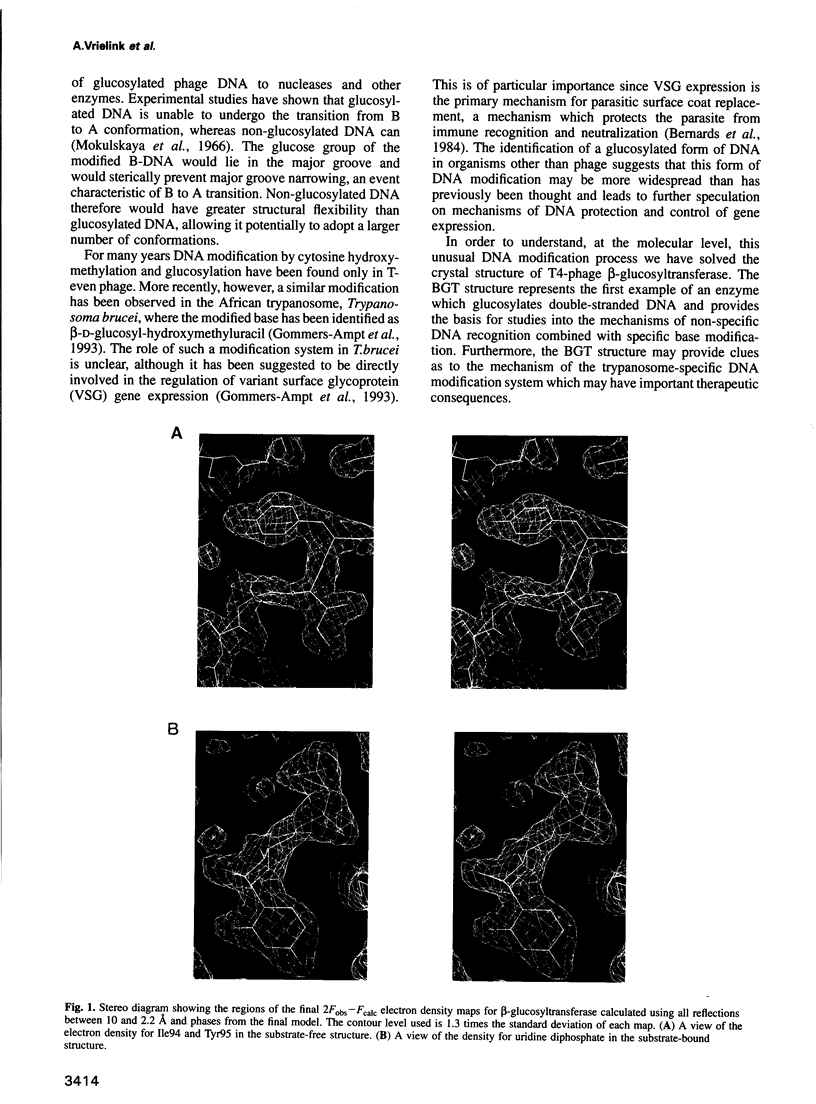
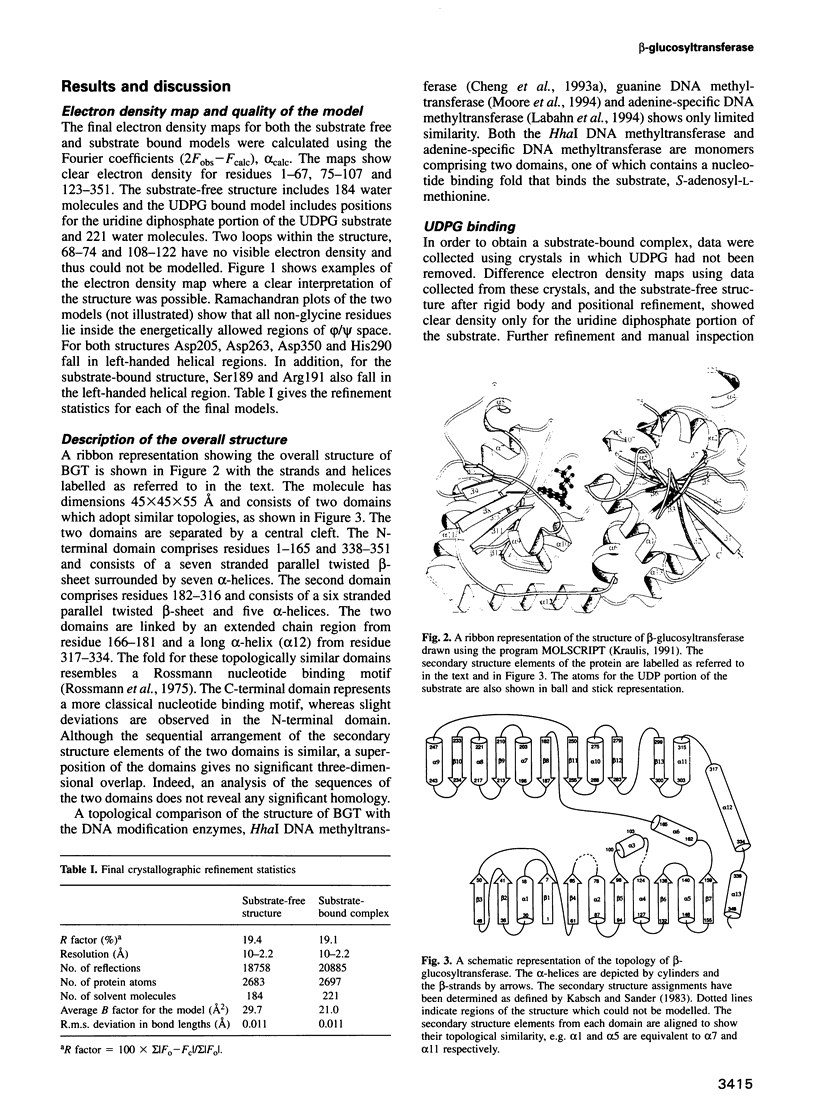
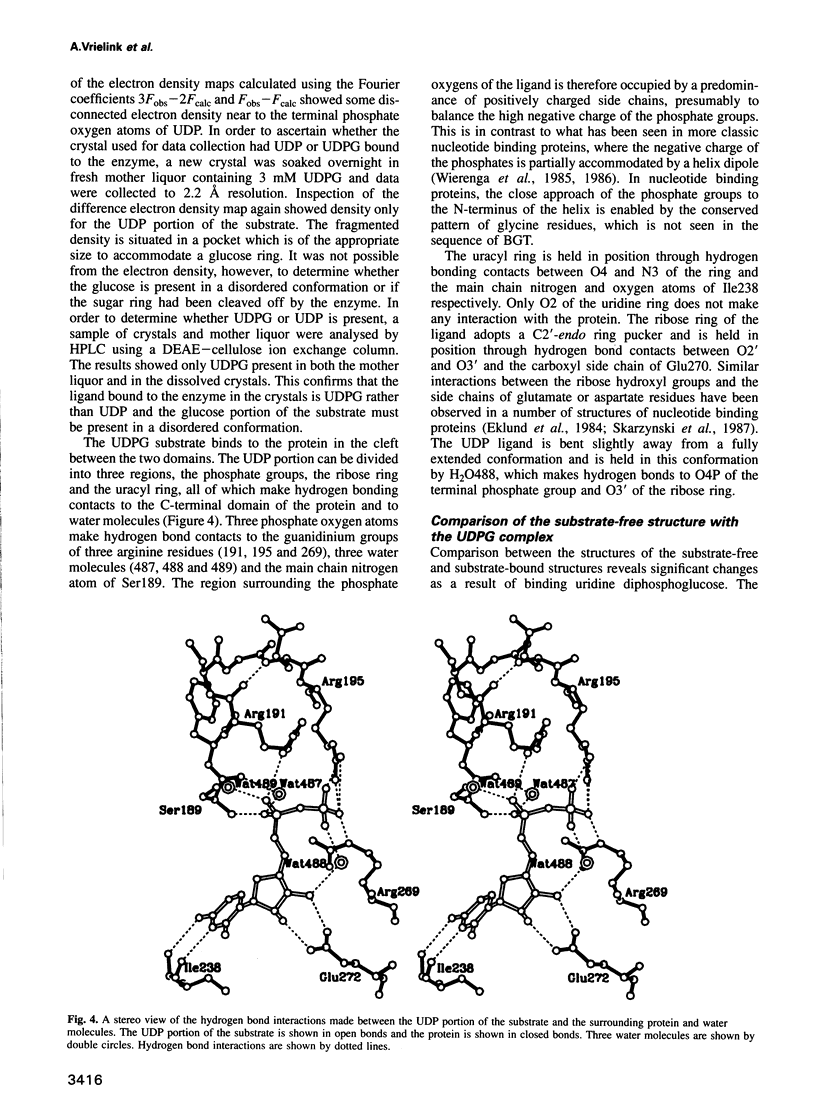
Abstract
Bacteriophage T4 beta-glucosyltransferase (EC 2.4.1.27) catalyses the transfer of glucose from uridine diphosphoglucose to hydroxymethyl groups of modified cytosine bases in T4 duplex DNA forming beta-glycosidic linkages. The enzyme forms part of a phage DNA protection system. We have solved and refined the crystal structure of recombinant beta-glucosyltransferase to 2.2 A resolution in the presence and absence of the substrate, uridine diphosphoglucose. The structure comprises two domains of similar topology, each reminiscent of a nucleotide binding fold. The two domains are separated by a central cleft which generates a concave surface along one side of the molecule. The substrate-bound complex reveals only clear electron density for the uridine diphosphate portion of the substrate. The UDPG is bound in a pocket at the bottom of the cleft between the two domains and makes extensive hydrogen bonding contacts with residues of the C-terminal domain only. The domains undergo a rigid body conformational change causing the structure to adopt a more closed conformation upon ligand binding. The movement of the domains is facilitated by a hinge region between residues 166 and 172. Electrostatic surface potential calculations reveal a large positive potential along the concave surface of the structure, suggesting a possible site for duplex DNA interaction.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adzhubei A. A., Sternberg M. J. Left-handed polyproline II helices commonly occur in globular proteins. J Mol Biol. 1993 Jan 20;229(2):472–493. doi: 10.1006/jmbi.1993.1047. [DOI] [PubMed] [Google Scholar]
- Anderson C. M., Stenkamp R. E., Steitz T. A. Sequencing a protein by x-ray crystallography. II. Refinement of yeast hexokinase B co-ordinates and sequence at 2.1 A resolution. J Mol Biol. 1978 Jul 25;123(1):15–33. doi: 10.1016/0022-2836(78)90374-1. [DOI] [PubMed] [Google Scholar]
