Abstract
The relationship between cancer progression and chronic inflammation is well documented but poorly understood. The innate immune system has long been recognized as the first line of defense against invading pathogens. More recently, endogenous molecules released from tissue matrix (Damage Associated Molecular Patterns [DAMPs]) following tissue injury or periods of active matrix remodeling have also been identified as regulators of innate immunity. DAMPs have been identified as ligands for Toll-like receptors (TLRs), a family of cell-surface proteins which regulate the immune response. TLRs have been identified on resident tissue cells as well as most tumor cells. Therefore, dysregulation of the innate immune response secondary to biochemical and mechanical driven changes in the extracellular matrix of the tumor microenvironment may be a critical component of the chronic inflammation associated with tumor progression. Here we review the role of extracellular matrix (ECM)-derived DAMPS in the activation of TLR4 signaling in the context of tumor progression. We also explore the various types of topographical changes that can lead to ECM-derived DAMPs and their contribution to TLR4 activation.
Keywords: extracellular matrix, toll-like receptor-4, fibronectin, DAMP
INTRODUCTION
The innate immune system can work efficiently to identify and destroy cancer cells ultimately leading to suppression of tumor growth. A vast amount of evidence from in vitro studies, animal models and clinical trials indicates that the host-protective action of the innate immune system is critical for the body's defense against cancer [1]. Unfortunately, during solid tumor progression the mechanisms that drive innate immunity can become dysregulated and chronic inflammation often ensues. Changes in cytokine profiles and the infiltration of a cancer-relevant population of immune cells unique to cancer stroma are drivers of tumor growth and spread [2, 3]. Changes in ECM mechanics and biochemistry are typically found to accompany these events in a variety of different cancers, and ECM remodeling is currently emerging as a major contributor to inflammation-based cancer progression [4, 5].
The conventional perspective of the ECM is that it exists to provide a physical support structure for cells. It is also viewed as an intermediary, regulating communication between the outside environment and intracellular signaling events [6]. However, alterations in ECM topography can drastically change the dynamic between the cell and its microenvironment. Cross-talk between tumor cells and surrounding stromal cells results in the restructuring of the ECM to benefit the overall homeostatic requirements of the tumor. Therefore, it is not surprising that there is continuous ECM remodeling during tumor progression. Alterations in the topography of the ECM arise through changes in the composition, structure and organization of ECM molecules [4]. These topographical changes are achieved through a combination of biochemical and mechanical mechanisms. In the context of tumor growth and invasion, proteolysis is a hallmark biochemical event that allows cells to break through the basement membrane and move throughout the tissue stroma. As tumors progress, the stiffness of the surrounding microenvironment increases [7, 8]. Mechanical remodeling is seen when increased physical stiffness of the ECM couples with increased cellular contractile forces to induce changes in protein structure and organization [9]. Three aspects of biochemical remodeling typically occur during solid tumor progression: 1) changes in the composition of ECM 2) generation of ECM fragments and 3) ECM crosslinking. These three events are emerging as drivers of dysfunctional inflammation that can result in tumor growth, metastasis and angiogenesis.
1. Changes in ECM topography drive dysregulated innate immunity
Constant remodeling of the ECM can be important for basic cellular processes and movement. However, the increased deposition of ECM components, as well as the synthesis of new proteins is a characteristic feature of fibrotic diseases, chronic inflammation and the desmoplastic reaction associated with tumor growth [4]. In fact, the extent of desmoplasia within the tumor stroma can be indicative of the aggressiveness of solid tumors. Type I collagen and fibronectin are the most highly expressed ECM components associated with tumor progression, leading to alterations in ECM structure, function and rigidity [8]. Additionally, a wide variety of other proteins and proteoglycans, such as tenascin-C and biglycan, can be deposited [10, 11]. The role of these ECM components in tumor progression, particularly the contribution of their biochemically and mechanically remodeled structures to dysregulated innate immunity and chronic inflammation, represents an understudied but important area of cancer research.
Solid tumor progression is accompanied by extensive remodeling that generates bioactive ECM fragments. Increased proteolysis is a major contributor to the generation of these fragments and is a major hallmark of invasive disease. When proteolysis of the ECM occurs, fragments are released into the microenvironment as discrete domains which maintain secondary structure and biological activity. These ECM domains, sometimes termed “matrikines”, act in a paracrine manner by functioning as ligands for receptors on the surface of cells within the microenvironment [12]. For instance, certain fragments can act as chemoattractants (i.e., recruit monocytes/macrophages), enhance phagocytic processes or promote increased inflammatory cell protease production [13]. Some of these domains can function as endogenous danger signals, DAMPs. DAMPs have been shown to influence a wide range of inflammatory events by serving as ligands for toll-like receptors, the critical regulators of the immune response [12, 14].
The recruitment of inflammatory cells in a tumor microenvironment is highly dependent on the production of pro-inflammatory cytokines [15]. In particular, the production of pro-inflammatory chemokines, a chemotactic subset of cytokines, can promote the infiltration of immune cells, tumor cell proliferation and metastatic dissemination [16, 17]. Numerous reports have documented that increased cytokine production not only increases the risk for tumor development, but can enhance the growth rate of tumors, aid in angiogenic processes and mediate metastatic invasion [18, 19]. A few examples of the cytokines commonly upregulated in tumors are IL-1, IL-6, IL-8, IL-23, and TNF-α, which can be produced by endothelial cells, tumor cells, fibroblasts and a wide range of immune cells [15, 20]. All of the above cytokines are controlled by the transcription factor, Nuclear Factor-kappa B (NF-κB), which is consistently linked to tumor initiation and inflammation-related tumor growth, EMT and invasion [21]. Furthermore, chronic activation of NF-κB in cancer-associated fibroblasts or other stromal cells is believed to provide tumors with a pro-inflammatory microenvironment that nurtures tumor growth and metastasis [22].
ECM fragments are directly linked to increased cytokine levels and activation of NF-κB. Fibronectin fragments in particular have been shown to increase the secretion of pro-inflammatory cytokines in a variety of cell types [23–25]. Elastin fragments were shown to induce NF-κB-dependent cytokine synthesis in cancer cells via the elastin receptor, S-Gal (spliced galactosidase) [26]. A peptide derived from a prominent basement membrane component, laminin-α5, was also implicated in pro-inflammatory responses, as it was shown to induce TNF-α secretion in neutrophils and macrophages [27]. Release of cytokines into a tumor microenvironment, whether it is by the tumor cells or surrounding stromal cells, will produce a pro-inflammatory feed-forward loop. For example, the NF-κB-mediated interleukin-8 (IL-8) production, as well as production of other pro-inflammatory chemokines, recruit neutrophils and other inflammatory cells to the site of tumor growth. At high levels, these tumor-associated neutrophils are associated with poor prognosis in cancer patients [28]. Furthermore, the tumor-associated neutrophils possess an arsenal of proteases that continue to alter the ECM topography as the cells infiltrate the tumor. Ultimately, the up-regulation of proteases causes the generation of more ECM fragments and the subsequent increase in cytokines, thus establishing a feed-forward inflammatory loop [29]. Deletion of tumor-associated neutrophils has long been known to inhibit tumor growth, metastasis and angiogenesis [30–32]. However, blocking the production of the cytokines that attract them to the site of tumor growth may provide an alternative therapeutic opportunity.
