Abstract
Tomato and lycopene (ψ, ψ-carotene) consumption is hypothesized to protect against nonalcoholic steatohepatitis and hepatocarcinogenesis, processes that may depend upon diet and gene interactions. To investigate the interaction of tomato or lycopene feeding with β-carotene-9′,10′-monooxygenase (Bco2) on hepatic metabolic and signaling pathways, male wild-type (WT) and Bco2−/− mice (3-wk-old; n = 36) were fed semi-purified control, 10% tomato powder–containing, or 0.25% lycopene beadlet–containing diets for 3 wk. Serum lycopene concentrations were higher in lycopene- and tomato-fed Bco2−/− mice compared with WT (P = 0.03). Tomato- and lycopene-fed mice had detectable hepatic apolipoprotein (apo)-6′-, apo-8′-, and apo-12′-lycopenal concentrations. Hepatic expression of β-carotene-15,15’-monooxygenase was increased in Bco2−/− mice compared with WT (P = 0.02), but not affected by diet. Evaluation of hepatic gene expression by focused quantitative reverse transcriptase-polymerase chain reaction arrays for nuclear receptors and coregulators (84 genes) and stress and metabolism (82 genes) genes indicates that tomato feeding affected 31 genes (≥1.5-fold, P < 0.05) and lycopene feeding affected 19 genes, 16 of which were affected by both diets. Lycopene down-regulation of 7 nuclear receptors and coregulators, estrogen-related receptor-α, histone deacetylase 3, nuclear receptor coactivator 4, RevErbA-β, glucocorticoid receptor, peroxisome proliferator-activated receptor (PPAR)-α, and PPAR-γ, coactivator 1 β was dependent upon interaction with Bco2 status. Lycopene and tomato feeding induced gene expression patterns consistent with decreased lipid uptake, decreased cell proliferation and mitosis, down-regulated aryl hydrocarbon receptor signaling, and decreased expression of genes involved in retinoid X receptor heterodimer activation. Tomato feeding also caused expression changes consistent with down-regulation of DNA synthesis and terpenoid metabolism. These data suggest tomato components, particularly lycopene, affect hepatic gene expression, potentially affecting hepatic responses to metabolic, infectious, or chemical stress.
Introduction
Emerging evidence suggests that consumption of tomato or its predominant red carotenoid, lycopene (ψ, ψ-carotene), may protect the liver from oxidative injury, nonalcoholic fatty liver disease (NAFLD)10, and NAFLD-driven precancerous lesions (1). NAFLD frequently coincides with metabolic syndrome, affecting as many as 30% of American adults and up to 90% of the morbidly obese, predisposing them to nonalcoholic steatohepatitis (NASH) and, consequently, hepatic failure or hepatocellular carcinoma (2). Lycopene is most concentrated in the liver (3), compared with other organs, and evidence suggests intact lycopene or its cleavage products may have protective properties against metabolic stressors.
Lycopene and tomato feeding were associated with protective activities in preclinical models of NASH (4) and hepatocarcinogenesis (5). In rodents fed a high-fat diet, lycopene feeding increased expression and activity of the antioxidant enzymes catalase and glutathione reductase, while decreasing lipid peroxidation (6) and hepatic steatosis (4). Both lycopene and tomato extract feeding also decreased the number of precancerous lesions, proinflammatory cytokine mRNA expression, and markers of hepatocyte proliferation in rats fed a high-fat diet with NASH-promoted, diethylnitrosamine-initiated hepatocarcinogenesis (5). Evidence suggests lycopene may affect transcriptional regulatory and cell signaling pathways independent of the antioxidant functions of lycopene (7). However, whether intact lycopene or its metabolites are responsible for these activities remains poorly understood.
Previously, we and others identified several lycopene cleavage products both in human plasma and in lycopene-containing foods, including raw tomato, processed tomato products, and watermelon (8, 9), suggesting these metabolites are absorbed and/or are endogenously produced. Emerging in vitro and in vivo evidence has documented that the eccentric carotenoid cleavage enzyme, β-carotene-9′,10′-oxygenase (BCO2), and the central carotenoid cleavage enzyme, β-carotene-15, 15′-monooxygenase (BCMO1), may be involved in lycopene metabolism (10–12). In particular, Bco2 ablation generally increases tissue and blood lycopene concentrations in mice, whereas ablation of Bcmo1 decreases hepatic lycopene concentrations (13–15), and gene polymorphisms of BCMO1 affect blood lycopene concentrations (16). Nonetheless, we are just beginning to understand the relation between these enzymes and tissue or blood lycopene concentrations.
It is well-known that the downstream products of β-carotene (β,β-carotene) central cleavage, all-trans and some cis-retinoic acid isomers, serve as activating ligands for homodimeric or heterodimeric nuclear receptor complexes such as retinoid X receptor (RXR)/RXR, retinoic acid receptor (RAR)/RXR, and RXR/PPARs (17), and it was recently discovered that several of the eccentric cleavage products of β-carotene antagonize RAR (18). In vitro and in vivo studies indicate that lycopene and/or its metabolites may act via nuclear receptors and transcription systems including PPARs, liver X receptor, and nuclear factor erythroid derived 2 (NRF2) to induce downstream physiologic changes (19–24). In particular, apo-8′-lycopenal acts via NRF2 to affect phase II enzyme expression in the liver (24), and apo-10′-lycopenoic acid activated luciferase activity associated with RAR binding of the retinoic acid response element (1). Moreover, the lycopene metabolite apo-10′-lycopenoic acid was recently shown to interrupt diethylnitrosamine-initiated, high-fat-diet–promoted hepatocarcinogenesis, further suggesting lycopene metabolites are bioactive in attenuating hepatic disease (12). However, whether lycopene or its BCO2-generated metabolites affect hepatic nuclear receptor expression, thus altering the orchestration of expression profiles (beyond those just mentioned) to change downstream biologic processes remains unknown.
To expand our understanding of the biologic processes in the liver affected by lycopene, BCO2-generated lycopene metabolites, as well as to compare and contrast lycopene alone with tomato feeding, we focused on hepatic lycopene metabolism and expression patterns of metabolism- and nuclear signaling–related genes. We first tested the hypothesis that the Bco2 genotype affects hepatic lycopene, lycopene isomers, and apo-lycopenal concentrations in tomato and lycopene-fed mice by examining Bco2−/− and wild-type (WT) mice. We further hypothesized that tomato and lycopene feeding would have many overlapping effects on gene expression, but because of its larger array of phytochemicals, tomato might have a broader effect than feeding lycopene alone. Using a gene network analysis tool, we integrated the observed expression changes to identify patterns consistent with altered metabolic and signaling pathways. We anticipate that these studies may provide insights into the mechanisms whereby BCO2 interacts with dietary tomato or lycopene to affect liver biology.
Materials and Methods
Animals, diets, and experimental design.
All animal protocols and procedures were reviewed and approved by the Ohio State University Institutional Animal Care and Use Committee to ensure the humane and ethical conduct of animal research. The generation and acquisition of Bco2−/− mice were previously described (25). The Bco2−/− (B6; 129S6-Bcdo2tm1Dnp) and WT mice were bred at the Ohio State University animal facility, and genotype was confirmed from genomic DNA with the Extract-N-Amp Tissue PCR kit (Sigma-Aldrich). A total of 66 3-wk-old Bco2−/− and WT male mice were randomly assigned to experimental powdered diets (n = 11/group; total of 6 groups): control (AIN-93G semi-purified diet; Research Diets) or diets supplemented with either 10% (w:w) tomato powder (Futureceuticals) or 0.25% (w:w) RediVivo (10% lycopene) beadlets (DSM). All diets contained placebo beadlets (DSM) (Supplemental Table 1). Tomato- and lycopene-supplemented diets were formulated to contain 250 mg of lycopene · kg diet−1 and delivered 384 and 462 mg of lycopene · kg−1 diet, respectively (Supplemental Table 1).The selected tomato powder diet amount was previously shown to be cancer-protective in rodents and to result in tissue and plasma concentrations achievable by humans (3, 26–28). This amount of tomato powder has not been previously shown to induce xenobiotic responses in rats (26), although some evidence suggests a stress response to carotenoids may depend on BCO2 status (25). Diets were stored in the dark at −20°C and were provided every other day. After 3 wk of feeding, mice were killed by CO2 and blood was collected by cardiac puncture. Liver was collected and either snap-frozen in liquid nitrogen and stored at −80°C for carotenoid and molecular analyses or prepared for histology.
