Seven of 15 different mouse formins localized in diverse patterns to cardiomyocyte sarcomeres. Four were required for proper organization of myofibrils, and two were critical for remodeling and repair of myofibril structure.
Abstract
Cardiac and skeletal muscle function depends on the proper formation of myofibrils, which are tandem arrays of highly organized actomyosin contractile units called sarcomeres. How the architecture of these colossal molecular assemblages is established during development and maintained over the lifetime of an animal is poorly understood. We investigate the potential roles in myofibril formation and repair of formin proteins, which are encoded by 15 different genes in mammals. Using quantitative real-time PCR analysis, we find that 13 formins are differentially expressed in mouse hearts during postnatal development. Seven formins immunolocalize to sarcomeres in diverse patterns, suggesting that they have a variety of functional roles. Using RNA interference silencing, we find that the formins mDia2, DAAM1, FMNL1, and FMNL2 are required nonredundantly for myofibrillogenesis. Knockdown phenotypes include global loss of myofibril organization and defective sarcomeric ultrastructure. Finally, our analysis reveals an unanticipated requirement specifically for FMNL1 and FMNL2 in the repair of damaged myofibrils. Together our data reveal an unexpectedly large number of formins, with diverse localization patterns and nonredundant roles, functioning in myofibril development and maintenance, and provide the first evidence of actin assembly factors being required to repair myofibrils.
INTRODUCTION
The physiological functions of cardiac and skeletal muscle require the ability to undergo coordinated contractions and generate large amounts of force. These functions in turn depend critically on the formation of highly ordered actomyosin arrays, or myofibrils. The basic contractile unit of muscle fibers is the sarcomere, which is immensely rich in its molecular composition and has a complex and organized architecture. Over the past 5 years, our understanding of the sarcomere has evolved substantially, as numerous studies have expanded the list of sarcomere-associated proteins and demonstrated a startling level of protein dynamics within sarcomeres (reviewed in Ono, 2010; Sanger et al., 2010). Along with our increasing appreciation of the complexity of the sarcomere has come an understanding that sarcomere composition and regulation can differ depending on cell type and developmental stage. Many of the components of cardiac sarcomeres have been linked to human cardiomyopathies, including desminopathy (Goldfarb and Dalakas, 2009), dilated cardiomyopathy (Arber et al., 1997), hypertrophic cardiomyopathy (Müller et al., 2012; Wooten et al., 2013), familial atrial septal defects (Posch et al., 2011), and others. Here we investigate the potential roles of the formin family of proteins in cardiomyocyte sarcomere development and repair.
Sarcomeres contain organized arrays of so-called thin (actin) filaments and interdigitating thick (myosin) filaments. Thin filaments have a uniform length within sarcomeres of the same muscle type. Despite the long list of actin filament regulators that localize to sarcomeres (reviewed in Ono, 2010), the specific mechanisms responsible for actin filament nucleation, elongation, bundling, and turnover dynamics during sarcomere development, maintenance, and repair are not well understood. Nebulin-N-WASP complex and Lmod were each shown to induce actin nucleation in muscle cell myofibrils (Chereau et al., 2008; Takano et al., 2010); however, the potential roles of formins in regulating sarcomeric actin assembly and organization have remained understudied. Formins are likely candidates for some of these roles based on their demonstrated abilities to nucleate and elongate linear actin filaments and antagonize capping proteins (Goode and Eck, 2007; Campellone and Welch, 2010; reviewed in Breitsprecher and Goode, 2013). In addition, many formins can bind to the sides of, and bundle, actin filaments in vitro (Michelot et al., 2005; Moseley and Goode, 2005; Harris et al., 2006; Vaillant et al., 2008; Schönichen et al., 2013), and there is growing evidence for the importance of filament bundling by formins in vivo (Jaiswal et al., 2013; Schönichen et al., 2013).
Formins in mammals are encoded by 15 different genes and classified into seven subfamilies (DIA, DAAM, FMN, INF, FHOD, FMNL/FRL, and Delphilin; Higgs and Peterson, 2005). Only two of the 15 mammalian formin proteins have been implicated in cardiac function, FHOD3 and DAAM1 (Kanaya et al., 2005; Taniguchi et al., 2009; Li et al., 2011; Kan-O et al., 2012a, b; Iskratsch et al., 2013; Wooten et al., 2013; see Discussion for details). Further, evidence suggests that neither of these formins is responsible for the initial polymerization of actin thin filaments during sarcomere formation. Thus the potential extent and diversity of formin functions in the heart has gone largely unexplored.
In the present study we cast a wide net to explore the possible roles of formins in mouse cardiomyocyte myofibril development. Using quantitative real-time PCR (qRT-PCR), we characterize the developmental expression patterns of all 15 formin genes in the heart and follow up with immunofluorescence staining to determine their subcellular localization patterns in primary cardiomyocytes. Using small interfering RNA (siRNA), we then knock down sarcomere-localized formins in primary cardiomyocytes to determine their effects on myofibril development and regeneration after damage. Our results show that an unexpectedly large number of formins are expressed in cardiomyocytes, where they exhibit intriguingly diverse localization patterns. Further, we find that members of multiple formin subfamilies perform nonredundant functional roles in sarcomere development, setting the stage for future investigations into the perplexing question of why so many actin-regulating formins are required to build a sarcomere.
RESULTS
Expression patterns of the 15 mammalian formin genes during heart development
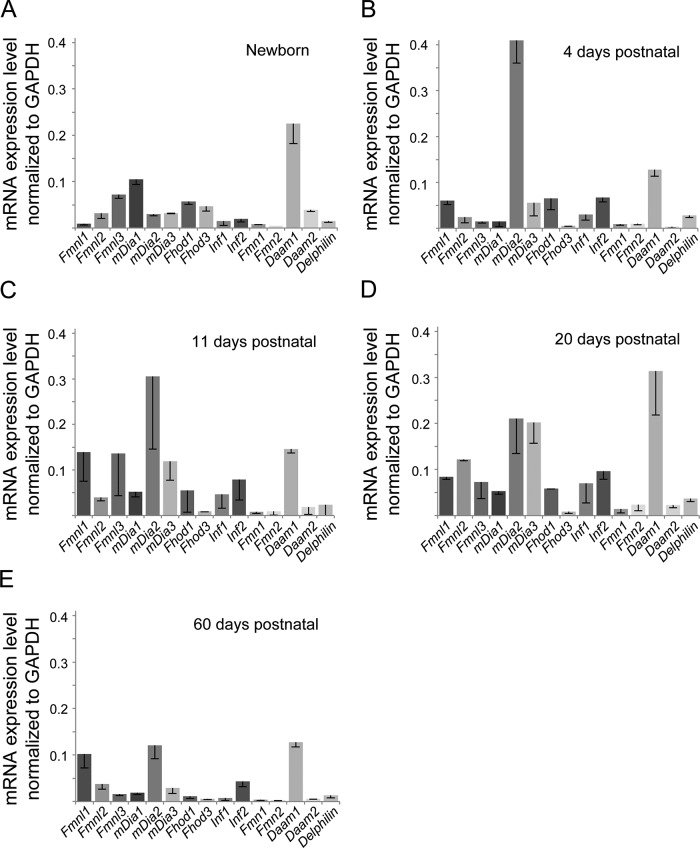
To begin to investigate which mammalian formins might play a role in cardiac muscle development, we used qRT-PCR to define the expression pattern of each of the 15 formin genes in mouse hearts at different developmental time points: newborn and 4, 11, 20, and 60 d (Figures 1 and 2 and Table 1). Although the heart is the first embryonic organ in the animal to take shape and begin functioning, it continues to grow and develop rapidly throughout the embryonic period and the first 14 d postnatal in mice (Oparil et al., 1984; Li et al., 1996; Leu et al., 2001). Mouse hearts reach adult size by ∼3 mo (Leu et al., 2001). Embryonic growth of the heart is hyperplastic, meaning that the heart grows primarily through cell division, whereas postnatal growth of the heart is hypertrophic, meaning that it grows primarily through enlargement of existing cells (Oparil et al., 1984; Li et al., 1996; Leu et al., 2001). By focusing on the postnatal period, any formin expression we detect is less likely to be due to a role in cell division and more likely to be due to a role in forming sarcomeres as the myofibrils expand during cell growth.
FIGURE 1:
Multiple formin genes are differentially expressed during mouse heart development. Comparative qRT-PCR analysis of the mRNA expression levels for the 15 mammalian formins in (A) newborn, (B) 4 d postnatal, (C) 11 d postnatal, (D) 20 d postnatal, and (E) 60 d postnatal mouse hearts. Formin mRNA values were normalized to GAPDH mRNA levels. The formin expression pattern was distinct at each time point. Each bar represents the average value of triplicate reactions from at least three independent experiments. Error bars are SEM.
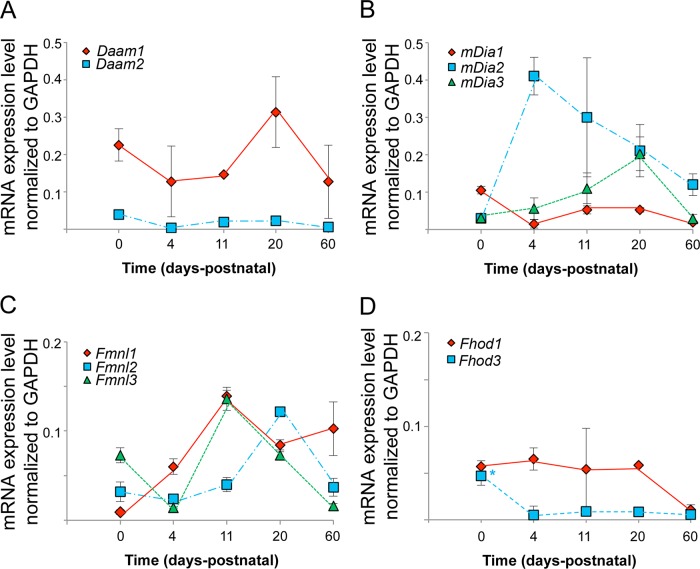
FIGURE 2:
Developmental changes in formin mRNA levels in mouse heart. Formin mRNA expression levels, as determined by qRT-PCR, throughout postnatal mouse heart development are graphed for members of the (A) DAAM, (B) DIA, (C) FMNL, and (D) FHOD subfamilies. Formin mRNA values were normalized to GAPDH mRNA levels.
TABLE 1:
Summary of formin gene expression in mammalian hearts.
| Expression at given number of days postnatal | |||||
|---|---|---|---|---|---|
| 0 | 4 | 11 | 20 | 60 | |
| Fmnl1 | + | ++ | ++ | ++ | ++ |
| Fmnl2 | + | + | + | +++ | + |
| Fmnl3 | ++ | + | + | + | + |
| mDia1 | ++ | – | + | + | + |
| mDia2 | + | +++ | +++ | +++ | ++ |
| mDia3 | + | + | ++ | +++ | + |
| Fhod1 | ++ | + | + | ++ | + |
| Fhod3 | + | – | + | – | – |
| Inf1 | – | + | – | + | – |
| Inf2 | + | ++ | – | ++ | + |
| Fmn1 | + | + | – | – | – |
| Fmn2 | – | + | – | + | – |
| Daam1 | +++ | +++ | + | +++ | +++ |
| Daam2 | + | – | – | + | – |
| Delphilin | + | + | – | + | + |
–, <0.5%; +, 0.6–5.0%; ++, 5.1–10.0%; +++, >10.1%. The data are shown in Figure 1.
