Abstract
The discovery of cancer stem cells in glioma has created a paradigm shift in our understanding of this deadly disease. Glioma stem cells exhibit sustained self-renewal and potent tumorigenic potential and differ from their more differentiated progeny in response to current therapies. Recurrent disease is likely derived from glioma stem cells or progeny reprogrammed to gain stem cell-like phenotypes, indicating that the stem cell phenotype is a crucial therapeutic target. While debate over cancer stem cell and clonal evolution models persists, important knowledge has been gained over the past decade from glioma stem cells investigation and clinical impact is expected.
Keywords: Glioma, cancer stem cells
Introduction
Malignant gliomas constitute a heterogeneous group of highly infiltrative primary brain tumors with distinct histopathological and molecular features. Each year in the United States, approximately 15,750 individuals are diagnosed with a malignant glioma and an estimated 12,740 patients succumb to this disease (1). These statistics highlight the particularly lethal nature of malignant gliomas and important need for enhanced therapeutic efficacy. Current classifications of glioma are based upon the seminal work of Bailey and Cushing, who in the 1920s named and divided glial tumors according to a putative cell type of origin and stage of cellular development (2). Likewise, efforts to provide more effective therapies continue to be driven by the studies of glioma cells of origin and underlying mechanisms of cellular development and growth. Paramount to these efforts is an evolving understanding of the cellular heterogeneity within gliomas. Thus, while the predominant cell type within an astrocytoma or oligodendroglioma may resemble an astrocyte or oligodendrocyte, respectively, each type of glioma is composed of morphologically, phenotypically and genetically heterogeneous cells.
In this review, two seemingly though not necessarily competing views of glioma heterogeneity are discussed, the stochastic and cancer stem cell models. How recent studies of microenvironmental cues, developmental signaling pathways, and treatment resistance inform our views of glioma heterogeneity, growth and therapy will also be reviewed.
Hierarchical organization vs. clonal evolution
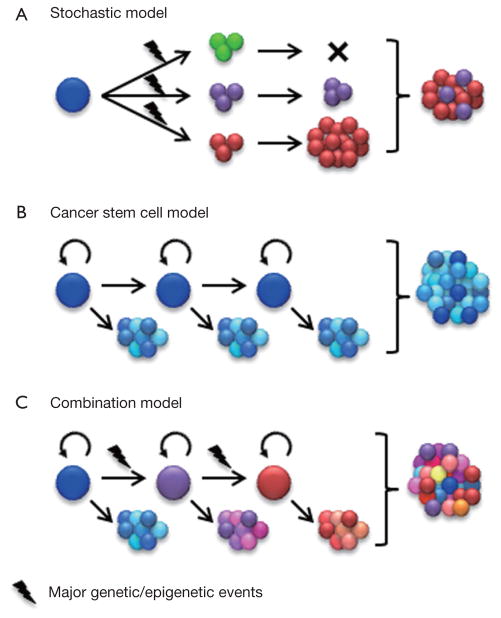
Although most cancers, including glioma, appear to be of monoclonal origin, at the time of diagnosis, tumors are composed of genetically and phenotypically heterogeneous clones (3–7). Intratumoral heterogeneity has traditionally been viewed according to a stochastic model outlined by Peter Nowell in 1976 (8). By this perspective, cancer heterogeneity and growth is an evolutionary process whereby neoplasms arise from a single cell of origin and tumor progression results from random accumulation of somatic mutations in genetically unstable cell populations with sequential selection of malignant subclones by environmental constraints (Figure 1A). In accordance with this model, it was proposed that each cancer may require individual-specific therapy due to the multitude of potentially random mutations that might drive tumor growth and emergence of treatment resistant subclones from neoplastic cells with roughly equal tumorigenic potential (8).
Figure 1.
Modeling cellular heterogeneity of cancer. (A) The stochastic model assumes that cancer cell phenotypes are primarily defined by intrinsic factors, in particular driver mutations. It indicates a clonal evolution of cancer. However, this model may not adequately address phenotypic variations within individual clones; (B) The cancer stem cell model assumes that cancer is organized in a hierarchical structure that, at least in part, resembles that of the tissue of origin. Tumorigenic potential is limited to the cancer stem cell subpopulation. In addition, cellular heterogeneity of the cancer is a product of multipotent cancer stem cells. However, the maintenance of coexisting genetically distinct clones in most late-stage cancers has not been adequately addressed by this model; (C) Emerging evidence suggests a combination of these two models in which cancers are driven by one or multiple dominating clones, some of which may be organized in a hierarchical manner. However, at the time of diagnosis, the original hierarchy may be altered due to acquisition of genetic or epigenetic events that promote tumorigenic capacity and impair differentiation.
Recent advances in genomic mapping have unambiguously demonstrated the complex genetic landscape in a wide variety of human tumors (9). Genetic heterogeneity in glioma was initially demonstrated by the presence of subclones with differing karyotype (10). More recent studies show that some glioblastoma tumors comprise subclones carrying amplifications of different receptor tyrosine kinases (RTKs, e.g., EGFR, MET, PDGRFA) in a mutually exclusive manner (11,12). The clonal heterogeneity RTK gene amplification in glioma suggests a mechanism for inherent resistance to agents that target only a single RTK. While clonal heterogeneity has been extensively documented in glioma and many other cancers, accumulating evidence suggests an additional level of functional heterogeneity based upon cellular differentiation.
Functional heterogeneity among cancer cells was documented decades ago by the demonstration that only a small subset of the cells within a tumor are capable of clonogenic growth in mice or soft agar (13,14). The cancer stem cell (CSC) model suggests a hierarchical organization of functional heterogeneity, with self-sustaining CSCs at the apex giving rise to heterogeneous transit-amplifying and differentiated cancer cell types (Figure 1B) (15). Two key outcomes of CSC divisions are differentiation into heterogeneous cancer cell types or self-renewal to sustain a cancer stem cell pool. Compelling evidence that the tumorigenic capacity may be restricted to cancer cells with stem cell phenotype was demonstrated by John Dick and colleagues in their seminal studies of human acute myelogenous leukemia (AML) (16). These landmark studies are supported in other tumor types by xenotransplantation assays showing that tumorigenic cells can be prospectively identified by selection for stem cell markers, whereas the remaining tumor cells are depleted of tumorigenic potential (15).
