Abstract
Complex developmental processes such as hematopoiesis require a series of precise and coordinated changes in cellular identity to ensure blood homeostasis. Epigenetic mechanisms help drive changes in gene expression that accompany the transition from hematopoietic stem cells to terminally differentiated blood cells. Genome-wide profiling technologies now provide valuable glimpses of epigenetic changes that occur during normal hematopoiesis, and genetic mouse models developed to investigate the in vivo functions of chromatin-modifying enzymes clearly demonstrate significant roles for these enzymes during embryonic and adult hematopoiesis. Here, we will review the basic science aspects of chromatin modifications and the enzymes that add, remove, and interpret these epigenetic marks. This overview will provide a framework for understanding the roles that these molecules play during normal hematopoiesis. Moreover, many chromatin-modifying enzymes are involved in hematologic malignancies, underscoring the importance of establishing and maintaining appropriate chromatin modification patterns to normal hematology.
Chromatin structure and epigenetic mechanisms
Hematopoiesis is an orderly process that involves the coordination of stem cell self-renewal, controlled expansion of progenitor cells, and timely differentiation. Developmental programs, such as hematopoiesis, are orchestrated by changes in patterns of gene expression. These patterns are directed by transcription factors, and the actions of these factors are strongly influenced by the chromatin structures of their target genes.
The basic repeating unit of chromatin is the nucleosome, which contains 2 copies of each core histone protein, H3, H4, H2A, and H2B and 146 base pairs of DNA spooled around the histone octamer. Histone H3 and histone H4 form a heterotetramer at the heart of the nucleosome, and 2 heterodimers of histones H2A and H2B associate with the tetramer to complete the octamer. Linker histones, typified by H1, bind to DNA entering into and exiting from the nucleosome, thereby providing further structural stability.
Chromatin that is in an open configuration, accessible to transcription factors and RNA polymerase, is often referred to as euchromatin, reflecting a less-condensed appearance in microscopic images of nuclear sections. Heterochromatin, in contrast, refers to more densely packed structures that are generally transcriptionally silent. Regions of the genome that include telomeres and centromeres are highly heterochromatic. Chromatin organization in other regions of the genome is more dynamic, reflecting changes in transcriptional competence and activity. Such plasticity is essential for regulation of various cellular processes such as cell division, gene transcription, DNA repair, replication, and recombination.
The term epigenetics is defined as inherited variation that occurs without changes in the DNA sequence.1 Epigenetic mechanisms allow a level of plasticity for the genetic information encoded by the DNA, which allows for the establishment of cell-specific expression programs. Modifications added to or removed from the chromatin template that carry epigenetic information include DNA methylation and histone posttranslational modifications (PTMs). Noncoding RNA molecules are also emerging as important regulators of gene expression during hematopoiesis.2,3 However, this review will highlight examples of the essential roles that chromatin-modifying enzymes play during hematopoietic cell development by regulating the dynamic nature of DNA and specific histone modifications, namely methylation and acetylation.
DNA modifications
Methylation of a cytosine residue in the context of a CpG dinucleotide represents a stable regulatory mark in the mammalian genome. Cytosine methylation (5-methyl cytosine [5mC]) is generally associated with heterochromatin formation and transcriptional repression. Conversely, stretches of unmethylated CpG dinucleotides, termed CpG islands, are often found in the promoters of actively transcribed genes and support a euchromatic environment. Additional functional groups were recently discovered to modify the 5-position of cytosine residues, namely hydroxymethyl (5hmC), formyl (5fC), and carboxyl (5caC), and the biological functions of these modifications throughout the genome are now being discovered.4
Covalent PTM of histone proteins
PTMs occurring on histone proteins regulate chromatin structure in a temporal and spatial manner. Histone modifications serve both as signaling mechanisms and as binding platforms to recruit other proteins. Covalent PTMs added to the core histone proteins include acetylation, methylation, phosphorylation, ubiquitination, sumoylation, adenosine 5'-diphosphate ribosylation, and biotinylation. The “writers” and “erasers” that govern the addition or removal of these PTMs often demonstrate site specificity; however, multiple enzymes are capable of modifying the same residue within a histone substrate.
Different patterns of histone PTMs have been proposed to act as a “code” in that specific patterns confer specific biological responses.5,6 The “histone code” hypothesis requires a way for effector molecules to interpret the information given by histone PTMs to mediate cellular processes outside of chromatin. Protein domains that recognize individual or combinatorial histone modifications have been termed reading or presenting domains.7 As more histone PTM reading domains are discovered, linking binding patterns to associated biological responses is becoming more complex. Accumulating evidence links deregulation of histone PTM interpretation with oncogenic transformation, highlighting the importance of histone reader proteins.8
Lysine acetylation and methylation occur on all 4 of the core histones, most prevalently on the N-terminal tails. The addition of an acetyl group neutralizes the positive charge of the lysine residue, which weakens interactions between the histones and DNA, facilitating more open and relaxed chromatin states. Acetylated histones are generally associated with transcriptionally active genes and euchromatic regions of the genome9 (Figure 1A). Lysine residues can accommodate up to 3 methyl groups (mono-, di-, or tri-), whereas arginine residues may be monomethylated or dimethylated, yielding monomethyl arginine, asymmetric dimethyl arginine, or symmetric dimethyl arginine. The degree of methylation at a particular lysine or arginine residue is just as important in influencing biological outcome as the target site (Figure 1).
Figure 1.
