The ε-subunit is essential for energy coupling in the mitochondrial ATP synthase. Moreover, mutations in the proton translocation domain of ATP synthase can obviate the requirement for the ε-subunit. The results help to explain the appearance of the ε-subunit with the evolution of mitochondrial ATP synthase.
Abstract
The central stalk of the ATP synthase is an elongated hetero-oligomeric structure providing a physical connection between the catalytic sites in F1 and the proton translocation channel in F0 for energy transduction between the two subdomains. The shape of the central stalk and relevance to energy coupling are essentially the same in ATP synthases from all forms of life, yet the protein composition of this domain changed during evolution of the mitochondrial enzyme from a two- to a three-subunit structure (γ, δ, ε). Whereas the mitochondrial γ- and δ-subunits are homologues of the bacterial central stalk proteins, the deliberate addition of subunit ε is poorly understood. Here we report that down-regulation of the gene (ATP15) encoding the ε-subunit rapidly leads to lethal F0-mediated proton leaks through the membrane because of the loss of stability of the ATP synthase. The ε-subunit is thus essential for oxidative phosphorylation. Moreover, mutations in F0 subunits a and c, which slow the proton translocation rate, are identified that prevent ε-deficient ATP synthases from dissipating the electrochemical potential. Cumulatively our data lead us to propose that the ε-subunit evolved to permit operation of the central stalk under the torque imposed at the normal speed of proton movement through mitochondrial F0.
INTRODUCTION
All members of the ATP synthase family of proteins are very similar at the structural level, regardless of evolutionary origin. Historically, the enzyme has been described in terms of an integral membrane domain (F0) and a peripheral domain (F1). The wealth of high-definition structural data now available (Devenish et al., 2008; Walker, 2013) reveals that F0 and F1 are divided further into distinct oligomeric substructures. The most hydrophobic F0 subunits associate in a proton-translocating complex, which in its simplest form is composed of one a-subunit adjacent to a ring of 8–15 c-subunits. The remaining F0 subunits assemble a stalk anchored at one end to the stationary a-subunit in the membrane and attached at the other end to the outer periphery of the soluble F1 domain. F1 is largely defined by a globular structure ([αβ]3 hexamer), which contains the catalytic sites, and two or three other proteins that constitute a central stalk, which makes contact with these sites at one end and the c-ring at the other. Together the c-ring and the central stalk comprise a functional “rotor” that turns relative to nonmoving parts to permit energy-coupling reactions catalyzed by ATP synthases. For example, in the direction of ATP synthesis, energy released by proton translocation through F0 activates the rotor, forcing conformational changes in the catalytic sites that effect release of ATP from the enzyme.
One of the least-understood aspects of the ATP synthase coupling mechanism relates to differences in the central stalk composition between bacterial and mitochondrial enzymes. In the mitochondrial enzyme, it is made of three subunits (γ, δ, and ε), whereas only two (γ and ε) are present in bacterial ATP synthase (Devenish et al., 2008; Walker, 2013). The γ-subunits of F1 are homologous, and mitochondrial subunit δ is homologous to bacterial subunit ε. However, mitochondrial subunit ε has no counterpart in the bacterial enzyme, identifying it as one of many so-called “supernumerary” ATP synthase subunits that most likely appeared with the establishment of mitochondria as internal organelles during evolution of the eukaryotic cell. (Unless otherwise stated, ε in the text and figures denotes the supernumerary subunit of mitochondrial F1.) The work reported here defines more clearly the importance of ε in energy coupling.
The F1 ε-subunit of Saccharomyces cerevisiae is encoded by the nuclear gene ATP15. Previous work (Guelin et al., 1993; Lai-Zhang et al., 1999) using different atp15-deletion (Δε) mutants reported that the yeast grew at poor to modest rates on nonfermentable substrates, which meant that cells had a variable level of respiratory competence. This finding was unexpected because the ε-subunit of mitochondrial F1 is not a vestigial element inherited from the prokaryotic progenitor but instead represents a new protein function acquired during evolution of the eukaryotic enzyme, and it was assumed to be indispensable for energy-coupled oxidative phosphorylation. However, it is important to note that all of the work thus far investigating the functional relevance of the ε-subunit in mitochondrial F1 used Δε strains, which are pleiotropic mutants. When cultured on fermentable carbons, upward of 70% of Δε cells fail to maintain mitochondrial DNA (mtDNA) and instead convert to respiratory-deficient ρ0/ρ− derivatives. Prompted by the notion that the functional relevance of mitochondrial F1 subunit ε would remain ambiguous until conditions were established under which the effect on respiration caused by the primary nuclear mutation (atp15-deletion allele) could be assessed in an otherwise respiratory wild-type (ρ+) background, we transformed Δε yeast with a plasmid for doxycycline-regulated expression of wild-type ATP15. Our results show that yeast cultures down-regulated for production of the ε-subunit retain partial respiratory activity as a direct function of suppressor mutations in a mitochondrial-encoded F0 protein (a- or c-subunit) that rescues the uncoupling defect imposed by the elimination of subunit ε. The effect of the ε-subunit on energy coupling and structure/function relationships in mitochondrial ATP synthase is discussed with respect to the more primitive (ε-less) bacterial enzyme.
