Abstract
Objective
Lipid-laden macrophages or foam cells are characterized by massive cytosolic lipid droplet (LD) deposition containing mostly cholesterol ester (CE) derived from the lipoproteins cleared from the arterial wall. Cholesterol efflux from foam cells is considered to be atheroprotective. Since cholesterol is effluxed as free cholesterol (FC), CE accumulation in LDs may limit FC efflux. Our objective was to identify proteins that regulate cholesterol trafficking through LDs.
Approach and results
In a proteomic analysis of the LD fraction of RAW 264.7 macrophages we identified an evolutionarily conserved protein with a canonical GXSXG lipase catalytic motif and a predicted α/β-hydrolase fold, the RIKEN cDNA 1110057K04 gene, which we named lipid droplet-associated hydrolase (LDAH). LDAH association to LDs was confirmed by immunoblotting and immunocytochemistry. LDAH was labeled with a probe specific for active serine hydrolases. LDAH showed relatively weak in vitro CE hydrolase activity. However, cholesterol measurements in intact cells supported a significant role of LDAH in CE homeostasis, since LDAH upregulation and downregulation decreased and increased, respectively, intracellular cholesterol and CE in HEK293 cells and RAW 264.7 macrophages. Mutation of the putative nucleophilic serine impaired active hydrolase probe binding, in vitro CE hydrolase activity, and the cholesterol lowering effect in cells, while this mutant still localized to the LD. LDAH upregulation increased CE hydrolysis and cholesterol efflux from macrophages and, interestingly, LDAH is highly expressed in macrophage-rich areas within mouse and human atherosclerotic lesions.
Conclusions
The data identify a candidate target to promote reverse cholesterol transport from atherosclerotic lesions.
Keywords: atherosclerosis, cholesterol trafficking, foam cell, lipid droplet
Introduction
The lipid-laden macrophage or foam cell is central to the initiation and progression of atherosclerosis.1 Foam cells are derived from circulating monocytes that migrate to the subendothelium and differentiate into macrophages in response to inflammatory stimuli triggered by, among other factors, the deposition of various forms of modified low-density lipoprotein (mLDL). Macrophages take up the mLDL by scavenger receptor (SR)-mediated endocytosis in an unfettered way, and consequently need to manage a large influx of cholesterol. After internalization, the mLDL-derived CE is hydrolyzed in the lysosomes by the acid lipase, and FC is exported to the cytoplasm. Surplus cytoplasmic FC is re-esterified in the endoplasmic reticulum (ER) by acetyl-CoA acetyltransferase 1 (ACAT1) and stored as CE in cytoplasmic lipid droplets (LDs).2, 3 The LD CE undergoes a continuous cycle of hydrolysis and re-esterification, and the FC generated by the hydrolytic arm of the cycle is known to constitute a suitable substrate for efflux.4 It is therefore conceivable that a low rate of CE hydrolysis may limit the amount of FC available for efflux and that, conversely, FC supply to efflux pathways could be increased by increasing the rate of CE hydrolysis, which in turn would promote reverse cholesterol transport from atherosclerotic lesions. Consequently, considerable efforts have been made to elucidate the identity of the enzyme(s) that hydrolyze the CE stored in LDs of foam cells, generically known as neutral cholesterol ester hydrolases (nCEH) because they function at the cytoplasmic neutral pH.
The most obvious candidate nCEH was hormone sensitive lipase (HSL), a robust esterase/lipase expressed mainly in adipose and steroidogenic tissues, where it associates to LDs and catalyzes the hydrolysis of triacylglycerol (TAG) and CE, respectively.5 However, lysates of HSL-knockout (HSL−/−) mouse peritoneal macrophages (MPM) displayed significantly reduced but not fully abolished CE hydrolase activity,6,7 and the mobilization of CE stores was similar in wild-type (WT) and HSL−/− MPM, indicating that other enzymes are able to catalyze CE hydrolysis in macrophages.7,8 Furthermore, while HSL is expressed in murine macrophages, studies in human monocytes, macrophages and human atherosclerotic lesions have reported very low or even undetectable levels of HSL expression.9 Two additional candidate nCEHs, that were named cholesteryl ester hydrolase (CEH) and neutral cholesterol ester hydrolase1 (NCEH1), were identified in RT-PCR screenings and in silico searches, respectively, that were aiming to uncover proteins with lipase-compatible sequences.10,11 CEH is identical to human hepatic carboxylesterase 1 (CES1), also known as triglyceride hydrolase (TGH).12 Carboxylesterases are primarily ER-associated proteins, with their catalytic domains facing the ER lumen12 although, interestingly, CEH was shown to remain in the cytosol and to associate to LDs in lipid-laden THP-1 macrophages, an obligatory step for the subsequent hydrolysis of CE present in the LDs.13 While CEH/CES1/TGH is expressed in human macrophages, its mouse ortholog, CES3/TGH, is not expressed in macrophages, suggesting that other enzymes are involved CE hydrolysis in HSL−/− murine macrophages.12 The third candidate, NCEH1, also named KIAA1363 or arylacetamide deacetylase-like 1 (AADACL1), is also an ER-associated protein with its active site situated in the ER lumen.12,14 Consistently with its intracellular localization, it was reported that CE hydrolysis was reduced in microsomes but not in soluble cytosolic fractions of NCEH1/KIAA1363/AADACL1-deficient macrophages,6 and the hydrolysis of preformed CE stores was similar MPM isolated from WT and NCEH1/KIAA1363/AADACL1-deficient mice.7
It is noteworthy that in foam cells the hydrolysis of the LD CE is not exclusively dependent upon nCEHs, since it was reported that LDs could be delivered to lysosomes via autophagy, where the acid lipase hydrolyzes CE to generate FC for efflux.15 Interestingly, inhibition of the neutral and lysosomal pathways of LD-associated CE hydrolysis increased macrophage CE mass in a cumulative fashion, and while inhibition of each individual pathway decreased CE hydrolysis, only the inhibition of both pathways totally abolished CE hydrolysis, indicating that the two pathways cannot fully compensate each other. Thus, while multiple enzymes may be able to hydrolyze CE in foam cells at different points of the CE cycle, the current evidence suggest that there may be additional nCEH(s) acting on LDs that have yet to be identified. Here we have performed a proteomic analysis of the LD fraction of RAW 264.7 macrophages in which we have identified a serine hydrolase we have named lipid droplet-associated hydrolase (LDAH). Results from biochemical cholesterol measurements and cholesterol trafficking experiments implicate a role of LDAH in CE turnover, and LDAH expression is enriched in macrophage-rich areas within mouse and human atherosclerotic lesions.
Materials and Methods
Materials and Methods are available in the online-only Data Supplement.