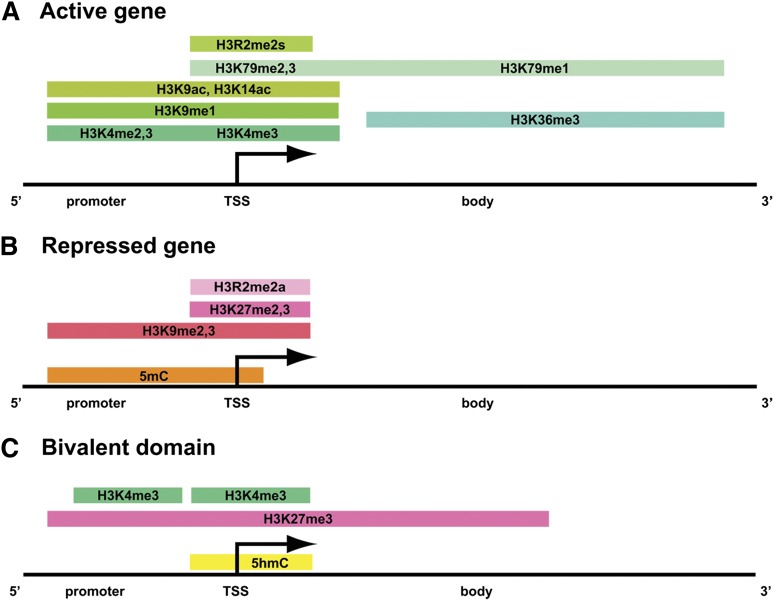
Histone modifications mark dynamically regulated genes. In all panels, shades of green indicate active marks, whereas shades of pink represent repressive marks. The orange and yellow colors mark regions of 5 mC and 5 hmC, respectively. (A) The distribution of histone methylation and acetylation marks, along with the degree of methylation, is illustrated across the promoter region, TSS (transcriptional start site), and gene body of a transcriptionally active gene. (B) Histone H3 methylation and DNA methylation are found in the promoter region and surrounding the TSS in transcriptionally repressed genes. (C) Bivalent chromatin domains consist of discrete pockets of histone H3K4me3 within large regions of histone H3K27me3. DNA 5hmC is also found in bivalent domains.
Chromatin modifications mark genomic regions
Many chromatin modifications are associated with either transcriptional activation or repression (Table 1); however, there are certainly exceptions to these assignments. The degree of a modification and where the modifications are located within genic regions affect the transcriptional outcome. Often, multiple enzymes modify a particular amino acid residue within a histone protein. The chromatin-modifying enzymes discussed in this review are listed in Table 1 along with their identified substrates, and Figure 1 serves as a general guide for matching chromatin modifications with transcriptional outcomes. Histone H3K4me2,3, H3R2me2s, H3K9ac, H3K14ac, K3K27ac, and H4K16ac mark euchromatic regions of the genome, particularly transcriptional enhancer and promoter regions10-12 (Figure 1A). Histone H3K36me3 is a mark associated with transcription elongation and is found in the body and 3′ ends of genes (Figure 1A). Methylation of histone H3K79 is largely associated with transcriptionally active genes, with dimethylation and trimethylation found near transcription start sites, and monomethylation occurring widely across active gene bodies.13
Table 1.
Chromatin-modifying enzymes
| Enzyme | Writer/eraser | Chromatin modification | Transcriptional outcome |
|---|---|---|---|
| MLL1, 2, 3, 4, 5 | Writer | H3K4me | Activation |
| DOT1L | Writer | H3K79me | Activation |
| PRMT1 | Writer | H4R3me2a | Activation |
| p300 | Writer | H3acH4ac | Activation |
| MOZ | Writer | H3ac | Activation |
| PCAF | Writer | H3ac | Activation |
| EZH2 | Writer | H3K27me | Repression |
| HDAC1, 2, 3 | Eraser | H3acH4ac | Repression |
| KDM1A (LSD1) | Eraser | H3K4meH3K9me | Repression |
| KDM5A (JARID1A) | Eraser | H3K4me | Repression |
| PRMT6 | Writer | H3R2me2a | Repression |
| DNMT1, 3a, 3b | Writer | 5mC | Repression |
| Tet1, 2, 3 | Writer/eraser | 5hmC/5mC | TBD |
| AID | Eraser | 5mC | TBD |
AID, activation-induced cytidine deaminase; TBD, to be determined.
Histone H3K9me3, H3K27me1,3, and H4K20me3, together with 5mC are marks of constitutive heterochromatin found in transcriptionally silenced regions of the genome, such as pericentromeric DNA and the inactive X chromosome14,15 (Figure 1B). Histone H3K9me2,3, H3K27me3, and H3R2me2a, as well as 5mC mark facultative heterochromatin and are found in transcriptionally repressed genes that are dynamically regulated16,17 (Figure 1B). The aforementioned repressive chromatin marks do not necessarily coexist at the same promoters. Furthermore, whereas methylated CpG dinucleotides in gene promoters and near transcription start sites often confer a repressive transcriptional state, CpG methylation within gene bodies does not interfere with transcription.18
Genomic regions that contain both transcriptional activating and repressing histone modifications are termed bivalent domains and are prevalent in undifferentiated embryonic stem cells. Bivalent domains consist of small patches of H3K4me3, near the transcriptional start site within larger regions of H3K27me319 (Figure 1C). In addition, 5hmC was recently identified as an additional epigenetic mark enriched at bivalent domains.20
In pluripotent embryonic stem cells, bivalent domains mark genes that are involved in cell fate determination and may help to maintain them in a repressed, yet potentially active, transcriptional state until the onset of differentiation.19 Genome-wide mapping of H3K4me3 and H3K27me3 in hematopoietic stem and progenitor cells revealed the presence of bivalent domains marking genes involved in lineage specification.21,22 Furthermore, the level of H3K4me3 in the progenitor cells correlated significantly with the expression of the marked genes in differentiated cell types.22 These results suggest that the plasticity of the chromatin landscape is crucial for the transition between multipotent and differentiated states.
Writers, erasers, and readers of chromatin modifications
Recent literature is replete with examples demonstrating roles for chromatin-modifying enzymes during hematopoiesis. Moreover, genes encoding these enzymes are frequently disrupted in hematologic malignancies (Table 2).23 Although genome-wide profiling methods provide snapshots of the chromatin landscape during hematopoiesis, genetic mouse models have afforded insights into the roles of chromatin-modifying enzymes during this process.
Table 2.
