RASSF1A is a microtubule-associated protein. This study provides evidence for RASSF1A regulating MT dynamics via segmental binding to provide local stabilization of the MT network, thus facilitating MT rescue. RASSF1A reconfigures the MT network through bundling of nearby MTs and provides a stable platform to maintain Golgi integrity.
Abstract
The tumor suppressor and microtubule-associated protein Ras association domain family 1A (RASSF1A) has a major effect on many cellular processes, such as cell cycle progression and apoptosis. RASSF1A expression is frequently silenced in cancer and is associated with increased metastasis. Therefore we tested the hypothesis that RASSF1A regulates microtubule organization and dynamics in interphase cells, as well as its effect on Golgi integrity and cell polarity. Our results show that RASSF1A uses a unique microtubule-binding pattern to promote site-specific microtubule rescues, and loss of RASSF1A leads to decreased microtubule stability. Furthermore, RASSF1A-associated stable microtubule segments are necessary to prevent Golgi fragmentation and dispersal in cancer cells and maintain a polarized cell front. These results indicate that RASSF1A is a key regulator in the fine tuning of microtubule dynamics in interphase cells and proper Golgi organization and cell polarity.
INTRODUCTION
Ras association domain family 1A (RASSF1A) is a tumor suppressor whose inactivation is believed to be responsible for 40 types of sporadic human cancers (van der Weyden and Adams, 2007). Recruitment of DNA methyltransferases to the RASSF1A promoter and subsequent promoter hypermethylation serves as the main mechanism of RASSF1A loss (Dammann et al., 2000; Burbee et al., 2001; Lee et al., 2001). Because RASSF1A lacks enzymatic function (Donninger et al., 2007), the currently accepted function is to serve as a scaffold for a number of essential signaling interactions (Donninger et al., 2007). Therefore RASSF1A can participate in a variety of processes that regulate apoptosis (Vos et al., 2000; Baksh et al., 2005) and cell cycle progress (Shivakumar et al., 2002), both of which likely impart the tumor-suppressive function of RASSF1A by decreasing cancer cell number. Besides the scaffolding function, RASSF1A also strongly binds microtubules (MTs) in vitro and in cells (Dallol et al., 2004). Such binding increases MT stability: in cells, RASSF1A overexpression induces MT hyperstabilization, whereas RASSF1A depletion decreases MT resistance against depolymerizing drugs (Liu et al., 2003; Dallol et al., 2004). The MT-binding ability of RASSF1A was proposed to be important for mitotic spindle dynamics and cell cycle progression (Rong et al., 2004; Vos et al., 2004). It is clear that RASSF1A regulates cell cycle progression by acting as an Aurora A–dependent scaffolding platform within a complex signaling mechanism (Rong et al., 2007). MT binding likely contributes to this mechanism via RASSF1A recruitment (Song et al., 2004; Rong et al., 2007), which may be important for the tumor-suppressive function of RASSF1A, as mutants deficient in MT binding were not as potent in cell cycle arrest (Dallol et al., 2004). However, there is no direct evidence for the importance of RASSF1A-dependent MT stabilization in mitosis.
Of importance, loss of RASSF1A expression in cancer cells is concomitant with the formation of a more invasive and metastatic cancer phenotype (Lee et al., 2001; Muller et al., 2003; Kang et al., 2004; Liu et al., 2005; Jo et al., 2006), which depends strongly on migratory rather than proliferative features of cancer cells. Indeed, RASSF1A depletion was shown to facilitate HeLa cell migration in vitro (Dallol et al., 2005). It has been hypothesized that the ability of RASSF1A to bind MTs is responsible for this phenotype (Dallol et al., 2004), which would have been a justified function for RASSF1A-dependent MT stabilization. Indeed, RASSF1A depletion affects the overall MT pattern (Dallol et al., 2005); however, the origin of this change has not been addressed, and whether and how RASSF1A regulates MT dynamic instability in motile cells are unknown.
Here we apply high-resolution confocal and total internal reflection fluorescence (TIRF) microscopy to determine RASSF1A-dependent MT regulation in motile cells. Because the function of known MT-binding proteins depends on their specific localization and dynamics within the MT network, we also investigate cellular localization of RASSF1A and potentially associated functions. We find that RASSF1A binds to MTs in a distinct segmental pattern and exerts its MT-stabilizing effect only within these segments. Such site-specific MT stabilization increases MT rescue frequency specifically at the RASSF1A-coated MT segments. RASSF1A-associated MT segments serve as sites for temporal bundling of short regions within MTs, which probably contributes to MT network organization. We also find that RASSF1A-associated MT segments are necessary for the Golgi complex integrity and positioning, which is a known essential determinant for polarized cell migration (Kupfer et al., 1983; Miller et al., 2009; Yadav et al., 2009). This study provides the first evidence of the role of MT-bound RASSF1A in MT dynamic instability regulation and the architecture of interphase cells.
RESULTS
RASSF1A localizes at discrete MT segments
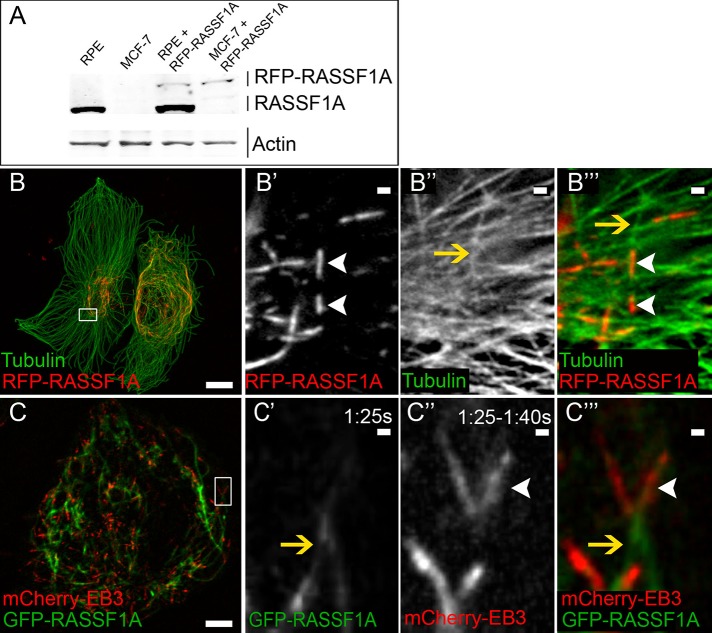
The goal of this study was to investigate functions for RASSF1A in interphase cells and determine whether loss of these functions is essential for phenotypes of cancer cells in which RASSF1A is silenced. To address this, we used two cell systems: normal retinal pigment epithelial cells (RPE1) and the epithelial breast cancer cell line Michigan Cancer Foundation-7 (MCF-7). To determine RASSF1A localization and dynamics, we ectopically expressed green fluorescent protein (GFP)– or red fluorescent protein (RFP)–fused RASSF1A in both cell types and examined expression levels by immunoblotting. We found that endogenous expression of RASSF1A was low in RPE1 and, as previously reported (Kellokumpu et al., 2002), completely absent in MCF-7. Because high expression of RASSF1A is known to overstabilize MTs (Liu et al., 2003; Dallol et al., 2004; Rong et al., 2004; Vos et al., 2004), ectopic expression of fluorescently fused RASSF1A in cells used for our experiments was maintained at near or slightly less than endogenous levels (Figure 1A). Under these conditions, RASSF1A was detected as short, linear segments (Figure 1, B′ and B′′′). Because previous reports showed that ectopically expressed RASSF1A can bind to the microtubule lattice (Rong et al., 2004), colocalization of these segments with MTs was tested by α-tubulin immunostaining of RFP-RASSF1A–expressing cells. We found that RASSF1A was localized linearly along the lattice of distinct MTs, highlighting a segmental pattern within the MT network (Figure 1, B–B′′′, left cell); however, when RFP-RASSF1A was overexpressed, this segmental pattern within the MT network was lost, and MT bundling occurred (Figure 1B, right cell). We hypothesize that this unusual binding pattern may impart an important function for MT organization.