Results
Identification of a novel LD-associated serine hydrolase in macrophages
Purified LD-associated proteins isolated from lipid-laden RAW 264.7 macrophages were identified by nano-LC–MS/MS. The LD-protein band pattern (Figure 1A) was consistent in various LD isolations. Forty proteins were identified based on three or more matching peptides, of which eleven (~28%) had not previously been found associated to LDs of mammalian cells (Table 1). Among them we identified a protein with an annotated esterase/lipase domain, the RIKEN cDNA 1110057K04 gene (GI: 55777092), the human homolog of which, named UPF0554 protein C2orf43 (GI: 11345458), had been classified as a member of the metabolic serine hydrolase family of proteins, which includes most known mammalian esterases and lipases.16 To simplify, we named this protein lipid droplet-associated hydrolase (LDAH), and we will refer to the mouse homolog as mLDAH and to the human homolog as hLDAH. While to our knowledge endogenous LDAH had not been identified in previous LD proteomics in mammalian cells, the LDAH Drosophila melanogaster homolog, named CG9186, had previously been identified in two proteomic analyses of LDs isolated from Drosophila embryos and late third instar larval fat bodies,17,18 and while this manuscript was in preparation Thiel et al. showed LD localization of both the fly and mouse overexpressed proteins.19
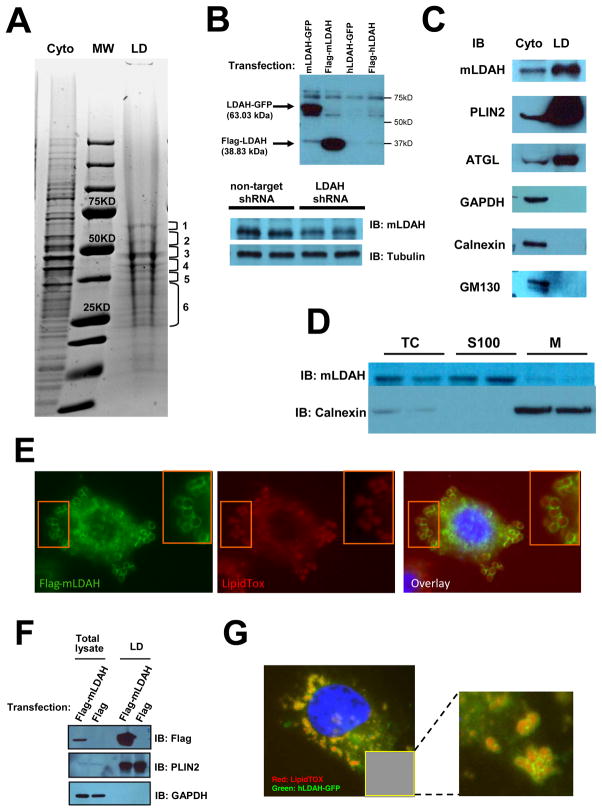
Figure 1. Identification of a novel candidate LD-associated CE hydrolase.
(A) LD proteins isolated from RAW 264.7 macrophages were resolved by SDS-PAGE and stained with Coomassie Blue. Six gel fragments containing most protein bands were analyzed by nano LC-MS/MS. Cyto= cytoplasmic fraction; LD= LD fraction; MW= molecular weight marker. (B) A custom anti-mLDAH antibody was generated, and its specificity was confirmed in HeLa cells transfected with mouse and human LDAH-GFP and flag-LDAH, and in RAW 264.7 macrophages with shRNA-mediated mLDAH downregulation. (C) Immunoblot with anti-mLDAH on the cytosolic and LD fractions that were used for the proteomic analysis. Immunoblots with antibodies against GAPDH (cytoplasmic protein), calnexin (ER protein), GM130 (Golgi protein), and PLIN2 and ATGL (LD proteins) verified the purity of the LD fraction. (D) Immunoblot with anti-mLDAH performed on total cytosol (TC), soluble 100,000 × g fractions (S100), and 100,000 × g membrane pellets (M) of RAW 264.7 macrophages. (E) Immunostaining with anti-flag (green) on acLDL-treated (50 μg/ml, 18h) RAW 264.7 macrophages transfected with flag-mLDAH. LipidTox™ (red) was used to stain LDs. (F) Immunoblot with anti-flag on total lysate and LD fractions of flag-mLDAH-transfected HeLa cells treated with oleic acid (360 μM, 18h). (G) Fluorescence microscopy on HeLa cells transfected with hLDAH-GFP (green) and treated with oleic acid (360 μM, 18h). LipidTOX™ was used to label LDs (Red).
Table 1.
LD associated proteins identified by LC-MS/MS.
| Fragment | Protein Name | MW (kDa) | GI |
|---|---|---|---|
| 1 | BIP* | 72.5 | 2598562 |
| Unnamed | 83.5 | 74147026 | |
| Plastin-2 | 70.7 | 31543113 | |
| UBXN4* | 56.8 | 30913398 | |
|
| |||
| 2 | Vimentin* | 51.6 | 2078001 |
| PLPL2 (ATGL)* | 54.5 | 81896337 | |
| Ubiquitin B* | 34.4 | 18044723 | |
| Prolyl-4-Hydrolase* | 57.4 | 42415475 | |
| CAP-1 | 51.9 | 729032 | |
| Unnamed | 58.8 | 12852157 | |
| Pyruvate kinase M* | 58.4 | 551295 | |
| Alpha-tubulin 8 | 50.7 | 8394493 | |
| PLCα* | 57.0 | 200397 | |
|
| |||
| 3 | PLIN2* | 46.9 | 116235489 |
| Enolase 1alpha* | 47.5 | 70794816 | |
| Protein disulfide-isomerase A3* | 57.1 | 112293264 | |
| Interferon gamma induced GTPase | 48.8 | 28261389 | |
| LysoPC acyltransferase-1* | 60.4 | 148747363 | |
|
| |||
| 4 | Gamma-actin | 41.3 | 809561 |
| Ancient ubiquitous protein* | 49.7 | 90403601 | |
| Unnamed | 38.9 | 74144652 | |
| VAT-1* | 43.3 | 33859662 | |
| ABHD5 (CGI-58)* | 39.5 | 13385690 | |
| IRGM-1* | 47.1 | 6680351 | |
|
| |||
| 5 | RIKEN cDNA 1110057K04 gene (LDAH) | 37.7 | 55777092 |
| Unnamed | 50.5 | 74142813 | |
| Annexin A1* | 39.0 | 124517663 | |
| Annexin A2* | 38.9 | 6996913 | |
| Unnamed | 50.8 | 74181454 | |
| Ribosomal protein, large, P0* | 34.3 | 13277927 | |
|
| |||
| 6 | RAB7* | 23.8 | 1050551 |
| DHRS-1* | 34.5 | 31980844 | |
| Diaphorase-1* | 34.3 | 19745150 | |
| RAB2A* | 23.7 | 10946940 | |
| RAB11B* | 24.6 | 6679583 | |
| RAB14* | 22.1 | 63087697 | |
| RAB5C* | 23.6 | 113866024 | |
| HSP70* | 71 | 309319 | |
| RAB18* | 23.3 | 30841008 | |
| RAP1B* | 21.0 | 7661678 | |
proteins found in previous LD proteomic analyses in mammalian cells. An extended table that includes references can be found in the Online Data Supplement (Table S2).
