Abstract
Until now, there was only serological evidence that hantaviruses were circulating in rodents and infecting humans from Madagascar. To assess the presence of a hantavirus on the island, between October, 2008, and March, 2010, we sampled 585 rodents belonging to seven species in the Anjozorobe-Angavo forest corridor, 70 km north from the capital city Antananarivo. A hantavirus was detected from organs of the ubiquist roof rat (Rattus rattus) and of the endemic Major's tufted-tailed rat (Eliurus majori). Amazingly, sequence analysis of the S (small), M (medium), and L (large) coding DNA sequence of this virus showed that the Anjozorobe strain (proposed name) was a new genetic variant of Thailand virus (THAIV) that comprises other variants found in Southeast Asia. Because THAIV is suspected of causing hemorrhagic fever with renal syndrome in humans, ongoing studies are addressing the risk of infection by this new variant in the Malagasy population.
Key Words: : Hantavirus, Madagascar, Rattus rattus, Eliurus majori
Introduction
Hantaviruses constitute one of the five genera in the family Bunyaviridae. The genome of these enveloped viruses is segmented and encompasses three molecules of negative-sense single-stranded RNA, designated S (small), M (medium), and L (large). These molecules encode the nucleoprotein (N), two external glycoproteins (Gn and Gc), and a RNA-dependent RNA polymerase (RdRp) or L protein, respectively. Twenty-three species are officially reported, but the number of described taxa is increasing exponentially (King et al. 2012). Each virus taxon is usually associated with one natural host species, including rodents (Order Rodentia) and insectivores (Order Soricomorpha) (Jonsson et al. 2010, Arai et al. 2012). Partial L sequences of new hantaviruses have also been recently detected in bats (Order Chiroptera) (Sumibcay et al. 2012, Weiss et al. 2012, Arai et al. 2013, Guo et al. 2013). Infection of the host species by their respective hantavirus remains generally unapparent. Transmission of the virus between individuals occurs through direct contact or through inhalation of excretions or secretions. Using these routes, some hantaviruses can be transmitted to humans and cause hemorrhagic fever with renal syndrome (HFRS) or cardiopulmonary syndrome. Human-to-human transmission is rare (Jonsson et al. 2010). There is no specific treatment of the diseases, although treatment with ribavirin showed some clinical benefit during Old World hantavirus infections. Inactivated vaccine against Hantaan and Seoul viruses are only available and licensed in China and South Korea (Krüger et al. 2011).
Although there was previous serological evidence of hantavirus circulation among rodents and humans in Africa, it is only since 2006 that hantaviruses have been identified in rodents, insectivores, and bats from this continent (Klempa et al. 2006, Klempa et al. 2007, Kang et al. 2011, Klempa et al. 2012, Meheretu et al. 2012, Sumibcay et al. 2012, Weiss et al. 2012). Similarly, in Madagascar, there were serological indications of hantavirus circulation in rodents (Rattus rattus and R. norvegicus) and humans (Rollin et al. 1986). We report here the first detection of a hantavirus associated with rodents in Madagascar.
Materials and Methods
Sample collection
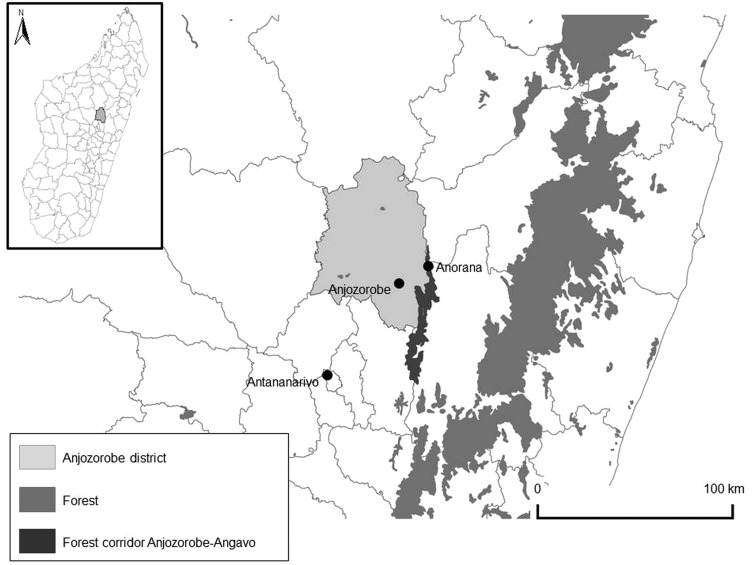
During a survey looking for hemorrhagic fever viruses associated with wild terrestrial small mammals in Madagascar (Olive et al. 2013), animals were captured in the Anjozorobe-Angavo forest corridor (Anjozorobe district) (18°18′41.9″ S, 48°00′57.6″ E, 70 km north from Antananarivo, the capital city) over four 3-week trapping sessions conducted in October, 2008, March and October, 2009, and March, 2010 (Fig. 1). Permits to capture and collect animals were obtained from the national authorities. Animals were collected twice a day, early in the morning and late in the afternoon, transferred to the site camp, anesthetized with chloroform, and euthanized by cardiac blood puncture. Species identification was based on characteristics outlined in Soarimalala and Goodman (2011). Organs were collected and stored in liquid nitrogen containers and then at −80°C upon arrival at the laboratory in Antananarivo.