FIGURE 1:
RASSF1A binds to the nondynamic portion of MTs. (A) Western blot analysis of endogenous RASSF1A (lanes 1 and 2) and expressed RFP-RASSF1A (lanes 3 and 4) in RPE1 and MCF-7 cells. (B) RPE1 cells expressing RFP-RASSF1A (red) fixed and stained for tubulin (green). Immunostaining. Bar, 5 μm. (B′–B′′′) RFP-RASSF1A (white arrowheads) localizes segmentally along MTs (yellow arrow) shown in B. Bar, 2 μm. (C) Detection of GFP-RASSF1A (green) and mCherry-EB3 (red) in time-lapse TIRFM movie of a MCF-7 cell (5 s/frame). Maximum intensity projection of four frames from mCherry-EB3 channel illustrates polymerizing MT plus end (C′′). Bar, 5 μm. (C′–C′′′) GFP-RASSF1A does not colocalize with mCherry-EB3 at the tip of MTs shown in C. Bar, 2 μm.
Binding of RASS1A locally stabilizes MT segments
Because localization of dynamic versus stable MTs defines the configuration of trafficking paths within a cell and thus is tightly connected with MT function, we addressed whether RASSF1A locally stabilizes MT segments. To determine whether RASSF1A-associated MT segments were dynamic, we applied live-cell TIRF imaging of cells coexpressing GFP-RASSF1A and a MT plus-tip marker, mCherry–end-binding protein 3 (EB3). Polymerizing MTs, detected by tracking of EB3 comets, were devoid of RASSF1A. In many cases, RASSF1A was found at the nondynamic portions of MTs, away from the plus ends undergoing dynamic instability (Figure 1, C–C′′′, and Supplemental Movie S1).
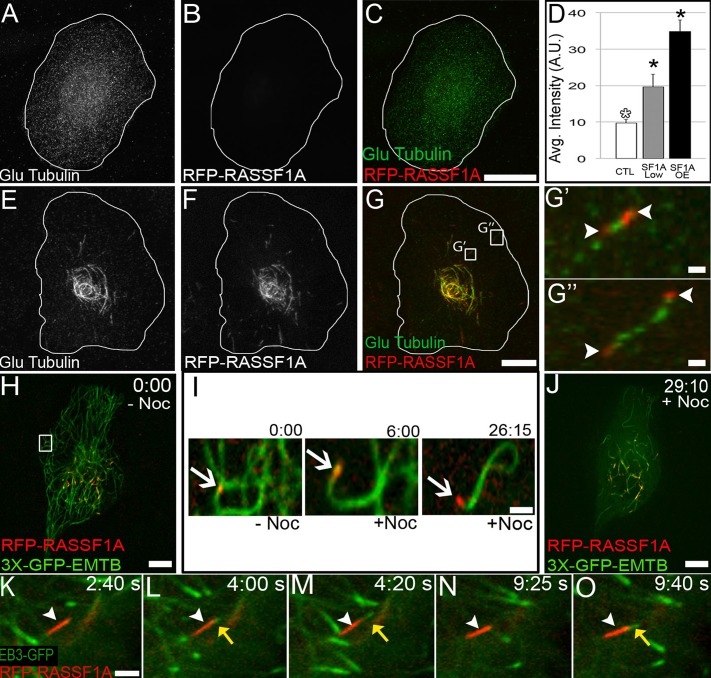
These data suggest that RASSF1A associates with specific, nondynamic regions of dynamic MTs away from the polymerizing plus ends. To test whether binding of RASSF1A extends lifetimes of distinct regions of MTs, we addressed whether these MT portions were posttranslationally modified. Because detyrosination (Glu tubulin) is an established marker for long-lived MTs, we first tested for this posttranslational modification (PTM) by immunostaining in RPE1 cells transiently expressing variable levels of RFP-RASSF1A. Nontransfected cells exhibited low Glu tubulin levels (Figure 2, A–D). We detected enhanced levels of Glu tubulin in cells expressing RFP-RASSF1A (Figure 2, E–G′′ and D), where these levels correlated with the level of RASSF1A expression (Figure 2D). On a single-MT level, Glu tubulin either was colocalized with RASSF1A or accumulated at MT regions flanked by RASSF1A-coated segments (Figure 2, G′–G′′), indicating that RASSF1A facilitated tubulin detyrosination indirectly by increasing MT lifetime. We also found that RASSF1A-associated MTs were often acetylated, although tubulin acetylation did not correlate with RASSF1A expression (unpublished data), indicating that although overstabilized MTs accumulate tubulin acetylation (Piperno et al., 1987; Westermann and Weber, 2003), other molecules are likely responsible for specific MT acetylation in cells. To examine whether RASSF1A-associated MT regions were indeed stabilized against depolymerization, we subjected RPE1 cells expressing RFP-RASSF1A and 3xGFP–ensconsin MT-binding domain (EMTB; Faire et al., 1999; Tanaka et al., 2009) to the MT-depolymerizing agent nocodazole. Confocal live-cell imaging revealed that MT depolymerization proceeds up to the point where RASSF1A is associated with the MT (Figure 2, H–J, and Supplemental Movies S2–S4), indicating that RASSF1A locally protects the MT from further depolymerization. Thus RASSF1A-coated MT segments might serve to regulate site-specific MT dynamics.
FIGURE 2:
RASSF1A binding stabilizes and protects MTs from depolymerization. (A–C) Nontransfected RPE1 cell does not exhibit detyrosinated (green) MTs in the absence of RFP-RASSF1A (red) expression. Immunostaining. (D) Intensity of detyrosinated (Glu) tubulin as correlated with RFP-RASSF1A expression. Representative examples out of 20 cells/condition. Error bars indicate SEM. (E–G) RFP-RASSF1A expression induces MT stabilization in RPE1 cells as detected by anti–detyrosinated (Glu) tubulin staining. Immunostaining. Bar, 5 μm. (G′–G′′) RFP-RASSF1A (red; white arrowheads) flanks portions of stable MTs as detected by anti–detyrosinated (Glu) tubulin staining shown in G. Immunostaining. Bar, 2 μm. (H, J) Video frames of RFP-RASSF1A– (red) and 3xGFP-EMTB (green)–expressing cells representing pre– and post–nocodazole treatment. Spinning-disk confocal (5 s/frames). (I) Time sequences pre– and post–nocodazole addition illustrates RFP-RASSF1A (white arrow) MT protection capacity. Bar, 5 μm. (K–O) RFP-RASSF1A (red; white arrowhead) expression in RPE1 cells can function as sites of MT rescue, as detected by EB3-GFP–labeled growing MT plus ends (green; yellow arrow). Spinning-disk confocal (5 s/frames). Bar, 5 μm.
RASSF1A localization defines sites of MT rescue
Because RASSF1A binds segmentally to the MT lattice, one can infer that the segment borders provide distinct points of stabilization, which could serve as platforms for switching of MTs from depolymerization to polymerization (MT rescue). The location and persistence of rescue sites at interphase MTs, which continuously undergo dynamic instability, is crucial for MT network configuration. However, although RASSF1A has been implicated in influencing MT dynamics in mitosis (Vos et al., 2004), the direct effects of RASSF1A on MT dynamics in interphase have not been addressed. To investigate the potential involvement of RASSF1A in regulating interphase MT dynamics, we performed steady-state analysis of RPE1 cells coexpressing low levels of RFP-RASSF1A and EB3-GFP. Cells were imaged with a spinning-disk confocal microscope to monitor the plus ends of growing MTs. RASSF1A-associated MTs acted as platforms for polymerizing MTs (Figure 2, K–O, and Supplemental Movies S5 and S6), which suggests that these sites can serve as points of rescue. To analyze the role of endogenous RASSF1A in overall MT dynamics parameters, we used TIRF microscopy to analyze peripheral MT dynamics in H1792 cells, a non–small cell lung carcinoma cell line, expressing either endogenous RASSF1A or short hairpin RNA (shRNA) depleted of RASSF1A (Figure 3A; Vos et al., 2006). Loss of RASSF1A via shRNA knockdown resulted in alterations in MT organization as compared with RASSF1A-expressing cells (Figure 3, B and C). Consistent with our model, we found that the number of rescue events (Figure 3D) and pause duration (Figure 3E) per MT were significantly decreased by RASSF1A depletion, indicating that this protein indeed functions as a rescue factor by stabilizing MT segments. In addition, our analyses revealed that RASSF1A depletion led to an increase of MT polymerization and depolymerization rates (Figure 3, G and H), as well as the number of catastrophes per MT (Figure 3F). Such enhancement of MT dynamicity could be a manifestation of a lack of short-term pauses and rescues and is characteristic for depletion of a rescue factor (Mimori-Kiyosue et al., 2005; Drabek et al., 2006). Collectively these alterations in dynamics strongly decreased MT lifetime after RASSF1A depletion (Figure 3, I–J).