Chromatin-modifying enzymes essential for hematopoiesis
| Enzyme | Epigenetic activity | Targeting strategy | Role in HSC self-renewal | Lineage-specific role | Ref no. |
|---|---|---|---|---|---|
| Moz | Histone acetylation | KO | Yes | No | 55 |
| KO | Yes | Myeloid | 56 | ||
| Mll1 | Histone methylation | KO | Yes | Myeloid | 67 |
| KO | Yes | No | 68 | ||
| CKO; Mx1-Cre,lysozyme M-Cre,CD19-Cre, lck-Cre | Yes | No | 69 | ||
| Ezh2 | Histone methylation | CKO; Mx1-Cre | — | Lymphoid | 75 |
| CKO; Tie2-Cre,Rosa26-Cre | No | Erythroid and lymphoid | 74 | ||
| Dot1L | Histone methylation | Inducible KO;Rosa26-Cre-ER | Yes | MPPs and myeloid | 91 |
| Inducible KO;Rosa26-Cre-ER | Yes | MPPs and myeloid | 90 | ||
| KO | — | Erythroid | 89 | ||
| Lsd1 | Histone demethylation | CKD; global | Yes | Myeloid | 99 |
| Dnmt1 | DNA 5mC | CKO; Mx1-Cre | Yes | Myeloid | 26 |
| CKO; Mx1-CreHypomorph | Yes | Lymphoid | 27 | ||
| Dnmt3a | DNA 5mC | CKO; Mx1-Cre | Yes | Long-term HSCs | 29 |
| Tet2 | DNA 5hmC | KO | Yes | Myeloid | 37 |
| KO | Yes | Myeloid | 36 | ||
| CKO; Vav-Cre,Ella-Cre, Mx1-Cre | Yes | Myeloid | 38 | ||
| KOCKO; Mx1-Cre | Yes | Lymphoid and myeloid | 39 |
CKO, conditional knockout; CKD, conditional knockdown; 5hmC, 5-hydroxymethyl cytosine; MPP, multipotent progenitor.
DNA methyltransferases (DNMTs) catalyze 5mC
In mammals, the principal enzymes that catalyze cytosine methylation are members of the DNMT1 and DNMT3 families. DNMT1 primarily serves as a maintenance enzyme by copying cytosine methylation patterns from a hemimethylated substrate after DNA replication.24 The DNMT3 family includes 2 active enzymes, DNMT3a and DNMT3b, each of which is further represented by several isoforms. DNMT3a and DNMT3b are referred to as de novo methyltransferases because of their preference to act on unmethylated DNA substrates.25
Conditional deletion of Dnmt1 in hematopoietic stem cells (HSCs) and progenitor cells revealed that Dnmt1 activity is required for early stages of hematopoiesis.26,27 Absence of Dnmt1 activity led to decreased self-renewal capacity26 and a rapid ablation of the HSC pool.27 Furthermore, mouse HSCs lacking Dnmt1 were unable to maintain a sufficient population of myeloid cells because of increased proliferative capacity and decreased differentiation potential of the myeloid progenitor pool.26
Deletion of Dnmt3a and Dnmt3b, either independently or together, in adult mouse HSCs indicated that these enzymes are required for self-renewal but not differentiation or lineage commitment.28 However, a long-term study investigating the role of Dnmt3a in mouse hematopoiesis demonstrated a requirement for Dnmt3a activity in maintaining the differentiation potential of HSCs after serial transplantation into wild-type recipient mice.29 Furthermore, a complex pattern of genomic cytosine methylation was observed in Dnmt3a-deficient HSCs, including hypermethylated regions, that might indicate compensation by other DNMTs.29 Mutations affecting the catalytic activity of DNMT3A have been identified in nearly one third of patients with acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS) and are associated with poor clinical outcome, indicating that the function of this enzyme is crucial to maintain normal hematopoiesis.30-32
DNMT1, DNMT3a, and DNMT3b physically and functionally interact with one another, thereby making it more difficult to ascribe the terms maintenance or de novo to any particular enzyme.25 Defining genomic targets of these enzymes will aid in assigning distinct functions. Taken together, these studies indicate that although the mechanisms are not yet clearly defined, appropriate expression and activity of DNA methyltransferases are essential during development and for adult blood homeostasis.
Ten-eleven translocation (TET) enzymes catalyze 5hmC
The TET family of enzymes, composed of TET1, TET2, and TET3, catalyze the oxidation of 5mC to 5hmC, which is an intermediate product of DNA demethylation mechanisms, and is proposed to have functional roles in stem cell biology.4 The TET enzymes have distinct expression patterns, with TET1 expressed in the fetal liver, TET2 in bone marrow, and both TET2 and TET3 in peripheral blood,33 suggesting unique functions for these enzymes during developmental and adult hematopoiesis. Recently, mutations within TET2 were identified as a prevalent occurrence in myeloid malignancies, prompting intense investigations to define the molecular function of TET2 during hematopoiesis.34,35
Mice lacking Tet2 are viable, but hematopoietic malignancies develop within 2 to 6 months.36-39 HSCs obtained from Tet2-deficient mice exhibit enhanced self-renewal capacity and achieve a greater contribution to peripheral blood production in transplanted recipients, compared with wild-type HSCs.36-39 The overall differentiation potential of HSCs was impeded in the absence of Tet2, with a bias toward expansion of monocyte and macrophage lineages.36-38 Disease progression resembling human chronic myelomonocytic leukemia was the most common phenotype observed in the Tet2-null mouse models.36,38,39 In addition, myeloid disorders similar to myeloproliferative disorder–like myeloid leukemia, myeloid leukemia with maturation, and MDS were reported to cause lethality in approximately one third of Tet2-null mice.36
Mutations that disrupt TET2 enzymatic activity and correlate with decreased global 5hmC levels are observed in patients with myeloid malignancies.40 TET2 mutations have also been detected in lymphoid malignancies, suggesting that mutations occur in early hematopoietic progenitors with myeloid and lymphoid potential.39 Moreover, TET2 mutations identified in patients are often monoallelic, indicating that haploinsufficiency contributes to transformation.34,38,39 Heterozygous gain-of-function mutations in the isocitrate dehydrogenase 1 and 2 enzymes, which are prevalent in AML, cause inhibition of TET2 catalytic activity and result in increased global 5mC levels, similar to the effect induced by loss of TET2 function.41
DNA mechanisms of demethylation
The passive loss of cytosine methylation after DNA replication is a widely accepted mechanism for the removal of CpG methylation. The direct removal of a methyl group from DNA is thermodynamically unfavorable.42 However, several pathways recently have been proposed to mediate indirect, yet active, removal of CpG methylation.