Eleven mLDAH matching peptides were identified in the LC-MS/MS analysis, the highest number in fragment five (Table SII), and these peptides aligned with six different fragments within the protein sequence (Figures SIA and SIB). To confirm that endogenous LDAH localizes to the LD, we generated an anti-mLDAH antibody that showed specificity in cell lysates with mLDAH overexpression and downregulation (Figure 1B). Immunoblots performed on the same LD and cytoplasmic protein fractions used for the proteomic analysis detected enriched mLDAH in the LD fraction (Figure 1C). The purity of this LD fraction was verified by immunoblotting against cytoplasmic (GAPDH), ER (Calnexin), Golgi (GM130), and LD [perilipin 2 (PLIN2) and adipose triglyceride lipase (ATGL)] markers (Figure 1C). Moreover, following subfractionation of cytoplasms at 100,000 × g, mLDAH was detected predominantly in the 100,000 × g supernatants, which is consistent with the fact that its sequence does not contain a signal peptide (Figure 1D). LDAH’s LD association was also confirmed by immunofluorescence in RAW 264.7 macrophages transfected with flag-mLDAH, which showed protein enrichment at the LD perimeter (Figure 1E). Interestingly, while Thiel et al. reported that LDAH induced LD clustering and in some cases fusion in non-monocytic cell lines that were treated with oleic acid,19 we did not observe changes in LD phenotype in cholesterol-laden macrophages with flag-mLDAH overexpression. LDAH was also found, by immunoblotting, in LD fractions isolated from flag-mLDAH transfected HeLa cells, and microscopy analysis of lipid-laden HeLa cells showed mLDAH colocalization with PLIN2 at the LD perimeter (Figure 1F and Figure SII). Furthermore, transfection with hLDAH-GFP also showed localization of the human homolog at the LD perimeter (Figure 1G). Collectivelly, these data support that LDAH is a genuine LD-associated protein.
LDAH has esterase/lipase features and plays a role in cholesterol homeostasis
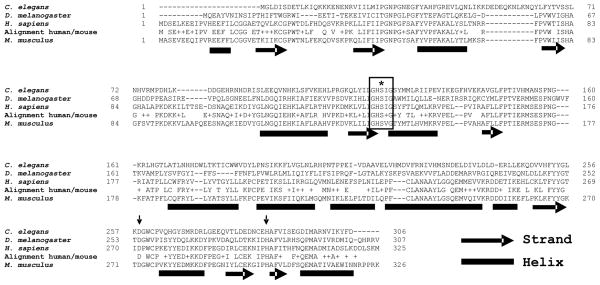
mLDAH is a 326-amino acid protein with a calculated molecular mass of ~36kD that is highly conserved through evolution (see examples in Figure 2). Esterases/lipases are typically built in an α/β-hydrolase fold structure, and in most cases their catalytic apparatus involves three residues that are responsible for the nucleophilic attack on the carbonyl carbon atom of the ester bond: a nucleophilic serine, an acid residue (glutamate or aspartate), and a histidine. Patatin domain-containing lipases such as ATGL are an exception, as they use serine-aspartate dyads for catalysis instead of catalytic triads.20 A highly conserved feature is that the nucleophilic serine is in a consensus GXSXG sequence, which is usually positioned in-between a β-strand and a α-helix, where the protein folds forming a sharp turn. mLDAH sequence analysis with the PSIPRED software21 predicted a secondary structure that was compatible with that expected from a protein containing an α/β-hydrolase fold (Figure 2). Furthermore, LDAH contains a central GXSXG motif harboring a putative nucleophilic serine (S140), and a conserved aspartate (D272) and a conserved histidine (H291) could complete the catalytic triad. These three candidate catalytic aminoacids are predicted to be positioned between strands and helixes (Figure 2).
Figure 2. LDAH has conserved esterase/lipase compatible features.
LDAH is evolutionarily conserved from rice to human. The figure displays the sequences of the C. elegans, Drosophila, human, and mouse LDAH homologs. The alignment between mouse and human LDAH is shown between their sequences. A putative active site nucleophilic serine (S, marked with an asterisk) is located within a conserved typical GXSXG pentapeptide lipase catalytic motif (boxed). A conserved aspartic acid (D) and a conserved histidine (H) are pointed by arrows. The bioinformatics prediction of mLDAH secondary structure is shown under the mouse sequence.
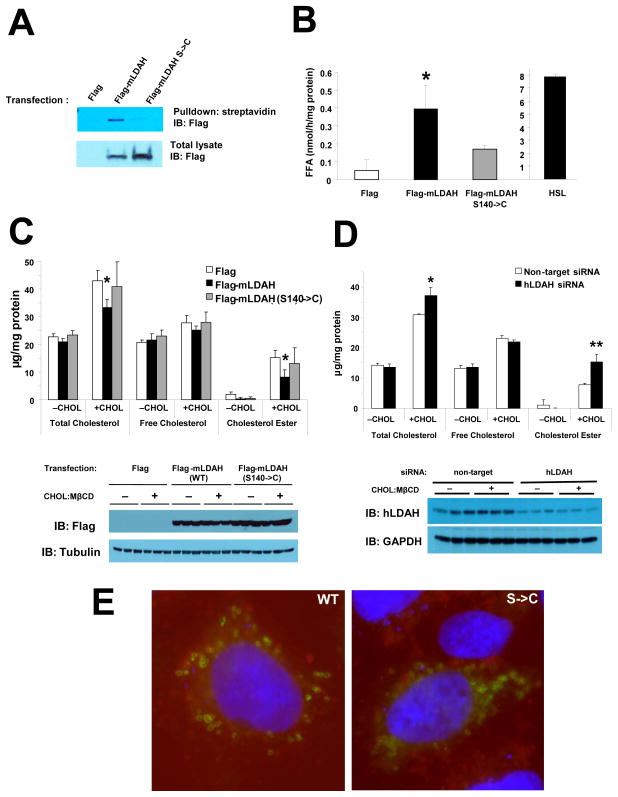
To test whether LDAH is an active serine hydrolase, we used a metabolic probe [Desthiobiotin-fluorophosphonate (DTB-FP)] that specifically and covalently binds to functionally active serine hydrolases and inhibits their activity.16 As seen in Figure 3A, WT mLDAH was labeled with DTB-FP, while a S140->C mutant, in which the predicted active-site serine was replaced by a cysteine, was not labeled. To test whether LDAH can hydrolyze CE, we carried out cholesterol esterase activity assays in which HeLa cell lysates were assayed against 14C-labeled CE emulsified with phospholipids.22 Data from these experiments showed increased CE hydrolysis in lysates from cells with mLDAH overexpression, which was inhibited by S140->C mutation or by preincubation with DTB-FP, although the activity was much weaker than the seen in extracts of cells transfected with HSL (Figure 3B and Figure SIIIA). Conversely, no activity against TAG was detected (Figure SIIIB). The results of experiments using cell-free extracts do not necessarily represent the protein’s role in intact cells. Since gene expression manipulations in macrophages are very challenging, to test whether LDAH plays a role in cholesterol homeostasis in intact cells initially we carried out LDAH gain- and loss-of-function experiments in HEK293 cells that remained untreated or were treated with cholesterol-methyl-β-cyclodextrin (CHOL:MβCD), a system that allows cholesterol loading in a SR-independent fashion.23 CE was readily detectable in CHOL:MβCD-treated cells, and as expected CE accumulation was paralleled by the inclusion of abundant cytoplasmic LDs (Figure 3C and Figure SIV). Interestingly, CE and total cholesterol levels were ~50% and ~25% lower, respectively, in cells transfected with mLDAH, while transfection with a S140->C mutant did not affect total cholesterol or CE levels (Figure 3C). Furthermore, knocking-down endogenous hLDAH by ~75% resulted in ~two-fold increase in intracellular CE and in ~25% increase in total cholesterol (Figure 3D). Of note, the S140->C mutation did not affect LDAH localization to the LD (Figure 3E). Thus, DTB-FP labeling confirmed the prediction that LDAH is an active serine hydrolase, and while experiments using lysates of cells transfected with mLDAH showed relatively weak in vitro activity, gain- and loss-of-function experiments in intact cells indicated that LDAH plays a significant role in cholesterol homeostasis.
