The isolated dynamin PH domain is an assembly-independent sensor of membrane curvature but not a curvature generator. In full-length dynamin, the PH alternates between two different orientations on the membrane surface during the GTP hydrolysis cycle, causing dramatic fluctuations in the diameter of dynamin polymers.
Abstract
The self-assembling GTPase dynamin catalyzes endocytic vesicle scission via membrane insertion of its pleckstrin homology (PH) domain. However, the molecular mechanisms underlying PH domain–dependent membrane fission remain obscure. Membrane-curvature–sensing and membrane-curvature–generating properties have been attributed, but it remains to be seen whether the PH domain is involved in either process independent of dynamin self-assembly. Here, using multiple fluorescence spectroscopic and microscopic techniques, we demonstrate that the isolated PH domain does not act to bend membranes but instead senses high membrane curvature through hydrophobic insertion into the membrane bilayer. Furthermore, we use a complementary set of short- and long-distance Förster resonance energy transfer approaches to distinguish PH-domain orientation from proximity at the membrane surface in full-length dynamin. We reveal, in addition to the GTP-sensitive “hydrophobic mode,” the presence of an alternate, GTP-insensitive “electrostatic mode” of PH domain–membrane interactions that retains dynamin on the membrane surface during the GTP hydrolysis cycle. Stabilization of this alternate orientation produces dramatic variations in the morphology of membrane-bound dynamin spirals, indicating that the PH domain regulates membrane fission through the control of dynamin polymer dynamics.
INTRODUCTION
Dynamin is a self-assembling GTPase that plays a key role in the scission of endocytic vesicles at the plasma membrane (Ramachandran, 2011; Schmid and Frolov, 2011; Ferguson and De Camilli, 2012). At the late stage of endocytic vesicle formation, dynamin assembles into a helical collar around the narrow, tubular membrane necks of deeply invaginated coated pits to direct vesicle scission. Membrane fission catalyzed by dynamin is believed to entail its dynamic helical polymerization around the highly curved membrane tether and GTP-dependent, mechanochemical conformational rearrangements in the polymer that promote rapid destabilization of the underlying membrane.
Mammalian cells encode three closely related but largely nonredundant isoforms of dynamin (Dyn1–3), each with distinct biochemical and biophysical properties (Barylko et al., 2010; Liu et al., 2011). Collectively referred to as classical dynamins, they share the presence of 1) an N-terminal GTPase (G) domain involved in basal and assembly-stimulated GTP hydrolysis, 2) a central pleckstrin homology (PH) domain that mediates lipid interactions at the membrane surface, 3) bridging middle and GTPase effector (GED) domains that together constitute a “stalk” to create interfaces for helical dynamin self-assembly, and 4) a C-terminal proline-rich domain (PRD) that interacts with the Src-homology 3 (SH3) domains of various accessory proteins to regulate membrane fission (Figure 1A; Faelber et al., 2012; Chappie and Dyda, 2013). Of these five functionally defined regions, the PH domain and the PRD are specific to classical dynamins and are not found in dynamin-related proteins that participate in other membrane-remodeling events in the cell (Heymann and Hinshaw, 2009).
