Abstract
A large body of evidence links a high dietary intake of n–3 (ω-3) polyunsaturated fatty acids (PUFAs) with improved cardiometabolic outcomes. Recent studies suggested that the biologic processes underlying the observed associations may involve epigenetic changes, specifically DNA methylation. To evaluate changes in methylation associated with n–3 PUFA intake, we conducted an epigenome-wide methylation association study of long-chain n–3 PUFA intake and tested associations between the diabetes- and cardiovascular disease–related traits. We assessed DNA methylation at ∼470,000 cytosine–phosphate–guanine (CpG) sites in a cross-sectional study of 185 Yup’ik Alaska Native individuals representing the top and bottom deciles of PUFA intake. Linear regression models were used to test for the associations of interest, adjusting for age, sex, and community group. We identified 27 differentially methylated CpG sites at biologically relevant regions that reached epigenome-wide significance (P < 1 × 10−7). Specifically, regions on chromosomes 3 (helicase-like transcription factor), 10 (actin α 2 smooth muscle/Fas cell surface death receptor), and 16 (protease serine 36/C16 open reading frame 67) each harbored 2 significant correlates of n–3 PUFA intake. In conclusion, we present promising evidence of association between several biologically relevant epigenetic markers and long-term intake of marine-derived n–3 PUFAs.
Introduction
Dietary n–3 PUFAs, specifically fish- and marine mammal–derived EPA and DHA, are inversely associated with the risk of cardiovascular disease (1). At very high intakes, such as the ones observed in Arctic populations, EPA and DHA are not only associated with improved dyslipidemia and reduced chronic inflammation (2) but also attenuate the effects of elevated BMI on markers of cardiometabolic dysfunction, such as elevated TGs of C-reactive protein (3). The underlying biologic mechanisms are incompletely understood but are thought to include altered eicosanoid metabolism and subsequent changes in cell signaling, transcription factor activity, and gene expression (4). An emerging body of evidence in animal models suggests that long-chain n–3 PUFAs may also influence global DNA methylation patterns because of their role in one-carbon metabolism (5). Such epigenetic changes likely contribute to the observed n–3 PUFA effects on gene expression, dyslipidemia, and inflammation and warrant additional investigation in the context of chronic disease.
To evaluate the contributions of dietary long-chain n–3 PUFAs to DNA methylation across the human genome, we analyzed data from a population-based sample of the Yup’ik people living in Southwest Alaska. Because of the traditional diet rich in fish and other marine foods, our study population has mean intakes of EPA and DHA that surpass mean intakes of the general U.S. population by >20-fold (6–8), offering an unprecedented opportunity to investigate connections between dietary PUFAs, DNA methylation patterns, and known markers of disease. In this epigenome-wide study of EPA and DHA biomarkers, we set out to identify genomic regions in which DNA methylation is correlated with habitual intake of long-chain marine n–3 PUFAs. In turn, we hypothesized that such differentially methylated genomic regions are associated with intermediate phenotypes, such as plasma lipids or inflammatory cytokines, presenting a comprehensive nutrigenomic paradigm of protection from chronic disease.
Participants and Methods
Participant recruitment and ethics.
Since 2003, the Center for Alaska Native Health Research has recruited 1518 participants aged ≥14 y in 11 Southwest Alaska communities to investigate obesity and chronic disease risk factors in the Yup’ik people. Details on participant recruitment and procedures can be found in previous Center for Alaska Native Health Research publications (9, 10). Briefly, all community residents are invited to participate, and the age distribution reflects that of all eligible participants as shown by the 2000 U.S. census data. Participants provided written informed consent using protocols approved by the University of Alaska Institutional Review Board, the National and Alaska Area Indian Health Service Institutional Review Boards, and the Yukon Kuskokwim Human Studies Committee.
Study sample.
From the 1518 participants of the main study, 1499 had available biomarker data on n–3 PUFA intake, measured using the nitrogen stable isotope ratio (δ15N)10 as described below. Of those, 450 were in the bottom 3 deciles of PUFA intake, whereas 446 were in the top 3 deciles. We chose individuals from the top 3 deciles and the bottom 3 deciles of the δ15N intake distribution to represent traditional Yup’ik and modern U.S. dietary patterns, respectively. This “extreme phenotypes” approach had the additional benefit of increasing statistical power (11).
All participants consented to participate in the convenience sample for the epigenetic analyses. Of all individuals in the top 3 and the bottom 3 deciles of PUFA intake, we selected 192 unrelated participants for the methylation assay based on their δ15N decile and age. We excluded individuals if they were in the lowest decile of age (<21 y), not of Yup’ik descent, or lacked information on clinical covariates. Of 192 individuals selected for the epigenetic study, methylation was successfully measured on 185. Of these 185 individuals, 93 were in the lowest 3 deciles of n–3 PUFA intake (32, 42, and 19 for deciles 1, 2, and 3, respectively), and 92 were in the top 3 (30, 38, and 24 in deciles 8, 9, and 10, respectively). More than 98% of the 185 individuals were unrelated, and <1% were closer than 4th-degree relatives. Given the small sample size and the sparseness of relations, we did not correct our analyses for familial correlation.