- Bennett W. S., Jr, Steitz T. A. Glucose-induced conformational change in yeast hexokinase. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4848–4852. doi: 10.1073/pnas.75.10.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett W. S., Jr, Steitz T. A. Structure of a complex between yeast hexokinase A and glucose. II. Detailed comparisons of conformation and active site configuration with the native hexokinase B monomer and dimer. J Mol Biol. 1980 Jun 25;140(2):211–230. doi: 10.1016/0022-2836(80)90103-5. [DOI] [PubMed] [Google Scholar]
- Bernards A., van Harten-Loosbroek N., Borst P. Modification of telomeric DNA in Trypanosoma brucei; a role in antigenic variation? Nucleic Acids Res. 1984 May 25;12(10):4153–4170. doi: 10.1093/nar/12.10.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Cheng X., Kumar S., Posfai J., Pflugrath J. W., Roberts R. J. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell. 1993 Jul 30;74(2):299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- Cox G. S., Conway T. W. Template properties of glucose-deficient T-even bacteriophage DNA. J Virol. 1973 Dec;12(6):1279–1287. doi: 10.1128/jvi.12.6.1279-1287.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmalingam K., Goldberg E. B. Restriction in vivo. III. General effects of glucosylation and restriction on phage T4 gene expression and replication. Virology. 1979 Jul 30;96(2):393–403. doi: 10.1016/0042-6822(79)90097-7. [DOI] [PubMed] [Google Scholar]
- Eklund H., Samama J. P., Jones T. A. Crystallographic investigations of nicotinamide adenine dinucleotide binding to horse liver alcohol dehydrogenase. Biochemistry. 1984 Dec 4;23(25):5982–5996. doi: 10.1021/bi00320a014. [DOI] [PubMed] [Google Scholar]
- Freemont P. S., Friedman J. M., Beese L. S., Sanderson M. R., Steitz T. A. Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont P. S., Rüger W. Crystallization and preliminary X-ray studies of T4 phage beta-glucosyltransferase. J Mol Biol. 1988 Sep 20;203(2):525–526. doi: 10.1016/0022-2836(88)90021-6. [DOI] [PubMed] [Google Scholar]
- Gommers-Ampt J. H., Van Leeuwen F., de Beer A. L., Vliegenthart J. F., Dizdaroglu M., Kowalak J. A., Crain P. F., Borst P. beta-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993 Dec 17;75(6):1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- Gram H., Rüger W. Genes 55, alpha gt, 47 and 46 of bacteriophage T4: the genomic organization as deduced by sequence analysis. EMBO J. 