Septal pores—the intercellular bridges of fungi—are open during interphase but closed at mitosis. The NIMA kinase mitotically regulates septal pore closing and opening potentially via mechanisms analogous to how it regulates mitotic nuclear pores. The findings explain how and why physically connected Aspergillus cells can maintain mitotic autonomy.
Abstract
Intercellular bridges are a conserved feature of multicellular organisms. In multicellular fungi, cells are connected directly via intercellular bridges called septal pores. Using Aspergillus nidulans, we demonstrate for the first time that septal pores are regulated to be opened during interphase but closed during mitosis. Septal pore–associated proteins display dynamic cell cycle–regulated locations at mature septa. Of importance, the mitotic NIMA kinase locates to forming septa and surprisingly then remains at septa throughout interphase. However, during mitosis, when NIMA transiently locates to nuclei to promote mitosis, its levels at septa drop. A model is proposed in which NIMA helps keep septal pores open during interphase and then closed when it is removed from them during mitosis. In support of this hypothesis, NIMA inactivation is shown to promote interphase septal pore closing. Because NIMA triggers nuclear pore complex opening during mitosis, our findings suggest that common cell cycle regulatory mechanisms might control septal pores and nuclear pores such that they are opened and closed out of phase to each other during cell cycle progression. The study provides insights into how and why cytoplasmically connected Aspergillus cells maintain mitotic autonomy.
INTRODUCTION
In multicellular organisms, cell-to-cell communication can occur indirectly via diffusible signaling molecules or more directly through stable intercellular bridges that allow cytoplasmic mixing between cells. Such direct communication occurs through plasmodesmata in plants, intercellular bridges and tunneling nanotubes in vertebrates and insects, and septal pores in filamentous fungi (Goodenough and Paul, 2009; Xu and Jackson, 2010; Jedd and Pieuchot, 2012; Bloemendal and Kuck, 2013). Fungal septal pores enable cell compartments that are actively growing at their cell tips to receive resources from subapical cells within the colony and vice versa. Fungal colonies therefore constitute connected cellular systems that enable different parts of the colony to support the growth of apical cell tips at far higher rates than could be supported in cells containing single nuclei. Foraging apical cells can then supply the main body of the colony resources for the complex developmental programs typical of filamentous fungi. This is a characteristic of saprophytic, symbiotic, and pathogenic fungi and enables a fast mode of integrated multicellular colony growth for foraging and invasion, leading to fungal development (Trinci, 1973).
Although fungal septal pores facilitate rapid growth and colony expansion into new growth substrates, they also possess a potential Achilles heel; if cell wall damage occurs, fungal colonies run the risk of uncontrolled cytoplasmic bleeding from connected colony compartments. However, septal pores are guarded by Woronin bodies in the Ascomycota—peroxisome-derived structures that are large enough to physically plug septal pores after cell damage and other forms of stress (Trinci and Collinge, 1973; Maruyama et al., 2005; Beck and Ebel, 2013), as first discovered in Neurospora crassa (Jedd and Chua, 2000; Tenney et al., 2000). This plugging prevents excessive cytoplasmic bleeding through septal pores after cell wall damage. Woronin bodies are built around a dense core of Hex1 protein, which is responsible for assembly of these structures (Jedd and Chua, 2000; Tenney et al., 2000). In addition to the Woronin body, the SO (SOFT) protein identified in N. crassa—a cytoplasmic protein that accumulates at septal pores in response to various stresses—is required for efficient plugging of septal pores (Fleissner and Glass, 2007). It has also been found that deletion of the so gene from Aspergillus oryzae allows some cytoplasmic leakage after cell wounding (Maruyama et al., 2010).
In Aspergillus nidulans, open septal pores pose another challenge related to how the cell cycle and mitoses are regulated in this model fungus. In A. nidulans all nuclei within a cellular compartment synchronously enter and exit mitosis, suggestive of a cytoplasmic level of mitotic regulation active upon all nuclei within a cell compartment (Rosenberger and Kessel, 1967; Sampson and Heath, 2005). In addition, only apical cells with actively growing tips transit the nuclear division cycle, whereas nuclei in subapical cells are arrested in G1 (Fiddy and Trinci, 1976; Momany, 2002; Nayak et al., 2010; Edgerton-Morgan and Oakley, 2012; Figure 1, D and E, red nuclei). The cell cycle arrest in subapical cells is maintained until a new growth axis is formed, which increases cell volume (Figure 1E, subapical cell compartment to the left) and promotes synchronous reentry into the cell cycle. How the mitotic cycles of nuclei in connected A. nidulans cells that share the same cytoplasm are independently regulated is unknown.
FIGURE 1:
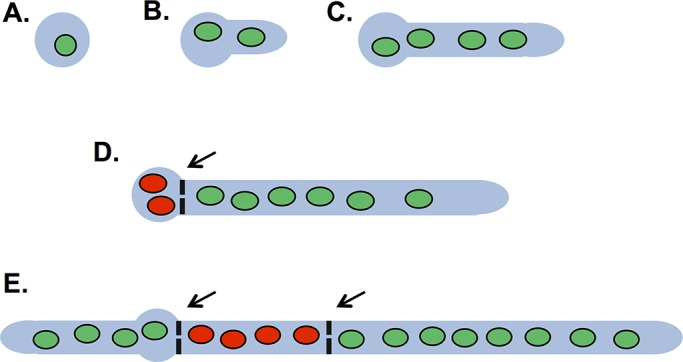
The cell cycle status of connected A. nidulans cells varies. (A) Cell growth is initiated from uninucleated asexual conidial spores. (B) Progression through the nuclear division cycle (green nuclei) is accompanied by highly polarized growth and germ tube development. (C) A second synchronous mitosis occurs, and the two nuclei divide into four daughter nuclei. (D) The first septum usually forms after three rounds of nuclear division and contains a central septal pore ∼50 nm in diameter. The left side (subapical) cellular compartment is removed from the cell cycle (red nuclei) until it forms a new axis of growth. The right (apical) cell compartment remains in the cell cycle, as it has a growing apical tip. (E) Growth of a second cell tip converts the cell to the left into an apical cell, which reenters the cell cycle. A second septum forms after the nuclei of the right apical cell undergo mitosis. This second septation event generates a subapical cell that is removed from the cell cycle until it forms a growth axis during cell branching (not shown). Arrows indicate septa that contain a central septal pore.
Septal pores pose an additional challenge to A. nidulans cells due to the semiopen nature of its mode of mitosis and the fact that connected cells are in different cell cycle states. During mitosis, partial nuclear pore complex (NPC) disassembly regulated by the NIMA kinase opens nuclear pores (De Souza et al., 2004; Osmani et al., 2006; Shen and Osmani, 2013; Laurell et al., 2011; Govindaraghavan et al., 2014), and nuclear proteins, including RNA polymerase II (Son and Osmani, 2009) and nucleolar proteins (Ukil et al., 2009), are released from nuclei into the cytoplasm. These nuclear proteins disperse throughout the cell until mitosis is complete, when they are reimported into the new, daughter G1 nuclei. The mitotic release of nuclear proteins creates a problem, as they could diffuse into subapical cells through septal pores and be imported into their interphase nuclei. This would severely compromise the functions of the apical cell's nuclei after mitosis because they would have to wait for resynthesis of nuclear proteins rather than being able to recycle existing nuclear proteins as occurs normally (De Souza et al., 2004; Osmani et al., 2006). How A. nidulans cells prevent leakage of cytoplasmic mitotic nuclear proteins into subapical cells is not understood but could involve the physical blocking of mitotic septal pores, as occurs after stress and cell wall damage, or some other form of cell cycle–regulated septal pore closing. Here we report that septal pores are subject to cell cycle regulation in a Woronin body and SO protein–independent manner, revealing a new level of cell-to-cell connectivity control linked to mitotic regulation via the mitotic NIMA kinase.