Anthropometric and clinical measurements.
Anthropometric measurements, which included height, weight, percentage body fat, and waist circumference, were obtained using the NHANES III Anthropometric Procedures Manual (12). Participants fasted overnight before collecting blood samples. We quantified metabolic traits, including plasma lipoproteins, fasting insulin and glucose, and glycosylated hemoglobin (HbA1c), as described in previous publications from our group (13–15).
n–3 PUFA measurements.
We assessed dietary intake of marine-derived long-chain n–3 PUFAs using the δ15N of RBCs as described previously (14, 15). The life of an RBC is ∼120 d; therefore, we can use this validated biomarker to objectively measure moderately long-term n–3 PUFA intake in our study population. Conventionally, δ15N are expressed in per mil (‰) abundance of 15N relative to atmospheric nitrogen (15N/14Natm = 0.0036765) and calculated using the following equation:
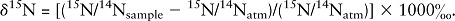 |
We tested analytical accuracy and precision by running laboratory reference materials (peptone, δ15N = 7.00); the resulting δ15N estimate was 7.0 ± 0.2‰ (mean ± SD). The range of isotopic variation in the data (9‰) was very large relative to analytical precision (0.2‰) (15).
DNA methylation measurements.
We quantified DNA methylation using the Infinium Human Methylation 450 array (Illumina, San Diego, CA). DNA was isolated using Puregene Blood Kits (Qiagen, Venlo, The Netherlands). Before the standard manufacturer protocol of amplification, hybridization, and imaging, we treated 500 ng of each DNA sample with sodium bisulfite (Zymo Research, Irvine, CA) (16). We used Illumina GenomeStudio software to estimate β scores, defined as the proportion of total signal from the methylation-specific probe or color channel, and detection P values, defined as the probability that the total intensity for a given probe falls within the background signal intensity. After measuring methylation on ∼470,000 autosomal cytosine–phosphate–guanine (CpG) sites, we removed any individual β scores with a detection P value > 0.01, all data from CpGs in which >10% of samples failed to yield adequate intensity, and all data from samples with >1.5% missing data points (16). Finally, we identified any CpGs in which the probe sequence mapped to either a location that did not match the annotation file or >1 locus and eliminated these markers from the analysis.
Statistical analyses.
All linear models that were fit included as covariates sex, age, and community group. The community group is a dichotomous variable that indicates proximity of the community to the coast as described previously (15). In addition, we included the microarray identification number (each array assays 12 samples) to account for possible batch effects at each CpG. Initial models used the logit-transformed methylation values similar to M-values as outcome variables (17) and included δ15N as a predictor and covariates described above to determine which loci were significantly associated with PUFA intake.
Additionally, the logit-transformed methylation values for significant loci obtained from the analysis above were then corrected for methylation principal components and batch effects in a linear model, and the residuals of these models were used as predictors in models for cardiovascular disease– and diabetes-related traits. We implemented stringent Bonferroni’s corrections to adjust for multiple testing, with the significance level set at 0.05/470,000 = 1.1 × 10−7 for the n–3 PUFA analyses and 0.05/27 = 0.002 for the chronic disease marker analyses.
Results
Demographic and clinical characteristics of the study population, stratified by intake of n–3 FAs, are summarized in Table 1. Community group was the strongest correlate of n–3 PUFA intake: compared with the inland communities, the coastal villages were more likely to be in the high δ15N group. Age and sex also exhibited statistically significant relations with marine-derived PUFA intake, with older individuals and women more likely to consume an n–3 PUFA-rich diet, although individuals in the high n–3 PUFA intake group had a marginally higher (P = 0.06) body fat percentage that did not translate to a difference in BMI, HbA1c, or fasting glucose. Individuals at the higher end of the δ15N distribution showed lower plasma TGs (P = 0.02) and higher HDL cholesterol (P = 0.003) concentrations but also borderline higher LDL (P = 0.07) and total (P = 0.06) cholesterol concentrations compared with individuals in the bottom 3 deciles of δ15N.
TABLE 1.
Demographic and clinical characteristics of the study sample by n–3 FA intake (n = 185)1
| High δ15N (n = 92) | Low δ15N (n = 93) | P value | |
| Age, y | 40 ± 1 | 36 ± 1 | 0.009 |
| Community group, n (%) | 4 × 10−12 | ||
| Coastal | 74 (80) | 28 (30) | |
| Upriver | 28 (20) | 65 (70) | |
| Male, n (%) | 37 (40) | 54 (58) | 0.02 |
| Current tobacco users, n (%) | 78 (85) | 77 (83) | 0.84 |
| BMI,2 kg/m2 | 28 ± 1 | 28 ± 1 | 0.57 |
| Body fat, % | 30 ± 2 | 27 ± 2 | 0.06 |
| Plasma TGs,3 mg/dL | 77 ± 4 | 103 ± 11 | 0.02 |
| Plasma LDL cholesterol, mg/dL | 146 ± 4 | 136 ± 4 | 0.07 |
| Plasma HDL cholesterol,2 mg/dL | 65 ± 2 | 58 ± 2 | 0.003 |
| Plasma total cholesterol, mg/dL | 227 ± 4 | 214 ± 5 | 0.06 |
| Plasma fasting glucose,3 mg/dL | 95 ± 1 | 93 ± 1 | 0.22 |
| Plasma glycosylated hemoglobin,3 % | 5.48 ± 0.03 | 5.45 ± 0.04 | 0.52 |
All values are means ± SEs or n (%). δ15N, nitrogen stable isotope ratio.
