Abstract
Background. Human immunodeficiency virus type 1 (HIV-1) dual infection (DI) has been associated with decreased CD4 T-cell counts and increased viral loads; however, the frequency of intrasubtype DI is poorly understood. We used ultradeep sequencing (UDS) to estimate the frequency of DI in a primary infection cohort of predominantly men who have sex with men (MSM).
Methods. HIV-1 genomes from longitudinal blood samples of recently infected, therapy-naive participants were interrogated with UDS. DI was confirmed when maximum sequence divergence was excessive and supported by phylogenetic analysis. Coinfection was defined as DI at baseline; superinfection was monoinfection at baseline and DI at a later time point.
Results. Of 118 participants, 7 were coinfected and 10 acquired superinfection. Superinfection incidence rate was 4.96 per 100 person-years (95% confidence interval [CI], 2.67–9.22); 6 occurred in the first year and 4 in the second. Overall cumulative prevalence of intrasubtype B DI was 14.4% (95% CI, 8.6%−22.1%). Primary HIV-1 incidence was 4.37 per 100 person-years (95% CI, 3.56–5.36).
Conclusions. Intrasubtype DI was frequent and comparable to primary infection rates among MSM in San Diego; however, superinfection rates declined over time. DI is likely an important component of the HIV epidemic dynamics, and development of stronger immune responses to the initial infection may protect from superinfection.
Keywords: intrasubtype HIV-1 dual infection, HIV-1 coinfection, HIV-1 superinfection, ultradeep sequencing, acute and early infection
Human immunodeficiency virus type 1 (HIV-1) dual infection (DI) is characterized by genetic evidence of 2 distinct viral subpopulations within the same host. DI cases can be divided into coinfections, namely, acquisition of at least 2 strains from different source partners simultaneously or within a brief period of time, and superinfections, namely, acquisition of a second strain after an immune response to the first infection has been established. DI can also be divided into intersubtype or intrasubtype. Intersubtype DI occurs when a person is infected by 2 HIV strains that are from different viral clades, and intrasubtype DI occurs when both strains are from the same viral clade [1]. In light of large genetic distances between clades, intersubtype DI is much easier to detect than intrasubtype DI with sequence analysis techniques, although intrasubtype DI may provide greater insight into local epidemic dynamics and be informative to vaccine design [1–5].
The first case of human HIV superinfection was described in 2002 [6], and several case reports and series followed in subsequent years [7–14]. Using standard genotyping and clonal sequencing, in 2004 our group estimated the incidence of intrasubtype superinfection in San Diego to be approximately 5% per year, similar to rates of initial infection among high-risk men who have sex with men (MSM) in the area [15]. The recent development of next-generation massively parallel sequencing modalities (eg, ultradeep pyrosequencing [UDS]) can identify DI more accurately and efficiently than previous methods [16]. Multiple platforms of UDS can detect circulating minority variants as low as 0.25% of the viral population [17, 18]. Optimization of high-throughput UDS protocols can further increase the efficacy of screening for evidence of DI. Using UDS, one group recently reported rates of superinfection comparable to those of primary HIV-1 infection in a heterosexual cohort in Rakai, Uganda [19]. Our group and others have also reported associations between DI and increased viral load [7, 10, 15, 20], faster decline in CD4+ T cells, and shorter time to AIDS diagnosis [21, 22]. A more accurate estimate of the frequency of DI may not only help paint a clearer picture of viral transmission dynamics within human populations but also assess the potential impact of DI on disease progression and correlates of protection from superinfection. To this end, we employed UDS to screen longitudinally collected blood plasma samples from recently infected individuals to determine the rates of coinfection and superinfection in San Diego, California.