TET enzymes catalyze sequential oxidation reactions that convert 5mC to 5hmC, 5fC, and finally to 5caC, which can be removed by DNA repair mechanisms.43 An alternative proposed active mechanism is deamination of 5mC by activation-induced cytidine deaminase, yielding a thymidine base that is subject to subsequent DNA mismatch repair and base excision repair mechanisms.42 Whereas these mechanisms do not directly return the methylated cytosine to an unmodified base, the 5mC functionality is no longer present. Defining DNA demethylation mechanisms is an active area of investigation that is likely to yield exciting results in the future.
DNA methylation readers
Methyl-binding domains (MBDs) recognize and bind to 5mC and are found in a conserved family of proteins that includes MBD1, MBD2, MBD3, MBD4, and MeCP2.44 MBD proteins are generally regarded as mediators of transcriptional repression by recruiting chromatin-modifying enzymes, such as histone deacetylases (HDACs), to regions of highly methylated DNA.44 The MBD proteins are not essential for embryonic development;44 however, transgenic mouse model studies have pointed to a role for Mbd2 in the silencing of embryonic globin gene expression in adult erythroid cells.45,46
The Set and RING finger associated domain within UHRF1, a DNMT1 interacting partner, and the MBD of MBD3 were recently reported as readers of 5hmC.47,48 The UHRF1 Set and RING finger associated domain was previously shown to bind hemimethylated DNA.49 It will be interesting to learn whether these molecular functions overlap or if they occur as distinct targeting mechanisms.
The specificity and affinity of MBD domains for 5mC have made them valuable reagents in defining patterns of global cytosine methylation.50 The MBD domain MBD2 was recently used to affinity-purify methylated DNA fragments from mouse HSCs, CMPs, and erythroblasts to determine changes in 5mC during myeloid differentiation. Using this method, a high level of 5mC was detected in mouse HSCs, with a dramatic loss of global methylation occurring during myeloid differentiation,51 similar to results obtained from a DNA methylation-profiling study performed during erythropoiesis using a bisulfite-based method.52 A similar approach using the MBD domain of MeCP2 was used to detect changes in DNA methylation during later stages of lymphoid differentiation; however, no dramatic change in 5mC patterns were noted,53 suggesting that global changes in DNA methylation accompany early differentiation events during hematopoiesis. Notably, the affinity capture approach can be applied to map the genomic locations of CpG islands using domains that recognize unmethylated CpG sites.54
Histone acetyltransferases (HATs) and deacetylases
A number of HAT enzymes have been identified as regulators of chromatin-templated processes, including gene transcription and DNA repair. Most HATs function within large multimeric complexes that include targeting subunits and additional histone-modifying activities. In mice, homozygous deletion of the HAT Moz leads to reduced numbers of hematopoietic progenitors, although all lineages are represented, indicating that differentiation and lineage commitment are not altered.55,56
Acetylation is a reversible reaction, and HDAC enzymes carry out deacetylation of histones as well as nonhistone substrates. The expression of HDACs 1,2,3 is low in hematopoietic progenitor cells but increases during differentiation. However, overexpression of HDAC1 was shown to inhibit myeloid differentiation.57 HDACs are notoriously linked with hematologic malignancies through their association with leukemogenic fusion proteins, and they mediate aberrant transcriptional repression of genes required for hematopoietic differentiation.58 Small-molecule HDAC inhibitors are used for leukemia therapy independently but are more successful when given in combination with other cytotoxic agents.59 Defining the genomic targets of HDACs will aid in developing inhibitors that specifically target those enzymes modifying genes involved in oncogenesis.
Acetylation readers
Many of the enzymes responsible for “writing” and “erasing” histone modifications contain domains that “read” modifications. Bromodomains are well-characterized modules that bind acetylated lysine residues.60 The bromodomain of the HAT p300/CBP-associated factor binds the acetylated N-terminal tail of H3 to promote transcriptional activation61 (Figure 2). Moreover, an example of a bromodomain indirectly promoting chromatin occupancy of a transcriptional regulator was recently reported. The bromodomain within the bromodomain extra terminal family member, BRD3, interacts with acetylated GATA1 to facilitate its recruitment to genomic targets, including genes involved in erythroid differentiation.62 Bromodomain-mediated interactions are amenable to small-molecule inhibition strategies.63 Several bromodomain extra terminal inhibitors show promise as therapeutic agents for AML through their effective inhibition of leukemia stem and progenitor cell proliferation and blocking of mixed lineage leukemia (MLL)–mediated transformation.64,65
Figure 2.
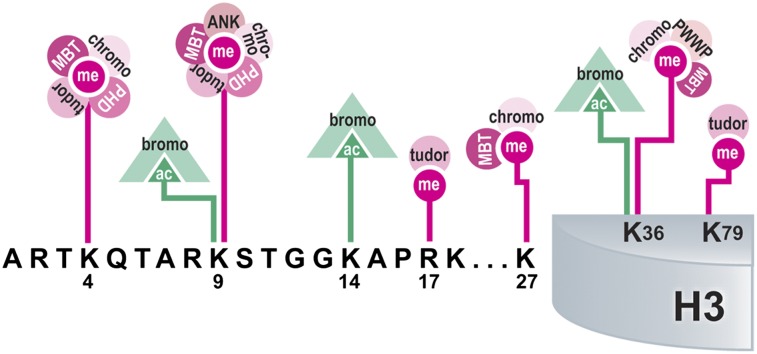
Modification-binding domains interpret histone PTMs. Domains from various histone-interacting proteins are depicted along with the PTMs that they bind on histone H3. Multiple reading domains can interact with a single modified site, as illustrated for histones H3K4, K9, K27, and K36. Interactions may also change depending on the modification present at the site, as shown for acetylation or methylation of histones H3K9 and K36. The colors represent different PTMs and associated binding domains, which are depicted as follows: pink, methylation; green, acetylation.