Carotenoid extraction and HPLC analysis.
Tomato carotenoids in tomato powder were quantitated as previously described (29). Serum and liver lycopene quantification was performed using HPLC as previously described (14).
Hepatic and dietary apo-lycopenal analysis.
Frozen hepatic tissue (500 mg) was pulverized (Cellcrusher), suspended in ethanol (5 mL) containing 0.1% (w/v) BHT, probe-sonicated, and centrifuged (300 × g, 5 min), and the supernatant was reserved. The tissue was extracted 3 times more with hexane:acetone (1:1, 5 mL), and extracts were pooled and mixed with deionized water (10 mL) and saturated NaCl (200 μL). The organic hexane layer was reserved and concentrated under nitrogen gas. Extracts were stored at −80°C for no more than 3 h before reconstitution in methanol:methyl tert-butyl ether (200 μL, 1:1) and centrifugation immediately prior to analysis by HPLC-tandem MS. Dietary apo-lycopenals were extracted in the same manner but without pulverization.
Hepatic and dietary apo-lycopenals were separated using an Acquity ultra-high–pressure liquid chromatography system (Waters) with a YMC C30 analytical column (4.6 × 150 mm, 5 μm; Waters) held at 40°C. Reconstituted samples were held at 27°C before 30-μL injection. A gradient elution method (1.8 mL/min) using mobile phase A [80:18:2 methanol:water:2% ammonium acetate (aq.) (v:v:v)] and mobile phase B [78:20:2 methyl tert-butyl ether:methanol:2% ammonium acetate (aq.)], increased linearly from 100% A to 100% B over 6 min, was held for 2 min and returned to 100% A over 2 min. The ultra-high–pressure liquid chromatography system was interfaced with a Quattro Ultima triple quadrupole mass spectrometer (Micromass UK) with atmospheric pressure chemical ionization operated in negative ion mode without flow splitting. Instrumental parameters included the following: corona current, 30 μA; cone voltage, 35 V; desolvation gas flow, 435 L/hr; source temperature, 110°C; and probe temperature, 500°C. Apo-lycopenals were identified and quantitated using the following selected tandem MS transitions: m/z 310.2 > 241.2 (apo-14′-lycopenal), 350.26 > 281.2 (apo-12′-lycopenal), 376.28 > 307.2 (apo-10′-lycopenal), 416.3 > 347.3 (apo-8′-lycopenal), and 442.32 > 373.3 (apo-6′-lycopenal). Apo-lycopenals were quantitated by external calibration curves generated from authentic standards (Carotenature) as previously described (8).
Gene expression analysis.
Total RNA was isolated from liver (n = 5) using the TRIzol Reagent (Invitrogen) method and RNeasy Mini Kit (Qiagen) per the manufacturer’s instructions, including an on-column DNase treatment. Two pathway-focused RT2 Profiler PCR arrays, the Stress and Toxicity PathwayFinder and the Nuclear Receptors and Coregulators PCR arrays (SABiosciences), examined 173 genes (including normalizer genes) (Supplemental Tables 2 and 3), using 400 ng of total RNA for the first strand cDNA synthesis reaction. RT-PCR was performed on an ABI 7900 HT (Applied Biosystems) using SYBR Green (universal cycling conditions: 95°C, 10 min; 95°C, 15 s; and 60°C, 1 min; repeated for 40 cycles). For data analysis, traditional housekeeping genes did not meet criteria to serve as biologic normalizers (they were changed by experimental treatment and/or had high within-group variability); therefore, normalizing genes were selected from the qRT-PCR array gene sets based upon the following criteria: expression amounts above the minimum limits of detection (35 cycles), low variability within treatment groups, and good stability across treatment groups (P > 0.05 by ANOVA). Briefly, ΔCt for each gene was calculated by normalizing to either Rxrb (nuclear receptor and co-regulator array) or the average of glutathione peroxidase 1 (Gpx1) and cyclin G1 (Ccng1) (stress and toxicity array). Fold-differences were calculated relative to the controls using the comparative Ct method. Single-gene, qRT-PCR (1 μg of total RNA) was performed using SYBR Green to quantify relative expression of Bcmo1 (NM_021486): forward-5′-ATGGAGATAATATTTGGCCAG-3′ and reverse-5′-AACTCAGACACCACGATTC-3′ Bco2 (NM_1332217): forward-5′-GTTATCTACTTCGAGTTGGACCTGG-3′ and reverse-5′-AAGCAACGCCATTCCATCA-3′. Single-gene qRT-PCR data were normalized to the geometric mean of Ccng1, Gpx1, and Rxrb.
Immunohistochemical analysis.
Liver tissue was immediately fixed in 10% neutral buffered formalin overnight, processed, and paraffin-embedded. Tissue sections were baked (1 h, 60°C) in the hybridization oven after deparaffinization, and rehydration antigen retrieval was performed (20 min, 1× CitraPlus Antigen Retrieval Solution; Biogenex). Tissue sections were incubated with PPARγ antibody (D69, Rabbit rabbit polyclonal, dilution diluted 1:300; Cell Signaling Technology), and immunohistochemically stained by endogenous H2O2 quenching for 10 min, primary antibody for 60 min, Envision + HRP-M (Dako) for 30 min, and 3,3′-diaminobenzidine staining for 4 min, and were counterstained with Mayer’s hematoxylin.
Liver TG analysis.
Briefly, liver was homogenized in 10× (v:w) ice-cold lysis buffer (20 nM Trizma base, 1% Triton ×100, 50 nM NaCl, 250 nM sucrose, 50 nM NaF, 5 nM Na4P2O7-10H2O). Homogenates were incubated at 4°C for 1 h prior to FA extraction, and liver TG concentrations were measured using the Triglyceride Assay Kit (Cayman Chemical Company) per the manufacturer’s instructions.
Statistical analysis.
Because of unequal variance between groups, hepatic lycopene concentrations were compared using the nonparametric Kruskal-Wallis test, and genotype within diet treatments and genotype effects across diet treatments were compared by the Mann-Whitney rank sum test. Serum lycopene concentrations, percentages of serum cis-lycopene, body weights, and gene expression ΔCt data were compared among 6 experimental groups (2 genotypes × 3 diets) by 2-factor ANOVA; post hoc comparisons were made by the Holm-Sidak test using SigmaPlot software (Systat Software) (α = 0.05). Gene expression data were log-transformed for analyses and the anti-logs taken on parameter estimates to provide estimates of fold-differences. Expression data that did not meet the assumption for homogeneity of variance after normalization and transformation were not analyzed further. The Ingenuity Pathway Analysis (IPA) (Ingenuity Systems) web-based application was used to simultaneously analyze gene expression fold-change and pair-wise P value (from post hoc Holm-Sidak analysis of ΔCt expression) data from the arrays to identify predicted pathways and biologic functions affected by lycopene or tomato feeding based on previously published relations. Pair-wise comparisons were made in IPA between lycopene vs. control feeding and tomato vs. control feeding and were considered biologically and statistically significant when ≥1.5-fold different and P ≤ 0.05. Canonical pathways affected by lycopene or tomato feeding that plausibly occur in the liver were viewed as significant at P ≤ 0.0004 (calculated as a Bonferroni-type correction for these data α/total number of canonical pathway = 0.05/116). Non-organ–specific biologic functions that can be inferred from hepatic gene transcription were considered significant when the Z-score was ≥2. To assess the contribution of diet, genotype, and/or hepatic lycopene concentrations on resultant hepatic apo-lycopenal concentrations, linear models were employed (α = 0.05), starting with 3 variables (genotype, diet, and hepatic lycopene concentration) and 3 interaction terms (genotype × diet, diet × hepatic lycopene, and genotype × hepatic lycopene). The linear model used does not require equal group sizes, which was important, because hepatic tissue availability for apo-lycopenal analyses was limited after completion of other assays. The full model was simplified using hierarchic elimination of nonsignificant factors (α = 0.05) to reach a final model, and apo-lycopenal data that did not meet assumptions for multiple linear regression were ln-transformed. The correlation between total lycopene and the percentage of cis-lycopene for serum and hepatic lycopene was measured using the Pearson’s product-moment correlation test.
Results
Body weights
Mouse body weight (final: 21.8 ± 0.3 g) was not substantially affected by either diets or genotype.