ECM fragmentation has also been linked to new blood vessel formation, particularly under conditions of inflammation-related tumor growth. Many pro-inflammatory cytokines, such as CXCL1,-2, IL-1, IL-6, IL-8, GM-CSF and TNF-α, have a dual function and are also considered to be pro-angiogenic factors. Tumor-derived TNF-α was shown to recruit monocytes to the site of cancer growth and induce their differentiation to a myeloid/endothelial phenotype in mouse models of melanoma, and lung and breast cancer [33]. TNF-α is also a well-documented activator of NF-κB, leading to continued upregulation of pro-inflammatory/pro-angiogenic cytokines [18]. Similarly, IL-1α and IL-1β up-regulate pro-angiogenic factors IL-8 and VEGF in cancer and inflammatory cells [34–36]. A variety of studies have demonstrated that tumor angiogenesis and increased levels of VEGF are, in fact, dependent on the presence of IL-1, underscoring the important contribution of these cytokines to tumor angiogenesis [37, 38]. Metastatic dissemination is also under the influence of pro-angiogenic cytokines. Inhibition of the NF-κB-dependent cytokines, CXCL1 and CXCL2, blocks infiltration of inflammatory cells and angiogenesis, which results in decreased breast and prostate cancer metastasis in vivo [39, 40]. Ultimately, the aberrant induction of cytokines by ECM fragments not only results in chronic pro-inflammatory responses but also drives angiogenic processes that feed tumor growth and facilitate metastasis. Furthermore, the growth of new blood vessels around a tumor is often associated with increased deposition of ECM and increased levels of angiogenesis-associated matrix metalloproteases (MMPs). Thus, MMP-mediated remodeling continues to generate bioactive ECM fragments that support angiogenic-associated inflammation and further tumor vascularization [41].
Mechanical remodeling specifically alters the physical relationship between cells and the surrounding ECM proteins. ECM crosslinking is an enzymatic event that changes compliance of the ECM and can have a significant impact on solid tumor growth [7, 42, 43]. Elevated levels of tissue stiffening agents, such as lysyl oxidase (LOX) and tissue transglutaminase, are often associated with increased inflammation [44–46]. Excessive exposure to pro-inflammatory cytokines such as TNF-α was shown to stimulate LOX expression in a variety of cells [47]. A more rigid tissue microenvironment can enhance Rho-dependent cytoskeletal tension, integrin clustering, disruption of adherens junctions and the promotion of cell growth [48]. This response to changes in tissue compliance results in a shift toward a more malignant phenotype, thus highlighting the dangers of tissue rigidity and its effect on cellular behavior. In an example of breast oncogenesis, collagen crosslinking via LOX induces integrin clustering, PI3Kinase activity, premetastatic niche formation and invasion of premalignant epithelium [7, 49]. Additionally, LOX activity was shown to control vascular permeability in vitro and in vivo, suggesting that tissue stiffening can synergize with inflammatory factors (IL-2, TNF-α) to drive inflammation [50]. However, a direct link between tissue stiffening and increased inflammatory cytokine production has not yet been demonstrated.
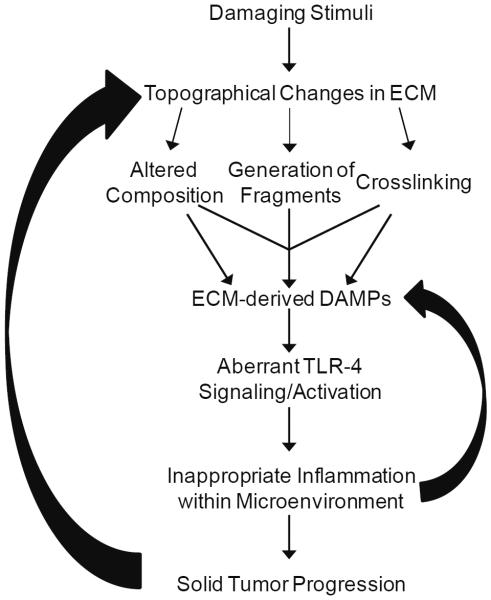
Due to the fact that innate immunity plays a crucial role in protecting hosts against cancer, scientists have begun looking at ways to block unwanted, while promoting protective, immune responses in tumors. The ability to selectively regulate positive and negative aspects of the immune response within tumors requires a further understanding of the mechanisms driving dysregulated immune responses and chronic inflammation. The endogenous danger signals that arise from tumor-associated ECM remodeling represent attractive therapeutic targets. Below we review the ECM-driven mechanisms that mediate dysfunctional innate immune signaling and cytokine production. By identifying and targeting the ECM components that drive immune dysfunction, it may be possible to break the feed-forward loop and intercede in inflammation-driven tumor growth (Figure 1).
Figure 1.
ECM-derived DAMPs modulate an inflammatory loop. Topographical changes within the ECM lead to the generation of ECM-derived DAMPs that drive TLR-4 signaling. Chronic activation of TLR-4 drives dysregulated innate immunity and inappropriate inflammation, characterized by excessive pro-inflammatory cytokine production. In turn, the pro-inflammatory mediators can generate more ECM-derived DAMPs, as well as directly contribute to solid tumor progression.
2. TLR4 signaling and activation in tumors
Toll-like receptors (TLRs) are an integral part of the cellular response to invading pathogens or endogenous danger signals that arise from damage and disease. They are a family of pattern recognition receptors on the surface of tumor cells, as well as resident stromal cells, that activate the major inflammatory pathways: NF-κB, mitogen-activated protein kinases (MAPKs) and interferon-regulatory factors 3, 5 and 7 (IRF3, 5, 7). Activation of these signaling pathways which lead to the release of inflammatory mediators occurs downstream of ligand binding to TLRs [51]. TLR4 was the first toll homologue to be discovered and since then its pro-inflammatory effects have been found to be wide ranging, leading to the activation of over 1000 genes [52]. Most recently, TLR4 has emerged as a principle regulator of the innate immune response in a variety of solid tumors [53]. TLR4 activation has been shown to promote the anti-cancer effects of chemo and radiotherapy, as TLR4 activation on the surface of immune cells boosted anti-tumor immunity [54–56]. In contrast, the expression and activation of TLR4 on tumor and/or stromal cells during the initiation and progression of tumor growth has been linked to increased evasion of immune surveillance [57]. TLR4 signaling in tumors can often arise from the presence of DAMPs that are generated during disease progression in response to ECM remodeling and tissue stiffening [58]. DAMPs are markers of sterile injury because they are non-microbial, while pathogen-associated molecular patterns (PAMPs) are markers of microbial, non-sterile injury. The consequences of sterile injury are quite similar to non-sterile inflammation: pro-inflammatory cytokines are produced and neutrophils and macrophages are recruited to the site of injury [59]. Our understanding of the TLR4 signaling pathway comes from studies on its activation by the canonical ligand, lipopolysaccharide (LPS). While LPS is recognized as a PAMP, the molecular mechanisms that drive DAMP-induced signaling are often compared to LPS' activation of TLR4 (Figure 2).