RESULTS
Aspergillus septal pores are closed during mitosis
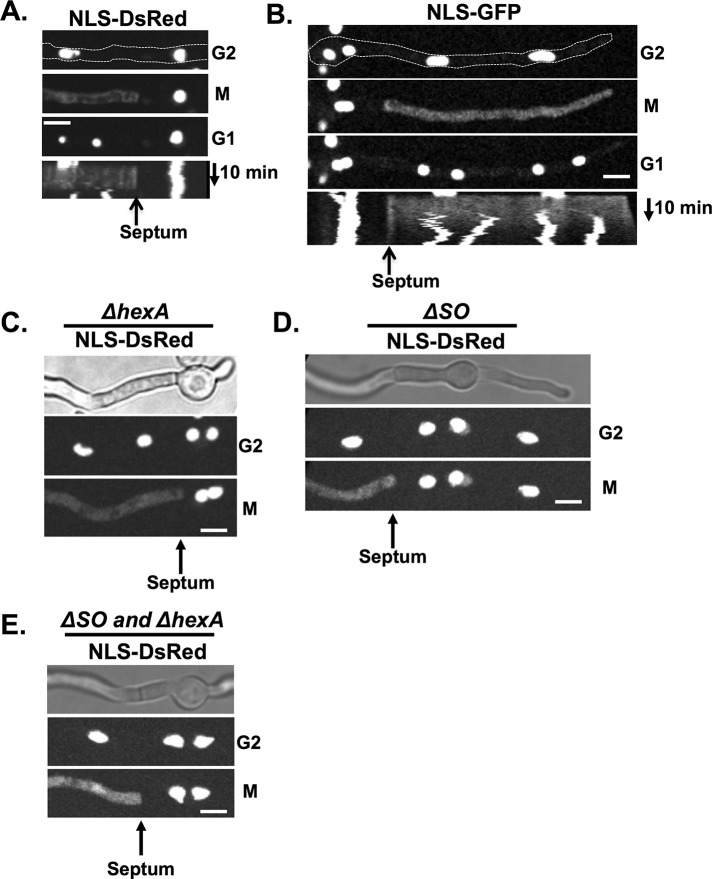
Vegetative fungal septal pores are believed to be in an open state unless hyphae are damaged (or subject to other stresses), after which septa are blocked by Woronin bodies to prevent excessive cytoplasmic bleeding in the Ascomycota (Tenney et al., 2000; Momany et al., 2002; Maruyama et al., 2005; Liu et al., 2008). However, A. nidulans apical cells function independently of subapical cells with regard to mitotic progression (Figure 1), suggesting that, even though septal pores allow cell-to-cell cytoplasmic continuity, individual cells maintain some autonomy, particularly during mitosis. One potential mechanism that might achieve mitotic cellular autonomy between connected cells would be regulated closing of septal pores during mitosis. To investigate this, we monitored the fate of a nuclear localization signal (NLS)–DsRed reporter construct during mitosis in septated cells. NLS-DsRed is transported into nuclei during interphase via the NLS of the stuA transcription factor (Suelmann et al., 1997) but disperses from nuclei during mitosis when nuclear pores are opened. This analysis revealed that septal pores adjacent to mitotic apical cells do not allow diffusion of NLS-DsRed from the mitotic cell into the subapical cell (Figure 2A). Kymographs generated from the live-cell imaging data demonstrate that cytoplasmic NLS-DsRed is unable to traverse the septal pore for the duration of mitosis and that NLS-DsRed is reimported into the G1 nuclei of the apical cell after mitosis (Figure 2A). During the same time period, nuclear NLS-DsRed in the subapical interphase cells remained unchanged (Figure 2A, nucleus to the right). Because DsRed undergoes oligomerization to form larger tetramers (Baird et al., 2000), we followed the fate of the smaller NLS–green fluorescent protein (GFP) marker protein during mitosis (Figure 2B). When NLS-GFP was released from nuclei at the onset of mitosis, it diffused throughout the cell and was also unable to pass through the septal pore into the subapical cell (Figure 2B). This reveals that septa provide a diffusion barrier between apical and subapical cells during mitosis.
FIGURE 2:
Septal pores are not open during mitosis. (A) NLS-DsRed (strain KF075) or (B) NLS-GFP (strain KF430) locates to nuclei in interphase and leaks into the cytoplasm during mitosis. Septa form a barrier preventing each protein from diffusing into adjacent cells, indicating that septal pores are closed during mitosis. Kymographs show a barrier exists between the cellular compartments throughout mitosis. (C) In hexA-null cells, which cannot form Woronin bodies (strain KF081), (D) in the absence of the SO protein (strain KF361), or (E) in hexA and so double-null mutants (strain KF394) septal pores are still closed during mitosis, stopping NLS-DsRed from leaking into neighboring cellular compartments. Bars, 5 μm.
We addressed the possibility that proteins known to be involved in septal pore occlusion might play a role in the mitotic closing of septal pores. We first observed the distribution of GFP-tagged HexA as a marker for Woronin bodies (Jedd and Chua, 2000; Tenney et al., 2000; Yuan et al., 2003) using live-cell imaging. A small percentage (∼5%) of completely immobile HexA foci were adjacent to mature septa. However, the majority (∼70%) moved short distances, with the remaining population (∼25%) displaying longer-range, faster motilities (Supplemental Figure S1A). The faster foci moved at a rate of 1.4 μm/s, similar to the rate previously determined for peroxisomes of 1.66 ± 0.77 μm/s for anterograde and 1.63 ± 0.87 μm/s for retrograde movements (Egan et al., 2012). The immobile HexA foci are therefore positioned near septa and could potentially help block septal pores during mitosis (Supplemental Figure S1B), as also recently found in Aspergillus fumigatus (Beck and Ebel, 2013).
Null alleles of hexA (Hynes et al., 2008) were generated to prevent Woronin body formation and ask whether they are required to block septal pores during mitosis. In the absence of HexA, NLS-DsRed was still unable to diffuse past septa during mitosis (Figure 2C). The SO protein, first identified in N. crassa (Fleissner and Glass, 2007), also locates to septal pores after stress in Aspergillus oryzae and is required for normal rates of septal pore closing after cellular damage (Maruyama et al., 2010). The SO protein is therefore a candidate that might help close septal pores during mitosis. However, neither HexA nor SO is required for blocking diffusion through septal pores during mitosis, as the single so and double so + hexA null mutants retain the mitotic septal barrier (Figure 2, C–E).
Septal pores are open in interphase and closed during mitosis
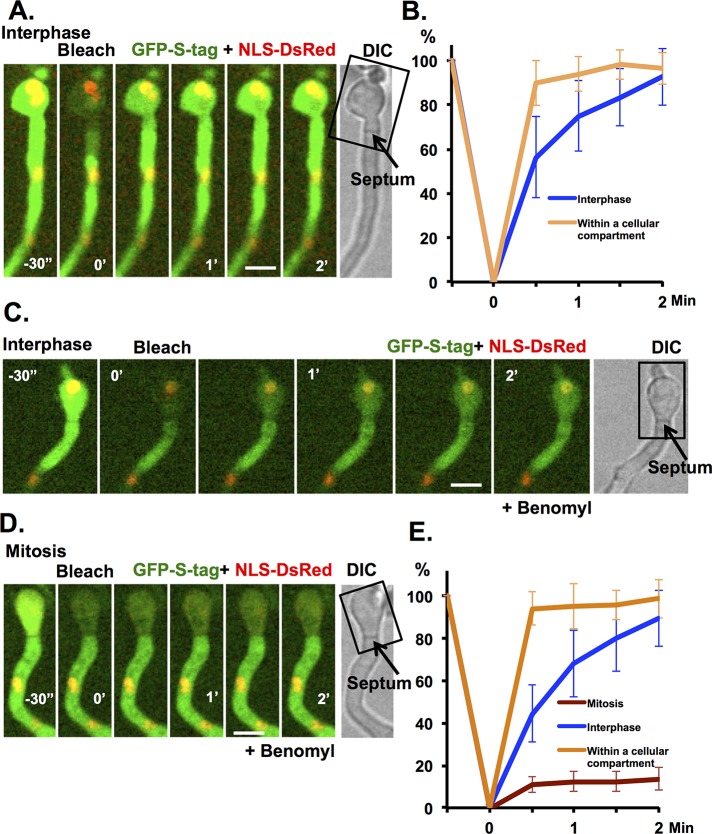
To monitor the state of septal pore opening and closing during the cell cycle, we developed a fluorescence recovery after photobleaching (FRAP)–based septal pore permeability assay. In this assay the cytoplasmic GFP signal in subapical cells of germlings (Figure 1D) is photobleached and fluorescence recovery quantified to determine the rate at which unbleached GFP diffuses through the septal pore from the adjacent cell. In the majority of cells analyzed, the GFP fluorescence signal rapidly equilibrated across the septal pore, with average fluorescence recovered being 85.8% with t1/2 < 30 s. Because most of the cell cycle time is devoted to interphase in A. nidulans, the cells analyzed are likely in interphase, which would suggest that during interphase, septal pores are in an open state (Figure 3, A and B), although they are closed during mitosis (Figure 2).
FIGURE 3:
A FRAP-based septal pore permeability assay reveals that septal pores are open in interphase and closed during mitosis. (A) FRAP analysis of cytoplasmic GFP-S-tag recovery across a septum when both cells are in interphase (NLS-DsRed is nuclear). The rectangle indicates the region that was photobleached. One prebleach and five postbleach images were captured at 30-s intervals. (B) The average fluorescence recovery profile of GFP-S-tag ±SD for recovery in subapical cells during interphase (septum crossing required; blue, n = 15) or in bleached portions of apical cell (no septum crossing required; orange, n = 10). (C) As for A but in the presence of benomyl, with cells on both sides of the septum in interphase (NLS-DsRed is nuclear) or (D) in a germling with the septum separating two cells, one in interphase and one in mitosis (NLS-DsRed dispersed from nuclei). (E) As in B when both cells were in interphase (blue, n = 12) and when one was in mitosis (brown, n = 10) or within a cell compartment (orange, n = 10) ± SD. Strain KF491 was used. Bars, 5 μm.
The rate of recovery of GFP fluorescence within the bleached area of the apical compartment was faster than that in the subapical compartment, which required crossing the septum (Figure 3B). Whether this is caused by the physical dimensions and/or biophysical properties of the septal pore remains to be determined. However, as described later, there are proteins that populate the septal pore region that might affect free diffusion through the septal pore.
To determine whether cells are in interphase or mitosis during the septal pore permeability assay, strains expressing NLS-DsRed as well as cytoplasmic GFP were used. Cells were scanned for those that had one cell compartment in mitosis (NLS-DsRed dispersed from nuclei) adjacent to one in interphase (nuclear NLS-DsRed) and the permeability assay completed. This approach proved difficult because mitosis is completed in ∼5 min, leaving minimal time to set up the analysis, although we did successfully collect data from one such cell that indicated that the mitotic septal pore is closed during mitosis (Supplemental Figure S2). To facilitate the experiment, we used benomyl to impose a mitotic spindle assembly checkpoint (SAC) mitotic arrest (De Souza et al., 2011; De Souza and Osmani, 2011), to provide more time to set up the assay. The resulting data indicate that there is a marked difference in permeability properties of septal pores during interphase and mitosis, with septa adjacent to mitotic cells forming a more effective barrier than septa that separate two cells in interphase (Figure 3, C–E).