Figure 3. LDAH is a serine hydrolase that plays a role in cholesterol homeostasis.
(A) HeLa cells were transfected with flag, flag-mLDAH, or flag-(S140->C)-mLDAH. Cell lysates were incubated with the ActivX® DTB-FP to label active serines in serine hydrolases. Proteins bound to the probe were pulled-down using streptavidin-agarose. The streptavidin-captured fraction and total cell lysates were immunoblotted with anti-flag. (B) In vitro CE hydrolase activity performed on protein extracts from HeLa cells transfected with flag, flag-mLDAH, or flag-(S140->C)-mLDAH. (n=3). (C) HEK293 cells were transfected with flag, flag-mLDAH, or flag-(S140->C)-mLDAH. The cells were cultured in DMEM-10% FBS and remained untreated (−CHOL), or were treated with CHOL:MβCD (10 μg/ml) for 24h (+CHOL). Total intracellular cholesterol, FC and CE levels were quantified and normalized to protein. (n=3). Immunoblots with anti-flag on protein lysates from the cells used for the experiments are shown under the chart. (D) Cholesterol levels in untreated or cholesterol-loaded HEK293 cells transfected with non-target or hLDAH siRNAs. (n=3). Immunoblots with anti-hLDAH on protein lysates from the cells used for the experiments are shown under the chart. (E) Immunofluorescence with an anti-flag antibody (green) on HEK293 cells transfected with flag-mLDAH and flag-(S140->C)-mLDAH; LipidTox™ (red) was used to stain LDs. All data are shown as mean ± SD. *p<0.05, **p<0.01 vs. flag-mLDAH or non-target-siRNA.
LDAH upregulation increases CE turnover and cholesterol efflux in macrophages
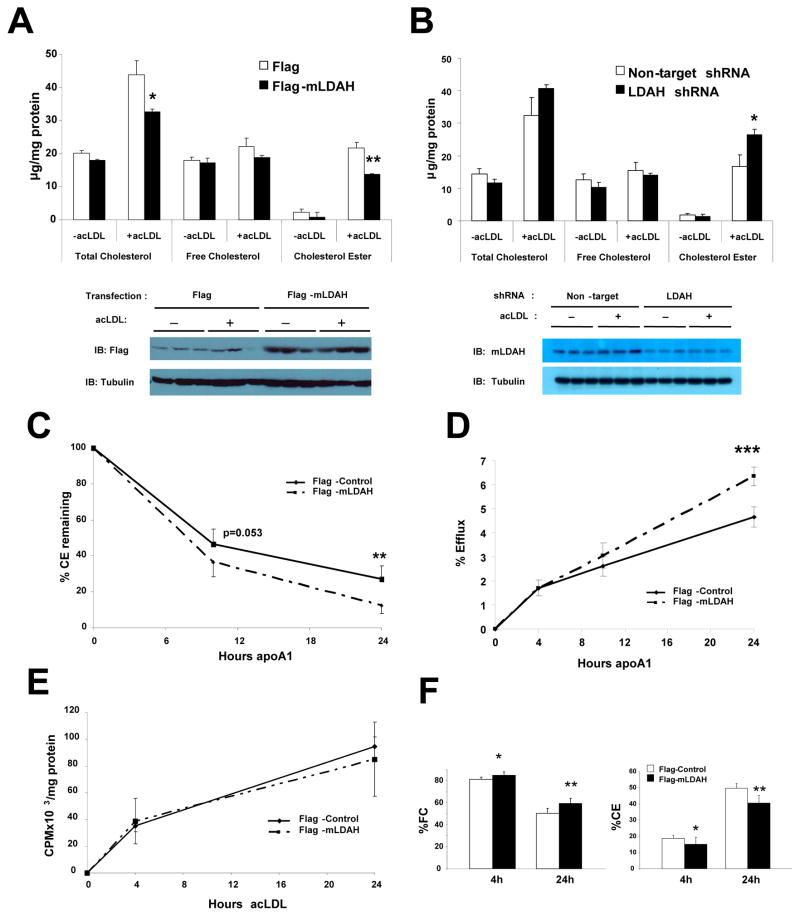
To test whether LDAH also plays role in CE accumulation in macrophages, we treated RAW 264.7 macrophages with acetylated (ac) LDL, a form of mLDL avidly internalized by macrophages. AcLDL loading increased CE content by ~10-fold and total cholesterol mass by ~2–3-fold. However, similar to the observations in HEK293 cells, these parameters were markedly affected by LDAH gain- and loss-of-function, which decreased and increased, respectively, intracellular total cholesterol and CE levels (Figures 4A and 4B). These data were consistent in several independent experiments. In addition, lentiviral vectors encoding three different shRNAs were tested for LDAH downregulation. Two of them reduced LDAH protein level by ~50%, which was associated with increased CE, while one of them did not affect either LDAH or CE levels (Figure SV). Furthermore, supporting that changes in cholesterol levels were actually caused by changes in LDAH levels, LDAH overexpression or knockdown did not affect the expression of HSL or NCEH1/KIAA1363/AADACL1 (Figure SVIA and SVIB), and CES3/TGH mRNA was undetectable in RAW 264.7 macrophages (Figure SVIC).
Figure 4. LDAH upregulation promotes cholesterol turnover from macrophages.
(A) RAW 264.7 macrophages were transfected with flag or flag-mLDAH plasmids. Cells were cultured for 18h in DMEM-1% FBS without or with acLDL (50 μg/ml). Cellular lipids were extracted, and total intracellular cholesterol, FC and CE levels were quantified and normalized to protein. (n= 3). Immunoblots on protein lysates of the cells used for the experiments are shown under the chart. (B) Total cholesterol, FC and CE levels in RAW 264.7 macrophages with lentiviral non-target shRNA or mLDAH shRNA-mediated knockdown. (n=3). Immunoblotts on protein lysates of RAW 264.7 macrophages with stable mLDAH knockdown are shown under the chart. (C) Time-course of CE hydrolysis in macrophages with and without mLDAH overexpression. RAW 264.7 macrophages were cholesterol-loaded with acLDL (50 μg/ml) for 20h in DMEM-0.2% BSA, washed, and pulsed with [3H] oleic acid (0.2 mM) in DMEM-0.4% BSA for 23h. Cells were then washed and pre-incubated with the ACAT1 inhibitor Sandoz 58–035 (10 μg/ml in DMEM-0.2% BSA) for 1h. ApoA1 (10 μg/ml) was added to the culture media that contained the ACAT1 inhibitor, and cells were harvested before and 6 and 24h after apoA1 was added. Intracellular lipids were resolved by TLC, and the CE at each timepoint was measured and normalized to protein. (n=6). (D) Effect of mLDAH overespression on cholesterol efflux. RAW 264.7 macrophages were loaded with [3H]-cholesterol-labeled acLDL (50 μg/ml in DMEM-0.2% BSA) for 18h, washed, equilibrated in DMEM-0.2% BSA for 1h, and cholesterol efflux was induced by adding apoA1 to the culture media. (n=5). (E and F) Time-course of cholesterol accumulation and distribution in the FC and CE fractions. RAW 264.7 macrophages were loaded with [3H]-cholesterol-labeled acLDL (50 μg/ml in DMEM-0.2% BSA) for 4 and 24h. Lipids were extracted and resolved by TLC, and FC and CE were quantified and normalized to protein. Total cholesterol (FC + CE) content is shown in panel (E), and the cholesterol distribution in the FC and CE fractions is shown in panel (F). (n=5). All data are shown as mean ± SD. *p<0.05, **p<0.01, ***p<0.001.