RT-PCR primers were carefully designed for specificity to each formin (see Materials and Methods). Formin expression levels were standardized to a housekeeping gene, glyceraldehyde 3-phosphate-dehydrogenase (GAPDH), which is constitutively expressed in neonatal and adult cardiomyocytes (Lam et al., 2002). Our results showed that 13 of the 15 formin genes were expressed in the heart during postnatal development, and at each developmental time point there was a distinct gene expression profile (Figure 1 and Table 1). Of interest, at least one representative from each formin subfamily was expressed at some point during development (Figure 1 and Table 1).
In newborn hearts, members of the FMNL, DIA, FHOD, and DAAM subfamilies were highly expressed, with Daam1 showing particularly high expression. At 4 d postnatal, members of the FMNL, DIA, FHOD, INF, and DAAM subfamilies were highly expressed, with mDia2 and Daam1 showing particularly high expression levels. At 11 and 20 d postnatal, FMNL, DIA, FHOD, INF, and DAAM subfamily members were still highly expressed, although the individual expression profiles differed (Figure 1). Consistent with our understanding of heart development, we observed the lowest diversity of formin expression in the heart at 60 d postnatal, which is a near-adult developmental time point. Thus formins that were highly expressed at 60 d (Fmnl1, Fmnl2, mDia2, mDia3, Inf2, Daam1) are likely to play roles in maintenance and/or repair rather than strictly in development.
Our results show that most formins are differentially expressed throughout heart development. In Figure 2, we graph the developmental expression levels for individual members of the DAAM, DIA, and FMNL subfamilies. Daam1 is the only formin expressed at both high and consistent levels throughout development (Figures 1 and 2). The DIA subfamily members have markedly different expression patterns (Figure 2). mDia1 expression increases during the postnatal period, peaks at 20 d (which is the approximate time that hypertrophic growth drops off), and then decreases to very low levels by 60 d. mDia2 is virtually unexpressed in the newborn heart, which is in its hyperplastic growth phase. mDia2 expression peaks at 4 d and then steadily drops, although expression is still relatively high at 60 d. mDia3 expression is consistently low throughout development.
Similar to members of the Dia subfamily, members of the FMNL subfamily show distinct expression patterns (Figure 2). Fmnl1 expression builds and then plateaus from 11 d onward. Fmnl2 expression peaks at 20 d and then falls off at 60 d, although at 60 d it is still expressed at significant levels. Fmnl3 is highly expressed in the newborn heart and exhibits a second peak of high expression at 11 d, finally dropping to low levels at 60 d. These striking differences in formin expression patterns hint at potentially diverse functional roles in development.
Formins are recruited to distinct locations within sarcomeres
Our qRT-PCR analysis showed that up to 13 different formin genes are expressed in the developing mouse heart, but some of this heterogeneity could arise from the variety of different cell types found in the heart. Therefore we used immunohistochemistry to visualize the presence and subcellular localization patterns of heart-expressed formins in primary cultured cardiomyocytes (Figures 3 and 4). Primary cardiomyocytes were harvested from newborn mice and grown for 11 d in vitro (DIV), allowing myofibrils to reform after plating, resulting in beating cells. Each sample was costained with a formin-recognizing antibody and an antibody against α-actinin to label the Z-bands of the sarcomeres (Figures 3A and 4A). We also determined that formins localized to sarcomeres in the intact heart by imaging mature cardiomyofibrils isolated from the hearts of 2-mo-old mice (Figure 3B and 4B). Fixation and immunostaining conditions were optimized for each antibody, and siRNA knockdowns in primary cardiomyocytes confirmed the specificity of staining for mDia2, DAAM1, FMNL1, and FMNL3 (see later discussion).
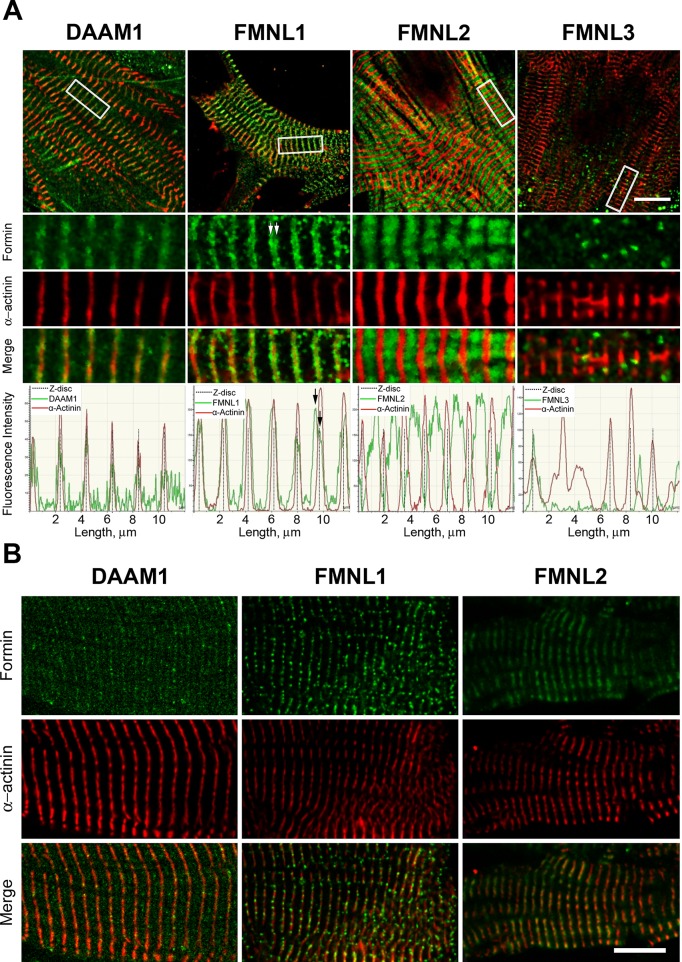
FIGURE 3:
Localization of DAAM and FMNL formin subfamily members to sarcomeres. (A) Immunohistochemical localization of the formins (green) DAAM1, FMNL1, FMNL2, and FMNL3 in relation to Z-bands (red, α-actinin) in neonatal primary mouse cardiomyocytes cultured 11 d in vitro. White boxes on cell overview (first row) indicate the zoom-in regions (rows 2–4). Graphs below depict fluorescence intensity in the two channels over 12 μm. Daam1 signal colocalized with α-actinin labeling of the Z-band. FMNL1 signal appeared as a broad band surrounding the α-actinin–labeled Z-band, with peaks on either side of the Z-band (white arrows in zoom; black arrows in graph). FMNL2 signal filled the area between α-actinin–labeled Z-bands. FMNL3 signal appeared as a few puncta with no regular sarcomere-related pattern. (B) Immunohistochemical localization of formin (green) and α-actinin (red) for DAAM1, FMNL1, and FMNL2 in mature myofibrils isolated from 2-mo-old mice. All three formins colocalize with α-actinin, indicating robust sarcomere localization. Scale bars, 10 μm.
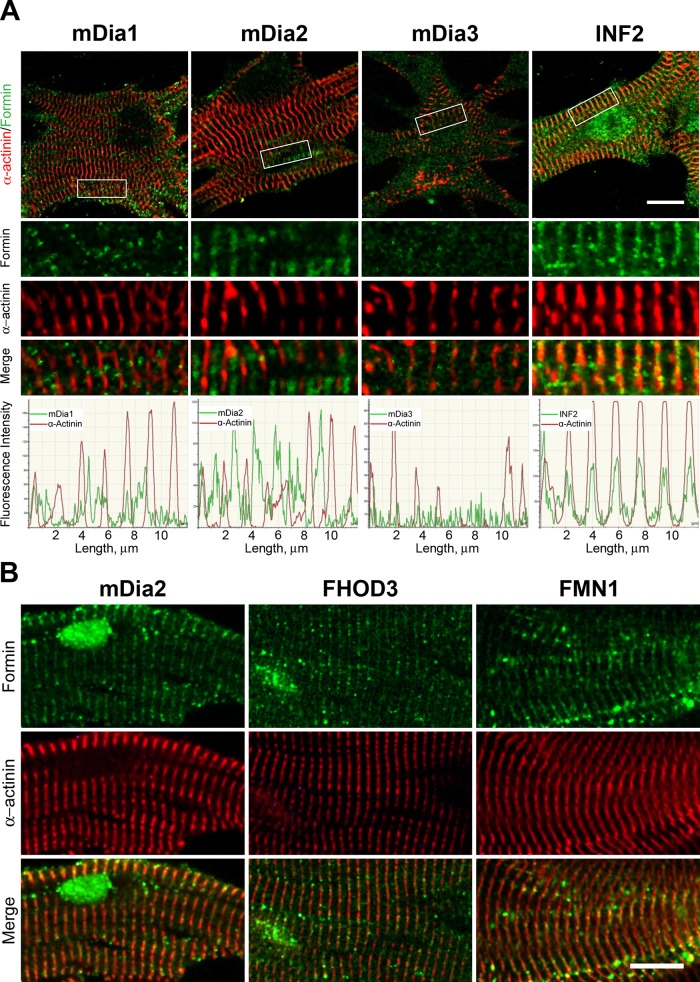
FIGURE 4:
Localization of DIA, INF, FHOD, and FMN formin subfamily members to sarcomeres. (A) Immunohistochemical localization of the formins (green) mDia1, mDia2, mDia3, and INF2 in relation to Z-bands (red, α-actinin) in neonatal primary mouse cardiomyocytes cultured 11 d in vitro. White boxes on cell overview (first row) indicate the zoom-in regions (rows 2–4). Graphs below depict fluorescence intensity in the two channels over 12 μm. mDia1 signal appeared as puncta with no clear sarcomere-related pattern. mDia2 localized to the M-band and Z-band, midway between the α-actinin labeled Z-bands. mDia3 signal was indistinguishable from noise. INF2 colocalized with α-actinin–labeled Z-bands. (B) Immunohistochemical localization of formins (green) and α-actinin (red) for mDia2, FHOD3, and FMN1 in mature myofibrils isolated from 2-mo-old male mice. All three formins colocalize with α-actinin, indicating robust sarcomere localization. Scale bars, 10 μm.
DAAM1 localized to Z-bands, as indicated by colocalization with α-actinin in both the primary cultured cardiomyocytes and mature cardiomyofibrils (Figure 3). These results are consistent with its previously reported role in maintaining Z-band integrity (Li et al., 2011). FMNL1 localized as a band with a double peak on either side of the Z-band in the primary cultures (Figure 3A) but as punctuated structures on the Z-band itself in the mature cardiomyofibrils (Figure 3B). FMNL2 localization in the primary culture cells filled the region between the Z-bands (Figure 3). In mature cardiomyofibrils, FMNL2 exhibited a broad banding pattern that overlaps with the Z-band (Figure 3). In contrast to FMNL1 and FMNL2, FMNL3 localized to puncta with no obvious sarcomere association (Figure 3A).
