieved within days of cold exposure whereas vernalization needs 4–6 weeks of cold exposure in Arabidopsis (Lang, 1965; Bond et al., 2011 ; Zografos and Sung, 2012). This is an adaptive feature of the vernalization response to ensure that plants respond to winter cold, but not to fluctuating temperatures.
THE VERNALIZATION REQUIREMENT
Variation in flowering time is commonly observed in many flowering plant species. One prominent variation in Arabidopsis accessions is the requirement of vernalization for accelerated flowering (Koornneef et al., 1991 ). Genetic studies on variations in the vernalization requirement using natural accessions of Arabidopsis demonstrate that this requirement is mainly due to two dominant genes, FRIGIDA (FRI: At4g00650) and FLC (Lee et al., 1993; Clarke and Dean, 1994; Lee et al., 1994; Michaels and Amasino, 1999; Johanson et al., 2000; Le Corre, 2005; Strange et al., 2011). Naturally occurring mutations in FRI are responsible for early flowering without vernalization in many accessions of Arabidopsis (Johanson et al., 2000; Strange et al., 2011). In the absence of active FRI allele, the level of FLC expression is reduced and plants do not require vernalization for accelerated flowering. In addition, there is natural variation in FLC alleles, which can result in low expression of FLC (Michaels et al., 2003; Strange et al., 2011). Natural accessions that contain both active FRI and FLC alleles require vernalization treatment for them to flower early. Therefore, the vernalization requirement in Arabidopsis is largely due to the level of FLC expression. There are also induced mutants in which FLC expression is elevated even in the absence of an active FRI allele. These mutants are collectively known as autonomous pathway mutants (Koornneef et al., 1991; Michaels and Amasino, 2001). Mutations in autonomous pathway genes result in high level of FLC expression and thus confer the vernalization requirement for early flowering in Arabidopsis. FLC encodes a MADS-box DNA binding protein that acts as a transcriptional repressor. FLC directly binds to downstream floral integrators, including FT, FD (At4g35900), and SOC1, to inhibit their transcription (Helliwell et al., 2006; Searle et al., 2006). Vernalization triggers mitotically-stable repression of FLC and thus allows plants to flower under the inductive photoperiod. Next, we describe molecular bases of these two genetic determinants that require plants to be vernalized to flower early in Arabidopsis.
FLC activation by a FRIGIDA (FRI) complex
FRI mainly acts to up-regulate FLC transcription and thus contributes to the vernalization requirement in winter-annual Arabidopsis accessions (Michaels and Amasino, 2001). Forward genetic approaches have been used to characterize the molecular mechanisms of FRI in the activation of FLC. Suppressor screenings identified a series of components that are required for the FRI-dependent activation of FLC. These include FRILIKE 1 (FRL1: At5g16320), FRI-LIKE 2 (FRL2: At1g31814), FRIGIDA ESSENTIAL 1 (FES1: At2g33835), SUPPRESSOR OF FRIGIDA 4 (SUF4: At1g30970), FLC EXPRESSOR (FLX: At2g30120), FLOWERING LOCUS C EXPRESSOR-LIKE 4 (FLL4: AT5g61920) (Michaels et al., 2004; Schmitz et al., 2005; Kim et al., 2006; Andersson et al., 2008; Ding et al., 2013; Lee and Amasino, 2013). Mutations in these genes commonly result in early flowering even in the presence of functional FRI allele, suggesting that these genes are required for the function of FRI. Interestingly, mutations in this group of genes have only a little effect on elevated levels of FLC expression caused by lesions in autonomous pathway genes. This suggests that FLC activation in mutations of the autonomous pathway genes is achieved independently of FRI. Biochemical purifications of FRI-containing complex (FRI-C) revealed that genetically identified components indeed form a large protein complex (Choi et al., 2011).
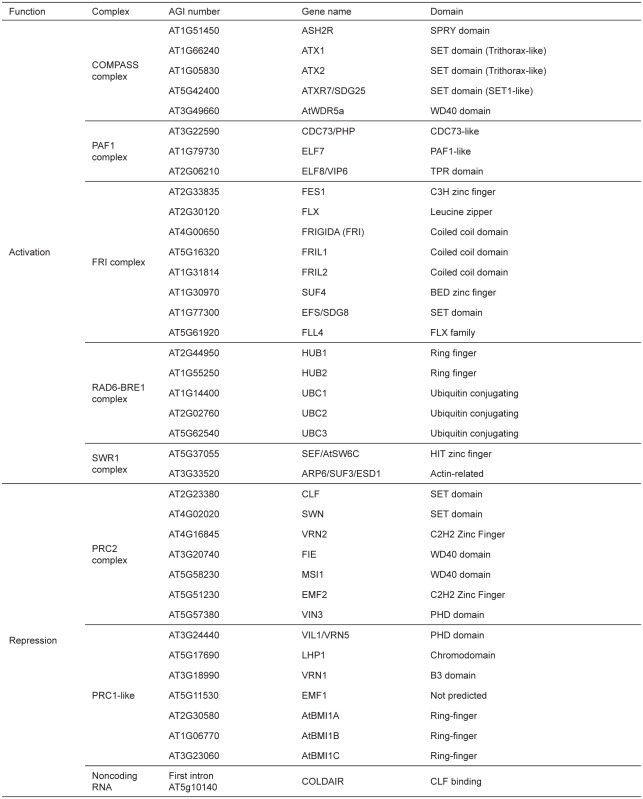
Among components of FRI-C, SUF4, a BED-type zinc finger protein, appears to recruit FRI-C to FLC through its binding to a 15 bp-sequence motif (-CCAAATTTTAAGTTT-) at the FLC promoter region (Choi et al., 2011). Leucine zipper-containing proteins, FLX and FLX4, shows transcriptional activator activity, which in part explains the transcriptional activation of FLC by FRIC C et al., 2013). However, the biochemical function of FRI or its related proteins, FRL1 and FRL2, are not known. FRI-C also shows strong association with components of chromatin remodeling complexes, including components of SWR1 complex: PHOTOPERIOD INDEPENDENT EARLY FLOWERING 1 (PIE1: At3g12810), ACTIN-RELATED PROTEIN 6 (ARP6)/ SUPPRESSOR OF FRI 3 (SUF3)/ESD1 (At3g33520), and SERRATED LEAVES AND EARLY FLOWERING (SEF)/AtSWC6 (At5g37055) (Choi et al., 2011). Trithorax-like SET domain protein, EARLY FLOWERING IN SHORT DAYS (EFS: At1g77300, also known as SDG8, ASHH2, and CCR1) is also intimately associated with FRI-C, indicating that chromatin modifications play a role in the activation of FLC by FRI-C (Choi et al., 2011). Unlike core components of FRI-C, these components of chromatin remodeling complexes are also required for the activation of FLC in autonomous pathway mutants as well. Therefore, chromatin remodeling complexes are more generally required for transcriptional activation of FLC. We will discuss the role of chromatin remodeling complexes in the activation of FLC later in this review. Components of FRI-C are listed in Table 1.
Table 1.
Genes involved in activation or repression of FLC in Arabidopsis
FLC activation by mutations in autonomous pathway genes
Historically, late flowering mutants were classified into two groups according to their flowering behavior under different environmental conditions (Koornneef et al., 1991). Although autonomous pathway mutants flower more rapidly under inductive long days than non-inductive short days, their flowering is markedly delayed under both conditions. In contrast, photoperiod pathway mutants show delayed flowering only under inductive photoperiod, suggesting that photoperiod pathway mutants are blind to inductive photoperiod. In addition, the flowering of autonomous pathway mutants is accelerated by vernalization treatment whereas flowering of photoperiod pathway mutants is not (Koornneef et al., 1991). Because autonomous pathway mutants remain responsive to both photoperiod and vernalization, the promotion of flowering by the autonomous pathway is considered to be independent of environmental stimuli (photoperiod and temperature).
Even though the autonomous pathway is independent of the vernalization pathway, all autonomous pathway genes function to repress the expression of FLC (Michaels and Amasino, 2001), indicating that FLC is a common target for both autonomous and vernalization pathways. Autonomous pathway genes include LUMINIDEPENDENS (LD: At4g02560), FCA (At4g16280), FPA (At2g43410), FY (At5g13480), FLOWERING LOCUS D (FLD: At3g 10390), FVE (At2g19520), FLOWERING LOCUS K (FLK: At3g04610) and RELATIVE OF EARLY FLOWERING 6 (REF6: At3g48430) (Macknight et al., 1997; Schomburg et al., 2001; Lim et al., 2004; Noh et al., 2004; Simpson, 2004). Largely, two classes of proteins are notable among the autonomous pathway proteins. Some of the autonomous proteins are implicated in RNA processing. For example, FCA, FPA, and FLK contain RNA binding motifs and FY is a homolog of the yeast 3′ processing factor Pfs2p (polyadenylation factor 1 subunit 2). Other autonomous pathway proteins are chromatin-modifying enzymes. FLD and REF6, encode two different types of histone demethylases (He et al., 2003; Noh et al., 2004; Jiang et al., 2007). The predicted biochemical functions of these autonomous pathway proteins are consistent with a model in which autonomous pathway proteins function through a coupling of RNA 3′ end processing and chromatin-modifying events (Kim et al., 2009; Michaels, 2009; Liu et al., 2010).