FIG. 1.
Study site in Madagascar—the Anorana camp, in the north of the forest corridor of Anjozorobe-Angavo.
RNA extraction from organs and hantavirus detection
About 50–100 mg of liver or liver and spleen from each individual were mixed and homogenized at a 1:10 dilution in culture medium containing 40% fetal bovine serum. After centrifugation, supernatants were collected and pooled by species (with a maximum of five individuals per pool). RNA was extracted from pooled supernatants using TRIzol LS reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
RNA was then reverse transcribed and cDNA amplified using a nested-PCR approach, according to Klempa et al., using sets of consensus primers previously designed to detect part of the L coding region of hantaviruses associated with rodents or insectivores (Klempa et al. 2006). Amplification products were sequenced on both strands by Cogenics (Essex, UK). Unverified sequences and chromatograms were compared and corrected when needed. A database search using the BLAST web server (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was performed to identify the hantavirus species.
Complete genome sequencing
Total RNA was extracted from liver and spleen tissues of some positive rodents using the NucliSENS® easyMAG™ bio-robot (bioMérieux, Marcy l'Etoile, France). cDNA was prepared with SuperScript® III Reverse Transcriptase (Invitrogen, Life Technologies, Inc.) and random hexamers (Roche, Mannheim, Germany). Overlapping PCR fragments for the different segments were obtained using consensus (degenerate) PCR primers. PCR products were cloned and sent for sequencing (Cogenics, Essex, UK). The generated sequences correspond to the consensus of three independent clones and have been deposited in GenBank under accession numbers KC490914–KC490918, KC490921, and KC490923. Strains from which the sequences were obtained were then named according to the following designation: <name of the district where the animals were sampled>, <host>, <country of sampling>, <year of sampling>, and <host specimen code>. For example, Anjozorobe/Em/MDG/2009/ATD-49 was the strain Anjozorobe (name of the district), obtained from the animal belonging to the Eliurus majori species, sampled in Madagascar in 2009 and coded ATD-49.
Sequence and phylogenetic analyses
Sequences from the same segment were compared when aligned, and a phylogenetic analysis was conducted using MEGA version 5 (Tamura et al. 2011). Sequences were aligned using ClustalW. The maximum likelihood method was used. According to the best-fit substitution model proposed by MEGA, analysis was performed applying the Tamura–Neil model using a Gamma distribution (+G) with five rate categories and by assuming that a certain fraction of the sites are evolutionarily invariable (+I). One thousand bootstrap replicates were generated.
Results
A total of 585 rodents belonging to seven species were sampled (Olive et al. 2013). Partial L amplification was positive in pools of organs from two species—the roof rat (R. rattus), the widely distributed species of the family Muridae introduced in Madagascar, and the Major's tufted-tailed rat (E. majori), a Malagasy endemic species of the family Nesomyidae. To confirm the results obtained from the pools, partial L detection was performed on supernatants of the individuals present in the positive pools. Among the 231 roof rats tested, constitutive of the positive pools, 92 were found positive. The unique ATD-49 individual, constitutive of the positive E. majori “pool,” was again found positive. PCR products from five pools and eight individuals of roof rats and from the Major's tufted-tailed rat ATD-49 were sequenced. Database BLAST searches using the sequences obtained confirmed the presence of a hantavirus. The sequences found matched especially to Serang virus, which is associated with the Oriental house rat Rattus tanezumi from Southeast Asia and which is member of the Thailand virus (THAIV) species (Johansson et al. 2010) (data not shown).