Similar to FMNL3, mDia1 localized to puncta in primary cultured cells, with no obvious sarcomere association. However, clusters of mDia1 signal were observed at sites near the cell periphery (Figure 4), where myofibrillogenesis occurs (reviewed in Sanger et al., 2000, 2010). In both primary culture and mature cardiomyocytes, examples could be found of mDia2 localizing to the Z-band and/or the M-band (Figures 4 and 5B). mDia3 staining in primary cultured cells was indistinguishable from background staining (Figure 4A). In addition, we observed Z-band staining of INF2 in primary culture cells (Figure 4A) and FMN1 in mature cardiomyofibrils (Figure 4B). Together these results demonstrate that multiple formins localize to cardiac sarcomeres and show distinct distribution patterns within these structures.
FIGURE 5:
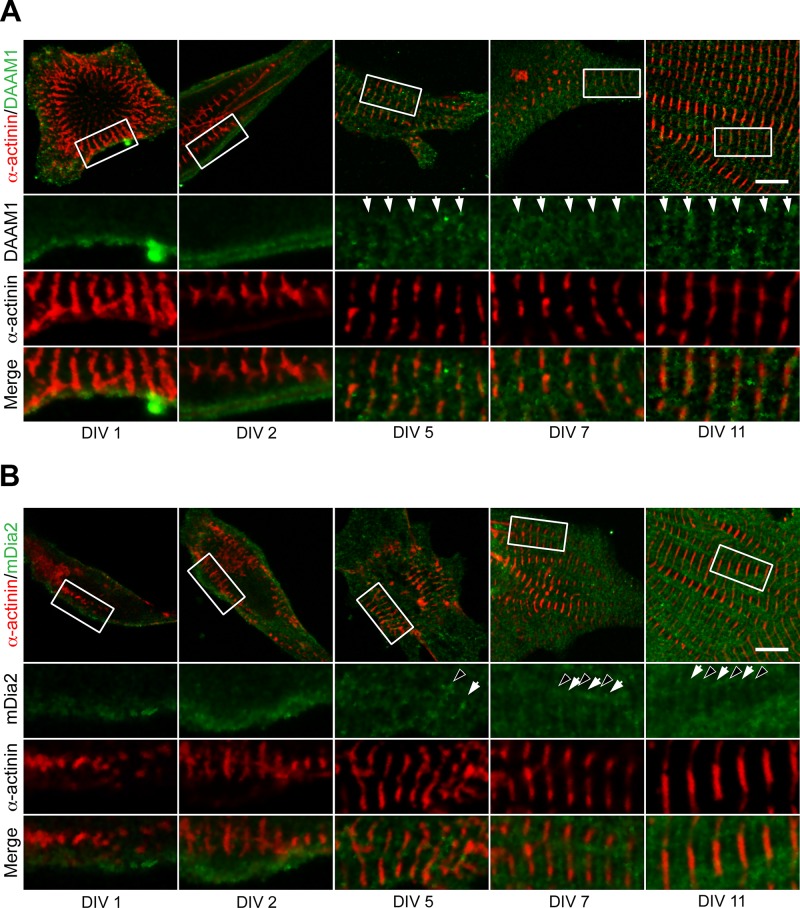
DAAM1 and mDia2 recruitment to sarcomeres during myofibrillogenesis. Immunohistochemistry was used to determine the localization of formins (green) and α-actinin (red) during myofibrillogenesis in cultured neonatal primary cardiomyocytes. Cells were imaged at 1, 2, 5, 7, and 11 DIV. White boxes on cell overview (first row) indicate zoom-in regions (rows 2–4). (A) DAAM1 localized to the cell cortex at DIV 1 and DIV 2, with no apparent sarcomere-related pattern. From DIV 5 onward, DAAM1 appeared in bands (white arrows) that colocalized with α-actinin–labeled Z-bands. (B) mDia2 also localized to the cell cortex at DIV 1 and DIV 2, with no apparent sarcomere-related pattern. At DIV 5, mDia2 began to appear as bands that correlated with the Z-bands (white arrows) or M-bands (black arrowheads). Z-band and M-band staining for mDia2 was robustly observed from DIV 7 onward. Scale bars, 10 μm.
Finally, we observed Z-band staining of FHOD3 in mature cardiomyocytes (Figure 4B). FHOD3 is one of the two formins that were previously demonstrated to function in heart development. Previous studies showed that FHOD3 localizes to the sarcomeres in rat and mouse cardiomyocytes, where it is required for normal sarcomere organization (Kanaya et al., 2005; Taniguchi et al., 2009; Kan-O et al., 2012a, b; Iskratsch et al., 2013). However, there were differing reports about the subcellular location of FHOD3, with some studies placing it at the I-band, adjacent to the M-band (Kan-O et al., 2012a, b) and others at the Z-band (Iskratsch et al., 2013). Our data agree more closely with the latter study.
Dynamic changes in formin localization during myofibrillogenesis
The results of our qRT-PCR analysis demonstrated that formin expression changes over the course of heart development. To determine whether formin subcellular localization in cardiomyocytes also changes during myofibrillogenesis, we followed DAAM1, mDia2, FMNL1, and FMNL2 localization in primary culture cells from DIV 1–20 (Figures 5 and 6 and Supplemental Figure S1). DAAM1 localized to the cell cortex at DIV 1 and 2 but did not appear to incorporate into the premyofibrils (Figure 5A). However, by DIV 5 and onward, DAAM1 localized to Z-bands (Figure 5A). Similarly, mDia2 started out diffusely localized at the cell cortex at DIV 1 and 2 but localized to Z-bands and/or M-bands from DIV 5 onward (Figure 5B and Supplemental Figure S1).
FIGURE 6:
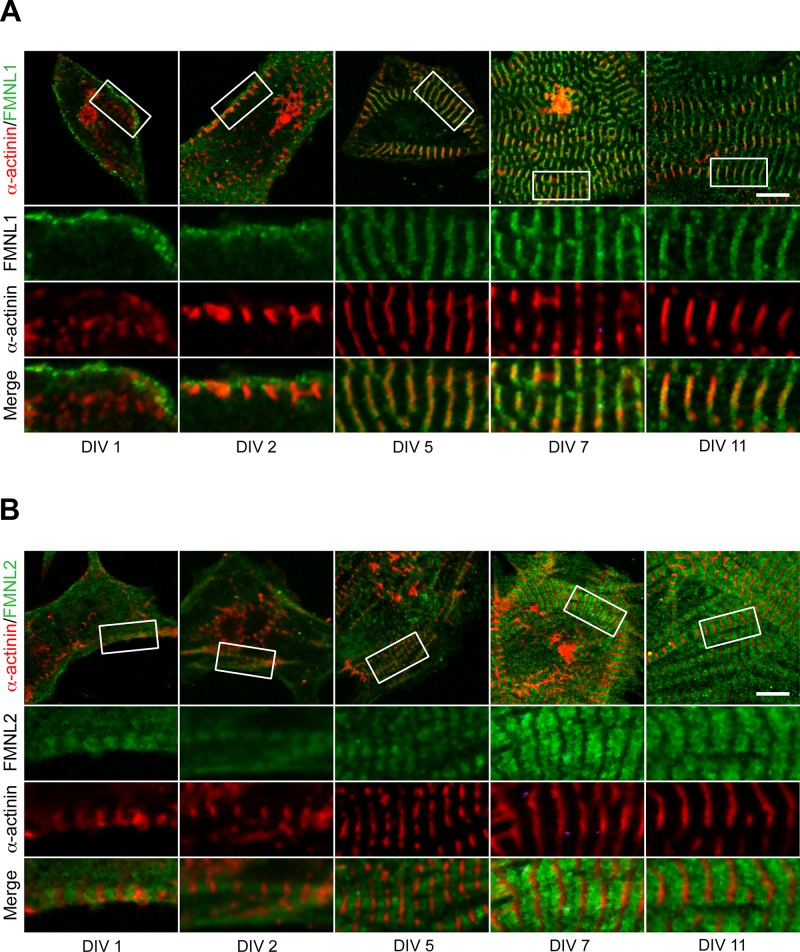
FMNL1 and FMNL2 recruitment to sarcomeres during myofibrillogenesis. Immunohistochemistry was used to determine the localization of formins (green) and α-actinin (red) during myofibrillogenesis in cultured neonatal primary cardiomyocytes. Cells were imaged at 1, 2, 5, 7, and 11 DIV. White boxes on cell overview (first row) indicate zoom-in regions (rows 2–4). (A) At DIV 1–2, FMNL1 localized to the cell cortex near the growing α-actinin–labeled Z-bodies/bands. From DIV 5 onward, FMNL1 localized to broad bands surrounding the Z-bands. (B) FMNL2 localized to clear sarcomere-associated bands between the α-actinin–labeled Z-bands as early as DIV 1. Scale bars, 10 μm.
At DIV 1, FMNL1 localized to the cell periphery (Figure 6). At DIV 2, FMNL1 signal surrounded the early Z-bodies/bands labeled by α-actinin. From DIV5 onward, FMNL1 localized to a broad band enveloping the Z-band (Figure 6A and Supplemental Figure S1). In contrast to other formins, within 24 h of culturing cells (DIV 1), FMNL2 localized in a banded pattern filling the space between Z-bodies/bands (Figure 6B). FMNL2’s localization pattern in relationship to α-actinin remained consistent through DIV 20 (Figure 6B and Supplemental Figure S1). From these results it is clear that the localization patterns of DAAM1, mDia2, FMNL1, and FMNL2 vary with the different stages of myofibrillogenesis, with FMNL2 recruited early to premyofibrils. Taken together, our results indicate that different formins are recruited to the sarcomere at different stages of cardiomyofibril development and that the localization pattern can change during the development.
Multiple formins are required for cardiomyofibril development
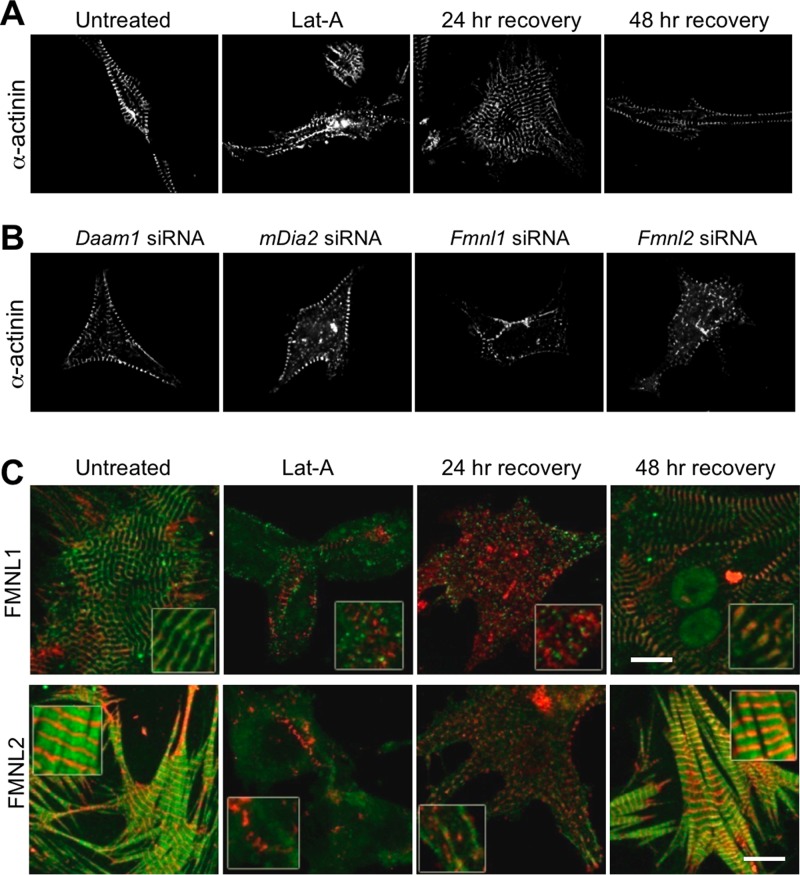
The formin localization patterns suggested that formins might function in the assembly and/or maintenance of sarcomere structures. To test this model, we individually knocked-down Daam1, mDia2, Fmnl1, and Fmnl2 gene expression in primary cultured cardiomyocytes. Cardiomyocytes were isolated from neonatal mice (0–3 d) and treated with siRNA oligos before plating. Cells were cotransfected at the same time with a plasmid expressing green fluorescent protein (GFP) to allow identification of successfully transfected and likely silenced cells. Control cells were transfected with a siRNA construct designed against the prokaryotic ampicillin gene. The cells were allowed to grow for 72 h posttransfection and then fixed and assessed for myofibril architecture by immunohistochemical labeling of α-actinin. Silencing of the targeted formin gene was confirmed by lack of immunofluorescence signal for the targeted formin in transfected (GFP-positive) cells (Supplemental Figure S2A).