METHODS
Study Participants and Clinical Data
This study included participants enrolled in the San Diego Primary Infection Cohort between January 1998 and January 2007 who were antiretroviral therapy (ART) naive, had deferred ART for at least 6 months after their estimated date of infection (EDI), and had at least 2 blood plasma samples available for sequencing. EDI was calculated at baseline for each participant using a well-established algorithm that characterizes stages of infection on the basis of serologic and virologic criteria [23]. Study participants were seen at weeks 2, 4, 8, 12, 16, 24, 48, and every 24 weeks thereafter for the duration of the study; and those who started ART as part of routine clinical care were followed every 8 weeks during ART for 24 weeks or until plasma viral load (VL) was ≤50 copies/mL. Upon collection, samples were aliquoted, frozen, and stored at −80°C. At all time points, CD4 T-cell counts (LabCorp), and blood plasma HIV-1 RNA levels (Amplicor HIV-1 Monitor Test, Roche Molecular Systems Inc.) were quantified. HIV-1 subtype was determined for each subject using HIV-1 pol sequence data generated by Viroseq 2.0 (Applied Biosystems) by the SCUEAL algorithm [24]. Demographics, clinical data, and standard laboratory values were also collected. Behavioral risk factors were assessed at baseline and longitudinally for all participants, as described elsewhere [25]. Similar assessments were conducted for individuals receiving HIV screening in the Early Test program, which includes antibody and nucleic acid testing [25].
RNA Extraction and Sequencing Methods
HIV-1 RNA was isolated from cryostored blood plasma (QIAamp Viral RNA Mini Kit, Qiagen, Hilden, Germany), and complementary DNA (cDNA) generated (RETROscript Kit, Applied Biosystems/Ambion, Austin, TX) according to manufacturers' instructions. As described previously [16, 20], 500 μL or 1000 μL of blood plasma were used for samples with viral loads above or below 20 000 copies/mL, respectively. Three coding regions—gag p24 (HXB2 coordinates 1366–1619), pol reverse transcriptase (RT, HXB2 coordinates 2708–3242), and env C2-V3 (HXB2 coordinates 6928–7344)—were amplified by polymerase chain reaction (PCR) with region-specific primers, and rubber gaskets were used to physically separate 16 concurrently sequenced samples on a single 454 GS FLX titanium picoliter plate (454 Life Sciences/Roche, Branford, CT) [16, 20, 26]. To avoid cross-contamination, extraction procedures, PCR and UDS library preparation of samples from the same cohort participants were performed on different days and in different sequencing runs. UDS data complemented prior single genome sequencing (SGS) data of env C2-V3 and pol RT generated for 49 cohort participants and published elsewhere [16, 20]. For each sample, the cDNA template input was calculated assuming 43% reverse transcription efficiency and was expressed as the number of templates (log10) present in the 10 μL reaction volume used for the first round of nested PCR (ie, cDNA input before any PCR amplification procedure). Average reverse transcription efficiency was validated by quantifying the input cDNA for 19 random samples from the total pool with real-time quantitative PCR (per) and comparing these results with the HIV RNA measurements, as described elsewhere [26].