A fundamental implication of the hierarchical model is that CSCs sustain and fuel tumor growth and that their eradication is crucial to effective therapy. Many aspects of the CSC model remain intensely debated, such as the cell of origin, cell surface markers, and the relative frequencies of CSCs. Nevertheless, several recent lineage-tracing studies provide crucial support of a hierarchical structure in some human cancers, including malignant glioma. These studies demonstrate that the majority of tumor cells have limited proliferative potential and are derived from a subpopulation of cancer cells that exhibit stem cell-like features (17–20). Luis Parada and colleagues crossed one strain of mice genetically engineered to develop gliomas with another strain expressing green fluorescent protein (GFP) under the control of the neural stem cell marker Nestin. In the resulting gliomas, the fraction of tumor cells expressing GFP were relatively quiescent and fulfilled key stem cell features. More importantly, following treatment with temozolomide, the recurrent tumors were derived from GFP-positive cells. Selective depletion of the GFP-positive subpopulation extended survival of experimental animals and improved tumor response to temozolomide (20). Hans Clevers and coworkers identified a minor population of cells expressing the intestinal stem cell marker Lgr5 as the stem cell fraction of intestinal adenoma (18). A cellular hierarchy resembling the organization of normal epidermis has also been revealed by lineage tracing studies in skin papilloma and squamous cell carcinoma (19). Interestingly, the genetically marked CSC pool expanded in malignant squamous cell carcinoma in comparison to benign papilloma (19), suggesting that hierarchical architecture may not be fixed during tumor progression. These results may explain some of the controversy regarding differences in CSC frequencies measured for the same tumor types by different investigators.
While the clonal evolution model and the CSC model interpret intratumoral heterogeneity in different ways, it is important to note that they are not mutually exclusive. Major genetic events continuously accumulate in CSCs and their progenies, which progressively gives rise to genetically distinguished new clones. These new clones may or may not be organized in a hierarchical structure. Therefore, the diversity in reported phenotypes of CSCs may in part reflect complexities of cancer genomes. Two recent studies analyzed the genetic diversity of CSCs in acute lymphoblastic leukemia driven by either BCR-ABL (breakpoint cluster region protein-Abelson murine leukemia viral oncogene homolog 1) or ETV6-RUNX1 (Ets Variant 6-Runt-related transcription factor 1) fusion genes (21,22). In these relatively less complex blood cancers, CSCs exhibited significant genetic diversity that reflects a branching clonal evolution. Importantly, xenografts and recurrent tumors were not always derived from the dominant clones, suggesting that CSCs of minor subclones may also be of clinical significance (21,22). On the other hand, CSCs isolated from different tumor types may share common phenotypes irrespective of their tissue background. For example CD133 expression is associated with CSCs from brain tumors and a wide range of other malignancies (23). Although direct experimental evidence is still emerging, the cellular complexity of many human cancers is likely the result of a combination of clonal genetic events and hierarchical differentiation (Figure 1C). The existence of genetically and phenotypically distinct cell populations within an individual’s tumor represents significant challenges to develop effective targeted therapies. Genetic diversity needs to be taken into account as we gain a better understanding of mechanisms commonly implicated in tumorigenic potential, therapeutic resistance, and other crucial phenotypes of CSCs.
Identity of glioma stem cells
A major advance in study of CSC is the ability to identify them among the rest of the tumor cells by phenotypic markers. Glioma stem cells (GSCs) were first identified by selection for the neural and hematopoietic stem cell marker, CD133 (prominin) (24,25). In these seminal studies, 100 CD133+ glioma cells were sufficient to develop xenografted tumors that recapitulated the heterogeneity of the original tumor, while CD133− tumor cells were effectively depleted for tumorigenic potential. Subsequent studies have either substantiated or challenged the specificity of CD133 as the GSC marker. Tumorigenic CD133− cells have been identified in different samples or even within the same tumors that contain CD133+ GSCs (26–29). These controversial observations can only be partially explained by a lack of technical consensus. Alternatively, the role of CD133 as the stem cell marker may vary in different molecular subtypes of glioma. Multiple groups have shown that gliomas driven by CD133+ cells exhibit transcription profiles resembling the proneural subtype, whereas CD133− GSCs may be associated with gliomas of the mesenchymal subtype (30–32). Further, Heidi Philips and colleagues have provided evidence for a more complex hierarchy driven by CD133− GSCs, which give rise to CD133+ intermediate progenitors and then CD133− differentiated progenies (33). In addition to CD133, a variety of other markers have been described, such as CD15, L1CAM, integrin α6, and A2B5 (28,34–36). Robust methods that can reproducibly identify and enrich for GSCs are of paramount importance to the field. However, it is conceivable that genetically and phenotypically diverse GSCs cannot be encompassed by a universal marker. With advances in high-throughput technologies for genomics and epigenomics, markers selective for subtypes of GSCs may be anticipated.
The crosstalk between glioma stem cells and their niche
Stem cell number and growth rate are tightly regulated by microenvironmental cues (a.k.a. niche). Like their normal counterparts, CSCs are affected by microenvironment factors. Therefore, disrupting the crosstalk between CSCs and their niches appear to be attractive therapeutic strategies. At least two types of niches have been identified for GSCs. Each is associated with a pathological hallmark of this disease, namely aberrant vascular proliferation and hypoxia-associated necrosis (37). Glioma is a highly angiogenic tumor, and GSCs are enriched in perivascular regions (38), where a variety of regional signals have been found to promote CSC phenotypes. Endothelial cells express Notch ligands, such as jagged1 (JAG1) and delta-like ligand 4 (DLL4) that activate Notch signaling in GSCs residing in perivascular region (39). The perivascular region is also enriched for extracellular matrix proteins that are capable of promoting proliferation, survival and migration of GSCs. For example, integrin α6 is highly expressed in perivascular GSCs and possibly functions as the receptor of laminin in the perivascular niche (35,40). GSCs are not passive residents of their niche, rather these cells play active roles in shaping tumor vasculature. GSCs produce high levels of pro-angiogenic factors, such as vascular endothelia growth factor (VEGF) (41). GSCs also produce differentiated progenies that exhibit features of endothelial cells and contribute to formation of cancer-specific vasculature (42–44). Similar observations have been made in other cancers, such as melanoma (45). More recently, Bao and colleagues demonstrate that GSCs are also a source of vascular pericytes (46). While it has been well documented that malignant cells are actively involved in cancer vasculature (a.k.a. vasculogenic mimicry) (47), these emerging results suggest that CSCs are key regulators of this process. Blood vessels formed by cancer cells may be pathologically important when proliferation of normal endothelial cells is not sufficient to sustain tumor growth or suppressed by factors such as anti-angiogenic therapy. The VEGF neutralizing antibody, bevacizumab, mitigates many symptoms in patients with recurrent glioblastoma (48). However, treatment with bevacizumab fails to improve patient survival. The ability of GSCs to generate functional blood vessels is one potential hurdle to anti-angiogenic therapy and suggests that it may need to be combined with other treatments that block the transdifferentiation of GSCs.