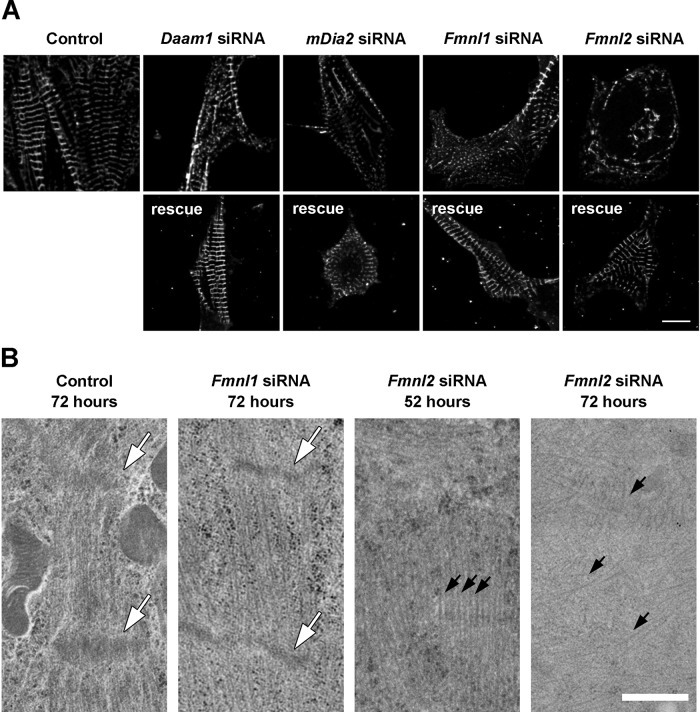
Silencing of Daam1, Fmnl1, and Fmnl2 each caused cellular defects that could be rescued by a siRNA-resistant copy of the targeted formin gene (Figure 7). After Daam1 siRNA treatment, disorganized α-actinin puncta were observed near the center of cells, although normal- appearing α-actinin bands were visible at the cell periphery (Figure 7A). In mDia2 siRNA cells, α-actinin was found only in small, regularly spaced puncta at the cell periphery, hinting that myofibrillogenesis may have been arrested at an early stage (Figure 7A). Silencing of Fmnl1 also significantly disrupted α-actinin banding, again with some regular banding at the cell periphery (Figure 7A). siRNA silencing of Fmnl2 completely disrupted α-actinin organization (Figure 7A). Staining cells with fluorescently labeled phalloidin gave similar results and revealed that there were no new large actin structures formed in the silenced cells (Supplemental Figure S2B).
FIGURE 7:
RNAi silencing of Daam1, mDia2, Fmnl1, or Fmnl2 in cardiomyocytes disrupts myofibril and sarcomere organization. siRNA constructs were used to individually silence Daam1, mDia2, Fmnl1, or Fmnl2 in cultured neonatal primary cardiomyocytes. (A) α-actinin staining of cardiomyocytes 72 h posttransfection revealed structural disruption of myofibrils (top). The absence of the targeted formin protein was confirmed by immunohistochemistry (Supplemental Figure S2). Specificity of the knockdowns was further demonstrated by successful rescues to wild-type phenotype with corresponding siRNA-resistant formin genes (bottom). Scale bar, 10 μm. (B) Electron microscopy imaging of control and siRNA-silenced cells at 52 and 72 h posttransfection. At 72 h posttransfection, siRNA Fmnl1 sarcomeres were longer on average and the Z-bands (white arrows) were thinner on average than in control cells (see Results). By 72 h posttransfection, siRNA Fmnl2 cells contained no structures resembling sarcomeres, although individual disorganized actin filaments were observed (black arrows). At 52 h posttransfection, siRNA Fmnl2 cells contained organized actin filaments but without Z-bands. Scale bar, 500 nm.
To further characterize the defects caused by silencing of Fmnl1 and Fmnl2, we investigated sarcomere ultrastructure by transmission electron microscopy at 72 and 52 h posttransfection (Figure 7B). Sarcomeres in Fmnl1-silenced cells showed several defects compared with control cells. First, sarcomeres in Fmnl1-silenced cells were longer than those in control cells (control, 1.19 ± 0.04 μm, n = 6; siRNA Fmnl1, 1.34 ± 0.04 μm, n = 6; p = 0.03). Second, Z-bands were abnormally narrow (control,125 ± 9 nm, n = 9; siRNA Fmnl1, 100 ± 6 nm, n = 9; p = 0.03). Third, actin thin filaments appeared more disorganized and less densely bundled than in control cells (Figure 7B). However, we could not discern from our images whether the changes in sarcomere length were due to a difference in contraction state or length of the thin filaments.
Consistent with our light microscopy analysis, at 72 h posttransfection, sarcomere ultrastructure in Fmnl2-silenced cells was more severely impaired than in Fmnl1-silenced cells. Fmnl2-silenced cells contained highly disorganized and scattered actin filaments, with nothing resembling sarcomere structures or Z-bands (Figure 7B). We also looked at Fmnl2-silenced cells at 52 h posttransfection, 20 h before the peak structural defects observed by light microscopy. At 52 h posttransfection, Fmnl2-silenced cells contained bundles with parallel actin filaments, but there was no evidence of Z-bands at the ends of these actin arrays and the bundles pointed in multiple directions, whereas actin structures in the control cells were oriented in the same direction (Figure 7B). Of importance, although no sarcomere structures were seen at either time point, a large amount of F-actin was still present. Together the siRNA results indicate that DAAM1, mDia2, FMNL1, and FMNL2 play critical and nonredundant roles in cardiomyofibril development.
Effects of actin assembly and myosin activity on formin localization within sarcomeres
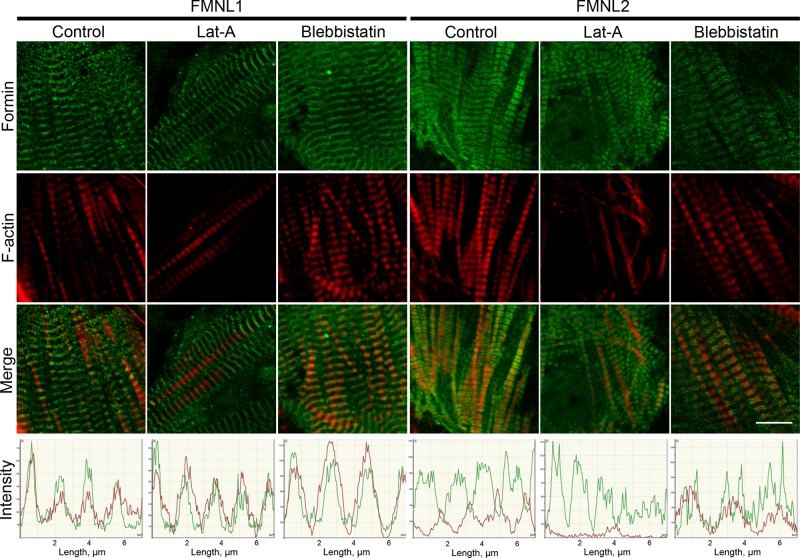
To determine whether DAAM1, mDia2, FMNL1, or FMNL2 sarcomere localization depended on either the availability of actin monomers or myosin activity, we examined formin localization in cardiomyocytes treated with latrunculin A (LatA) or blebbistatin, respectively. LatA binds and sequesters actin monomers, inhibiting new polymerization; its effects are transient and can be reversed by washing out the drug from the growth media (Spector et al., 1989; Ayscough et al., 1997). Previous studies showed that treatment of mouse primary cardiomyocytes with low concentrations of LatA (0.2–0.5 μM) for 15–30 min stops new actin polymerization without disassembling mature, existing myofibrils (Wang et al., 2005). Blebbistatin is a drug that traps the myosin II in a state with low affinity for actin filaments, inhibiting actomyosin contraction (Straight et al., 2003; Kovács et al., 2004). We grew primary cardiomyocytes for 6 d in vitro, allowing them to develop mature myofibrils, and then treated them with either LatA for 30 min or blebbistatin for 15 min. LatA treatment caused no obvious change in localization of DAAM1, mDia2, FMNL1, or FMNL2 (Figure 8 and Supplemental Figure S3). FMNL1 and mDia2 localization were also unaffected by blebbistatin treatment (Figure 8 and Supplemental Figure S3). However, the FMNL2 localization changed dramatically after blebbistatin treatment, collapsing from the broad band between Z-bands seen in control cells to two narrow peaks located on either side of the Z-bands (Figure 8). These findings indicate that FMNL2 localization to the zones with actin–myosin overlap (A-bands) requires myosin binding and/or activity.
FIGURE 8:
FMNL1 and FMNL2 localization in cardiomyocytes after LatA or blebbistatin treatment. After 6 d in culture, neonatal mouse cardiomyocytes were treated with LatA (30 min) or blebbistatin (15 min), fixed, and stained with antibodies. Localization of FMNL1 (green) and F-actin (red) was unaffected by LatA or blebbistatin treatment. Localization of FMNL2 (green) bands was unaffected by LatA treatment. However, blebbistatin caused the broad FMNL2 bands between Z-bands to collapse into narrow bands on either side of the Z-bands. Graphs show fluorescence intensity in the two channels. Scale bars, 10 μm.