Coupling of RNA 3′ end processing and chromatin modifications may happen during 3′ end processing of a group of antisense RNAs (Liu et al., 2007; Hornyik et al., 2010; Liu et al., 2010). These antisense noncoding RNAs (ncRNAs; collectively known as COOLAIR) are transcribed from the 3′ end of FLC locus. Although these ncRNAs may be responsible for the accumulation of siRNA that targets a downstream region of FLC 3′ end (Swiezewski et al., 2007), overall transcription level of COOLAIR is not correlated with that of sense FLC mRNA. For example, a mutant in which COOLAIR is expressed higher than the wild type results in elevated level of FLC and thus late-flowering (Sun et al., 2013). Rather, a proposed model suggests that differential 3′ end processing and polyadenylation result in repression of FLC (Liu et al., 2007; De Lucia and Dean, 2011). COOLAIR transcripts are preferentially processed at the proximal end in the presence of active FCA, FPA and FY. Given that FLD is required for the function of FCA in the repression of FLC, FCA-mediated 3′ end processing of COOLAIR appears to be a part of mechanism in which FLD mediates the demethylation of methylated Histone H3 Lys 4 (H3K4) at FLC chromatin (Liu et al., 2007; De Lucia and Dean, 2011). However, how these coordinated processes are eventually merged to repress FLC transcription remains to be addressed.
FLC activation through chromatin modifications
Although there is a clear difference between the FRI-mediated activation of FLC and the activation of FLC in autonomous pathway mutants, a group of genes are commonly required for the activation of FLC in both cases. For example, EFS was isolated as a component of FRI-C (Choi et al., 2011). However, EFS is also required for the activation of FLC in autonomous pathway mutants (Kim et al., 2005). EFS encodes a histone methyltransferase that has a dual function for di- and tri-methylation of both histone H3 Lys 4 (H3K4) and H3 Lys 36 (H3K36) at FLC chromatin (Kim et al., 2005; Zhao et al., 2005; Xu et al., 2008; Ko et al., 2010). Methylations of H3K4 and H3K26 are hallmarks of actively transcribed chromatin and are required for the activation of FLC transcription. Similar to EFS, additional genes are required for transcriptional activation of FLC. This group of genes shares two interesting features. First, most of them function to modify and/or remodel chromatin. In addition, mutations in these genes cause pleiotropic effects on other developmental processes. For example, mutations in EFS result in not only early flowering but also other developmental defects, including reduced organ size and shoot branching, carotenoid accumulation, and seed fertility (Kim et al., 2005; Zhao et al., 2005; Dong et al., 2008; Xu et al., 2008; Cazzonelli et al., 2009; Grini et al., 2009).
H3K4me3 is a representative active histone mark in eukaryotes (Schneider et al., 2004) and is associated with high levels of FLC expression. Methylation of H3K4 is catalyzed by SETdomain containing proteins. In yeast, two SET-domain containing proteins, SET1 and SET2, are responsible for the methylation of H3K4 and H3K36, respectively. Yeast SET1 is an essential component of a complex named COMPASS (Complex proteins associated with SET1) (Krogan et al., 2003). Another protein complex, PAF1 (RNA polymerase II associated factor 1) complex, is also necessary for H3K4me3 enrichments in eukaryotes. COMPASS and PAF1 complexes are physically associated to coordinate transcriptional activation of target genes by linking H3K4 histone methylation and transcriptional activation (Krogan et al., 2003). PAF1 complex connects COMPASS to RNA polymerase II machinery and thus leads to active transcription of target genes (Betz et al., 2002).
Two Arabidopsis homologs of yeast PAF1 complex components, EARLY FLOWERING 7 (ELF7: At1g79730) and EARLY FLOWERING 8 (ELF8: At2g06210; also known as VERNALIZATION INDEPENDENCE 6, VIP6) were isolated as essential components for FLC activation. Both ELF7 and ELF8 are required for H3K4me3 enrichment at FLC chromatin and ELF7 and ELF8 encode homologs of yeast PAF1 and CTR9 (a component of yeast PAF1-complex), respectively (He et al., 2004). Other Arabidopsis homologs encoding members of yeast PAF1-COMPASS complexes have also been isolated as required components for the activation of FLC. These include VERNALIZATION INDEPENDENCE 3 (VIP3: At4g29830: Arabidopsis homolog of human hSki8), VERNALIZATION INDEPENDENCE 4 (VIP4: At5g61150: Arabidopsis homolog of yeast Leo1), VERNALIZATION INDEPENDENCE 5 (VIP5: At1g61040: Arabidopsis homolog of yeast Rtf1), ARABIDOPSIS TRITHORAX-LIKE 1 (ATX1: At2g31650) and ARABIDOPSIS TRITHORAX LIKE 2 (ATX2: At1g05830) (Baumbusch et al., 2001 ; Zhang and van Nocker, 2002; AlvarezVenegas et al., 2003; He et al., 2004; Oh et al., 2004; Pien et al., 2008; Saleh et al.,2008). Lesions in these PAF 1-associated complex components commonly result in decreased H3K4me3 enrichment at FLC chromatin, and thus lead to early flowering in the presence of active FRI allele or autonomous pathway mutations.
A group of SET domain proteins, including ATX1, ATX2, ARABIDOPSIS TRITHORAX-RELATED 3 (ATXR3: At4g15180), and ARABIDOPSIS TRITHORAX-RELATED 7 (ATXR7: At5g42400), are redundantly responsible for H3K4 methylations at FLC chromatin through their histone methyltransferase activities (Pien et al., 2008; Tamada et al., 2009; Berr et al., 2010; Guo et al., 2010; Yun et al., 2012). Therefore, the COMPASS-PAF1 complex establishes the vernalization requirement in Arabidopsis through the activation of FLC. Genetically characterized components of the COMPASS-PAF1 complex involved in FLC activation in Arabidopsis are listed in Table 1.
Mono-ubiquitination of histone H2B (H2Bub1), together with H3K4me3, is associated with gene activation in eukaryotes (Wood et al., 2003; Zhang, 2003). In yeast, RAD6 (E2-ubiquitin conjugating enzyme) and BRE1 (E3-ubiquitin ligase enzyme) are responsible for H2Bub1 for certain targets (Jentsch et al., 1987; Robzyk et al., 2000; Yamashita et al., 2004). Similar to COMPASS, PAF1 is necessary for RAD6-BRE1 containing complexmediated H2Bub1. In addition, H2Bub1 is also required for proper H3K4me3 deposition (Wood et al., 2003). In Arabidopsis, there are three RAD6 homologs [UBUQUITIN-CONJUGATING ENZYME 1 (AtUBCI : At1 g14400, AtUBC2: At2g02760, and AtUBC3: At5g62540)] and two BRE1 homologs [HISTONE MONOUBIQUITlNATION 1 (HUB1 : At2g44950 and HUB2: AT1g55250)] (Cao et al., 2008). AtUBC1 and AtUBC2 are redundantly involved in flowering time regulation through enrichment of H2Bub1 at FLC chromatin, whereas AtUBC3 is dispensable for activation of FLC (Xu et al., 2009). Each hubl and hub2 single mutant shows early flowering which results from defects in the enrichments of H2Bub1 and H3K4me3 at FLC chromatin (Cao et al., 2008), suggesting that HUB1 and HUB2 function non-redundantly at FLC chromatin. Known components of RAD6-BRE1 complex involved in FLC activation in Arabidopsis are listed in Table 1.
VERNALIZATION AS AN EPIGENETIC CHANGE
Unlike other biological responses, the vernalization response (accelerated flowering) is not immediately triggered by the stimulus (low temperatures). Rather, accelerated flowering happens when the original stimulus (low temperatures) is removed (warm temperatures in the following spring). This epigenetic nature of vernalization predicts that low temperatures during winter establish stable changes that last until the following spring to promote the floral transition (Lang, 1965). In Arabidopsis, a major stable change by vernalization is the stable repression of a floral repressor, FLC. Prior to vernalization, high levels of FLC block the floral transition in winter-annual strains of Arabidopsis (Figure 2). Prolonged exposure to low temperatures results in epigenetic repression of FLC. The repressed state of FLC triggered by low temperatures is stably maintained throughout subsequent mitotic cell divisions even when plants are returned to warm growth temperatures. Therefore, stable repression of FLC by vernalization allows the floral transition to occur when inductive day length activates the photoperiod pathway in the spring (Figure 1).
Figure 2.
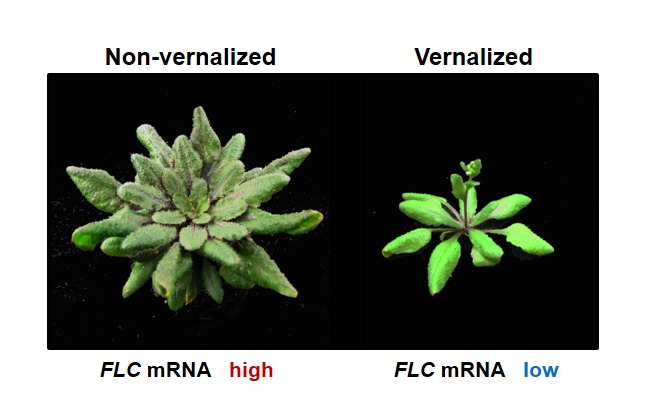
Vernalization-mediated acceleration of flowering.