Malignant glioma, and in particular glioblastoma, is associated with widespread hypoxia (49). Necrosis is a histologic hallmark of glioblastoma and predictor of poor prognosis (50), suggesting that necrosis may promote tumor progression and therapeutic resistance. The necrotic regions of glioblastoma are characterized by severe hypoxia. Low oxygen tension has been shown to promote self-renewal for various types of normal stem cells (51). Emerging evidence further suggests that hypoxia promotes stem cell-like phenotypes in glioma, thus hypoxic regions function as an important niche factor for GSCs (52). The hypoxia inducible factors (HIF) family of transcription factors is a central regulator of tumor response to hypoxia. In particular, HIF2α appears to be a key player in maintenance of stem cells in glioma (53,54). GSCs express higher levels of HIF2α in comparison to non-stem glioma cells and normal neural progenitors (54). Knockdown of HIF2α specifically compromise proliferation and survival of GSCs, while HIF1α may have important functions in non-stem glioma cells as well (54). Prolonged hypoxic stresses not only stimulate expansion of CD133+ GSCs but also exhibit the potential to reprogram non-stem glioma cells to a CSC-like phenotype (55,56). However, these results should not be over-interpreted, as HIF may directly activate expression of CD133 (57,58). The ability of GSCs to expand under hypoxic condition imposes additional challenges to anti-angiogenic therapy, as reduction in vascular supply may have limited impact on these cells. In fact, compromised tumor vasculature may mobilize GSCs, as a number of anti-angiogenic therapies have been found to stimulate cancer invasion and metastasis (59,60). The plasticity of GSCs to co-opt drastically different tumor microenvironments, again, suggests that combinatorial approaches will be required to effectively disrupt their putative niches.
GSCs and pathways that direct cell fate determination
Vogelstein and colleagues analyzed the seemingly innumerable genetic alterations identified by comprehensive genomic studies in human cancer to identify those that significantly promote or “drive” tumorigenesis. Through this effort, they distilled driver genes into 12 signaling pathways that regulate core cellular processes of cell fate determination, proliferation, survival, and genome maintenance. Key among the signaling pathways regulating cell fate are Notch, Hedgehog, and Wnt (61). These three pathways are instrumental in embryonic development and adult tissue homeostasis. Aberrant activation of these pathways promotes stem cell-like phenotypes in cancer and dampens CSC differentiation (62). Although components of these three pathways are not frequently mutated in glioma, they appear to be crucial GSCs niche factors and thus attractive therapeutic targets.
Notch signaling pathway
The Notch signaling pathway is an evolutionarily conserved mechanism that regulates cell fate determination across many tissue types (63). Notch receptors are activated by ligands expressed on the surface of neighboring cells. The intracellular domains of activated Notch receptors are proteolytically released by the γ-secretase complex, translocate into nucleus, and subsequently activate transcription of Notch-responsive target genes. During embryonic development, Notch signaling plays critical roles to maintain neural stem cell proliferation, survival and self-renewal (64). In contrast, EGFR activation leads to expansion of progenitor cells (65). Crosstalk between the Notch and EGFR pathways regulates a crucial balance between neural stem and progenitor cells. Disruption of Notch signaling in the embryonic mouse brain by knockout of the DNA binding subunit RBPJ/CBF1 (recombining binding protein suppressor of hairless) or the catalytic subunit of γ-secretase (presenilin-1) promotes premature neuronal differentiation with profound consequences on neural development (66,67). Notch signaling also regulates stem cells in adult tissue homeostasis, including the brain (68), and aberrant Notch signaling has been widely implicated in cancer.
The oncogenic function of Notch pathway activation is best exemplified by the presence of activating mutations in Notch1 in more than half of human T-cell acute lymphoblastic leukemias (69). While genetic alterations of the Notch pathway are rare in glioma, Notch signaling can be activated by a variety of microenvironmental cues. In xenotransplantation assays, the addition of human brain microvascular endothelial cells improves the tumorigenic potential of glioblastoma sphere cells, and the effect is abolished upon knockdown of Notch ligands, JAG1 or DLL4, in endothelial cells (39). In colorectal cancer, endothelial cells produce a soluble form of JAG1 that promotes CSC phenotypes (70). In addition to the expression of Notch ligands, endothelial cells also produce nitric oxide that induces Notch activation in perivascular glioma cells (71). Blockade of Notch signaling by γ-secretase inhibitors (GSIs) reduces proliferation, neurosphere formation, and tumorigenic potential of GSCs (72). In addition to these effects on GSC proliferation, it is worth noting that Notch inhibition compromises the ability of GSCs to resist radiation as well as temozolomide (73,74).
Multiple GSIs have entered clinical trials. The first clinical evidence for potential efficacy came from a recent phase I clinical trial of a Merck GSI, MK-0752, in patients with advanced solid tumors that included glioma. Stable disease was reported in about 24% of patients with advanced glioma in addition to a complete response in one patient with a grade III glioma (75). Of particular interest in this trial, clinical response of MK-0752 was predominantly found in adult patients with glioma. However, the efficacy of MK-0752 in refractory pediatric brain cancers was not significant (76). Another GSI, RO4929097 (Roche), also exhibited moderate clinical efficacy in a phase I trial, though the trial was not focused on glioma (77). Trials combining GSIs with other therapeutic modality are currently ongoing in glioblastoma and other cancers (e.g., clinicaltrials.gov identifier-NCT01119599 and NCT01098344).
Wnt signaling pathway
The Wnt family of secreted signaling proteins and their receptors have important functions in embryonic development, particularly in tissue patterning (78). The central player of the canonical Wnt signaling is the cytoplasmic protein β-catenin. In the absence of Wnt pathway stimulation, β-catenin is constitutively degraded by a destruction complex and Wnt target gene expression is repressed by DNA-bound TCF/LEF (T-cell factor and lymphoid enhancing factor) transcription factors. Binding of Wnt ligands to the Frizzled family of receptors inhibits kinase activity of the destruction complex, leading to stabilization and nuclear translocation of β-catenin. Nuclear β-catenin converts TCF/LEF into a transcription activator of Wnt target genes that induce downstream signaling (79). The function of the Wnt pathway in tumorigenesis is best documented in colorectal cancer. Mutational inactivation of Adenomatous polyposis coli (APC), a scaffolding protein of the destruction complex, plays a key role in development of hereditary and sporadic colorectal cancer (80,81). Lgr5, a Wnt receptor, is preferentially expressed in both normal and malignant stem cells of colon and intestine (18,82). Though the roles of Wnt signaling in glioma are less understood, β-catenin is essential for neuronal progenitor cell proliferation (83) and the expression of Wnt1, β-catenin, and the downstream target cyclin D1 has been demonstrated in a considerable percentage of gliomas and correlate with increasing World Health Organization tumor grade (84). PLAGL2 (pleomorphic adenoma gene-like 2) is a recently identified proto-oncogene that is amplified in glioma and promotes proliferation and self-renewal of GSCs (85). The ability of PLAGL2 to regulate “stemness” in glioma and normal neural tissues is attributable in part to activation of the Wnt pathway (85). Like PLAGL2, additional transcription factors overexpressed in glioblastoma, such as Forkhead box protein M1 (FOXM1) and Achaete-scute homolog 1 (ASCL1), also cooperate with the Wnt/β-catenin pathway to regulate stemness of GSCs (86,87). However, controversial results have been reported. In primary glioblastoma cells, activation of Wnt signaling promotes neuronal differentiation and compromises malignant phenotypes of CD133+ glioblastoma cells, particularly in a hypoxic microenvironment (88). These effects appear to be mediated, at least in part, via antagonism of Notch signaling (88). The different roles of Wnt signaling in glioma may reflect the genetic heterogeneity of this disease. A recent study showed that the Wnt pathway is a downstream target of MET in glioblastoma stem cells (89). MET is a tyrosine receptor kinase frequently amplified in glioblastoma and associated with the mesenchymal subtype (90,91). Therefore, the functions of Wnt signaling may vary in different molecular subtype of glioma, which demands further investigation.