The variable was logit-transformed to achieve normality.
The variable underwent a Box-Cox transformation to achieve normality.
The results from the epigenome-wide analysis of δ15N are illustrated by the Manhattan plot in Supplemental Fig. 1. Of ∼470,000 autosomal markers that were tested, 27 have reached the Bonferroni’s threshold for epigenome-wide significance and are listed in Table 2. Despite the limited sample size, our findings show robust statistical significance, with P < 1 × 10−10 for 10 markers. Regions that harbored >1 statistically significant CpG were located on chromosomes 3 [helicase-like transcription factor (HLTF)], 10 [actin α 2 smooth muscle/Fas cell surface death receptor (ACTA2/FAS)], and 16 [protease serine 36 (PRSS36)/C16 open reading frame 67 (C16orf67)]. Interestingly, the direction of association within these 3 genomic regions was concordant across CpG sites, with n–3 PUFA intake mostly associated with increased methylation (78% of significant associations) (Table 2). Deviations from the expected distribution were assessed using a quantile–quantile plot (Supplemental Fig. 2). Sensitivity analyses that included information on tobacco use, an established predictor of methylation status, did not yield substantially different results.
TABLE 2.
Top cytosine–phosphate–guanine methylation sites associated with n–3 PUFA intake in the study population (n = 185)1
| Chromosome | Position | Marker | Gene | Genomic region | Direction of association | P |
| 1 | 201,625,499 | cg07999988 | NAV1 | — | + | <1 × 10−16 |
| 16 | 57,437,864 | cg02050165 | CCL17 | — | + | 8 × 10−16 |
| 10 | 124,905,105 | cg05144067 | — | North Shore | − | 5 × 10−15 |
| 3 | 148,804,275 | cg21926402 | HLTF | Island | − | 2 × 10−14 |
| 12 | 133,345,809 | cg07249343 | GOLGA3 | North Shelf | + | 8 × 10−14 |
| 1 | 12,633,465 | cg05230551 | DHRS3 | — | + | 1 × 10−12 |
| 3 | 148,804,272 | cg04836786 | HLTF | Island | − | 1 × 10−11 |
| 17 | 29,159,594 | cg05952098 | ATAD5 | South Shore | + | 2 × 10−11 |
| 11 | 64,794,859 | cg17101226 | SNX15 | Island | + | 3 × 10−11 |
| 6 | 7,313,623 | cg00520393 | SSR1 | Island | + | 5 × 10−11 |
| 5 | 434,998 | cg22951524 | AHRR | — | + | 1 × 10−10 |
| 2 | 171,571,716 | cg06272543 | SP5/LOC440925 | Island | − | 2 × 10−10 |
| 1 | 182,759,062 | cg15293611 | — | Island | + | 2 × 10−10 |
| 15 | 34,635,452 | cg22686731 | NOP10 | — | + | 3 × 10−10 |
| 1 | 150,601,613 | cg23912435 | ENSA | North Shore | + | 3 × 10−9 |
| 10 | 90,749,255 | cg06450397 | ACTA2/FAS | North Shore | + | 4 × 10−9 |
| 2 | 26,101,348 | cg19084174 | ASXL2 | Island | + | 5 × 10−9 |
| 3 | 148,804,477 | cg24041269 | HLTF | Island | − | 1 × 10−8 |
| 16 | 31,711,872 | cg16585380 | C16orf67 | Island | + | 1 × 10−8 |
| 15 | 40,226,279 | cg21830797 | EIF2AK4 | Island | + | 2 × 10−8 |
| 9 | 13,284,899 | cg21186560 | — | — | + | 2 × 10−8 |
| 20 | 331,827 | cg24601011 | NRSN2 | South Shelf | − | 4 × 10−8 |
| 10 | 90,750,580 | cg03672854 | ACTA2/FAS | Island | + | 5 × 10−8 |
| 6 | 21,666,569 | cg10086104 | FLJ22536 | North Shore | + | 6 × 10−8 |
| 3 | 51,705,097 | cg03855276 | TEX264 | Island | + | 6 × 10−8 |
| 1 | 43,855,450 | cg05704639 | C1orf84/MED8 | Island | + | 6 × 10−8 |
| 16 | 31,153,871 | cg07065683 | PRSS36 | Island | + | 8 × 10−8 |
ACTA2, actin α 2 smooth muscle; AHRR, aryl-hydrocarbon receptor repressor; ASXL2, additional sex combs-like 2; ATAD5, ATPase family AAA domain containing 5; C16orf67, C16 open reading frame 67; C1orf84, C1 open reading frame 84; CCL17, chemokine ligand 17; DHRS3, dehydrogenase/reductase SDR family member 3; EIF2AK4, eukaryotic translation initiation factor 2 α kinase 4; ENSA, endosulfine α FAS, Fas cell surface death receptor; FLJ22536, hypothetical locus LOC401237; GOLGA3, golgin A3; HLTF, helicase-like transcription factor; LOC440925, uncharacterized LOC440925; MED8, mediator complex subunit 8; NAV1, neuron navigator 1; NOP10, NOP10 ribonucleoprotein; NRSN2, neurensin 2; PRSS36, protease serine 36; SNX15, sorting nexin 15; SP5, SP5 transcription factor; SSR1, signal sequence receptor α TEX264, testis expressed 264; —, not available.