Sequence Analysis and Dual Infection Confirmation
UDS data were analyzed using the 454 UDS bioinformatics pipeline, available as part of the HyPhy software package [27] and the Datamonkey web server [24]. The methodology has been described in detail elsewhere [20]. Briefly, high-quality reads were retained and aligned to HXB2 library reference genes for the 3 coding regions using an iterative codon-based alignment procedure, and the maximum divergence (MDI) was calculated for each coding region as the maximum likelihood divergence using the HKY85 substitution model in sliding windows of 150 nucleotides (50-nucleotide shifts, minimum site coverage 500 reads). The output alignments of all UDS datasets have been uploaded to the Los Alamos National Laboratory HIV-DB Next Generation Sequence Archive (http://www.hiv.lanl.gov/content/sequence/HIV/NextGenArchive) and are also available upon request from the corresponding author. Putative DI was suggested when the MDI was >5% in env and >2.5% in gag and pol. If the MDI exceeded these thresholds in at least one coding region, standard phylogenetic analyses (neighbor-joining and maximum likelihood tree reconstruction and bootstrapping) were performed and highlighter plots were created using 500 randomly selected reads, and if 2 clusters were separated by a branch with a bootstrap value >90%, the sample was preliminarily classified as DI. Representative haplotypes were then input into BLAST against a database of locally generated reads to assess for contamination, and if none was found, a different plasma aliquot from the same time point (or within-3-months) underwent repeat RNA extraction, cDNA generation, and UDS to confirm DI. If DI was not observed in the repeat sample, the participant was classified as monoinfected. Longitudinal viral sequences for each coding region from a participant were also assembled into a single phylogeny to further assess for evidence of DI that could be missed by focusing solely on intrasample genetic relatedness (ie, superinfection with viral replacement). Coinfection was defined as DI at baseline, and superinfection was defined as monoinfection at baseline and DI at a later time point. The algorithm for the selection of time point sampling was described elsewhere [20]. Timing of superinfection was calculated as the midway point between the last time point with monoinfection and the first time point with DI.
Incidence of Initial HIV-1 Infection and Superinfection
All participants were enrolled in the San Diego Primary Infection Cohort after being identified with recent HIV-1 infection in a local HIV testing and counseling program (Early Test) targeting high-risk MSM [25]. An HIV incidence rate was calculated among Early Test participants who initially tested negative in the program, but upon repeat testing, subsequently tested positive. All incidence rates were calculated per person-years of follow-up. The incidence of superinfection was derived by dividing the total number of superinfection cases by the total number of person-years contributed by monoinfection participants and superinfection cases (coinfection cases were removed from this measure). The person-years of follow-up was calculated by adding the time to last study time point sampled for all monoinfected participants plus the estimated time to superinfection event (timing of superinfection as described above) for all superinfected cases. The last study time point for monoinfected participants was the time point before study withdrawal or ART initiation, whichever happened first.
RESULTS
At study baseline, participants (n = 118) were infected with HIV-1 subtype B and were predominantly white men (82%) with a median age of 32 years (IQR: 26–38 years) who reported sex with other men as their main HIV risk factor (Table 1). At enrollment, the median CD4+ T-cell count was 544 cells/mL (IQR: 460–709 cells/mL), the median time from EDI was 71 days (IQR: 70–133 days), and the median blood plasma level of HIV-1 RNA was 4.81 log10 copies/mL (IQR: 3.96–5.38 log10 copies/mL). Further breakdown of CD4+ T-cell count, plasma HIV-1 RNA, and other clinical markers according to dual infection status is outlined in Table 1. The median time from EDI to ART initiation in the group was 16.2 months (IQR: 6.7–33.2 months). When using UDS to screen for DI, the median calculated cDNA input in the first-round nested PCR was 3.42 log10copies/10 μL (IQR: 3.13–3.90 log10 copies); the median number of UDS reads generated passing quality filters for partial env was 3877 (IQR: 1771–6417) with a median nucleotide coverage of 2032 (IQR: 944–3492), the median number of reads generated for partial gag was 8263 (IQR: 3189–12 911) with a median coverage of 3018 (IQR: 1093–4927), and for partial pol, the median read number was 4268 (IQR: 1911–8082) with a median per site nucleotide coverage of 1320 (IQR: 603–2619; Table 2).
Table 1.