Winter-annual strains of Arabidopsis flower late without vernalization (Left). Flowering of winter-annual strains of Arabidopsis is accelerated by vernalization (Right).
Genetic characterization of the vernalization pathway
To understand the molecular mechanism underlying vernalization-mediated FLC repression, genetic approaches have been used (Chandler et al., 1996; Gendall et al., 2001; Levy et al., 2002; Sung and Amasino, 2004; Greb et al., 2007). The first component characterized from genetic screens was VERNALIZATION (VRN2: At4g16845) (Gendall et al., 2001). VRN2 is a homolog of the Polycomb-group zinc finger protein, SUPPRESSOR OF ZESTE-12 (Su(Z)12), and lesions in VRN2 results in de-repression of FLC. Polycomb-group genes are classically known to be essential for gene repressions in higher eukaryotes (Ringrose and Paro, 2004). Biochemical purification and subsequent characterization revealed that Su(Z)12 is a component of a histone methyltransferase complex, Polycomb Repression Complex 2 (PRC2). PRC2 mediates methylation at H3K27 through one of its components, ENHANCER OF ZESTE (E(z)), a SET-domain containing methyltransferase (Cao et al., 2002; Kuzmichev et al., 2002; Muller et al., 2002). Additional components of the vernalization pathway include VERNALIZATION1 (VRN1: At3g18990), VERNALIZATION INSENSITIVE 3 (VIN3: At5g57380) and VIN3-LIKE 1 (VIL1)IVERNALIZATION 5 (VRN5) (At3g24440), which are non-redundantly necessary for vernalization-mediated re pression of FLC (Levy et al., 2002; Sung and Amasino, 2004; Sung et al., 2006a; Greb et al., 2007). VRN1 belongs to a small family of plant-specific B3 DNA-binding proteins and VIN3 and VIL1/VRN5 are plant-specific Plant Homeo Domain (PHD) motifcontaining proteins. PHD motifs recognize and bind a wide variety of modified histones in eukaryotes (Musselman and Kutateladze, 2011). VRN1, VRN2, VIL1/VRN5 are constitutively expressed regardless of vernalization. In contrast, VIN3 is only induced when plants are kept under prolonged periods of cold temperature. When plants are returned to warm growth temperatures, transcription of VIN3 quickly decreases (Sung and Amasino, 2004; Kim and Sung, 2013). Therefore, VIN3 is a cold-specific component in the vernalization pathway in Arabidopsis.
Changes in histone modifications at FLC by vernalization
Identification of VRN2 and VIN3 as essential components in vernalization-mediated FLC repression implicated that histone modifications play roles in the process. Indeed, the chromatin context of FLC undergoes a series of changes by vernalization. During and after vernalization, levels of histone modifications associated with gene activation are reduced (Bastow et al., 2004; Sung and Amasino, 2004). By contrast, repressive histone modifications (i.e. H3K9me2 and H3K27me3) are substantially increased at FLC chromatin by vernalization (Bastow et al., 2004; Sung and Amasino, 2004; Schubert et al., 2006; Sung et al., 2006a; Greb et al., 2007). VIN3 co-purifies with components of PRC2, including VRN2 (Wood et al., 2006; De Lucia et al., 2008). Both VIN3 and VRN2 are required for methylation of H3K27 at FLC chromatin by vernalization (Bastow et al., 2004; Sung and Amasino, 2004; Kim and Sung, 2013).
VIN3 is a member of a plant-specific small gene family together with VIL1/VRN5, VIN3-LIKE 2 (VIL2)/VRN5-LIKE 1 (VEL1) (At4g30200) and VIN3-LIKE 3 (VIL3) /VRN5-LIKE 2 (VEL2) (At2g18880). VIL1 and VIL2 biochemically co-purify together with VIN3-containing PRC2 complex (De Lucia et al., 2008), suggesting that they share a common biochemical function in the vernalization response. Although FLC is a major target for repression by vernalization, other FLC-related genes are also repressed by vernalization (Kim and Sung, 2013). In the presence of functional FRI allele, FLC is the main contributor for the vernalization requirement. FLC-related genes include FLOWERING LOCUS M (FLM)IMADS AFFECTING FLOWERING 1 (MAF1) (At1g77080), MADS AFFECTING FLOWERING 2 (MAF2: At5g65050), MADS AFFECTING FLOWERING 3 (MAF3: At5g65060), MADS AFFECTING FLOWERING 4 (MAF4: At5g65070), and MADS AFFECTING FLOWERING 5 (MAF5: At5g65080). They commonly act as floral repressors (Ratcliffe et al., 2001; Scortecci et al., 2001 ; Ratcliffe et al., 2003; Gu et al., 2013; Kim and Sung, 2013). The chromatin of all FLC and FLC-related loci become enriched with repressive histone marks (H3K9me2 and H3K27me3) as a result of vernalization (Kim and Sung, 2013).
Roles of VIN3 family of proteins in vernalization
Although all members of VIN3 gene family function in vernalization, their contributions differ in “timing” during the course of vernalization. VIN3 and VIL2/VEL1 act during cold exposure, whereas VIL1/ VRN5 and VIL3 are predominantly involved after cold (Sung et al., 2006a; Greb et al., 2007; De Lucia et al., 2008; Kim and Sung, 2013). Each member of VIN3 family of proteins is associated with certain FLC gene family chromatin to exert their repressive activities on their respective targets. VIN3 is required for repression of all members of FLC gene family, indicating that VIN3 is a master regulator for vernalization. Other members of the VIN3 family of proteins are necessary for a subset of FLC-related genes (Sung et al., 2006a; Kim and Sung, 2010, 2013). VIL1/VRN5 is necessary for the repression of FLC and FLM whereas VIL2/VEL1 is necessary for the repression of MAF4 and MAF5 by vernalization. VIL3/VEL2 is enriched at MAF2 ~ MAF5 chromatin and necessary for proper repression of MAF2 ~ MAF5. Given that VIN3, VIL1 and VIL2 can be found together with PRC2 complex, it is likely that VIN3 family of proteins function through alternative complexes with the core components of PRC2 at their target chromatin (De Lucia et al., 2008; Kim and Sung, 2013). Although the VIN3 family of proteins can directly interact with one and another (i.e. direct interaction between VIN3 and VIL1 though their VID motifs) (Sung et al., 2006a; Greb et al., 2007), no direct interaction between members of the VIN3 family of proteins and core components of PRC2 is known.
All VIN3 family proteins contain a PHD finger, a motif known to bind a wide range of modified histones (Musselman and Kutateladze, 2011). Indeed, all VIN3 family proteins preferentially bind to H3K9me2 peptides in vitro through PHD motifs (Figure 3). H3K-9me2 mark is enriched at FLC gene family chromatin by vernalization and the VIN3 gene family is necessary for the enrichment of H3K9me2 at their respective target chromatin, supporting the biological significance of such binding activities by VIN3 family of proteins (Kim and Sung, 2013). Mutations in the VIN3 gene family also impair vernalization-mediated enrichment of H3K27me3 at FLC gene family chromatin. Therefore, it appears that the preferential binding to H3K9me2 by VIN3 family proteins may reinforce the activity of PRC2, H3K27 methylation, at target chromatin (Figure 3) (Kim and Sung, 2013).
Figure 3.
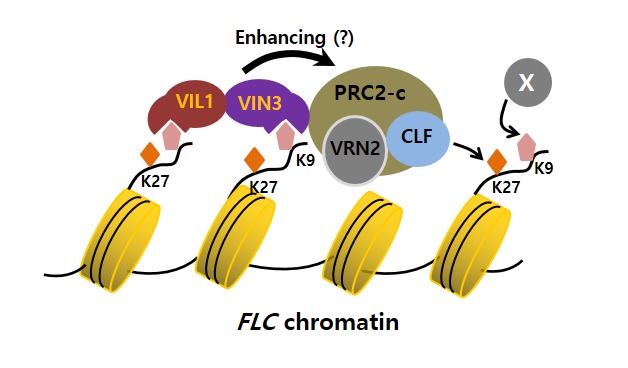
Cooperative activity of VIN3 and PRC2 for the repression of FLC by vernalization.PRC2 mediates tri-methylation of H3K27 at FLC chromatin. H3K9me2 at FLC chromatin can be recognized by VIN3 and VIL1/VRN5 through their PHD motifs. VIN3 and VIL1/VRN5 physically associate with PRC2 and enhance the H3K27 methylation activity of PRC2.