We then tested for associations between the 27 CpG sites that emerged as the statistically significant correlates of δ15N and a number of chronic disease risk factors, shown in Table 3. After adjusting for multiple testing, none of these associations reached statistical significance. However, a CpG in the aryl-hydrocarbon receptor repressor (AHRR) was suggestively negatively associated with fasting glucose (P = 0.004), and CpGs in the ACTA2/FAS and additional sex combs-like 2 (ASXL2) loci reached or approached nominal significance for LDL cholesterol (P = 0.07, 0.01, and 0.01, respectively) (Table 3). Specifically, the methylation status of both CpGs in ACTA2/FAS was positively associated with the lipid concentrations, whereas methylation of a CpG site in ASXL2 had an inverse association (Table 3). Additional adjustment for δ15N strengthened the associations with the methylation of ACTA2/FAS CpGs (P = 0.01 and 0.06), decreased the association between AHRR methylation and fasting glucose (P = 0.009), and did not appreciably alter the association between ASXL2 methylation and LDL cholesterol (P = 0.01); however, after implementing the Bonferroni’s correction, none of the δ15N-adjusted associations were significant (Table 3).
TABLE 3.
Associations between epigenetic correlates of n–3 PUFA intake and selected disease-related traits (n = 185)1
| HDL cholesterol |
LDL cholesterol |
Total cholesterol |
TGs |
HbA1c |
Fasting glucose |
||||||||
| CpG site | Gene | Adj-P2 | P3 | Adj-P | P | Adj-P | P | Adj-P | P | Adj-P | P | Adj-P | P |
| cg00520393 | SSR1 | 0.88 | 0.64 | 0.32 | 0.41 | 0.65 | 0.76 | 0.31 | 0.43 | 0.91 | 0.95 | 0.28 | 0.21 |
| cg02050165 | CCL17 | 0.36 | 0.28 | 0.30 | 0.35 | 0.74 | 0.81 | 0.73 | 0.63 | 0.96 | 0.98 | 0.05 | 0.04 |
| cg03672854 | ACTA2/FAS | 0.74 | 0.60 | 0.01 | 0.01 | 0.09 | 0.10 | 0.77 | 0.67 | 0.57 | 0.56 | 0.12 | 0.09 |
| cg03855276 | TEX264 | 0.66 | 0.75 | 0.22 | 0.25 | 0.35 | 0.38 | 0.72 | 0.66 | 0.94 | 0.94 | 0.02 | 0.02 |
| cg04836786 | HLTF | 0.45 | 0.62 | 0.26 | 0.31 | 0.40 | 0.47 | 0.55 | 0.67 | 0.41 | 0.43 | 0.83 | 0.94 |
| cg05144067 | — | 0.44 | 0.32 | 0.33 | 0.28 | 0.19 | 0.16 | 0.77 | 0.90 | 0.49 | 0.52 | 0.23 | 0.18 |
| cg05230551 | DHRS3 | 0.92 | 0.59 | 0.67 | 0.52 | 0.66 | 0.53 | 0.36 | 0.57 | 0.99 | 0.93 | 0.92 | 0.72 |
| cg05704639 | C1orf84/MED8 | 0.31 | 0.24 | 0.54 | 0.60 | 0.88 | 0.94 | 0.45 | 0.38 | 0.92 | 0.94 | 0.09 | 0.07 |
| cg05952098 | ATAD5 | 0.47 | 0.30 | 0.53 | 0.66 | 0.67 | 0.79 | 0.62 | 0.48 | 0.42 | 0.45 | 0.41 | 0.32 |
| cg06272543 | SP5/LOC440925 | 0.44 | 0.72 | 0.97 | 0.88 | 0.93 | 0.93 | 0.08 | 0.15 | 0.92 | 0.88 | 0.64 | 0.83 |
| cg06450397 | ACTA2/FAS | 0.74 | 0.88 | 0.06 | 0.07 | 0.85 | 0.90 | 0.86 | 0.95 | 0.65 | 0.64 | 0.12 | 0.10 |
| cg07065683 | PRSS36 | 0.82 | 0.75 | 0.43 | 0.40 | 0.35 | 0.33 | 0.54 | 0.60 | 0.20 | 0.20 | 0.30 | 0.29 |
| cg07249343 | GOLGA3 | 0.47 | 0.31 | 0.58 | 0.69 | 0.95 | 0.94 | 0.60 | 0.47 | 0.49 | 0.48 | 0.09 | 0.06 |