Septal pore–associated proteins
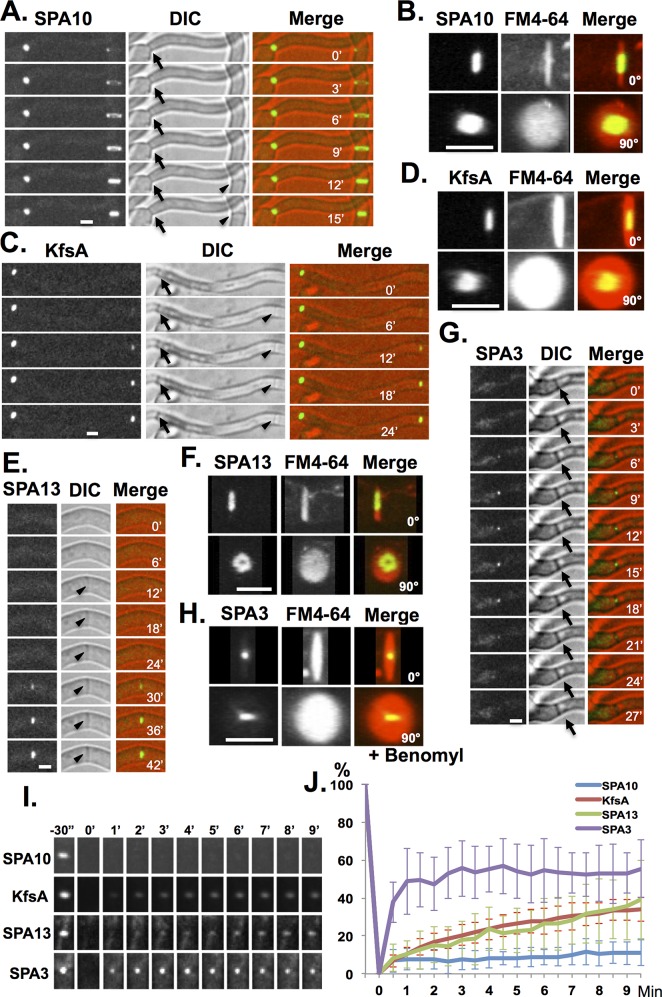
Our data indicate the diffusion properties and hence potentially the protein composition of septal pores might be regulated during the cell cycle. A recent N. crassa study identified 18 septal pore–associated (SPA) proteins, which possess long, intrinsically disordered domains able to form aggregates at septal pores (Lai et al., 2012). We identified and endogenously tagged three putative SPA homologues with GFP (AN4698-SPA3, AN1948-SPA10, and AN4484-SPA13) and monitored their subcellular locations through the cell cycle. We included the KfsA protein kinase (AN0822; kinase for septation), as it also contains long, intrinsically disordered domains and fulfils sequence characteristics of a SPA protein (Lai et al., 2012) as defined (Chuan Hock Koh, personal communication) using Sirius PSB software (Koh et al., 2009). KfsA was also previously shown to locate to septa (Takeshita et al., 2007).
The SPA proteins of N. crassa locate to mature septa, and we wanted to establish whether they similarly locate to mature septa in A. nidulans. During septum formation, SPA10-GFP located to a closing ring structure (Figure 4A) reminiscent of the contractile actomyosin ring (CAR) that drives cytokinesis (Seiler and Justa-Schuch, 2010; Lee et al., 2012). The dynamic ring of SPA10-GFP preceded septum formation that was visible by bright-field microscopy (Figure 4A). However, unlike other CAR-associated proteins, such as actin and myosin II, upon completion of septation, SPA10-GFP remained associated with mature septa. High-resolution through Z confocal imaging and FM4-64 costaining (Fischer-Parton et al., 2000; Penalva, 2005) of septal wall membrane (Figure 4B) revealed SPA10-GFP located as a central disk structure in the vicinity of the septal pore. No redistribution of SPA10-GFP was apparent during cell cycle progression or during mitotic SAC arrest. SPA10-GFP was stably associated with septa with minimal recovery during FRAP analysis over a 9-min recovery period (Figure 4, I and J).
FIGURE 4:
Analysis of GFP-tagged septal pore–associated proteins during cell cycle progression. Live-cell imaging of (A) SPA10-GFP with (B) FM4-64 using strain SO1306, (C) KfsA-GFP with (D) FM4-64 using strain SO1316, (E) SPA13-GFP with (F) FM4-64 using strain SO1310, and (G) SPA3-GFP with (H) FM4-64 using strain SO1302. Cells were stained with 5 μM FM4-64 for 5 min before imaging. The GFP + FM4-64 images were captured with 0.2 μm Z-sections, and three-dimensional images were reconstructed using ImageJ, showing 0 and 90° rotations. (I) Images collected during FRAP analysis of the indicated GFP-tagged SPA proteins at mature septa. (J) FRAP analysis of SPA-GFP proteins at septa (SPA10-GFP, n = 15; KfsA-GFP, n = 11; SPA13-GFP, n = 17; SPA3-GFP, n = 10). The average fluorescence recovery profile of each is shown ± SD as indicated. Arrows indicate mature septa, and arrowheads indicate forming septa. Bars, 2.5 μm.
N-terminally tagged GFP-KfsA expressed from the regulatable alcA promoter was found to locate to the cell cortex, as well as to mature septa (Takeshita et al., 2007). The endogenously C-terminally tagged KfsA-GFP we generated did not locate to the cell cortex but located both to forming septa as they became visible by bright-field microscopy and at mature septa (Figure 4C). Unlike SPA10-GFP (Figure 4A), we did not detect evidence of KfsA-GFP associating with a CAR-like structure during septum formation. Instead KfsA-GFP accumulated as a disk structure centered in the septal pore region (Figure 4D) as septa formed (Figure 4C). No redistribution of KfsA-GFP was apparent during cell cycle progression or during mitotic SAC arrest. FRAP analysis indicated that KfsA-GFP was a relatively stable component of the septal pore region, although not as stable as SPA10-GFP (Figure 4, I and J).
SPA13-GFP located to septa after they were visible by light microscopy—noticeably at a later time than KfsA-GFP (Figure 4E). In addition, instead of forming a disk structure, three-dimensional rotations revealed that SPA13-GFP displayed a washer-like ring structure around the cell wall of the septal pore (Figure 4F). No redistribution of SPA13-GFP occurred during cell cycle progression or during mitotic SAC arrest. FRAP analysis showed that SPA13-GFP was a relatively stable component of the septal pore region, although, again, not as stable as SPA10-GFP (Figure 4, I and J).
Initial live-cell imaging of SPA3-GFP found that it did not locate to forming septa or at all mature septa. Instead, SPA3-GFP transiently located as a defined dot in the center of a subpopulation of septa, suggestive of regulated location at the septal pore (Figure 4, G and H). The percentage of septa to which SPA3-GFP was located increased from 29 to 85% after benomyl addition, which causes SAC-mediated mitotic arrest. This indicates that SPA3-GFP locates to the center region of septal pores in a regulated manner during mitosis. The SPA3-GFP at septal pores was not stably associated and showed rapid, yet incomplete recovery during FRAP analysis (Figure 4, I and J).