Polycomb-mediated FLC repression by vernalization
Polycomb group proteins maintain gene expression pattern in a wide variety of cells during development by regulating chromatin structure. Core components of PRC2 are well conserved in higher eukaryotes, including Arabidopsis (Hsieh et al., 2003). PRC2 core components consist of Su(z)12, E(z), EED and RbAp46/48 in mammals. Among Arabidopsis components of PRC2, two homologs of E(z), CURLY LEAF (CLF: At2g23380) and SWINGER (SWN: At4g02020) and a homolog of Su(z)12, VRN2, are involved in the repression of FLC (Chanvivattana et al., 2004). The enrichment of PRC2 increases at FLC chromatin by vernalization. PRC2 contributes to the repression of genes mainly through its H3K27 methyltranseferase activity (Cao et al., 2002; Kuzmichev et al., 2002; Muller et al., 2002). Components of PRC2 complex involved in FLC repression in Arabidopsis are listed in Table 1.
Another major Polycomb group complex, POLYCOMB REPRESSIVE COMPLEX 1 (PRC1), exerts another histone modification activity, mono-ubiquitination of histone H2A (H2Aub1, a repressive histone mark) (Margueron and Reinberg, 2011). PRC1 is also involved in chromatin compaction through the binding to H3K27me3 mark by Polycomb protein in Drosophila and mammals. In Arabidopsis, LIKE-HETEROCHROMATIN PROTEIN 1 (LHP1: At5g17690), binds to H3K27me3 in vitro (Zhang et al., 2007) and accumulates at FLC chromatin by vernalization (Mylne et al., 2006; Sung et al., 2006b; Turck et al., 2007). In Ihp1 mutants, vernalization-mediated repression of FLC is not stable, indicating its essential role in the maintenance of repressed chromatin. EMBRYONIC FLOWERING 1 (EMF1: At5g11530) encodes a plantspecific protein with motifs found in transcriptional regulators and may function as a component of PRC1 -like complex in Arabidopsis (Aubert et al., 2001; Calonje et al., 2008; Kim et al., 2010; Kim et al., 2012). In mammals, PRC1 stabilizes repressed state of H3K-27me3-enriched target chromatin through its H2A mono-ubiquitination activity (Margueron and Reinberg, 2011). Similarly, two Arabidopsis RING-finger proteins, AtBMI1A (At2g30580) and AtBMI1B (At1g06770), function to mediate the formation of H2Aub1 at chromatin in Arabidopsis (Bratzel et al., 2010; Yang et al., 2013). atbmi1a/atbmi1b double mutants result in de-repression of Polycomb target genes and thus display similar phenotypes to those of PRC2 component mutants. AtBMI1A and AtBMI1B show H2Aub1 activity in vitro and interact with other Arabidopsis PRC1-like components, LHP1 and EMF1. Another Arabidopsis RING finger protein AtBMI1C (At3g23060) was also reported to be involved in repression of FLC through its H2Aub1 activity (Li et al., 2011). Therefore, Arabidopsis PRC1-like complex consists of at least LHP1, EMF1, AtBMI1A, AtBMI1B, and AtBMI1C and contributes to Polycombmediated repression of FLC (Figure 4).
Figure 4.
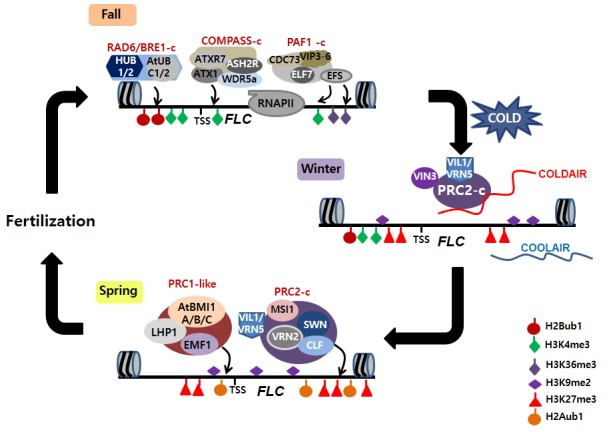
Schematic representation of mechanisms underlying FLC activation and repression.
A) Prior to vernalization (fall), FLC is highly expressed by activation chromatin-remodeling complexes, PAF1-C, COMPASS-C and RAD6-BRE1-C. B) During winter, a long ncRNA, COLDAIR, is transcribed from the first intron of FLC and functions to recruit PRC2. COOLAIR and VIN3 is also transiently induced by cold and PRC2 together with PHD finger proteins, VIN3 and VIL1/VRN5, becomes associated with FLC chromatin. Level of FLC mRNA decreases during cold exposure. C) After cold (Spring), the repressed state of FLC is stably maintained through combinatorial activities of PRC2 and PRC1-like complex.
Physical alteration of higher order structure is closely correlated with gene expression in eukaryotes (Fraser and Bickmore, 2007; Hubner et al., 2013). Conformational changes of higher order chromatin structure are also reported for Polycomb-mediated silenced loci in Drosophila and mammals, known as polycomb bodies within nucleus (Lanzuolo et al., 2007; Bantignies and Cavalli, 2011; Nora et al., 2012). Similarly in plants, it has been observed that FLC chromatin is repositioned in nucleus by vernalization (Rosa et al., 2013). Therefore, Polycomb-mediated silencing of FLC by vernalization may also involve physical repositioning of chromatin.
Role of noncoding RNAs in vernalization
Polycomb group complexes regulate a wide range of genes in eukaryotes. However, how these complexes are recruited to certain target genes is not well understood. Recent studies show that noncoding RNAs (ncRNAs) are becoming recognized as a part of PRC2 recruitment machinery. In mammals, several long ncRNAs, such as ANRIL, HOTAIR, Xist and Kcnq1ot1, physically interact with components of PRC2 and direct PRC2 to target chromatin (Pandey et al., 2008; Zhao et al., 2008; Tsai et al., 2010; Kotake et al., 2011). The involvement of ncRNAs is not restricted to PRC2. A number of ncRNAs have been co-purified with various types of chromatin modifying complexes, indicating that ncRNAs function in various gene expression regulations in eukaryotes (Tsai et al., 2010; Guttman et al., 2011; Spitale et al., 2011; Guttman and Rinn, 2012). In the regulation of FLC, two different types of long ncRNAs, COOLAIR and COLDAIR, appear to have regulatory function (Liu et al., 2007; Swiezewski et al., 2009; Hornyik et al., 2010; Liu et al., 2010; Heo and Sung, 2011).
First, a group of antisense transcripts are detectable from FLC locus. These antisense ncRNAs are largely grouped into two classes based on different polyadenylation sites, proximal and distal. A total of 6 splicing variants also exist among these two classes of antisense ncRNAs (Hornyik et al., 2010). It has been proposed that the proximal polyadenylation by components of autonomous pathway, including FCA, FY and FPA, triggers the repression of FLC through the recruitment of FLD (De Lucia and Dean, 2011). Transcriptional activity of these antisense RNAs transiently increases during the course of vernalization. Therefore, these antisense ncRNAs, known as COOLAIR, are also implicated in the vernalization-mediated FLC repression (Swiezewski et al., 2009). Both proximally and distally polyadenylated antisense transcripts increase during early time periods (~ 2 weeks) of vernalization treatment, but eventually decrease to the basal level at later periods of vernalization treatment (Swiezewski et al., 2009). Alternative polyadenylation of COOLAIR does not play a role in vernalization since all known autonomous pathway mutants are responsive to vernalization treatment. Several T-DNA insertion lines, in which COOLAIR transcription is largely impaired, are responsive to vernalization (Helliwell et al., 2011). In addition, a mutant in which COOLAIR is up-regulated exhibits derepression of FLC (Sun et al., 2013). Therefore, increased levels of COOLAIR transcripts do not trigger FLC repression. An alternative model has been proposed to have a “co-transcriptional” regulation circuitry (De Lucia and Dean, 2011). In this model, the antisense transcription “read-through” may interfere with sense transcription of FLC, therefore contributes to initial transcriptional repression of FLC during the course of vernalization. Mechanistic details of this model remain to be elucidated.
Another long ncRNA, known as COLDAIR (COLD ASSISTED INTRONIC NON-CODING RNA), is also transcribed from the FLC locus. COLDAIR is transcribed from the first intron of FLC in a sense direction compared to the FLC transcript (Heo and Sung, 2011). Similar to COOLAIR, COLDAIR is also transiently induced by vernalization. COLDAIR transcripts (about 1.1 kb long) physically interact with CLF, one of PRC2 components. Reduced expression of COLDAIR using RNAi impairs the vernalizationmediated enrichment of PRC2 at FLC chromatin. Reduced enrichment of PRC2 at FLC chromatin results in decreased enrichment of H3K27me3 (Heo and Sung, 2011). Taken together, COLDAIR is a part of the machinery that recruits PRC2 to FLC chromatin by vernalization. Biochemical properties of COLDAIR are similar to those of PRC2 associated ncRNAs in mammals (Rinn et al., 2007; Tsai et al., 2010; Guttman et al., 2011; Spitale et al., 2011; Guttman and Rinn, 2012). Therefore, ncRNA-mediated recruitment of PRC2 may be an evolutionally conserved mechanism in eukaryotes. Transiently increased transcription of COLDAIR indicates that COLDAIR may function in initial recruitment of PRC2 to FLC by vernalization. However, it is not yet clear whether the same mechanism also plays a role in the maintenance of PRC2 recruitment to FLC after the cold treatment. PRC2 recruitment by HOTAIR includes a conserved sequence motif of HOTAIR appears to be necessary for the recruitment of PRC2, perhaps through RNADNA sequence recognition (Chu et al., 2011). However, it remains to be addressed how the COLDAIR targets PRC2 to FLC locus.