FMNL formins are required for myofibril repair after injury
Next we investigated the potential involvement of formins in cardiomyofibril repair after damage induced by prolonged treatment with LatA. To characterize normal recovery from this damage, we grew primary cardiomyocytes to DIV 3–4, until cells had mature myofibrils, and then added LatA to the growth medium for 24 h. This longer (24 h) exposure to LatA disrupted Z-band organization as determined by α-actinin staining (Figure 9A). We then washed LatA from the growth medium and checked myofibril staining after 24 and 48 h of recovery. At 24 h of recovery, myofibrillogenesis had begun at the cell periphery, and by 48 h of recovery, mature myofibril organization was fully restored and cells appeared normal in regard to both morphology (Figure 9A) and beating. To assess whether specific formins were required for the regeneration of myofibrils after the same damage, we repeated these assays in cells silenced for Daam1, mDia2, Fmnl1, or Fmnl2 (Figure 9B). Whereas Daam1- and mDia2-silenced cells recovered normally, Fmnl1- and Fmnl2-silenced cells failed to recover properly and lacked normal myofibril architecture even after 48 h of recovery (Figure 9B). Finally, we examined localization of FMNL1 and FMNL2 during myofibril repair. FMNL2 (but not FMNL1) was associated with the early myofibril structures at 24 h, and normal localization of both formins was restored by 48 h (merged images in Figure 9C; individual images in Supplemental Figure S4). These exciting findings point to specific role(s) for FMNL subfamily members in repairing damaged cardiomyofibrils.
FIGURE 9:
FMNL1 and FMNL2 are required for repair of damaged myofibrils. (A) Time course of Z-band repair after injury. To induce myofibril damage, neonatal mouse cardiomyocytes were grown in culture for 48 h, then treated with LatA for 24 h and allowed to recover for 24 or 48 h. α-Actinin banding patterns returned to normal by 24 h of recovery from LatA. (B) Formin requirement in myofibril repair. LatA was used to induce myofibril damage in neonatal primary culture cardiomyocytes in which either Daam1, mDia2, Fmnl1, or Fmnl2 was silenced using siRNA. Images of the injured siRNA cells after 48 h of recovery from LatA treatment. Note that Fmnl1 and Fmnl2 siRNA cells fail to reestablish organized sarcomeric structures, even after 48 h of recovery, whereas Daam1 and mDia2 siRNA cells recovered normally within 24 h. (C) Localization of FMNL1 or FMNL2 (green) and α-actinin (red) in control (untreated) cells, in cells treated with LatA for 24 h, and in cells allowed to recover for 24 or 48 h after LatA treatment. Merged images are shown (individual images are shown in Supplemental Figure S4). Scale bars, 10 μm.
DISCUSSION
In this study, we asked the broad question of how many of the 15 mammalian formin proteins participate in sarcomere development and/or maintenance. We determined that 13 different formins are expressed at some point during postnatal heart development (Figures 1 and 2 and Table 1) and that at least seven formins (DAAM1, mDia2, FMNL1, FMNL2, INF2, FHOD3, FMN1) localize to sarcomeres in either primary cultured cardiomyocyte cells or mature cardiomyofibrils isolated from the heart (Figures 3 and 4). Because only two formins (DAAM1 and FHOD3) had previously been shown to be important for cardiac function, our observations greatly expand and diversify the roles that formins play in this process.
Focusing on the four formins with the most distinct and robust localization patterns (DAAM1, mDia2, FMNL1, FMNL2), we demonstrated that they are recruited in distinct localization patterns to the sarcomere at different times during development (Figures 5 and 6). Through siRNA silencing of the individual genes in primary cardiomyocytes, we further showed that each of these four formins is essential for myofibrillogenesis in a nonredundant manner (Figure 7). Moreover, our siRNA analysis revealed that myofibril repair after LatA-induced damage specifically requires FMNL1 and FMNL2 but not DAAM1 or mDia2. This differs from myofibrillogenesis, which requires all four of these formins.
Formin gene expression in different stages of mammalian heart development
Our qRT-PCR expression profile was based on total RNA from whole hearts (including multiple cell types) isolated at different stages of postnatal mouse development. We found that 13 different formins are expressed in at least one stage examined during the postnatal period (Figure 1). The distinct expression profiles found at each time point suggests that different formins are involved in hyperplastic growth (newborn), hypertrophic growth (4 d postnatal), the transition from hypertrophic growth to mature heart cells (11 and 20 d postnatal), and maintenance of mature heart cells (60 d postnatal). Although formin expression profiles change through development, it is interesting to note that at least one member of the FMNL, DIA, FHOD, INF, and DAAM families is expressed at each time point (Figure 1). This may point to each formin subfamily having distinct functional roles.
Because our immediate interest was in the roles formins play in cardiomyocyte sarcomere formation and maintenance, we followed up on our qRT-PCR results by determining each formin's expression and subcellular localization in primary culture cardiomyocytes. Of the 12 formins for which we had antibodies, we found that seven (DAAM1, mDia2, FMNL1, FMNL2, INF2, FHOD3, FMN1) localized to the sarcomere. This does not preclude the localization of other formins to cardiomyocyte sarcomeres, as we could not check for each of the 13 formins at every developmental time point and because only a single antibody was used in each case. Further, there are additional roles that formins may play in the development of the heart besides the formation of sarcomeres, including assembly of stress fibers, maintenance of cell shape, mediation of adhesion junctions, and roles in cell division. Our qRT-PCR data provide surprising information about the abundance and diversity of formin expression during heart development, which we hope will aid researchers in further investigating the molecular basis for regulation of actin dynamics and organization in the heart.
Formin localization during cardiomyocyte myofibrillogenesis
The best-accepted model for myofibrillogenesis proposes that myofibrils are assembled in stages. In this model, premyofibrils with α-actinin containing Z-bodies and nonmuscle myosin II form at the cell periphery and later mature into myofibrils with α-actinin–containing Z-bands and muscle myosin II (Rhee et al., 1994; Sanger et al., 2002, 2005; Golson et al., 2004). This suggests the potential for multiple roles for formins during myofibrillogenesis, including initial nucleation and polymerization of thin filaments at the cell periphery, repair of damaged thin filaments in mature sarcomeres, facilitation of dynamic turnover of thin filaments, potential capping/regulation of the barbed ends of thin filaments, and polymerization of actin filaments that anchor Z-bands to desmin filament networks or to costameres that connect sarcomeres to the cell membrane. For many of these roles, we might expect the formin to localize to the Z-band where the barbed (fast-growing) ends of thin filaments are found, where anchoring of sarcomeres to other structures occurs, and where chaperones involved in sarcomere repair reside until damage occurs (Etard et al., 2008). Indeed, although several of the formins show distinct localization patterns, we frequently observed signals at the Z-band, and in the cases of INF2, FHOD3, DAAM1, and FMN1, we detected the formin exclusively at the Z-band (Figures 3 and 4).
Based on the biochemical characterization of mammalian formins, including mDia1, mDia2, DAAM1, FMNL2, and INF2, it appears that each formin has a distinct set of effects on actin filament nucleation, elongation, bundling, capping, severing, and/or depolymerization (reviewed in Goode and Eck, 2007; Breitsprecher and Goode, 2013; Chesarone et al., 2010; Schonichen and Geyer, 2010). These biochemical differences are likely to support the distinct cellular tasks performed by each formin at their particular locations within the sarcomere. In the following we discuss in more detail what our findings, combined with those of previous studies, suggest about the specific roles of DAAM1, mDia2, FMNL1, and FMNL2 in myofibrillogenesis.
DAAM1
Daam1 was detected in our qRT-PCR analysis as one of two formin genes that are highly expressed throughout heart development (Figures 1 and 2). A previous study showed that DAAM1 is expressed in the heart during mouse embryogenesis and that Daam1−/− mutant mice have defective adherens junctions and Z-disks, as well as ventricular noncompaction (Li et al., 2011). The phenotypes of the DAAM1 mutant suggest that it associates with the Z-bands; however, its subcellular localization was not previously determined. We found that DAAM1 localizes to the cell periphery early in myofibrillogenesis and then is recruited to the Z-bands of mature myofibrils (Figure 5). Consistent with the studies of Daam1−/− mice, we found that myofibril organization is disrupted by Daam1 silencing (Figure 7A). The phenotypes of Daam1−/− mice indicate that this formin is necessary for proper formation of both adherens junctions and Z-bands in cardiomyocytes (Li et al., 2011), and our localization data place DAAM1 exactly where it needs to be to directly participate in these functions. It is worth noting that although DAAM1 appears to be a component of mature Z-bands, it does not colocalize with Z-bodies earlier in myofibrillogenesis (Figure 5). Thus DAAM1 may not participate in the initial nucleation/polymerization of thin filaments. However, the wide and disorganized Z-bands of Daam1−/− mutant mice (Li et al., 2011) suggest a role for DAAM1 in regulating the organization rather than polymerization of thin filaments in mature sarcomeres. Consistent with this view, in nonmuscle cells DAAM1 serves an actin-filament bundling role that is required for the structural integrity and protrusion of filopodial shafts (Jaiswal et al., 2013).
mDia2
mDia2 is best known for its actin-polymerization activities in filopodia and lamellipodia (Pellegrin and Mellor, 2005; Schirenbeck et al., 2005; Yang et al., 2007), but our qRT-PCR results indicate that mDia2 is also highly expressed throughout postnatal heart development (Figures 1 and 2). In our analysis of cardiomyocyte myofibrillogenesis, mDia2 was found diffusely located at the cell periphery until it was recruited to the Z-bands and M-bands of mature myofibrils (Figure 5B). Knockdown of mDia2 disrupted myofibrillogenesis such that α-actinin was only found in small and regularly spaced puncta at the cell periphery (Figure 7A), suggesting that premyofibrils fail to mature into bona fide myofibrils. This observation is interesting, given that cardiomyofibril maturation requires the exchange of nonmuscle myosin II for muscle myosin II (reviewed in Sanger et al., 2010). Dia2 is already known to contribute to myosin II recruitment during the assembly of contractile stress fibers in nonmuscle cells (Tojkander et al., 2011). Given that myofibrils and stress fibers are believed to develop through similar mechanisms (reviewed in Sparrow and Schöck, 2009), it is possible that Dia2 plays a similar function in myofibril and stress fiber assembly. Thus one possibility is that M-line–localized mDia2 we observed in cardiomyocytes plays a role in stabilizing muscle myosin II integration into maturing nascent myofibrils. mDia2 also bundles actin filaments in vitro (Harris et al., 2006), so another possibility is that its bundling activity is required at the Z-band.
The Z-band and M-band localization pattern of mDia2 in mature myofibrils is reminiscent of ankyrin-B localization (Mohler et al., 2003; Cunha and Mohler, 2008). Ankyrins are adaptor proteins that link cytoskeletal components such as actin filaments to membrane proteins. Whether ankyrin-B is mediating such connections at the Z-bands and M-bands is not yet clear (reviewed in Cunha and Mohler, 2009), but ankyrin-B is necessary for the recruitment of at least one protein (PP2A) to the M-band (Cunha and Mohler, 2008), and a future goal should be to test whether mDia2 localization also depends on ankyrin-B.
FMNL1
FMNL1 expression in the postnatal heart increases through the hypertrophic phase of growth and then plateaus as the heart nears maturity (Figure 2). Fmnl1a is also expressed in zebrafish heart during development (Santos-Ledo et al., 2013), suggesting that FMNL1’s functional role(s) in the heart may be evolutionarily conserved. Early in myofibrillogenesis (DIV 2), FMNL1 signal surrounds the premyofibrils (Figure 6A). In mature myofibrils, FMNL1 is present as a double-peaked band enveloping the Z-band (Figures 3 and 6A), that is, it localizes to the I-band region closest to the Z-band. On silencing of Fmnl1, α-actinin organization is disrupted, and sarcomeres become longer, contain fewer thin filaments, and have narrower Z-bands (Figure 7). However, it is not clear whether the increased sarcomere length is due to a change in the contraction state or the length of the thin filaments. FMNL1 also plays an essential role in myofibril repair after LatA-induced damage (Figure 9).