1985 Jan;4(1):257–264. doi: 10.1002/j.1460-2075.1985.tb02344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOSSE J., KORNBERG A. Glucosylation of deoxyribonucleic acid. III. alpha- and beta-Glucosyl transferases from T4-infected Escherichia coli. J Biol Chem. 1962 Jun;237:1968–1976. [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- KORNBERG S. R., ZIMMERMAN S. B., KORNBERG A. Glucosylation of deoxyribonucleic acid by enzymes from bacteriophage-infected Escherichia coli. J Biol Chem. 1961 May;236:1487–1493. [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kang C. H., Shin W. C., Yamagata Y., Gokcen S., Ames G. F., Kim S. H. Crystal structure of the lysine-, arginine-, ornithine-binding protein (LAO) from Salmonella typhimurium at 2.7-A resolution. J Biol Chem. 1991 Dec 15;266(35):23893–23899. [PubMed] [Google Scholar]
- LEHMAN I. R., PRATT E. A. On the structure of the glucosylated hydroxymethylcytosine nucleotides of coliphages T2, T4, and T6. J Biol Chem. 1960 Nov;235:3254–3259. [PubMed] [Google Scholar]
- Lamm N., Wang Y., Mathews C. K., Rüger W. Deoxycytidylate hydroxymethylase gene of bacteriophage T4. Nucleotide sequence determination and over-expression of the gene. Eur J Biochem. 1988 Mar 15;172(3):553–563. doi: 10.1111/j.1432-1033.1988.tb13925.x. [DOI] [PubMed] [Google Scholar]
- Mokul'skaia T. D., Gorlenko Zh M., Zamchuk L. A., Bogdanova E. S., Mokul'skii M. A., Gol'dfarb D. M., Khesin R. B. Nekotorye svoistva negliukozilirovannoi DNK fega T2. Biokhimiia. 1966 Jul-Aug;31(4):749–759. [PubMed] [Google Scholar]
- Moore M. H., Gulbis J. M., Dodson E. J., Demple B., Moody P. C. Crystal structure of a suicidal DNA repair protein: the Ada O6-methylguanine-DNA methyltransferase from E. coli. EMBO J. 1994 Apr 1;13(7):1495–1501. doi: 10.1002/j.1460-2075.1994.tb06410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh B. H., Pandit J., Kang C. H., Nikaido K., Gokcen S., Ames G. F., Kim S. H. Three-dimensional structures of the periplasmic lysine/arginine/ornithine-binding protein with and without a ligand. J Biol Chem. 1993 May 25;268(15):11348–11355. [PubMed] [Google Scholar]