Re-activation of FLC in the next generation
Prior to vernalization, FLC is highly expressed and its chromatin is enriched with H3K4me3, an active histone mark. By vernalization, active histone marks at FLC chromatin decrease. The reduction of active histone marks at FLC chromatin is accompanied by decreased enrichment of active chromatin modifying complex components, including ATXR7 and EFS (Kim and Sung, 2013). Instead, repressive chromatin modifying complexes, including PRC2, become predominantly associated with FLC chromatin, which result in the enrichment of repressive histone marks, such as H3K27me3. Therefore, vernalization triggers changes in chromatin landscape at FLC (Figure 4). This repression of FLC is stable even after the cold exposure. However, this repression is only stable throughout mitosis and FLC is re-activated in the next generation. This is an adaptive feature of the vernalization response to ensure that each generation of Arabidopsis plants re-achieve the vernalization requirement. FLC appears to be reactivated during the gametogenesis and early embryogenesis after fertilization (Sheldon et al., 2008; Choi et al., 2009). At this stage, FLC chromatin must undergo reprogramming of chromatin context from repressive to active states. Active chromatin modifying complex components, particularly components of FRI-C and PAF1, are necessary for the reactivation of FLC (Yun et al., 2011). Mechanisms of how these active chromatin modifying complexes are recruited to FLC chromatin and how repressive chromatin modifying complexes are excluded from FLC chromatin are not known.
VERNALIZATION IN OTHER FLOWERING PLANTS
Although Arabidopsis has served as an excellent model system to understand molecular mechanism of the vernalization response, other vernalization-required species use different gene regulatory circuitries. Here, we briefly describe current understanding on molecular circuitries of the vernalization response in other flowering species.
Arabis alpina
Arabis alpina, a perennial relative of Arabidopsis, is distinctive in the vernalization response compared to annual/biennial Arabidopsis accessions (Koch et al., 2006; Ansell et al., 2008). Annual plants initiate the floral transition in all apical meristems at the same time during their life cycle, known as monocarpy. In contrast, perennial plants bloom in spring and summer seasons but arrest flowering later. Perennial plants resume vegetative growth in fall and repeatedly undergo vernalization. Therefore, perennial plants flower and set seeds many times in their life cycle (known as polycarpy). Arabis plants repeat the cycle of vegetative and reproductive growth phases. Similar to Arabidopsis, an ortholog of FLC (PERPETUAL FLOWERING 1 (PEP1)) acts as a major floral repressor in Arabis (Wang et al., 2009). PEP1 is repressed by vernalizing cold and thus allow plants to bloom. Unlike Arabidopsis, however, PEP1 is re-activated when plants are returned to warm growth temperature. This transient nature of the repression of PEP1 confers polycarpic flowering behavior in Arabis. Consistent with fluctuating expressions of PEP1, a repressive histone mark, H3K27me3, is accumulated at PEP1 chromatin during the cold exposure, but depleted when plants are returned to warm temperature.
An APETALA2-type transcription factor, PERPETUAL FLOWERING 2 (PEP2, an Arabis ortholog of Arabidopsis AP2) also function to repress flowering in Arabis (Bergonzi et al., 2013). PEP2 acts to up-regulate PEP1 to prevent flowering prior to vernalization. Interestingly, Arabis plants respond vernalization only when plants reach to a certain mature age. This age-dependent response to vernalization is achieved via a microRNA, miRNA156. When plants are young, miRNA156 is abundant and prevents flowering through blocking expression of floral activators SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL). However, as plants age, miRNA156 levels decline, resulting in an increase of floral activator, SPL. Therefore, it is likely that PEP2-PEP1 and miRNA156 act in parallel to ensure that Arabis plants become competent to flower only when they have reached appropriate vegetative stage and have been exposed to vernalization (Figure 5).
Figure 5.
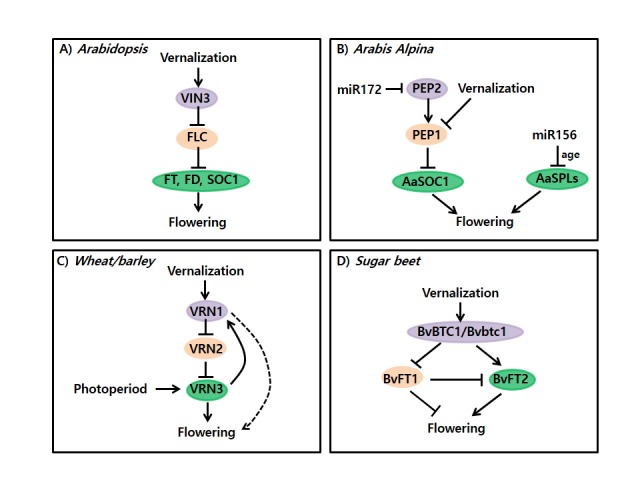
Models of flowering time regulation by vernalization in various flowering plants.
Green: floral activator, Pink: floral repressor, Violet: upstream repressor of floral repressor.
Cereals (wheat and barley)
In temperate cereals, such as wheat and barley, genetic analysis between winter and spring cultivars of the crops has identified several genes involved in the vernalization requirement (Trevaskis et al., 2007; Distelfeld et al., 2009). Three loci, VERNALIZATION1 (VRN1), VRN2, and VRN3 (also known as HvFT1 in barley) are important in determining vernalization requirement and flowering time regulation in temperate cereals (Danyluk et al., 2003; Trevaskis et al., 2003; Yan et al., 2003; Yan et al., 2004a; Yan et al., 2004b). VRN1 and VRN2 from cereals are not related to Arabidopsis VRN1 and VRN2. VRN1 encodes a MADS-box transcription factor that promotes flowering in cereals, while VRN2 encodes a CCT-domain protein and acts as a floral repressor by blocking VRN3 expression (Yan et al., 2004b; Yan et al., 2006). There is no apparent homolog of Arabidopsis FLC in cereals. Instead, a floral repressor, VRN2, acts as a floral repressor in cereals, similar to FLC in Arabidopsis. For example, vernalization results in stable repression of VRN2 (Figure 5). In winter cultivars, VRN1 is induced by vernalizing cold treatment and is required for the repression of VRN2. It is interesting to note that VRN1 chromatin context is subjected to histone modification change by vernalization. Vernalization results in decreased level of enrichment of H3K27me3 and increased level of enrichment of H3K4me3 at VRN1 chromatin. On the other hand, there are no significant changes in histone modifications at VRN2 and VRN3 chromatins (Oliver et al., 2009). Taken together, changes of chromatin structure at VRN1 locus appear to take part in the epigenetic mechanism of the vernalization response in cereals.
Sugar beet (Beta vulgaris)
In sugar beet, a pair of FT homologs (BvFT1 and BvFT2), which encode phosphatidylethanolamine-binding protein, acts antagonistically in the floral transition. BvFT1 acts as a floral repressor whereas and BvFT2 promotes flowering (Pin et al., 2010). In addition, vernalization results in down-regulation of BvFT1. Vernalization-induced repression of BvFT1 is stably maintained even after plants are returned to warm growth temperatures, indicating that BvFT1 functions similarly to FLC. Vernalization requirement in sugar beet is mainly conferred by a dominant allele named BvBTC1 through its regulation of BvFT1 and BvFT2 (Pin et al., 2012). In annual sugar beet, expression of a dominant BvBTC1 allele is increased by long days. This results in the floral transition through the repression of BvFT1 and the activation of BvFT2 under long day conditions. Therefore, annual sugar beet plants with a dominant BvBTC1 allele do not need vernalization for early flowering. In contrast, biennial sugar beet plants carry a partial loss-of-function allele of Bvbtcl. Bvbtc1 is not significantly induced even under long days without vernalization treatment. Bvbtc1 allele can be gradually activated by vernalization treatment to the level sufficient to repress BvFT1 and activate BvFT2 (Figure 5).
Divergent regulatory circuitries of vernalization pathway in flowering plants suggest that plants have independently evolved systems to mediate the vernalization response. Despite clear differences in components of flowering regulatory circuits, one basic theme is conserved; vernalization commonly results in competence to flower through ‘repression of floral repressor’ (Figure 5).
CONCLUSION
Studies using Arabidopsis shed light on our understanding on molecular mechanisms of the vernalization response. Mechanisms underlying vernalization involves various modes of gene expression regulation, from histone modifications to noncoding RNAs. Therefore, what we learn from vernalization studies contributes to our understanding of gene expression. The inducible nature of gene expression makes vernalization one of the best model systems to study mechanistic details of gene expression changes by environmental stimuli in eukaryotes. Combined with a rich genetic resource and recent technological advances, vernalization study using Arabidopsis and other flowering plants continue to provide insights on our understanding of gene regulation in eukaryotes.