FIGURE 1:
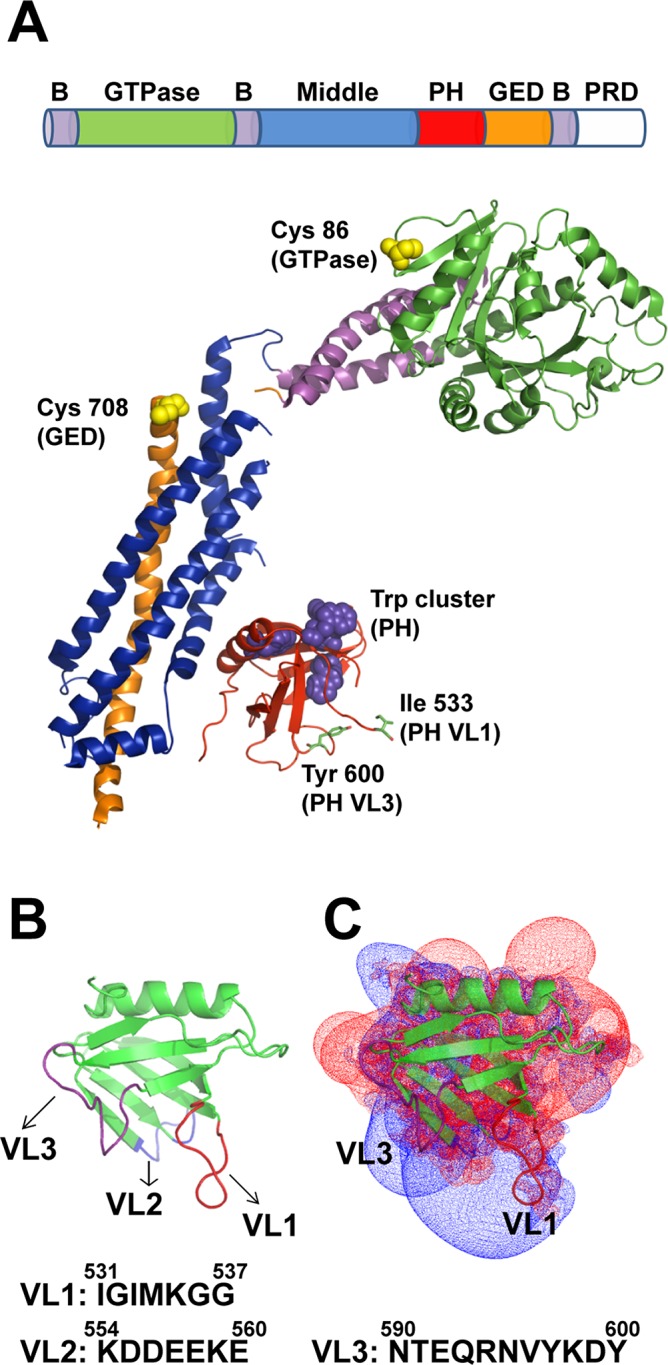
Dynamin structure and structural elements. (A) Domain arrangement in the primary structure of dynamin and color-coded ribbon representation of the Dyn1ΔPRD crystal structure (Faelber et al., 2011; Protein Data Bank [PDB] ID 3SNH). Residues Ile-533 in VL1 and Tyr-600 in VL3 are shown in stick representations. Trp residues located in the PH domain (magenta) and solvent-exposed Cys-708 (GED) and Cys-86 (GTPase) residues fluorescently labeled in this study (yellow) are shown in space-filled representations. B, constituent helices of the three-helix bundle signaling element (BSE); GED, GTPase effector domain; PH, pleckstrin homology domain; PRD, proline-rich domain. (B) Ribbon representation of Dyn1PH (PDB ID 1DYN) with the variable loops (VL1–3) highlighted in color. Membrane-proximal amino acid residue sequences in VL1–3 are shown underneath. (C) Electrostatic potential map of Dyn1PH in the same orientation as in B contoured at −1.5kT (red; negative) and +1.5kT (blue; positive) as originally shown in Ferguson et al. (1994) and Lemmon and Ferguson (2000). Inset, ribbon representation of the Dyn1PH backbone. Calculations using GRASP in Swiss-PdbViewer.
The PH domain, long considered to be a membrane adaptor for classical dynamins, has recently garnered attention for its purported mechanical role in the mechanism of membrane fission. Critical membrane interactions of its variable loops have been suggested to play a direct role in the membrane-remodeling events leading to membrane fission (Liu et al., 2011; Shnyrova et al., 2013). A membrane-bending role has been described for the Dyn1 PH domain based on the inability of a PH domain mutant defective in membrane insertion, Dyn1 I533A, to generate curvature from planar membrane templates in vitro (Ramachandran et al., 2009). Dyn1 I533A is consequently believed to fail in mediating endocytic vesicle scission in vivo (Ramachandran et al., 2009). Alternatively, a curvature-sensing role has been proposed for the Dyn2 PH domain based on the selective fission of precurved membrane tethers by full-length Dyn2 in vitro (Liu et al., 2011). Although the differential membrane-remodeling activities of the two dynamin isoforms in vitro map unambiguously to their respective PH domains (Liu et al., 2011), the molecular mechanisms underlying these functional differences remain unknown.