Consequently, S coding DNA sequences (CDS) were recovered from four roof rats (ATD-9, ATD-56, ATD-108, and ATD-261) and the Major's tufted-tailed rat ATD-49. The CDS were found to be 1290 nucleotides long, encoding a 429 amino-acid-long N protein. All nucleotide sequences were very close (92.9–100% within the roof rat virus sequences and 92.5–96.7% between themselves and the Anjozorobe/Em/MDG/2009/ATD-49 sequence). In addition, sequences exhibited 100% amino acid identity between themselves, except for the R. rattus sequence of Anjozorobe/Rr/MDG/2009/ATD-261, which exhibited a V69I mutation. Alignment and comparison of one of these five sequences (strain Anjozorobe/Rr/MDG/2009/ATD-56) with the 1290-nucleotide-long S CDS of hantavirus taxa showed a high percentage of identity between this sequence and those from THAIV and its variants Jurong and Serang viruses (97.0–98.1% amino acid identity) (Table 1). Then, M and L CDS were recovered from the Anjozorobe/Rr/MDG/2009/ATD-56 strain. M CDS was 3402 nucleotides long, encoding a 1133-amino-acid-long glycoprotein precursor (GPC). The WAASA motif corresponding to the GPC cleavage site was identified at positions 642–646. As for the S CDS, when compared to sequences of the same batch of hantaviruses, the M CDS was also found to be close to M CDS of THAIV and its variants (91.7–93.6% amino acid identity) (Table 1). L CDS was 6456 nucleotides long, encoding a 2151-amino-acid-long RdRp. Nevertheless, because complete sequences from THAIV and its variants were not available, comparisons were based on the available 1560-, 411-, and 316-nucleotide-long partial L sequences of Jurong, Serang, and Thailand viruses, respectively. Nucleotide identities with Anjozorobe/Rr/MDG/2009/ATD-56 L sequence ranged from 75.0% (THAIV) to 80.8% (Serang virus) and amino acid identities ranged from 93.3% (THAIV) to 96.3% (Jurong virus).
Table 1.
Identities (%) of the Entire Coding DNA Sequences of the S (Small), M (Medium), and L (Large) Segments of Anjozorobe/Rr/2009/MDG/ATD-56 Hantavirus Strain and Selected Hantaviruses Harbored by Rodents and Soricomorphs
| S CDS | M CDS | L CDS | ||||
|---|---|---|---|---|---|---|
| Virus (strain) | 1290 nt | 429 aa | 3402 nt | 1133 aa | 6456 nt | 2151 aa |
| Hantaviruses associated with Murinae | ||||||
| Jurong (TJK/O6 [RT49]) | 84.7 | 97.4 | 79.7 | 93.6 | NA | NA |
| Jurong (TJK/O6 [RT50]) | 84.7 | 97.4 | 79.7 | 93.6 | NA | NA |
| Serang (Rt60/2000) | 83.9 | 98.1 | NA | NA | NA | NA |
| Thailand (741 for S; 749 for M and L) | 83.0 | 97.0 | 79.7 | 91.7 | NA | NA |
| Thailand (N-R/Bi0017/2004) | 82.7 | 97.0 | NA | NA | NA | NA |
| Seoul (HR 80-39) | 75.3 | 86.5 | 74.6 | 83.1 | 76.3 | 88.9 |
| Hantaan (76–118) | 75.0 | 84.6 | 71.1 | 77.8 | 74.4 | 84.9 |
| Da-Bie-Shang (NC167) | 75.0 | 84.8 | 71.0 | 76.6 | 74.9 | 85.4 |
| Amur (Khekhtsir/AP209/2005) | 75.5 | 85.3 | 70.6 | 77.8 | 75.2 | 85.5 |
| Soochong (SC-1) | 74.7 | 85.1 | 71.1 | 77.7 | 74.5 | 85.3 |
| Dobrava-Belgrade (GRW/Aa) | 73.3 | 84.4 | 71.4 | 77.1 | 75.0 | 85.9 |
| Sangassou (SA14) | 73.1 | 83.0 | 71.2 | 78.2 | 74.6 | 84.4 |
| Saaremaa (160V) | 72.9 | 82.8 | 70.8 | 77.1 | 75.2 | 86.3 |
| Hantavirus associated with Arvicolinae | ||||||
| Tula (Moravia/5302Ma/94) | 62.4 | 61.8 | 58.6 | 54.4 | 66.5 | 68.6 |
| Malacky (Ma32/94) | 62.3 | 62.2 | NA | NA | NA | NA |
| Hantavirus associated with soricomorphs | ||||||
| Jemez Springs (MSB144475) | 63.9 | 61.8 | NA | NA | NA | NA |
| Seewis (mp70) | 62.1 | 59.4 | NA | NA | NA | NA |
| Qiandao-Lake (YN05-284) | 64.8 | 61.3 | NA | NA | NA | NA |
| Kenkeme (MSB148794) | 61.7 | 59.0 | NA | NA | NA | NA |
CDS, coding DNA sequences; nt, nucleotides; aa, amino acids; NA, corresponding complete sequence not available.