ACKNOWLEDGEMENTS
Sung Lab is supported by the University of Texas at Austin, National Science Foundation (IOS-0950785) and National Institute of Health (R01GM100108).
Footnotes
Citation: Dong-Hwan Kim and Sibum Sung. (2014) Genetic and Epigenetic Mechanisms Underlying Vernalization. The Arabidopsis Book 11:e0171. doi:10.1199/tab.0171
elocation-id: e0171
First published on February 12, 2014: e0171. doi: 10.1199/tab.0171
REFERENCES
- Alvarez-Venegas R., Pien S., Sadder M., Witmer X., Grossniklaus U., Avramova Z. ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr. Biol. 2003;13:627–637. doi: 10.1016/s0960-9822(03)00243-4. [DOI] [PubMed] [Google Scholar]
- Andersson C.R., Helliwell C.A., Bagnall D.J., Hughes T.P., Finnegan E.J., Peacock W.J., Dennis E.S. The FLX gene of Arabidopsis is required for FRI-dependent activation of FLC expression. Plant Cell Physiol. 2008;49:191–200. doi: 10.1093/pcp/pcm176. [DOI] [PubMed] [Google Scholar]
- Ansell S.W., Grundmann M., Russell S.J., Schneider H., Vogel J.C. Genetic discontinuity, breeding-system change and population history of Arabis alpina in the Italian Peninsula and adjacent Alps. Mol. Ecol. 2008;17:2245–2257. doi: 10.1111/j.1365-294X.2008.03739.x. [DOI] [PubMed] [Google Scholar]
- Aubert D., Chen L., Moon Y.H., Martin D., Castle L.A., Yang C.H., Sung Z.R. EMF1, a novel protein involved in the control of shoot architecture and flowering in Arabidopsis. Plant Cell. 2001;13:1865–1875. doi: 10.1105/TPC.010094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies F., Cavalli G. Polycomb group proteins: repression in 3D. Trends Genet. 2011;27:454–464. doi: 10.1016/j.tig.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Bastow R., Mylne J.S., Lister C., Lippman Z., Martienssen R.A., Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- Baumbusch L.O., Thorstensen T., Krauss V., Fischer A., Naumann K., Assalkhou R., Schulz I., Reuter G., Aalen R.B. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergonzi S., Albani M.C., Ver Loren van Themaat E., Nordstrom K.J., Wang R., Schneeberger K., Moerland P.D., Coupland G. Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science. 2013;340:1094–1097. doi: 10.1126/science.1234116. [DOI] [PubMed] [Google Scholar]
- Bernier G., Kinet J.-M., Sachs R.M. The physiology of flowering. Boca Raton, Fla: CRC Press; 1981. [Google Scholar]
- Berr A., Shafiq S., Shen W.H. Histone modifications in transcriptional activation during plant development. Biochim. Biophys. Acta. 2010;1809:567–576. doi: 10.1016/j.bbagrm.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Betz J.L., Chang M., Washburn T.M., Porter S.E., Mueller C.L., Jaehning J.A. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics. 2002;268:272–285. doi: 10.1007/s00438-002-0752-8. [DOI] [PubMed] [Google Scholar]
- Bond D.M., Dennis E.S., Finnegan E.J. The low temperature response pathways for cold acclimation and vernalization are independent. Plant Cell Environ. 2011;34:1737–48. doi: 10.1111/j.1365-3040.2011.02370.x. [DOI] [PubMed] [Google Scholar]
- Bratzel F., Lopez-Torrejon G., Koch M., Del Pozo J.C., Calonje M. Keeping cell identity in arabidopsis requires PRC1 RINGfinger homologs that catalyze H2A monoubiquitination. Curr. Biol. 2010;20:1853–1859. doi: 10.1016/j.cub.2010.09.046. [DOI] [PubMed] [Google Scholar]
- Calonje M., Sanchez R., Chen L., Sung Z.R. EMBRYONIC FLOWER1 participates in polycomb group-mediated AG gene silencing in Arabidopsis. Plant Cell. 2008;20:277–291. doi: 10.1105/tpc.106.049957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cao Y., Dai Y., Cui S., Ma L. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell. 2008;20:2586–2602. doi: 10.1105/tpc.108.062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli C.I., Cuttriss A.J., Cossetto S.B., Pye W., Crisp P., Whelan J., Finnegan E.J., Turnbull C., Pogson B.J. Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell. 2009;21:39–53. doi: 10.1105/tpc.108.063131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J., Wilson A., Dean C. Arabidopsis mutants showing an altered response to vernalization. Plant J. 1996;10:637–644. doi: 10.1046/j.1365-313x.1996.10040637.x. [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y., Bishopp A., Schubert D., Stock C., Moon Y.H., Sung Z.R., Goodrich J. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development. 2004;131:5263–5276. doi: 10.1242/dev.01400. [DOI] [PubMed] [Google Scholar]
- Chu C., Qu K., Zhong F.L., Artandi S.E., Chang H.Y. Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Mol. Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J., Zhu J.K. Cold stress regulation of gene expression in plants. Trends Plant Sei. 2007;12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Choi J., Hyun Y., Kang M.J., In Yun H., Yun J.Y., Lister C., Dean C., Amasino R.M., Noh B., Noh Y.S., Choi Y. Resetting and regulation of FLOWERING LOCUS C expression during Arabidopsis reproductive development. Plant J. 2009;23:289–303. doi: 10.1111/j.1365-313X.2008.03776.x. [DOI] [PubMed] [Google Scholar]
- Choi K., Kim J., Hwang H.J., Kim S., Park C., Kim S.Y., Lee I. The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell. 2011;23:289–303. doi: 10.1105/tpc.110.075911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouard P. Vernalization and its relations to dormancy. Annu Rev Plant Physiol. 1960;11:191–238. [Google Scholar]
- Clarke J.H., Dean C. Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Mol. Gen. Genet. 1994;242:81–89. doi: 10.1007/BF00277351. [DOI] [PubMed] [Google Scholar]
- Danyluk J., Kane N.A., Breton G., Limin A.E., Fowler D.B., Sarhan F. TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol. 2003;132:1849–1860. doi: 10.1104/pp.103.023523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F., Dean C. Long non-coding RNAs and chromatin regulation. Curr. Opin. Plant Biol. 2011;14:168–173. doi: 10.1016/j.pbi.2010.11.006. [DOI] [PubMed] [Google Scholar]
- De Lucia F., Crevillen P., Jones A.M., Greb T., Dean C. A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. U S A. 2008;105:16831–16836. doi: 10.1073/pnas.0808687105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Kim S.Y., Michaels S.D. FLOWERING LOCUS C EXPRESSOR Family Proteins Regulate FLOWERING LOCUS C Expression in Both Winter-Annual and Rapid-Cycling Arabidopsis. Plant Physiol. 2013;163:243–252. doi: 10.1104/pp.113.223958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A., Li C., Dubcovsky J. Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 2009;12:178–184. doi: 10.1016/j.pbi.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Dong G., Ma D.P., Li J. The histone methyltransferase SDG8 regulates shoot branching in Arabidopsis. Biochem. Biophys. Res. Commun. 2008;373:659–664. doi: 10.1016/j.bbrc.2008.06.096. [DOI] [PubMed] [Google Scholar]
- Fraser P., Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Gendall A.R., Levy Y.Y., Wilson A., Dean C. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell. 2001;107:525–535. doi: 10.1016/s0092-8674(01)00573-6. [DOI] [PubMed] [Google Scholar]
- Greb T., Mylne J.S., Crevillen P., Geraldo N., An H., Gendall A.R., Dean C. The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr. Biol. 2007;17:73–78. doi: 10.1016/j.cub.2006.11.052. [DOI] [PubMed] [Google Scholar]
- Grini P.E., Thorstensen T., Alm V., Vizcay-Barrena G., Windju S.S., Jorstad T.S., Wilson Z.A., Aalen R.B. The ASH1 HOMOLOG 2 (ASHH2) histone H3 methyltransferase is required for ovule and anther development in Arabidopsis. PLoS One. 2009;4:e7817. doi: 10.1371/journal.pone.0007817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Le C., Wang Y., Li Z., Jiang D., He Y. Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 2013;4:1947. doi: 10.1038/ncomms2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Yu Y., Law J.A., Zhang X. SET DOMAIN GROUP2 is the major histone H3 lysine 4 trimethyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2010;107:18557–18562. doi: 10.1073/pnas.1010478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L., Yang X., Amit I., Meissner A., Regev A., Rinn J.L., Root D.E., Lander E.S. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Michaels S.D., Amasino R.M. Regulation of flowering time by histone acetylation in Arabidopsis. Science. 2003;302:1751–1754. doi: 10.1126/science.1091109. [DOI] [PubMed] [Google Scholar]
- He Y., Doyle M.R., Amasino R.M. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 2004;18:2774–2784. doi: 10.1101/gad.1244504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C.A., Wood C.C., Robertson M., James Peacock W., Dennis E.S. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecularweight protein complex. Plant J. 2006;46:183–192. doi: 10.1111/j.1365-313X.2006.02686.x. [DOI] [PubMed] [Google Scholar]
- Helliwell C.A., Robertson M., Finnegan E.J., Buzas D.M., Dennis E.S. Vernalization-Repression of Arabidopsis FLC Requires Promoter Sequences but Not Antisense Transcripts. PLoS One. 2011:6. doi: 10.1371/journal.pone.0021513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Dean C. Control of Arabidopsis flowering: the chill before the bloom. Development. 2004;131:3829–3838. doi: 10.1242/dev.01294. [DOI] [PubMed] [Google Scholar]