RESULTS
A 25% depletion in subunit ε leads to a total uncoupling of mitochondria
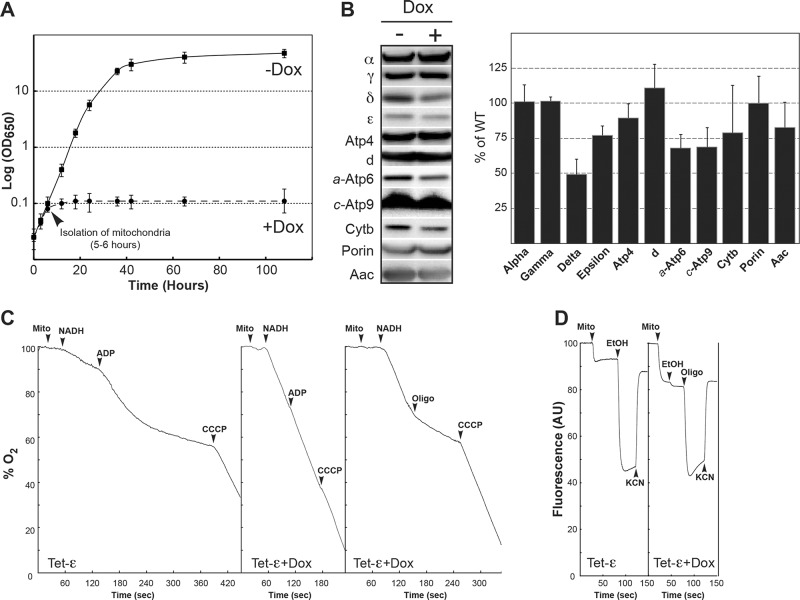
We constructed yeast strain YE1 (henceforth referred to by its acronym Tet-ε), which has a null allele in place of the ε-subunit gene (ATP15) in the chromosome and carries a doxycycline-repressible form of the gene on a low-copy plasmid (see Materials and Methods and genotypes in Table 1). Without the drug, Tet-ε grew in nonfermentable glycerol/ethanol media like its parental strain (SDC22) and displayed normal subunit ε levels. Down-regulation of ATP15 expression was achieved using doxycycline at a concentration (10 μM) known to not affect the respiratory capacity of wild-type yeast (Duvezin-Caubet et al., 2003). The respiratory growth of Tet-ε remained normal for 6 h after the addition of doxycycline to the culture medium and then stopped abruptly (Figure 1A, arrowhead). Samples were removed at this time point from both the –Dox and +Dox Tet-ε cultures, and mitochondria were isolated. Western blots revealed that the amount of the ε-subunit decreased by ∼25% at the time of doxycycline-induced growth arrest compared with control mitochondria from Tet-ε cells grown in the absence of doxycycline (Figure 1B). The amount of the δ subunit was reduced by ∼50% in mitochondria from growth-arrested +Dox cells, whereas the levels of other nuclear gene products evaluated (subunits α, γ, Atp4, d, porin, Aac) were quite similar for the two samples. The three mitochondrial-encoded proteins a-Atp6, c-Atp9, and cytochrome b were reduced in samples from +Dox versus –Dox cells, but the effect was less severe than with the δ-subunit.
TABLE 1:
Genotypes of yeast strains.
| Strain (acronym) | Nuclear genotype | mtDNA | Source |
|---|---|---|---|
| SDC22 (WT) | Mat α ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 arg8::HIS3 | ρ+Arg8m | Duvezin-Caubet et al. (2003) |
| SDC6 (Tet-δ) | Mat α ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 arg8::HIS3 atp16::KanMX + pCM189-ATP16 | ρ+Arg8m | Duvezin-Caubet et al. (2003) |
| YG1 (Tet-γ) | Mat α ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 arg8::HIS3 atp3::KanMX + pCM189-ATP3 | ρ+Arg8m | This study |
| YE1 (Tet-ε) | Mat α ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 arg8::HIS3 atp15::KanMX + pCM189-ATP15 | ρ+Arg8m | This study |
| S1 (Δε+c-L57F) | Mat α ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 arg8::HIS3 atp15::KanMX | ρ+Arg8m + c-L57F | This study |
| S1+ε (ε+c-L57F) | Mat α ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 arg8::HIS3 atp15::KanMX + pCM189-ATP15 | ρ+Arg8m + c-L57F | This study |
| S2 (Δε+a-A120V) | Mat α ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 arg8::HIS3 atp15::KanMX | ρ+Arg8m + a-A120V | This study |
FIGURE 1:
A block in ATP synthase subunit ε expression is rapidly followed by F0-mediated proton leaks that dissipate the mitochondrial membrane potential. The consequences of a block in subunit ε expression were followed using a strain (Tet-ε) in which this protein is under the control of a doxycycline-repressible promoter. (A) Growth curves of Tet-ε in rich glycerol/ethanol medium at 28°C in the presence (+Dox) or absence (–Dox) of doxycycline. Mitochondria were extracted from the two cultures at the time indicated by the arrowhead and used in the experiments shown in the subsequent panels. (B) Mitochondrial proteins were separated via SDS–PAGE and probed with antibodies against the indicated proteins. Setting the amount of each protein in Tet-ε cells grown in the absence of doxycycline at 100%, the relative levels of the proteins in Tet-ε cultured in the presence of the drug (+Dox) are plotted in the bar graph. The results shown are means of three experiments, and the vertical lines denote the SD in the data. (C) Oxygen consumption. The additions were 0.15 mg/ml mitochondrial proteins (mito), 4 mM NADH, 400 μM ADP, 3 μg/ml oligomycin (oligo), and 4 μM CCCP. (D) Energization of the mitochondrial inner membrane. Variations in ΔΨ were monitored by the fluorescence quenching of rhodamine 123. The additions were 0.5 μg/ml rhodamine 123, 0.15 mg/ml proteins (mito), 10 μl of ethanol (EtOH), 4 μg/ml oligomycin (oligo), 2 mM potassium cyanide (KCN), and 4 μM CCCP. Oxygen consumption and fluorescence traces are representative of three to five experimental trials.
Respiratory responses to metabolites and drugs measured for mitochondria from –Dox cells were within normal range (Figure 1C and Table 2). Compared to the maximal respiratory activity, which was measured in the presence of an uncoupling agent (carbonyl cyanide m-chlorophenyl hydrazone [CCCP]), the rate of mitochondrial oxygen consumption was 17% with NADH alone (state 4 respiration) and increased to 53% after addition of ADP (state 3 respiration). Instead, with NADH alone, the rate of oxygen consumption in mitochondria from +Dox cells was essentially equal to the CCCP-stimulated rate. Oligomycin, which blocks proton translocation through the F0, restored mitochondria to the slower state 4 respiration rate expected under conditions in which the respiratory substrate (NADH) is available but not the phosphorylation substrate (ADP). Hence the partial depletion of subunit ε, which was induced by doxycycline, correlated with 100% mitochondrial uncoupling due specifically to proton leakage through F0. This finding was corroborated in experiments that used rhodamine 123 as a reporter of mitochondrial membrane potential (ΔΨ; Figure 1D). In mitochondria from untreated Tet-ε cells, supplying the respiratory chain with electrons from ethanol produced a large fluorescent quenching of the dye that collapsed after subsequent addition of potassium cyanide. In contrast, the mitochondria partially depleted of subunit ε could not be energized with ethanol until the proton leak in F0 was blocked by the addition of oligomycin to the reaction sample. The severity of the uncoupling defect observed for doxycycline-treated Tet-ε cells was not unexpected because the steady-state level of the δ-subunit was reduced by half in these cells (Figure 1B; see earlier discussion), and we knew from previous work (Duvezin-Caubet et al., 2003) that decreasing the amount of F1 δ by 50% was sufficient to uncouple yeast mitochondria completely. Assays of oligomycin-sensitive mitochondrial ATPase activity in the two samples showed that although comparable rates were obtained in the absence of the inhibitor, the sample from +Dox cells was less sensitive than normal to oligomycin (77 vs. 89% inhibition; Table 3). The latter finding suggests that there is a higher percentage of F1 in the free, oligomycin-insensitive state in mitochondria isolated from +Dox yeast, which was predicted given the fact that these cells are partially depleted for two proteins (subunits δ and ε) that are known to be critical factors in mitochondria for physically coupling F1 to F0.