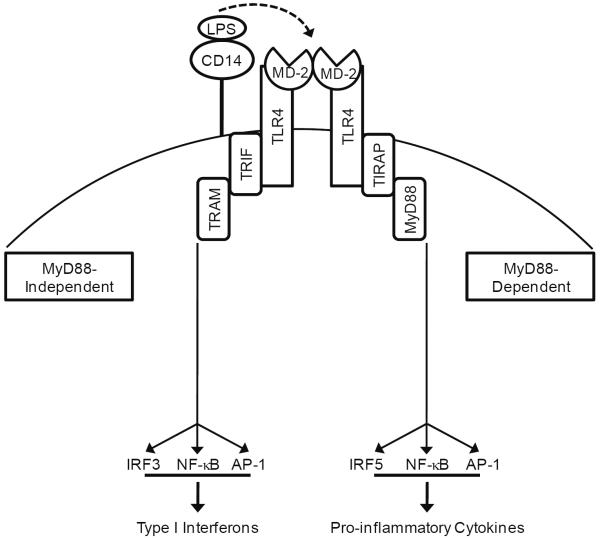
Figure 2.
TLR4 Signaling Pathway. LPS-induced activation of TLR4 is mediated by CD14. CD14 delivers LPS to TLR4/MD-2 complex and downstream signaling is divided into MyD88-dependent and independent signaling. MyD88-dependent signaling results in the rapid activation of NF-κB and the induction of pro-inflammatory cytokines, while MyD88-independent signaling results in late activation of NF-κB and induction of type I interferons.
The TLR4 signaling cascade begins in a somewhat complex and unique manner. Signaling, initiated in response to LPS, is facilitated by the accessory molecule, CD14, which transfers LPS from the bacterial membrane to a TLR4/MD-2 complex. MD-2, myeloid differentiation factor-2, is the ligand-binding constituent of the receptor complex [60]. Crystallography studies show that when LPS is presented to MD-2 by CD14, two copies of the TLR-4/MD-2 complex unite symmetrically at the cell surface. LPS interacts with the hydrophobic pocket in MD-2 and links the two TLR-4/MD-2 units, resulting in dimerization [61]. When LPS recognition occurs, the cytoplasmic TIR (Toll-interleukin-1 receptor) domain of TLR4 initiates two distinct signaling cascades: MyD88-dependent and MyD88-independent cascades. In MyD88-dependent signaling, MyD88 and TIRAP (TIR-domain containing adaptor protein) recruit death domain (DD)-containing proteins, IRAKs (IL-1 receptor-associated kinases), to the TLR4 scaffold, resulting in the activation of several transcription factors: NF-κB, AP-1 and IRF5. The MyD88-mediated activation of NF-κB results in rapid pro-inflammatory cytokine gene expression [62]. Conversely, MyD88-independent signaling results in induction of type I interferons and interferon-inducible genes, which are mainly responsible for eliciting an anti-viral response [63]. For the MyD88-independent signaling to begin, the adaptor proteins TRIF (TIR-domain containing adaptor protein inducing interferon β) and TRAM (TRIF-related adaptor molecule) are recruited. This event leads to the activation of IRF3, as well as AP-1 and late-phase activation of NF-κB [64]. Interestingly, recent evidence has also implicated IRF3-mediated gene expression in the late and sustained activation of NF-κB. The MyD88-dependent cascade acts to rapidly activate NF-κB and initiate a pro-inflammatory cytokine response, while the sustained activation of NF-κB can be maintained through the MyD88-independent cascade. The activation of the TLR4-NF-kB signaling axis is an example of an enhanced transcriptional program that utilizes two different signaling cascades to sustain gene transcription. The adaptor proteins that are recruited to the dimerized TLR4 complex facilitate dual activation of NF-κB in a MyD88-dependent- and independent-manner, contributing κB in a MyD88-to the chronic expression of cytokines [65].
3. ECM-derived DAMPs as emerging activators of aberrant TLR4 signaling
The DAMPs that are generated within the tumor microenvironment are emerging as potent activators of TLR4 signaling [66]. ECM-derived DAMPS develop early in tumor progression in response to increased proteolysis and changes in matrix composition. These DAMPs lead to activation of pro-inflammatory signaling pathways, cytokine production and ultimately inflammation-associated tumor growth. The matrix components that are typically altered in structure and function fall into two groups: proteins/peptides and proteoglycans/glycosaminoglycans [5]. Here we focus on these two groups and their role as ECM-derived DAMPs that arise during cancer progression. We examine their potential interactions with TLR4 and the co-receptors that dictate downstream signaling.
3.1. Proteoglycans/GAGs
Biglycan is a proteoglycan which binds to type I and II collagen. It is ubiquitously expressed in most tissue microenvironments. However, when cleaved from the matrix during remodeling or expressed during infiltration of immune cells, biglycan behaves as a DAMP, ultimately influencing inflammation-related tumor growth [67–69]. Biglycan was found to interact with TLR4 and MD-2 via CD14, resulting in the activation of the pro-inflammatory pathways p38, ERK and NF-κB and the induction of pro-inflammatory factors such as TNF-α, CXCL2 and CXCL13 [70, 71]. These inflammatory factors function as chemo attractants, which further recruit macrophages and neutrophils, thus perpetuating the inflammatory response, as these cells carry out de novo synthesis of biglycan [72]. Increased expression of biglycan corresponds with the development of inflammatory disorders which often accompany tumorigenesis [73, 74] and correlates with enhanced malignant phenotypes in colorectal and gastric cancer patients [10, 68]. Recent data now suggests that the role of biglycan as a DAMP is context-sensitive; when biglycan is bound to the ECM it does not appear to behave as a DAMP [69]. It functions to activate TLR4 signaling only when it is in a soluble form, indicating that increased tissue remodeling will boost soluble levels of this ECM-derived DAMP as it is freed from the matrix.
Similarly, the proteoglycan decorin functions as a TLR4-activating DAMP when presented to cells from solution phase. Decorin does not elicit an inflammatory response when present as an integral part of the ECM, suggesting that proteolytic cleavage of decorin from the matrix regulates the activation of TLR4. Decorin induced the TLR4-dependent activation of p38, ERK and NF-κB, leading to the induction of an array of pro-inflammatory mediators in macrophages. Interestingly, and in contrast to biglycan, the induction of these mediators resulted in the decreased growth of established tumor xenografts in mice [75]. This evidence would suggest that the presence of decorin mediates anti-cancer inflammation, as opposed to inflammation that drives tumor growth. Accordingly, several reports have also documented an inhibitory effect of decorin on tumor growth in models of squamous cell carcinoma and mammary carcinoma [76–78]. The molecular basis for the pro- and anti-tumorigenic functions of these proteoglycan ligands for TLR4 is not known. However, the identification of possible co-receptors involved in the regulation of TLR4 signaling by decorin and biglycan may provide some important clues.