Serum and liver intact carotenoid concentrations
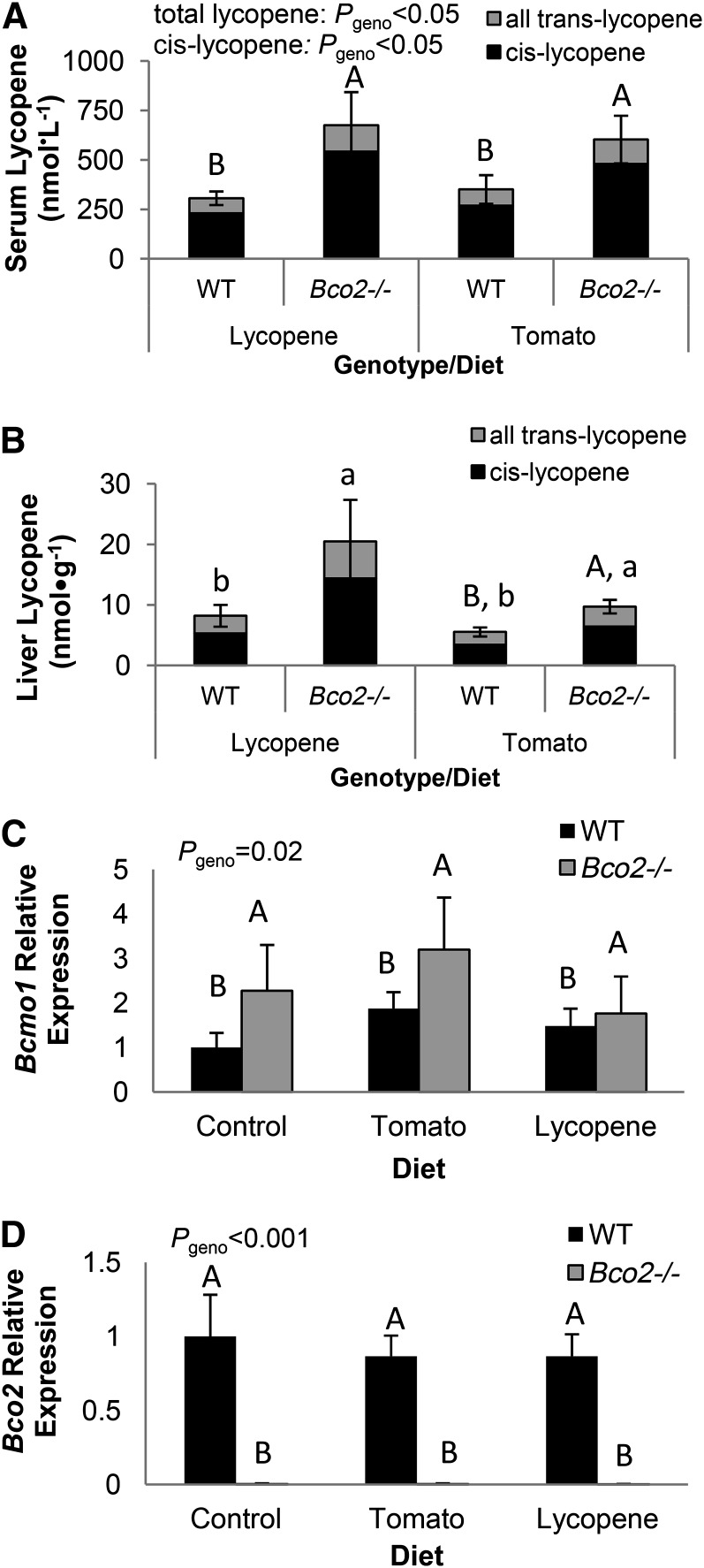
Lycopene was not detected in the serum of control diet–fed mice but was present in both tomato- and lycopene-fed WT mice, with substantially higher concentrations in Bco2−/− mice (Fig. 1A). Hepatic total lycopene was substantially higher in Bco2−/− mice fed either tomato- or lycopene-containing diets compared with WT mice fed the same respective diets (Fig. 1B), and none was detected in control-fed mice. Although the relative percentages of hepatic cis-lycopene were significantly higher in Bco2−/− mice (68 ± 1%) than in WT mice (63 ± 1%) (P = 0.004), the proportion of cis-lycopene was significantly correlated with total lycopene (r = 0.67, P = 0.002). Serum cis-lycopene was substantially higher in lycopene-fed Bco2−/− mice compared with other tomato- and lycopene-fed Bco2−/− and WT mice (Fig. 1A), and the percentage of cis-lycopene (74 ± 2%) was higher in Bco2−/− mice than in WT mice (68 ± 1%) (P = 0.05). However, the percentages of cis-lycopene significantly correlated with total lycopene (r = 0.63, P = 0.03). Phytoene, phytofluene, β-carotene, and ζ-carotene were detected only in tomato-fed mouse livers. Hepatic phytofluene was not altered by the absence of BCO2, and concentrations of phytoene, β-carotene, and ζ-carotene were below those necessary for precise quantification and evaluation of genotype impact (data not shown).
FIGURE 1.
Serum (A) and hepatic (B) lycopene concentrations and hepatic relative mRNA expression of Bcmo1 (C) and Bco2 (D) in tomato- or lycopene-fed WT and Bco2−/− mice. Bars represent means ± SEMs, n = 3 (A) or 5 (B–D). Differing capital letters indicate significant differences by genotype (A). Hepatic lycopene concentrations differed by genotype within tomato-fed groups (P < 0.02) (indicated by differing capital letters), and hepatic lycopene was greater in Bco2−/− vs. WT mice (P < 0.025) across lycopene- and tomato-fed groups (indicated by differing lowercase letters) (B). Bcmo1 and Bco2 expressions are shown as fold-change (2-ΔΔ Ct) compared with the WT, control-fed group, and differences by genotype are indicated by differing capital letters (C and D). Bcmo1, β-carotene-15, 15′-monooxygenase; Bco2, β-carotene-9′,10′-oxygenase; geno, genotype; WT, wild-type.
Hepatic Bcmo1 and Bco2 gene expression
Bco2−/− mice expressed higher hepatic Bcmo1 than WT mice, but diet did not substantially alter Bcmo1 expression (Fig. 1C). The absence of Bco2 expression in Bco2−/− mice was confirmed (Fig. 1D). Bco2 mRNA expression was unchanged in WT mice by diet.
Tomato bioactive components and genotype modulate hepatic nuclear receptor and coactivator and stress- and metabolism-related gene expression
Gene expression altered by genotype.
In these healthy mice, subjected to neither dietary nor environmental stressors, few of the 175 genes assayed were affected by Bco2 genotype. However, interleukin-18 (Il18) was strongly down-regulated [P < 0.001; relative expression (RE) = 0.41] in Bco2−/− mice, and crystalline-α B (Cryab) (P = 0.02, RE = 0.72) and estrogen-related receptor-γ (Esrrg) (P = 0.05, RE = 0.74) were modestly down-regulated. Expression was modestly up-regulated for RevErbA-β (Nr1d2) (P = 0.03, RE = 1.19) and histone deacetylase (Hdac) 5 (P = 0.03, RE = 1.17) in Bco2−/− mice compared with WT.
Gene expression altered by diet.
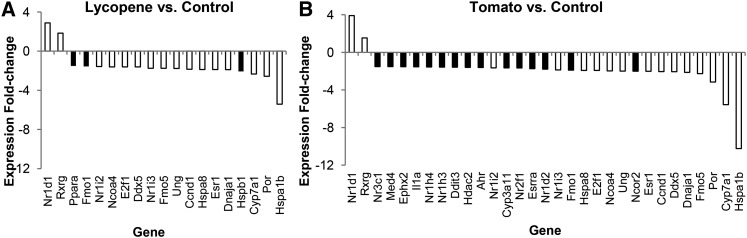
Dietary treatment affected 29 nuclear receptor–related genes and 30 stress-related genes; tomato and lycopene feeding down-regulated many of the same genes (Supplemental Table 4). Compared with control feeding, tomato biologically affected 31 genes, whereas lycopene affected 19 genes, with 16 of these genes being affected by both lycopene and tomato (Fig. 2).
FIGURE 2.
The effect of (A) lycopene vs. control diet feeding and (B) tomato vs. control diet feeding in WT and Bco2−/− mice on hepatic stress– and nuclear receptor–related gene expression. Data are significant fold-changes (>1.5-fold, P < 0.05); black bars represent gene expression significantly changed by either tomato or lycopene feeding, and white bars represent genes changed by both tomato and lycopene feeding. Values are means, n = 9–10. Bco2, β-carotene-9′,10′-oxygenase; WT, wild-type. Refer to Supplemental Tables 2 and 3 for definitions of gene symbols in Fig. 2.