Lysine methyltransferases (KMTs) and demethylases (KDMs)
The majority of mammalian KMTs characterized to date function within multisubunit complexes and target the N-terminal tail of histone H3, where they coordinate with additional histone-modifying activities to drive either activation or repression of transcription. The MLL methyltransferases carry out methylation of histone H3K4 and are members of the trithorax group family, whereas enzymes that methylate H3K27 belong to the polycomb group (PcG) family. The functional antagonism between these groups of proteins regulates transcription of developmental genes, including the Hox gene cluster, and establishes patterning during embryonic development in vertebrates.
MLL1 is required for viability, as Mll1−/− embryos die by E10.5.66 Yolk sacs derived from Mll1−/− embryos produce fewer and smaller myeloid colonies than yolk sacs derived from Mll1+/+ embryos, indicating that MLL1 is critical for primitive hematopoiesis.67 Mll1 is required for HSC and early progenitor self-renewal during hematopoiesis in the fetal liver68 as well as in the adult bone marrow.69 The human MLL1 gene is often disrupted by chromosomal translocations observed in acute leukemias,70 and defining the molecular mechanisms underlying transformation remains an active area of investigation.71,72 Furthermore, inactivating mutations in other MLL proteins, including MLL2 and MLL3, are observed in various cancers.23
There are 2 well-characterized PcG complexes that incorporate multiple histone-modifying activities to mediate transcriptional repression: polycomb repressive complex 1 and 2 (PRC1 and PRC2). The enhancer of zeste homolog 2 (EZH2) enzyme functions within the PRC2 complex to catalyze dimethylation and trimethylation of histone H3K27, which is then bound by the PRC1 complex to maintain transcriptional repression of PcG target genes. EZH2 silences genes required for cell fate decisions, thus promoting stem cell self-renewal. Fitting with this role, overexpression of Ezh2 enhances the long-term repopulating potential of HSCs after serial transplantations into mice.73 Ezh2 is required for fetal HSC proliferation and erythropoiesis in the developing embryo, but loss of Ezh2 has only a trivial effect on the function of HSCs in the adult bone marrow, primarily their ability to contribute to lymphopoiesis.74
Consistently, Ezh2 activity was previously found to be essential during the early stages of B-cell development but was dispensable for the maturation and activation of pro-B cells.75 Notably, mutations abrogating the catalytic function of EZH2 were recently identified in acute and chronic myeloid malignancies,76-79 whereas gain-of function mutations were identified in B-cell lymphomas80,81,23. Inhibiting the methyltransferase activity of EZH2 in patients with lymphoma carrying activating mutations might be a promising therapeutic endeavor. Treatment with an EZH2-specific small-molecule inhibitor resulted in decreased lymphoid cell proliferation in vitro and in mouse xenograft models.82
A number of point mutations in the polycomb-associated gene addition of sex combs-like 1 (ASXL1) have been identified in patients with MDS or AML and correlate with a poor prognosis.83-86 ASXL1 mutations promote transformation by decreasing the recruitment of PRC2 to leukemogenic target genes, resulting in loss of histone H3K27 methylation and transcriptional repression of those genes.87 Given that gain- or loss-of-function mutations affects various components of the PRC2 complex suggests that altering PRC2 activity and/or H3K27 methylation levels in either direction are detrimental to normal hematopoiesis.
The disruptor of the telomeric silencing 1-like (DOT1L) enzyme methylates the globular domain of histone H3 at K79, which correlates with active transcription.88 Mice deficient for Dot1L exhibit severe anemia and die between E10.5-13.5.89 Erythroid development was specifically inhibited in Dot1L-null mice, and altered H3K79 methylation status and expression of the erythroid regulatory genes, Gata2 and Pu1, were observed.89 Conditional targeting strategies revealed a role for Dot1L in maintaining adult hematopoiesis as well.90,91 Notably, aberrant recruitment of DOT1L by several MLL fusion proteins to genes involved in cellular transformation is a common mechanism underlying MLL-mediated leukemogenesis.13,92 Moreover, DOT1L is a promising therapeutic target in mixed myeloid leukemias in which MLL and/or AF10 oncogenic fusion proteins are expressed.93,94
Histone lysine methylation is dynamic and is regulated by KDM enzymes. Two classes of KDMs acting on histones have been identified: amine oxidases and jumonji C domain–containing enzymes. Lysine-specific demethylase 1 (LSD1) was the first histone KDM identified and exhibits dual substrate specificity for histones H3K4 and H3K9.95,96 LSD1 interacts with transcriptional repressors and demethylates histone H3K4me1,2 yielding an unmodified lysine residue. LSD1 and HDACs1 and 2 interact with the T-cell acute lymphocytic leukemia 1 transcription factor in erythroleukemia cell lines and negatively regulate T-cell acute lymphocytic leukemia 1 target genes to inhibit the onset of erythroid differentiation.97 LSD1 also cooperates with the Gfi-1 transcriptional repressors in a lineage-specific manner to regulate megakaryocytic, granulocytic, and erythroid differentiation.98 Furthermore, depletion of Lsd1 in mice leads to enhanced HSC and progenitor cell proliferation and deficient erythroid differentiation.99 Regulatory roles for LSD1 in defining hematopoietic lineage commitment highlight the importance of restricting histone H3K4 methylation patterns to establish and maintain differentiation programs.
Arginine methyltransferases
Methylation of arginine residues within histone and nonhistone proteins is catalyzed by protein arginine (R) methyltransferases (PRMTs). There are 9 identified enzymes in the mammalian PRMT family (PRMT1-9). They are generally classified as type 1 enzymes (PRMTs 1, 3, 4, 6, and 8), which catalyze asymmetric dimethylation; or type 2 enzymes (PRMTs 5 and 7), which catalyze symmetric dimethylation.100 Methylated arginine residues identified in histones H3 and H4 directly and indirectly regulate transcription.