Based on its localization to the LD surface and on its effects on CE levels, we hypothesized that LDAH promotes the turnover of the CE stored in LDs. Following internalization by macrophages, the CE carried by the lipoproteins is hydrolyzed to FC in the lysosomes, and excess cytoplasmic FC is re-esterified in the ER and stored as CE in LDs. To specifically assess the turnover of the LD CE depots, RAW 264.7 macrophages were incubated with cold-acLDL and pulsed with 3H-labeled oleic acid to label the newly synthesized CE as 3H-oleic-cholesterol. This was followed by a chase period in which cholesterol turnover was promoted by adding the cholesterol acceptor apolipoprotein A1 (apoA1) to the culture media, while re-esterification was blocked with an ACAT1 inhibitor. As seen in Figure 4C, cells transfected with flag-mLDAH displayed a higher rate of 3H-oleic-cholesterol hydrolysis than flag-transfected control cells. Next, we asked whether LDAH upregulation also results in increased cholesterol efflux. The two main transporters involved in cholesterol efflux are ATP-binding cassette A1 and G1 (ABCA1 and ABCG1), which promote efflux to lipid poor apoA1 and HDL particles, respectively. ABCA1 and ABCG1 can act sequentially with ABCA1 initiating the formation of nascent HDL particles, which then acquire additional lipids via ABCG1.24 Therefore, to take into consideration both the ABCA1 and ABCG1-mediated efflux, RAW 264.7 macrophages were cholesterol-loaded with 3H-cholesterol-labeled acLDL, and, after washing and equilibration, apoA1 was added to the culture media to induce efflux. As seen in Figure 4D, while early efflux was similar in macrophages with and without LDAH overexpression, the amount of cholesterol effluxed increased over time at a higher rate in cells transfected with flag-mLDAH than in flag-transfected controls. Interestingly, these results are consistent with those of other interventions at the LD level, that also showed a significant response on later rather than earlier efflux.3,15,25 The changes in efflux seen in cells with mLDAH overexpression were not due to increased expression of ABCA1 or ABCG1 (Figure SVII).
To rule out the possibility that the differences seen in the previous experiments were due to changes in cholesterol uptake, RAW 264.7 macrophages were loaded with 3H-cholesterol-labeled acLDL in the absence of serum for 4 and 24h, and the 3H-cholesterol in the FC and CE fractions was quantified. As seen in Figure 4E, the total amount of cholesterol (free + esterified) was similar between flag-mLDAH and flag- transfected macrophages, indicating that LDAH upregulation did not affect cholesterol uptake. At the earlier timepoint most cholesterol (~80%) still remained as FC, while at the 24h timepoint ~40–50% of the cholesterol was incorporated into the CE pool. However, at both timepoints the % of acLDL-derived cholesterol in the form of FC and CE were higher and lower, respectively, in macrophages with LDAH overexpression (Figure 4F). Thus, LDAH upregulation did not affect the rate of cholesterol uptake by macrophages, but it increased the hydrolysis of re-esterified CE, and this was associated with increased cholesterol efflux.
LDAH is expressed in mouse and human atherosclerotic lesions
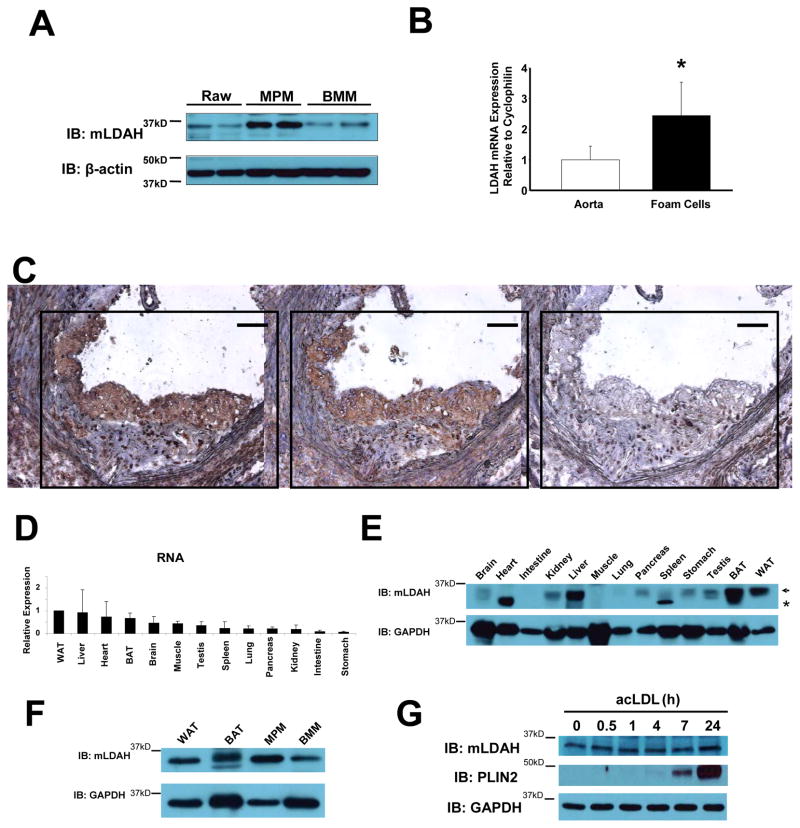
LDAH expression was not restricted to the RAW 264.7 cell line, but it was also expressed in primary MPM and bone marrow-derived macrophages (BMM) (Figure 5A). Regarding atherosclerotic lesions, quantitative real-time PCR (qPCR) analysis detected ~2.5-fold higher mLDAH mRNA in RNA isolated by laser capture microdissection (LCM) from lesional macrophages/foam cells than in RNA isolated from apoE−/− mice whole aortas (Figure 5B). At the protein level, immunohistological analyses readily detected immunoreactive LDAH predominantly in macrophage-rich areas within the lesions (Figure 5C). With respect to other tissues, LDAH mRNA was ubiquitously detected, with higher levels found in white adipose tissue (WAT), liver, heart and brown adipose tissue (BAT) (Figure 5D). Protein levels were high in liver and white and brown adipose tissues, but lower levels were also observed in testis, stomach, brain, kidney and pancreas. While LDAH mRNA levels were detected in heart and muscle, no protein band of the expected size was observed in these tissues (Figure 5E). Interestingly, LDAH levels in mouse macrophages were relatively high, comparable to the levels seen in the tissues with higher LDAH expression such as WAT and BAT (Figure 5F). Given its role in cholesterol metabolism and the fact that LDAH is expressed by foam cells, we speculated that LDAH expression could be regulated by cholesterol loading. However, as seen in Figure 5G, LDAH expression was not affected by treatment with acLDL, while the expression of the LD-associated protein PLIN2 was highly induced.