Multiple actin isoforms are expressed in cardiomyocytes, although the thin filaments in mature cardiomyofibrils appear to be primarily composed of α-cardiac actin (Ruzicka and Schwarz, 1988; von Arx et al., 1995). In addition to the thin filament network, skeletal myofibrils also contain a network of nonmuscle γ-actin and nonmuscle γ-tropomyosin that connects sarcomeres to costameres (Craig and Pardo, 1983; Rybakova et al., 2000; Nakata et al., 2001; Kee et al., 2004; Papponen et al., 2009). The localization of the γ-actin and γ-tropomyosin with respect to α-actinin (Kee et al., 2004) is strikingly similar to our observation of FMNL1 in cardiomyocytes (Figures 3 and 6A). To our knowledge, no group has tested for the presence of the γ-actin filament network in these cardiac cells; however, costameres are present (Danowski et al., 1992), linked to the Z-bands (Danowski et al., 1992) and essential for proper cardiomyofibrillogenesis (reviewed in Ervasti, 2003; Zemljic-Harpf et al., 2009). FMNL1’s localization suggests that it may act as a nucleator for a Z-band/costamere–linking actin network. If FMNL1 is regulating a costamere-linked γ-actin network, this could account for the phenotypes observed in the Fmnl1 siRNA cells. It is easy to imagine how the absence of the network would lead to a loss of Z-band proteins, particularly α-actinin, which is associated with costameres (reviewed in Ervasti, 2003; Zemljic-Harpf et al., 2009). This would explain both the abnormal α-actinin staining and the narrow Z-bands in Fmnl1 siRNA cells. It is also well established that force transduction (involving costameres) and adhesion to the substrate are necessary for proper myofibrillogenesis, possibly explaining the other observed sarcomeric defects (less dense and more disorganized actin filaments).
FMNL2
FMNL2 was expressed throughout postnatal heart development at varying levels and was localized throughout the sarcomere, excluding Z-bands. Like FMNL1, FMNL2 isoforms are expressed in the developing zebrafish heart (Santos-Ledo et al., 2013), suggesting that FMNL2’s role in the heart may be conserved. FMNL2 was the only formin studied here that localized to the premyofibrils at DIV1, suggesting that FMNL2 may play a role in the initial polymerization and/or organization of actin filaments. Indeed, in Fmnl2 siRNA–silenced cells, actin filaments were disorganized and sarcomere structures failed to form (Figure 7).
Studies suggest that only a fraction of the thin filaments in mature myofibrils are dynamic and that they are likely to be those undergoing repair (Littlefield et al., 2001; Skwarek-Maruszewska et al., 2009). The persistence of dynamic thin filaments appears to depend on contraction, as the dynamic population disappears with blebbistatin treatment (Skwarek-Maruszewska et al., 2009). In our cultured cardiomyocytes, inhibiting myosin II with blebbistatin caused the FMNL2 signal to collapse to two narrow peaks in the I-band region flanking the Z-band (Figure 8), indicating that the normal localization pattern of FMNL2 is dependent on myosin binding and/or sarcomere contraction. To test whether FMNL2 plays a role in repairing damaged sarcomeres, we treated cardiomyocytes with LatA for 24 h and then washed out the drug and observed the recovery of myofibril architecture. In the absence of FMNL2, cells did not recover proper myofibril architecture even after 48 h (Figure 9). Although myofibril repair is poorly understood, a study of the myosin chaperones Unc45b and Hsp90a in striated muscle demonstrated that the chaperones reside at the Z-band until injury (Etard et al., 2008). After injury, such as heat, cold, membrane puncture, or exposure to paraformaldehyde, these chaperones spread across the sarcomere in a pattern not unlike that of FMNL2 in cells fixed with paraformaldehyde (Etard et al., 2008; Figure 3). Thus future live-cell imaging studies should be performed to determine whether FMNL2 also dynamically relocates in response to myofibril injury. In addition to its actin-nucleating activities, FMNL2 bundles actin filaments in vitro (Vaillant et al., 2008), which could be highly relevant to its function in myofibrillogenesis or myofibril repair.
Conclusion
Our examination of the expression and localization patterns of mammalian formins in cardiomyocytes revealed that a surprising large number of formins localize to the sarcomere. Their distinct patterns of expression, developmental recruitment into the sarcomere, and subsarcomeric localization patterns suggest diverse functional roles for them in the development and repair of myofibrils. Although we have begun to identify some of the unique features of these formins in this study, much work remains to be done to determine the specific functional and mechanistic roles of each of these formins within the sarcomere.
MATERIALS AND METHODS
Analysis of gene expression by quantitative real-time PCR
Total RNA from whole mouse hearts at different ages was isolated using TRIzol reagent (Life Technologies, Grand Island, NY) according to manufacturer's instructions. cDNA was generated from the RNA (Superscript II; Life Technologies) and served as a template for qRT-PCR using SYBR Green PCR master mix (Bio-Rad, Hercules, CA) and a DNA engine chromo 4 real-time PCR detector (Bio-Rad). Internal housekeeping gene GAPDH, forward 5′- AGTCCCTGCCCTTTGTACACA-3′, and reverse, 5′-CCGAGGGCCTCACTAAACC-3′. Special attention was given to the design of the oligonucleotides used for RT-PCR amplification of the formin genes. We avoided splice junctions between introns and exons, as well as targeting splice isoform variant regions predicted by DNA analysis software. Five different oligonucleotide primer sets were designed for each formin and tested by PCR cycling analysis. Oligonucleotide primer sets that resulted in amplification of the desired product size were selected, and oligonucleotide primer specificity was verified by sequencing the PCR products. Primer locations for each formin gene were as follows, with the identity of the mouse gene used in parentheses after the formin name; numbering refers to nucleotide positions in the open reading frame: Daam1 (NM_025102) forward (488–508) and reverse (776–756), Daam2 (NM_001008231) forward (686–706) and reverse (1018–998), Fmnl1 (NM_019679) forward (2499–2519) and reverse (2993–2973), Fmnl2 (NM_172409) forward (1316–1336) and reverse (1454–1434), Fmnl3 (NM_011711) forward (897–916) and reverse (1257–1238), Inf1 (EDL15405) forward (3049–3069) and reverse (3497–3477), Inf2 (NM_198411) forward (3330–3349) and reverse (3588–3569), Fmn1 (NM_010230) forward (2553–2573) and reverse (3111–3091), Fmn2 (NM_019445) forward (3537–3557) and reverse (3982–3962), Delphilin/Grid2ip (NM_133355) forward (3143–3163) and reverse (3200–3180), Fhod1 (NM_177699) forward (771–791) and reverse (917–897), Fhos2/Fhod3 (NM_175276) forward (941–961) and reverse (1288–1268), mDia1/Diap1 (NM_007858) forward (644–664) and reverse (919–899), mDia2/Diap3 (NM_019670) forward (4056–4076) and reverse (4164–4144), and mDia3/Diap2 (NM_172493) forward (32–51) and reverse (291–271). Cycling parameters were 95°C for 3 min, followed by 40 cycles of 95°C for 30 s, 55°C for 45 s, and 72°C for 45 s. Fluorescence intensities were plotted versus the number of cycles using an algorithm provided by the manufacturer (Bio-Rad). mRNA values were normalized to those from GAPDH. Data are presented as mean ± SEM of at least three independent mRNA isolation experiments, with qRT-PCR reaction run in duplicate. Significant changes in mRNA expression levels during developmental stages were determined by analysis of variance single-way factor analysis with p < 0.05, followed by t test analysis.
Isolation and culture of primary cardiomyocytes
Primary cardiomyocytes were isolated and cultured from neonatal to 3-d-old C57BL/6J mice as previously described (Maass and Buvoli, 2007). All animal procedures were conducted in accordance with National Institutes of Health guidelines and were approved by the Brandeis University Animal Care and Use Committee. Briefly, cardiac tissue was dissected and digested enzymatically using 2 mg/ml trypsin (Sigma-Aldrich, St. Louis, MO) and 1 mg/ml collagenase type II (Worthington, Lakewood, NJ) in calcium-free buffered 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid solution at room temperature with gentle shaking (60 rpm). After digestion, cells were centrifuged at room temperature for 15 min at 800 × g and resuspended in growth medium (D-MEM supplemented with 10% calf serum, 2 mM l-glutamine, nonessential amino acid mixture [Invitrogen], 5 mg/ml insulin [Sigma-Aldrich], 5 mg/ml transferrin [Sigma-Aldrich], 0.1 mM norepinephrine [Sigma-Aldrich], and 75 μg/ml ampicillin [Invitrogen]). Cells were preplated for 30 min to remove cardiac fibroblasts. Cardiac myocytes where then replated at a cell density of 300,000 cells/ml on Falcon six-well culture dishes on 0.1% gelatin (Sigma-Aldrich)-coated coverslips. After 24 h, growth medium was changed to maintain cells.
Immunofluorescence microscopy
Cells were fixed for 20 min in 4% paraformaldehyde (Sigma-Aldrich) in phosphate-buffered saline (PBS) and permeabilized with 0.2% Triton X-100 in PBS for 3 min, followed by blocking incubation in PBS and 0.5% bovine serum albumin) for 20 min at room temperature. Immunofluorescence was carried out with antibodies and dilutions as follows. Three different fixing methods (paraformaldehyde, glutaraldehyde, and a mixture of alcohols [ethanol and methanol]) were tested to find the one that best preserved sarcomeric structures as determined by staining with antibodies to sarcomeric proteins. For each antibody, a series of antibody concentrations was tested to determine what dilution and incubation times resulted in clean staining with minimal nonspecific background. siRNA knockdowns confirmed antibody specificity for mDia2, DAAM1, FMNL1, and FMNL3 (see Results) and for FMNL2, mDia1, and mDia3 (M.R. and B.L.G., unpublished data). Primary antibodies were as follows: 1:1000 anti–α-Actinin (Sigma-Aldrich), 1:1000 anti–α-sarcomeric Actin (Sigma-Aldrich), 1:800 anti-titin (9D10; Developmental Studies Hybridoma Bank, Iowa City, IA), and 1:800 anti-myosin (MF-20; Developmental Studies Hybridoma Bank). Formin antibodies (Santa Cruz Biotechnology, Dallas, TX) were all used at 1:100: anti-Dia1, anti-Dia2, anti-Dia3, anti-DAAM1, anti-DAAM2, anti-FMNL1, anti-FMNL2, anti-FMNL3, anti-FMN1, anti-FMN2, and anti-FHOD3. Anti-INF2 antibody was used at 1:25 (Sigma-Aldrich). Secondary antibodies were all used at 1:1000: anti-goat immunoglobulin G (IgG)–fluorescein isothiocyanate (Santa Cruz Biotechnology), anti-goat IgG-TR (Santa Cruz Biotechnology), and Alexa Fluor 633 F(ab′)2 rabbit anti-mouse IgG (H+L; Life Technologies). F-actin was visualized using Alexa Fluor 633–phalloidin (dilution 1:50; Life Technologies). Images were acquired using a Leica TCS SP5 DM6000 confocal microscope (Leica Microsystems, Buffalo Grove, IL) with a 100× HCX Plan APO 1.40 oil-immersion objective lens and LCS software (Leica Microsystems). Images were processed using Photoshop Element 3 (Adobe, San Jose, CA).