Heparan sulfate (HS) molecules are glycosaminoglycan (GAG) chains that can be cleaved from proteoglycans, such as syndecans and glypicans, and float freely through the extracellular environment. The release of HS into the microenvironment can be regulated by the enzymes, heparanase and elastase. Heparanase is secreted during inflammatory responses, trauma and tumor progression [79, 80]. Elastases are also upregulated in tissues that are chronically inflamed or have undergone some type of trauma [81]. Thus, it is not surprising that the presence of free floating or “shed” HS is increased in inflammation-related cancers and corresponds to poor patient outcome [82, 83]. Johnson et al. [84] demonstrated that HS can interact with TLR4 and mediate dendritic cell maturation. Later it was demonstrated that HS induced the expression of pancreatic pro-inflammatory mediators in vivo in a TLR4/MD-2-dependent manner. In fact, cells treated with elastase exhibited increased activation of TLR4 signaling, most likely as a result of increased levels of soluble HS [85]. However, it remains unclear whether HS can directly bind to TLR4/MD-2 or whether a co-receptor molecule is required for HS signaling [86]. Ultimately, it is emerging that HS functions as a DAMP in a soluble form to activate TLR4 signaling.
Hyaluronan or hyaluronic acid (HA) is a prevalent, linear, high MW GAG within the ECM. During the course of ECM remodeling, HA is broken down into smaller forms through the action of hyaluronidases resulting in the release of low MW, soluble HA fragments (LMWHA) into the microenvironment. During the progression of inflammation-related disease, the levels of hyaluronidases (HYAL1/2) become increased [87, 88]. LMWHA, specifically HA that is < 80 kDa, can trigger inflammatory responses in a variety of cells via activation of TLR4 [89–92]. A pro-inflammatory response by unfragmented, high MW HA has not been documented. The activation of TLR4 signaling by HA appears to be similar to that of decorin and biglycan in that only the soluble LMW form of HA acts as a DAMP to promote inflammation. LMWHA has been shown to activate the TLR4-dependent signaling pathways (i.e., p38, ERK, NF-κB) in dendritic cells leading to the induction of the pro-inflammatory cytokine, TNF-α [90]. Later studies demonstrated that there were differences in the cytokine profile induced by LMWHA vs. LPS, suggesting that the mechanisms for receptor activation may be different. Consistent with this idea, Taylor et al. [93] demonstrated that LMWHA activates TLR4 signaling via a novel co-receptor complex involving TLR4, MD-2 and CD44. The use of CD44 as a TLR4 co-receptor to mediate LMWHA signaling is in contrast to LPS signaling, where CD14 functions as the co-receptor.
3.2. Proteins/peptides
The presence of fibrinogen (FBG), a plasma glycoprotein, has been identified in breast cancer tissue, metastatic brain tumors and prostate cancer [94–96]. Similar to other ECM proteins, FBG can incorporate into the ECM when plasma FBG exudes from the vasculature. FBG can also be locally synthesized and secreted by a variety of different cells to act as support or provisional structure for growth factor and cytokine binding [94]. The presence of FBG has been shown to enhance inflammation-related tumor growth, migration and invasion into surrounding tissues [97, 98]. Addition of FBG to macrophages results in the release of macrophage inflammatory proteins, MIP1α, MIP1β and MIP2, as well as a monocyte chemoattractant protein, MCP1, in a TLR4-dependent manner [99]. A later study demonstrated that the mutations in TLR4, D229G and T399I, which normally render cells LPS-unresponsive, had no effect on FBG signaling through TLR4 [100]. This finding suggests that fibrinogen and LPS may bind to different sites on the TLR4 receptor complex, or possibly utilize distinct co-receptor molecules. The molecular mechanisms underlying the differential response initiated by LPS and FBG are yet to be elucidated, but these findings do indicate that the TLR4 receptor complexes discriminate among various ligands.
Tenascin-C is an ECM glycoprotein that is expressed only under conditions of tissue remodeling or wound healing. It is found in most solid tumors and its presence has most recently been correlated with increased invasion and migration of tumor cells [101, 102]. Cancer cells themselves can produce large amounts of tenascin-C, as can myofibroblasts and other stromal cells [11]. The multimodular structure of tenascin-C, consisting of an assembly domain, a series of EGF-like repeats, a series of FNIII domains, and a C-terminal fibrinogen globe, provides numerous cellular-interaction sites [103]. Tenascin-C was identified as an endogenous activator of TLR4 on macrophages and synovial fibroblasts, leading to the induction of pro-inflammatory cytokines. Tenascin-C interacts with TLR4 through its fibrinogen globe and the induction of cytokines does not require CD14 or MD2 [104]. These data point to the involvement of distinct co-receptors in the TLR4 signaling initiated by tenascin-C and LPS. More recently, tenascin-C was shown to promote an LPS-response in an experimental model of sepsis by post-transcriptionally controlling TNF-α levels indicating an alternative mode of pro-inflammatory action by tenascin-C [105]. Thus, tenascin-C within a tumor microenvironment acts both directly and indirectly to regulate TLR4 signaling by direct binding to the TLR4 receptor complex and by priming immune cells to TLR4 activation by LPS. This type of cellular-sensitization to TLR4-ligands would be expected to enhance inappropriate inflammation within tumors.
Fibronectin is one of the most abundant ECM proteins within the tumor microenvironment and it has wide ranging effects on almost all aspects of cellular behavior. Structurally, the protein is organized into different domains, identified as the type I, II and III. These different regions can bind to various ECM molecules or act as ligands for receptors on the surface of cells [106]. During tumor progression, fibronectin undergoes biochemical changes as additional type III repeats, extra domain A (EDA) and extra domain B (EDB) are induced in the molecule by alternative splicing [107]. Cleavage of fibronectin within the tumor stroma also occurs as fibronectin is highly sensitive to a variety of proteases [108]. Furthermore, changes in tissue compliance lead to alterations in fibronectin secondary structure. The type III domains, which unlike the type I and II domains are not stabilized by disulfide bonds, easily unfold under cell-generated forces in response to increased tissue stiffening [109].
Two regions of fibronectin have been identified as activators of TLR4 signaling (Figure 3). Fn-EDA was identified as an endogenous ligand for TLR4 [110]. Activation of TLR4 by Fn-EDA led to the nuclear translocation of NF-κB, which was shown to be dependent on the presence of MD-2. Recently Fn-EDA was shown to induce inflammatory brain damage in vivo by upregulating pro-inflammatory mediators such as COX-2 and TNF-α, via TLR4 [111]. Fn-EDA has recently been identified as a TLR4 activator in mesenchymal uterine cells, resulting in the release of pro-inflammatory mediators and the induction of pre-term labor [112]. Furthermore, the presence of the EDA domain in tumor-associated fibronectin has a strong correlation with metastatic spread, neo-vascularization and sustained cancer stem cell populations [113, 114]. Although Fn-EDA is highly expressed in tumor stroma, its contribution to tumor inflammation and metastatic progression is unknown.