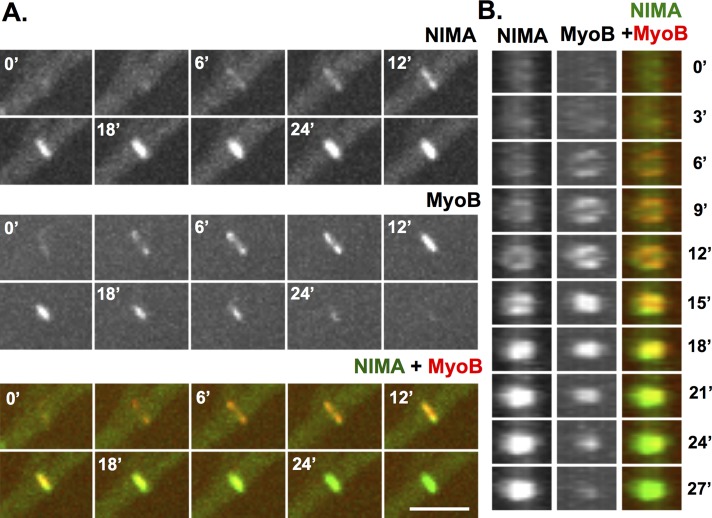
NIMA colocalizes with MyoB during early stages of septum formation
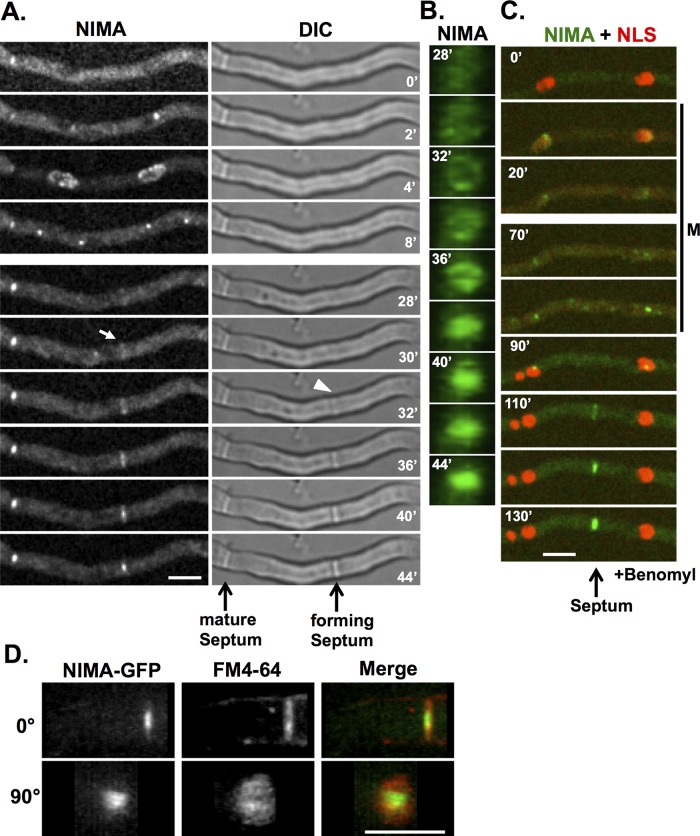
We recently endogenously GFP-tagged NIMA to generate a functional chimera and used live imaging to follow its locations during mitosis (Shen and Osmani, 2013). During these studies, in addition to its mitotic nuclear locations, NIMA-GFP was also seen to appear with a closing ring structure during septum formation (Figure 5, A and B) reminiscent of the CAR. SepA, a formin, and MyoB, a type II myosin, are components and therefore markers for the CAR (Sharpless and Harris, 2002; Taheri-Talesh et al., 2012; Supplemental Figure S3, A and B). It was recently shown that, before locating to the CAR, MyoB forms string-like structures that coalesce to form the CAR (Taheri-Talesh et al., 2012). Using GFP-tagged MyoB, we also observed this behavior. However, when using the mCherry variant of red fluorescent protein (chRFP)–tagged version of MyoB, as for our studies, the MyoB string structures were not easily detected (Supplemental Figure S3). As soon as we were able to detect MyoB-chRFP concentrating at the forming CAR, we also detected NIMA-GFP starting to accumulate at the same locations (Figure 5, A and B). However, a marked difference in the locations of MyoB and NIMA occurred during the later stages of the CAR constriction. During CAR constriction the level of MyoB-chRFP decreased and was no longer enriched at mature septa. In contrast, as the levels of MyoB-chRFP decreased, the levels of NIMA-GFP at septa increased, and NIMA-GFP remained associated with mature septa after MyoB-chRFP was no longer detectable (Figure 5, A and B). At mature septa (Figure 5B), NIMA-GFP formed a central disk structure near the septal pore region. Costaining with FM4-64 confirmed that NIMA-GFP (Figure 6D) locates at septal pores. These location studies indicate NIMA has the potential to play roles during septum formation and at mature septa in addition to its known roles at nuclei during mitosis.
FIGURE 5:
NIMA colocalizes with the MyoB during septum formation. (A) A NIMA-GFP strain (KF244) with MyoB-chRFP was imaged during septation. (B) The septation region was rotated 90° (using ImageJ software) for an end-on view of septum formation. Both NIMA-GFP and MyoB-chRFP appear at forming septa as rings that contract and fill. Once a septum is formed, NIMA-GFP persists at the septum, whereas MyoB-chRFP does not. Bar, 5 μm.
FIGURE 6:
NIMA locates to forming septa and mature septa. (A) After first locating to SPB and the nuclear periphery during mitosis, NIMA-GFP (strain KF005) appears at the forming septa before septa become visible by differential interference contrast. The arrow indicates when NIMA-GFP first becomes visible at the forming septa, and the arrowhead indicates when a septum starts to become visible. (B) The region of NIMA-GFP at the forming septa in A was rotated 90° (using ImageJ software) for an end-on view of the NIMA-GFP ring converting to a disk. (C) In the presence of benomyl, cells (strain KF033) enter and arrest at the mitotic SAC arrest point, during which some NIMA-GFP localizes to the kinetochore region, as previously shown (Shen and Osmani, 2013), and NLS-DsRed remains dispersed from nuclei. After cells exit the mitotic SAC arrest, NLS-DsRed is reimported into the G1 nuclei, and after a delay of ∼15 min, NIMA-GFP locates to forming septa, as occurs also after normal mitosis. (D) A NIMA-GFP strain (KF005) was stained with FM4-64 and images were captured at 0.2-μm Z-sections. Three-dimensional images of NIMA-GFP and FM4-64 were reconstructed using ImageJ, and 0° and 90° rotations are shown. Bars, 5 μm.
Insights into septum formation using NIMA-GFP as a marker for mitosis and septation
During mitotic entry, NIMA-GFP first located to spindle pole bodies (SPBs) and then spread around the nuclear periphery (Figure 6A, 0–4′) as previously described (De Souza et al., 2000, 2004; Shen and Osmani, 2013). During mitotic exit, NIMA-GFP located back to the separated SPBs (Figure 6A, 4′–8′), and for a period after mitosis (typically 20–30 min; Supplemental Figure S4), NIMA-GFP did not locate to specific locations but then located to forming septa as they became visible by light microscopy (Figure 6A and rotation in Supplemental Movie S1). The position of septum formation was suggested to be marked by the location of a mitotic nucleus (Harris, 2001; Seiler and Justa-Schuch, 2010). Because we could follow the site of mitosis and subsequent location of the septa using NIMA-GFP, we were able to test this idea. We found that the majority of septa do not form at a position dictated by a previous mitotic nucleus. Instead, the majority of septa were formed at a position corresponding to a location in between two previous mitotic nuclei (Figure 6A). Of 20 cells tracked from mitosis to septation, 18 formed septa at a position located between two previous mitotic nuclei. It was been reported, in studies using fixed cell samples (Momany and Hamer, 1997), that microtubules are required for the initiation and progression of septation in A. nidulans. To investigate this using live-cell imaging, we treated cells expressing NIMA-GFP and the nuclear transport marker NLS-DsRed (Suelmann et al., 1997) with benomyl to depolymerize microtubules and monitored for mitosis and septation. Cells without microtubule functions traverse interphase and become mitotically arrested due to activation of the SAC (De Souza et al., 2011). During entry into mitotic SAC arrest (Figure 6C), NIMA-GFP locates to mitotic nuclei, and NLS-DsRED is dispersed from nuclei because nuclear transport is suspended during A. nidulans mitotic arrest (De Souza et al., 2011), as occurs during normal mitosis. During mitotic SAC arrest, some NIMA-GFP remained associated with the SPB region of mitotic nuclei (Shen and Osmani, 2013). However, as previously shown (De Souza et al., 2011), the mitotic arrest is not permanent, and cells exit the mitotic state into G1 after the SAC is turned off. NLS-DsRED was transported back into nuclei during mitotic exit (Figure 6C, 90′). This process has been termed spindle-independent mitotic exit (SIME; De Souza et al., 2011). When cells exit mitotic SAC arrest there is a lag period, similar to that after completion of normal mitoses, only after which does NIMA-GFP appear as a constricting ring at the site of septation (Figure 6C, 110′). The data show that NIMA-GFP can locate to forming septa in the absence of microtubules, indicating that microtubules are not required for initiation or completion of septation (Figure 6C). Septa were predominantly formed between previous mitotic nuclei and not at a location corresponding to where a mitotic nucleus had been located (Figure 6C; 13 of 14 cells had septation occur between previous mitotic nuclei). We asked whether microtubule depolymerization affects the timing of the start, or the time to complete, septation (Supplemental Figure S4). We found that in the absence of microtubules the delay between exit from mitosis and septation becomes shorter, whereas the time to complete septation becomes slightly longer (Supplemental Figure S4).