Hedgehog signaling pathway
Hedgehog secreted signaling proteins are critical for embryonic tissue development (patterning) and postnatal tissue homeostasis (92,93). Aberrant activation of the Hedgehog pathway has been implicated in the growth of many malignancies in a role that is largely attributed to action of the pathway on stem or progenitor cells (94). Cellular responses to Hedgehog signaling are regulated through the primary cilium by the transmembrane proteins Patched-1 (PTCH1) and Smoothened (SMOH) (95). PTCH1 functions to suppress the activity of Smoothened. Hedgehog ligand binding to PTCH1 inhibits this function to activate the GLI family of transcription factors. PTCH1 and GLI1 are transcriptional gene targets of Hedgehog signaling, and in the proper context their expression levels can be used to monitor Hedgehog pathway activity in malignancies.
A role for aberrant Hedgehog signaling in tumorigenesis was first appreciated by the series of discoveries that mutations in the Hedgehog signal transduction components PTCH1 and SMOH may confer ligand-independent pathway activation in heritable (Gorlin or basal cell nevus syndrome) and sporadic forms of medulloblastoma and basal cell carcinoma (BCC) (96–99). Shortly afterwards, studies of an acquired form of cyclopia (100,101) identified the teratogen cyclopamine as a potent inhibitor of Hedgehog signal reception through direct binding and antagonism of SMOH (102–104). Cyclopamine is a plant-derived alkaloid and several synthetic SMOH antagonists have since been identified that appear to bind the same sight as cyclopamine but with enhanced efficacy for inhibiting SMOH bearing oncogenic mutations (105). Some of the SMOH antagonists have progressed into clinical trial and one, vismodegib (GDC-0449; Genentech), has received approval by the FDA for treatment of adults with metastatic BCC or locally advanced disease who are not candidates for surgery or radiation (106). Enthusiasm for dramatic initial response to GDC-0449 in a patient with metastatic medulloblastoma was dampened by the emergence of treatment resistance with disease relapse (107). Gene sequencing of the recurrent disease, however, identified acquisition of a SMOH missense mutation that decreased GDC-0449 binding affinity (108), demonstrating the critical importance of Hedgehog pathway activation for tumor growth and offering hope for the efficacy of other mechanistically diverse Hedgehog inhibitors.
In contrast to medulloblastoma and BCC in which the Hedgehog pathway is constitutively activated by pathway component mutation, ligand-dependent activation of the Hedgehog pathway in the absence of mutation has been identified in a broader array of malignancies (109). In these tumors, the Hedgehog pathway appears to be activated in a small population of cells that have been proposed to have stem or progenitor-like features (92). Although the Hedgehog transcription factor GLI1 was first discovered (and named) as a gene that was amplified in a glioblastoma cell line (110), GLI1 gene amplification or other genomic alterations in Hedgehog pathway components are generally absent in gliomas (91,111). Instead, the Hedgehog pathway is activated by a ligand-dependent mechanism in gliomas (112–114). Activation of the pathway in GSCs regulates tumor growth and inhibition of the pathway in preclinical animal models confers a significant survival advantage (112,113,115).
In contrast to Wnt signaling, where pathway component expression levels correlate with tumor grade, Hedgehog component and gene target expression is higher among grades II and III gliomas than in grade IV gliomas (113,114). Further, the Hedgehog pathway is not operant in all malignant gliomas (114,115) and thus the clinical utility of targeting this pathway could be enhanced by clear identification of Hedgehog-responsive glioma subtypes. Somatic mutations in the isocitrate dehydrogenase (IDH) gene have recently emerged as a surrogate marker for identifying gliomas with an operant Hedgehog pathway (116). In adult gliomas, IDH mutations occur in more than 70% of diffuse astrocytomas, oligodendrogliomas, oligoastrocytomas and secondary glioblastomas that evolve from lower grade astrocytomas (117,118). Conversely, IDH mutation occurs in less than 7% of primary glioblastomas, which occur without evidence or antecedent disease and represent greater than 95% of glioblastomas. Increasing evidence suggests that IDH mutation is an early genetic alteration in a common cell of origin for astrocytic or oligodendroglial tumors that is distinct from the cellular origin for primary glioblastoma (119,120). The Hedgehog pathway is frequently activated in secondary glioblastoma and lower-grade gliomas carrying IDH mutations. Taken together, these observations suggest an interesting model whereby lower grade infiltrating gliomas and secondary glioblastoma arise from Hedgehogdependent cell types and primary glioblastoma from cell types that are not Hedgehog responsive (116).