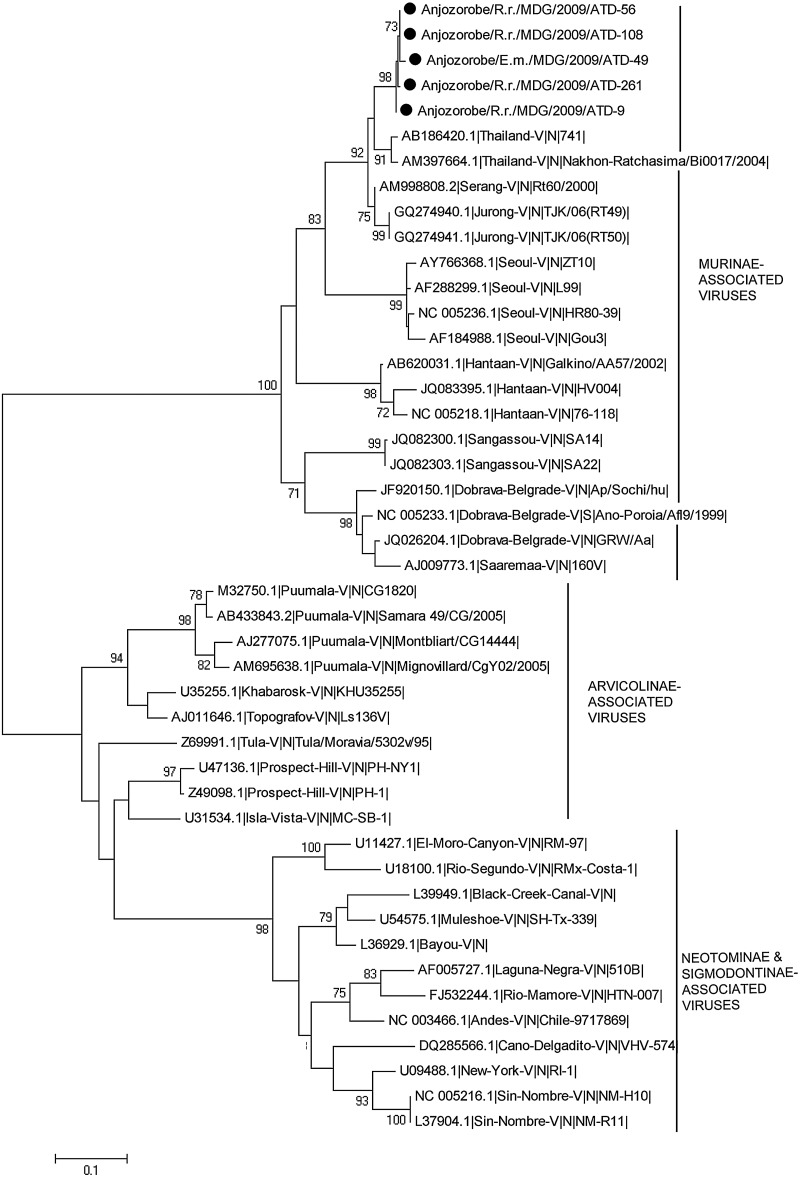
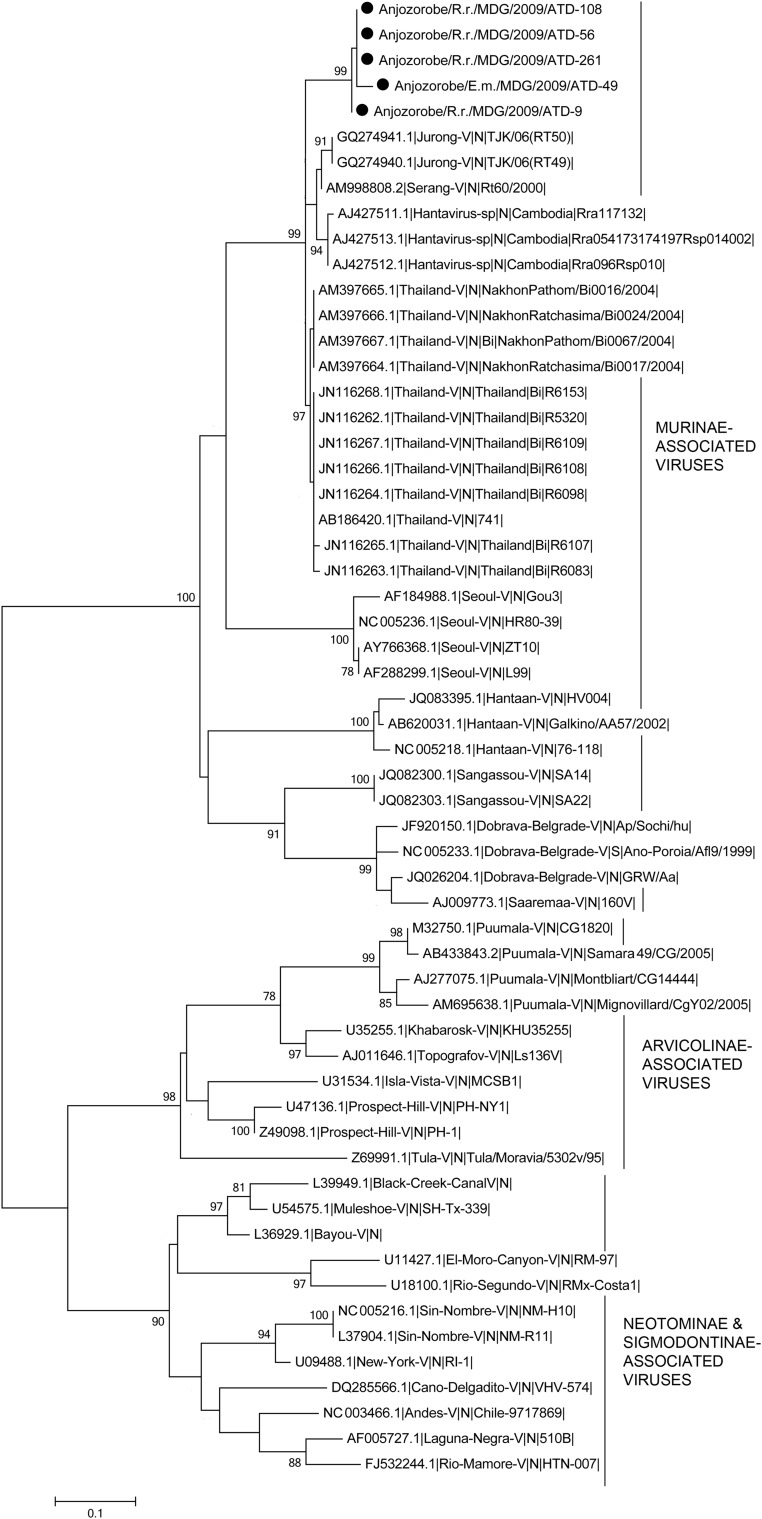
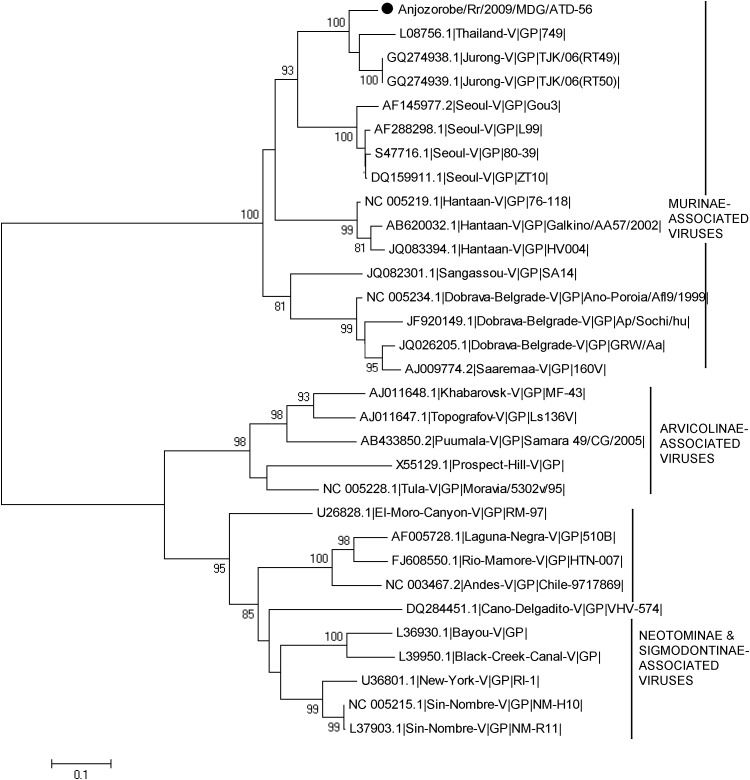
Phylogenetic analysis based on entire S CDS showed that Anjozorobe strains belonged to the THAIV species and were much closer to THAIV than to Jurong and Serang variants that are also members of the THAIV species (Fig. 2). This phylogenetic analysis was also performed with a reduction of the S CDS dataset to 519 nucleotides (positions 364–882). This allowed the inclusion of the sequences of Cambodian strains close to THAIV, and of other THAIV sequences (Reynes et al. 2003, Hugot et al. 2006, Blasdell et al. 2011). Although the tree topology changed slightly and the statistical support was lower, all sequences grouped together (Fig. 3). The clustering of the Anjozorobe variant with THAIV and its other variants was also confirmed by phylogenetic analyses of the entire M CDS (Fig. 4) and partial L (286 nucleotides) CDS (THAIV entire CDS not yet available) (data not shown).