| cg07999988 | NAV1 | 0.36 | 0.20 | 0.27 | 0.37 | 0.82 | 0.96 | 0.98 | 0.78 | 0.31 | 0.33 | 0.72 | 0.89 |
| cg10086104 | FLJ22536 | 0.43 | 0.56 | 0.45 | 0.40 | 0.48 | 0.43 | 0.07 | 0.10 | 0.45 | 0.44 | 0.91 | 0.83 |
| cg15293611 | — | 0.22 | 0.11 | 0.67 | 0.53 | 0.54 | 0.43 | 0.41 | 0.27 | 0.66 | 0.70 | 0.70 | 0.92 |
| cg16585380 | C16orf67 | 0.39 | 0.41 | 0.34 | 0.33 | 0.60 | 0.59 | 0.31 | 0.32 | 0.72 | 0.72 | 0.63 | 0.61 |
| cg17101226 | SNX15 | 0.60 | 0.49 | 0.39 | 0.45 | 0.52 | 0.58 | 0.37 | 0.31 | 0.52 | 0.54 | 0.86 | 0.76 |
| cg19084174 | ASXL2 | 0.28 | 0.17 | 0.01 | 0.01 | 0.01 | 0.01 | 0.61 | 0.47 | 0.83 | 0.85 | 0.52 | 0.45 |
| cg21186560 | — | 0.85 | 0.66 | 0.49 | 0.58 | 0.50 | 0.58 | 0.63 | 0.52 | 0.82 | 0.79 | 0.23 | 0.30 |
| cg21830797 | EIF2AK4 | 0.58 | 0.49 | 0.66 | 0.71 | 0.88 | 0.93 | 0.97 | 0.89 | 0.64 | 0.66 | 0.43 | 0.38 |
| cg21926402 | HLTF | 0.49 | 0.31 | 0.22 | 0.30 | 0.66 | 0.78 | 0.96 | 0.85 | 0.08 | 0.09 | 0.76 | 0.93 |
| cg22686731 | NOP10 | 0.47 | 0.35 | 0.46 | 0.40 | 0.22 | 0.19 | 0.52 | 0.63 | 0.42 | 0.40 | 0.09 | 0.06 |
| cg22951524 | AHRR | 0.29 | 0.15 | 0.23 | 0.16 | 0.13 | 0.09 | 0.69 | 0.50 | 0.98 | 0.98 | 0.009 | 0.004 |
| cg23912435 | ENSA | 0.20 | 0.37 | 0.70 | 0.58 | 0.85 | 0.74 | 0.39 | 0.55 | 0.89 | 0.92 | 0.11 | 0.08 |
| cg24041269 | HLTF | 0.47 | 0.68 | 0.10 | 0.14 | 0.26 | 0.33 | 0.17 | 0.24 | 0.09 | 0.11 | 0.88 | 0.97 |
| cg24601011 | NRSN2 | 0.97 | 0.65 | 0.17 | 0.12 | 0.16 | 0.12 | 0.07 | 0.14 | 0.28 | 0.31 | 0.44 | 0.34 |
ACTA2, actin α 2 smooth muscle; Adj, adjusted; AHRR, aryl-hydrocarbon receptor repressor; ASXL2, additional sex combs-like 2; ATAD5, ATPase family AAA domain containing 5; C16orf67, C16 open reading frame 67; C1orf84, C1 open reading frame 84; CCL17, chemokine ligand 17; CpG, cytosine–phosphate–guanine; DHRS3, dehydrogenase/reductase SDR family member 3; EIF2AK4, eukaryotic translation initiation factor 2 α kinase 4; ENSA, endosulfine α FAS, Fas cell surface death receptor; FLJ22536, hypothetical locus LOC401237; GOLGA3, golgin A3; HbA1c, glycosylated hemoglobin; HLTF, helicase-like transcription factor; LOC440925, uncharacterized LOC440925; MED8, mediator complex subunit 8; NAV1, neuron navigator 1; NOP10, NOP10 ribonucleoprotein; NRSN2, neurensin 2; PRSS36, protease serine 36; SNX15, sorting nexin 15; SP5, SP5 transcription factor; SSR1, signal sequence receptor α TEX264, testis expressed 264; —, not available.
For all Adj-P columns, correlation adjusted for n–3 PUFA intake.
For all P columns, correlation unadjusted for n–3 PUFA intake.
Discussion
We present an epigenome-wide association study of n–3 PUFA intake, conducted in a Yup’ik population characterized by unique dietary PUFA variability. Our study links DNA methylation patterns in several genomic regions to n–3 PUFA intake, as well as fasting plasma glucose and LDL cholesterol. Thus, we provide preliminary evidence in support of the emerging hypothesis of nutritional regulation of epigenetic changes that may be involved in protection from metabolic and cardiovascular disease.