The level of NIMA at mature septa is under cell cycle control
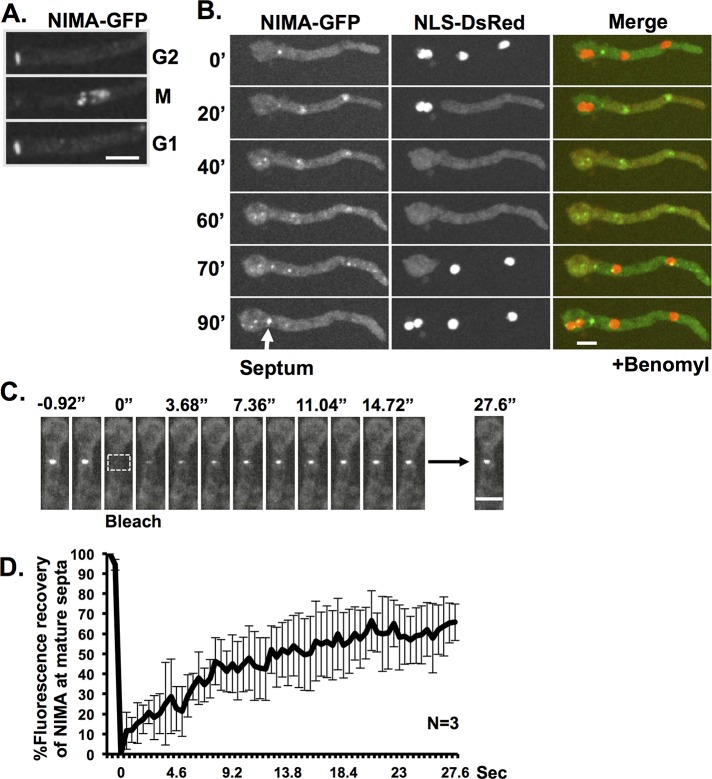
The cell cycle–regulated characteristics of septal pores, as revealed here, indicates that mitotic regulators might influence septa during mitosis. In this regard, after completion of septation, some NIMA-GFP remains associated with the mature septa during interphase. However, we noticed that the level of NIMA-GFP at septa is reduced when cells enter mitosis and returns to higher interphase levels upon completion of mitosis (Figures 6A and 7A). This pattern was observed in 31 of 32 mitotic cells with septa. Similar behavior occurred during mitotic SAC arrest, with NIMA-GFP only returning to its higher interphase septal levels after nuclei had undergone SIME and entered interphase (Figure 7B). These observations indicate that septal NIMA is under cell cycle regulation such that during interphase its levels are high but during mitosis its levels at septa are decreased. The cell cycle–regulated septal levels of NIMA-GFP indicate that it might not be stably bound to septa, and we investigated this using FRAP analysis. After photobleaching, the fluorescence of septal NIMA-GFP recovered rapidly (Figure 7C), with the average fluorescence recovering to 65.8% and t1/2 of only 6.4 s (Figure 7D). This indicates that NIMA is weakly associated at mature septa during interphase.
FIGURE 7:
NIMA is a dynamic protein at mature septa. (A) NIMA-GFP locates at mature septa during interphase but is removed during mitosis, when NIMA appears at nuclei to promote mitosis. NIMA-GFP is then visible again at septa after mitosis when cells enter G1. (B) In benomyl-treated cells, NIMA-GFP behaves similarly, but because cells remain in an extended mitotic SAC arrest, NIMA-GFP is removed from septa during this mitotic period but returns when both cells exit mitotic SAC arrest and reimport NLS-DsRed to their nuclei. (C) FRAP analysis of NIMA-GFP at mature septa. The rectangle indicates the region that was photobleached. Images were taken every 0.46 s. (D) The average fluorescence recovery profile of NIMA-GFP at septa (n = 3). The average is shown ± SD. Strain KF005 was used in A and C. Strain KF033 was used in B. Bars, 5 μm.
NIMA activity is required to maintain septal pore opening
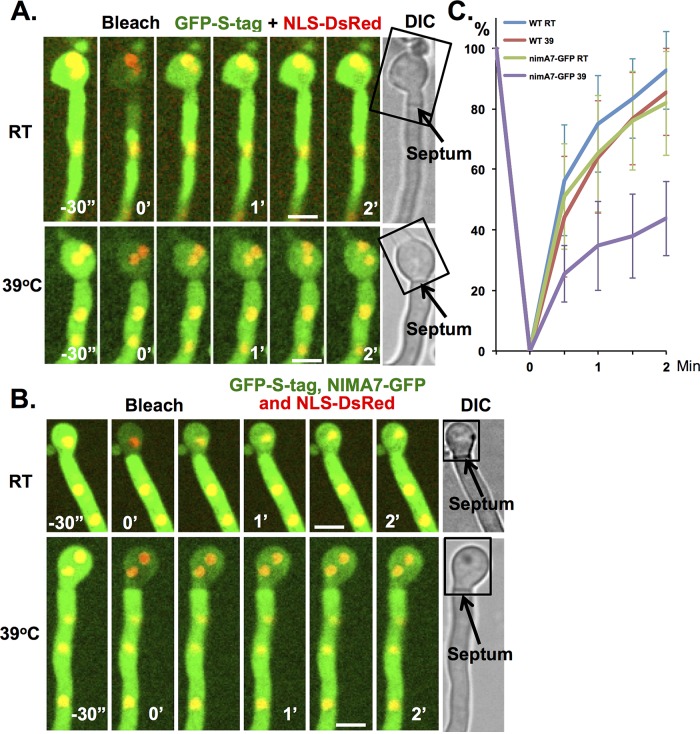
The dynamic associations of NIMA-GFP with septa and their closing during mitosis suggested the following. During interphase, NIMA at mature septa helps to maintain them in an open state, and its removal at mitosis promotes septal pore closing. To test this idea, we generated strains into which the temperature-sensitive nimA7 mutation was incorporated. This allowed us to investigate the effects of NIMA inactivation during interphase on septal pore permeability. The strain also carried cytoplasmic GFP and NLS-DsRed, enabling assay of septal pore permeability. We compared the rate of diffusion across the septal pore between a wild-type strain and the NIMA7-GFP strain at 23 and 39°C (restrictive for nimA7) during interphase. The results indicate that inactivation of NIMA caused a marked decrease in septal pore permeability (Figure 8, A–C), revealing a functional role for NIMA in keeping septal pores in an open state during interphase.
FIGURE 8:
NIMA is required for opening septal pores in interphase. FRAP analysis in (A) wild-type strain (KF491) and (B) nimA7-GFP strain (KF486). (A) The fast recovery of GFP-S-tag at room temperature and 39°C in wild-type cells indicates that septal pores are open in interphase and temperature does not affect the opening of septal pores. (B) The fast recovery of GFP-S-tag at room temperature in nimA7-GFP cells indicates that septal pores are open in the presence of uninhibited NIMA function. The slower recovery of GFP-S-tag at 39°C in nimA7-GFP cells indicates that septal pores are more closed in interphase when NIMA function is impaired. The rectangles in A and B indicate the region that was photobleached. One prebleach and five postbleach images were captured at 30-s intervals. (C) The average fluorescence recovery profile at room temperature or 39°C in wild-type (n = 15 for room temperature and 39°C) or nimA7-GFP cells (n = 12 for room temperature and 39°C). Average ± SD. Bars, 5 μm.
DISCUSSION
Septal pore permeability is subject to cell cycle regulation
It has been known for several decades that nuclei within tip cells of A. nidulans progress synchronously through the cell cycle, whereas nuclei within connected subapical cells are removed from the cell cycle (Kaminskyj and Hamer, 1998) and were more recently shown to be arrested in G1 (Nayak et al., 2010; Edgerton-Morgan and Oakley, 2012). How apical cells can enter a mitotic state without affecting mitosis of nuclei in adjacent connected cells was not understood. Our studies reveal for the first time that the permeability properties of A. nidulans septal pores are subject to cell cycle regulation to ensure that during mitosis they are closed. This likely explains how the mitotic state of apical cells does not affect the nuclei in cells previously assumed to be directly connected throughout the cell cycle via open septal pores. The mitotic closing of septal pores also prevents nuclear proteins released during mitosis from moving into adjacent connected cells. This would ensure that the mitotically released nuclear proteins in apical cells are available for reimport and normal nuclear G1 functions when mitosis is completed rather than being sequestered in the G1 nuclei of subapical cells during mitosis.
Given the key roles that Woronin bodies play during fungal septal pore closing, we investigated their potential roles during mitotic pore closing. We found that neither the formation of Woronin bodies nor that of SO proteins was required to close septal pores at mitosis. This suggests that additional proteins might be involved in mitotic septal pore closing. Prime candidates are a recently discovered class of proteins called SPA proteins identified in N. crassa. Seventeen such proteins have been identified that locate to septa as ring structures that line the septal pore or have central or peripheral septal pore locations. The SPA proteins are characterized by long, intrinsically disordered domains and as such are poorly conserved at the primary sequence level. We were able to identify three SPA proteins that locate to A. nidulans septal pores, each in a distinctive manner. The time at which SPA proteins locate to septal pores has not been reported in N. crassa. We found that SPA10 locates to the forming septum in a manner initially analogous to the contractile ring of the CAR. Unlike CAR proteins, which are removed from septa once they are formed, SPA10-GFP remained as a disk structure associated near the septal pore of mature septa.
FRAP analysis showed SPA10-GFP to be a stable component of the septal pore region. The early location and stability of SPA10 at septal pores indicate that the pore is not free of obstruction, and recent electron microscope analysis further supports the presence of potential barrier material within the septal pore of A. nidulans (Griffith et al., 2011) as well as N. crassa (Lai et al., 2012). This suggests that septal pores of N. crassa and A. nidulans are not completely open conduits that provide free mixing of cytoplasm between interphase cells. The stable presence of proteins at septal pores might explain why movement of proteins throughout the cytoplasm is significantly faster than that through the septal pores between cells. This likelihood becomes more plausible when the biophysical properties of SPA proteins are considered, as they are capable of forming hydrogel-like structures that can form pore-occluding plugs (Lai et al., 2012). Our findings show that pore occlusion in A. nidulans is under cell cycle regulation, and we identified one SPA protein, SPA3, that locates specifically to septal pores only during mitosis and could potentially participate in septal pore closing during mitosis. However, initial deletion analysis indicates that septal pores are closed during mitosis in the absence of SPA3. One reason for this might be functional redundancies with other SPA proteins. Such redundancy has been shown for the intrinsically disordered FG-repeat proteins that occupy the central transport channel of nuclear pore complexes, which can also form hydrogels. For example, Saccharomyces cerevisiae cells can tolerate deletion of several of its FG-repeat Nup genes representing half its total mass of FG-repeat domains without loss of the NPC permeability barrier or viability (Strawn et al., 2004). Future identification and deletion analysis of additional SPA proteins will allow their roles in mitotic septal pore occlusion to be investigated further.