PRMT1 catalyzes asymmetric dimethylation of histone H4R3 (H4R3me2a), which promotes p300-mediated acetylation of H4K8 and K12, resulting in transcriptional activation.101 PRMT1 interacts with a leukemogenic MLL fusion protein (MLL-EEN) to promote transformation of primary hematopoietic progenitor cells. The oncogenic mechanism is dependent on PRMT1-mediated methylation of histone H4R3 and transcriptional activation of MLL target genes.102 Furthermore, cross-talk occurring between PRMTs and KMTs is necessary to establish appropriate patterns of histone methylation. For instance, PRMT6-mediated methylation of histone H3R2me2a indirectly represses transcription by inhibiting MLL-mediated activity toward histone H3K4,17,103 and by blocking the association of histone H3K4-methyl–binding proteins.104
PRMTs influence transcription via methylation of histone and nonhistone substrates, including transcription factors. In human hematopoietic progenitor cells, PRMT6 physically interacts with runt-related transcription factor 1/AML1 at the promoters of megakaryocytic genes and maintains a chromatin environment that is repressed but primed for transcriptional activation.105 Moreover, PRMT1-mediated methylation of runt-related transcription factor 1/AML1 itself enhances its ability to activate transcription of several target genes during hematopoietic differentiation by inhibiting its association with the SIN3A corepressor complex.106 PRMT1 also interacts with an alternatively spliced version of the AML1-ETO leukemogenic fusion protein, AE9a, and activates transcription of AE9a target genes to enhance proliferation of hematopoietic progenitor cells.107
Lysine and arginine methylation readers
Methyl lysine-binding domains include the plant homeodomain (PHD) finger, chromo, tudor, malignant brain tumor, PWWP, and the ankyrin repeat. Multiple binding domains show an affinity for particular lysine or arginine residues within histone tails; however, binding by different effector molecules depends on whether methylation or acetylation is present (Figure 2). In addition, effector binding also depends on the degree of the modification present on a particular residue, such as me1, me2, me3. These requirements suggest temporal regulation of binding events to ensure that the appropriate interactions occur between the histone substrate and the binding effector molecule.
Histone-modifying enzymes often contain domains that bind modifications. For instance, the histone demethylase JARID1A contains a C-terminal PHD finger that binds its substrate, H3K4me2,3 (Figure 2). Translocations occurring in AML create a JARID1A-NUP98 fusion protein that uses this PHD finger to aberrantly bind PcG-silenced Hox genes and maintain them in a transcriptionally active state, resulting in an arrest of hematopoietic differentiation.108 Furthermore, replacing the JARID1A PHD domain with a PHD domain that reads unmethylated histone H3K4 fails to induce transformation, supporting the idea that induced transcriptional activation of temporally regulated genes is crucial to the leukemogenic mechanism.108 This example demonstrates that aberrant genomic targeting of a histone modification–reading domain is sufficient to drive transformation.
The conserved PHD domains within MLL1 are lost after leukemogenic translocations.109 It was recently shown that MLL1 PHD3 binds histone H3K4me2/3 and is required for localization of wild-type MLL1 to target gene promoters to activate transcription.110,111 Introduction of the third PHD finger (PHD3) into the MLL-ENL fusion protein blocks aberrant HoxC8 gene activation in an HDAC-dependent manner and subsequently inhibits hematopoietic progenitor transformation.112 Similarly, adding multiple PHD fingers to the MLL-AF9 fusion protein inhibits association with the HoxA9 locus and immortalization of hematopoietic progenitors.109 Taken together, it is evident that loss of the PHD fingers is a common contributing factor to MLL1-mediated leukemogenesis.
Epigenetic mechanisms in normal hematology and in hematologic malignancies
Genomic loci encoding chromatin-modifying enzymes are frequently disrupted in human cancers, and these alterations are especially prevalent in hematologic disorders. It is noteworthy that many modification readers promote transformation by cooperating with oncogenic enzymes. Loss or gain of conserved catalytic domains and/or modification-reading domains results in deregulated enzyme activity toward histone and nonhistone substrates that ultimately affects chromatin structure and function to promote leukemogenesis. In addition, fusion proteins created by translocations often lead to altered protein-protein interactions that target histone-modifying activities to inappropriate genomic targets. One significant point that remains is how these enzymes are targeted to specific genomic loci. Patterns of DNA methylation and histone modification are frequently interdependent because of functional interactions between the enzymes that modify DNA and histones. It is critical to define common DNA methylation and histone modification patterns, in addition to downstream gene targets affected by genetic alterations in the writers, erasers, and readers of chromatin modifications as we look toward the future design of molecular-based therapies to treat hematologic malignancies.
Acknowledgments
The authors thank Elizabeth McIvor for helpful comments and discussion on the manuscript. The authors also thank Leisa McCord for assistance with the artwork.
This work was supported by the Cancer Prevention Research Institute of Texas (CPRIT) RP100429 to S.Y.R.D.
Authorship
Contribution: J.S.B. and S.Y.R.D. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sharon Dent, Department of Molecular Carcinogenesis, The University of Texas MD Anderson Cancer Center Science Park, PO Box 389, Smithville, TX 78957; e-mail sroth@mdanderson.org.