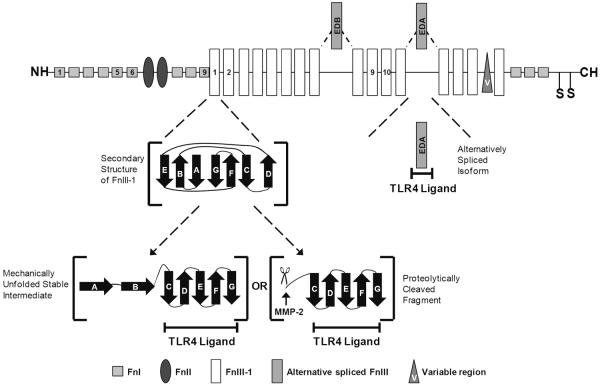
Figure 3.
Schematic of the TLR4 ligands within the fibronectin monomer. The fibronectin type III domains are organized into a beta-sandwich structure consisting of seven beta strands (A–G) that are arranged in two anti-parallel beta sheets consisting of the AGE and BFCD strands. The first type III (FnIII-1) domain is of interest because it has been shown to unfold in response to mechanical force into a stable intermediate structure consisting of strands c-g. The A and B strands can also be proteolytically cleaved from FnIII-1 by MMP-2 to generate the same structure, termed FnIII-1c. Extra Domain A (EDA) is a type III module which is alternatively spliced into the fibronectin during periods of active matrix remodeling. It is very prominent in the stroma of most solid tumors. Both FnIII-1c and EDA are activators of TLR4.
Another region of fibronectin, the first type III domain (FnIII-1), has a well established role in regulating fibronectin polymerization due to its ability to unfold in response to cell-generated tension [115–117]. A peptide, FnIII-1c or anastellin, derived from the FnIII1 domain was shown to activate inflammatory gene expression in a TLR4-dependent manner in human dermal fibroblasts [118]. Steered molecular dynamics and atomic force microscopy studies predict that the FnIII1 domain can partially unfold in response to cell-mediated tension to form a stable intermediate structure similar to FnIII-1c [109]. Forced unfolding experiments predict that the intermediate FnIII-1c structure remains after the first two β-strands (A and B) completely unfold from the remaining protein [119, 120]. Interestingly, recent proteolytic studies identify a functional MMP-2 cleavage site within the FnIII-1 domain, 631XNAPQ [121]. Cleavage at this site will remove the A and B strands from FnIII-1, thereby biochemically releasing fragments containing the FnIII-1c structure from the ECM. In this context, FnIII-1c functions as a DAMP to activate TLR4-dependent inflammatory responses. These findings suggest that fibronectin has two separate domains which can interact with TLR4 to generate an immune response. This activity may be cryptic within the matrix until exposed through the action of proteases, changes in alternative splicing or force-induced unfolding.
The signaling mechanisms downstream of TLR4 are fundamentally understood. However, the field has yet to fully identify all of the TLR4 co-receptors that control TLR4 activation and signaling in response to ECM-derived DAMPs (Table 1). The identification and classification of these co-receptor molecules may help to delineate the molecular events leading to ligand and/or cell-type specific responses to ECM-derived DAMPs. Tenascin-C, for instance, induces a fairly different pro-inflammatory gene signature in human macrophages compared with synovial fibroblasts. While tenascin-C induced the synthesis of TNF-α, IL-6 and IL-8 in macrophages, it only induced the synthesis of IL-6 in the fibroblasts [104]. Conversely, FnIII-1c induced expression of both TNF-α and IL-8, but not IL-6, in human fibroblasts [118]. The variance in pro-inflammatory gene signature may result from changes in ligand binding to the TLR4 complex due to co-receptor specificity or availability. Identification of specific co-receptor-ligand pairs will provide the foundation for future pre-clinical studies designed to target these coreceptors to control dysfunctional immune responses without sacrificing host defense. TLR4 co-receptors are expected to be attractive therapeutic targets for the design of reagents to treat dysregulated inflammation.
Table 1.
TLR4 co-receptors and accessory molecules that mediate ECM-denved DAMP recognition and signaling
| ECM-derived DAMPs | TLR4 co-receptors or accessory molecules | Source |
|---|---|---|
| Fn-EDA | MD-2 | 110 |
| Biglycan | MD-2, CD14 | 70 |
| HA fragments | MD-2, CD44 | 93 |
| Heparan sulfate fragments | MD-2 | 85 |
| Decorin | MD-2 | 75 |
| FnIII-1c | Unknown | |
| Fibrinogen | Unknown | |
| Tenascin-C | Unknown |
Footnotes
CONFLICT OF INTEREST STATEMENT The authors have no conflicts of interest.
REFERENCES
- 1.Vaseley MD, Kershaw MH, Schreiber RD, Smyth MJ. Annu. Rev. Immunol. 2011;29:235. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- 2.Lin W, Karin M. J. Clin. Invest. 2007;117:1175. doi: 10.1172/JCI31537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinarello CA. Cancer Metastasis Rev. 2006;25:307. doi: 10.1007/s10555-006-9000-8. [DOI] [PubMed] [Google Scholar]
- 4.Cox TR, Erler JT. Dis. Model Mech. 2011;4:165. doi: 10.1242/dmm.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu P, Weaver VM, Werb Z. J. Cell Biol. 2012;196:395. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim SH, Turnbull J, Guimond S. J. Endocrinol. 2011;209:139. doi: 10.1530/JOE-10-0377. [DOI] [PubMed] [Google Scholar]
- 7.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM. Cell. 2009;139:891. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butcher DT, Alliston T, Weaver VM. Nature. 2009;9:108. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu H, Mouw JK, Weaver VM. Trends Cell Biol. 2011;21:47. doi: 10.1016/j.tcb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang B, Li GX, Zhang SG, Wang Q, Wen YG, Tang HM, Zhou CZ, Xing AY, Fan JW, Yan DW, Qiu GQ, Yu ZH, Peng ZH. Exp. Biol. Med. 2011;236:1247. doi: 10.1258/ebm.2011.011124. [DOI] [PubMed] [Google Scholar]