Isolation of cardiac myofibrils
Myofibrils were isolated from 2-mo-old C57BL/6J mice. All steps were performed at 4°C. Whole-heart tissue was excised and placed in cold muscle extraction buffer (75 mM KCl, 10 mM imidazole, pH 7.2, 2 mM MgCl2, 2 mM ethylene glycol tetraacetic acid, 1 mM NaN3), made in 50% glycerol (vol/vol), with 1 mM (final) dithiothreitol. Slightly stretched heart muscle was tied to applicator sticks and submerged in muscle extraction buffer plus 0.5% Triton X-100 (vol/vol) and 0.1 mM phenylmethylsulfonyl fluoride at 4°C for 24 h and then transferred to −20°C for 1 wk. The attached ends of the muscle bundles were removed, and the tissue was homogenized by applying 10–15 strokes with a Dounce homogenizer using loose-fitting pestle. The suspension was placed on lysine-coated slides and fixed with a 3% formaldehyde solution in muscle extraction buffer for 10–15 min on the microscope slides. Fixed cardiac myofibrils on slides were then prepared for immunofluorescence and imaged as described.
Depletion of formins by siRNA
siRNA-mediated gene silencing was performed using synthetic siRNA duplex against specific formin genes: mDia2/Diap3 (NM_019670), siRNA mDiaP3-1 sense 5′-AGTGAAGTCTTGAAGCTTTTT-3′ (Miki et al., 2008); Fmnl1 (NM_010230), siRNA Fmnl1-1 sense 5′-CTGAAGAGCTACCTGGATATT-3′ and siRNA Fmnl1-2 sense 5′-TGGAGGAGACAGAGACGAATT-3′; Fmnl2 (NM_172409), siRNA Fmnl2-1 sense 5′-CAACTTATGCAGAGGAATAATT-3′ and siRNA Fmnl2-2 sense 5′-GAACCTACCTCCTGACAAATT-3′; and Daam1 (NM_025102), siRNA Daam1-1 sense 5′-GGAAAGAGCAGGCAGAGAATT-3′ and siRNA Daam1-2 sense 5′-GAAGAGGAGGAGAGGAGTATT-3′. Control siRNA was designed against prokaryotic ampicillin gene, a validated nonsilencing siRNA. siRNA oligonucleotide duplexes along with reporter plasmid pEGFP-C3 were electroporated into freshly isolated neonatal mouse cardiomyocytes using a Nucleofector machine, according to the manufacturer's instructions (Amaxa, Allendale, NJ). Cells were plated for culture, and 72 h postelectroporation, cells were fixed and prepared for immunofluorescence microscopy as described. Rescues were performed by cotransfecting cells with a plasmid (pEGFP-C3) that expresses an RNAi-resistant copy of the full-length formin. For mouse mDia2/Diap3, the RNAi-resistant gene construct was kindly provided by Shuh Narumiya (Kyoto University). For Daam1, Fmnl1, and Fmnl2 rescues, we used plasmids expressing full-length human cDNAs (OriGene, Rockville, MD), which were RNAi resistant to the siRNA oligos used.
Treatment of cardiomyocytes with blebbistatin and latrunculin A
Neonatal primary mouse cardiomyocytes were grown for 6 d in culture before treatment. Cardiomyocyte cells grown for 6 d are well spread and exhibit mature myofibrils. For inhibition of myosin type II contraction, cells were treated with 100 mM blebbistatin (Sigma-Aldrich) or control dimethyl sulfoxide in the culture medium for 15 min until muscle contraction ceased. Cells were fixed and immunostained. Actin monomer sequestration was carried out by treatment of cells with 25 mM LatA (Sigma-Aldrich) in the medium for 30 min (Coué et al., 1987). Of importance, this brief treatment, which blocks new actin polymerization, did not produce any changes in the visual beating pattern of cells. This indicated that existing sarcomeric structure and function remained intact, which was further confirmed by immunohistochemistry.
Correlative light and electron microscopy
Primary cadiomyocytes were isolated from 2- or 3-d-old C57BL/6J mice and immediately transfected with siRNA as described. Cells were grown on 1.5-mm-diameter Aclar disks punched out of Aclar film (50426-10; EMS, Fort Washington, PA) and mounted on a Lab-Tek II Chambered Cover glass (155379; Nalge Nunc International, Rochester, NY). The disks were previously marked with a pattern that allowed us to track the cells of interest throughout light microscopy and electron microscopy sample preparation. Cells were imaged using bright-field and fluorescent microscopy on an Evos FL epifluorescence microscope (Advanced Microscopy Group, Life Technologies), and transfected cells were identified by enhanced GFP signal. Aclar disks containing the cells of interest were transferred into aluminum platelets for high-pressure freezing (Wohlwend, Sennwald, Switzerland), covered with 150 mM sucrose (as cryoprotectant) in D-MEM, and covered with another platelet, thus enclosing the cells in a cavity of 0.1-mm depth. Samples were rapidly frozen using a Leica HPM-100 high-pressure freezer (Leica Microsystems, Vienna, Austria). The frozen cells were freeze substituted at low temperatures (starting at −90°C) over 3 d in a solution containing 1% osmium tetroxide (EMS), 0.5% anhydrous glutaraldehyde (EMS), and 2% water in anhydrous acetone (AC32680-0010; Fisher Scientific, Pittsburgh, PA) using a Leica AFS-2 automatic freeze-substitution device. Afterward, the temperature was raised to 4°C, and cells on disks were infiltrated and embedded in EMbed 812/Araldite 502-Resin (EMS). Guided by the light microscopy imaging and the pattern on the Aclar disk, we trimmed the resin block so that only the quadrant containing the cell(s) of interest remained for ultrathin sectioning. The ultrathin sections (70 nm) were collected on slot grids covered with Formvar support film and poststained with 2% uranyl acetate and Reynold lead citrate. Transmission electron microscopy was performed using either an FEI Morgagni 228 TEM or Tecnai F30 intermediate-voltage TEM (FEI, Hillsboro, OR) with a 4k charge-coupled device camera (Gatan, Pleasanton, CA). For overviews of cells at medium magnification we acquired montages in an automated manner using the microscope control software SerialEM (Mastronarde, 2005).
Myofibril repair after injury
Primary mouse cardiomyocytes isolated from neonatal mice were allowed to spread and grow in culture for 7 d. Cells containing mature myofibrils were then treated with 10 μM LatA in the culture medium. Cells were closely monitored for viability, and after 16–24 h, LatA was removed from the medium by washing five times with prewarmed medium. Cells were allowed to continue to grow for an additional 24 or 48 h posttreatment before being fixed and prepared for immunofluorescence. Myofibril architecture was determined at each of the two time points by staining with α-actinin antibodies. Localization of DAAM1, mDia2, FMNL1, and FMNL2 were also determined by immunofluorescence. For myofibril repair after injury, the cardiomyocytes were grown for 48 h before treatment with 10 μM LatA for 24 h, then transfected with siRNA oligonucleotide duplexes as described using the calcium phosphate method (Kingston et al., 2001). At 48 h post siRNA treatment, cells were fixed for immunofluorescence microscopy.
Supplementary Material
Acknowledgments
We are grateful to Irina Krykbaeva and Alexandra Pizzi for technical assistance, Ed Dougherty and Chen Xu for light and electron microscopy training, and Shuh Namuriya for providing the RNAi-resistant pEGFP-mDia2 construct. We thank Melissa Chesarone-Cataldo and Avital Rodal for critical reading of the manuscript. M.R. received support from National Institutes of Health T32 NS07292, and C.F.B was supported by a fellowship from the Leukemia and Lymphoma Society and in part by National Institutes of Health P30 NS45713. Research was supported by National Institutes of Health NS066977 to S.B., National Science Foundation 0722582 to D.N., and National Science Foundation DMR-MRSEC-0820492 and National Institutes of Health GM083137 to B.G.
Abbreviations used:
- DIV
days in vitro
- GAPDH
glyceraldehyde 3-phosphate-dehydrogenase
- GFP
green fluorescent protein
- Lat A
latrunculin A
- PBS
phosphate-buffered saline
- qRT-PCR
quantitative real-time PCR
- siRNA
small interfering RNA
- SEM
standard error of the mean
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-08-0443) on January 15, 2014.
*These authors contributed equally.