FIGURE 3:
RASSF1A depletion influences MT dynamics. (A) Western blot analysis of RASSF1A in vector-expressing (lane 1) and RASSF1A shRNA–depleted (lane 2) H1792 cells. (B, C) H1792 cells expressing vector (B) or RASSF1A shRNA depleted (C) fixed and immunostained for tubulin. Immunostaining. Bar, 5 μm. (D–J) Analysis of various MT dynamics parameters in H1792 RASSF1A shRNA–depleted cells reveals an increase in MT dynamicity. p < 0.05. Error bars indicate SEM. (I, J), Examples of MT life history plots in control H1792 cells and cells depleted of RASSF1A. Plots reveal decreased MT lifetimes upon RASSF1A depletion. Representative examples out of 20 cells/condition.
Taken together, our data suggest a critical role for RASSF1A in fine tuning interphase MT network dynamics.
RASSF1A bundles neighboring MTs
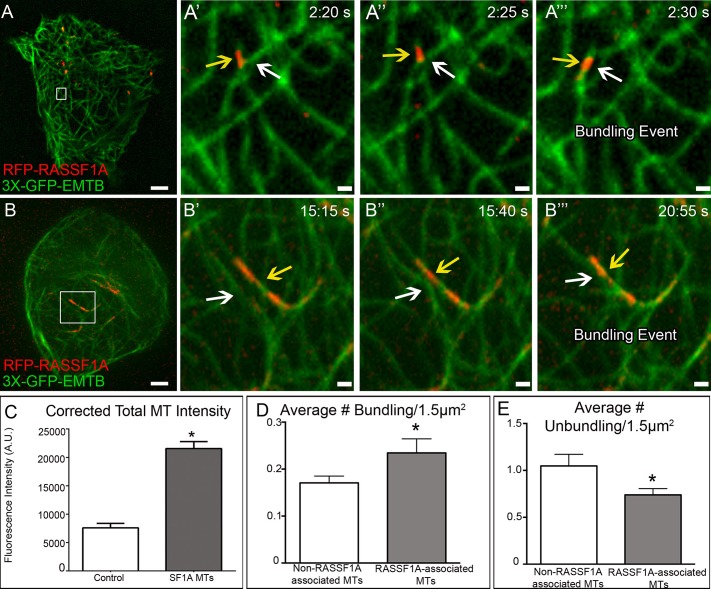
Because RASSF1A-associated MT segments clearly bear a specialized function in local MT behavior, we next addressed their positioning and integration within the whole MT network. We hypothesized that if RASSF1A were stably bound to MTs, addition of nocodazole would not increase cytoplasmic RFP-RASSF1A levels. Confocal live-cell imaging under these conditions revealed no appreciable increase in cytoplasmic RFP-RASSF1A levels, and instead RFP-RASSF1A remained associated with MT fragments (compare a prenocodazole cytoplasmic fluorescence intensity of 176,952 a.u. to a postnocodazole intensity of 183,031 a.u.; no significance). FRAP analysis also revealed that RFP-RASS1A is nondynamic, suggesting that it is stably associated with a MT (Supplemental Figure S1). Further analysis of live-cell imaging sequences showed that RASSF1A could promote bundling of preexisting steady-state MTs (Figure 4, A–B′′′, Supplemental Figure S2, and Supplemental Movies S7 and S8). MT bundling often resulted in a change in MT directionality from random to parallel and coincided with the formation of thin MT bundles. Fluorescence intensity analysis of 3xGFP-EMTB–labeled MTs revealed that on average there is a threefold increase in EMTB signal for RASSF1A-associated segments as compared with single MTs not bound by RASSF1A (Figure 4C), suggesting that RASSF1A can bundle up to three MTs. Quantification of non–RASSF1A-associated MT bundling events in 1.5-μm2 squares (similar to the size of RASSF1A segments) and RASSF1A-associated MTs revealed that the majority of RASSF1A-coated MT segments facilitate bundling within the MT network (Figure 4D). In addition, quantification of non–RASSF1A-associated MT unbundling reveals a significantly higher number of unbundling events as than with RASSF1A-associated MTs (Figure 4E). Thus RASSF1A stabilizes MT–MT interactions, which could provide a significant influence in altering the MT network configuration. These properties can be used for specific MT functions required at distinct cellular locations. For example, centrally located RASSF1A-associated segments may facilitate reliable Golgi complex assembly (Ryan et al., 2012) by maintaining the Golgi complex integrity (Cole et al., 1996).
FIGURE 4:
RASSF1A induces local bundling to alter MT network configuration. (A, B) RPE1 cells expressing RFP-RASSF1A and 3xGFP-EMTB. Area in box is enlarged to the right. Spinning-disk confocal. Bar, 5 μm (A′–A′′′ and B′–B′′′). RFP-RASSF1A (yellow arrow) locally bundles neighboring MTs (white arrow). Bar, 2 μm. (C) Analysis of fluorescence intensity in single vs. RASSF1A-associated MTs reveals RASSF1A can bundle up to three MTs at one time. Representative examples out of 15 cells/condition. Error bars indicate SEM. (D) Average number of bundling events per 1.5 μm2 reveals a significant increase in these events with RASSF1A-associated vs. non–RASSF1A-associated MTs. (E) Average number of unbundling events per 1.5 μm2 reveals a significant decrease in unbundling events in RASSF1A-associated vs. non–RASSF1A-associated MTs. Representative examples out of 10 cells.