Bco2 genotype–dependent dietary regulation of stress- and nuclear receptor signaling–related gene expression.
A number of important interactions between diet and BCO2 were detected in stress- and metabolism-related genes (Table 1). Nuclear receptor and coregulator gene expression was affected by lycopene and tomato feeding in a genotype-specific manner. Tomato-induced up-regulation of androgen receptor (Ar) was dependent on Bco2 presence and was the only instance of Bco2-dependent up-regulation of gene expression by diet. Lycopene-induced down-regulation of estrogen-related receptor-α (Esrra), Hdac3, nuclear receptor coactivator (Ncoa) 4, Nr1d2, peroxisome proliferator-activated receptor-α (Ppara), Ppar-γ, coactivator 1 β (Ppargc1b), and glucocorticoid receptor (Nr3c1) was dependent on the presence of Bco2, whereas tomato decreased expression of these 7 genes independently of Bco2 status.
TABLE 1.
Genotype-specific dietary regulation of nuclear receptor and stress- and toxicity-related gene expression in livers of −/− and WT male mice fed control, tomato powder, or lycopene beadlet diets for 3 wk1
| Control |
Tomato |
Lycopene |
|||||
| Gene | Overall P value for diet × genotype interaction | WT | Bco2–/– | WT | Bco2–/– | WT | Bco2–/– |
| Fold of Control-WT | Fold of Control-WT | Fold of Control-WT | |||||
| Nuclear receptors and coregulators | |||||||
| Ar | 0.048 | 1 | 1.26 | 1.24a | 0.59b | 0.61 | 0.77 |
| Esrra | 0.009 | 1 | 0.94 | 0.6 | 0.52 | 0.54d | 0.89c |
| Hdac3 | 0.033 | 1 | 0.94 | 0.67 | 0.71 | 0.67d | 1.06c |
| Ncoa4 | 0.045 | 1 | 0.89 | 0.48 | 0.48 | 0.44d | 0.71c |
| Nr1d2 | 0.003 | 1 | 0.96 | 0.56 | 0.53 | 0.41f | 0.93e |
| Nr3c1 | 0.031 | 1 | 0.82 | 0.6 | 0.61 | 0.54d | 0.87c |
| Ppara | 0.019 | 1 | 0.99 | 0.71 | 0.63 | 0.51d | 0.78c |
| Ppargc1b | 0.003 | 1 | 0.82 | 1.11 | 0.91 | 0.56f | 1.26e |
| Stress- and toxicity-related | |||||||
| Cryab | 0.016 | 1 | 0.75 | 0.98 | 1 | 1.26e | 0.58f |
| Hspa5 | 0.003 | 1 | 1.24 | 1 | 1.11 | 1.39c | 0.73d |
| Cdkn1a | 0.014 | 1 | 1.55 | 0.57 | 0.69 | 2.12c | 0.51d |
| Cyp1b1 | 0.009 | 1 | 0.99 | 0.70b | 1.50a | 1.15a | 0.43b |
| Egr1 | 0.026 | 1 | 0.38 | 0.37b | 1.24a | 0.4 | 0.4 |
| Por | 0.004 | 1 | 1.12 | 0.48a | 0.19b | 0.19b | 0.47a |
| Ung | 0.003 | 1 | 0.92 | 0.48 | 0.48 | 0.29f | 0.67e |
Values are means, n = 4–5. Genotype interaction, P < 0.05, and significant genotype within diet group expression difference (>50%). Labeled means in a row without a common letter differ: a,b, P < 0.05; c,d, P < 0.01; and e,f, P < 0.001. Ar, androgen receptor; Bco2, β-carotene-9′,10′-oxygenase; Cdkn1a, cyclin-dependent kinase inhibitor 1a; Cryab, crystalline-α B; Cyp1b1, cytochrome P450 1b1; Egr1, early growth response 1; Esrra, estrogen-related receptor-α Hdac3, histone deacetylase 3; Hspa5, heat shock protein 70kDa protein 5; Ncoa4, nuclear receptor coactivator 4; Nr1d2, RevErbA-β Nr3c1, glucocorticoid receptor; Por, P450 (cytochrome) oxidoreductase; Ppara, peroxisome proliferator-activated receptor-α Ppargc1b, peroxisome proliferator-activated receptor-γ, coactivator 1 β Ung, uracil DNA glycosylase; WT, wild-type.
Changes in gene expression in response to diet were genotype-dependent for 10 genes encoding liver metabolic- and stress-related genes (Table 1). Lycopene feeding up-regulated the expression of cyclin-dependent kinase inhibitor 1a (Cdkn1a), Cryab, and heat shock protein 70kDa protein 5 (Hspa5) in WT mice but not in Bco2−/− mice. The expression of uracil DNA glycosylase (Ung) in response to lycopene feeding and early growth response 1 (Egr1) to tomato feeding were also dependent on BCO2. Lycopene and tomato feeding had divergent effects in the gene expression of cytochrome P450 (Cyp) 1b1 and P450 (cytochrome) oxidoreductase (Por) in response to diet. As anticipated, the qRT-PCR array results were in agreement with single-gene qRT-PCR analyses for several test genes with representative expression patterns, including Rev-ErbA-α (Nr1d1), Cyp7a1, and Por (data not shown) because the qRT-PCR array is simply simultaneous analysis of multiple genes using traditional qRT-PCR technology.
IPA of dietary effects
Predicted canonical pathways substantially affected by diet.
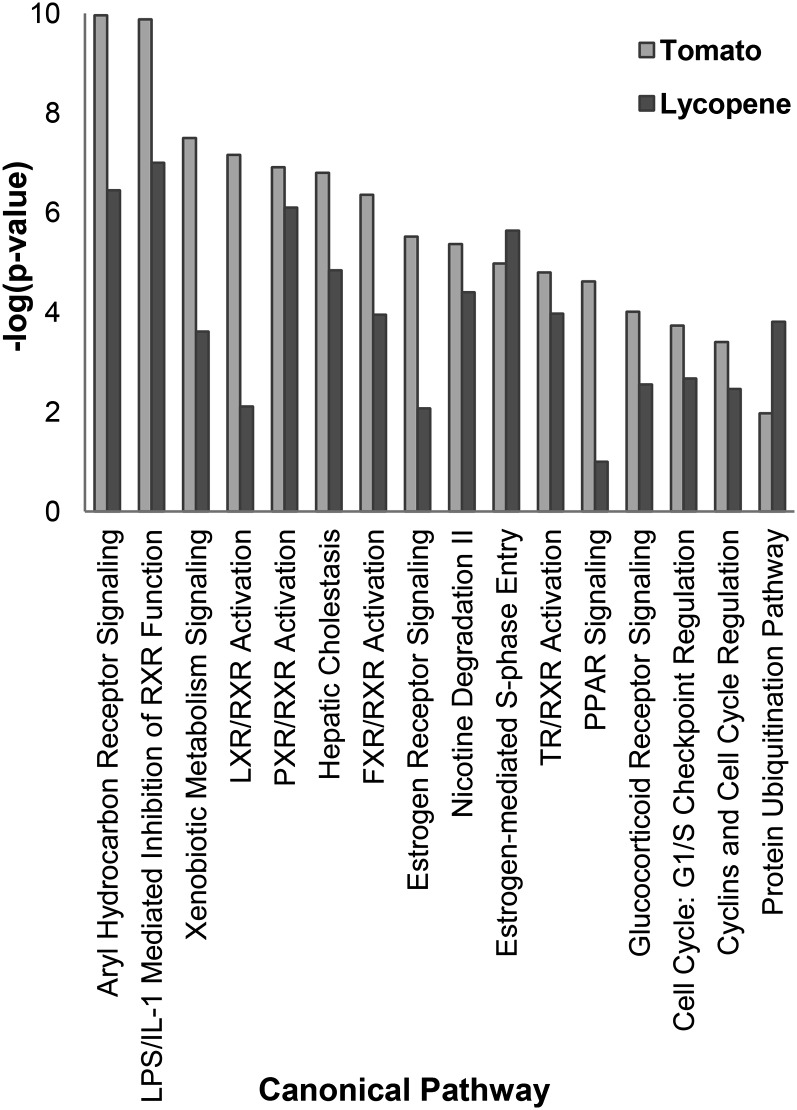
Diet-induced changes in gene expression patterns integrated by IPA showed that 16 of 116 calculated canonical pathways were regulated by both tomato and/or lycopene feeding (Fig. 3; Supplemental Table 5). For 14 of 16 (88%), P values indicated that tomato feeding was more strongly associated with a change in the pathway than lycopene feeding.