NIMA at forming septa
NIMA-GFP was found to locate to forming septa as a constricting CAR-like ring structure. Formation of the CAR is a key feature of septation and guides deposition of septal wall material. NIMA-GFP colocalizes with markers of the CAR during the early stages of septum formation, indicating that NIMA has the potential to play a role in regulating septation. However, unlike other CAR components, NIMA remained at mature septa in a manner similar to SPA10, as described. Unlike SPA10, however, NIMA is not a stable component of the septal pore, but instead dynamically associates with them, suggesting a more dynamic regulatory function.
There are several potential reasons that NIMA might locate to the septum formation apparatus in addition to perhaps helping regulate septation itself. Unlike most other types of cytokinesis, the final stages of abscission are incomplete in filamentous fungi, leaving a stable septal pore structure, which in A. nidulans is typically ∼50 nm in diameter. Little is known about how septation is halted to generate septal pores. However, other types of intercellular bridges are formed via a process that involves arresting completion of abscission, leading to incomplete cytokinesis and generation of pores of defined sizes (Haglund et al., 2011; Bloemendal and Kuck, 2013). Because NIMA remains in the vicinity of the forming septal pore during septation, it could perhaps help regulate the arrest of cytokinesis, leading to the subsequent formation of septal pores. Of further interest, NIMA-related kinases (Nek kinases) have been implicated in the regulation of cytokinesis, and Nek2, Nek6, and Nek7 locate to the midbody during mitotic exit (Fletcher et al., 2005; O'Regan and Fry, 2009), and Nek7 deletion in mice causes cytokinesis defects and polyploidy (Salem et al., 2010). Moreover, Drosophila Nek2 locates to the midbody during mitotic exit, and ectopic expression of Nek2 causes a failure in cytokinesis, indicating that it plays an important role in cell division (Prigent et al., 2005). Collectively, with our findings, the data suggest a conserved role for NIMA-related kinases during cytokinesis. It will also be interesting to investigate the potential roles of this family of kinase in the formation of intercellular bridges that form via incomplete cytokinesis.
The role of NIMA at mature septa
One function for NIMA at septa for which we provide experimental evidence is modulation of their gating properties. During normal cell cycle progression, NIMA-GFP locates to mature septa when they are in an open state but is removed from them when they are closed during mitosis. This pattern is consistent with the idea that NIMA could help opening of septal pores during interphase and then allow them to close during mitosis when NIMA locates to nuclei to promote mitosis. Further supporting this model, we found that inactivation of NIMA during interphase affects septal pore permeability, promoting their closing as occurs during mitosis.
When NIMA promotes mitosis it does so by transiently locating to NPCs, triggering their partial disassembly. This involves NIMA promoting the removal of all peripheral NPC proteins, including the FG-repeat Nups that provide the central diffusion barrier of the NPC. In this manner NPCs are opened during mitosis to allow diffusion in and out of mitotic nuclei. The SPA proteins, like FG-repeat Nups, are intrinsically disordered proteins that can self-assemble to form higher-ordered hydrogels (Lai et al., 2012). SPA proteins therefore share several similarities with FG-Nups, including their assembly at biologically important pores, the presence of long, intrinsically disordered domains, and their ability to form hydrogels in vitro (Denning et al., 2003; Frey et al., 2006). It is therefore possible that there are mechanisms to regulate the assembly and disassembly of SPA protein aggregates that are similar to the mechanisms that regulate the assembly and disassembly of FG-Nups to open and close nuclear pores. In such a scenario NIMA would play a role in regulating the assembly/disassembly of SPA protein aggregates to induce the opening and closing of septal pores. From these considerations we propose the following working model: NIMA is removed from septal pores during mitosis to initiate the closing of septal pores and subsequently locates to NPCs to trigger the opening of nuclear pores. During mitotic exit, NIMA is removed from NPCs to allow NPC reassembly and locates back to septal pores to prompt their opening. Although further experimentation will be required to define and test this model, it is important that we have shown that at least one SPA protein is kept from septal pores during interphase but resides at them during mitosis. In addition, experimental inactivation of NIMA was shown to cause closing of septal pores during interphase. These two findings fit well with the model and provide impetus and direction to further define how the fungal septal pore class of intercellular bridges are opened and closed during cell cycle progression.
MATERIALS AND METHODS
General techniques and strains
Media and general techniques for genetic analysis of A. nidulans, gene deletion, endogenous fluorescent protein tagging, transformation, and confocal microscopy have been described (Pontecorvo et al., 1953; Yang et al., 2004; Nayak et al., 2006; Szewczyk et al., 2006; Liu et al., 2009; De Souza et al., 2013). Strains used in this study are listed in Table 1 and primers in Supplemental Table S1. All GFP- and chRFP-tagged proteins are the only copy of each protein expressed in the cells and are expressed from their own promoter, except GFP-S-Tag (De Souza et al., 2014), which is expressed under the promoter of AN1553. To express GFP-S-Tag under the control of the AN1553 promoter, a GFP-S-Tag targeting construct containing the AN1553 promoter region, GFP-S-Tag-pyrGAF, and the 5′ region of AN1553 was generated by fusion PCR and transformed into strain AY02. To C-terminally tag HexA with GFP-PTS1 (peroxisome-targeting signal 1), a two-step strategy was used. In the first, a HexA-GFP targeting construct containing the region upstream of the stop codon of HexA, then GFP-pyrGAF, and then the 3′-untranslated region of hexA was generated by fusion PCR and transformed into strain KF075 generating a HexA-GFP strain (KF078). Because the C-terminal GFP fusion interferes with the C-terminal PTS1 sequence (S-R-L) required for HexA targeting to Woronin bodies (Gould et al., 1989; Keller et al., 1991; Jedd and Chua, 2000), the HexA-GFP failed to target to Woronin bodies. To generate a HexA-GFP-PTS1 strain (KF095), a HexA-GFP-PTS1 targeting fragment was made by fusion PCR incorporating a primer to insert the PTS1 amino acid sequence for S-R-L at the C-terminus of HexA-GFP. The construct was landed at the HexA-GFP-pyrGAF locus using 5-fluoroorotic acid counterselection in strain KF078.
TABLE 1:
Strains used.
| Strain | Genotype |
|---|---|
| Ay02 | pyrG89; pyroA4; ΔyA:NLS-DsRed; ΔnKuA::argB; (argB2) |
| KF005 | nimA-GFP::pyrGAF; (pyrG89); pyroA4; ΔnKuA:: argB; (argB2); sE15; nirA14; wA3; fwA1; chaA1 |
| KF033 | nimA-GFP::pyrGAF; pyroA4; ΔyA:NLS-DsRed; ΔnKuA::argB; (argB2); (nirA14?); (sE15?) |
| KF075 | pyrG89; pyroA4; nirA14(?);ΔyA::NLS-DsRed; ΔnKuA::argB; (argB2?) |
| KF078 | hexA-GFP::pyrGAF; (pyrG89); pyroA4; nirA14(?);ΔyA::NLS-DsRed; ΔnKuA::argB; (argB2?) |
| KF081 | ΔhexA::pyrGAF; (pyrG89); pyroA4; (nirA14?); ΔyA::NLS-DsRed; ΔnKuA::argB; (argB2?) |
| KF095 | hexA-GFP-PTS1; pyrG89; pyroA4; nirA14(?);ΔyA::NLS-DsRed; ΔnKuA::argB (argB2?) |
| KF244 | nimA-GFP::pyrGAF; (pyrG89); myoB-chRFP::pyroAAF; (pyroA4); nirA14; sE15; ΔnKuA:: argB; (argB2); wA3; fwA1; chaA1 |
| KF298 | sepA-GFP::pyrGAF; (pyrG89); myoB-chRFP::pyroAAF; (pyroA4); ΔnKuA::argB (argB2); nirA14; sE15; wA3 |
| KF361 | ∆SO::pyrGAF; ΔyA::NLS-DsRed; ∆nKuA :: argB; (argB2); pyrG89; pyroA4 |
| KF394 | ∆SO::pyroAAF; (pyroA4); ∆hex1::pyrGAF; (pyrG89); ΔyA::NLS-DsRed; ∆nKuA:: argB; (argB2) |
| KF430 | NLS-GFP::pyr4; (pyrG89); pyroA4; sE15; nirA14; ΔnKuA::argB; (argB2); fwA1; chaA1; wA3 |
| KF486 | nimA7-GFP::pyrGAF; GFP-S-tag::pyrGAF; argB2; gcp3-chRFP::riboAF?; ΔyA::NLS-DsRed; nirA14?; sE15? |
| KF491 | GFP-S-tag::pyrGAF; ΔnKuA::argB; (argB2); pyroA4; ΔyA::NLS-DsRed; nirA14?; sE15? |
| SO1302 | An-spa3-GFP::pyrGAF; (pyrG89); pyroA4; ΔnKuA:: argB; (argB2); sE15; nirA14; wA3; fwA1; chaA1 |
| SO1306 | An-spa10-GFP::pyrGAF; (pyrG89); pyroA4; ΔnKuA:: argB; (argB2); sE15; nirA14; wA3; fwA1; chaA1 |
| SO1310 | An-spa13-GFP::pyrGAF; (pyrG89); pyroA4; ΔnKuA:: argB; (argB2); sE15; nirA14; wA3; fwA1; chaA1 |
| SO1316 | kfsA-GFP::pyrGAF; (pyrG89); pyroA4; ΔnKuA:: argB; (argB2); sE15; nirA14; wA3; fwA1; chaA1 |
A question mark indicates that the marker may or may not be present in the strain.