RASSF1A-associated MT segments are required for Golgi integrity
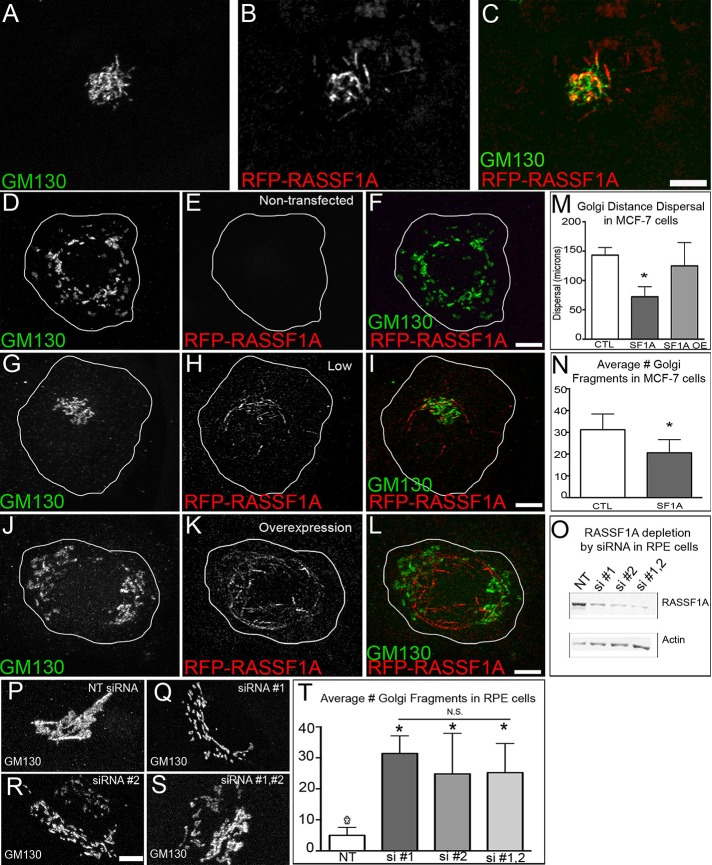
Analysis of the cellular localization of RASSF1A-associated MT segments revealed that they were accumulated predominantly in the central cell region. Costaining with a Golgi marker, Golgi matrix protein 130 (GM130), revealed the close proximity of these segments with the Golgi apparatus (Figure 5, A–C). Such specific localization may indicate that MTs in the Golgi area possess higher RASSF1A-binding properties because, for example, a cofactor needed for this segmental binding is enriched at the Golgi membrane or in the perinuclear region (e.g., the nuclear transport regulator small GTPase Ran, which was identified as a possible cofactor for MT binding of RASSF1A; Dallol et al., 2004, 2009). Another attractive possibility is that such location reflects specific functions of RASSF1A-associated MTs in Golgi organization or positioning. We hypothesize that RASSF1A localization at the Golgi suggests a functional relationship by which RASSF1A-induced MT stabilization provides a scaffold for Golgi assembly. Indeed, in MCF-7 breast cancers cells, which lack endogenous expression of RASSF1A, Golgi stacks are dispersed throughout the cell (Kellokumpu et al., 2002; Figure 5, D–F, and M). To determine the role of RASSF1A in Golgi positioning, we examined the effects of altering RASSF1A protein levels on the cellular distribution of the Golgi, visualized by GM130 immunostaining. First, we used reexpression of RFP-RASSF1A in MCF-7 cells at low levels, which we consider close to physiological expression in noncancerous cells (Figure 1A). Of interest, we found that under these low-expression conditions, the typically dispersed Golgi fragments were collected to the cell center (Figure 5, G–I and M), and the number of Golgi fragments was significantly reduced compared with nontransfected cells (Figure 5N). However, when RFP-RASSF1A was overexpressed at high levels in MCF-7 cells, resulting in coating of the entire MT network, the Golgi became redistributed throughout the cell, resembling nontransfected controls (Figure 5, J–M). This result suggests that low levels of RASSF1A, when it functions to stabilize discrete MT segments, are necessary for proper Golgi assembly. To further test this hypothesis, we addressed whether RASSF1A depletion in noncancerous cells also affects Golgi structure. In RPE1 cells treated with either small interfering RNA (siRNA) against human RASSF1A or control scrambled siRNA (Figure 5O), we determined the effect of RASSF1A depletion on Golgi structure by examining individual cells stained with GM130. In scramble control cells, the Golgi showed a compact morphology (Figure 5P). In contrast, RASSF1A-depleted cells showed aberrant Golgi features, including fragmentation of the Golgi (Figure 5, Q–S). Of interest, the level of Golgi fragmentation in RASSF1A siRNA–depleted RPE1 cells was similar to levels observed in nontransfected or RFP-RASSF1A–overexpressing MCF-7 cells (Figure 5, N and T). Therefore we conclude that centrally located RASSF1A is essential for proper Golgi complex organization.
FIGURE 5:
RASSF1A expression alters Golgi morphology. (A–C) MCF-7 cell expressing low levels of RFP-RASSF1A, fixed and stained for Golgi protein, GM130 (green). Immunostaining. RFP-RASSF1A localizes at MTs associated with the Golgi and at the cell periphery. (D–L) MCF-7 cells expressing various levels of RFP-RASSF1A (red) and immunostained for Golgi protein, GM130 (green). (D–F) Nontransfected MCF-7 cell exhibits a dispersed and fragmented Golgi. (G–I) MCF-7 cell expressing low levels of RFP-RASSF1A exhibits a compact Golgi. (J–L) MCF-7 cell overexpressing RFP-RASSF1A exhibits a dispersed and fragmented Golgi. Bar, 5 μm. (M) Analysis of Golgi dispersal under various conditions of RFP-RASSF1A expression. Representative examples out of 10 cells/condition. (N) Analysis of Golgi fragmentation in nontransfected and low RFP-RASSF1A–expressing MCF-7 cells. Representative examples out of 10 cells/condition. (O) Western blot analysis of RASSF1A siRNA depletion in RPE1 cells. Lane 1, nontargeted (NT) RASSF1A siRNA; lanes 2–4, RASSF1A siRNA combinations. (P–S) RPE1 cells treated with RASSF1A siRNA and immunostained for Golgi protein, GM130. (T) Analysis of Golgi fragmentation in RPE1 cells after RASSF1A siRNA depletion. Bar, 5 μm (L, MT). Representative examples out of 10 cells/condition. Error bars indicate SEM.
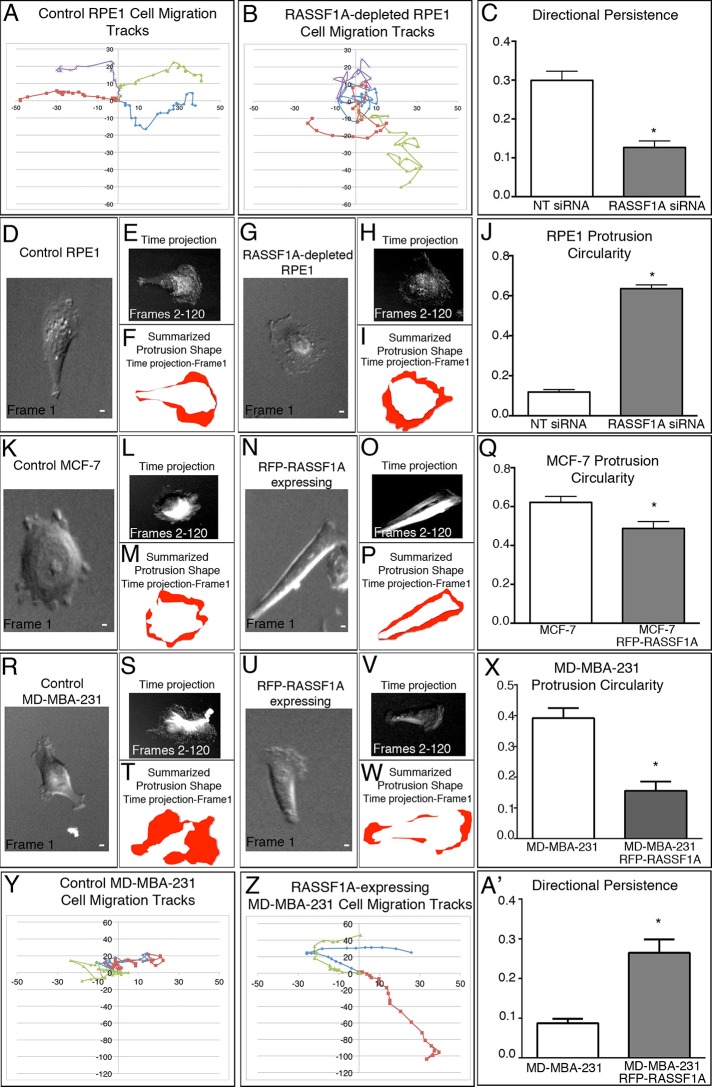
RASSF1A depletion disrupts cell polarity and migration
Because loss of RASSF1A alters MT dynamics and Golgi organization, one can hypothesize that this loss would also affect cell migration. Previous reports found that RASSF1A overexpression can suppress cell migration (Dallol et al., 2005; Jung et al., 2013); however, the ability of low levels of RASSF1A expression to influence directionality of cell migration or polarization of migrating cells has yet to be examined. To address the effect on directional persistence of migration, we examined control or RASSF1A-depleted RPE1 cells by differential interference contrast (DIC) microscopy and subjected them to persistence analysis. Depletion of RASSF1A significantly decreased the directional persistence of RPE1 cells (Figure 6, A–C), indicating that RASSF1A, likely through modulation of Golgi integrity, promotes persistent migration. Cell polarity was analyzed by assessing the direction of cell protrusions formed throughout a time sequence. Area taken by all the protrusions was combined, and the circularity of the resulting shape was quantified. In this analysis, if protrusions were formed equally around the cell perimeter (lack of polarity), the overlaid protrusion area formed a donut shape (circularity equals 1). If protrusions form only at one side of a cell (high polarity), the overlaid area is significantly asymmetric (circularity approaches 0). This analysis reveals a significant increase in protrusion circularity when cells were depleted of RASSF1A (Figure 6, D–J). MCF-7 cells also exhibited high measures of protrusion circularity similar to siRNA RASSF1A–depleted RPE1 cells (Figure 6, K–M and Q). Reexpression of RFP-RASSF1A in MCF-7 cells significantly reduced protrusion circularity (Figure 6, N–P and Q, and Supplemental Figure S3) but was unable to completely restore polarity in these cells, which may require additional molecular players that are inactive in these immobile cells. In addition, reexpression of RFP-RASSF1A did not appear to influence the overall migratory ability of MCF-7 cells. Furthermore, upon reexpression of RFP-RASSF1A in MD-MBA-231 cells, a highly motile cell type, polarity was restored to levels seen in control RPE1 cells (Figure 6, R–X, and Supplemental Figure S3), and these cells exhibited increased directional persistence (Figure 6, Y and A′). Thus these data suggest that the variation in RASSF1A regulation of cell polarity may greatly depend on the inherent motile capacity within cell types. Collectively these data suggest that RASSF1A regulation of cell polarity can potentially occur through modulation of Golgi integrity and organization, as well as via regulation of MT dynamics.