Figure 5. LDAH expression in mouse tissues.
(A) Immunoblot with anti-mLDAH in lysates of RAW 264.7 macrophages, thioglycollate elicited MPM, and mouse BMM. (B) qPCR analysis comparing LDAH levels in RNA isolated by LCM from macrophage/foam cell rich-areas within atherosclerotic lesions and whole aorta RNA of apoE−/− mice. Data are shown as mean ± SD. (n=5). *p<0.05. (C) Immunoperoxidase staining with mLDAH antisera (left panel, brown color) and anti-Lamp-2 (macrophage marker, middle panel, brown color) in consecutive sections of the aortic sinus of apoE−/− mice. Preimmune serum was used as negative control (right panel). Bar= 100 μM. (D) qPCR analysis of LDAH RNA levels in male mouse tissues. (n=3). (E) Immunoblotting with anti-mLDAH in mouse tissues. The arrow indicates the specific mLDAH band. *non-specific band. (F) Immunoblotting with anti-mLDAH comparing mLDAH levels between WAT, BAT, MPM and BMM. (G) LDAH levels were not affected by cholesterol loading with acLDL (50 μg/ml in DMEM-1% FBS). The LD-associated protein PLIN2 was highly induced by the treatment.
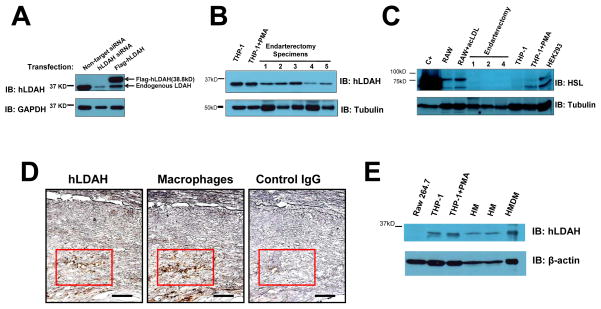
To test whether LDAH is also expressed in human atherosclerotic lesions, we generated and anti-hLDAH antibody that specifically recognized hLDAH (Figure 6A). By immunoblotting, hLDAH was detected in THP-1 macrophages, a human monocytic cell line, and more importantly, in human carotid endarterectomy specimens (Figure 6B). On the contrary, in agreement with previous reports,8, 26 HSL levels were sensibly lower in THP-1 macrophages than in RAW 264.7 macrophages, and HSL was undetectable in endarterectomy specimens (Figure 6C). In addition, in line with the mouse data, immunohistological analyses of endarterectomy specimens detected hLDAH predominantly in macrophage-rich areas within the human lesions (Figure 6D). Furthermore, LDAH was also detected in lysates of human monocytes and monocyte-derived macrophages (Figure 6E). Thus, LDAH is highly expressed in mouse and human macrophages of different sources, and the protein is expressed in mouse and human atheroma, predominantly in foam cell-rich areas within the lesions.
Figure 6. LDAH is expressed in foam cells within human atherosclerotic lesions.
(A) The specificity of the antibody generated against hLDAH was tested in HEK293 cells transfected with hLDAH siRNA and overexpressing flag-hLDAH. (B) 15 μg of THP-1 lysates and 30 μg of human endarterectomy lysates were used to assess hLDAH expression by immunoblotting. The figure displays five representative examples from a total of thirteen endarterectomy specimens analyzed; hLDAH was detected in all the samples tested. (C) Immunoblotting with anti-HSL on 70 μg of protein lysates from mouse RAW 264.7 macrophages, human THP-1 macrophages, HEK 293 cells, and endarterectomy specimens. HEK293 cells transfected with HSL were used as a positive control (C+). (D) Immunoperoxidase staining with anti-hLDAH (left panel, brown color) and anti-CD68 (macrophage marker, middle panel, brown color) in consecutive sections of human endarterectomy specimens. Rabbit IgG was used as negative control (right panel). Bar= 100 μM. (E) Immunoblotting with anti-hLDAH on 7.5 μg of protein lysates from RAW 264.7 macrophages, THP-1 macrophages, human monocytes (HM) and human monocyte-derived macrophages (HMDM).
Discussion
The lipid-laden macrophage or foam cell is a hallmark of atherosclerotic lesions, and there is compelling evidence that enhancing reverse cholesterol transport from foam cells is an effective antiatherogenic strategy.27 In macrophages, cholesterol is stored as CE but effluxed as FC.2 Thus, theoretically the hydrolysis of the CE accrued within LDs could be considered as a preliminary and perhaps limiting step for cholesterol efflux and reverse cholesterol transport from atherosclerotic lesions, and proteins controlling the process could become pharmacological targets to ameliorate atherogenesis. Unfortunately, the identity of the proteins that regulate the hydrolysis of the CE stored in LDs of foam cells is still far from clear. Most known lipases and esterases belong to the subfamily of metabolic serine hydrolases, and therefore it is likely that the enzymes involved in CE hydrolysis in macrophages are among the ~115 members of this family, and probably among the ~50% family members with unknown function.16 Interestingly, our macrophage LD proteomics led to the identification of LDAH, an evolutionarily conserved protein that had been classified as a metabolic serine hydrolase.16
To our knowledge, the only functional characterization of any of the LDAH homologs was very recently reported by Thiel et al., who found that overexpression of CG9186 or mLDAH fused to GFP caused LD clustering in cells cultured with oleic acid, which was abrogated by deleting the 106 C-terminal aminoacids, but not by mutating the putative nucleophilic serine, suggesting that this phenotype is not related to the protein’s hydrolytic activity. In addition, neither the Drosophila or mouse homologs displayed activity towards TAG, diacylglycerol or monoacylglycerol, nor did CG9186 overexpression change TAG content in Drosophila Kc167 cells, while CG9186 knockdown actually decreased TAG in flies, facts that are at odds with the protein playing a role in TAG hydrolysis. Thus, to date no enzymatic activity for this protein had been demonstrated, and one possibility to take into consideration was that LDAH is not an active hydrolase. For example, comparative gene identification-58 (CGI-58) is an α/β-hydrolase fold protein that associates to LDs and is considered to be an activator of ATGL, a rate limiting enzyme in TAG hydrolysis. However, CGI-58 contains a pseudo-catalytic triad in which the nucleophilic serine in the GXSXG motif is replaced by an asparagine, and therefore is very unlikely that it exhibits hydrolase activity.28 While LDAH contains the actual GXSXG motif, it was important to demonstrate that the protein is an active hydrolase. This was shown by labeling experiments with DTB-TP, a probe that when is hydrolyzed by serine hydrolases remains covalently bound to the nucleophilic serine. With respect to the LDAH substrate(s), in agreement with the data by Thiel et al.,19 our results did not show increased TAG hydrolase activity in lysates of HeLa cells with LDAH overexpression. Interestingly, extracts of mLDAH-transfectd HeLa cells showed significantly increased CE hydrolase activity, although the in vitro activity was much weaker than the seen in lysates of HSL overexpressing HeLa cells. In vitro conditions that work well for one enzyme may not work as well or even be detrimental to others. However, we assayed CE hydrolase activity in cell lysates under different conditions including PC/PI micelles with or without taurocholate, PC alone or PI of different sources, and the CE hydrolase activity in lysates of cells transfected with HSL was consistently higher than that of lysates of cells transfected with LDAH (data not shown). This is not surprising, since HSL is a very robust CE hydrolase that is highly expressed in tissues where adequate FC supply is critical for proper esteroidogenesis, while the high accumulation of CE seen in foam cells suggests that the CE may be hydrolyzed by enzymes with lower catalytic rates.29, 30 On the other hand, the results of in vitro activity assays may not adequately reflect the protein’s physiological relevance, for example because of lack other LD-associated proteins or cofactors that may modulate the protein’s activity in vivo, or because protein-protein interactions at the LD surface are disrupted upon cell lysis. Experiments using intact cells provided stronger evidence that LDAH plays a role in cholesterol homeostasis, since cellular CE content decreased when LDAH was overexpressed and increased when LDAH was downregulated, both in HEK293 and RAW 264.7 cells. Interestingly, mutation of the nucleophilic serine impaired active hydrolase probe binding, in vitro lipase activity, and the cholesterol lowering effect in cells, while this mutant still localized to the LD. In addition, cholesterol trafficking experiments indicated that LDAH may be a promising candidate target for stimulating reverse cholesterol transport from foam cells, since macrophages with LDAH overexpression showed increased CE turnover and efflux. Furthermore, LDAH’s relatively high expression in macrophage-rich areas within atherosclerotic lesions also indicates that LDAH could be a suitable target to promote reverse cholesterol transport from the lesions.