Study Cohort Baseline Characteristics
| Factor | Groups |
||
|---|---|---|---|
| Monoinfection (N = 101) | Coinfection (N = 7) | Superinfection (N = 10) | |
| Age at enrollment: median years (IQR) | 31 (27–39) | 34 (27–39) | 31 (24–34) |
| Sex, no. (%) | |||
| Male | 98 (97) | 7 (100) | 10 (100) |
| Female | 3 (3) | 0 (0) | 0 (0) |
| Race, no. (%) | |||
| White | 84 (83) | 4 (57) | 8 (80) |
| Black | 7 (7) | 0 (0) | 1 (10) |
| Native American | 6 (6) | 0 (0) | 1 (10) |
| Hawaiian or Pacific Islander | 1 (1) | 2 (29) | 0 (0) |
| Asian | 1 (1) | 0 (0) | 0 (0) |
| Unknown | 2 (2) | 1 (14) | 0 (0) |
| Ethnicity, no. (%) | |||
| Hispanic or Latino | 17 (17) | 2 (29) | 2 (20) |
| Not Hispanic or Latino | 32 (32) | 3 (43) | 4 (40) |
| Unknown | 52 (51) | 2 (29) | 4 (40) |
| HIV risk factors, no. (%) | |||
| MSM | 98 (97) | 7 (100) | 10 (100) |
| Injection drug use | 4 (4) | 0 (0) | 0 (0) |
| HIV viral load: median log10 copies/mL (IQR) | 4.84 (4.13–5.38) | 4.58 (4.21–5.11) | 4.68 (3.16–4.85) |
| CD4 T-cell count: median cells/μL (IQR) | 542 (460–706) | 546 (421–706) | 543 (476–685) |
| cDNA input: median log10 copies/mL (IQR) | 3.51 (3.02–4.04) | 3.24 (3.02–3.77) | 3.34 (2.12–3.51) |
Abbreviations: cDNA, complementary DNA; HIV, human immunodeficiency virus; IQR, interquartile range; MSM, men who have sex with men.
Table 2.
Characteristics of UDS Reads Generated for Each HIV-1 Coding Region
| Coding Region | No. of Reads: Median (IQR) | Read Length: Mean (IQR) | Nucleotide Coverage: Median (IQR) |
|---|---|---|---|
| env (C2-V3) | 3877 (1771–6417) | 296 (269–329) | 2032 (944–3492) |
| gag (p24) | 8263 (3189–12 911) | 233 (226–244) | 3018 (1093–4927) |
| pol (RT) | 4268 (1911–8082) | 223 (194–242) | 1320 (603–2619) |
Abbreviations: HIV-1, human immunodeficiency virus type 1; IQR, interquartile range; RT, reverse transcriptase; UDS, ultradeep sequencing.
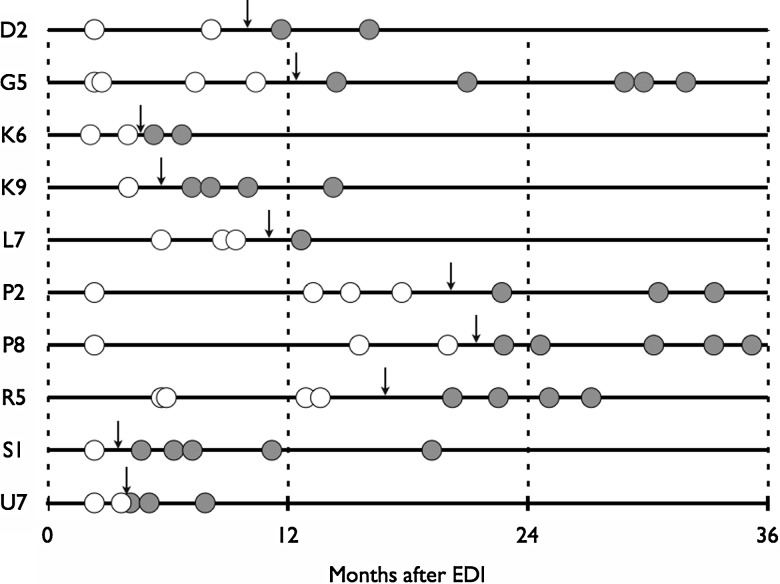
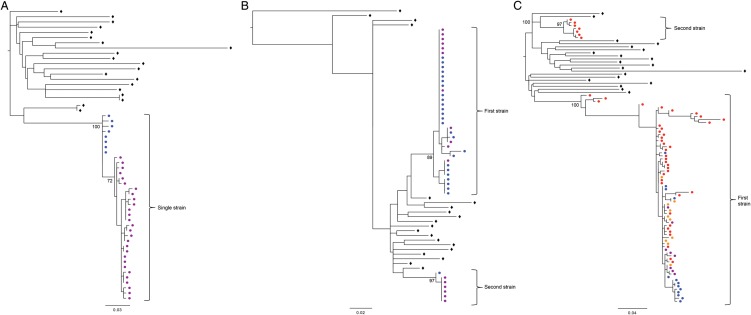
At baseline, 7 cases of coinfection were identified at a median time from EDI of 2.8 months (IQR: 2.3–3.2 months after EDI), and a coinfection prevalence of 5.9% (95% CI, 2.4%–11.8%). During follow-up, 10 participants were identified with superinfection over 201.6 person-years of follow-up. This resulted in an overall superinfection incidence rate of 4.96 per 100 person-years (95% CI, 2.67–9.22). Six superinfection cases occurred in the first year after EDI, 4 within the first 6 months (first-year incidence rate of 29.20 per 100 person-years, 95% CI, 1.13–6.50), and 4 within the second year (second-year incidence rate of 9.25 per 100 person-years, 95% CI, 3.47–24.64; Figure 1). The median time to superinfection was 10.5 months after EDI (IQR: 5.2–15.6 months). Together, the cumulative prevalence of dual infection (including both coinfections and superinfections) was 14.4% (95% CI, 8.6%–22.1%). All DI cases were infected with 2 subtype B viruses. Phylogenetic trees built using PHYML from longitudinal sequences chosen to represent HIV-1 monoinfection, coinfection, and superinfection cases are shown in Figure 2. Sampling of time points after estimated date of infection for all cohort participants is also listed (Supplementary Table 1).