Glioma stem cells and resistance to radiation
Adult gliomas are highly infiltrative and cannot be completely removed by surgery. Radiation and temozolomide are the primary adjuvant therapies for malignant gliomas. Response to chemoradiotherapy in malignant glioma is generally short-lived, and the almost universal recurrence suggests inadequate eradication of tumorigenic cells (121). Identification of CSCs in glioma provides fresh mechanistic insights into intrinsic resistance to radiation and chemotherapy. GSCs appear to substantially differ in response to radiation in comparison to differentiated cancer cells, although some controversy persists (122). The percentage of CD133+ cells within malignant gliomas markedly increases following radiotherapy (123). CD133+ cells are also enriched in glioma tumorsphere cultures and orthotopic tumors following radiation, potentially due an inherent capacity of GSCs to more effectively repair radiation-induced DNA damage (122). In addition, the CD133+ subpopulation is preferentially protected in a physiologically relevant microenvironment, associated with fewer phosphorylated histone H2AX (γH2AX) and TP53 binding protein 1 (53BP1) loci than CD133− cells within the same tumor (124). Ropolo and colleagues found no difference in DNA base excision, single-strand break repair, or γH2AX foci resolution between patient-derived glioblastoma spheroid cultures and differentiated cells or established cell lines grown in serum-containing medium (125). However, enhanced basal activation of DNA damage checkpoint kinases Chk1 and Chk2 in CD133+ cells may protect these cells against radiation (125). In addition to DNA damage repair mechanisms, a variety of signaling pathways that are preferentially activated in GSCs may protect these cells from radiation-induced toxicity. Ionizing radiation activates Notch in GSCs, and Notch signaling confers protection from radiation via an Akt-mediated mechanism (73). Wnt and MET also exhibits radioprotective functions in GSCs (126,127) as well as breast CSCs (128). Conflicting results have been reported comparing the radiation response between GSCs and non-stem glioma cells. For example, McCord and colleagues observed that CD133+ GSCs derived from surgical specimens were more radiosensitive than established glioma cell lines based on clonogenic assays (129). The CD133+ primary cultures actually exhibited a defective DNA damage checkpoint (129). Pallini and colleagues found significantly higher percentage of CD133+ cells in recurrent specimens based on examination of 37 paired glioblastoma samples (130). Interestingly, the percentage of CD133+ cells correlated with longer survival after tumor recurrence. Further studies suggested that a significant portion of the CD133+ cells in recurrent glioblastoma specimens were normal neural stem cells with potential antineoplastic activity (130). Discrepancies with regard to the radiation responses of GSCs may reflect technical difficulties of identifying GSCs, thus highlighting significant challenges in this field of study. Genetic heterogeneity in samples generates additional difficulties in interpreting these results. Lineage tracing assays provide an alternative strategy to interrogate therapeutic response of specific cellular subpopulations.
Conclusions and outlooks
While not all cancers may contain hierarchical organization, the existence of CSCs in glioma has been extensively documented and validated by rigorous measures including serial transplantation and in vivo lineage tracing assays (20,25). Introduction of the CSC concept into brain cancer research has led to a paradigm shift and significant advances in the field. For example, it has been convincingly demonstrated that the genomic integrity and cellular heterogeneity of patient tumors cannot be maintained in widely used established glioma cell lines (131). In contrast, culture conditions designed for normal neural progenitors and GSCs preserve the phenotypes and genotypes of patient tumors and thus represent a more physiologically relevant in vitro model (132). The ability of GSCs to repopulate the original tumor following treatment and their inherent potentials for conferring treatment resistance indicate that GSCs are crucial targets for novel therapeutics. Over the past decade, a rapidly growing list of novel targets has been identified by interrogating the biology of GSCs, including the developmental signaling pathways, Notch, Wnt and Hedgehog. Other targets in GSCs have emerged, such as the epigenetic regulators-EZH2 (133,134), kinases-bone marrow X-linked (BMX) and maternal embryonic leucine-zipper kinase (MELK) (135,136), and transcription factors like STAT3, REST, and MYC (137–139). Notably, and potentially introducing another layer of complexity, in the majority of these studies, the glioma molecular subtypes are not determined, and thus the roles of these novel targets among gliomas of different molecular subtypes remain unclear. Future studies of GSC-targeted therapy will need to establish links with glioma molecular subtypes in order to design more selective and effective clinical trials.
Lastly, it is important to note that while GSCs represent crucial therapeutic targets, differentiated glioma cells are not merely bystanders. Although there is still a lack of direct experimental evidence, the hierarchical structure of glioma may not be as strict as that of normal tissues. Both mature astrocytes and neurons can be genetically reprogramed to confer stem and glioma-initiating properties (140–142). Reprogramming might also be induced by environmental factors, such as low pH, hypoxia and even stem cell culture condition (56,143,144). Dedifferentiation may even be accelerated by treatments that change the microenvironment and increase mutations rates.
Recent advances in the study of the cancer genome and epigenome are rapidly transforming research in the field of neuro-oncology. Studies of GSCs in malignant gliomas with defined genetic backgrounds will likely offer greater success in identifying important drug targets that are tailored for individual glioma subtypes. In addition, targeting the microenvironment may more broadly impact GSCs irrespectively of genetic background and possibly reduce the rate of dedifferentiation. To considerably improve outcomes of clinical trials, combinations of multiple therapeutic modalities that target GSCs as well as their microenvironment appear to be essential.
Acknowledgments
Disclosure: This work is supported in part by the National Institutes of Health grant 1R01CA166492 (to J.W.) and the Department of Veterans Affairs grant 1I01BX000744-01 (to M. K. C.). The contents do not represent the views of the Department of Veterans Affairs or the United States Government.
References
- 1.Dolecek TA, Propp JM, Stroup NE, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. 2012;14:v1–49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey P, Cushing H. A classification of the tumors of the glioma group on a histogenetic basis with a correlated study of prognosis. Philadelphia: Lippincott; 1926. [Google Scholar]
- 3.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell PJ, Pleasance ED, Stephens PJ, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci U S A. 2008;105:13081–6. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding L, Ellis MJ, Li S, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805:105–17. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 9.Aparicio S, Caldas C. The implications of clonal genome evolution for cancer medicine. N Engl J Med. 2013;368:842–51. doi: 10.1056/NEJMra1204892. [DOI] [PubMed] [Google Scholar]
- 10.Shapiro JR, Yung WK, Shapiro WR. Isolation, karyotype, and clonal growth of heterogeneous subpopulations of human malignant gliomas. Cancer Res. 1981;41:2349–59. [PubMed] [Google Scholar]