While LD proteomic analyses may be technically challenging, they have the advantage over indirect screening methods that they allow circumventing ambiguity with the proteins’ cellular localization. The intracellular localization of the proteins that regulate the CE cycle is not a trivial question. Part of the cycle takes place in the ER, where FC is re-esterified by ACAT1 and, therefore, it could be argued that the ER could also be a potential site for therapeutic intervention. However, studies on ACAT1 inhibition have shown that raising FC levels inside the ER can give rise to adverse side effects. Compared to other cell membranes, the ER membrane is relatively poor in cholesterol and it is well established that FC accumulation at the ER membrane, such as seen under ACAT1 inhibition, results in ER stress-mediated apoptosis, increased synthesis of inflammatory mediators and, in vivo, in accelerated atherosclerosis with grossly necrotic lesions.31, 32 On the other hand, FC accumulation in other cell compartments, such as the plasma membrane, is not as harmful to the macrophage,33 and macrophages can tolerate PLIN2 deficiency, which hampers their ability to accumulate cytoplasmic LDs, much better than ACAT1 inhibition.34 Presumably, the ER FC levels would also increase if CE hydrolysis were to be mediated by ER lumen-resident enzymes. Thus, considering that the safety of a molecular intervention with potential therapeutic use may be as important as its efficiency, current evidence indicate that targeting proteins that mediate CE hydrolysis at the LD level is safer than targeting ER-resident proteins.
In conclusion, this study identifies a novel LD-associated serine hydrolase in macrophages and uncovers a role of the protein in cholesterol mobilization. Interestingly, in addition to foam cells, high LDAH levels are also seen in important metabolic tissues such as WAT, BAT and liver. Atherosclerosis development is closely related to a number of co-morbidities such as obesity, dyslipidemia, insulin resistance and type 2 diabetes, which are also caused or aggravated by excessive lipid accumulation in tissues and, accordingly, multiple lines of evidence have linked LD biology to their pathogenesis.35 Thus, LDAH could be a significant player not only in atherogenesis, but also in other lipid-related metabolic derangements.
Supplementary Material
Significance.
The hydrolysis of the CE accrued within LDs of foam cells is considered a limiting step for cholesterol efflux and reverse cholesterol transport from atherosclerotic lesions. Unfortunately, to date the identity of the enzymes responsible for CE hydrolysis at the LD level, generically known as neutral cholesterol ester hydrolases, remains unclear. In a proteomic analysis of the LD fraction of RAW 264.7 macrophages we have identified a novel serine hydrolase which we have named LDAH. Gain- and loss-of-function experiments in cell culture showed an inverse relationship between LDAH levels and intracellular cholesterol and CE levels, and LDAH upregulation promoted the hydrolysis of the CE stored in LDs, which was associated with increased cholesterol efflux. In vivo, LDAH is highly expressed in foam cell-rich areas within mouse and human atherosclerotic lesions. The data identify a potential new target to promote reverse cholesterol transport from atherosclerotic lesions.
Acknowledgments
We thank the Vascular Surgery Group at Albany Medical Center for providing the endarterectomy samples.
Sources of Funding
This study was supported by a National Institutes of Health R01 grant HL104251 (to A.P.).