Figure 1.
Timing of superinfection (dark arrows) in 10 individuals was estimated as the halfway point between the last sampled time point with monoinfection and the first sampled time point with dual infection. White circles: sampled time points with monoinfection; gray circles: sampled time points with dual infection. Abbreviation: EDI: estimated date of initial infection.
Figure 2.
Phylogenetic trees using PHYML were generated from longitudinal env sequences for participants I4, U1, and L7 illustrating examples of HIV-1 (A) monoinfection, (B) co-infection, and (C) superinfection, respectively. Solid triangles and circles represent haplotypes from different viral strains, open diamonds indicate background sequences, and brackets delineate different timepoints (T1: timepoint 1; T2: timepoint 2; T3: timepoint 3; T4: timepoint 4). The same set of background sequences were used in all trees.
HIV prevalence in the Early Test program was 2.5% (95% CI, 2.0%–3.1%) among first time testers [25]; and among repeat clients, 92 seroconverted over 21 065 person-years, resulting in an HIV incidence of 4.37 per 100 person-years (95% CI, 3.56–5.36). Behavioral risk factors, including acquisition of sexually transmitted infections, were not significantly different among the MSM identified with primary infection, or DI when assessed longitudinally, although the presence of an STI infection was based on self-report (data not shown).
DISCUSSION
Our study used UDS to investigate the incidence and prevalence of intrasubtype DI in a predominantly high-risk cohort of individuals enrolled during primary infection. All identified cases of DI were intrasubtype B, and the cumulative prevalence of DI (both coinfections and superinfections) was high at 14.4%. Of the 17 total cases of DI, 10 superinfections were observed over 215 person-years, giving an incidence of HIV-1 superinfection of 4.96 per 100 person-years. The majority of these superinfection cases occurred in the first year after initial infection, consistent with our group's prior report [15]. On the other hand, the incidence of primary HIV in our screening program was 4.37 per 100 person-years (among clients who had repeat testing). Although the incidence rates of primary infection and superinfection were not directly compared, they seemed similar. This is consistent with recent reports of the incidence of superinfection in a cohort with different HIV risk factors [19]. The rate of superinfection declined from 29.20 per 100 person-years in the first year to 9.25 per 100 person-years in the second year, which might suggest some protection against superinfection over time; the small number of cases and shorter follow-up from access to ART may bias this observation.