- 11.Little SE, Popov S, Jury A, et al. Receptor tyrosine kinase genes amplified in glioblastoma exhibit a mutual exclusivity in variable proportions reflective of individual tumor heterogeneity. Cancer Res. 2012;72:1614–20. doi: 10.1158/0008-5472.CAN-11-4069. [DOI] [PubMed] [Google Scholar]
- 12.Snuderl M, Fazlollahi L, Le LP, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–7. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Bruce WR, Van Der Gaag H. A quantitative assay for the number of murine lymphoma cells capable of proliferation in vivo. Nature. 1963;199:79–80. doi: 10.1038/199079a0. [DOI] [PubMed] [Google Scholar]
- 14.Hamburger AW, Salmon SE. Primary bioassay of human tumor stem cells. Science. 1977;197:461–3. doi: 10.1126/science.560061. [DOI] [PubMed] [Google Scholar]
- 15.Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–44. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 16.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 17.Humphries A, Cereser B, Gay LJ, et al. Lineage tracing reveals multipotent stem cells maintain human adenomas and the pattern of clonal expansion in tumor evolution. Proc Natl Acad Sci U S A. 2013;110:E2490–9. doi: 10.1073/pnas.1220353110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schepers AG, Snippert HJ, Stange DE, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–5. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 19.Driessens G, Beck B, Caauwe A, et al. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–30. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–6. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson K, Lutz C, van Delft FW, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–61. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 22.Notta F, Mullighan CG, Wang JC, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–7. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 23.Mizrak D, Brittan M, Alison M. CD133: molecule of the moment. J Pathol. 2008;214:3–9. doi: 10.1002/path.2283. [DOI] [PubMed] [Google Scholar]
- 24.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–8. [PubMed] [Google Scholar]
- 25.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 26.Beier D, Hau P, Proescholdt M, et al. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–5. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 27.Nishide K, Nakatani Y, Kiyonari H, et al. Glioblastoma formation from cell population depleted of Prominin1-expressing cells. PLoS One. 2009;4:e6869. doi: 10.1371/journal.pone.0006869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogden AT, Waziri AE, Lochhead RA, et al. Identification of A2B5+CD133− tumor-initiating cells in adult human gliomas. Neurosurgery. 2008;62:505–14. doi: 10.1227/01.neu.0000316019.28421.95. discussion 514–5. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Sakariassen PØ, Tsinkalovsky O, et al. CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int J Cancer. 2008;122:761–8. doi: 10.1002/ijc.23130. [DOI] [PubMed] [Google Scholar]
- 30.Lottaz C, Beier D, Meyer K, et al. Transcriptional profiles of CD133+ and CD133− glioblastoma-derived cancer stem cell lines suggest different cells of origin. Cancer Res. 2010;70:2030–40. doi: 10.1158/0008-5472.CAN-09-1707. [DOI] [PubMed] [Google Scholar]
- 31.Joo KM, Kim SY, Jin X, et al. Clinical and biological implications of CD133-positive and CD133-negative cells in glioblastomas. Lab Invest. 2008;88:808–15. doi: 10.1038/labinvest.2008.57. [DOI] [PubMed] [Google Scholar]
- 32.Mao P, Joshi K, Li J, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A. 2013;110:8644–9. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen R, Nishimura MC, Bumbaca SM, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17:362–75. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 34.Son MJ, Woolard K, Nam DH, et al. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–52. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lathia JD, Gallagher J, Heddleston JM, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–32. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bao S, Wu Q, Li Z, et al. Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res. 2008;68:6043–8. doi: 10.1158/0008-5472.CAN-08-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Filatova A, Acker T, Garvalov BK. The cancer stem cell niche(s): the crosstalk between glioma stem cells and their microenvironment. Biochim Biophys Acta. 2013;1830:2496–508. doi: 10.1016/j.bbagen.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 38.Calabrese C, Poppleton H, Kocak M, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 39.Zhu TS, Costello MA, Talsma CE, et al. Endothelial cells create a stem cell niche in glioblastoma by providing NOTCH ligands that nurture self-renewal of cancer stem-like cells. Cancer Res. 2011;71:6061–72. doi: 10.1158/0008-5472.CAN-10-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lathia JD, Li M, Hall PE, et al. Laminin alpha 2 enables glioblastoma stem cell growth. Ann Neurol. 2012;72:766–78. doi: 10.1002/ana.23674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bao S, Wu Q, Sathornsumetee S, et al. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006;66:7843–8. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- 42.Ricci-Vitiani L, Pallini R, Biffoni M, et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468:824–8. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- 43.Soda Y, Marumoto T, Friedmann-Morvinski D, et al. Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proc Natl Acad Sci U S A. 2011;108:4274–80. doi: 10.1073/pnas.1016030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang R, Chadalavada K, Wilshire J, et al. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468:829–33. doi: 10.1038/nature09624. [DOI] [PubMed] [Google Scholar]
- 45.Lai CY, Schwartz BE, Hsu MY. CD133+ melanoma subpopulations contribute to perivascular niche morphogenesis and tumorigenicity through vasculogenic mimicry. Cancer Res. 2012;72:5111–8. doi: 10.1158/0008-5472.CAN-12-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng L, Huang Z, Zhou W, et al. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 2013;153:139–52. doi: 10.1016/j.cell.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Folberg R, Hendrix MJ, Maniotis AJ. Vasculogenic mimicry and tumor angiogenesis. Am J Pathol. 2000;156:361–81. doi: 10.1016/S0002-9440(10)64739-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kunnakkat S, Narayana A. Bevacizumab in the treatment of high-grade gliomas: an overview. Angiogenesis. 2011;14:423–30. doi: 10.1007/s10456-011-9232-2. [DOI] [PubMed] [Google Scholar]
- 49.Evans SM, Judy KD, Dunphy I, et al. Hypoxia is important in the biology and aggression of human glial brain tumors. Clin Cancer Res. 2004;10:8177–84. doi: 10.1158/1078-0432.CCR-04-1081. [DOI] [PubMed] [Google Scholar]
- 50.Barker FG, 2nd, Davis RL, Chang SM, et al. Necrosis as a prognostic factor in glioblastoma multiforme. Cancer. 1996;77:1161–6. doi: 10.1002/(sici)1097-0142(19960315)77:6<1161::aid-cncr24>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 51.Davy P, Allsopp R. Hypoxia: are stem cells in it for the long run? Cell Cycle. 2011;10:206–11. doi: 10.4161/cc.10.2.14535. [DOI] [PubMed] [Google Scholar]
- 52.Li Z, Rich JN. Hypoxia and hypoxia inducible factors in cancer stem cell maintenance. Curr Top Microbiol Immunol. 2010;345:21–30. doi: 10.1007/82_2010_75. [DOI] [PubMed] [Google Scholar]
- 53.Seidel S, Garvalov BK, Wirta V, et al. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2 alpha. Brain. 2010;133:983–95. doi: 10.1093/brain/awq042. [DOI] [PubMed] [Google Scholar]