TABLE 2:
Oxygen consumption of mitochondria.
| Strain (acronym) | Dox | Asc/TMPD + CCCP (nAtO2/min/mg) | NADH + CCCP (nAtO2/min/mg) | NADH + ADP (nAtO2/min/mg) | NADH + oligo (nAtO2/min/mg) | NADH (nAtO2/min/mg) | ρ−/0 (%) |
|---|---|---|---|---|---|---|---|
| YE1 (Tet-ε) | – | 1617 ± 325 | 1031 ± 169 | 546 ± 107 | 217 ± 79 | 181 ± 55 | 2 ± 1 |
| YE1 (Tet-ε) | + | 1708 ± 203 | 1182 ± 70 | 975 ± 94 | 195 ± 31 | 966 ± 84 | 4 ± 1 |
| S1 (Δε+c-L57F) | – | 2180 ± 7 | 1240 ± 179 | 792 ± 109 | 216 ± 32 | 753 ± 86 | 12 ± 2 |
| S1+ε (c-L57F) | – | 1430 ± 302 | 912 ± 260 | 444 ± 55 | 221 ± 55 | 202 ± 56 | 3 ± 1 |
The strains were grown in rich glycerol/ethanol medium in the presence/absence of 10 μM doxycycline (Dox). Oxygen consumption was measured using isolated mitochondria. The additions were 0.15 mg/ml mitochondrial proteins, 4 mM NADH, 150 μM ADP, 12.5 mM ascorbate (Asc), 1.4 mM N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD), 3 μg/ml oligomycin (oligo), and 4 μM CCCP. n = 3–7.
TABLE 3:
ATP synthesis/hydrolysis activities of mitochondria.
| Strain (acronym) | Dox | ATP synthase (nmol/ATP/min/mg) | ATPase (nmol Pi/min/mg) | Inhibition (%) | ρ−/0 (%) | |
|---|---|---|---|---|---|---|
| –Oligo | +Oligo | |||||
| YE1 (Tet-ε) | – | 697 ± 141 | 3030 ± 163 | 341 ± 17 | 89 ± 1 | 2 ± 1 |
| YE1 (Tet-ε) | + | ND | 3063 ± 216 | 706 ± 66 | 77 ± 2 | 4 ± 1 |
| S1 (Δε+c-L57F) | – | 188 ± 22 | 1133 ± 191 | 1010 ± 179 | 11 ± 2 | 12 ± 2 |
| S1+ε (c-L57F) | – | 488 ± 96 | 3102 ± 77 | 323 ± 9 | 90 ± 1 | 3 ± 1 |
The strains were grown in rich glycerol/ethanol medium in the presence/absence of 10 μM doxycycline (Dox). ATP synthesis was measured in freshly isolated mitochondria using 0.15 mg/ml mitochondrial proteins, 4 mM NADH, and 1 mM ADP. For the ATPase assays, mitochondria kept at −80°C were thawed and the reaction performed in absence of osmotic protection and at pH 8.4 in the absence or presence of 6 μg/ml oligomycin (Oligo). The percentage of inhibition of ATPase activity by oligomycin is indicated.
Differential effects on mtDNA stability in Δε versus Δγ or Δδ null mutants
The oligomycin-sensitive uncoupling defect we observed in response to partial depletion of the ε-subunit is similar to the phenotype others reported for yeast in which the cellular level of γ- or δ- subunit protein was effectively reduced, by either dilution through the creation of heterozygous diploids (Δγ/γ or Δδ/δ; Xiao et al., 2000) or the use of a repressible promoter (Duvezin-Caubet et al., 2003). Hence a partial deficit in any one of the three rotor subunits can interfere with the mitochondrial ΔΨ by allowing protons to leak through F0. The circumstances are exacerbated in yeast null mutants that are completely deleted for one or another rotor subunit protein. Pertinent to our work, previous characterization of such strains revealed a dichotomy in mtDNA phenotypes that could not be explained. On one hand, Δγ and Δδ cells rapidly lost mtDNA and converted 100% to the ρ−/ρ0 state (Giraud and Velours, 1997; Lai-Zhang et al., 1999). Instead, cultures of Δε mutants harboring one (Guelin et al., 1993) or another (Lai-Zhang et al., 1999) null allele in place of ATP15 contained a significant subpopulation (∼30%) of ρ+ cells. We investigated the discrepancy in mtDNA phenotypes in Δε versus Δγ or Δδ yeast in experiments that monitored, side by side, the conversion of ρ+ cells to the ρ−/ρ0 state as each of the three rotor subunits became limiting. Three of the four yeast strains used in this study (Tet-γ, Tet-δ, and Tet-ε) produced one of the rotor subunits from a plasmid-borne, doxycycline-repressible gene; the fourth corresponded to the parental strain (SDC22), in which the rotor protein genes are all under control of their respective native promoter in the chromosome. All four strains were grown in rich galactose supplemented with 10 μM doxycycline, conditions under which ρ−/ρ0 cells can survive and divide. At the time doxycycline was added, ρ−/ρ0 levels were <2% for each strain (Figure 2). After five to seven doublings of the cell density, the Tet-δ and Tet-γ cultures were already almost completely invaded by ρ−/ρ0 cells (90–95%), whereas 70% of Tet-ε cells were still scored ρ+. After 20 generations, the Tet-δ and Tet-γ cultures were composed exclusively of ρ−/ρ0 cells, whereas the Tet-ε cultures still contained a substantial subpopulation of ρ+ cells, with a 50% peak after 12–15 generations, followed by a progressive decline to 25% after 40 generations. Conversely, the ρ−/ρ0 level remained <5% in SDC22 cultures during the course of the entire experiment.
FIGURE 2:
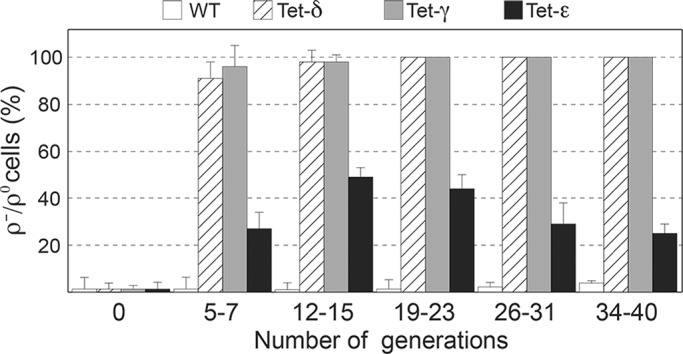
Kinetics of ρ−/ρ0 cell production. The Tet-γ, Tet-δ, and Tet-ε strains and their parental strain SDC22 (WT) were grown in rich galactose in the presence of 10 μM doxycycline. The cultures were refreshed several times with the same medium to produce a total of 40 generations, which was estimated by measuring the turbidity at 650 nm. The contents in ρ−/ρ0 cells were determined at the indicated number of generations.