Using a more rigorous disease progression analysis, our group previously reported that HIV-1 superinfection was associated with more rapid rates of viral load increase over time but not with the rates CD4 T-cell count decline [20]. As more cases of DI are identified, more specific immunologic, clinical, and virologic correlates of DI can be determined. One limitation of this study is that while UDS is very sensitive at detecting DI, it is subject to experimental and reproducibility biases, especially when preceded by PCR amplification [28]. Despite increased sensitivity to detect DI in a blood sample, the absence of detected DI does not mean that it did not occur; it could still have been missed by not sampling at the right time, viral recombination, or lack of sensitivity of the UDS platform. The classification of DI in the present study was rigorous. Confirmation of DI at a sampled time point required meeting a series of stringent criteria: excessive sequence divergence, phylogenetic trees consistent with DI, and maintenance of the same phylogenetic relationships in the resampled time point. Although this helped control for sampling biases by the sequencing method and bioinformatics alignment artifacts, the DI call was conservative, and some DI cases may have been misclassified as monoinfections.
The initiation of ART in the study cohort has the potential to modulate some of the conclusions. First, it limited our ability to assess the incidence of superinfection over longer durations of follow-up. Second, because only ART-naive individuals were considered in the calculation of the incidence of superinfection, this could lead to a selection bias if those who were more (or less) susceptible to superinfection were more (or less) likely to initiate ART. For example, if those who had riskier sex behaviors chose to start ART sooner than those who remained treatment naive, then our sample would have been biased to consist of a group which was less likely to become superinfected. The opposite scenario could likewise be true, and other unmeasured confounders could have influenced one's decision to start or not start ART, which would result in unmeasured bias.
Another important caveat is that the group screened for evidence of DI did not comprise HIV-negative individuals wherein at-risk individuals could be followed for estimates of incidence of initial infection. Therefore, a direct comparison of the incidence of primary infection to superinfection in these groups would not be appropriate because the denominators (ie, populations at risk) were different. Because the risks for superinfection are likely the same as those for primary infection, the superinfection group is likely riskier because the person time is for those with one infection already, whereas the person time for initial HIV infection is among all those at risk for infection. Despite these limitations, however, the improvement of our understanding regarding the incidence of intrasubtype HIV superinfection may provide important insight into the correlates of protection necessary to prevent against initial HIV infection, which could inform preventative vaccine design.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We are grateful to all the participants in the San Diego Primary Infection Cohort, to Mehdi Bouhaddou for his assistance with sequence data organization and processing, to Homero Vazquez for bioinformatics assistance, to Christy Anderson for invaluable statistical advice and cohort database assistance, to Susanna Var for assistance with data processing and management, to Ying Shi for assistance with sequence processing, to Stephen Espitia for assistance with sequence data analysis, to Caroline Ignacio for technical support, and to Demetrius Dela Cruz for his administrative assistance.
Author Contributions. G. A. W. participated in the study design, performed the laboratory experiments, participated in the data analyses for this study and wrote the primary version of the manuscript, M. E. P. participated in the study design, performed the laboratory experiments, participated in the data analyses for this study, and assisted in the writing of the article, G. C. performed the laboratory experiments, S. J .L. designed and implemented the protocol of the study cohort, and assisted in the writing of the article, S. K. P. designed and implemented the bioinformatics analysis pipeline, and assisted in the writing of the article, A. C. participated in data organization and analysis, A. E. R. participated in data analysis and in the editing of the article, S. R. M. participated in data analysis and assisted in the writing of the article, D. D. R. assisted in the writing of the article, D. M. S. conceived the study design, assisted in the collection, analysis and interpretation of data, and in the writing of the article.