References
- 1.Zhu B, Reinberg D. Epigenetic inheritance: uncontested? Cell Res. 2011;21(3):435–441. doi: 10.1038/cr.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paralkar VR, Weiss MJ. A new ‘Linc’ between noncoding RNAs and blood development. Genes Dev. 2011;25(24):2555–2558. doi: 10.1101/gad.183020.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Connell RM, Baltimore D. MicroRNAs and hematopoietic cell development. Curr Top Dev Biol. 2012;99:145–174. doi: 10.1016/B978-0-12-387038-4.00006-9. [DOI] [PubMed] [Google Scholar]
- 4.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2012;13(1):7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Gardner KE, Allis CD, Strahl BD. Operating on chromatin, a colorful language where context matters. J Mol Biol. 2011;409(1):36–46. doi: 10.1016/j.jmb.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25(1):15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 8.Chi P, Allis CD, Wang GG. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10(7):457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Migliori V, Müller J, Phalke S, et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19(2):136–144. doi: 10.1038/nsmb.2209. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein BE, Kamal M, Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120(2):169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Kim TH, Barrera LO, Zheng M, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436(7052):876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steger DJ, Lefterova MI, Ying L, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28(8):2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schotta G, Lachner M, Sarma K, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18(11):1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plath K, Fang J, Mlynarczyk-Evans SK, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300(5616):131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 16.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Guccione E, Bassi C, Casadio F, et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449(7164):933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 18.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 20.Pastor WA, Pape UJ, Huang Y, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473(7347):394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui K, Zang C, Roh TY, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4(1):80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adli M, Zhu J, Bernstein BE. Genome-wide chromatin maps derived from limited numbers of hematopoietic progenitors. Nat Methods. 2010;7(8):615–618. doi: 10.1038/nmeth.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butler JS, Koutelou E, Schibler AC, et al. Histone-modifying enzymes: regulators of developmental decisions and drivers of human disease. Epigenomics. 2012;4(2):163–177. doi: 10.2217/epi.12.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song J, Rechkoblit O, Bestor TH, et al. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science. 2011;331(6020):1036–1040. doi: 10.1126/science.1195380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ooi SK, O’Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J Cell Sci. 2009;122(Pt 16):2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trowbridge JJ, Snow JW, Kim J, et al. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5(4):442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bröske AM, Vockentanz L, Kharazi S, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41(11):1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 28.Tadokoro Y, Ema H, Okano M, et al. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J Exp Med. 2007;204(4):715–722. doi: 10.1084/jem.20060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Challen GA, Sun D, Jeong M, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2011;44(1):23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan XJ, Xu J, Gu ZH, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43(4):309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 32.Yamashita Y, Yuan J, Suetake I, et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29(25):3723–3731. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- 33.Lorsbach RB, Moore J, Mathew S, et al. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia. 2003;17(3):637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 34.Abdel-Wahab O, Mullally A, Hedvat C, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114(1):144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ko M, Rao A. TET2: epigenetic safeguard for HSC. Blood. 2011;118(17):4501–4503. doi: 10.1182/blood-2011-08-373357. [DOI] [PubMed] [Google Scholar]
- 36.Li Z, Cai X, Cai CL, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118(17):4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ko M, Bandukwala HS, An J, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA. 2011;108(35):14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moran-Crusio K, Reavie L, Shih A, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quivoron C, Couronné L, Della Valle V, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20(1):25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Ko M, Huang Y, Jankowska AM, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franchini DM, Schmitz KM, Petersen-Mahrt SK. 5-Methylcytosine DNA demethylation: more than losing a methyl group. Annu Rev Genet. 2012;46:419–441. doi: 10.1146/annurev-genet-110711-155451. [DOI] [PubMed] [Google Scholar]
- 43.He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bogdanović O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118(5):549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rupon JW, Wang SZ, Gaensler K, et al. Methyl binding domain protein 2 mediates gamma-globin gene silencing in adult human betaYAC transgenic mice. Proc Natl Acad Sci USA. 2006;103(17):6617–6622. doi: 10.1073/pnas.0509322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rupon JW, Wang SZ, Gnanapragasam M, et al. MBD2 contributes to developmental silencing of the human ε-globin gene. Blood Cells Mol Dis. 2011;46(3):212–219. doi: 10.1016/j.bcmd.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frauer C, Hoffmann T, Bultmann S, et al. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS ONE. 2011;6(6):e21306. doi: 10.1371/journal.pone.0021306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yildirim O, Li R, Hung JH, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147(7):1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bostick M, Kim JK, Estève PO, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317(5845):1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 50.Cross SH, Charlton JA, Nan X, et al. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6(3):236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- 51.Hogart A, Lichtenberg J, Ajay SS, et al. NIH Intramural Sequencing Center. Genome-wide DNA methylation profiles in hematopoietic stem and progenitor cells reveal overrepresentation of ETS transcription factor binding sites. Genome Res. 2012;22(8):1407–1418. doi: 10.1101/gr.132878.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shearstone JR, Pop R, Bock C, et al. Global DNA demethylation during mouse erythropoiesis in vivo. Science. 2011;334(6057):799–802. doi: 10.1126/science.1207306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deaton AM, Webb S, Kerr AR, et al. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011;21(7):1074–1086. doi: 10.1101/gr.118703.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Illingworth R, Kerr A, Desousa D, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6(1):e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas T, Corcoran LM, Gugasyan R, et al. Monocytic leukemia zinc finger protein is essential for the development of long-term reconstituting hematopoietic stem cells. Genes Dev. 2006;20(9):1175–1186. doi: 10.1101/gad.1382606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katsumoto T, Aikawa Y, Iwama A, et al. MOZ is essential for maintenance of hematopoietic stem cells. Genes Dev. 2006;20(10):1321–1330. doi: 10.1101/gad.1393106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wada T, Kikuchi J, Nishimura N, et al. Expression levels of histone deacetylases determine the cell fate of hematopoietic progenitors. J Biol Chem. 2009;284(44):30673–30683. doi: 10.1074/jbc.M109.042242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26(37):5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 59.Quintás-Cardama A, Santos FP, Garcia-Manero G. Histone deacetylase inhibitors for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Leukemia. 2011;25(2):226–235. doi: 10.1038/leu.2010.276. [DOI] [PubMed] [Google Scholar]