Mutations in F0-subunit genes enable yeast to maintain mtDNA in the absence of the F1 ε-subunit
Samples of the ρ+ Tet-ε cells that had accumulated in galactose media after 40 generations in the presence of doxycycline (Figure 2) formed either medium-sized (S1) or small (S2) colonies on rich glycerol/ethanol medium (Figure 3A), and both types of clones failed to grow on glucose plates lacking uracil (Figure 3B). The latter condition indicated that strains S1 and S2 had lost the URA3-plasmid (pCM189-ATP15) bearing the doxycycline-repressible gene for wild-type subunit ε and were, in fact, ε-subunit null (Δε) strains. Missense mutations were identified in F0 subunits c (c-L57F) and a (a-A120V) of strains S1 and S2, respectively (see Materials and Methods). S1 (Δε+c-L57F) was notably impaired for growth in liquid glycerol/ethanol medium in comparison with the plasmid-bearing Tet-ε cells from which it originated but could be restored to the control level after reintroduction of pCM189-ATP15 (Figure 3C, S1+ε).
FIGURE 3:
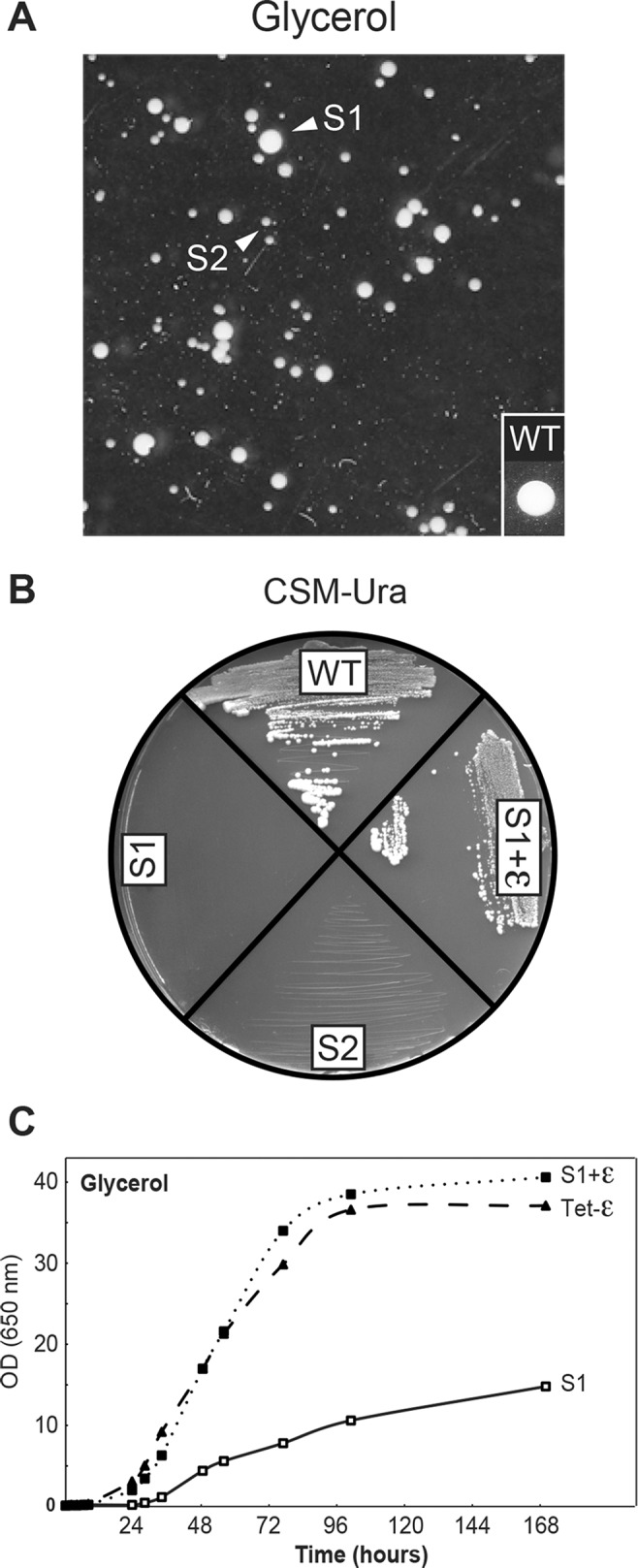
Mutations in F0 a- and c-subunits can bypass the need for subunit ε. (A) Samples of the Tet-ε galactose cultures after 40-generation growth in the presence of doxycycline (Figure 2) produce colonies that grow on rich glycerol/ethanol medium. Arrowheads indicate medium-size (S1) and small (S2) clones that carry mutations in subunit c (c-L57F) and subunit a (a-A128V), respectively. (B, C) Growth of Tet-ε, S1, and S1 retransformed with the Tet-ε gene on glucose plates lacking uracil (B) and in liquid glycerol/ethanol medium (C).
The c-L57F mutation slows F0 activity
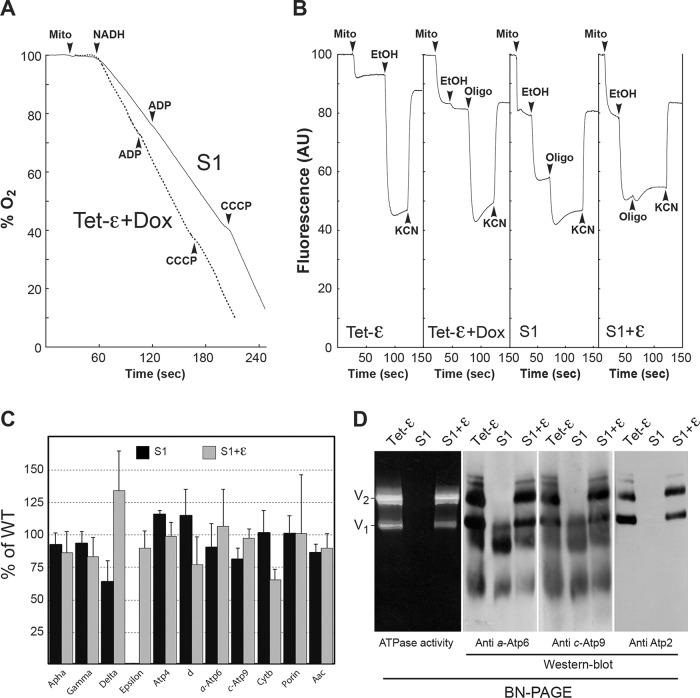
Respiration in the S1 (Δε+c-L57F) mitochondria was only partially uncoupled, as evidenced by a significant stimulation by both ADP and CCCP (Figure 4A and Table 2). These mitochondria could sustain a significant ΔΨ with ethanol without adding oligomycin (Figure 4B), and 27% the control level of ATP synthesis activity was observed (Table 3). These data provide evidence that the c-L57F mutation enables partial recovery of the physical and functional coupling of F1 to F0 defect imposed by the absence of subunit ε. However, although Western blots revealed the presence of near-normal to modestly reduced amounts of ATP synthase subunits α, γ, δ, Atp4, d, a, and c in S1 mitochondria (Figure 4C), F1F0 complexes minus the ε-subunit could not detected by blue native PAGE (BN-PAGE) by in-gel activity assays or Western blotting with antibody against the β subunit of F1 (Figure 4D). Instead, Western blots showed evidence of high–molecular weight oligomers of the hydrophobic F0 proteins, subunits a and c. Such findings indicate that the S1 variant produces an ε-minus form of the ATP synthase that is fragile and dissociates in response to detergent extraction and/or gel electrophoresis.