The identified CpG sites in or near neuron navigator 1 (NAV1), chemokine ligand 17 (CCL17), ACTA2/FAS, and AHRR are of particular biologic salience. The CpG site with the strongest association to δ15N is located proximally to a gene encoding the inactivation no afterpotential D-like protein (INADL) and contains a genetic variant estimated previously to explain 3% of the variance in body weight and body composition in Hispanic children (18). However, in our sample, the methylation of that site was not associated with BMI, body fat percentage (results not shown), or any other chronic disease marker, potentially suggesting actions of yet unknown compensatory mechanisms and warranting future investigation. The second most robustly associated CpG site is located in CCL17, which encodes a chemokine pivotal to regulating T-cell homeostasis that had been linked to atherosclerosis and other inflammatory conditions (19). Two studies of infants whose mothers were administered n–3 PUFA supplementation during pregnancy (20, 21) found that long-chain PUFAs reduce plasma Ccl17, attenuating the inflammatory response and providing a plausible biologic mechanism for our observed association. However, it is worth noting that the observed associations between CCL17 methylation and δ15N also did not translate into changes in chronic disease risk factors, possibly because of the limited sample size. In light of these findings and other studies from our group (3), future investigations in larger cohorts should consider including measures of C-reactive protein or other inflammatory cytokines to further elucidate the interplay between n–3 PUFA intake and CCL17 methylation in human health.
Two markers in the ACTA2/FAS cluster were significantly correlated with δ15N (P = 4 × 10−9 and 5 × 10−8) and suggestively associated with LDL cholesterol (P = 0.01 and 0.07). FAS is crucial to controlling apoptosis in the immune system. The Fas death receptor is expressed on T-cells within atherosclerotic lesions; in mice, Fas deficiency has been shown to promote systemic inflammation (22). A polymorphism located in the FAS promoter region has been linked to lipodystrophy in the context of HIV (23). It is possible that DNA methylation in that region, perhaps in concert with sequence variations, reduces FAS expression and regulates lipid metabolism through the apoptotic pathway. Future investigations may consider integrating genetic, epigenetic, and transcriptional data to further disentangle the observed relation.
Another finding of biologic interest emerged from a CpG site in AHRR, the differential methylation and expression of which have been linked recently to smoking status (24). However, our sensitivity analyses that adjusted for tobacco use (data not shown) did not appreciably change the estimate of association, suggesting involvement of alternative mechanisms. AHRR encodes the aryl-hydrocarbon receptor, a pleiotropic protein also implicated in carcinogenesis and oxidative stress pathways (25). In addition to its association with n–3 PUFA intake (P = 5 × 10−10), AHRR methylation was suggestively associated (P = 0.004) with fasting glucose in our data set. The positive effects of n–3 PUFA intake on insulin sensitivity and glucose tolerance have been well established, including in circumpolar populations (26). Our findings suggest that the underlying mechanisms may involve the oxidative stress pathway, in particular AHRR methylation and consequent changes in expression. Another finding linking carcinogenesis and n–3 PUFA intake is the differential methylation of 2 markers in HLTF, a tumor suppressor protein silenced in a variety of digestive tract cancers (27). Although in our population the methylation status of 1 of the HLTF sites was only suggestively associated (P = 0.09) with disease markers—particularly HbA1c—further studies of that region may produce valuable insights into the shared inflammatory mechanisms relevant to both cancer and metabolic disease.
Finally, we observed an association of the methylation status of a CpG site in ASXL2 with δ15N (P = 5 × 10−9), as well as marginal associations with LDL (P = 0.01) and total (P = 0.01) cholesterol. ASXL2 is located under the linkage peak for serum lipid concentrations identified in the Amish Family Study (28), and the observed association is likely because of the proximity to well-known lipid loci in apolipoprotein B (APOB) and ATP-binding cassette subfamily G member 8 (ABCG8). Because several studies, including 1 from our group (29), demonstrated that DNA methylation status is often influenced by the underlying sequence, future interrogations of that genomic region may consider genetic and epigenetic variation jointly to produce a more complete picture of the association observed in our study population.
The main strengths of this study are the use of a validated biomarker for EPA and DHA intake, the unparalleled range of marine-derived n–3 PUFA consumption among the Yup’ik people, the epigenome-wide coverage of the methylation array, and the biologically relevant nature of our most significant findings. Nevertheless, our results must be interpreted in light of several considerations. First, our study is cross-sectional and thus cannot be used to infer causality, because the temporal order of the relation between δ15N and DNA methylation is yet to be determined. Second, we do not have genotype or transcription information at the differentially methylated loci and are thus limited in our understanding of the underlying biologic mechanisms. Because our study represents preliminary data, future investigations based on our findings should include replication in independent populations and functional analyses of the top CpG hits and technical validation of the DNA methylation array in this study population. Third, because this is an investigation involving only a small sample of individuals, it is possible that some of the association findings did not reach statistical significance because of insufficient statistical power. However, the magnitude of the observed effects remains impressive despite the limited sample size, and our findings merit follow-up in larger, well-characterized cohorts. Fourth, a recent study from our group showed that n–3 PUFA consumption in the Yup’ik people is part of the larger subsistence dietary pattern (7, 30, 31), and thus our observed associations could be due to confounding by other dietary or lifestyle variables. Although we have taken careful steps to statistically account for this possibility, residual confounding remains a plausible explanation for our findings. Finally, because our study represents a hypothesis-free research approach, our findings must be replicated in an independent population to ensure validity.
In conclusion, we conducted an epigenome-wide search for CpG methylation sites associated with marine-derived EPA and DHA intake and discovered several biologically relevant regions with robust associations. By highlighting the most promising regions of the genome, our study lays the groundwork for future hypothesis-driven investigations of the role of n–3 PUFAs in epigenetic processes and metabolic health in human populations.