FIG. 2.
Phylogenetic tree based on the complete nucleoprotein coding sequences from the Anjozorobe virus (indicated by ●), from the 22 hantavirus species associated with rodents and listed by the International Committee on Taxonomy of Viruses, from strains related to Thailand virus, and from Sangassou hantavirus recently described in Africa. Bootstrap percentages ≥70% (from 1000 resamplings) are indicated at each node. Scale bar indicates nucleotide substitution per site.
FIG. 3.
Phylogenetic tree based on the partial nucleoprotein coding sequences (nucleotides 364–882) from the Anjozorobe virus (indicated by ●), from the 22 hantavirus species associated with rodents and listed by the International Committee on Taxonomy of Viruses, from strains related to Thailand virus, and from the Sangassou hantavirus recently described in Africa. Bootstrap percentages ≥70% (from 1000 resamplings) are indicated at each node. Scale bar indicates nucleotide substitution per site.
FIG. 4.
Phylogenetic tree based on the complete glycoproteins coding sequences from the Anjozorobe virus (indicated by ●), from 18 of the 22 hantavirus species associated with rodents and listed by the International Committee on Taxonomy of Viruses, from strains related to the Thailand virus, and from the Sangassou hantavirus recently described in Africa. Bootstrap percentages ≥70% (from 1000 resamplings) are indicated at each node. Scale bar indicates nucleotide substitution per site.
Discussion
Our study demonstrates the presence of a hantavirus in Madagascar (proposed name Anjozorobe, according to the name of the district where the rodents were sampled). This hantavirus, associated with the introduced R. rattus species and the endemic E. majori species, belongs to the THAIV species, because the level of amino acid divergence between Anjozorobe strains and viruses of the THAIV species is less than 10% and 12% on the complete S and M CDS, respectively (Maes et al. 2009).
Thailand virus species include THAIV associated with the rodent Bandicota indica in Thailand, as well as Serang and Jurong variants and an unnamed variant, all associated with R. tanezumi in Indonesia, Singapore, and Cambodia, respectively (Elwell et al. 1985, Reynes et al. 2003, Hugot et al. 2006, Plyusnina et al. 2009, Johansson et al. 2010). The presence of a THAIV variant in Madagascar, amazingly far from South and Southeast Asia, is probably explained by the origin of R. rattus, which is native to the Indian Peninsula, and has been introduced to Madagascar, through the Arabic Peninsula, about 2000–3000 years ago on the coast, and later in the highlands, with the arrival of the first human migrants and the colonization of the island (Tollenaere et al. 2010). R. rattus and B. indica are sympatric species in South Asia and in the western part (Myanmar) of Southeast Asia (Wilson and Reeder 2005). This presupposes the presence of THAIV or of a THAIV variant in this area. This occurrence had been suspected but not yet demonstrated and requires further investigation (Chandy et al. 2013).
Two other rodent species introduced by humans are recorded in Madagascar—the brown rat (R. norvegicus) and the house mouse (Mus musculus) (Soarimalala and Goodman 2011). The brown rat and the roof rat are known to host Seoul virus, whereas there is no report of a hantavirus associated with M. musculus. During our study, we did not detect any Seoul virus. However, our sampling area was limited to a secondary and primary forest and, consequently, no brown rat has been sampled. A survey exploring the association between hantavirus and rodents from urban environments could assess the presence of Seoul virus within the country.