REFERENCES
- Arber S, Hunter JJ, Ross J, Hongo M, Sansig G, Borg J, Perriard JC, Chien KR, Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Goode BL. Formins at a glance. J Cell Sci. 2013;126:1–7. doi: 10.1242/jcs.107250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D, Boczkowska M, Skwarek-Maruszewska A, Fujiwara I, Hayes DB, Rebowski G, Lappalainen P, Pollard TD, Dominguez R. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320:239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- Coué M, Brenner SL, Spector I, Korn ED. Inhibition of action polymerization by latrunculin A. FEBS Lett. 1987;213:316–318. doi: 10.1016/0014-5793(87)81513-2. [DOI] [PubMed] [Google Scholar]
- Craig SW, Pardo JV. Gamma actin, spectrin, and intermediate filament proteins colocalize with vinculin at costameres, myofibril-to-sarcolemma attachment sites. Cell Motil. 1983;3:449–462. doi: 10.1002/cm.970030513. [DOI] [PubMed] [Google Scholar]
- Cunha SR, Mohler PJ. Obscurin targets ankyrin-B and protein phosphatase 2A to the cardiac M-line. J Biol Chem. 2008;283:31968–31980. doi: 10.1074/jbc.M806050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha SR, Mohler PJ. Ankyrin protein networks in membrane formation and stabilization. J Cell Mol Med. 2009;13:4364–4376. doi: 10.1111/j.1582-4934.2009.00943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danowski BA, Imanaka-Yoshida K, Sanger JM, Sanger JW. Costameres are sites of force transmission to the substratum in adult rat cardiomyocytes. J Cell Biol. 1992;118:1411–1420. doi: 10.1083/jcb.118.6.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etard C, Roostalu U, Strähle U. Shuttling of the chaperones Unc45b and Hsp90a between the A band and the Z line of the myofibril. J Cell Biol. 2008;180:1163–1175. doi: 10.1083/jcb.200709128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ervasti JM. Costameres: the Achilles’ heel of Herculean muscle. J Biol Chem. 2003;278:13591–13594. doi: 10.1074/jbc.R200021200. [DOI] [PubMed] [Google Scholar]
- Goldfarb LG, Dalakas MC. Tragedy in a heartbeat: malfunctioning desmin causes skeletal and cardiac muscle disease. Clin J Invest. 2009;119:1806–1813. doi: 10.1172/JCI38027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golson ML, Sanger JM, Sanger JW. Inhibitors arrest myofibrillogenesis in skeletal muscle cells at early stages of assembly. Cell Motil Cytoskeleton. 2004;59:1–16. doi: 10.1002/cm.20017. [DOI] [PubMed] [Google Scholar]
- Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Harris ES, Rouiller I, Hanein D, Higgs HN. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J Biol Chem. 2006;281:14383–14392. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Peterson KJ. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell. 2005;16:1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskratsch T, Reijntjes S, Dwyer J, Toselli P, Dégano IR, Dominguez I, Ehler E. Two distinct phosphorylation events govern the function of muscle FHOD3. Cell Mol Life Sci. 2013;70:893–908. doi: 10.1007/s00018-012-1154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal R, Breitsprecher D, Collins A, Correa IR, Xu M-Q, Goode BL. The formin Daam1 and fascin directly collaborate to promote filopodia formation. Curr Biol. 2013;23:1373–1379. doi: 10.1016/j.cub.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya H, Takeya R, Takeuchi K, Watanabe N, Jing N, Sumimoto H. Fhos2, a novel formin-related actin-organizing protein, probably associates with the nestin intermediate filament. Genes Cells. 2005;10:665–678. doi: 10.1111/j.1365-2443.2005.00867.x. [DOI] [PubMed] [Google Scholar]
- Kan-O M, Takeya R, Abe T, Kitajima N, Nishida M, Tominaga R, Kurose H, Sumimoto H. Mammalian formin Fhod3 plays an essential role in cardiogenesis by organizing myofibrillogenesis. Biol Open. 2012a;1:889–896. doi: 10.1242/bio.20121370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan-O M, Takeya R, Taniguchi K, Tanoue Y, Tominaga R, Sumimoto H, Weed SA. Expression and subcellular localization of mammalian formin Fhod3 in the embryonic and adult heart. PLoS One. 2012b;7:e34765. doi: 10.1371/journal.pone.0034765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee AJ, et al. Sorting of a nonmuscle tropomyosin to a novel cytoskeletal compartment in skeletal muscle results in muscular dystrophy. J Cell Biol. 2004;166:685–696. doi: 10.1083/jcb.200406181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Chen CA, Okayama H. Calcium phosphate transfection. Curr Protoc Immunol. 2001;10:10.13.1–10.13.9. doi: 10.1002/0471142735.im1013s31. [DOI] [PubMed] [Google Scholar]
- Kovács M, Tóth J, Hetényi C, Málnási-Csizmadia A, Sellers JR. Mechanism of blebbistatin inhibition of myosin II. J Biol Chem. 2004;279:35557–35563. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- Lam ML, Bartoli M, Claycomb WC. The 21-day postnatal rat ventricular cardiac muscle cell in culture as an experimental model to study adult cardiomyocyte gene expression. Mol Cell Biochem. 2002;229:51–62. doi: 10.1023/a:1017999216277. [DOI] [PubMed] [Google Scholar]
- Leu M, Ehler E, Perriard JC. Characterization of postnatal growth of the murine heart. Anat Embryol. 2001;204:217–224. doi: 10.1007/s004290100206. [DOI] [PubMed] [Google Scholar]
- Li D, et al. Dishevelled-associated activator of morphogenesis 1 (Daam1) is required for heart morphogenesis. Development. 2011;138:303–315. doi: 10.1242/dev.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. Mol J Cell Cardiol. 1996;28:1737–1746. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- Littlefield RS, Almenar-Queralt A, Fowler VM. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat Cell Biol. 2001;3:544–551. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
- Maass AH, Buvoli M. Cardiomyocyte preparation, culture, and gene transfer. Methods Mol Biol. 2007;366:321–330. doi: 10.1007/978-1-59745-030-0_18. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Michelot A, Guerin C, Huang S, Ingouff M, Richard S, Rodiuc N, Staiger CJ, Blanchoin L. The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell. 2005;17:2296–2313. doi: 10.1105/tpc.105.030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Okawa K, Sekimoto T, Yoneda Y, Watanabe S, Ishizaki T, Narumiya S. mDia2 shuttles between the nucleus and the cytoplasm through the importin α/β and CRM1 -mediated nuclear transport mechanism. J Biol Chem. 2008;284:5753–5762. doi: 10.1074/jbc.M806191200. [DOI] [PubMed] [Google Scholar]
- Mohler PJ, et al. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature. 2003;421:634–639. doi: 10.1038/nature01335. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Goode BL. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J Biol Chem. 2005;280:28023–28033. doi: 10.1074/jbc.M503094200. [DOI] [PubMed] [Google Scholar]
- Müller M, et al. Functional characterization of the human α-cardiac actin mutations Y166C and M305L involved in hypertrophic cardiomyopathy. Cell Mol Life Sci. 2012;69:3457–3579. doi: 10.1007/s00018-012-1030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata T, Nishina Y, Yorifuji H. Cytoplasmic gamma actin as a Z-disc protein. Biochem Biophys Res Commun. 2001;286:156–163. doi: 10.1006/bbrc.2001.5353. [DOI] [PubMed] [Google Scholar]
- Ono S. Dynamic regulation of sarcomeric actin filaments in striated muscle. Cytoskeleton. 2010;67:677–692. doi: 10.1002/cm.20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparil S, Bishop SP, Clubb FJ., Jr Myocardial cell hypertrophy or hyperplasia. Hypertension. 1984;6:38–43. doi: 10.1161/01.hyp.6.6_pt_2.iii38. [DOI] [PubMed] [Google Scholar]
- Papponen H, Kaisto T, Leinonen S, Kaakinen M, Metsikko K. Evidence for gamma-actin as a Z disc component in skeletal myofibers. Exp Cell Res. 2009;315:218–225. doi: 10.1016/j.yexcr.2008.10.021. [DOI] [PubMed] [Google Scholar]
- Pellegrin S, Mellor H. The Rho family GTPase Rif induces filopodia through mDia2. Curr Biol. 2005;15:129–133. doi: 10.1016/j.cub.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Posch MG, et al. Cardiac alpha-myosin (MYH6) is the predominant sarcomeric disease gene for familial atrial septal defects. PLoS One. 2011;6:e28872. doi: 10.1371/journal.pone.0028872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee D, Sanger JM, Sanger JW. The premyofibril: evidence for its role in myofibrillogenesis. Cell Motil Cytoskeleton. 1994;28:1–24. doi: 10.1002/cm.970280102. [DOI] [PubMed] [Google Scholar]
- Ruzicka DL, Schwartz RJ. Sequential activation of alpha-actin genes during avian cardiogenesis: vascular smooth muscle alpha-actin gene transcripts mark the onset of cardiomyocyte differentiation. J Cell Biol. 1988;107:2575–2586. doi: 10.1083/jcb.107.6.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybakova IN, Patel JR, Ervasti JM. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J Cell Biol. 2000;150:1209–1214. doi: 10.1083/jcb.150.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger JW, Ayoob JC, Chowrashi P, Zurawski D, Sanger JM. Assembly of myofibrils in cardiac muscle cells. Adv Exp Med Biol. 2000;481:89–102. doi: 10.1007/978-1-4615-4267-4_6. [DOI] [PubMed] [Google Scholar]
- Sanger JW, Chowrashi P, Shaner NC, Spalthoff S, Wang J, Freeman NL, Sanger JM. Myofibrillogenesis in skeletal muscle cells. Clin Orthop Relat Res. 2002;403(Suppl):S153–S162. doi: 10.1097/00003086-200210001-00018. [DOI] [PubMed] [Google Scholar]
- Sanger JW, Kang S, Siebrands CC, Freeman N, Du A, Wang J, Stout AL, Sanger JM. How to build a myofibril. J Muscle Res Cell Motil. 2005;26:343–354. doi: 10.1007/s10974-005-9016-7. [DOI] [PubMed] [Google Scholar]
- Sanger JW, Wang J, Fan Y, White J, Sanger JM. Assembly and dynamics of myofibrils. J Biomed Biotechnol. 2010;2010:858606. doi: 10.1155/2010/858606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Ledo A, Jenny A, Marlow FL. Comparative gene expression analysis of the FMNL family of formins during zebrafish development and implications for tissue specific functions. Gene Expr Patterns. 2013;13:30–37. doi: 10.1016/j.gep.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirenbeck A, Bretschneider T, Arasada R, Schleicher M, Faix J. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat Cell Biol. 2005;7:619–625. doi: 10.1038/ncb1266. [DOI] [PubMed] [Google Scholar]
- Schonichen A, Geyer M. Fifteen formins for an actin filament: a molecular view on the regulation of human formins. Biochim Biophys Acta. 2010;1803:152–163. doi: 10.1016/j.bbamcr.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Schönichen A, et al. FHOD1 is a combined actin filament capping and bundling factor that selectively associates with actin arcs and stress fibers. J Cell Sci. 2013;26:1891–1901. doi: 10.1242/jcs.126706. [DOI] [PubMed] [Google Scholar]
- Skwarek-Maruszewska A, Hotulainen P, Mattila PK, Lappalainen P. Contractility-dependent actin dynamics in cardiomyocyte sarcomeres. J Cell Sci. 2009;122:2119–2126. doi: 10.1242/jcs.046805. [DOI] [PubMed] [Google Scholar]
- Sparrow JC, Schöck F. The initial steps of myofibril assembly: integrins pave the way. Nat Rev Mol Cell Biol. 2009;10:293–298. doi: 10.1038/nrm2634. [DOI] [PubMed] [Google Scholar]
- Spector I, Shochet NR, Blasberger D, Kashman Y. Latrunculins—novel marine macrolides that disrupt microfilament organization and affect cell growth: I. comparison with cytochalasin D. Cell Motil Cytoskeleton. 1989;13:127–144. doi: 10.1002/cm.970130302. [DOI] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- Takano K, Watanabe-Takano H, Suetsugu S, Kurita S, Tsujita K, Kimura S, Karatsu T, Takenawa T, Endo T. Nebulin and N-WASP cooperate to cause IGF-1-induced sarcomeric actin filament formation. Science. 2010;330:1536–1540. doi: 10.1126/science.1197767. [DOI] [PubMed] [Google Scholar]
- Taniguchi K, Takeya R, Suetsugu S, Kan-O M, Narusawa M, Shiose A, Tominaga R, Sumimoto H. Mammalian formin fhod3 regulates actin assembly and sarcomere organization in striated muscles. J Biol Chem. 2009;284:29873–29881. doi: 10.1074/jbc.M109.059303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojkander S, Gateva G, Schevzov G, Hotulainen P, Naumanen P, Martin C, Gunning PW, Lappalainen P. A molecular pathway for myosin II recruitment to stress fibers. Curr Biol. 2011;21:539–550. doi: 10.1016/j.cub.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Vaillant DC, Copeland SJ, Davis C, Thurston SF, Abdennur N, Copeland JW. Interaction of the N- and C-terminal autoregulatory domains of FRL2 does not inhibit FRL2 activity. J Biol Chem. 2008;283:33750–33762. doi: 10.1074/jbc.M803156200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arx P, Bantle S, Soldati T, Perriard JC. Dominant negative effect of cytoplasmic actin isoproteins on cardiomyocyte cytoarchitecture and function. J Cell Biol. 1995;131:1759–1773. doi: 10.1083/jcb.131.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sanger JM, Sanger JW. Differential effects of latrunculin-A on myofibrils in cultures of skeletal muscle cells: insights into mechanisms of myofibrillogenesis. Cell Motil Cytoskeleton. 2005;62:35–47. doi: 10.1002/cm.20083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten EC, et al. Formin homology 2 domain containing 3 (FHOD3) variants associated with hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2013;6:10–18. doi: 10.1161/CIRCGENETICS.112.965277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemljic-Harpf A, Manso AM, Ross RS. Vinculin and talin: focus on the myocardium. Invest J Med. 2009;57:849–855. doi: 10.231/JIM.0b013e3181c5e074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.