Acknowledgments
H.K.T., B.B.B., and D.M.A. designed the research; P.J.H., K.L.S., D.M.O., S.E.H., B.B.B., and D.M.A. conducted the research; D.M.A. and H.W.W. analyzed the data; S.A. wrote the paper; S.A., D.M.A., B.B.B., and H.K.T. had primary responsibility for the final content. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: ABCG8, ATP-binding cassette subfamily G member 8; ACTA2, actin α 2 smooth muscle; AHRR, aryl-hydrocarbon receptor repressor; APOB, apolipoprotein B; ASXL2, additional sex combs-like 2; C16orf67, C16 open reading frame 67; CCL17, chemokine ligand 17; CpG, cytosine–phosphate–guanine; FAS, Fas cell surface death receptor; HbA1c, glycosylated hemoglobin; HLTF, helicase-like transcription factor; INADL, gene encoding the inactivation no afterpotential D-like protein; NAV1, neuron navigator 1; PRSS36, protease serine 36; δ15N, nitrogen stable isotope ratio.
Literature Cited
- 1.Wang C, Harris WS, Chung M, Lichtenstein AH, Balk EM, Kupelnick B, Jordan HS, Lau J. n-3 Fatty acids from fish or fish-oil supplements, but not alpha-linolenic acid, benefit cardiovascular disease outcomes in primary- and secondary-prevention studies: a systematic review. Am J Clin Nutr. 2006;84:5–17 [DOI] [PubMed] [Google Scholar]
- 2.Makhoul Z, Kristal AR, Gulati R, Luick B, Bersamin A, Boyer B, Mohatt GV. Associations of very high intakes of eicosapentaenoic and docosahexaenois acids with biomarkers of chronic disease risk among Yup’ik Eskimos. Am J Clin Nutr. 2010;91:777–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makhoul Z, Kristal AR, Gulati R, Luick B, Bersamin A, O’Brien D, Hopkins SE, Stephensen CB, Stanhope KL, Havel PH, et al. Associations of obesity with triglycerides and C-reactive protein are attenuated in adults with high red blood cell eicosapentaenoic and docosahexaenoic acids. Eur J Clin Nutr. 2011;65:808–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simopoulos AP. Omega-3 fatty acids in inflammation and autoimmune diseases. J Am Coll Nutr. 2002;21:495–505 [DOI] [PubMed] [Google Scholar]
- 5.Kulkarni A, Dangat K, Kale A, Sable P, Chavan-Gautam P, Joshi S. Effects of altered maternal folic acid, vitamin B12 and docosahexaenoic acid on placental global DNA methylation patterns in Wistar rats. PLoS One `2011;10;6:e17706. [DOI] [PMC free article] [PubMed]
- 6.Johnson JS, Nobmann ED, Asay E, Lanier AP. Dietary intake of Alaska native people in two regions and implications for health: the Alaska Native Dietary and Subsistence Food Assessment Project. Int J Circumpolar Health. 2009;68:109–22 [DOI] [PubMed] [Google Scholar]
- 7.Bersamin A, Luick BR, King IB, Stern JS, Zidenberg-Cherr S. Westernizing diets influence fat intake, red blood cell fatty acid composition, and health in remote Alaskan Native Communities in the Center for Alaska Native Health Study. J Am Diet Assoc. 2008;108:266–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Brien DM, Kristal AR, Jeannet MA, Wilkinson MJ, Bersamin A, Luick B. Red blood cell delta15N: a novel biomarker of dietary eicosapentaenoic acid and docosahexaenoic acid intake. Am J Clin Nutr. 2009;89:913–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer BB, Mohatt GV, Lardon C, Plaetke R, Luick BR, Hutchison SH, Mayolo GA, Ruppert E, Bersamin A. Building a community-based participatory research center to investigate obesity and diabetes in Alaska Natives. Int J Circumpolar Health. 2005;64:281–90 [DOI] [PubMed] [Google Scholar]
- 10.Mohatt GV, Plaetke R, Klejka J, Luick BR, Lardon C, Bersamin A, Hopkins SE, Dondanville M, Herron J, Boyer BB. The Center for Alaska Native Health Research Study: a community-based participatory research study of obesity and chronic disease-related protective and risk factors. Int J Circumpolar Health. 2007;66:8–18 [DOI] [PubMed] [Google Scholar]
- 11.Barnett IJ, Lee S, Lin X. Detecting rare variant effects using extreme phenotype sampling in sequencing association studies. Genet Epidemiol. 2013;37:142–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lohman TG, Roche AF. Anthropometric standardization reference manual. Champaign, IL: Human Kinetics Books; 1988.