One endemic rodent species, the Major's tufted-tailed rat (E. majori) was found to be infected, carrying the Anjozorobe variant. This nocturnal, arboreal, and frugivorous species is distributed primarily in humid forest in the east of the island (Soarimalala and Goodman 2011). This rodent was sympatric with roof rats in the study area, where about 20% of the captured roof rats were infected by this virus. Molecular identification of the species using mitochondrial cytochrome b DNA sequence obtained from the liver (from which the virus was detected twice in two independent assays) confirmed species identification and allowed us to exclude a sampling error (data not shown). Therefore, we suspected a spillover event among rodents from the same superfamily Muroidea. Because only one E. majori individual was found to be infected, the association between this rodent and Anjozorobe variant needs to be confirmed, particularly if the Major's tufted-tailed rat may act as a reservoir for this variant.
All rodent species, except for R. rattus, R. norvegicus, and M. musculus, present in Madagascar are endemic species belonging to the Nesomyinae subfamily, a monophyletic phylum originating from Africa and for which 27 species are now reported (Soarimalala and Goodman 2011). Insectivorous mammals present in Madagascar include tenrecs, about 32 endemic species (Family Tenrecidae, Afrosoricida) originating from Africa, and two introduced species—the pygmy shrew (Suncus etruscus, Soricomorpha) and the Asian house shrew (Suncus murinus, Soricomorpha), an ubiquist species that, in Asia, hosts the Thottapalayam virus (Poux et al. 2005, Omar et al. 2011, Soarimalala and Goodman 2011). In our study, rodents from only six endemic species were sampled, with a restricted number of individuals sampled due to conservation practices. In addition, insectivorous species have not been investigated. Extensive studies among these animals should probably allow the identification of viruses related to hantaviruses recently described in Africa (Klempa et al. 2007, Kang et al. 2011, Klempa et al. 2012, Meheretu et al. 2012, Sumibcay et al. 2012, Weiss et al. 2012).
THAIV and its variants Jurong and Serang have only ever been detected in rodents and not in humans. However, because serological evidence suggests that THAIV is capable of infecting humans and causing HFRS (Pattamadilok et al. 2006, Gamage et al. 2011), the Anjozorobe variant, belonging to the THAIV species, may be a causative agent of HFRS in Madagascar. We have not yet succeeded in isolating the Anjozorobe variant on Vero E6 cells, and serological tools are still unavailable. Nevertheless, molecular investigations are ongoing to evaluate the presence of the Anjozorobe variant not only in human samples presenting with symptoms compatible with hantavirus infection, but also in roof rats living in forested areas and close to human settlements in several regions from Madagascar to determine the risk for human infection.
Acknowledgments
We thank Benoît de Thoisy and Damien Donato for their technical support and Philippe Marianneau for providing the Puumala virus RNA positive control. We also thank Philip Lawrence for his contribution as native English speaker. This work received financial support from the International Division of the Institut Pasteur in Paris (“Actions Concertées Inter-Pasteurienne A17/2009”).
Author Disclosure Statement
No competing financial interests exist.
References
- Arai S, Gu SH, Baek LJ, Tabara J, et al. Divergent ancestral lineages of newfound hantaviruses harbored by phylogenetically related crocidurine shrew species in Korea. Virology 2012; 424:99–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S, Nguyen ST, Boldgiv B, Fukui D, et al. Novel bat-borne hantavirus, Vietnam. Emerg Infect Dis 2013; 19:1159–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasdell K, Cosson JF, Chaval Y, Herbreteau V, et al. Rodent-borne hantaviruses in Cambodia, Lao PDR, and Thailand. Ecohealth 2011; 8:432–443 [DOI] [PubMed] [Google Scholar]
- Chandy S, Ulrich RG, Schlegel M, Petraityte R, et al. Hantavirus infection among wild small mammals in Vellore, South India. Zoonoses Public Health 2013; 60:336–340 [DOI] [PubMed] [Google Scholar]
- Elwell MR, Ward GS, Tingpalapong M, LeDuc JW. Serologic evidence of Hantaan-like virus in rodents and man in Thailand. Southeast Asian J Trop Med Public Health 1985; 16:349–354 [PubMed] [Google Scholar]
- Gamage CD, Yasuda SP, Nishio S, Kularatne SA, et al. Serological evidence of Thailand virus-related hantavirus infection among suspected leptospirosis patients in Kandy, Sri Lanka. Jpn J Infect Dis 2011; 64:72–75 [PubMed] [Google Scholar]