- 54.Li Z, Bao S, Wu Q, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–13. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soeda A, Park M, Lee D, et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha. Oncogene. 2009;28:3949–59. doi: 10.1038/onc.2009.252. [DOI] [PubMed] [Google Scholar]
- 56.Heddleston JM, Li Z, McLendon RE, et al. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 2009;8:3274–84. doi: 10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohnishi S, Maehara O, Nakagawa K, et al. Hypoxia-Inducible Factors Activate CD133 Promoter through ETS Family Transcription Factors. PLoS One. 2013;8:e66255. doi: 10.1371/journal.pone.0066255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Griguer CE, Oliva CR, Gobin E, et al. CD133 is a marker of bioenergetic stress in human glioma. PLoS One. 2008;3:e3655. doi: 10.1371/journal.pone.0003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pàez-Ribes M, Allen E, Hudock J, et al. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15:220–31. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ebos JM, Lee CR, Cruz-Munoz W, et al. Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer Cell. 2009;15:232–9. doi: 10.1016/j.ccr.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takebe N, Ivy SP. Controversies in cancer stem cells: targeting embryonic signaling pathways. Clin Cancer Res. 2010;16:3106–12. doi: 10.1158/1078-0432.CCR-09-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bolós V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev. 2007;28:339–63. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- 64.Lathia JD, Mattson MP, Cheng A. Notch: from neural development to neurological disorders. J Neurochem. 2008;107:1471–81. doi: 10.1111/j.1471-4159.2008.05715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aguirre A, Rubio ME, Gallo V. Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature. 2010;467:323–7. doi: 10.1038/nature09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Handler M, Yang X, Shen J. Presenilin-1 regulates neuronal differentiation during neurogenesis. Development. 2000;127:2593–606. doi: 10.1242/dev.127.12.2593. [DOI] [PubMed] [Google Scholar]
- 67.Imayoshi I, Sakamoto M, Yamaguchi M, et al. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. 2010;30:3489–98. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ables JL, Breunig JJ, Eisch AJ, et al. Not(ch) just development: Notch signalling in the adult brain. Nat Rev Neurosci. 2011;12:269–83. doi: 10.1038/nrn3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 70.Lu J, Ye X, Fan F, et al. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell. 2013;23:171–85. doi: 10.1016/j.ccr.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Charles N, Ozawa T, Squatrito M, et al. Perivascular nitric oxide activates notch signaling and promotes stem-like character in PDGF-induced glioma cells. Cell Stem Cell. 2010;6:141–52. doi: 10.1016/j.stem.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fan X, Khaki L, Zhu TS, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang J, Wakeman TP, Lathia JD, et al. Notch promotes radioresistance of glioma stem cells. Stem Cells. 2010;28:17–28. doi: 10.1002/stem.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gilbert CA, Daou MC, Moser RP, et al. Gamma-secretase inhibitors enhance temozolomide treatment of human gliomas by inhibiting neurosphere repopulation and xenograft recurrence. Cancer Res. 2010;70:6870–9. doi: 10.1158/0008-5472.CAN-10-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Krop I, Demuth T, Guthrie T, et al. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J Clin Oncol. 2012;30:2307–13. doi: 10.1200/JCO.2011.39.1540. [DOI] [PubMed] [Google Scholar]
- 76.Fouladi M, Stewart CF, Olson J, et al. Phase I trial of MK-0752 in children with refractory CNS malignancies: a pediatric brain tumor consortium study. J Clin Oncol. 2011;29:3529–34. doi: 10.1200/JCO.2011.35.7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tolcher AW, Messersmith WA, Mikulski SM, et al. Phase I study of RO4929097, a gamma secretase inhibitor of Notch signaling, in patients with refractory metastatic or locally advanced solid tumors. J Clin Oncol. 2012;30:2348–53. doi: 10.1200/JCO.2011.36.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 79.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–50. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 80.Fodde R. The APC gene in colorectal cancer. Eur J Cancer. 2002;38:867–71. doi: 10.1016/s0959-8049(02)00040-0. [DOI] [PubMed] [Google Scholar]
- 81.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 82.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 83.Zechner D, Fujita Y, Hülsken J, et al. beta-Catenin signals regulate cell growth and the balance between progenitor cell expansion and differentiation in the nervous system. Dev Biol. 2003;258:406–18. doi: 10.1016/s0012-1606(03)00123-4. [DOI] [PubMed] [Google Scholar]
- 84.Liu C, Tu Y, Sun X, et al. Wnt/beta-Catenin pathway in human glioma: expression pattern and clinical/prognostic correlations. Clin Exp Med. 2011;11:105–12. doi: 10.1007/s10238-010-0110-9. [DOI] [PubMed] [Google Scholar]
- 85.Zheng H, Ying H, Wiedemeyer R, et al. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell. 2010;17:497–509. doi: 10.1016/j.ccr.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rheinbay E, Suvà ML, Gillespie SM, et al. An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma. Cell Rep. 2013;3:1567–79. doi: 10.1016/j.celrep.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang N, Wei P, Gong A, et al. FoxM1 promotes β-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell. 2011;20:427–42. doi: 10.1016/j.ccr.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rampazzo E, Persano L, Pistollato F, et al. Wnt activation promotes neuronal differentiation of glioblastoma. Cell Death Dis. 2013;4:e500. doi: 10.1038/cddis.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim KH, Seol HJ, Kim EH, et al. Wnt/β-catenin signaling is a key downstream mediator of MET signaling in glioblastoma stem cells. Neuro Oncol. 2013;15:161–71. doi: 10.1093/neuonc/nos299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 91.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–31. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 93.Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–72. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 94.Barakat MT, Humke EW, Scott MP. Learning from Jekyll to control Hyde: Hedgehog signaling in development and cancer. Trends Mol Med. 2010;16:337–48. doi: 10.1016/j.molmed.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rohatgi R, Scott MP. Patching the gaps in Hedgehog signalling. Nat Cell Biol. 2007;9:1005–9. doi: 10.1038/ncb435. [DOI] [PubMed] [Google Scholar]
- 96.Aszterbaum M, Rothman A, Johnson RL, et al. Identification of mutations in the human PATCHED gene in sporadic basal cell carcinomas and in patients with the basal cell nevus syndrome. J Invest Dermatol. 1998;110:885–8. doi: 10.1046/j.1523-1747.1998.00222.x. [DOI] [PubMed] [Google Scholar]
- 97.Hahn H, Wicking C, Zaphiropoulous PG, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–51. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 98.Johnson RL, Rothman AL, Xie J, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–71. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 99.Reifenberger J, Wolter M, Weber RG, et al. Missense mutations in SMOH in sporadic basal cell carcinomas of the skin and primitive neuroectodermal tumors of the central nervous system. Cancer Res. 1998;58:1798–803. [PubMed] [Google Scholar]
- 100.Keeler RF, Binns W. Teratogenic compounds of Veratrum californicum (Durand). I. Preparation and characterization of fractions and alkaloids for biologic testing. Can J Biochem. 1966;44:819–28. doi: 10.1139/o66-100. [DOI] [PubMed] [Google Scholar]