FRAP analysis
FRAP analysis was performed by using a photokinesis unit on an UltraVIEW ERS confocal system (PerkinElmer, Waltham, MA). Regions were selected for photobleaching, and the fluorescence recovery rates of GFP-tagged proteins were recorded by time-lapse imaging. To examine the fluorescence recovery of septal NIMA-GFP, a single confocal slice was captured at 0.46-s intervals. Two prebleach images and 60 postbleach images were recorded. To monitor the fluorescence recovery of SPA proteins at mature septa, 1 prebleach image and 20 postbleach images were captured at 30-s intervals. To determine the opening and closing of septal pores, a FRAP-based septal pore permeability assay was developed. Cells were grown at 24°C for 22 h to generate cells containing septa before analysis. In the presence or absence of 2.4 μg/ml benomyl, one cellular compartment was selected for FRAP analysis. Previously published data indicate that the plugging of septal pores might occur under various stress conditions, including laser treatment (Maruyama et al., 2010). To avoid plugging of septal pores due to laser exposure, images were collected as single confocal slices and captured at 30-s intervals. One prebleach image and 5 postbleach images were recorded for monitoring the recovery of GFP-S-tag. Data were processed according to Hames et al. (2005). For each time point, the fluorescence intensity of the photobleached region of interest (ROI; P1) was determined using ImageJ software (National Institutes of Health, Bethesda, MD). The fluorescence intensity of an unbleached ROI in the cell was determined (U2), and a normalized intensity (In) was calculated using P1/U2. The background fluorescence intensity (Bk) was set as the first frame after photobleaching, and this value was subtracted from all frames (In-Bk). The fluorescence intensity of the frame (In-Bk) before photobleaching was normalized to 100% and used to normalize (In-Bk) at the other time points (Hames et al., 2005). To determine a role of NIMA in opening septal pores in interphase, cells (wild type or nimA7-GFP) were grown at 24°C for 21 h and then either continued to grow at 24°C or shifted to 39°C for 1 h before FRAP analysis at 24 or 39°C.
Identification of SPA proteins in A. nidulans
Potential orthologues of N. crassa SPA proteins were identified using BLASTP analysis at AspGD (Arnaud et al., 2012) with the N. crassa SPA protein sequences. Reciprocal BLASP analysis at the National Center for Biotechnology Information and comparison of the relative distribution and levels of disordered unstructured domains versus predicted structured regions (IUPred; Dosztanyi et al., 2005) and the presence of coiled-coiled domains (Lupas et al., 1991) were further used to define the potential orthologues as previously described (Lai et al., 2012).
Supplementary Material
Acknowledgments
We thank all members of the Osmani laboratory for help and input to this work and Peter Punt for the suggestion to use the AN1553 promoter to constitutively express GFP-Stag. We also thank Colin P. De Souza for helpful reading of the manuscript and use of the GFP-S-Tag construct he developed, as well as Chuan Hock Koh for a bioinformatics search for potential SPA proteins in A. nidulans. We are also indebted to Greg Jedd for insightful comments regarding SPA proteins and septal pore biology. This work was supported by a grant from the National Institutes of Health (GM042564) to S.A.O.
Abbreviations used:
- chRFP
mCherry variant of red fluorescent protein
- Nek
NIMA-related kinase
- NPC
nuclear pore complex
- SAC
spindle assembly checkpoint
- SPA
septal pore associated
- SPB
spindle pole body
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-12-0718) on January 22, 2014.
REFERENCES
- Arnaud MB, et al. The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 2012;40:D653–D659. doi: 10.1093/nar/gkr875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird GS, Zacharias DA, Tsien RY. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J, Ebel F. Characterization of the major Woronin body protein HexA of the human pathogenic mold Aspergillus fumigatus. Int J Med Microbiol. 2013;303:90–97. doi: 10.1016/j.ijmm.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Bloemendal S, Kuck U. Cell-to-cell communication in plants, animals, and fungi: a comparative review. Naturwissenschaften. 2013;100:3–19. doi: 10.1007/s00114-012-0988-z. [DOI] [PubMed] [Google Scholar]
- Denning DP, Patel SS, Uversky V, Fink AL, Rexach M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci USA. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Hashmi SB, Osmani AH, Andrews P, Ringelberg CS, Dunlap JC, Osmani SA. Functional analysis of the Aspergillus nidulans kinome. PLoS One. 2013;8:e58008. doi: 10.1371/journal.pone.0058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Hashmi SB, Osmani AH, Osmani SA. Application of a new dual localization affinity purification tag reveals novel aspects of protein kinase biology in Aspergillus nidulans. PLoS ONE. 2014 doi: 10.1371/journal.pone.0090911. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Hashmi SB, Yang X, Osmani SA. Regulated inactivation of the spindle assembly checkpoint without functional mitotic spindles. EMBO J. 2011;30:2648–2661. doi: 10.1038/emboj.2011.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Osmani SA. A new level of spindle assembly checkpoint inactivation that functions without mitotic spindles. Cell Cycle. 2011;10:3805–3806. doi: 10.4161/cc.10.22.18187. [DOI] [PubMed] [Google Scholar]
- De Souza CP, Osmani AH, Hashmi SB, Osmani SA. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr Biol. 2004;14:1973–1984. doi: 10.1016/j.cub.2004.10.050. [DOI] [PubMed] [Google Scholar]
- De Souza CP, Osmani AH, Wu LP, Spotts JL, Osmani SA. Mitotic histone H3 phosphorylation by the NIMA kinase in Aspergillus nidulans. Cell. 2000;102:293–302. doi: 10.1016/s0092-8674(00)00035-0. [DOI] [PubMed] [Google Scholar]
- Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- Edgerton-Morgan H, Oakley BR. gamma-Tubulin plays a key role in inactivating APC/C(Cdh1) at the G(1)-S boundary. J Cell Biol. 2012;198:785–791. doi: 10.1083/jcb.201203115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MJ, Tan K, Reck-Peterson SL. Lis1 is an initiation factor for dynein-driven organelle transport. J Cell Biol. 2012;197:971–982. doi: 10.1083/jcb.201112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiddy C, Trinci AP. Mitosis, septation, branching and the duplication cycle in Aspergillus nidulans. J Gen Microbiol. 1976;97:169–184. doi: 10.1099/00221287-97-2-169. [DOI] [PubMed] [Google Scholar]
- Fischer-Parton S, Parton RM, Hickey PC, Dijksterhuis J, Atkinson HA, Read ND. Confocal microscopy of FM4-64 as a tool for analysing endocytosis and vesicle trafficking in living fungal hyphae. J Microsc. 2000;198:246–259. doi: 10.1046/j.1365-2818.2000.00708.x. [DOI] [PubMed] [Google Scholar]
- Fleissner A, Glass NL. SO, a protein involved in hyphal fusion in Neurospora crassa, localizes to septal plugs. Eukaryot Cell. 2007;6:84–94. doi: 10.1128/EC.00268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher L, Cerniglia GJ, Yen TJ, Muschel RJ. Live cell imaging reveals distinct roles in cell cycle regulation for Nek2A and Nek2B. Biochim Biophys Acta. 2005;1744:89–92. doi: 10.1016/j.bbamcr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Frey S, Richter RP, Gorlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- Goodenough DA, Paul DL. Gap junctions. Cold Spring Harb Perspect Biol. 2009;1:a002576. doi: 10.1101/cshperspect.a002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Keller GA, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindaraghavan M, Lad AA, Osmani SA. The NIMA kinase is required to execute stage specific mitotic functions after initiation of mitosis. Eukaryot Cell. 2014;13:99–109. doi: 10.1128/EC.00231-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J, Penalva MA, Reggiori F. Adaptation of the Tokuyasu method for the ultrastructural study and immunogold labelling of filamentous fungi. J Electron Microsc (Tokyo) 2011;60:211–216. doi: 10.1093/jmicro/dfr026. [DOI] [PubMed] [Google Scholar]
- Haglund K, Nezis IP, Stenmark H. Structure and functions of stable intercellular bridges formed by incomplete cytokinesis during development. Commun Integr Biol. 2011;4:1–9. doi: 10.4161/cib.4.1.13550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames RS, Crookes RE, Straatman KR, Merdes A, Hayes MJ, Faragher AJ, Fry AM. Dynamic recruitment of Nek2 kinase to the centrosome involves microtubules, PCM-1, and localized proteasomal degradation. Mol Biol Cell. 2005;16:1711–1724. doi: 10.1091/mbc.E04-08-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SD. Septum formation in Aspergillus nidulans. Curr Opin Microbiol. 2001;4:736–739. doi: 10.1016/s1369-5274(01)00276-4. [DOI] [PubMed] [Google Scholar]