- Guo WP, Lin XD, Wang W, Tian JH, et al. Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog 2013; 9:1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Plyusnina A, Herbreteau V, Nemirov K, et al. Genetic analysis of Thailand hantavirus in Bandicota indica trapped in Thailand. Virol J 2006; 5:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson P, Yap G, Low HT, Siew CC, et al. Molecular characterization of two hantavirus strains from different rattus species in Singapore. Virol J 2010; 7:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson CB, Figueiredo LT, Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev 2010; 23:412–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kadjo B, Dubey S, Jacquet F, et al. Molecular evolution of Azagny virus, a newfound hantavirus harbored by the West African pygmy shrew (Crocidura obscurior) in Côte d'Ivoire. Virol J 2011; 8:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. VirusT. Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses. New York: Elsevier Academic Press, 2012 [Google Scholar]
- Klempa B, Fichet-Calvet E, Lecompte E, Auste B, et al. Hantavirus in African wood mouse, Guinea. Emerg Infect Dis 2006; 12:838–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempa B, Fichet-Calvet E, Lecompte E, Auste B, et al. Novel hantavirus sequences in Shrew, Guinea. Emerg Infect Dis 2007; 13:520–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempa B, Witkowski PT, Popugaeva E, Auste B, et al. Sangassou virus, the first hantavirus isolate from Africa, displays genetic and functional properties distinct from those of other murinae-associated hantaviruses. J Virol 2012; 86:3819–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger DH, Schönrich G, Klempa B. Human pathogenic hantaviruses and prevention of infection. Hum Vaccin 2011; 7:685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes P, Klempa B, Clement J, Matthijnssens J, et al. A proposal for new criteria for the classification of hantaviruses, based on S and M segment protein sequences. Infect Genet Evol 2009; 9:813–820 [DOI] [PubMed] [Google Scholar]
- Meheretu Y, Cížková D, Těšíková J, Welegerima K, et al. High diversity of RNA viruses in rodents, Ethiopia. Emerg Infect Dis 2012; 18:2047–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive MM, Razafindralambo N, Barivelo TA, Rafisandratantsoa JT, et al. Absence of Rift Valley fever virus in wild small mammals, Madagascar. Emerg Infect Dis 2013;19:1025–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar H, Adamson HA S, Bhassu S, Goodman SM, et al. Phylogenetic relationships of Malayan and Malagasy pygmy shrews of the genus Suncus (Soricomorpha: Soricidae) inferred from mitochondrial cytochrome b gene sequences. Raff Bull Zoo 2011; 59:237–243 [Google Scholar]
- Pattamadilok S, Lee BH, Kumperasart S, Yoshimatsu K, et al. Geographical distribution of hantaviruses in Thailand and potential human health significance of Thailand virus. Am J Trop Med Hyg 2006; 75:994–1002 [PubMed] [Google Scholar]
- Plyusnina A, Ibrahim IN, Plyusnin A. A newly recognized hantavirus in the Asian house rat (Rattus tanezumi) in Indonesia. J Gen Virol 2009; 90:205–209 [DOI] [PubMed] [Google Scholar]
- Poux C, Madsen O, Marquard E, Vieites DR, et al. Asynchronous colonization of Madagascar by the four endemic clades of primates, tenrecs, carnivores, and rodents as inferred from nuclear genes. Syst Biol 2005; 54 :719–730 [DOI] [PubMed] [Google Scholar]
- Reynes JM, Soares JL, Hüe T, Bouloy M, et al. Evidence of the presence of Seoul virus in Cambodia. Microbes Infect 2003; 5:769–773 [DOI] [PubMed] [Google Scholar]
- Rollin PE, Mathiot C, Nawrocka E, Ravaoalimalala VE, et al. [Hemorrhagic fever with renal syndrome in Madagascar. First seroepidemiologic survey of rat populations]. Arch Inst Pasteur Madagascar 1986; 52:181–186. [in French] [PubMed] [Google Scholar]
- Soarimalala V, Goodman SM. Les petits mammifères de Madagascar. Antananarivo (Madagascar): Association Vahatra, 2011 [Google Scholar]
- Sumibcay L, Kadjo B, Gu SH, Kang HJ, et al. Divergent lineage of a novel hantavirus in the banana pipistrelle (Neoromicia nanus) in Côte d'Ivoire. Virol J 2012; 9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollenaere C, Brouat C, Duplantier JM, Rahalison L, et al. Phylogeography of the introduced species Rattus rattus in the western Indian Ocean, with special emphasis on the colonization history of Madagascar. J Biogeogr 2010; 37:398–410 [Google Scholar]
- Weiss S, Witkowski PT, Auste B, Nowak K, et al. Hantavirus in bat, Sierra Leone. Emerg Infect Dis 2012; 18:159–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DE, Reeder DM. Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd ed. Baltimore: Johns Hopkins University Press, 2005, 2142 pp [Google Scholar]