- 101.Keeler RF, Binns W. Teratogenic compounds of Veratrum californicum (Durand). V. Comparison of cyclopian effects of steroidal alkaloids from the plant and structurally related compounds from other sources. Teratology. 1968;1:5–10. doi: 10.1002/tera.1420010103. [DOI] [PubMed] [Google Scholar]
- 102.Chen JK, Taipale J, Cooper MK, et al. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–8. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cooper MK, Porter JA, Young KE, et al. Teratogen-mediated inhibition of target tissue response to Shh signaling. Science. 1998;280:1603–7. doi: 10.1126/science.280.5369.1603. [DOI] [PubMed] [Google Scholar]
- 104.Taipale J, Chen JK, Cooper MK, et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–9. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 105.Lin TL, Matsui W. Hedgehog pathway as a drug target: Smoothened inhibitors in development. Onco Targets Ther. 2012;5:47–58. doi: 10.2147/OTT.S21957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Axelson M, Liu K, Jiang X, et al. U.S. Food and Drug Administration approval: vismodegib for recurrent, locally advanced, or metastatic basal cell carcinoma. Clin Cancer Res. 2013;19:2289–93. doi: 10.1158/1078-0432.CCR-12-1956. [DOI] [PubMed] [Google Scholar]
- 107.Rudin CM, Hann CL, Laterra J, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173–8. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yauch RL, Dijkgraaf GJ, Alicke B, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–4. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov. 2006;5:1026–33. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- 110.Kinzler KW, Bigner SH, Bigner DD, et al. Identification of an amplified, highly expressed gene in a human glioma. Science. 1987;236:70–3. doi: 10.1126/science.3563490. [DOI] [PubMed] [Google Scholar]
- 111.Bigner SH, Wong AJ, Mark J, et al. Relationship between gene amplification and chromosomal deviations in malignant human gliomas. Cancer Genet Cytogenet. 1987;29:165–70. doi: 10.1016/0165-4608(87)90045-8. [DOI] [PubMed] [Google Scholar]
- 112.Bar EE, Chaudhry A, Lin A, et al. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25:2524–33. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Clement V, Sanchez P, de Tribolet N, et al. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165–72. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ehtesham M, Sarangi A, Valadez JG, et al. Ligand-dependent activation of the hedgehog pathway in glioma progenitor cells. Oncogene. 2007;26:5752–61. doi: 10.1038/sj.onc.1210359. [DOI] [PubMed] [Google Scholar]
- 115.Sarangi A, Valadez JG, Rush S, et al. Targeted inhibition of the Hedgehog pathway in established malignant glioma xenografts enhances survival. Oncogene. 2009;28:3468–76. doi: 10.1038/onc.2009.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gerardo Valadez J, Grover VK, Carter MD, et al. Identification of Hedgehog pathway responsive glioblastomas by isocitrate dehydrogenase mutation. Cancer Lett. 2013;328:297–306. doi: 10.1016/j.canlet.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–74. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 118.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Watanabe T, Nobusawa S, Kleihues P, et al. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–53. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lai A, Kharbanda S, Pope WB, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol. 2011;29:4482–90. doi: 10.1200/JCO.2010.33.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 122.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 123.Tamura K, Aoyagi M, Wakimoto H, et al. Accumulation of CD133-positive glioma cells after high-dose irradiation by Gamma Knife surgery plus external beam radiation. J Neurosurg. 2010;113:310–8. doi: 10.3171/2010.2.JNS091607. [DOI] [PubMed] [Google Scholar]
- 124.Jamal M, Rath BH, Tsang PS, et al. The brain microenvironment preferentially enhances the radioresistance of CD133(+) glioblastoma stem-like cells. Neoplasia. 2012;14:150–8. doi: 10.1593/neo.111794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ropolo M, Daga A, Griffero F, et al. Comparative analysis of DNA repair in stem and nonstem glioma cell cultures. Mol Cancer Res. 2009;7:383–92. doi: 10.1158/1541-7786.MCR-08-0409. [DOI] [PubMed] [Google Scholar]
- 126.Joo KM, Jin J, Kim E, et al. MET signaling regulates glioblastoma stem cells. Cancer Res. 2012;72:3828–38. doi: 10.1158/0008-5472.CAN-11-3760. [DOI] [PubMed] [Google Scholar]
- 127.Kim Y, Kim KH, Lee J, et al. Wnt activation is implicated in glioblastoma radioresistance. Lab Invest. 2012;92:466–73. doi: 10.1038/labinvest.2011.161. [DOI] [PubMed] [Google Scholar]
- 128.Woodward WA, Chen MS, Behbod F, et al. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci U S A. 2007;104:618–23. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McCord AM, Jamal M, Williams ES, et al. CD133+ glioblastoma stem-like cells are radiosensitive with a defective DNA damage response compared with established cell lines. Clin Cancer Res. 2009;15:5145–53. doi: 10.1158/1078-0432.CCR-09-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pallini R, Ricci-Vitiani L, Montano N, et al. Expression of the stem cell marker CD133 in recurrent glioblastoma and its value for prognosis. Cancer. 2011;117:162–74. doi: 10.1002/cncr.25581. [DOI] [PubMed] [Google Scholar]
- 131.Li A, Walling J, Kotliarov Y, et al. Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol Cancer Res. 2008;6:21–30. doi: 10.1158/1541-7786.MCR-07-0280. [DOI] [PubMed] [Google Scholar]
- 132.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 133.Kim E, Kim M, Woo DH, et al. Phosphorylation of EZH2 Activates STAT3 Signaling via STAT3 Methylation and Promotes Tumorigenicity of Glioblastoma Stem-like Cells. Cancer Cell. 2013;23:839–52. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Suvà ML, Riggi N, Janiszewska M, et al. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009;69:9211–8. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- 135.Gu C, Banasavadi-Siddegowda YK, Joshi K, et al. Tumor-specific activation of the C-JUN/MELK pathway regulates glioma stem cell growth in a p53-dependent manner. Stem Cells. 2013;31:870–81. doi: 10.1002/stem.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Guryanova OA, Wu Q, Cheng L, et al. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer Cell. 2011;19:498–511. doi: 10.1016/j.ccr.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kamal MM, Sathyan P, Singh SK, et al. REST regulates oncogenic properties of glioblastoma stem cells. Stem Cells. 2012;30:405–14. doi: 10.1002/stem.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sherry MM, Reeves A, Wu JK, et al. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27:2383–92. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang J, Wang H, Li Z, et al. c-Myc is required for maintenance of glioma cancer stem cells. PLoS One. 2008;3:e3769. doi: 10.1371/journal.pone.0003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Friedmann-Morvinski D, Bushong EA, Ke E, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–4. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bachoo RM, Maher EA, Ligon KL, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–77. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 142.Dai C, Celestino JC, Okada Y, et al. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 2001;15:1913–25. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kang SK, Park JB, Cha SH. Multipotent, dedifferentiated cancer stem-like cells from brain gliomas. Stem Cells Dev. 2006;15:423–35. doi: 10.1089/scd.2006.15.423. [DOI] [PubMed] [Google Scholar]
- 144.Hjelmeland AB, Wu Q, Heddleston JM, et al. Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ. 2011;18:829–40. doi: 10.1038/cdd.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]