FIGURE 4:
Properties of the c-L57F mutation. All of the experiments shown were performed using mitochondria prepared from cells grown in rich glycerol/ethanol medium. (A) Oxygen consumption. The additions were 0.15 mg/ml mitochondrial proteins (mito), 4 mM NADH, 150 μM ADP, and 4 μM CCCP. (B) Energization of the mitochondrial inner membrane. Variations in ΔΨ were monitored by the fluorescence quenching of rhodamine 123. The additions were 0.5 μg/ml rhodamine 123, 0.15 mg/ml proteins (mito), 10 μl ethanol (EtOH), 4 μg/ml oligomycin (oligo), and 2 mM potassium cyanide (KCN). For comparison, we include the experiment shown in Figure 1, which was performed with mitochondria prepared from Tet-ε cells grown in the presence of doxycycline. Oxygen consumption and fluorescence traces are representative of three to five experimental trials. (C) Steady-state levels of different mitochondrial proteins in S1. Mitochondrial proteins were separated via SDS–PAGE and probed with antibodies against the indicated proteins. The level of each protein in S1 relative to the Tet-ε control is represented by the bars. The results are representative of at least three experiments. The lines indicate standard deviations. (D) Mitochondrial proteins were extracted with digitonin (2 g/g), separated by BN-PAGE, and either assayed for in-gel ATPase activity or transferred to membranes for Western blotting with the indicated antibodies. V1 and V2, monomeric and dimeric species of ATP synthase (complex V), respectively.
The nuclear and mitochondrial chromosomal genotypes of the strains S1 and S1+ε are identical (Δε+c-L57F), the difference being that the latter strain is transformed with plasmid pCM189-ATP15, in which the wild-type gene for subunit ε is subject to down-regulation by doxycycline. In the absence of drug, S1+ε yeast are replete with the ε-subunit and provide a cell model to study the c-L57F substitution as a stand-alone mutation. Strain S1+ε grew as well as the control Tet-ε cells in rich glycerol/ethanol medium (Figure 3C). State 4 respiration was also normal in S1+ε mitochondria and was stimulated efficiently (4.5-fold) by CCCP (Table 2). However, state 3 respiration (Table 2) and ATP synthesis (Table 3) were both diminished, by ∼20–30%, compared with the control, notwithstanding the fact that BN-PAGE (Figure 4D) revealed control amounts of fully assembled ATP synthase in the S1+ε strain. These data suggest that, by itself, the c-L57F mutation reduces the rate of proton translocation through F0 without any effect on F1F0 coupling and assembly.
DISCUSSION
Energy coupling in the ATP synthase occurs by means of an elongated hetero-oligomeric structure called the central stalk, which makes contact at one end with the catalytic sites in the soluble F1 domain and at the other end with the proton-translocating unit in the membrane (Devenish et al., 2008; Walker, 2013). In the mitochondrial enzyme, the central stalk is made of three subunits (γ, δ, and ε) instead of two as found in bacterial ATP synthase (γ and ε). Bacterial subunit ε is homologous to mitochondrial subunit δ, whereas the mitochondrial ε-subunit has no counterpart in bacteria. Our investigation into the functional relevance of mitochondrial ε provides information that helps explain the appearance of this novel protein with the evolution of mitochondrial ATP synthase. The results also offer valuable insight into previous publications in which the phenotype of yeast strains lacking the chromosomal gene encoding the ε-subunit (Δε) was described but could not be explained fully.
Deleting the yeast gene for one of the two conserved, central stalk proteins (γ or δ) destroys oxidative phosphorylation in the cell (Paul et al., 1994; Giraud and Velours, 1997; Lai-Zhang et al., 1999). Instead, poor to modest growth on nonfermentable substrates has been reported for yeast Δε strains (Guelin et al., 1993; Lai-Zhang et al., 1999). Another complicating feature is that primary mutations in any of the three central stalk proteins promote the loss of mtDNA. The mtDNA instability is most severe in Δγ and Δδ yeast, which convert 100% to ρ−/ρ0 cells (Giraud and Velours, 1997; Lai-Zhang et al., 1999; Duvezin-Caubet et al., 2003, 2006; this study). This phenomenon is best described as a survival tactic based on the fact the proton-translocating subunits of F0 are mitochondrial gene products, and the seal provided by the central stalk can be compromised if the structure of this element in incomplete (Lai-Zhang et al., 1999; Duvezin-Caubet et al., 2006; Godard et al., 2011). By eliminating mtDNA, the ρ−/ρ0 derivatives of Δγ and Δδ yeast lose the capacity to synthesize an integral membrane F0 domain that can collapse the ΔΨ by leaking protons.
In contrast, Δε yeast convert only partially to cytoplasmic petite derivatives such that the constitution of stable cultures is a heterogeneous mixture of ρ+ and ρ−/ρ0 cells (∼30:70 split). The degree of mtDNA instability makes it impossible to distinguish the effects on respiration attributed solely to the missing ε-subunit. For this reason we used a transformed Δε yeast strain (Tet-ε) for our experiments that carried a doxycycline-repressible form of the ε-subunit gene, ATP15, on a plasmid. In the absence of drug, Tet-ε is ρ+ and behaves just like the parental control strain (SDC22), in which ATP15 is expressed from its native promoter in the chromosome. Instead, a doxycycline-induced block in subunit ε production was rapidly followed by a growth arrest in nonfermentable media (Figure 1A). At the point of growth arrest, the mitochondria were only partially depleted of subunit ε (25% depletion; Figure 1B), had a normal content of mtDNA, but were totally uncoupled due to F0-mediated proton leaks (Figure 1, C and D, and Table 1).