- Hynes MJ, Murray SL, Khew GS, Davis MA. Genetic analysis of the role of peroxisomes in the utilization of acetate and fatty acids in Aspergillus nidulans. Genetics. 2008;178:1355–1369. doi: 10.1534/genetics.107.085795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd G, Chua NH. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat Cell Biol. 2000;2:226–231. doi: 10.1038/35008652. [DOI] [PubMed] [Google Scholar]
- Jedd G, Pieuchot L. Multiple modes for gatekeeping at fungal cell-to-cell channels. Mol Microbiol. 2012;86:1291–1294. doi: 10.1111/mmi.12074. [DOI] [PubMed] [Google Scholar]
- Kaminskyj SG, Hamer JE. hyp loci control cell pattern formation in the vegetative mycelium of Aspergillus nidulans. Genetics. 1998;148:669–680. doi: 10.1093/genetics/148.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller GA, Krisans S, Gould SJ, Sommer JM, Wang CC, Schliebs W, Kunau W, Brody S, Subramani S. Evolutionary conservation of a microbody targeting signal that targets proteins to peroxisomes, glyoxysomes, and glycosomes. J Cell Biol. 1991;114:893–904. doi: 10.1083/jcb.114.5.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CH, Lin S, Jedd G, Wong L. Sirius PSB: a generic system for analysis of biological sequences. J Bioinform Comput Biol. 2009;7:973–990. doi: 10.1142/s0219720009004436. [DOI] [PubMed] [Google Scholar]
- Lai J, Koh CH, Tjota M, Pieuchot L, Raman V, Chandrababu KB, Yang D, Wong L, Jedd G. Intrinsically disordered proteins aggregate at fungal cell-to-cell channels and regulate intercellular connectivity. Proc Natl Acad Sci USA. 2012;109:15781–15786. doi: 10.1073/pnas.1207467109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, Kutay U. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 2011;144:539–550. doi: 10.1016/j.cell.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Lee IJ, Coffman VC, Wu JQ. Contractile-ring assembly in fission yeast cytokinesis: recent advances and new perspectives. Cytoskeleton (Hoboken) 2012;69:751–763. doi: 10.1002/cm.21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HL, De Souza CP, Osmani AH, Osmani SA. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex. Mol Biol Cell. 2009;20:616–630. doi: 10.1091/mbc.E08-06-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Ng SK, Lu Y, Low W, Lai J, Jedd G. Making two organelles from one: Woronin body biogenesis by peroxisomal protein sorting. J Cell Biol. 2008;180:325–339. doi: 10.1083/jcb.200705049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Maruyama J, Escano CS, Kitamoto K. AoSO protein accumulates at the septal pore in response to various stresses in the filamentous fungus Aspergillus oryzae. Biochem Biophys Res Commun. 2010;391:868–873. doi: 10.1016/j.bbrc.2009.11.154. [DOI] [PubMed] [Google Scholar]
- Maruyama J, Juvvadi PR, Ishi K, Kitamoto K. Three-dimensional image analysis of plugging at the septal pore by Woronin body during hypotonic shock inducing hyphal tip bursting in the filamentous fungus Aspergillus oryzae. Biochem Biophys Res Commun. 2005;331:1081–1088. doi: 10.1016/j.bbrc.2005.03.233. [DOI] [PubMed] [Google Scholar]
- Momany M. Polarity in filamentous fungi: establishment, maintenance and new axes. Curr Opin Microbiol. 2002;5:580–585. doi: 10.1016/s1369-5274(02)00368-5. [DOI] [PubMed] [Google Scholar]
- Momany M, Hamer JE. Relationship of actin, microtubules, and crosswall synthesis during septation in Aspergillus nidulans. Cell Motil Cytoskeleton. 1997;38:373–384. doi: 10.1002/(SICI)1097-0169(1997)38:4<373::AID-CM7>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Momany M, Richardson EA, Van Sickle C, Jedd G. Mapping Woronin body position in Aspergillus nidulans. Mycologia. 2002;94:260–266. [PubMed] [Google Scholar]
- Nayak T, Edgerton-Morgan H, Horio T, Xiong Y, De Souza CP, Osmani SA, Oakley BR. Gamma-tubulin regulates the anaphase-promoting complex/cyclosome during interphase. J Cell Biol. 2010;190:317–330. doi: 10.1083/jcb.201002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Regan L, Fry AM. The Nek6 and Nek7 protein kinases are required for robust mitotic spindle formation and cytokinesis. Mol Cell Biol. 2009;29:3975–3990. doi: 10.1128/MCB.01867-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani AH, Davies J, Liu HL, Nile A, Osmani SA. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol Biol Cell. 2006;17:4946–4961. doi: 10.1091/mbc.E06-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalva MA. Tracing the endocytic pathway of Aspergillus nidulans with FM4-64. Fungal Genet Biol. 2005;42:963–975. doi: 10.1016/j.fgb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- Prigent C, Glover DM, Giet R. Drosophila Nek2 protein kinase knockdown leads to centrosome maturation defects while overexpression causes centrosome fragmentation and cytokinesis failure. Exp Cell Res. 2005;303:1–13. doi: 10.1016/j.yexcr.2004.04.052. [DOI] [PubMed] [Google Scholar]
- Rosenberger RF, Kessel M. Synchrony of nuclear replication in individual hyphae of Aspergillus nidulans. J Bacteriol. 1967;94:1464–1469. doi: 10.1128/jb.94.5.1464-1469.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem H, Rachmin I, Yissachar N, Cohen S, Amiel A, Haffner R, Lavi L, Motro B. Nek7 kinase targeting leads to early mortality, cytokinesis disturbance and polyploidy. Oncogene. 2010;29:4046–4057. doi: 10.1038/onc.2010.162. [DOI] [PubMed] [Google Scholar]
- Sampson K, Heath IB. The dynamic behaviour of microtubules and their contributions to hyphal tip growth in Aspergillus nidulans. Microbiology. 2005;151:1543–1555. doi: 10.1099/mic.0.27750-0. [DOI] [PubMed] [Google Scholar]
- Seiler S, Justa-Schuch D. Conserved components, but distinct mechanisms for the placement and assembly of the cell division machinery in unicellular and filamentous ascomycetes. Mol Microbiol. 2010;78:1058–1076. doi: 10.1111/j.1365-2958.2010.07392.x. [DOI] [PubMed] [Google Scholar]
- Sharpless KE, Harris SD. Functional characterization and localization of the Aspergillus nidulans formin SEPA. Mol Biol Cell. 2002;13:469–479. doi: 10.1091/mbc.01-07-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen KF, Osmani SA. Regulation of mitosis by the NIMA kinase involves TINA and its newly discovered partner, An-WDR8, at spindle pole bodies. Mol Biol Cell. 2013;24:3842–3856. doi: 10.1091/mbc.E13-07-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S, Osmani SA. Analysis of all protein phosphatase genes in Aspergillus nidulans identifies a new mitotic regulator, fcp1. Eukaryot Cell. 2009;8:573–585. doi: 10.1128/EC.00346-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn LA, Shen T, Shulga N, Goldfarb DS, Wente SR. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat Cell Biol. 2004;6:197–206. doi: 10.1038/ncb1097. [DOI] [PubMed] [Google Scholar]
- Suelmann R, Sievers N, Fischer R. Nuclear traffic in fungal hyphae: in vivo study of nuclear migration and positioning in Aspergillus nidulans. Mol Microbiol. 1997;25:757–769. doi: 10.1046/j.1365-2958.1997.5131873.x. [DOI] [PubMed] [Google Scholar]
- Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Taheri-Talesh N, Xiong Y, Oakley BR. The functions of myosin II and myosin V homologs in tip growth and septation in Aspergillus nidulans. PLoS One. 2012;7:e31218. doi: 10.1371/journal.pone.0031218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita N, Vienken K, Rolbetzki A, Fischer R. The Aspergillus nidulans putative kinase, KfsA (kinase for septation), plays a role in septation and is required for efficient asexual spore formation. Fungal Genet Biol. 2007;44:1205–1214. doi: 10.1016/j.fgb.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Tenney K, Hunt I, Sweigard J, Pounder JI, McClain C, Bowman EJ, Bowman BJ. Hex-1, a gene unique to filamentous fungi, encodes the major protein of the Woronin body and functions as a plug for septal pores. Fungal Genet Biol. 2000;31:205–217. doi: 10.1006/fgbi.2000.1230. [DOI] [PubMed] [Google Scholar]
- Trinci AP. Growth of wild type and spreading colonial mutants of Neurospora crassa in batch culture and on agar medium. Arch Mikrobiol. 1973;91:113–126. doi: 10.1007/BF00424755. [DOI] [PubMed] [Google Scholar]
- Trinci AP, Collinge AJ. Structure and plugging of septa of wild type and spreading colonial mutants of Neurospora crassa. Arch Mikrobiol. 1973;91:355–364. doi: 10.1007/BF00425054. [DOI] [PubMed] [Google Scholar]
- Ukil L, De Souza CP, Liu HL, Osmani SA. Nucleolar separation from chromosomes during Aspergillus nidulans mitosis can occur without spindle forces. Mol Biol Cell. 2009;20:2132–2145. doi: 10.1091/mbc.E08-10-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Jackson D. Lights at the end of the tunnel: new views of plasmodesmal structure and function. Curr Opin Plant Biol. 2010;13:684–692. doi: 10.1016/j.pbi.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Yang L, Ukil L, Osmani A, Nahm F, Davies J, De Souza CP, Dou X, Perez-Balaguer A, Osmani SA. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot Cell. 2004;3:1359–1362. doi: 10.1128/EC.3.5.1359-1362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Jedd G, Kumaran D, Swaminathan S, Shio H, Hewitt D, Chua NH, Swaminathan K. A HEX-1 crystal lattice required for Woronin body function in Neurospora crassa. Nat Struct Biol. 2003;10:264–270. doi: 10.1038/nsb910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.