FIGURE 3.
Top 16 IPA-calculated canonical pathway liver gene expression changes imputed to tomato or lycopene feeding [α = 0.0004 or −log(P value) = 3.4] in Bco2−/− and WT mice. Bco2, β-carotene-9′,10′-oxygenase; FXR, farnesoid X receptor; IPA, Ingenuity Pathway Analysis; LXR, liver X receptor; PXR, pregnane X receptor; RXR, retinoid X receptor; TR, thyroid hormone receptor; WT, wild-type.
Biologic functions predicted to be changed by diet.
Several biologic functions were identified by IPA as likely downstream effects of tomato and/or lycopene feeding. Tomato feeding induced expression changes associated with decreased terpenoid metabolism, typically mediated through the 3-hydroxy-3-methylglutaryl-coenzymeA reductase pathway, and decreased steroid metabolism (P = 6.6 × 10−12; Z = −2.49 and P = 3.4 × 10−11; Z = −2.324, respectively). Tomato feeding was also associated with decreased mitosis (P = 3.4 × 10−4; Z = −2.22), decreased epithelial cell proliferation (P = 4.7 × 10−5; Z = −2.18), and decreased DNA synthesis (P = 1.8 × 10−5; Z = −2.02). Lycopene feeding was correlated with decreased lipid uptake (P = 8.3 × 10−7; Z = −2.15), decreased liver and general cell proliferation (P = 1.6 × 10−6, Z = −2.20; P = 1.1 × 10−4, Z = −2.23, respectively), and decreased mitosis (P = 2.3 × 10−4; Z = −2.22). The Bco2 genotype affected too few genes to impute changes in canonical pathways or biologic functions.
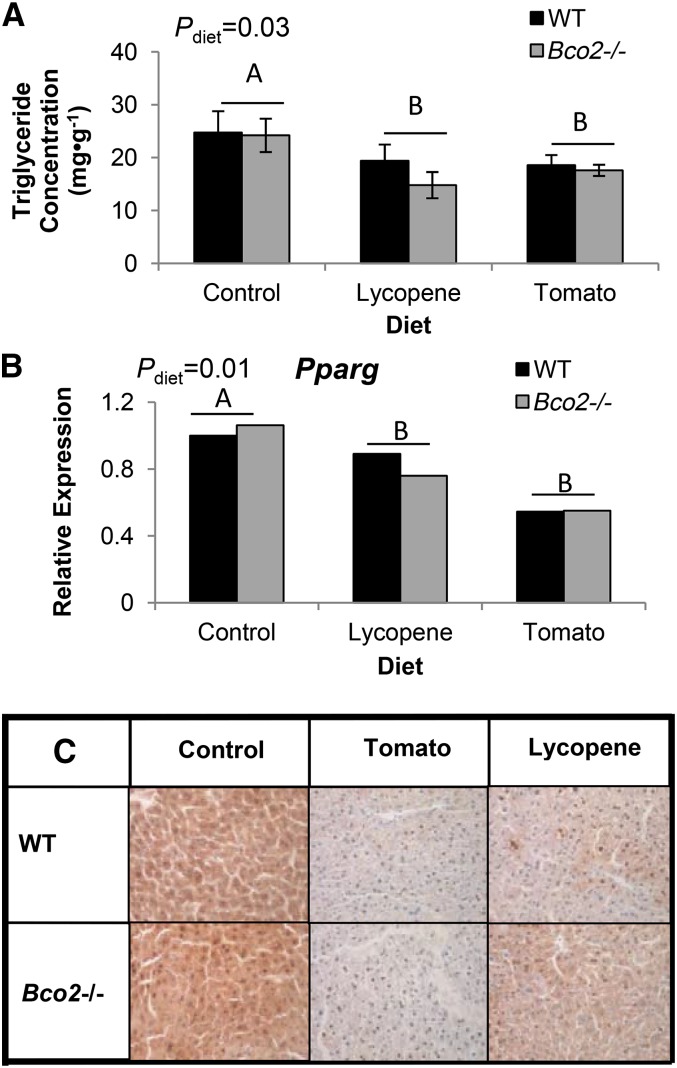
Hepatic TG concentrations and PPARγ protein and mRNA expression
Hepatic TG concentrations were lower in tomato- and lycopene-fed mice compared with control-fed mice, regardless of genotype (Fig. 4A). Tomato and lycopene feeding decreased both Pparg expression and cytoplasmic staining of PPARγ in tomato- and lycopene-fed mice compared with control-fed mice (Fig. 4B, C).
FIGURE 4.
Hepatic triglyceride concentrations (A), Pparg expression (B), and PPARγ staining (C, 400× magnification) in Bco2−/− and WT male mice fed control, tomato powder, or lycopene beadlet diets for 3 wk. Bars represent means ± SEMs, n = 3–5. Significant diet main effects on hepatic triglyceride concentrations and Pparg expression are indicated by differing capital letters (A and B). Bco2, β-carotene-9′,10′-oxygenase; Pparg, peroxisome proliferator-activated receptor-γ WT, wild-type.
Dietary and hepatic apo-lycopenals
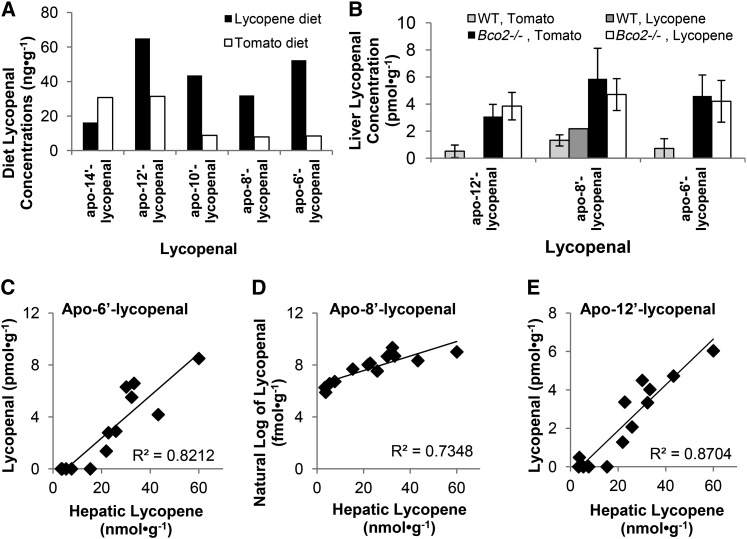
Several apo-lycopenals, from apo-14′- to apo-6′-lycopenal were detected in both the tomato- and lycopene-containing diets. The concentrations and percentages varied between the diets (Fig. 5), with higher percentages of the longer-chained lycopenals in the lycopene diet. Apo-12′-, apo-8′-, and apo-6′-lycopenals were detected in the livers of tomato-fed WT and Bco2−/− mice, as well as lycopene-fed Bco2−/− mice, whereas only apo-8′-lycopenal was detected in a lycopene-fed WT mouse. Apo-lycopenal concentrations were generally higher in Bco2−/− mice (Fig. 5), and apo-6′-lycopenal and apo-12′-lycopenal were correlated with hepatic lycopene concentrations with a weaker correlation between ln transformed apo-8′-lycopenal and hepatic lycopene concentration. Based on our linear model, hepatic intact lycopene was an important predictor for concentrations of all 3 apo-lycopenals. In addition, diet type influenced apo-6′-lycopenal, a diet × hepatic lycopene interaction influenced apo-8′-lycopenal, and genotype-influenced apo-12′-lycopenal (Table 2).
FIGURE 5.
Dietary (A) and hepatic (B) apo-lycopenal concentrations, and the linear relations between hepatic apo-lycopenal and intact lycopene concentrations (C–E) in Bco2−/− and WT male mice fed tomato powder or lycopene beadlet diets for 3 wk. Values are the means of duplicate analyses (A). Values are means ± SDs, n = 4, except WT, lycopene-fed mice, where n = 1 (B). Bco2, β-carotene-9′,10′-oxygenase; WT, wild-type.
TABLE 2.