Financial support. This work was supported by the U.S. Department of Veterans Affairs; the National Institutes of Health (AI090970, AI100665, AI080353, MH097520, DA034978, MH83552, AI36214, MH62512, MH81482, AI43638, AI74621, TW008908, AI69432, AI096113, AI47745, GM093939); the International AIDS Vaccine Initiative; and the James B. Pendleton Charitable Trust. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the article.
Potential conflicts of interest. D. D. R. has served as a consultant for Biota, Chimerix, Gen-Probe, Gilead Sciences, Merck & Co, Monogram Biosciences, and Tobira Therapeutics. D. M. S. has received research support from ViiV Pharmaceuticals and has served as a consultant to Gen-Probe and Testing Talent Services. S. K. P. has served as a consultant to Monogram Biosciences and Gen-Probe. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Smith DM, Richman DD, Little SJ. HIV superinfection. J Infect Dis. 2005;192:438–44. doi: 10.1086/431682. [DOI] [PubMed] [Google Scholar]
- 2.Smith DM, Strain MC, Frost SD, et al. Lack of neutralizing antibody response to HIV-1 predisposes to superinfection. Virology. 2006;355:1–5. doi: 10.1016/j.virol.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Blish CA, Dogan OC, Derby NR, et al. Human immunodeficiency virus type 1 superinfection occurs despite relatively robust neutralizing antibody responses. J Virol. 2008;82:12094–103. doi: 10.1128/JVI.01730-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chohan BH, Piantadosi A, Overbaugh J. HIV-1 superinfection and its implications for vaccine design. Curr HIV Res. 2010;8:596–601. doi: 10.2174/157016210794088218. [DOI] [PubMed] [Google Scholar]
- 5.Gross KL, Porco TC, Grant RM. HIV-1 superinfection and viral diversity 1. AIDS. 2004;18:1513–1520. doi: 10.1097/01.aids.0000131361.75328.47. [DOI] [PubMed] [Google Scholar]
- 6.Ramos A, Hu DJ, Nguyen L, et al. Intersubtype human immunodeficiency virus type 1 superinfection following seroconversion to primary infection in two injection drug users. J Virol. 2002;76:7444–52. doi: 10.1128/JVI.76.15.7444-7452.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altfeld M, Allen TM, Yu XG, et al. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature. 2002;420:434–9. doi: 10.1038/nature01200. [DOI] [PubMed] [Google Scholar]
- 8.Brenner B, Routy JP, Quan YD, et al. Persistence of multidrug-resistant HIV-1 in primary infection leading to superinfection (vol 18, pg 1653, 1660) 1. AIDS. 2004;18:2107–7. doi: 10.1097/01.aids.0000131377.28694.04. [DOI] [PubMed] [Google Scholar]
- 9.Chohan B, Lavreys L, Rainwater SMJ, Overbaugh J. Evidence for frequent reinfection with human immunodeficiency virus type 1 of a different subtype. J Virol. 2005;79:10701–8. doi: 10.1128/JVI.79.16.10701-10708.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jost S, Bernard MC, Kaiser L, et al. A patient with HIV-1 superinfection. N Engl J Med. 2002;347:731–6. doi: 10.1056/NEJMoa020263. [DOI] [PubMed] [Google Scholar]
- 11.Koelsch KK, Smith DM, Little SJ, et al. Clade B HIV-1 superinfection with wild-type virus after primary infection with drug-resistant clade B virus. AIDS. 2003;17:F11–6. doi: 10.1097/00002030-200305020-00001. [DOI] [PubMed] [Google Scholar]