FIGURE 6:
RASSF1A expression and depletion disrupt directional persistence and cell polarity. (A, B) Representative migration tracks from control and RASSF1A-depleted RPE1. (C) Analysis of directional persistence in control and RASSF1A-depleted RPE1 cells reveals that depleted cells fail to migrate directionally and exhibit more-random movement. Representative examples out of 10 cells/condition. (D–I) RASSF1A-depleted RPE1 cells demonstrate a lack of polarity as compared with control RPE1. (J) Analysis of protrusion circularity in control and RASSF1A-depleted RPE1 cells. Representative examples out of 12 cells/condition. (K–P) Reexpression of RFP-RASSF1A in MCF-7 cells partially rescues cell polarity defects. (Q) Analysis of protrusion circularity in control and RFP-RASSF1A–expressing MCF-7 cells. Representative examples out of 12 cells/condition. (R–W) Reexpression of RFP-RASSF1A in MD-MBA-231 cells significantly reduces the protrusion circularity. (X) Analysis of protrusion circularity in control and RFP-RASSF1A–expressing MD-MBA-231. Representative examples out of 12 cells/condition. (Y–Z) Migration tracks from control and RFP-RASSF1A–expressing MD-MBA-231 cells indicate an increase in directional persistence upon RASSF1A reexpression. (A′) Analysis of directional persistence in MD-MBA-231 cells. Representative examples out of 12 cells/condition.
DISCUSSION
In this study, we detected segmental binding of RASSF1A at MTs, which is an unusual pattern for a MT-associated protein. The mechanism by which RASSSF1A accumulates at selected regions of the MT lattice may be a consequence of cooperative binding of RASSF1A molecules to a MT. Indeed, previously described ability of RASSF1A for self-association (Ortiz-Vega et al., 2002), as well as for heterodimerization with other RASSF family members (Ortiz-Vega et al., 2002; Praskova et al., 2004; Guo et al., 2007), may promote the cooperative binding. Alternatively, a preferred RASSF1A-binding area may exist within a MT, either due to accumulation of a RASSF1A-binding partner (e.g., MAP1B; Dallol et al., 2005) or if modification of tubulin within the MT lattice favors association with RASSF1A. The latter possibility is less likely, as indicated by our data (Figure 2). Any of these mechanisms likely leads to a high concentration of RASSF1A molecules on the MT lattice. Thus these short regions of MT likely exhibit properties similar to those of extremely stable MTs described in cells overexpressing RASSF1A (Liu et al., 2003; van der Weyden et al., 2005). On one hand, the capacity to stabilize MTs as the result of MT lattice-binding proteins is very common; many MT-associated proteins (MAPs), such as neuronal Tau (Tanaka et al., 2009) or MAP2 (Takemura et al., 1992) and MAP4 (Nguyen et al., 1997), stabilize MTs to build long, reliable tracks for MT-dependent transport. On the other hand, the unusual segmental pattern observed with RASSF1A-driven stabilization of short MT portions provides a mechanism for distinguishing two contrasting dynamic behaviors within one MT and serves to regulate site-specific MT dynamics.
Furthermore, our data on MT bundling indicate that massive MT bundling previously observed upon RASSF1A overexpression (Liu et al., 2003; El-Kalla et al., 2010) is not a random artifact but an exaggerated manifestation of a bona fide function. This evidence places RASSF1A within a subset of MAPs that contain MT-bundling abilities (e.g., PRC1; Mollinari et al., 2002; and synapsin I; Baines and Bennett, 1986). Such properties likely indicate that RASSF1A is capable of remodeling the interphase MT network via local bundling and connection of short fragments among two or three MTs. RASSF1A-associated segments found close to the cell periphery, which can be revealed by TIRF microscopy (and thus proximal to the ventral cell surface), might be involved in the formation of bundled MT arrays, which may serve for directional post-Golgi trafficking toward the cell front in motile cells (Rahkila et al., 1997; Schmoranzer and Simon, 2003; Lee et al., 2009).
At the same time, we show that centrally located RASSF1A-associated segments facilitate Golgi complex assembly and integrity. We hypothesize that RASSF1A defines the sites of preferred Golgi interaction with MTs and that association of RASSF1A with MT fragments in the cell center promotes Golgi stack fusion and Golgi complex integrity. In this scenario, extending RASSF1A association over the whole MT network results in scattering of these sites throughout the cell. The connection between the Golgi and MTs may be mediated either directly through RASSF1A (or some unidentified linker protein) binding to the Golgi components or by creating MT sites capable of stronger molecular motor binding. Alternatively, it is also possible that RASSF1A affects Golgi organization indirectly via MT stabilization specifically within the Golgi region, providing a long-lived platform for Golgi assembly, or by facilitation of molecular motor binding (Cai et al., 2009). Furthermore, it was previously proposed that acetylation, a robust tubulin PTM found within the Golgi region, is important for MT-dependent Golgi assembly (Thyberg and Moskalewski, 1993; Ryan et al., 2012). MT acetylation may contribute to a RASSF1A-dependent mechanism of Golgi assembly, but is not likely to be the whole mechanism, because tubulin acetylation was found, in many cases, to be independent of RASSF1A binding to MTs (unpublished data). Ultimately, we can infer that a disruption in the Golgi structure, by loss of RASSF1A, specifically interferes with other MT-dependent processes at the Golgi, such as post-Golgi trafficking.
In addition, we show that loss of RASSF1A leads to defects in cell migration and polarity; reexpression of RASSF1A can partially rescue polarization defects. We hypothesize that loss of RASSF1A at the Golgi significantly compromises the integrity and organization of the Golgi structure, both of which are required for directional cell migration and polarization (Bershadsky and Futerman, 1994; Yadav et al., 2009). With this idea, loss of RASSF1A would lead to scattering of Golgi membranes throughout the cell and the inability to form a single protrusion, potentially due to lack of directed secretion (Yadav et al., 2009). Moreover, lack of the bundling capacity exerted by RASSF1A may fail to provide the necessary stable MT tracks needed to establish a polarized front (Manneville et al., 2010; Chen et al., 2013). Alternatively, or additionally, RASSF1A-mediated regulation of cell migration and polarity may be a result of altered MT dynamics and organization. Ultimately, we can infer that perturbations to RASSF1A levels can result in significant changes in cell morphology and behavior.