- 11.Oskarsson T, Acharyya S, Zhang Z, Vanharanta S, Tavazoie SF, Morris PG, Downey RJ, Manova-Todorova K, Brogi E, Massague J. Nat. Med. 2011;17:867. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marquart FX, Pasco S, Ramont L, Hornebeck W, Monboisse JC. Crit. Rev. Oncol. Hematol. 2004;49:199. doi: 10.1016/j.critrevonc.2003.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Adair-Kirk TL, Senior RM. Int. J. Biochem. Cell Biol. 2008;40:1101. doi: 10.1016/j.biocel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newton K, Dixit VM. Cold Spring Harb. Perspect. Biol. 2012;4(3) doi: 10.1101/cshperspect.a006049. pii: a006049, doi: 10.1101/cshperspect.a00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Carcinogenesis. 2009;30:1073. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 16.Andjilani M, Droz JP, Benahmed M, Tabone E. Int. J. Cancer. 2005;117:68. doi: 10.1002/ijc.21144. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Yao Y, Gong C, Yu F, Su S, Chen J, Liu B, Deng H, Wang F, Lin L, Yao H, Su F, Anderson KS, Liu Q, Ewan ME, Yao X, Song E. Cancer Cell. 2011;19:541. doi: 10.1016/j.ccr.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Multhoff G, Molls M, Radons J. Front. Immunol. 2012;2:98. doi: 10.3389/fimmu.2011.00098. doi:10.3389/fimmu.2011.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aggarwal BB, Gehlot P. Curr. Opin. Pharmacol. 2009;9:351. doi: 10.1016/j.coph.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheu BC, Chang WC, Cheng CY, Lin HH, Chang DY, Huang SC. Front. Biosci. 2008;13:6255. doi: 10.2741/3152. [DOI] [PubMed] [Google Scholar]
- 21.Lin Y, Bai L, Chen W, Xu S. Expert Opin. Ther. Targets. 2010;14:45. doi: 10.1517/14728220903431069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karin M. Nature. 2006;441:431. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 23.Austin BA, Liu B, Li Z, Nussenblatt RB. Invest. Ophthalmol. Vis. Sci. 2009;50:2896. doi: 10.1167/iovs.08-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beezhold DH, Personius C. J. Leukoc. Biol. 1992;51:59. doi: 10.1002/jlb.51.1.59. [DOI] [PubMed] [Google Scholar]
- 25.Loeser RF, Forsyth CB, Samarel AM, Im HJ. J. Biol. Chem. 2003;278:24577. doi: 10.1074/jbc.M304530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Debret R, Le Naour RR, Sallenave JM, Deshorgue A, Hornebeck WG, Guenounou M, Bernard P, Antonicelli FD. J. Invest. Dermatol. 2006;126:1860. doi: 10.1038/sj.jid.5700337. [DOI] [PubMed] [Google Scholar]
- 27.Adair-Kirk TL, Atkinson JJ, Kelley DG, Arch RH, Miner JH, Senior RM. J. Immunol. 2005;174:1621. doi: 10.4049/jimmunol.174.3.1621. [DOI] [PubMed] [Google Scholar]
- 28.Jensen HK, Donskov F, Marcussen N, Nordsmark M, Lundbeck F, von der Masse H. J. Clin. Oncol. 2009;27:4709. doi: 10.1200/JCO.2008.18.9498. [DOI] [PubMed] [Google Scholar]
- 29.Pham CTM. Nat. Rev. Immunol. 2006;6:541. doi: 10.1038/nri1841. [DOI] [PubMed] [Google Scholar]
- 30.Sparmann A, Bar-Sagi D. Cancer Cell. 2004;6:447. doi: 10.1016/j.ccr.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 31.Pekarek LA, Starr BA, Toledano AY, Schreiber H. J. Exp. Med. 1995;181:435. doi: 10.1084/jem.181.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tazawa H, Okada F, Kobayashi T, Tada M, Mori Y, Une Y, Sendo F, Kobayshi M, Hosokawa M. Am. J. Pathol. 2003;163:2221. doi: 10.1016/S0002-9440(10)63580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B, Vincent A, Cates J, Brantley-Sieders DM, Polk DB, Young PP. Cancer Res. 2009;69:338. doi: 10.1158/0008-5472.CAN-08-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung YD, Liu W, Reinmuth N, Ahmad SA, Fan F, Gallick GE, Ellis LM. Angiogenesis. 2001;4:155. doi: 10.1023/a:1012291524723. [DOI] [PubMed] [Google Scholar]
- 35.Salven P, Hattori K, Heissig B, Rafii S. FASEB J. 2002;16:1471. doi: 10.1096/fj.02-0134fje. [DOI] [PubMed] [Google Scholar]
- 36.Akagi Y, Liu W, Xie K, Zebrowski B, Shaheen RM, Ellis LM. Brit. J. Cancer. 1999;80:1506. doi: 10.1038/sj.bjc.6690553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song X, Voronov E, Dvorkin T, Fima E, Cagnano E, Benharroch D, Shendler Y, Bjorkdahl O, Segal S, Dinarello CA, Apte RN. J. Immunol. 2003;171:6448. doi: 10.4049/jimmunol.171.12.6448. [DOI] [PubMed] [Google Scholar]
- 38.Elaraj DM, Weinreich DM, Varghese S, Puhlmann M, Hewitt SM, Carroll NM, Feldman ED, Turner EM, Alexander HR. Clin. Cancer Res. 2006;12:1088. doi: 10.1158/1078-0432.CCR-05-1603. [DOI] [PubMed] [Google Scholar]
- 39.Killian PH, Kronski E, Michalik KM, Barbieri O, Astigiano S, Sommerhoff CP, Pfeffer U, Nerlich AG, Bachmeier BE. Carcinogenesis. 2012;33:2507. doi: 10.1093/carcin/bgs312. [DOI] [PubMed] [Google Scholar]
- 40.Bachmeier BE, Mohrenz IV, Mirisola V, Schleicher E, Romeo F, Hohneke C, Jochum M, Nerlich AG, Pfeffer U. Carcinogenesis. 2008;29:779. doi: 10.1093/carcin/bgm248. [DOI] [PubMed] [Google Scholar]
- 41.Mott JD, Werb Z. Curr. Opin. Cell Biol. 2004;16:558. doi: 10.1016/j.ceb.2004.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cox TR, Bird D, Baker AM, Barker HE, Ho MW, Lang G, Erler JT. Cancer Res. [Epub ahead of print], Mar. 2013:6. doi: 10.1158/0008-5472.CAN-12-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erler JT, Weaver VM. Clin. Exp. Metast. 2009;26:35. doi: 10.1007/s10585-008-9209-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lazarus HM, Cruikshank WW, Narasimhan N, Kagan HM, Center DM. Matrix Biol. 1995;14:727. doi: 10.1016/s0945-053x(05)80015-0. [DOI] [PubMed] [Google Scholar]
- 45.Cexus ONF, Labrousse D, Luciani A, Elliott T, Maiuri L, Quaratina S. J. Immunol. 2009;182(48):12. (abstract) [Google Scholar]
- 46.Trackman PC, Graham RJ, Bittner HK, Carnes DL, Gilles JA, Graves DT. Histochem. Cell Biol. 1998;110:9. doi: 10.1007/s004180050259. [DOI] [PubMed] [Google Scholar]
- 47.Xiao Q, Ge G. Cancer Microenviron. 2012 doi: 10.1007/s12307-012-0105-z. April 13 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Cancer Cell. 2005;8:241. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 49.Erler JT, Bennewith KL, Cox TR, Lang G, Bird D, Koong A, Le Q-T, Giaccia AJ. Cancer Cell. 2009;15:35. doi: 10.1016/j.ccr.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mammoto A, Mammoto T, Kanapathipillai M, Yung CW, Jiang E, Jiang A, Lofgren K, Gee EPS, Ingber DE. Nat. Commun. 2013;4:1759. doi: 10.1038/ncomms2774. doi: 10.1038/ncomms2774. [DOI] [PubMed] [Google Scholar]