- Remington S., Wiegand G., Huber R. Crystallographic refinement and atomic models of two different forms of citrate synthase at 2.7 and 1.7 A resolution. J Mol Biol. 1982 Jun 15;158(1):111–152. doi: 10.1016/0022-2836(82)90452-1. [DOI] [PubMed] [Google Scholar]
- Rüger W. Transcription of bacteriophage T4 DNA in vitro: selective initiation with dinucleotides. Eur J Biochem. 1978 Jul 17;88(1):109–117. doi: 10.1111/j.1432-1033.1978.tb12427.x. [DOI] [PubMed] [Google Scholar]
- Sack J. S., Saper M. A., Quiocho F. A. Periplasmic binding protein structure and function. Refined X-ray structures of the leucine/isoleucine/valine-binding protein and its complex with leucine. J Mol Biol. 1989 Mar 5;206(1):171–191. doi: 10.1016/0022-2836(89)90531-7. [DOI] [PubMed] [Google Scholar]
- Sack J. S., Trakhanov S. D., Tsigannik I. H., Quiocho F. A. Structure of the L-leucine-binding protein refined at 2.4 A resolution and comparison with the Leu/Ile/Val-binding protein structure. J Mol Biol. 1989 Mar 5;206(1):193–207. doi: 10.1016/0022-2836(89)90532-9. [DOI] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Sharff A. J., Rodseth L. E., Spurlino J. C., Quiocho F. A. Crystallographic evidence of a large ligand-induced hinge-twist motion between the two domains of the maltodextrin binding protein involved in active transport and chemotaxis. Biochemistry. 1992 Nov 10;31(44):10657–10663. doi: 10.1021/bi00159a003. [DOI] [PubMed] [Google Scholar]
- Skarzyński T., Moody P. C., Wonacott A. J. Structure of holo-glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus at 1.8 A resolution. J Mol Biol. 1987 Jan 5;193(1):171–187. doi: 10.1016/0022-2836(87)90635-8. [DOI] [PubMed] [Google Scholar]
- Spurlino J. C., Lu G. Y., Quiocho F. A. The 2.3-A resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J Biol Chem. 1991 Mar 15;266(8):5202–5219. doi: 10.2210/pdb1mbp/pdb. [DOI] [PubMed] [Google Scholar]
- Tomaschewski J., Gram H., Crabb J. W., Rüger W. T4-induced alpha- and beta-glucosyltransferase: cloning of the genes and a comparison of their products based on sequencing data. Nucleic Acids Res. 1985 Nov 11;13(21):7551–7568. doi: 10.1093/nar/13.21.7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WYATT G. R., COHEN S. S. A new pyrimidine base from bacteriophage nucleic acids. Nature. 1952 Dec 20;170(4338):1072–1073. doi: 10.1038/1701072a0. [DOI] [PubMed] [Google Scholar]