- 12.McCutchan FE, Hoelscher M, Tovanabutra S, et al. In-depth analysis of a heterosexually acquired human immunodeficiency virus type 1 superinfection: evolution, temporal fluctuation, and intercompartment dynamics from the seronegative window period through 30 months postinfection. J Virol. 2005;79:11693–704. doi: 10.1128/JVI.79.18.11693-11704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang OO, Daar ES, Jamieson BD, et al. Human immunodeficiency virus type 1 clade B superinfection: evidence for differential immune containment of distinct clade B strains. J Virol. 2005;79:860–8. doi: 10.1128/JVI.79.2.860-868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yerly S, Jost S, Monnat M, et al. HIV-1 co/super-infection in intravenous drug users 1. AIDS. 2004;18:1413–21. doi: 10.1097/01.aids.0000131330.28762.0c. [DOI] [PubMed] [Google Scholar]
- 15.Smith DM, Wong JK, Hightower GK, et al. Incidence of HIV superinfection following primary infection. JAMA. 2004;292:1177–8. doi: 10.1001/jama.292.10.1177. [DOI] [PubMed] [Google Scholar]
- 16.Pacold M, Smith D, Little S, et al. Comparison of methods to detect HIV dual infection. AIDS Res Hum Retroviruses. 2010;26:1291–8. doi: 10.1089/aid.2010.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Archer J, Weber J, Henry K, et al. Use of four next-generation sequencing platforms to determine HIV-1 coreceptor tropism. PLoS One. 2012;7:e49602. doi: 10.1371/journal.pone.0049602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Redd AD, Collinson-Streng A, Martens C, et al. Identification of HIV superinfection in seroconcordant couples in Rakai, Uganda, by use of next-generation deep sequencing. J Clin Microbiol. 2011;49:2859–67. doi: 10.1128/JCM.00804-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redd AD, Mullis CE, Serwadda D, et al. The rates of HIV superinfection and primary HIV incidence in a general population in Rakai, Uganda. J Infect Dis. 2012;206:267–74. doi: 10.1093/infdis/jis325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pacold ME, Pond SL, Wagner GA, et al. Clinical, virologic, and immunologic correlates of HIV-1 intraclade B dual infection among men who have sex with men. AIDS. 2012;26:157–65. doi: 10.1097/QAD.0b013e32834dcd26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cornelissen M, Pasternak AO, Grijsen ML, et al. HIV-1 dual infection is associated with faster CD4+ T-cell decline in a cohort of men with primary HIV infection. Clin Infect Dis. 2012;54:539–47. doi: 10.1093/cid/cir849. [DOI] [PubMed] [Google Scholar]
- 22.Gottlieb GS, Nickle DC, Jensen MA, et al. Dual HIV-1 infection associated with rapid disease progression. Lancet. 2004;363:619–22. doi: 10.1016/S0140-6736(04)15596-7. [DOI] [PubMed] [Google Scholar]
- 23.Le T, Wright EJ, Smith DM, et al. Enhanced CD4+ T-cell recovery with earlier HIV-1 antiretroviral therapy. N Engl J Med. 2013;368:218–30. doi: 10.1056/NEJMoa1110187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kosakovsky Pond SL, Posada D, Stawiski E, et al. An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comput Biol. 2009;5:e1000581. doi: 10.1371/journal.pcbi.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris SR, Little SJ, Cunningham T, et al. Evaluation of an HIV nucleic acid testing program with automated internet and voicemail systems to deliver results. Ann Intern Med. 2010;152:778–85. doi: 10.1059/0003-4819-152-12-201006150-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gianella S, Delport W, Pacold ME, et al. Detection of minority resistance during early HIV-1 infection: natural variation and spurious detection rather than transmission and evolution of multiple viral variants. J Virol. 2011;85:8359–67. doi: 10.1128/JVI.02582-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosakovsky Pond SL, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–9. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 28.Liu SL, Rodrigo AG, Shankarappa R, et al. HIV quasispecies and resampling. Science. 1996;273:415–6. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.