Footnotes
Disclosures
None
References
- 1.Glass CK, Witztum JL. Atherosclerosis: The Road Ahead. Cell. 2001;104:503–16. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 2.Brown MS, Ho YK, Goldstein JL. The cholesteryl ester cycle in macrophage foam cells. Continual hydrolysis and re-esterification of cytoplasmic cholesteryl esters. J Biol Chem. 1980;255:9344–9352. [PubMed] [Google Scholar]
- 3.Paul A, Chang BH, Li L, Yechoor VK, Chan L. Deficiency of adipose differentiation-related protein impairs foam cell formation and protects against atherosclerosis. Circ Res. 2008;102:1492–1501. doi: 10.1161/CIRCRESAHA.107.168070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown MS, Goldstein JL, Krieger M, Ho YK, Anderson RG. Reversible accumulation of cholesteryl esters in macrophages incubated with acetylated lipoproteins. J Cell Biol. 1979;82:597–613. doi: 10.1083/jcb.82.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeaman SJ. Hormone-sensitive lipase--a multipurpose enzyme in lipid metabolism. Biochim Biophys Acta. 1990;1052:128–132. doi: 10.1016/0167-4889(90)90067-n. [DOI] [PubMed] [Google Scholar]
- 6.Sekiya M, Osuga Ji, Nagashima S, et al. Ablation of Neutral Cholesterol Ester Hydrolase 1 Accelerates Atherosclerosis. Cell Metabolism. 2009;10:219–228. doi: 10.1016/j.cmet.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Buchebner M, Pfeifer T, Rathke N, et al. Cholesteryl ester hydrolase activity is abolished in HSL−/− macrophages but unchanged in macrophages lacking KIAA1363. J Lipid Res. 2010;51:2896–2908. doi: 10.1194/jlr.M004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Contreras JA. Hormone-sensitive lipase is not required for cholesteryl ester hydrolysis in macrophages. Biochem Biophys Res Commun. 2002;292:900–903. doi: 10.1006/bbrc.2002.6757. [DOI] [PubMed] [Google Scholar]
- 9.Kraemer FB, Shen WJ. Hormone-sensitive lipase: control of intracellular tri-(di-)acylglycerol and cholesteryl ester hydrolysis. J Lipid Res. 2002;43:1585–1594. doi: 10.1194/jlr.r200009-jlr200. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh S. Cholesteryl ester hydrolase in human monocyte/macrophage: cloning, sequencing, and expression of full-length cDNA. Physiol Genomics. 2000;2:1–8. doi: 10.1152/physiolgenomics.2000.2.1.1. [DOI] [PubMed] [Google Scholar]
- 11.Okazaki H, Igarashi M, Nishi M, et al. Identification of neutral cholesterol ester hydrolase, a key enzyme removing cholesterol from macrophages. J Biol Chem. 2008;283:33357–33364. doi: 10.1074/jbc.M802686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quiroga AD, Lehner R. Role of endoplasmic reticulum neutral lipid hydrolases. Trends in Endocrinology & Metabolism. 2011;22:218–225. doi: 10.1016/j.tem.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Zhao B, Fisher BJ, Clair RWS, Rudel LL, Ghosh S. Redistribution of macrophage cholesteryl ester hydrolase from cytoplasm to lipid droplets upon lipid loading. J Lipid Res. 2005;46:2114–2121. doi: 10.1194/jlr.M500207-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Igarashi M, Osuga J, Isshiki M, Sekiya M, Okazaki H, Takase S, Takanashi M, Ohta K, Kumagai M, Nishi M, Fujita T, Nagai R, Kadowaki T, Ishibashi S. Targeting of neutral cholesterol ester hydrolase to the endoplasmic reticulum via its N-terminal sequence. J Lipid Res. 2010;51:274–285. doi: 10.1194/jlr.M900201-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13:655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon GM, Cravatt BF. Activity-based Proteomics of Enzyme Superfamilies: Serine Hydrolases as a Case Study. J Biol Chem. 2010;285:11051–5. doi: 10.1074/jbc.R109.097600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 18.Beller M, Riedel D, Jansch L, Dieterich G, Wehland J, Jackle H, Kuhnlein RP. Characterization of the Drosophila lipid droplet subproteome. Mol Cell Proteomics. 2006;5:1082–1094. doi: 10.1074/mcp.M600011-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Thiel K, Heier C, Haberl V, Thul PJ, Oberer M, Lass A, Jackle H, Beller M. The evolutionarily conserved protein CG9186 is associated with lipid droplets, required for their positioning and for fat storage. J Cell Sci. 2013;126:2198–2212. doi: 10.1242/jcs.120493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachovchin DA, Cravatt BF. The pharmacological landscape and therapeutic potential of serine hydrolases. Nat Rev Drug Discov. 2012;11:52–68. doi: 10.1038/nrd3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchan DW, Ward SM, Lobley AE, Nugent TC, Bryson K, Jones DT. Protein annotation and modelling servers at University College London. Nucleic Acids Res. 2010;38:W563–W568. doi: 10.1093/nar/gkq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm C, Osterlund T. Hormone-sensitive lipase and neutral cholesteryl ester lipase. Methods Mol Biol. 1999;109:109–121. doi: 10.1385/1-59259-581-2:109. [DOI] [PubMed] [Google Scholar]
- 23.Rong JX, Shapiro M, Trogan E, Fisher EA. Transdifferentiation of mouse aortic smooth muscle cells to a macrophage-like state after cholesterol loading. Proc Natl Acad Sci U S A. 2003;100:13531–13536. doi: 10.1073/pnas.1735526100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tall AR. Cholesterol efflux pathways and other potential mechanisms involved in the athero-protective effect of high density lipoproteins. J Intern Med. 2008;263:256–273. doi: 10.1111/j.1365-2796.2007.01898.x. [DOI] [PubMed] [Google Scholar]
- 25.Larigauderie G, Furman C, Jaye M, Lasselin C, Copin C, Fruchart JC, Castro G, Rouis M. Adipophilin Enhances Lipid Accumulation and Prevents Lipid Efflux From THP-1 Macrophages: Potential Role in Atherogenesis. Arterioscler Thromb Vasc Biol. 2004;24:504–510. doi: 10.1161/01.ATV.0000115638.27381.97. [DOI] [PubMed] [Google Scholar]
- 26.Igarashi M, Osuga J, Uozaki H, et al. The critical role of neutral cholesterol ester hydrolase 1 in cholesterol removal from human macrophages. Circ Res. 2010;107:1387–1395. doi: 10.1161/CIRCRESAHA.110.226613. [DOI] [PubMed] [Google Scholar]
- 27.Ouimet M, Marcel YL. Regulation of Lipid Droplet Cholesterol Efflux From Macrophage Foam Cells. Arterioscler Thromb Vasc Biol. 2012;32:575–581. doi: 10.1161/ATVBAHA.111.240705. [DOI] [PubMed] [Google Scholar]
- 28.Schweiger M, Lass A, Zimmermann R, Eichmann TO, Zechner R. Neutral lipid storage disease: genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ABHD5. Am J Physiol Endocrinol Metab. 2009;297:E289–E296. doi: 10.1152/ajpendo.00099.2009. [DOI] [PubMed] [Google Scholar]
- 29.Contreras JA, Lasuncion MA. Essential differences in cholesteryl ester metabolism between human monocyte-derived and J774 macrophages. Evidence against the presence of hormone-sensitive lipase in human macrophages. Arterioscler Thromb. 1994;14:443–452. doi: 10.1161/01.atv.14.3.443. [DOI] [PubMed] [Google Scholar]
- 30.Harte RA, Hulten LM, Lindmark H, Reue K, Schotz MC, Khoo J, Rosenfeld ME. Low level expression of hormone-sensitive lipase in arterial macrophage-derived foam cells: potential explanation for low rates of cholesteryl ester hydrolysis. Atherosclerosis. 2000;149:343–350. doi: 10.1016/s0021-9150(99)00345-7. [DOI] [PubMed] [Google Scholar]
- 31.Tabas I. The Role of Endoplasmic Reticulum Stress in the Progression of Atherosclerosis. Circ Res. 2010;107:839–850. doi: 10.1161/CIRCRESAHA.110.224766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fazio S, Linton M. Failure of ACAT inhibition to retard atherosclerosis. N Engl J Med. 2006;354:1307–1309. doi: 10.1056/NEJMe068012. [DOI] [PubMed] [Google Scholar]
- 33.Feng B, Yao PM, Li Y, Devlin CM, Zhang D, Harding HP, Sweeney M, Rong JX, Kuriakose G, Fisher EA, Marks AR, Ron D, Tabas I. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol. 2003;5:781–792. doi: 10.1038/ncb1035. [DOI] [PubMed] [Google Scholar]
- 34.Son SH, Goo YH, Chang BH, Paul A. Perilipin 2 (PLIN2)-Deficiency Does Not Increase Cholesterol-Induced Toxicity in Macrophages. PLoS ONE. 2012;7:e33063. doi: 10.1371/journal.pone.0033063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le LS, Dugail I. Connecting lipid droplet biology and the metabolic syndrome. Prog Lipid Res. 2009;48:191–195. doi: 10.1016/j.plipres.2009.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.