Collectively our results strongly suggest a role for RASSF1A in influencing interphase MT dynamics and Golgi positioning (Figure 7A). These functions probably contribute to preventing such cancer phenotype features as disorganized cell migration, which was shown to arise from the loss of RASSF1A (Dallol et al., 2005). In particular, precise spatial regulation of MT dynamics in migrating cells is necessary for maintenance of persistent lamella at the cell front (Waterman-Storer et al., 1999), contact inhibition of migration (Kadir et al., 2011), and asymmetric focal adhesion turnover (Kaverina et al., 1999). Moreover, an integral polarized Golgi complex in the cell center is necessary for directional post-Golgi trafficking to the cell front and directional cell migration (Yadav et al., 2009). Our findings allow us to hypothesize that a dependence of directional cell migration on RASSF1A arises from spatial regulation of the MT network and Golgi organization by RASSF1A-associated MT segments (Figure 7B).
FIGURE 7:

Models of RASSF1A contribution to MT functions and Golgi organization. (A) Model of RASSF1A functions in interphase cells. (B) Model of the role of RASSF1A in directional cell migration.
MATERIALS AND METHODS
Cells
Immortalized human retinal pigment epithelial cells, hTert-RPE1 (Clontech), were maintained in DMEM/F12 (Gibco Media, Grand Island, NY) with 10% fetal bovine serum (Atlanta Biological, Flowery Branch, GA). H1792 human tumor cell lines (a gift of Geoffrey J. Clark) were grown in RPMI (Mediatech, Manassas, VA) with 10% fetal calf serum. MCF-7 breast cancer cells (American Type Culture Collection, Manassas, VA) and HeLa GFP-tubulin cells (a kind gift of Paul Chang, MIT, Cambridge, MA) were grown in DMEM with 10% fetal bovine serum. Cells were grown in 5% CO2 at 37°C. Cells were plated on fibronectin-coated glass coverslips 24 h before experiments. In all live-cell experiments, cells were maintained on the microscope stage at 37°C under mineral oil for medium equilibrium maintenance.
Treatments
For MT depolymerization during live-cell imaging, cells were initially imaged in media lacking nocodazole (Sigma, St. Louis, MO). In the subsequent frames, the media was aspirated off and replaced with cell culture media containing nocodazole (2.5 μg/ml) while imaging.
shRNA, siRNA, and expression constructs
RASSF1A shRNA was previously described (Vos et al., 2006). Single or a combination of single stealth siRNA oligos against RASSF1A (HSS174151, HSS117377; Life Technologies, Invitrogen, Darmstadt, Germany) were transfected into RPE1 cells using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The RASSF1HSS174151 siRNA–targeted sequence was 5′-GGGACGCCUUCAGCAUGCCUGAACU-3′, and the RASSFHSS117377 siRNA–targeted sequence was 5′-ACGCACAAGGGCACGUGAAGUCAUU-3′. Experiments were conducted 72 h posttransfection, as at this time minimal protein levels were detected. Nontargeting siRNA (Dharmacon, Thermo Scientific, Pittsburgh, PA) was used for controls. GFP-RASSF1A and RFP-RASSF1A were described by Vos et al. (2004) and Vos et al. (2006), respectively. mCherry-EB3 (a gift from J. V. Small, Institute of Molecular Biology, Vienna, Austria), EGFP-EB3 (a gift from A. Akhmanova, Utrecht University, Utrecht, Netherlands), and 3xGFP-EMTB (a gift from J. C. Bulinski, Columbia University, New York, NY) were used for MT plus-tip and MT visualization. RPE1 and MCF-7 cells were transfected with Fugene6 (Roche, Indianapolis, IN) according to manufacturer's protocols.
Antibodies and immunofluorescence details
For Golgi identification, mouse monoclonal antibody against GM130 (1:300; BD Transduction Laboratories, San Jose, CA) was used. MTs were stained with anti–α-tubulin rabbit polyclonal antibody (1:1000; Abcam, Cambridge, MA). For detyrosinated tubulin detection, a rabbit polyclonal antibody was used (1:500; Millipore, Billerica, MA). For MT and Golgi staining, cells were fixed (15 min at room temperature) in 4% paraformaldehyde, 0.025% glutaraldehyde, and 0.3% Triton in cytoskeleton buffer (10 mM 2-(N-morpholino)ethanesulfonic acid, 150 mM NaCl, 5 mM ethylene glycol tetraacetic acid, 5 mM glucose, and 5 mM MgCl2, pH 6.1). Alexa 488–conjugated highly cross-absorbed goat anti-mouse immunoglobulin G (IgG) antibodies and Alexa 488–conjugated goat anti-rabbit IgG antibodies (1:500; Molecular Probes, Invitrogen, Eugene, OR) were used as secondary antibodies.
Confocal and live-cell imaging
A Leica TCS SP5 confocal laser scanning microscope with an HCX PL APO 100× oil lens, numerical aperture (NA) 1.47, was used for taking confocal stacks of fixed cells and photobleaching of living cells.
TIRFM live-cell videos were acquired on a Nikon TE2000E microscope with a Nikon TIRF2 system using a TIRFM 100×/1.49 NA oil lens and Cascade 512B camera (Photometrics) and IPLab software (Scanalytics).
Live cells plated on MatTek glass-bottom dishes were maintained at 37°C by heated stage (Warner Instruments). Single-plane confocal video sequences were taken were taken by a Yokogawa QLC-100/CSU-10 spinning-disk head (Visitec assembled by Vashaw) attached to a Nikon TE2000E microscope using a CFI PLAN APO VC 100× oil lens, NA 1.4, with 1.5× intermediate magnification and a back-illuminated electron-multiplying charge-coupled device camera, Cascade 512B (Photometrics), driven by IPLab software (Scanalytics). A 75-mW 488/568 krypton-argon laser (Melles Griot) with acousto-optical tunable filter was used for two-color excitation. Custom double dichroic mirror and filters (Chroma) in a filter wheel (Ludl) were used in the emission light path.
Western blot analysis
Western blotting was performed with the Protein Electrophoresis and Western Blotting System (Bio-Rad, Hercules, CA). For Western blotting, a mouse polyclonal antibody against actin (1:1000; pan-Ab-5; Thermo Scientific, Pittsburgh, PA) and a mouse anti-human antibody against RASSF1A (1:500; EBioscience, San Diego, CA) were used. Nitrocellulose membrane was incubated with primary and then secondary antibody (IRDye 800 and 700, LI-COR Bioscience, Lincoln, NE) diluted in Odyssey Blocking Buffer with 0.2% Tween-20 to lower background. An Odyssey Infrared Imaging System (LI-COR Bioscience) was used for membrane scanning.
FRAP
FRAP was performed using a Leica TCS SP5 confocal laser scanning microscope. Regions of interest (ROIs) were introduced in cells expressing GFP-tubulin and RFP-RASSF1A within RFP-RASSF1A segments. Photobleaching of these ROIs was carried out with 100% laser power of a 561-nm laser. Cells were then imaged for 15 min with 30-s intervals to monitor RFP-RASSF1A recovery.
Quantitative analyses
MT dynamics parameters.
To analyze MT dynamic parameters in nontargeted (NT) control cells and RASSF1A-depleted cells, we used 5-min single-channel TIRFM sequences (3 s/frame) of cells expressing 3xGFP-EMTB. Individual MTs were manually tracked using the MTrackJ plug-in of ImageJ (National Institutes of Health, Bethesda, MD) by following movements of the MT tip as visualized by 3xGFP-EMTB expression. Cells expressing low levels of 3xGFP-EMTB were used to avoid analysis of cells with dampened MT dynamics. We analyzed 20 MTs per condition (NT control or RASSF1A depleted). Average numbers of events (rescue, catastrophe, pause, and pause duration) were manually calculated per MT. This was then used to calculate the frequency (rescue and catastrophe) per MT/min. Average MT growth and shrinkage rates were calculated from instantaneous velocity measurements obtained from the MTrackJ plug-in of ImageJ.
Distance dispersal.
Distance dispersal analysis was first performed with ImageJ. Golgi particles were subjected to thresholding and x, y coordinates obtained using the Analyze Particles function of ImageJ. The x, y coordinates were then analyzed using a custom program written in MATLAB to calculate average distance between all particles.
Golgi fragmentation.