- 51.Ahmed A, Redmond HP, Wang JH. Oncoimmunology. 2013;2:e22945. doi: 10.4161/onci.22945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oblak A, Jerala R. Clin Dev Immunol. 2011;2011:609579. doi: 10.1155/2011/609579. doi: 10.1155/2011/609579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weng H, Deng Y, Xie Y, Liu H, Gong F. BMC Cancer. 2013;13:311. doi: 10.1186/1471-2407-13-311. doi:10.1186/1471-2407-13-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiui MC, Ullrich E, Saulnier P, Yang H, Amigorena S, Ryffel B, Barrat FJ, Saftig P, Levi F, Lidereau R, Nogues C, Mira JP, Chompret A, Joulin V, Clavel-Chapelon F, Bourhis J, Andre F, Delaloge S, Tursz T, Kroemer G, Zitvogel L. Nat. Med. 2007;13:1050. [Google Scholar]
- 55.Zhang Y, Wang Y, Yuan J, Qin W, Liu F, Wang F, Zhang G, Yang X. Cell Biol. Toxicol. 2012;28:269. doi: 10.1007/s10565-012-9221-2. [DOI] [PubMed] [Google Scholar]
- 56.Rajput S, Volk-Draper LD, Ran S. Mol. Cancer Ther. 2013;12:1676. doi: 10.1158/1535-7163.MCT-12-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang B, Zhao J, Li H, He KL, Chen Y, Mayer L, Unkeless JC, Xiong H. Cancer Res. 2005;65:5009. doi: 10.1158/0008-5472.CAN-05-0784. [DOI] [PubMed] [Google Scholar]
- 58.Srikrishna G, Freeze HH. Neoplasia. 2009;11:615. doi: 10.1593/neo.09284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen GY, Nunez G. Nat. Rev. Immunol. 2010;10:826. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miyake K. J. Endotoxin Res. 2006;12:195. doi: 10.1179/096805106X118807. [DOI] [PubMed] [Google Scholar]
- 61.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. Nature. 2009;458:1191. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 62.Suzuki N, Suzuki S, Duncan GS, Millar DG, Wada T, Mirtsos C, Takada H, Wakeham A, Itie A, Li S, Penninger JM, Wesche H, Ohashi PS, Mak TW, Yeh W-C. Nature. 2002;416:750. doi: 10.1038/nature736. [DOI] [PubMed] [Google Scholar]
- 63.Lu YC, Yeh W-C, Ohashi PS. Cytokine. 2008;42:145. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 64.Takeda K, Akira S. Semin. Immunol. 2003;16:3. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 65.Covert MW, Leung TH, Gaston JE, Baltimore D. Science. 2005;309:1854. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
- 66.Piccinini AM, Midwood KS. Mediators of Inflamm. 2010;2010 doi: 10.1155/2010/672395. article ID 672395, Epub 2010 Jul 13, 21 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boivin WA, Shackleford M, Vanden Hoeck A, Zhao H, Hackett TL, Knight DA, Granville DJ. PLoS ONE. 2012;7:e33163. doi: 10.1371/journal.pone.0033163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gu X, Ma Y, Xiao J, Zheng H, Song C, Gong Y, Xing X. Clin. Exp. Med. 2012;12:195. doi: 10.1007/s10238-011-0155-4. [DOI] [PubMed] [Google Scholar]
- 69.Nastase MV, Young MF, Schaefer L. J. Histochem. Cytochem. 2012;60:963. doi: 10.1369/0022155412456380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schaefer L, Babelova A, Kiss E, Hausser H-J, Baliova M, Crzyzankova M, Marsche G, Young MF, Mihalik D, Gotte M, Malle E, Schaefer RM, Grone H-J. J. Clin. Invest. 2005;115:2223. doi: 10.1172/JCI23755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moreth K, Brodbeck R, Babelova A, Gretz N, Spieker T, Zeng-Brouwers J, Pfeilschifter J, Young MF, Schaefer RM, Schaefer L. J. Clin. Invest. 2010;120:4251. doi: 10.1172/JCI42213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moreth K, Iozzo RV, Schaefer L. Cell Cycle. 2012;11:2084. doi: 10.4161/cc.20316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Westergren-Thorsson G, Hernnas J, Sarnstrand B, Oldberg A, Heinegard D, Malmstrom A. J. Clin. Invest. 2003;92:632. doi: 10.1172/JCI116631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Kluijver J, Schrumpf JA, Evertse CE, Sont JK, Roughley PJ, Rabe KF, Hiemstra PF, Mauad T, Sterk PJ. Clin. Exp. Allergy. 2005;35:1361. doi: 10.1111/j.1365-2222.2005.02334.x. [DOI] [PubMed] [Google Scholar]
- 75.Merline R, Moreth K, Beckmann J, Nastase MV, Zeng-Brouwers J, Tralhao JG, Lemarchand P, Pfeilschifter J, Schaefer RM, Iozzo RV, Schaefer L. Sci. Signal. 2011;4(199):ra75. doi: 10.1126/scisignal.2001868. doi:10.1126/scisignal.2001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goldoni S, Seidler DG, Heath J, Fassan M, Baffa R, Thakur ML, Owens RT, McQuillan DJ, Iozzo RV. Am. J. Pathol. 2008;173:844. doi: 10.2353/ajpath.2008.080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Iozzo RV, Sanderson RD. J. Cell Mol. Med. 2011;15:1013. doi: 10.1111/j.1582-4934.2010.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seidler DG, Goldoni S, Agnew C, Cardi C, Thakur ML, Owens RT, McQuillan DJ, Iozzo RV. J. Biol. Chem. 2006;281:28408. doi: 10.1074/jbc.M602853200. [DOI] [PubMed] [Google Scholar]
- 79.Ihrcke NS, Parker W, Reissner KJ, Platt JL. J. Cell Physiol. 1998;175:255. doi: 10.1002/(SICI)1097-4652(199806)175:3<255::AID-JCP3>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 80.Hulett MD, Freeman C, Hamdorf BJ, Baker RT, Harris MJ, Parish CR. Nat. Med. 1999;5:803. doi: 10.1038/10525. [DOI] [PubMed] [Google Scholar]
- 81.Döring G. Am. J. Respir. Crit. Care Med. 1994;150:S114–S117. doi: 10.1164/ajrccm/150.6_Pt_2.S114. [DOI] [PubMed] [Google Scholar]
- 82.Joensuu H, Anttonen A, Eriksson M, Makitaro R, Alfthan H, Kinnula V, Leppa S. Cancer Res. 2002;62:5210. [PubMed] [Google Scholar]
- 83.Seidel C, Sundan A, Hjorth M, Turesson I, Dahl IM, Abildgaard N, Waage A, Borset M. Blood. 2000;95:388. [PubMed] [Google Scholar]
- 84.Johnson GB, Brunn GJ, Kodaira Y, Platt JL. J. Immunol. 2002;168:5233. doi: 10.4049/jimmunol.168.10.5233. [DOI] [PubMed] [Google Scholar]