The correlation of 25% reduction in the ε-subunit with 100% mitochondrial uncoupling in growth-arrested Tet-ε cells supports the idea that a full complement of ε protein is required to support energy coupling in the organelle. However, this conclusion is difficult to reconcile with the phenotype of simple Δε strains, which lack completely the ε-subunit but retain partial respiratory competence (Guelin et al., 1993; Lai-Zhang et al., 1999). A solution to the puzzle was revealed from experiments in which Tet-ε cells were cultured for 40 generations with a fermentable carbon source (galactose) in a rich medium that also contained doxycycline (Figure 2). Because rich galactose media can support the growth of respiratory-deficient yeast that are auxotrophic for uracil, it was no surprise to find that many cells from the original culture had lost the URA3-linked plasmid, which rendered them equivalent to untransformed Δε yeast that were incapable of producing any ε-subunit (Figure 3B). Remarkably, whereas after two or three generations ATP15 plasmid-replete Tet-ε cells stopped growing on respiratory substrates in media that contained 10 μM doxycycline (Figure 1A), the cured cells that were recovered after extensive culturing in a rich fermentable medium supplemented with the drug formed two populations (S1 and S2) of respiring colonies on glycerol/ethanol plates (Figure 3A).
Suppressor mutations were identified in the mitochondrial genes coding for subunits of the proton translocation machinery in strains S1 and S2. For S1, a missense mutation was found in ATP9 that resulted in phenylalanine substitution for Leu-57 in the c-subunit, and for S2 a genetic lesion was located to ATP6 that caused an A120V mutation in subunit a. Because the respiratory function of S2 was much worse relative to S1, only the latter strain was investigated in detail (Figure 4 and Tables 2 and 3). Experiments with S1 that queried the characteristics of respiration and energy coupling in the ATP synthase revealed significant levels of coupled enzyme activities that indicated the assembly in mitochondria of an ε-deficient F1F0 complex that did not leak protons. The fact such complex was not detected by BN-PAGE analysis of digitonin-extracted mitochondria (Figure 4D) suggests that the physical association of F1 with F0 is compromised significantly in S1 yeast. Under conditions in which the c-L57F mutation could be investigated in the context of an otherwise normal F1F0 structure (S1+ε strain), the results showed that in this configuration the ATP synthase is fully coupled but reduced in capacity to translocate protons across the membrane (Tables 2 and 3).
Having shown that combining either the c-L57F or a-A120V mutation with the null allele for subunit ε enables yeast to retain mtDNA, we suggest that similar mutations may have occurred in the genetic background of the Δε strains that have been described (Guelin et al., 1993; Lai-Zhang et al., 1999), which permitted a subpopulation of ρ+ cells to persist. In contrast, it has not been possible to recover ρ+ derivatives of Δγ and Δδ that are respiratory competent. Previously we showed that the mtDNA instability in Δδ yeast can be suppressed by mutations in the mitochondrial genes for subunits c and a, but whereas these maintain the mitochondrial inner membrane in a proton-impermeable state, such mutant ATP synthases fail to ensure proton-linked energy-transducing activities (Godard et al., 2011).
Information on the atomic structure of ATP synthases supports an explanation for phenotypic differences between Δε and Δδ yeast strains. Partial structures of the enzyme show that most of the contacts between the central stalk and the proton translocation apparatus in the membrane involve the δ-subunit (Stock et al., 1999; Dautant et al., 2010). Hence transduction of the proton motive force to the F1 catalytic sites largely depends on subunit δ. However, within the central stalk, the ε-subunit provides more than half of the atomic contacts for the δ-subunit (Figure 5, blue vs. red spheres). It follows that the stability of δ in the protein structure is likely compromised under conditions in which the ε-subunit is missing (e.g., Δε). Accelerated degradation of unbound subunit δ in doxycycline-treated Tet-ε is supported by Western analysis, which shows a reduction in the levels of this protein in comparison with mitochondria from control cells (Figure 1B).
FIGURE 5:
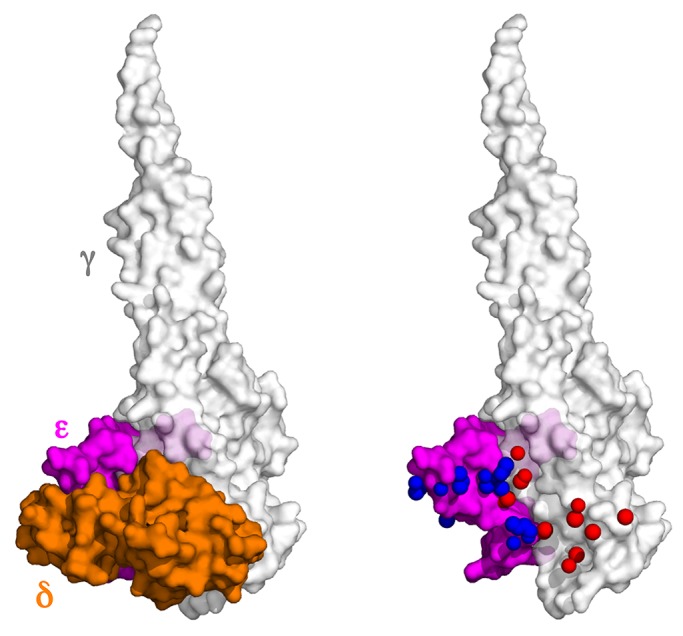
Contact zones between the different subunits of the central stalk in yeast F1. Left, surface-rendered model of the yeast F1 showing only γ (light gray, rendered at 20% transparency), δ (orange), and ε (magenta) subunits. Right, the surface of the δ-subunit was removed and in its place the red and blue dots show the positions of atoms in δ that are within 3.5 Å of the γ- and ε-subunits, respectively. PyMOL was used to calculate the atomic distances and make the Figure with the coordinates from the x-ray structure of yeast F1C10 ATP synthase (2WPD.pdb).
We propose that the ε-subunit evolved as a means of securing the contact between γ and δ, which otherwise might be compromised by the torque imposed on subunit δ when the c-ring rotates. Consistent with this idea is that the c-L57F mutation acquired by Δε yeast to sustain coupled oxidative phosphorylation did so by slowing the rate of proton-linked activities in the enzyme, as this may have been necessary to reduce the amount of strain on the structure.