Regression coefficients for factors affecting hepatic apo-lycopenal concentrations in −/− and WT male mice fed control, tomato powder, or lycopene beadlet diets for 3 wk1
| Model | β (SE) | P |
| Apo-6′-lycopenal2 (pmol · g−1) | ||
| Constant | −1.15 (0.62) | 0.095 |
| Hepatic lycopene (nmol · g−1) | 0.16 (0.02) | <0.001 |
| Genotype | 0.61 (0.38) | 0.145 |
| Diet | −0.83 (0.33) | 0.035 |
| Apo-8′-lycopenal3 (ln of fmol · g−1) | ||
| Constant | 1.56 (0.25) | <0.001 |
| Hepatic lycopene (nmol · g−1) | 0.06 (0.01) | <0.001 |
| Genotype | 0.13 (0.15) | 0.441 |
| Diet | 0.48 (0.24) | 0.080 |
| Hepatic lycopene × diet interaction | −0.03 (0.01) | 0.013 |
| Apo-12′-lycopenal4 (pmol · g−1) | ||
| Constant | −0.28 (0.40) | 0.491 |
| Hepatic lycopene (nmol · g−1) | 0.10 (0.02) | <0.001 |
| Genotype | 0.58 (0.25) | 0.041 |
| Diet | −0.17 (0.21) | 0.441 |
Bco2, β-carotene-9′,10′-oxygenase; WT, wild type.
Adjusted r2 = 0.87.
Adjusted r2 = 0.85.
Adjusted r2 = 0.90.
Discussion
In this study of Bco2−/− and WT mice fed either lycopene- or tomato-containing diets or a control diet for 3 wk, we report that tomato and lycopene feeding have widespread effects on hepatic metabolic, stress, nuclear receptor, and nuclear coregulator gene expression, of which some are substantially dependent upon the Bco2 genotype. Integration of the gene expression data identified a number of pathways and possible downstream biologic functions that are related to hepatic disease prevention and are very consistent with emerging themes derived from previous in vitro and in vivo studies. Furthermore, we found the percentage of cis-lycopene and apo-6′-, apo-8′-, and apo-12′-lycopenal concentrations in liver tissue to be in proportion to total hepatic lycopene concentrations. This may suggest that hepatic lycopene may be isomerized and cleaved to yield these specific lycopenals either non-enzymatically or by enzymes other than BCO2, which should be the subject of future mechanistic studies.
The first objective of the study was to determine if the BCO2 genotype affects hepatic lycopene, lycopene isomer, or apo-lycopenal concentrations. The absence of BCO2 resulted in increased serum and liver lycopene concentrations with no change in phytofluene concentrations, which we have previously seen (14, 15). Serum and liver lycopene concentrations may be elevated in Bco2−/− mice because of slower lycopene metabolism and clearance; however, differences in absorption and tissue distribution cannot be completely excluded. It has been suggested that cis-lycopene isomers are the preferred substrates for BCO2 (1). In this study, the percentages of serum and hepatic lycopene present as cis-isomers were higher in Bco2−/− mice than WT mice, yet the percentage of cis-lycopene was also significantly correlated with total lycopene concentrations. Therefore, it cannot be definitively determined whether Bco2−/− mice have higher cis-lycopene because of decreased BCO2 cleavage of cis-isomers or as a consequence of BCO2-independent isomerization processes associated with higher concentrations of lycopene.
Several observations suggested that apo-lycopenals might be generated via pathways independent of BCO2. First, apo-lycopenals were found in the livers of both Bco2−/− and WT mice. Second, hepatic lycopene concentration but not genotype was the most important predictive factor (Table 2) of apo-lycopenal concentrations. Third, apo-10′-lycopenal, a proposed product of BCO2 cleavage (10) was not detectable in liver tissue of WT mice, suggesting this may not be a major BCO2 product in the liver. In addition to non-enzymatic or enzymatic production of apo-lycopenals, they may be absorbed from the diet; indeed small amounts of lycopenals are spontaneously formed in food products and were found in the lycopene-containing diets fed to our mice. Regression analyses indicated that hepatic apo-6′- and apo-8′-lycopenal concentrations were influenced by diet type, suggesting that the food may be one source in addition to in vivo production. Recently, it was reported that BCMO1 may centrally cleave lycopene to yield acycloretinal (apo-15′-lycopenal) (11). In our study, Bcmo1 expression was higher in Bco2−/− mice, and because we did not measure the central-cleavage product of lycopene in the liver, this analysis should be undertaken in the future. We did observe that the apo-lycopenal distribution differed between the diets, possibly because of compound stability differences in the 2 diet matrices. Differences in apo-lycopenal profiles between those found in the diet versus in liver, however, suggest that absorption, production, or clearance rates of these apo-lycopenals are contributory. Consistent with our previous findings (14), it does not appear that tissue lycopene or apo-lycopenal concentrations may influence the expression of either Bcmo1 or Bco2 (Fig. 1C, D).
Next, we examined the effects of tomato and lycopene on gene expression (Fig. 2), with tomato affecting expression of more genes than lycopene alone. However, the substantial overlap for gene expression patterns between lycopene- and tomato-fed mice indicates that a major proportion of the biologic effect of tomato feeding is due to lycopene, but certainly other phytochemicals contribute as well. IPA indicated tomato- and lycopene-driven expression patterns consistent with altered canonical pathway signaling (Fig. 3), with marked predicted impacts on aryl hydrocarbon signaling and RXR heterodimer activation pathways. Both lycopene- and tomato-induced expression changes were highly consistent with depressed aryl hydrocarbon signaling. A recent study indicated that aryl hydrocarbon receptor (AHR) activation promotes NASH in mice exposed to NASH-promoting diets (30); thus, lycopene or tomato feeding may act via AHR signaling to reduce NASH progression. Tomato and/or lycopene feeding also substantially decreased gene expression in the RXR heterodimer activation pathways as well as in the LPS-IL-1 mediated inhibition of RXR function pathway. Additional molecular studies will be necessary to determine if lycopene derivatives agonize or antagonize RXR function or perturb other steps of RXR homo- or heterodimer–regulated pathways, as proposed by Sharoni et al. (7). Lastly, tomato and/or lycopene feeding also induced expression changes consistent with decreased mitosis, DNA synthesis, and cell proliferation, which are all supported by previous reports as well (27, 28, 31–34). Future studies should determine the effects of tomato and lycopene on these specific canonical pathways to define the implications for hepatic disease risk.
Expression changes induced by lycopene and tomato feeding in nuclear receptor–related and stress- and metabolism-related genes were consistent with downstream changes in lipid metabolism/homeostasis-related biologic functions, which align with previous reports. First, in the current study, tomato feeding down-regulated expression of genes encoding proteins that inhibit the PPAR signaling canonical pathway, an avenue of active investigation (19, 20, 35). The down-regulation of the PPAR inhibitors may have resulted from decreased Pparg expression and PPARγ protein expression because of tomato feeding (Fig. 4B, C). Previously, β-carotene feeding also was found to down-regulate PPARγ expression and protein activity, which can contribute to both increased lipolysis and fat mobilization (35). Second, lycopene feeding was correlated with decreased lipid uptake, which we confirmed by hepatic TG analysis, and has also been previously observed with β-carotene feeding (35). Decreased lipid uptake may play a role in attenuating NASH. Therefore, further investigation of the interface between carotenoid metabolites, PPARγ gene expression, lipid metabolism, and hepatic steatosis is warranted.
Nuclear receptor signaling controls adaptive responses to dietary variation and to stressors (36). Our results suggest that the effect of diet on nuclear receptor expression was affected by Bco2 genotype. For example, the effect of tomato feeding on Ar expression was dependent on Bco2 genotype such that Ar expression was higher in tomato-fed WT mice and lower in tomato-fed Bco2−/− mice compared with WT, control-fed mice. Increased hepatic Ar expression has been correlated with lower hepatocellular carcinoma risk (37), suggesting a role for BCO2-mediated maintenance of hepatic Ar expression by dietary tomato. The expression of 7 other nuclear receptors and coactivators was down-regulated by lycopene only when BCO2 was present, whereas tomato feeding down-regulated the expression of these 7 genes independent of Bco2 status. This pattern may indicate that the interaction between Bco2 and lycopene reduces expression of these factors, whereas the complex mixture of phytochemicals present in tomato reduces their expression independent of Bco2 status. The impacted nuclear receptors are involved in diverse roles including circadian liver metabolic regulation (Esrra) (38), mitochondrial metabolism (Ppargc1b) (39), inflammation and immunity (Nr3c1) (40), carcinogenesis (Nr3c1, Esrra) (41), mediation of steroid receptor–driven gene transcription (Ncoa2 and Ncoa4) (National Center for Biotechnology Information gene database), heme-mediated transcription repression (Nr1d2) (42), and gene transcription silencing by histone deacetylation (Hdac3). The effect of lycopene and tomato feeding on several stress- and metabolism-related genes was also Bco2-dependent (Table 1), but not in the consistent pattern seen with the nuclear receptors and coactivators. These novel observations warrant the investigation of how lycopene interacts with BCO2 to affect nuclear receptor and coactivator expression.