- 60.Filippakopoulos P, Knapp S. The bromodomain interaction module. FEBS Lett. 2012;586(17):2692–2704. doi: 10.1016/j.febslet.2012.04.045. [DOI] [PubMed] [Google Scholar]
- 61.Dhalluin C, Carlson JE, Zeng L, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399(6735):491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 62.Lamonica JM, Deng W, Kadauke S, et al. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc Natl Acad Sci USA. 2011;108(22):E159–E168. doi: 10.1073/pnas.1102140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vidler LR, Brown N, Knapp S, et al. Druggability analysis and structural classification of bromodomain acetyl-lysine binding sites. J Med Chem. 2012;55(17):7346–7359. doi: 10.1021/jm300346w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dawson MA, Prinjha RK, Dittmann A, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zuber J, Shi J, Wang E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu BD, Hess JL, Horning SE, et al. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378(6556):505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 67.Hess JL, Yu BD, Li B, et al. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood. 1997;90(5):1799–1806. [PubMed] [Google Scholar]
- 68.Yagi H, Deguchi K, Aono A, et al. Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood. 1998;92(1):108–117. [PubMed] [Google Scholar]
- 69.Jude CD, Climer L, Xu D, et al. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1(3):324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71(4):691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 71.Bernt KM, Armstrong SA. Targeting epigenetic programs in MLL-rearranged leukemias. Hematology Am Soc Hematol Educ Program. 2011;2011:354-360. [DOI] [PubMed]
- 72.Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25(7):661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kamminga LM, Bystrykh LV, de Boer A, et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107(5):2170–2179. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mochizuki-Kashio M, Mishima Y, Miyagi S, et al. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood. 2011;118(25):6553–6561. doi: 10.1182/blood-2011-03-340554. [DOI] [PubMed] [Google Scholar]
- 75.Su IH, Basavaraj A, Krutchinsky AN, et al. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4(2):124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 76.Nikoloski G, Langemeijer SM, Kuiper RP, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42(8):665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 77.Makishima H, Jankowska AM, Tiu RV, et al. Novel homo- and hemizygous mutations in EZH2 in myeloid malignancies. Leukemia. 2010;24(10):1799–1804. doi: 10.1038/leu.2010.167. [DOI] [PubMed] [Google Scholar]
- 78.Ernst T, Chase AJ, Score J, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42(8):722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 79.Jankowska AM, Makishima H, Tiu RV, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 2011;118(14):3932–3941. doi: 10.1182/blood-2010-10-311019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17(9):2613-2618. [DOI] [PubMed]
- 82.McCabe MT, Ott HM, Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 83.Gelsi-Boyer V, Trouplin V, Adélaïde J, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145(6):788–800. doi: 10.1111/j.1365-2141.2009.07697.x. [DOI] [PubMed] [Google Scholar]
- 84.Bejar R, Stevenson K, Abdel-Wahab O, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364(26):2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Metzeler KH, Becker H, Maharry K, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood. 2011;118(26):6920–6929. doi: 10.1182/blood-2011-08-368225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gelsi-Boyer V, Brecqueville M, Devillier R, Murati A, Mozziconacci MJ, Birnbaum D. Mutations in ASXL1 are associated with poor prognosis across the spectrum of malignant myeloid diseases. J Hematol Oncol. 2012;5:12. [DOI] [PMC free article] [PubMed]
- 87.Abdel-Wahab O, Adli M, LaFave LM, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22(2):180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Feng Q, Wang H, Ng HH, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12(12):1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 89.Feng Y, Yang Y, Ortega MM, et al. Early mammalian erythropoiesis requires the Dot1L methyltransferase. Blood. 2010;116(22):4483–4491. doi: 10.1182/blood-2010-03-276501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jo SY, Granowicz EM, Maillard I, et al. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117(18):4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nguyen AT, He J, Taranova O, et al. Essential role of DOT1L in maintaining normal adult hematopoiesis. Cell Res. 2011;21(9):1370–1373. doi: 10.1038/cr.2011.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Okada Y, Feng Q, Lin Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121(2):167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 93.Daigle SR, Olhava EJ, Therkelsen CA, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20(1):53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen L, Deshpande AJ, Banka D, et al. Abrogation of MLL-AF10 and CALM-AF10-mediated transformation through genetic inactivation or pharmacological inhibition of the H3K79 methyltransferase Dot1l. Leukemia. 2012 Nov 9. doi: 10.1038/leu.2012.327. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 96.Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437(7057):436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 97.Hu X, Li X, Valverde K, et al. LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. Proc Natl Acad Sci USA. 2009;106(25):10141–10146. doi: 10.1073/pnas.0900437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Saleque S, Kim J, Rooke HM, et al. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell. 2007;27(4):562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 99.Sprüssel A, Schulte JH, Weber S, et al. Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation. Leukemia. 2012;26(9):2039–2051. doi: 10.1038/leu.2012.157. [DOI] [PubMed] [Google Scholar]
- 100.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33(1):1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang H, Huang ZQ, Xia L, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293(5531):853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 102.Cheung N, Chan LC, Thompson A, et al. Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9(10):1208–1215. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- 103.Hyllus D, Stein C, Schnabel K, et al. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21(24):3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Iberg AN, Espejo A, Cheng D, et al. Arginine methylation of the histone H3 tail impedes effector binding. J Biol Chem. 2008;283(6):3006–3010. doi: 10.1074/jbc.C700192200. [DOI] [PubMed] [Google Scholar]
- 105.Herglotz J, Kuvardina ON, Kolodziej S, et al. Histone arginine methylation keeps RUNX1 target genes in an intermediate state. Oncogene. 2012 doi: 10.1038/onc.2012.274. Jul 9. doi: 10.1038/onc.2012.274. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 106.Zhao X, Jankovic V, Gural A, et al. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008;22(5):640–653. doi: 10.1101/gad.1632608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shia WJ, Okumura AJ, Yan M, et al. PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. Blood. 2012;119(21):4953–4962. doi: 10.1182/blood-2011-04-347476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang GG, Song J, Wang Z, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459(7248):847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Muntean AG, Giannola D, Udager AM, et al. The PHD fingers of MLL block MLL fusion protein-mediated transformation. Blood. 2008;112(12):4690–4693. doi: 10.1182/blood-2008-01-134056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Milne TA, Kim J, Wang GG, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell. 2010;38(6):853–863. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chang PY, Hom RA, Musselman CA, et al. Binding of the MLL PHD3 finger to histone H3K4me3 is required for MLL-dependent gene transcription. J Mol Biol. 2010;400(2):137–144. doi: 10.1016/j.jmb.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen J, Santillan DA, Koonce M, et al. Loss of MLL PHD finger 3 is necessary for MLL-ENL-induced hematopoietic stem cell immortalization. Cancer Res. 2008;68(15):6199–6207. doi: 10.1158/0008-5472.CAN-07-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]