- Heo J.B., Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- Hornyik C., Terzi L.C., Simpson G.G. The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev. Cell. 2010;18:203–213. doi: 10.1016/j.devcel.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Hubner M.R., Eckersley-Maslin M.A., Spector D.L. Chromatin organization and transcriptional regulation. Curr. Opin. Genet. Dev. 2013;23:89–95. doi: 10.1016/j.gde.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., McGrath J.P., Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987;329:131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- Jiang D., Yang W., He Y., Amasino R.M. Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell. 2007;19:2975–2987. doi: 10.1105/tpc.107.052373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U., West J., Lister C., Michaels S., Amasino R., Dean C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000;290:344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Sung S. The Plant Homeo Domain finger protein, VIN3-LIKE 2, is necessary for photoperiod-mediated epigenetic regulation of the floral repressor, MAF5. Proc. Natl. Acad. Sci. U S A. 2010;107:17029–17034. doi: 10.1073/pnas.1010834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Sung S. Coordination of the Vernalization Response through a VIN3 and FLC Gene Family Regulatory Network in Arabidopsis. Plant Cell. 2013;25:454–469. doi: 10.1105/tpc.112.104760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Doyle M.R., Sung S., Amasino R.M. Vernalization: winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 2009;25:277–299. doi: 10.1146/annurev.cellbio.042308.113411. [DOI] [PubMed] [Google Scholar]
- Kim S., Choi K., Park C., Hwang H.J., Lee I. SUPPRESSOR OF FRIGIDA4, encoding a C2H2-Type zinc finger protein, represses flowering by transcriptional activation of Arabidopsis FLOWERING LOCUS C. Plant Cell. 2006;18:2985–2998. doi: 10.1105/tpc.106.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., Zhu T., Sung Z.R. Epigenetic regulation of gene programs by EMF1 and EMF2 in Arabidopsis. Plant Physiol. 2010;152:516–528. doi: 10.1104/pp.109.143495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., Lee J., Eshed-Williams L., Zilberman D., Sung Z.R. EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet. 2012;8:e1002512. doi: 10.1371/journal.pgen.1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., He Y., Jacob Y., Noh Y.S., Michaels S., Amasino R. Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell. 2005;17:3301–3310. doi: 10.1105/tpc.105.034645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J.H., Mitina I., Tamada Y., Hyun Y., Choi Y., Amasino R.M., Noh B., Noh Y.S. Growth habit determination by the balance of histone methylation activities in Arabidopsis. EMBO J. 2010;29:3208–3215. doi: 10.1038/emboj.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M.A., Kiefer C., Ehrich D., Vogel J., Brochmann C., Mummenhoff K. Three times out of Asia Minor: the phylogeography of Arabis alpina L. (Brassicaceae). Mol. Ecol. 2006;15:825–839. doi: 10.1111/j.1365-294X.2005.02848.x. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C.J., van der Veen J.H. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Kotake Y., Nakagawa T., Kitagawa K., Suzuki S., Liu N., Kitagawa M., Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Dover J., Wood A., Schneider J., Heidt J., Boateng M.A., Dean K., Ryan O.W., Golshani A., Johnston M., Greenblatt J.F., Shilatifard A. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A. Physiology of flower initiation. Berlin: Springer-Verlag; 1965. [Google Scholar]
- Lanzuolo C., Roure V., Dekker J., Bantignies F., Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat. Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- Le Corre V. Variation at two flowering time genes within and among populations of Arabidopsis thaliana: comparison with markers and traits. Mol. Ecol. 2005;14:4181–92. doi: 10.1111/j.1365-294X.2005.02722.x. [DOI] [PubMed] [Google Scholar]
- Lee I., Bleecker A., Amasino R. Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol. Gen. Genet. 1993;237:171–176. doi: 10.1007/BF00282798. [DOI] [PubMed] [Google Scholar]
- Lee I., Michaels S.D., Masshardt A.S., Amasino R.M. The late-flowering phenotype of FRIGIDA and mutations in LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J. 1994;6:903–909. [Google Scholar]
- Lee J., Amasino R.M. Two FLX family members are nonredundantly required to establish the vernalization requirement in Arabidopsis. Nat. Commun. 2013;4:2186. doi: 10.1038/ncomms3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy Y.Y., Mesnage S., Mylne J.S., Gendall A.R., Dean C. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science. 2002;297:243–246. doi: 10.1126/science.1072147. [DOI] [PubMed] [Google Scholar]
- Li W., Wang Z., Li J., Yang H., Cui S., Wang X., Ma L. Overexpression of AtBMl1C, a polycomb group protein gene, accelerates flowering in Arabidopsis. PLoS One. 2011;6:e21364. doi: 10.1371/journal.pone.0021364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M.H., Kim J., Kim Y.S., Chung K.S., Seo Y.H., Lee I., Kim J., Hong C.B., Kim H.J., Park C.M. A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell. 2004;16:731–740. doi: 10.1105/tpc.019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Marquardt S., Lister C., Swiezewski S., Dean C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327:94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- Liu F., Quesada V., Crevillen P., Baurle I., Swiezewski S., Dean C. The Arabidopsis RNA-binding protein FCA requires a lysinespecific demethylase 1 homolog to downregulate FLC. Mol. Cell. 2007;28:398–407. doi: 10.1016/j.molcel.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Macknight R., Bancroft I., Page T., Lister C., Schmidt R., Love K., Westphal L., Murphy G., Sherson S., Cobbett C., Dean C. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell. 1997;89:737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- Margueron R., Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D. Flowering time regulation produces much fruit. Curr. Opin. Plant Biol. 2009;12:75–80. doi: 10.1016/j.pbi.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell. 2001;13:935–941. doi: 10.1105/tpc.13.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Bezerra I.C., Amasino R.M. FRIGIDArelated genes are required for the winter-annual habit in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2004;101:3281–3285. doi: 10.1073/pnas.0306778101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., He Y., Scortecci K.C., Amasino R.M. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sei. U S A. 2003;100:10102–10107. doi: 10.1073/pnas.1531467100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J., Hart C.M., Francis N.J., Vargas M.L., Sengupta A., Wild B., Miller E.L., O'Connor M.B., Kingston R.E., Simon J.A. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Musselman C.A., Kutateladze T.G. Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res. 2011;39:9061–9071. doi: 10.1093/nar/gkr613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylne J.S., Barrett L., Tessadori F., Mesnage S., Johnson L., Bernatavichute Y.V., Jacobsen S.E., Fransz P., Dean C. LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc. Natl. Acad. Sci. U S A. 2006;103:5012–5017. doi: 10.1073/pnas.0507427103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B., Lee S.H., Kim H.J., Yi G., Shin E.A., Lee M., Jung K.J., Doyle M.R., Amasino R.M., Noh Y.S. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell. 2004;16:2601–2613. doi: 10.1105/tpc.104.025353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J., Gribnau J., Barillot E., Bluthgen N., Dekker J., Heard E. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S., Zhang H., Ludwig P., van Nocker S. A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell. 2004;16:2940–2953. doi: 10.1105/tpc.104.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S.N., Finnegan E.J., Dennis E.S., Peacock W.J., Trevaskis B. Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. Proc. Natl. Acad. Sci. U S A. 2009;106:8386–8391. doi: 10.1073/pnas.0903566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R.R., Mondal T., Mohammad F., Enroth S., Redrup L., Komorowski J., Nagano T., Mancini-Dinardo D., Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Pien S., Fleury D., Mylne J.S., Crevillen P., Inze D., Avramova Z., Dean C., Grossniklaus U. ARABIDOPSIS TRITHORAX 1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell. 2008;20:580–588. doi: 10.1105/tpc.108.058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin P.A., Benlloch R., Bonnet D., Wremerth-Weich E., Kraft T., Gielen J.J., Nilsson O. An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science. 2010;330:1397–1400. doi: 10.1126/science.1197004. [DOI] [PubMed] [Google Scholar]