MATERIALS AND METHODS
Strains and media
Escherichia coli XL1-Blue strain (Stratagene, Santa Clara, CA) was used for the cloning and propagation of plasmids. The S. cerevisiae strains used and their genotypes are listed in Table 1. The following rich media were used for the growth of yeast: 1% (wt/vol) yeast extract, 1% (wt/vol) peptone, 40 mg/l adenine, 2% (wt/vol) glucose, 2% (wt/vol) galactose, or 2% (wt/vol) glycerol. The glycerol medium was buffered at pH 6.2 with 50 mM potassium phosphate, and 2% (wt/vol) ethanol was added after sterilization. We also used complete synthetic medium (CSM; 0.17% [wt/vol] yeast nitrogen base without amino acids and ammonium sulfate, 0.5% [wt/vol] ammonium sulfate, 2% [wt/vol] glucose, and 0.8% [wt/vol] of a mixture of amino acids and bases from ForMedium, Norfolk, UK). The solid media contained 2% (wt/vol) agar.
Construction of a yeast strain expressing the subunit ε under the control of a doxycycline-repressible promoter
The coding sequence of the subunit ε gene (ATP15) was amplified by PCR using DNA from strain W303-1B as a template and the primers ATP15tet1 (5′-cgcggatccATGTCTGCCTGGAGGAAAGCTG-3′) for the sense strand and ATP15tet2 (5′-ataagaatgcggccgcCTATTTTGTTATTGGAGTGGGTTC-3′) for the antisense strand. The PCR product was digested with BamHI-NotI and ligated into the vector pCM189 (Duvezin-Caubet et al., 2003) to produce plasmid pZVT1/13. The cloned gene was verified by DNA sequencing. The SDC22 strain (Duvezin-Caubet et al., 2003) was transformed with pZVT1/13 and selected on synthetic complete medium lacking uracil. SDC22 containing pZVT1/13 was transformed with the deletion cassette of ATP15 obtained by PCR amplification using DNA from strain YPL271w (Δatp15 from the European Saccharomyces cerevisiae Archive for Functional Analysis, Frankfurt, Germany) as a template and the primers ATP15-300 (5′-AGCATTGTACTAGTTCTCTCG-3′) for the sense strand and ATP15+300 (5′-AGAAGGGGTGACCAAAGACCG-3′) for the antisense strand according to a previously described procedure (Wach et al., 1994). The transformants were selected on YPGA (yeast extract, peptone, glucose, adenine) supplemented with 200 μg/ml G418 and analyzed by PCR analysis. One clone, called YE1, carrying the expected deletion in the chromosomal ATP15 gene, in a ρ+ state and containing pZVT1/13, was retained for further analysis.
Construction of a yeast strain expressing the subunit γ under the control of a doxycycline-repressible promoter
The coding sequence of the γ-subunit gene (ATP3) was amplified by PCR using DNA from strain W303-1B as template and primers ATP3tet1 (5′-ataggatccATGTTGTCAAGAATTGTATCAAACAATG-3′) for the sense strand and ATP3tet2 (5′-atagcggccgcTCATCCCAAAGAGGAAGCACCAGTAATAATATC-3′) for the antisense strand. The PCR product was digested with BamHI-NotI and ligated into the vector pCM189 (Duvezin-Caubet et al., 2003) to produce plasmid pSE1. The 3′ untranslated region of ATP3 was amplified by PCR using DNA from strain W303-1B as template and primers GUTR1 (5′-catcctgcagTAAAAAAATCACCCTGCATTGCC-3′) for the sense strand and GUTR2 (5′- gatcaagcttCAAATCATTGAGATTGCGACC-3′) for the antisense strand. The PCR product was digested with PstI-HindIII and ligated into the vector pSE1 to produce plasmid pTE1. The cloned gene was verified by DNA sequencing. The SDC22 strain was transformed with pTE1 and selected on synthetic complete medium lacking uracil. SDC22 containing pTE1 was transformed with the deletion cassette of ATP3 obtained by the PCR amplification of the pUG6 plasmid containing the KanMX4 module (Wach et al., 1994) as template and primers ATP3/Kan/Pro (5′-aggtggaaacaattgaagacgagcagtaaacattattttatttagtagtcCATAGGCCACTAGTGGATCT-3′) for the sense strand and ATP3/Kan/Ter (5′-ttctacaaaaacaacgtcaaataaagaggcaatgcagggtgatttttttaCAGCTGAAGCTTCGTACGC-3′) for the antisense strand according to a previously described procedure. Lowercase letters refer to regions of homology to ATP3, and capital letters refer to regions of KanMX4 homology. The transformants were selected on yeast extract/peptone/dextrose supplemented with 200 μg/ml G418 and analyzed by PCR analysis. One clone, called YG1, carrying the expected deletion in the chromosomal ATP3 gene, in a ρ+ state and containing pTE1, was retained for further analysis.
Miscellaneous procedures
Isolated mitochondria were prepared by the enzymatic method (Guerin et al., 1979). The protein amounts were determined by the Lowry method (Lowry et al., 1951) in the presence of 5% SDS. Oxygen consumption rates were measured with a Clark electrode in the respiration buffer (0.65 M mannitol, 0.36 mM ethylene glycol tetraacetic acid, 5 mM Tris-phosphate, 10 mM Tris-maleate, pH 6.8) as previously described (Rigoulet and Guerin, 1979). For ATP synthesis rate measurements, mitochondria (0.3 mg/ml) were placed in a 2-ml thermostatically controlled chamber at 28°C in respiration buffer. The reaction was started by the addition of 4 mM NADH and 1 mM ADP and stopped by 3.5% perchloric acid and 12.5 mM EDTA. The samples were then neutralized to pH 6.5 by the addition of KOH and 0.3 M 3-(N-morpholino)propanesulfonic acid. ATP was quantified in a luciferin/luciferase assay (Perkin Elmer) with an LKB bioluminometer. The specific ATPase activity was measured at pH 8.4 by using a previously described procedure (Somlo, 1968). The participation of the F1F0-ATP synthase in ATP production or hydrolysis was assessed by addition of oligomycin (20 μg/mg of protein). Variations in ΔΨ were evaluated in the same buffer by measuring the fluorescence quenching of rhodamine 123 with a SAFAS (Monte Carlo, Monaco) fluorescence spectrophotometer (Emaus et al., 1986). SDS–PAGE was conducted as described previously (Laemmli, 1970; Schagger and von Jagow, 1987). BN-PAGE and clear native-PAGE experiments were carried out as described previously (Schagger and von Jagow, 1991). Briefly, mitochondrial extracts solubilized with digitonin to a protein ratio of 2 g/g were separated in a 3–13% acrylamide continuous gradient gel. After electrophoresis, the gel was either stained with Coomassie blue or incubated in a solution of 5 mM ATP, 5 mM MgCl2, 0.05% lead acetate, 50 mM glycine-NaOH, pH 8.4, to detect the ATPase activity (Grandier-Vazeille and Guerin, 1996) or transferred to poly(vinylidene difluoride) membranes and analyzed by Western blotting. Western blot analyses were performed as previously described (Arselin et al., 1996). Polyclonal antibodies raised against yeast ATP synthase were used at a dilution of 1:50,000 for subunit α; 1:10,000 for subunits γ, Atp4, a-Atp6, d, c-Atp9, ε, and δ; 1:10,000 for Aac2; and 1:5000 for cytochrome b. Monoclonal antibodies against yeast porin (from Molecular Probes) were used at a dilution of 1:5000. Nitrocellulose membranes were incubated with peroxidase-labeled antibodies at a 1:5000 dilution (Promega), and the blot visualization was conducted with an electrochemiluminescence reagent.