- 13.Boyer BB, Mohatt GV, Plaetke R, Herron J, Stanhope KL, Stephensen C, Havel PJ; CANHR Project Team Metabolic syndrome in Yup’ik Eskimos: the Center for Alaska Native Health Research (CANHR) Study. Obesity (Silver Spring). 2007;15:2535–40 [DOI] [PubMed] [Google Scholar]
- 14.Lemas DJ, Wiener HW, O’Brien DM, Hopkins S, Stanhope KL, Havel PJ, Allison DB, Fernandez JR, Tiwari HK, Boyer BB. Genetic polymorphisms in carnitine palmitoyltransferase 1A gene are associated with variation in body composition and fasting lipid traits in Yup’ik Eskimos. J Lipid Res. 2012;53:175–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aslibekyan S, Vaughan LK, Wiener HW, Lemas DJ, Klimentidis YC, Havel PJ, Stanhope KL, O’Brien DM, Hopkins SE, Boyer BB, et al. doi: 10.1002/ajhb.22429. Evidence for novel genetic loci associated with metabolic traits in Yup’ik people. Am J Hum Biol. 25:673-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Absher DM, Li X, Waite LL, Gibson A, Roberts K, Edberg J, Chatham WW, Kimberly RP. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and remodeling of CD4+ T-cell populations. PLoS Genet. 2013;9:e1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA, Butte NF. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One. 2012;7:e51954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber C, Meiler S, Doring Y, Koch M, Drechsler M, Megens RT, Rowinska Z, Bidzhekov K, Fecher C, Ribechini E, et al. CCL17-expressing dendritic cells drive atherosclerosis by restraining regulatory T cell homeostasis in mice. J Clin Invest. 2011;121:2898–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romero VC, Somers EC, Stolberg V, Clinton C, Chensue S, Djuric Z, Berman DR, Treadwell MC, Vahratian AM, Mozurkewich E. Developmental programming for allergy: a secondary analysis of the Mothers, Omega-3, and Mental Health Study. Am J Obstet Gynecol. 2013;208:e1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furuhjelm C, Jenmalm MC, Falth-Magnusson K, Duchen K. Th1 and Th2 chemokines, vaccine-induced immunity, and allergic disease in infants after maternal ω-3 fatty acid supplementation during pregnancy and lactation. Pediatr Res. 2011;69:259–64 [DOI] [PubMed] [Google Scholar]
- 22.de Claro RA, Zhu X, Tang J, Morgan-Stevenson V, Schwartz BR, Iwata A, Liles WC, Raines EW, Harlan JM. Hematopoietic Fas deficiency does not affect experimental atherosclerotic lesion formation despite inducing a proatherogenic state. Am J Pathol. 2011;178:2931–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zanone Poma B, Riva A, Nasi M, Cicconi P, Broggini V, Lepri AC, Mologni D, Mazzotta F, Monforte AD, Mussini C, et al. Genetic polymorphisms differently influencing the emergence of atrophy and fat accumulation in HIV-related lipodystrophy. AIDS. 2008;22:1769–78 [DOI] [PubMed] [Google Scholar]
- 24.Shenker NS, Polidoro S, van Veldhoven K, Sacerdote C, Ricceri F, Birrell MA, Belvisi MG, Brown R, Vineis P, Flanagan JM. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identified novel genetic loci associated with smoking. Hum Mol Genet. 2013;22:843–851 [DOI] [PubMed] [Google Scholar]
- 25.Cheng YH, Huang SC, Lin CJ, Cheng LC, Li LA. Aryl hydrocarbon receptor protects lung adenocarcinoma cells against cigarette sidestream smoke particulates-induced oxidative stress. Toxicol Appl Pharmacol. 2012;259:293–301 [DOI] [PubMed] [Google Scholar]
- 26.Ebbesson SO, Risica PM, Ebbesson LO, Kennish JM, Tejero ME. Omega-3 fatty acids improve glucose tolerance and components of the metabolic syndrome in Alaskan Eskimos: the Alaska Siberia Project. Int J Circumpolar Health. 2005;64:396–408 [DOI] [PubMed] [Google Scholar]
- 27.Hibi K, Nakayama H, Kanyama Y, Kodera Y, Ito K, Akiyama S, Nakao A. Methylation pattern of HLTF gene in digestive tract cancers. Int J Cancer. 2003;104:433–436 [DOI] [PubMed] [Google Scholar]
- 28.Pollin TI, Hsueh WC, Steinle NI, Snitker S, Shuldiner AR, Mitchell BD. A genome-wide scan of serum lipid levels in the Old Order Amish. Atherosclerosis. 2004;173:89–96 [DOI] [PubMed] [Google Scholar]
- 29.Zhi D, Aslibekyan S, Irvin MR, Claas SA, Borecki IB, Ordovas JM, Absher DM, Arnett DK. doi: 10.4161/epi.25501. SNPs located at CpG sites modulate genome-epigenome interaction. Epigenetics. 2013;8:802-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bersamin A, Zidenberg-Cherr S, Stern JS, Luick BR. Nutrient intakes are associated with adherence to a traditional diet among Yup’ik Eskimos living in remote Alaska Native communities: the CANHR study. Int J Circumpolar Health. 2007;66:62–70 [DOI] [PubMed] [Google Scholar]
- 31.Nash SH, Bersamin A, Kristal AR, Hopkins SE, Church RS, Pasker RL, Luick BR, Mohatt GV, Boyer BB, O’Brien DM. Stable nitrogen and carbon isotope ratios indicated traditional and market food intake in an indigenous circumpolar population. J Nutr. 2012;142:84–90 [DOI] [PMC free article] [PubMed] [Google Scholar]