Unexpectedly, 2 marked, diet-independent effects of Bco2 ablation were observed. First, interestingly, Bco2−/− mice expressed substantially lower Il18; a previous genome-wide association scan found several single nucleotide polymorphisms in Bco2 to significantly correlate with circulating IL-18 (30). Second, Bco2 ablation led to substantially higher Bcmo1 expression, as previously reported (15), supporting a cross-regulatory relation between Bcmo1 and Bco2. These findings are in addition to the known BCO2 roles related to carotenoid metabolism, which include preventing xanthophyll-induced mitochondrial stress (25) and controlling carotene deposition in the fat, milk, and tissues of animals (14, 43).
The current study’s strengths include the use of the Bco2−/− mice to identify changes in hepatic gene expression in response to tomato and lycopene feeding and the use of focused qRT-PCR arrays in combination with literature-based functional and network analysis to more broadly compare the effects to tomato, lycopene, and BCO2 genotype on expression of genes in hypothesized functional gene groups than single-gene qRT-PCR allows. More information can be gained in the future with bigger group sizes for apo-lycopenal analyses and use of animal models of NASH and NAFLD. Although our hepatic apo-lycopenal post hoc analyses cannot unequivocally determine the role of BCO2 in apo-lycopenal generation because of limited group sizes, it does seem that factors other than BCO2, including hepatic intact lycopene concentrations and diet type, contribute to resultant hepatic apo-lycopenal concentrations. Furthermore, the current study was conducted in healthy mice, and future studies to investigate the interactions between BCO2 and tomato or lycopene in a model of NASH and NAFLD should be pursued.
In summary, we found that the impact of lycopene or tomato feeding on multiple biologic signaling pathways was largely overlapping; however, instances where tomato feeding had stronger biologic impacts than lycopene alone were dominant, a finding supported by clinical and preclinical trials (27, 44, 45). A number of gene expression changes related to hepatic stress and nuclear signaling were affected by tomato and lycopene feeding, and some of these changes were dependent on Bco2. The Bco2-dependence of lycopene and independence of tomato on down-regulation of nuclear factor expression provides support for future intervention trials and public health recommendations focused on whole foods compared with single pharmacologic agents. Finally, we found that total lycopene, the proportion of cis-lycopene, and apo-lycopenals were higher in Bco2−/− mice, but the underlying mechanisms of these differences warrant further investigation. Biologic pathways predicted to be affected by tomato or lycopene feeding and/or BCO2-generated metabolites should be studied to define how tomato or lycopene feeding are protective in the context of NAFLD and NASH.
Acknowledgments
The authors thank Johannes von Lintig (Case Western Reserve) for providing the Bco2−/− breeding pairs and Chureeporn (Julie) Chitchumroonchokchai [Ohio State University (OSU)] for assisting in TG analyses. Lycopene metabolite analyses were conducted at the OSU Comprehensive Cancer Center (OSU-CCC) Nutrient and Phytochemical Analytic Shared Resource and qRT-PCR samples were analyzed at the OSU-CCC Nucleic Acid Shared Resource (directed by S.J.S. and co-directed by K.M.R.). H.-L.T., N.E.M., J.M.T.-A., K.M.R., M.J.C., S.J.S., J.W.E., and S.K.C. designed the research; H.-L.T., N.E.M., M.J.C., J.M.T.-A., and S.K.C. conducted the research; and N.E.M., H.-L.T., J.M.T.-A., D.K.P., M.J.C., and S.K.C. analyzed the data and prepared the manuscript. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: AHR, aryl hydrocarbon receptor; Ar, androgen receptor; BCMO1, β-carotene-15, 15′-monooxygenase; BCO2, β-carotene-9′,10′-oxygenase; Ccng1, cyclin G1; Cdkn1a, cyclin-dependent kinase inhibitor 1a; Cryab, crystalline-α B; Cyp, cytochrome P450; Egr1, early growth response 1; Esrra, estrogen-related receptor-α Esrrg, estrogen-related receptor-γ Gpx1, glutathione peroxidase 1; Hdac, histone deacetylase; Hspa5, heat shock protein 70kDa protein 5; Il18, interleukin-18; IPA, Ingenuity Pathway Analysis; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; Ncoa, nuclear receptor coactivator; Nr1d1, Rev-ErbA-α Nr1d2, RevErbA-β NRF2, nuclear factor erythroid derived 2; Por, P450 (cytochrome) oxidoreductase; Ppar, peroxisome proliferator-activated receptor; RAR, retinoic acid receptor; RE, relative expression; RXR, retinoid X receptor; Ung, uracil DNA glycosylase; WT, wild-type.
Literature Cited
- 1.Ip BC, Wang XD. Non-alcoholic steatohepatitis and hepatocellular carcinoma: implications for lycopene intervention. Nutrients. 2013;6:124–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Starley BQ, Calcagno CJ, Harrison SA. Nonalcoholic fatty liver disease and hepatocellular carcinoma: a weighty connection. Hepatology. 2010;51:1820–32 [DOI] [PubMed] [Google Scholar]
- 3.Moran NE, Erdman JW, Jr, Clinton SK. Complex interactions between dietary and genetic factors impact lycopene metabolism and distribution. Arch Biochem Biophys. 2013;539:171–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn J, Lee H, Jung CH, Ha T. Lycopene inhibits hepatic steatosis via microRNA-21-induced downregulation of fatty acid-binding protein 7 in mice fed a high-fat diet. Mol Nutr Food Res. 2012;56:1665–74 [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Ausman LM, Greenberg AS, Russell RM, Wang XD. Dietary lycopene and tomato extract supplementations inhibit nonalcoholic steatohepatitis-promoted hepatocarcinogenesis in rats. Int J Cancer. 2010;126:1788–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi SK, Seo JS. Lycopene supplementation suppresses oxidative stress induced by a high fat diet in gerbils. Nutr Res Pract. 2013;7:26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharoni Y, Linnewiel-Hermoni K, Zango G, Khanin M, Salman H, Veprik A, Danilenko M, Levy J. The role of lycopene and its derivatives in the regulation of transcription systems: implications for cancer prevention. Am J Clin Nutr. 2012;96:1173S–8S [DOI] [PubMed] [Google Scholar]
- 8.Kopec RE, Riedl KM, Harrison EH, Curley RW, Jr, Hruszkewycz DP, Clinton SK, Schwartz SJ. Identification and quantification of apo-lycopenals in fruits, vegetables, and human plasma. J Agric Food Chem. 2010;58:3290–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez EB, Rodriguez-Amaya DB. Lycopene epoxides and apo-lycopenals formed by chemical reactions and autoxidation in model systems and processed foods. J Food Sci. 2009;74:C674–82 [DOI] [PubMed] [Google Scholar]
- 10.Hu KQ, Liu C, Ernst H, Krinsky NI, Russell RM, Wang XD. The biochemical characterization of ferret carotene-9′,10'-monooxygenase catalyzing cleavage of carotenoids in vitro and in vivo. J Biol Chem. 2006;281:19327–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dela Seña C, Narayanasamy S, Riedl KM, Curley RW, Jr, Schwartz SJ, Harrison EH. Substrate specificity of purified recombinant human beta-carotene 15,15'-oxygenase (BCO1). J Biol Chem. 2013;288:37094–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ip BC, Hu KQ, Liu C, Smith DE, Obin MS, Ausman LM, Wang XD. Lycopene metabolite, apo-10'-lycopenoic acid, inhibits diethylnitrosamine-initiated, high fat diet-promoted hepatic inflammation and tumorigenesis in mice. Cancer Prev Res (Phila). 2013;6:1304–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindshield BL, King JL, Wyss A, Goralczyk R, Lu CH, Ford NA, Erdman JW., Jr Lycopene biodistribution is altered in 15,15'-carotenoid monooxygenase knockout mice. J Nutr. 2008;138:2367–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ford NA, Clinton SK, von Lintig J, Wyss A, Erdman JW., Jr Loss of carotene-9′,10'-monooxygenase expression increases serum and tissue lycopene concentrations in lycopene-fed mice. J Nutr. 2010;140:2134–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ford NA, Elsen AC, Erdman JW., Jr Genetic ablation of carotene oxygenases and consumption of lycopene or tomato powder diets modulate carotenoid and lipid metabolism in mice. Nutr Res. 2013;33:733–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang TT, Edwards AJ, Clevidence BA. Strong and weak plasma response to dietary carotenoids identified by cluster analysis and linked to beta-carotene 15,15'-monooxygenase 1 single nucleotide polymorphisms. J Nutr Biochem. 2013;24:1538–46 [DOI] [PubMed] [Google Scholar]