- 85.Brunn GJ, Bungum MK, Johnson GB, Platt JL. FASEB J. 2005;19:7872. doi: 10.1096/fj.04-3211fje. [DOI] [PubMed] [Google Scholar]
- 86.Akbarshahi H, Axelsson J, Said K, Malmstrom A, Fischer H, Andersson R. J. Transl. Med. 2011;9:219. doi: 10.1186/1479-5876-9-219. doi: 10.1186/1479-5876-9-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tan JX, Wang XY, Su XL, Li HY, Shi Y, Wang L, Ren GS. PLoS ONE. 2011;6:e22836. doi: 10.1371/journal.pone.0022836. doi: 10.1371/journal.pone.0022836. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 88.Siiskonen H, Poukka M, Tynnela-Korhonen K, Sironen R, Pasonen-Seppänen S. BMC Cancer. 2013;13:181. doi: 10.1186/1471-2407-13-181. doi: 10.11/86/1471-2407-13-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hill DR, Kessler SP, Rho HK, Cowman MK, de la Motte CA. J. Biol. Chem. 2012;287:30610. doi: 10.1074/jbc.M112.356238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Termeer C, Benedix F, Sleeman J, Fieber C, Voith U, Ahrens T, Miyake K, Freudenberg M, Galanos C, Simon JC. J. Exp. Med. 2002;195:99. doi: 10.1084/jem.20001858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Taylor KR, Trowbridge JM, Rudisill JA, Termeer CC, Simon JC, Gallo RL. J. Biol. Chem. 2004;279:17079. doi: 10.1074/jbc.M310859200. [DOI] [PubMed] [Google Scholar]
- 92.Voelcker V, Gebhardt C, Averbeck M, Saalbach A, Wolf V, Weih F, Sleeman J, Anderegg U, Simon J. Exp. Dermatol. 2008;17:100. doi: 10.1111/j.1600-0625.2007.00638.x. [DOI] [PubMed] [Google Scholar]
- 93.Taylor KR, Yamasaki K, Radek KA, DiNardo A, Goodarzi H, Golenbock D, Beutler B, Gallo RL. J. Biol. Chem. 2007;282:18265. doi: 10.1074/jbc.M606352200. [DOI] [PubMed] [Google Scholar]
- 94.Simpson-Haidaris PJ, Rybarczyk B. Ann. N. Y. Acad. Sci. 2006;936:406. [PubMed] [Google Scholar]
- 95.Bárdos H, Molnár P, Csécsei G, Adány R. Blood Coagul. Fibrinolysis. 1996;7:536. [PubMed] [Google Scholar]
- 96.Wojtukiewicz MZ, Zacharski LR, Memoli VA, Kisiel W, Kudryk BJ, Moritz TE, Rousseau SM, Stump DC. Cancer. 1991;67:1377. doi: 10.1002/1097-0142(19910301)67:5<1377::aid-cncr2820670517>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 97.Davalos D, Akassoglou K. Semin. Immunopathol. 2012;34:43. doi: 10.1007/s00281-011-0290-8. [DOI] [PubMed] [Google Scholar]
- 98.dos Santos Silva I, De Stavola BL, Pizzi C, Meade TW. Int. J. Epidemiol. 2010;39:699. doi: 10.1093/ije/dyq012. [DOI] [PubMed] [Google Scholar]
- 99.Smiley ST, King JA, Hancock WW. J. Immunol. 2001;167:2887. doi: 10.4049/jimmunol.167.5.2887. [DOI] [PubMed] [Google Scholar]
- 100.Hodgkinson CP, Patel K, Ye S. Thromb. Haemost. 2008;100:301. [PubMed] [Google Scholar]
- 101.Grahovac J, Becker D, Wells A. J. Invest. Dermatol. 2013;133:210. doi: 10.1038/jid.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goh FG, Piccinini AM, Krausgruber T, Udalova IA, Midwood KS. J. Immunol. 2010;184:2655. doi: 10.4049/jimmunol.0903359. [DOI] [PubMed] [Google Scholar]
- 103.Midwood KS, Hussenet T, Langlois B, Orend G. Cell Mol. Life Sci. 2011;68:3175. doi: 10.1007/s00018-011-0783-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Midwood K, Sacre S, Piccinini AM, Inglis J, Trebaul A, Chan E, Drexler S, Sofat N, Kashiwagi M, Orend G, Brennan F, Foxwelll B. Nature, Med. 2009;15:774. doi: 10.1038/nm.1987. [DOI] [PubMed] [Google Scholar]
- 105.Piccinini AM, Midwood KS. Cell Rep. 2012;2:914. doi: 10.1016/j.celrep.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ruoslahti E. Ann. Rev. Biochem. 1988;57:375. doi: 10.1146/annurev.bi.57.070188.002111. [DOI] [PubMed] [Google Scholar]
- 107.Kaczmarek J, Castellani P, Nicolo G, Spina B, Allemanni G, Zardi L. Int. J. Cancer. 1994;59:11. doi: 10.1002/ijc.2910590104. [DOI] [PubMed] [Google Scholar]
- 108.To WS, Midwood KS. Fibrogenesis Tissue Repair. 2011;4:21. doi: 10.1186/1755-1536-4-21. doi: 10.1186/1755-1536-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gao M, Craig D, Lequin O, Campbell ID, Vogel V, Schulten K. Proc. Natl. Acad. Sci. 2003;100:14784. doi: 10.1073/pnas.2334390100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Okamura Y, Watari M, Jerud ES, Young DW, Ishizaka ST, Rose J, Chow JC, Strauss JR., 3rd J. Biol. Chem. 2001;276:10229. doi: 10.1074/jbc.M100099200. [DOI] [PubMed] [Google Scholar]
- 111.Khan MM, Gandhi C, Chauhan N, Stevens JW, Motto DG, Lentz SR, Chauhan AK. Stroke. 2012;43:1376. doi: 10.1161/STROKEAHA.111.635516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mogami H, Kishore AH, Shi H, Keller PW, Akgul Y, Word RA. J. Biol. Chem. 2013;288:1953. doi: 10.1074/jbc.M112.424366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ou J, Deng J, Wei X, Xie G, Zhou R, Yu L, Liang H. Stem Cell Res. 2013;11:820. doi: 10.1016/j.scr.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rybak JN, Roesli C, Kaspar M, Villa A, Neri D. Cancer Res. 2007;67:10948. doi: 10.1158/0008-5472.CAN-07-1436. [DOI] [PubMed] [Google Scholar]
- 115.Morla A, Zhang Z, Ruoslahti E. Nature. 1994;367:193. doi: 10.1038/367193a0. [DOI] [PubMed] [Google Scholar]
- 116.Hocking DC, Sottile J, McKeown-Longo PJ. J. Biol. Chem. 1994;269:19183. [PubMed] [Google Scholar]
- 117.Zhong C, Chrzanowska-Wodnicka M, Brown J, Shaub A, Belkin AM, Burridge K. J. Cell Biol. 1998;141:539. doi: 10.1083/jcb.141.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.You R, Zheng M, McKeown-Longo PJ. J. Biol. Chem. 2010;285:36255. doi: 10.1074/jbc.C110.176990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Morla A, Ruoslahti E. J. Cell Biol. 1992;118:421. doi: 10.1083/jcb.118.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Briknarova K, Akerman ME, Hoyt DW, Ruoslahti E, Ely KR. J. Mol. Biol. 2003;332:205. doi: 10.1016/s0022-2836(03)00890-8. [DOI] [PubMed] [Google Scholar]
- 121.Doucet A, Overall CM. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.003533. M110.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]