Golgi fragmentation analysis was performed with the Analyze Particles function of ImageJ. Golgi particles were subjected to thresholding, and the number of particles was automatically calculated.
Fluorescence intensity.
Intensity in cells expressing various levels of RFP-RASSF1A was measured using ImageJ software. Cells of interest were outlined with a selection tool, and measurements were set (area, integrated density, and mean gray value). Three background measurements were taken. To correct for background, the following formula was used: corrected total cell fluorescence = integrated density − (area × mean fluorescence of background readings). Similar methods were used to determine fluorescence intensity of RFP-RASSF1A–associated MTs and single, nonassociated MTs.
Bundling and unbundling.
Bundling analysis of non–RASSF1A-associated MTs was restricted to 1.5-μm2 ROIs (similar to the size of RASSF1A segments). The total number of ROIs analyzed was equal to the total number of RASSF1A segments in the cell. Bundling and unbundling events were quantified as those lasting two frames (10 s) or more. Events per ROI were divided by the total number of MTs within the ROI to determine the “per-MT” contribution to bundling (or unbundling). RASSF1A-associated MTs were quantified per RASSF1A fragment.
Cell polarity.
To analyze cell polarity, we used 6-h DIC sequences (180 s/frame) of cells depleted (RPE1) or expressing (MCF-7 and MD-MBA-231) RASSF1A. Cell polarity was analyzed by assessing the direction of cell protrusions formed throughout the time sequence. The initial area of the cell (frame 1) was subtracted from the combined area taken by all protrusions in subsequent frames. The circularity of the resulting shape was quantified by ImageJ Circularity analysis. In this analysis, if protrusions were formed equally around the cell perimeter (lack of polarity), the overlaid protrusion area formed a donut shape (circularity equals 1). If protrusions form only at one side of a cell (high polarity), the overlaid area is significantly asymmetric (circularity approaches 0).
Statistical analysis
Statistical significance was determined by Student's t-test (two-tailed, unpaired).
Supplementary Material
Acknowledgments
We thank Ashley Grimaldi for providing technical and proofreading expertise. This work was supported by National Institutes of Health Integrated Biological Systems Training in Oncology Training Grant 5T32CA119925-03 to C.A., American Heart Predoctoral Fellowship 12PRE12040218 to C.A, National Institutes of Health Grant R01-GM078373 to I.K., and American Heart Association Grant-in-Aid 13GRNT16980096 to I.K.
Abbreviations used:
- EB3
end-binding protein 3
- EMTB
ensconsin microtubule-binding domain
- GFP
green fluorescent protein
- GM130
Golgi matrix protein 130
- MCF-7
Michigan Cancer Foundation-7
- MT
microtubule
- RASSF1A
Ras association domain family 1A
- RFP
red fluorescent protein
- RPE1
retinal pigment epithelial cell
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-07-0374) on January 29, 2014.
*Present address: Department of Biology, University of Pennsylvania, Philadelphia, PA 19104.
REFERENCES
- Baines AJ, Bennett V. Synapsin I is a microtubule-bundling protein. Nature. 1986;319:145–147. doi: 10.1038/319145a0. [DOI] [PubMed] [Google Scholar]
- Baksh S, Tommasi S, Fenton S, Yu VC, Martins LM, Pfeifer GP, Latif F, Downward J, Neel BG. The tumor suppressor RASSF1A and MAP-1 link death receptor signaling to Bax conformational change and cell death. Mol Cell. 2005;18:637–650. doi: 10.1016/j.molcel.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Bershadsky AD, Futerman AH. Disruption of the Golgi apparatus by brefeldin A blocks cell polarization and inhibits directed cell migration. Proc Natl Acad Sci USA. 1994;91:5686–5689. doi: 10.1073/pnas.91.12.5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbee DG, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, McEwen DP, Martens JR, Meyhofer E, Verhey KJ. Single molecule imaging reveals differences in microtubule track selection between kinesin motors. PLoS Biol. 2009;7:e1000216. doi: 10.1371/journal.pbio.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Chen J, Shi H, Wei M, Castaneda-Castellanos DR, Bultje RS, Pei X, Kriegstein AR, Zhang M, Shi SH. Regulation of microtubule stability and organization by mammalian Par3 in specifying neuronal polarity. Dev Cell. 2013;24:26–40. doi: 10.1016/j.devcel.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NB, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol Biol Cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallol A, Agathanggelou A, Fenton SL, Ahmed-Choudhury J, Hesson L, Vos MD, Clark GJ, Downward J, Maher ER, Latif F. RASSF1A interacts with microtubule-associated proteins and modulates microtubule dynamics. Cancer Res. 2004;64:4112–4116. doi: 10.1158/0008-5472.CAN-04-0267. [DOI] [PubMed] [Google Scholar]
- Dallol A, Agathanggelou A, Tommasi S, Pfeifer GP, Maher ER, Latif F. Involvement of the RASSF1A tumor suppressor gene in controlling cell migration. Cancer Res. 2005;65:7653–7659. doi: 10.1158/0008-5472.CAN-05-0247. [DOI] [PubMed] [Google Scholar]
- Dallol A, Hesson LB, Matallanas D, Cooper WN, O'Neill E, Maher ER, Kolch W, Latif F. RAN GTPase is a RASSF1A effector involved in controlling microtubule organization. Curr Biol. 2009;19:1227–1232. doi: 10.1016/j.cub.2009.05.064. [DOI] [PubMed] [Google Scholar]
- Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120:3163–3172. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- Drabek K, et al. Role of CLASP2 in microtubule stabilization and the regulation of persistent motility. Curr Biol. 2006;16:2259–2264. doi: 10.1016/j.cub.2006.09.065. [DOI] [PubMed] [Google Scholar]
- El-Kalla M, Onyskiw C, Baksh S. Functional importance of RASSF1A microtubule localization and polymorphisms. Oncogene. 2010;29:5729–5740. doi: 10.1038/onc.2010.316. [DOI] [PubMed] [Google Scholar]
- Faire K, Waterman-Storer CM, Gruber D, Masson D, Salmon ED, Bulinski JC. E-MAP-115 (ensconsin) associates dynamically with microtubules in vivo and is not a physiological modulator of microtubule dynamics. J Cell Sci. 1999;112:4243–4255. doi: 10.1242/jcs.112.23.4243. [DOI] [PubMed] [Google Scholar]
- Guo C, Tommasi S, Liu L, Yee JK, Dammann R, Pfeifer GP. RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr Biol. 2007;17:700–705. doi: 10.1016/j.cub.2007.02.055. [DOI] [PubMed] [Google Scholar]
- Jo H, Kim JW, Kang GH, Park NH, Song YS, Kang SB, Lee HP. Association of promoter hypermethylation of the RASSF1A gene with prognostic parameters in endometrial cancer. Oncol Res. 2006;16:205–209. doi: 10.3727/000000006783981125. [DOI] [PubMed] [Google Scholar]
- Jung HY, Jung JS, Whang YM, Kim YH. RASSF1A suppresses cell migration through inactivation of HDAC6 and increase of acetylated alpha-tubulin. Cancer Res Treat. 2013;45:134–144. doi: 10.4143/crt.2013.45.2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadir S, Astin JW, Tahtamouni L, Martin P, Nobes CD. Microtubule remodelling is required for the front-rear polarity switch during contact inhibition of locomotion. J Cell Sci. 2011;124:2642–2653. doi: 10.1242/jcs.087965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang GH, Lee S, Lee HJ, Hwang KS. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. J Pathol. 2004;202:233–240. doi: 10.1002/path.1503. [DOI] [PubMed] [Google Scholar]