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique, the Université Bordeaux Segalen, the Conseil Régional d'Aquitaine, and the Agence Nationale de la Recherche (program Chloro/mito CES) to J.P.d.R., as well as by funding from the Office of the Vice President for Research at Wayne State University to S.H.A. We particularly thank G. Lauquin for providing the immune serum against the Aac2 transporter and J. Velours and T. Langer for yeast ATP synthase and cytochrome b antibodies.
Abbreviations used:
- BN-PAGE
blue native polyacrylamide gel electrophoresis
- Dox
doxycycline
- mtDNA
mitochondrial DNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-02-0112) on January 22, 2014.
E.T. and J.-P.d.R. designed the study; E.T., F.G., and M.-F.G. conducted the research; E.T., F.G., M.-F.G, S.H.A., and J.-P.d.R. analyzed data; and E.T., S.H.A., and J.-P.d.R. wrote the article.
REFERENCES
- Arselin G, Vaillier J, Graves PV, Velours J. ATP synthase of yeast mitochondria. Isolation of the subunit h and disruption of the ATP14 gene. J Biol Chem. 1996;271:20284–20290. doi: 10.1074/jbc.271.34.20284. [DOI] [PubMed] [Google Scholar]
- Dautant A, Velours J, Giraud MF. Crystal structure of the Mg.ADP-inhibited state of the yeast F1c10 ATP synthase. J Biol Chem. 2010;285:29502–29510. doi: 10.1074/jbc.M110.124529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devenish RJ, Prescott M, Rodgers AJ. The structure and function of mitochondrial F1F0-ATP synthases. Int Rev Cell Mol Biol. 2008;267:1–58. doi: 10.1016/S1937-6448(08)00601-1. [DOI] [PubMed] [Google Scholar]
- Duvezin-Caubet S, Caron M, Giraud MF, Velours J, di Rago JP. The two rotor components of yeast mitochondrial ATP synthase are mechanically coupled by subunit delta. Proc Natl Acad Sci USA. 2003;100:13235–13240. doi: 10.1073/pnas.2135169100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvezin-Caubet S, Rak M, Lefebvre-Legendre L, Tetaud E, Bonnefoy N, di Rago JP. A “petite obligate” mutant of Saccharomyces cerevisiae: functional mtDNA is lethal in cells lacking the delta subunit of mitochondrial F1-ATPase. J Biol Chem. 2006;281:16305–16313. doi: 10.1074/jbc.M513805200. [DOI] [PubMed] [Google Scholar]
- Emaus RK, Grunwald R, Lemasters JJ. Rhodamine 123 as a probe of transmembrane potential in isolated rat-liver mitochondria: spectral and metabolic properties. Biochim Biophys Acta. 1986;850:436–448. doi: 10.1016/0005-2728(86)90112-x. [DOI] [PubMed] [Google Scholar]
- Giraud MF, Velours J. The absence of the mitochondrial ATP synthase delta subunit promotes a slow growth phenotype of rho-yeast cells by a lack of assembly of the catalytic sector F1. Eur J Biochem. 1997;245:813–818. doi: 10.1111/j.1432-1033.1997.00813.x. [DOI] [PubMed] [Google Scholar]
- Godard F, Tetaud E, Duvezin-Caubet S, di Rago JP. A genetic screen targeted on the FO component of mitochondrial ATP synthase in Saccharomyces cerevisiae. J Biol Chem. 2011;286:18181–18189. doi: 10.1074/jbc.M110.214825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandier-Vazeille X, Guerin M. Separation by blue native and colorless native polyacrylamide gel electrophoresis of the oxidative phosphorylation complexes of yeast mitochondria solubilized by different detergents: specific staining of the different complexes. Anal Biochem. 1996;242:248–254. doi: 10.1006/abio.1996.0460. [DOI] [PubMed] [Google Scholar]
- Guelin E, Chevallier J, Rigoulet M, Guerin B, Velours J. ATP synthase of yeast mitochondria. Isolation and disruption of the ATP epsilon gene. J Biol Chem. 1993;268:161–167. [PubMed] [Google Scholar]
- Guerin B, Labbe P, Somlo M. Preparation of yeast mitochondria (Saccharomyces cerevisiae) with good P/O and respiratory control ratios. Methods Enzymol. 1979;55:149–159. doi: 10.1016/0076-6879(79)55021-6. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lai-Zhang J, Xiao Y, Mueller DM. Epistatic interactions of deletion mutants in the genes encoding the F1-ATPase in yeast Saccharomyces cerevisiae. EMBO J. 1999;18:58–64. doi: 10.1093/emboj/18.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Paul MF, Ackerman S, Yue J, Arselin G, Velours J, Tzagolof A, Ackermann S. Cloning of the yeast ATP3 gene coding for the gamma-subunit of F1 and characterization of atp3 mutants. J Biol Chem. 1994;269:26158–26164. [PubMed] [Google Scholar]
- Rigoulet M, Guerin B. Phosphate transport and ATP synthesis in yeast mitochondria: effect of a new inhibitor: the tribenzylphosphate. FEBS Lett. 1979;102:18–22. doi: 10.1016/0014-5793(79)80919-9. [DOI] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Somlo M. Induction and repression of mitochondrial ATPase in yeast. Eur J Biochem. 1968;52:276–284. doi: 10.1111/j.1432-1033.1968.tb00368.x. [DOI] [PubMed] [Google Scholar]
- Stock D, Leslie AG, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Walker JE. The ATP synthase: the understood, the uncertain and the unknown. Biochem Soc Trans. 2013;41:1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Metzl M, Mueller DM. Partial uncoupling of the mitochondrial membrane by a heterozygous null mutation in the gene encoding the gamma- or delta-subunit of the yeast mitochondrial ATPase. J Biol Chem. 2000;275:6963–6968. doi: 10.1074/jbc.275.10.6963. [DOI] [PubMed] [Google Scholar]