- Pin P.A., Zhang W., Vogt S.H., Dally N., Buttner B., Schulze-Buxloh G., Jelly N.S., Chia T.Y., Mutasa-Gottgens E.S., Dohm J.C., Himmelbauer H., Weisshaar B., Kraus J., Gielen J.J., Lommei M., Weyens G., Wahl B., Schechert A., Nilsson O., Jung C., Kraft T., Muller A.E. The role of a pseudo-response regulator gene in life cycle adaptation and domestication of beet. Curr. Biol. 2012;22:1095–1101. doi: 10.1016/j.cub.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Ratcliffe O.J., Nadzan G.C., Reuber T.L., Riechmann J.L. Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 2001;126:122–132. doi: 10.1104/pp.126.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe O.J., Kumimoto R.W., Wong B.J., Riechmann J.L. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell. 2003;15:1159–1169. doi: 10.1105/tpc.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L., Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., Chang H.Y. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robzyk K., Recht J., Osley M.A. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- Rosa S., De Lucia F., Mylne J.S., Zhu D., Ohmido N., Pendle A., Kato N., Shaw P., Dean C. Physical clustering of FLC alleles during Polycomb-mediated epigenetic silencing in vernalization. Genes Dev. 2013;27:1845–1850. doi: 10.1101/gad.221713.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Yilmaz M., Le O., Hou G., Sadder M., Al-Abdallat A., Xia Y., Lu G., Ladunga I., Avramova Z. The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell. 2008;20:568–579. doi: 10.1105/tpc.107.056614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R.J., Hong L., Michaels S., Amasino R.M. FRIGIDA-ESSENTIAL 1 interacts genetically with FRIGIDA and FRIGIDA-LIKE 1 to promote the winter-annual habit of Arabidopsis thaliana. Development. 2005;132:5471–5478. doi: 10.1242/dev.02170. [DOI] [PubMed] [Google Scholar]
- Schneider R., Bannister A.J., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Schomburg F.M., Patton D.A., Meinke D.W., Amasino R.M. FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell. 2001;13:1427–1436. doi: 10.1105/tpc.13.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D., Orimavesi L., Bishopp A., Roberts G., Doonan J., Jenuwein T., Goodrich J. Silencing by plant Polycombgroup genes requires dispersed trimethylation of histone H3 at lysine 27. EM BO J. 2006;25:4638–49. doi: 10.1038/sj.emboj.7601311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortecci K.C., Michaels S.D., Amasino R.M. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 2001;26:229–236. doi: 10.1046/j.1365-313x.2001.01024.x. [DOI] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Krober S., Amasino R.A., Coupland G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C.C., Burn J.E., Perez P.P., Metzger J., Edwards J.A., Peacock W.J., Dennis E.S. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11:445–58. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C.C., Hills M.J., Lister C., Dean C., Dennis E.S., Peacock W.J. Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc. Natl. Acad. Sci. U S A. 2008;105:2214–2219. doi: 10.1073/pnas.0711453105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G.G. The autonomous pathway: epigenetic and posttranscriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant Biol. 2004;7:570–574. doi: 10.1016/j.pbi.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Spitale R.C., Tsai M.C., Chang H.Y. RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6:539–543. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange A., Li P., C Lister., Anderson J., Warthmann N., Shindo C., Irwin J., Nordborg M., Dean C. Major-effect alleles at relatively few loci underlie distinct vernalization and flowering variation in Arabidopsis accessions. PLoS One. 2011;6:e19949. doi: 10.1371/journal.pone.0019949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Csorba T., Skourti-Stathaki K., Proudfoot N.J., Dean C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science. 2013;340:619–621. doi: 10.1126/science.1234848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S., Amasino R.M. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature. 2004;427:159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- Sung S., Amasino R.M. Molecular genetic studies of the memory of winter. J. Exp. Bot. 2006;57:3369–3377. doi: 10.1093/jxb/erl105. [DOI] [PubMed] [Google Scholar]
- Sung S., Schmitz R.J., Amasino R.M. A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev. 2006a;20:3244–3248. doi: 10.1101/gad.1493306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S., He Y., Eshoo T.W., Tamada Y., Johnson L., Nakahigashi K., Goto K., Jacobsen S.E., Amasino R.M. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat. Genet. 2006b;38:706–710. doi: 10.1038/ng1795. [DOI] [PubMed] [Google Scholar]
- Swiezewski S., Liu F., Magusin A., Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- Swiezewski S., Crevillen P., Liu F., Ecker J.R., Jerzmanowski A., Dean C. Small RNA-mediated chromatin silencing directed to the 3′ region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc. Natl. Acad. Sci. U S A. 2007;104:3633–3638. doi: 10.1073/pnas.0611459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada Y., Yun J.Y., Woo S.C., Amasino R.M. ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell. 2009;21:3257–3269. doi: 10.1105/tpc.109.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M.F. So what–s new in the field of plant cold acclimation? Lots! Plant Physiol. 2001;125:89–93. doi: 10.1104/pp.125.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B., Hemming M.N., Dennis E.S., Peacock W.J. The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 2007;12:352–357. doi: 10.1016/j.tplants.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Trevaskis B., Bagnall D.J., Ellis M.H., Peacock W.J., Dennis E.S. MADS box genes control vernalization-induced flowering in cereals. Proc. Natl. Acad. Sci. U S A. 2003;100:13099–13104. doi: 10.1073/pnas.1635053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F., Roudier F., Farrona S., Martin-Magniette M.L., Guillaume E., Buisine N., Gagnot S., Martienssen R.A., Couplant G., Colot V. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:e86. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Farrona S., Vincent C., Joecker A., Schoof H., Turck F., Alonso-Blanco C., Coupland G., Albani M.C. PEP1 regulates perennial flowering in Arabis alpina. Nature. 2009;459:423–427. doi: 10.1038/nature07988. [DOI] [PubMed] [Google Scholar]
- Wellensiek S.J. Dividing Cells as the Prerequisite for Vernalization. Plant Physiol. 1964;39:832–835. doi: 10.1104/pp.39.5.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A., Schneider J., Dover J., Johnston M., Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- Wood C.C., Robertson M., Tanner G., Peacock W.J., Dennis E.S., Helliwell C.A. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc. Natl. Acad. Sci. U S A. 2006;103:14631–14636. doi: 10.1073/pnas.0606385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Zhao Z., Dong A., Soubigou-Taconnat L., Renou J.P., Steinmetz A., Shen W.H. Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell Biol. 2008;28:1348–1360. doi: 10.1128/MCB.01607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Menard R., Berr A., Fuchs J., Cognât V., Meyer D., Shen W.H. The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J. 2009;57:279–288. doi: 10.1111/j.1365-313X.2008.03684.x. [DOI] [PubMed] [Google Scholar]
- Yamashita K., Shinohara M., Shinohara A. Rad6-Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis. Proc. Natl. Acad. Sci. U S A. 2004;101:11380–11385. doi: 10.1073/pnas.0400078101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Tranquilli G., Helguera M., Fahima T., Dubcovsky J. Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. U S A. 2003;100:6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Helguera M., Kato K., Fukuyama S., Sherman J., Dubcovsky J. Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor. Appl. Genet. 2004a;109:1677–1686. doi: 10.1007/s00122-004-1796-4. [DOI] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Blechl A., Tranquilli G., Ramakrishna W., SanMiguel P., Bennetzen J.L., Echenique V., Dubcovsky J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004b;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Fu D., Li C., Blechl A., Tranquilli G., Bonafede M., Sanchez A., Valarik M., Yasuda S., Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. U S A. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Bratzel F., Hohmann K., Koch M., Turck F., Calonje M. VAL- and AtBMl1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 2013;23:1324–1329. doi: 10.1016/j.cub.2013.05.050. [DOI] [PubMed] [Google Scholar]
- Yun H., Hyun Y., Kan M.J., Noh Y.S., Noh B., Choi Y. Identification of regulators required for the reactivation of FLOWERING LOCUS C during Arabidopsis reproduction. Planta. 2011;234:1237–1250. doi: 10.1007/s00425-011-1484-y. [DOI] [PubMed] [Google Scholar]
- Yun J.Y., Tamada Y., Kang Y.E., Amasino R.M. ARABIDOPSIS TRITHORAX-RELATED3/SET DOMAIN GROUP2 is Required for the Winter-Annual Habit of Arabidopsis thaliana. Plant Cell Physiol. 2012;53:834–846. doi: 10.1093/pcp/pcs021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart J.A.D. Physiology of flower formation. Annu Rev Plant Physiol. 1976;27:321–348. [Google Scholar]
- Zhang H., van Nocker S. The VERNALIZATION INDEPENDENCE 4 gene encodes a novel regulator of FLOWERING LOCUS C. Plant J. 2002;31:663–673. doi: 10.1046/j.1365-313x.2002.01380.x. [DOI] [PubMed] [Google Scholar]
- Zhang X., Germann S., Blus B.J., Khorasanizadeh S., Gaudin V., Jacobsen S.E. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 2007;14:869–871. doi: 10.1038/nsmb1283. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003;17:2733–2740. doi: 10.1101/gad.1156403. [DOI] [PubMed] [Google Scholar]
- Zhao J., Sun B.K., Erwin J.A., Song J.J., Lee J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Yu Y., Meyer D., Wu C., Shen W.H. Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat. Cell Biol. 2005;7:1256–1260. doi: 10.1038/ncb1329. [DOI] [PubMed] [Google Scholar]
- Zografos B.R., Sung S. Vernalization-mediated chromatin changes. J. Exp. Bot. 2012;63:4343–4348. doi: 10.1093/jxb/ers157. [DOI] [PubMed] [Google Scholar]