- Kaverina I, Krylyshkina O, Small JV. Microtubule targeting of substrate contacts promotes their relaxation and dissociation. J Cell Biol. 1999;146:1033–1044. doi: 10.1083/jcb.146.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellokumpu S, Sormunen R, Kellokumpu I. Abnormal glycosylation and altered Golgi structure in colorectal cancer: dependence on intra-Golgi pH. FEBS Lett. 2002;516:217–224. doi: 10.1016/s0014-5793(02)02535-8. [DOI] [PubMed] [Google Scholar]
- Kupfer A, Dennert G, Singer SJ. Polarization of the Golgi apparatus and the microtubule-organizing center within cloned natural killer cells bound to their targets. Proc Natl Acad Sci USA. 1983;80:7224–7228. doi: 10.1073/pnas.80.23.7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Kim HY, Byun DS, Lee SJ, Lee CH, Kim JI, Chang SG, Chi SG. Frequent epigenetic inactivation of RASSF1A in human bladder carcinoma. Cancer Res. 2001;61:6688–6692. [PubMed] [Google Scholar]
- Lee S, Han JW, Leeper L, Gruver JS, Chung CY. Regulation of the formation and trafficking of vesicles from Golgi by PCH family proteins during chemotaxis. Biochim Biophys Acta. 2009;1793:1199–1209. doi: 10.1016/j.bbamcr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Broaddus RR, Yao JC, Xie S, White JA, Wu TT, Hamilton SR, Rashid A. Epigenetic alterations in neuroendocrine tumors: methylation of RAS-association domain family 1, isoform A and p16 genes are associated with metastasis. Mod Pathol. 2005;18:1632–1640. doi: 10.1038/modpathol.3800490. [DOI] [PubMed] [Google Scholar]
- Liu L, Tommasi S, Lee DH, Dammann R, Pfeifer GP. Control of microtubule stability by the RASSF1A tumor suppressor. Oncogene. 2003;22:8125–8136. doi: 10.1038/sj.onc.1206984. [DOI] [PubMed] [Google Scholar]
- Manneville JB, Jehanno M, Etienne-Manneville S. Dlg1 binds GKAP to control dynein association with microtubules, centrosome positioning, and cell polarity. J Cell Biol. 2010;191:585–598. doi: 10.1083/jcb.201002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller PM, Folkmann AW, Maia AR, Efimova N, Efimov A, Kaverina I. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat Cell Biol. 2009;11:1069–1080. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinari C, Kleman JP, Jiang W, Schoehn G, Hunter T, Margolis RL. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol. 2002;157:1175–1186. doi: 10.1083/jcb.200111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HM, Widschwendter A, Fiegl H, Ivarsson L, Goebel G, Perkmann E, Marth C, Widschwendter M. DNA methylation in serum of breast cancer patients: an independent prognostic marker. Cancer Res. 2003;63:7641–7645. [PubMed] [Google Scholar]
- Nguyen HL, Chari S, Gruber D, Lue CM, Chapin SJ, Bulinski JC. Overexpression of full- or partial-length MAP4 stabilizes microtubules and alters cell growth. J Cell Sci. 1997;110:281–294. doi: 10.1242/jcs.110.2.281. [DOI] [PubMed] [Google Scholar]
- Ortiz-Vega S, Khokhlatchev A, Nedwidek M, Zhang XF, Dammann R, Pfeifer GP, Avruch J. The putative tumor suppressor RASSF1A homodimerizes and heterodimerizes with the Ras-GTP binding protein Nore1. Oncogene. 2002;21:1381–1390. doi: 10.1038/sj.onc.1205192. [DOI] [PubMed] [Google Scholar]
- Piperno G, LeDizet M, Chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praskova M, Khoklatchev A, Ortiz-Vega S, Avruch J. Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem J. 2004;381:453–462. doi: 10.1042/BJ20040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahkila P, Vaananen K, Saraste J, Metsikko K. Endoplasmic reticulum to Golgi trafficking in multinucleated skeletal muscle fibers. Exp Cell Res. 1997;234:452–464. doi: 10.1006/excr.1997.3633. [DOI] [PubMed] [Google Scholar]
- Rong R, Jiang LY, Sheikh MS, Huang Y. Mitotic kinase Aurora-A phosphorylates RASSF1A and modulates RASSF1A-mediated microtubule interaction and M-phase cell cycle regulation. Oncogene. 2007;26:7700–7708. doi: 10.1038/sj.onc.1210575. [DOI] [PubMed] [Google Scholar]
- Rong R, Jin W, Zhang J, Sheikh MS, Huang Y. Tumor suppressor RASSF1A is a microtubule-binding protein that stabilizes microtubules and induces G2/M arrest. Oncogene. 2004;23:8216–8230. doi: 10.1038/sj.onc.1207901. [DOI] [PubMed] [Google Scholar]
- Ryan SD, Bhanot K, Ferrier A, De Repentigny Y, Chu A, Blais A, Kothary R. Microtubule stability, Golgi organization, and transport flux require dystonin-a2-MAP1B interaction. J Cell Biol. 2012;196:727–742. doi: 10.1083/jcb.201107096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoranzer J, Simon SM. Role of microtubules in fusion of post-Golgi vesicles to the plasma membrane. Mol Biol Cell. 2003;14:1558–1569. doi: 10.1091/mbc.E02-08-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol. 2002;22:4309–4318. doi: 10.1128/MCB.22.12.4309-4318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, et al. The tumour suppressor RASSF1A regulates mitosis by inhibiting the APC-Cdc20 complex. Nat Cell Biol. 2004;6:129–137. doi: 10.1038/ncb1091. [DOI] [PubMed] [Google Scholar]
- Takemura R, Okabe S, Umeyama T, Kanai Y, Cowan NJ, Hirokawa N. Increased microtubule stability and alpha tubulin acetylation in cells transfected with microtubule-associated proteins MAP1B, MAP2 or tau. J Cell Sci. 1992;103:953–964. doi: 10.1242/jcs.103.4.953. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Nohara T, Iwamoto M, Sumiyoshi K, Kimura K, Takahashi Y, Tanigawa N. Tau expression and efficacy of paclitaxel treatment in metastatic breast cancer. Cancer Chemother Pharmacol. 2009;64:341–346. doi: 10.1007/s00280-008-0877-5. [DOI] [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S. Relationship between the Golgi complex and microtubules enriched in detyrosinated or acetylated alpha-tubulin: studies on cells recovering from nocodazole and cells in the terminal phase of cytokinesis. Cell Tissue Res. 1993;273:457–466. doi: 10.1007/BF00333700. [DOI] [PubMed] [Google Scholar]
- van der Weyden L, Adams DJ. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim Biophys Acta. 2007;1776:58–85. doi: 10.1016/j.bbcan.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weyden L, Tachibana KK, Gonzalez MA, Adams DJ, Ng BL, Petty R, Venkitaraman AR, Arends MJ, Bradley A. The RASSF1A isoform of RASSF1 promotes microtubule stability and suppresses tumorigenesis. Mol Cell Biol. 2005;25:8356–8367. doi: 10.1128/MCB.25.18.8356-8367.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos MD, Dallol A, Eckfeld K, Allen NP, Donninger H, Hesson LB, Calvisi D, Latif F, Clark GJ. The RASSF1A tumor suppressor activates Bax via MOAP-1. J Biol Chem. 2006;281:4557–4563. doi: 10.1074/jbc.M512128200. [DOI] [PubMed] [Google Scholar]
- Vos MD, Ellis CA, Bell A, Birrer MJ, Clark GJ. Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J Biol Chem. 2000;275:35669–35672. doi: 10.1074/jbc.C000463200. [DOI] [PubMed] [Google Scholar]
- Vos MD, Martinez A, Elam C, Dallol A, Taylor BJ, Latif F, Clark GJ. A role for the RASSF1A tumor suppressor in the regulation of tubulin polymerization and genomic stability. Cancer Res. 2004;64:4244–4250. doi: 10.1158/0008-5472.CAN-04-0339. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Worthylake RA, Liu BP, Burridge K, Salmon ED. Microtubule growth activates Rac1 to promote lamellipodial protrusion in fibroblasts. Nat Cell Biol. 1999;1:45–50. doi: 10.1038/9018. [DOI] [PubMed] [Google Scholar]
- Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol. 2003;4:938–947. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- Yadav S, Puri S, Linstedt AD. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol Biol Cell. 2009;20:1728–1736. doi: 10.1091/mbc.E08-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.