- Wang B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- Warner H. R., Snustad P., Jorgensen S. E., Koerner J. F. Isolation of bacteriophage T4 mutants defective in the ability to degrade host deoxyribonucleic acid. J Virol. 1970 Jun;5(6):700–708. doi: 10.1128/jvi.5.6.700-708.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwicker J., Watson H. C. Calculation of the electric potential in the active site cleft due to alpha-helix dipoles. J Mol Biol. 1982 Jun 5;157(4):671–679. doi: 10.1016/0022-2836(82)90505-8. [DOI] [PubMed] [Google Scholar]
- Westbrook E. M. Crystal density measurements using aqueous ficoll solutions. Methods Enzymol. 1985;114:187–196. doi: 10.1016/0076-6879(85)14019-x. [DOI] [PubMed] [Google Scholar]
- Wiegand G., Remington S., Deisenhofer J., Huber R. Crystal structure analysis and molecular model of a complex of citrate synthase with oxaloacetate and S-acetonyl-coenzyme A. J Mol Biol. 1984 Mar 25;174(1):205–219. doi: 10.1016/0022-2836(84)90373-5. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Terpstra P., Hol W. G. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986 Jan 5;187(1):101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- Winkler F. K., Banner D. W., Oefner C., Tsernoglou D., Brown R. S., Heathman S. P., Bryan R. K., Martin P. D., Petratos K., Wilson K. S. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993 May;12(5):1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M., Rüger W. Cloning and sequencing of the genes of beta-glucosyl-HMC-alpha-glucosyl-transferases of bacteriophages T2 and T6. Nucleic Acids Res. 1993 Mar 25;21(6):1500–1500. doi: 10.1093/nar/21.6.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Geiduschek E. P. The role of replication proteins in the regulation of bacteriophage T4 transcription. II. Gene 45 and late transcription uncoupled from replication. J Mol Biol. 1975 Aug 25;96(4):539–562. doi: 10.1016/0022-2836(75)90138-2. [DOI] [PubMed] [Google Scholar]
- Zou J. Y., Flocco M. M., Mowbray S. L. The 1.7 A refined X-ray structure of the periplasmic glucose/galactose receptor from Salmonella typhimurium. J Mol Biol. 1993 Oct 20;233(4):739–752. doi: 10.1006/jmbi.1993.1549. [DOI] [PubMed] [Google Scholar]