- 17.Al Tanoury Z, Piskunov A, Rochette-Egly C. Vitamin A and retinoid signaling: genomic and nongenomic effects. J Lipid Res. 2013;54:1761–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eroglu A, Hruszkewycz DP, dela Sena C, Narayanasamy S, Riedl KM, Kopec RE, Schwartz SJ, Curley RW, Jr, Harrison EH. Naturally occurring eccentric cleavage products of provitamin A beta-carotene function as antagonists of retinoic acid receptors. J Biol Chem. 2012;287:15886–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaripheh S, Nara TY, Nakamura MT, Erdman JW., Jr Dietary lycopene downregulates carotenoid 15,15'-monooxygenase and PPAR-gamma in selected rat tissues. J Nutr. 2006;136:932–8 [DOI] [PubMed] [Google Scholar]
- 20.Yang CM, Lu IH, Chen HY, Hu ML. Lycopene inhibits the proliferation of androgen-dependent human prostate tumor cells through activation of PPARγ-LXRα-ABCA1 pathway. J Nutr Biochem. 2012;23:8–17 [DOI] [PubMed] [Google Scholar]
- 21.Lian F, Wang XD. Enzymatic metabolites of lycopene induce Nrf2-mediated expression of phase II detoxifying/antioxidant enzymes in human bronchial epithelial cells. Int J Cancer. 2008;123:1262–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ben-Dor A, Steiner M, Gheber L, Danilenko M, Dubi N, Linnewiel K, Zick A, Sharoni Y, Levy J. Carotenoids activate the antioxidant response element transcription system. Mol Cancer Ther. 2005;4:177–86 [PubMed] [Google Scholar]
- 23.Sahin K, Tuzcu M, Sahin N, Ali S, Kucuk O. Nrf2/HO-1 signaling pathway may be the prime target for chemoprevention of cisplatin-induced nephrotoxicity by lycopene. Food Chem Toxicol. 2010;48:2670–4 [DOI] [PubMed] [Google Scholar]
- 24.Chung J, Koo K, Lian F, Hu KQ, Ernst H, Wang XD. Apo-10'-lycopenoic acid, a lycopene metabolite, increases sirtuin 1 mRNA and protein levels and decreases hepatic fat accumulation in ob/ob mice. J Nutr. 2012;142:405–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amengual J, Lobo GP, Golczak M, Li HN, Klimova T, Hoppel CL, Wyss A, Palczewski K, von Lintig J. A mitochondrial enzyme degrades carotenoids and protects against oxidative stress. FASEB J. 2011;25:948–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell JK, Stroud CK, Nakamura MT, Lila MA, Erdman JW., Jr Serum testosterone is reduced following short-term phytofluene, lycopene, or tomato powder consumption in F344 rats. J Nutr. 2006;136:2813–9 [DOI] [PubMed] [Google Scholar]
- 27.Canene-Adams K, Lindshield BL, Wang S, Jeffery EH, Clinton SK, Erdman JW., Jr Combinations of tomato and broccoli enhance antitumor activity in dunning r3327-h prostate adenocarcinomas. Cancer Res. 2007;67:836–43 [DOI] [PubMed] [Google Scholar]
- 28.Zuniga KE, Clinton SK, Erdman JW., Jr The interactions of dietary tomato powder and soy germ on prostate carcinogenesis in the TRAMP model. Cancer Prev Res (Phila). 2013;6:548–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moran NE, Clinton SK, Erdman JW., Jr Differential bioavailability, clearance, and tissue distribution of the acyclic tomato carotenoids, lycopene and phytoene, in mongolian gerbils. J Nutr. 2013;143:1920–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He M, Cornelis MC, Kraft P, van Dam RM, Sun Q, Laurie CC, Mirel DB, Chasman DI, Ridker PM, Hunter DJ, et al. Genome-wide association study identifies variants at the IL18–BCO2 locus associated with interleukin-18 levels. Arterioscler Thromb Vasc Biol. 2010;30:885–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agca CA, Tuzcu M, Gencoglu H, Akdemir F, Ali S, Sahin K, Kucuk O. Lycopene counteracts the hepatic response to 7,12-dimethylbenz[a]anthracene by altering the expression of Bax, Bcl-2, caspases, and oxidative stress biomarkers. Pharm Biol. 2012;50:1513–8 [DOI] [PubMed] [Google Scholar]
- 32.Gupta P, Bansal MP, Koul A. Evaluating the effect of lycopene from Lycopersicum esculentum on apoptosis during NDEA induced hepatocarcinogenesis. Biochem Biophys Res Commun. 2013;434:479–85 [DOI] [PubMed] [Google Scholar]
- 33.Rafi MM, Kanakasabai S, Reyes MD, Bright JJ. Lycopene modulates growth and survival associated genes in prostate cancer. J Nutr Biochem. 2013;24:1724–34 [DOI] [PubMed] [Google Scholar]
- 34.Ford NA, Elsen AC, Zuniga K, Lindshield BL, Erdman JW., Jr Lycopene and apo-12'-lycopenal reduce cell proliferation and alter cell cycle progression in human prostate cancer cells. Nutr Cancer. 2011;63:256–63 [DOI] [PubMed] [Google Scholar]
- 35.Lobo GP, Amengual J, Li HN, Golczak M, Bonet ML, Palczewski K, von Lintig J. Beta,beta-carotene decreases peroxisome proliferator receptor gamma activity and reduces lipid storage capacity of adipocytes in a beta,beta-carotene oxygenase 1-dependent manner. J Biol Chem. 2010;285:27891–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vacca M, Degirolamo C, Massafra V, Polimeno L, Mariani-Costantini R, Palasciano G, Moschetta A. Nuclear receptors in regenerating liver and hepatocellular carcinoma. Mol Cell Endocrinol. 2013;368:108–19 [DOI] [PubMed] [Google Scholar]
- 37.Chang C, Lee SO, Yeh S, Chang TM. Androgen receptor (AR) differential roles in hormone-related tumors including prostate, bladder, kidney, lung, breast and liver. Oncogene. 2013;Jul 22 (Epub ahead of print; DOI:10.1038/onc.2013.274). [DOI] [PubMed] [Google Scholar]
- 38.Dufour CR, Levasseur MP, Pham NH, Eichner LJ, Wilson BJ, Charest-Marcotte A, Duguay D, Poirier-Heon JF, Cermakian N, Giguere V. Genomic convergence among ERRalpha, PROX1, and BMAL1 in the control of metabolic clock outputs. PLoS Genet. 2011;7:e1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–35 [DOI] [PubMed] [Google Scholar]
- 40.Ramamoorthy S, Cidlowski JA. Exploring the molecular mechanisms of glucocorticoid receptor action from sensitivity to resistance. Endocr Dev. 2013;24:41–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang CY, Kazmin D, Jasper JS, Kunder R, Zuercher WJ, McDonnell DP. The metabolic regulator ERRalpha, a downstream target of HER2/IGF-1R, as a therapeutic target in breast cancer. Cancer Cell. 2011;20:500–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burris TP. Nuclear hormone receptors for heme: REV-ERBalpha and REV-ERBbeta are ligand-regulated components of the mammalian clock. Mol Endocrinol. 2008;22:1509–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartonova P, Vrtkova I, Kaplanova K, Urban T. Association between CSN3 and BCO2 gene polymorphisms and milk performance traits in the Czech Fleckvieh cattle breed. Genet Mol Res. 2012;11:1058–63 [DOI] [PubMed] [Google Scholar]
- 44.Basu A, Imrhan V. Tomatoes versus lycopene in oxidative stress and carcinogenesis: conclusions from clinical trials. Eur J Clin Nutr. 2007;61:295–303 [DOI] [PubMed] [Google Scholar]
- 45.Boileau TW, Liao Z, Kim S, Lemeshow S, Erdman JW, Jr, Clinton SK. Prostate carcinogenesis in N-methyl-N-nitrosourea (NMU)-testosterone-treated rats fed tomato powder, lycopene, or energy-restricted diets. J Natl Cancer Inst. 2003;95:1578–86 [DOI] [PubMed] [Google Scholar]