Early biochemical experiments with the isolated, monomeric PH domains of Dyn1 and Dyn2 did not reveal any significant binding to negatively charged phosphoinositides in vitro, including phosphatidylinositol-4,5-bisphosphate (PIP2), essential for dynamin function in vivo (Ferguson et al., 1994; Klein et al., 1998). In contrast, the respective PH domains, when dimerized through glutathione S-transferase fusion, bound PIP2 with significantly greater affinity, suggesting that stable PH domain–membrane association in full-length dynamin requires the high-avidity membrane interactions facilitated by dynamin oligomerization (Salim et al., 1996; Klein et al., 1998). These early studies, performed with liposomes of undefined curvature, did not incorporate the stringent roles of membrane curvature and/or membrane geometry in the stabilization of dynamin–membrane interactions described since (Ramachandran and Schmid, 2008; Ramachandran et al., 2009; Roux et al., 2010). Interestingly, however, the monomeric PH domain of Dyn1 was found to bind PIP2, but only when the lipid was solubilized in highly curved detergent micelles (Zheng et al., 1996), suggesting that 1) high membrane curvature rather than high avidity plays a role in stabilizing PH domain–membrane interactions and 2) the differential activities of the Dyn1 and Dyn2, both in vitro and in vivo, may be related to significant differences in the nature of their respective PH domain–membrane interactions. These early observations, as well as recent findings, prompted us to reinvestigate and reevaluate the role of the PH domain in dynamin function.
Recent x-ray structures of assembly-defective but nearly full-length variants of Dyn1 position the globular PH domain (residues 518–630) near the base of the dynamin stalk (Faelber et al., 2011; Ford et al., 2011). Capped by a stalk-interfacing amphipathic α-helix at one end, the domain's β-sandwich core is lined at the other by three variable loops (VL1, VL2, and VL3) that create a binding pocket for PIP2 (Ferguson et al., 1994; Lemmon and Ferguson, 2000; Figure 1, A and B). Of these three variable loops, the hydrophobic tip of VL1 has been shown to penetrate the acyl-chain hydrocarbon core of a target lipid bilayer in vitro (Ramachandran and Schmid, 2008). Furthermore, hydrophobic character–disrupting mutations in VL1 (I533A, M534C) impair membrane fission in vivo (Ramachandran et al., 2009). The residues of VL2 and VL3, on the other hand, are polar and/or charged, making excursions into the nonpolar membrane core energetically unfavorable and highly unlikely (Ferguson et al., 1994; Zheng et al., 1996; Figure 1B). Yet a mutation in Dyn1 PH domain VL3 (Y600L) has been shown to inhibit dynamin's fission activity in vitro (Liu et al., 2011). As observed for PH domains in other proteins, clusters of positively charged residues (Lys and Arg) localized around the variable loops and of negatively charged residues (Asp and Glu) on the opposite face of the domain render the dynamin PH domain electrostatically polarized (Ferguson et al., 1994; Lemmon and Ferguson, 2000; Figure 1C). Of interest, the calculated positive field potentials for the domain have a distinctly bilobal distribution that coincides approximately with the relative positions of VL1 and VL3, suggesting that the PH domain may associate with the membrane through loop-specific electrostatic interactions (Rebecchi and Scarlata, 1998; Lemmon and Ferguson, 2000; Figure 1C). Point mutations of select Lys (K) residues in either loop considerably weaken or even completely abrogate dynamin membrane binding in vitro (Salim et al., 1996; Zheng et al., 1996; Achiriloaie et al., 1999; Vallis et al., 1999).
To better understand the role of the PH domain in dynamin function and gain new insight into the mechanisms underlying PH domain–dependent, dynamin-mediated membrane fission, we reexamined the biochemical and biophysical properties of the Dyn1 PH domain both in isolation and in the context of the full-length molecule. We demonstrate that the isolated PH domain is an exquisite sensor of membrane curvature independent of dynamin self-assembly. The PH domain, however, cannot generate membrane curvature on its own, indicating that dynamin-mediated membrane remodeling requires concomitant dynamin self-assembly. Furthermore, using a complementary set of Förster resonance energy transfer (FRET) approaches, we show that the dynamin PH domain can adopt alternate orientations on the membrane surface that directly control dynamin polymer morphology and, indirectly, membrane fission. Our studies thus provide critical new insight into the versatile role of the dynamin PH domain in the regulation of membrane fission.
RESULTS
Dyn1PH is an assembly-independent membrane-curvature sensor
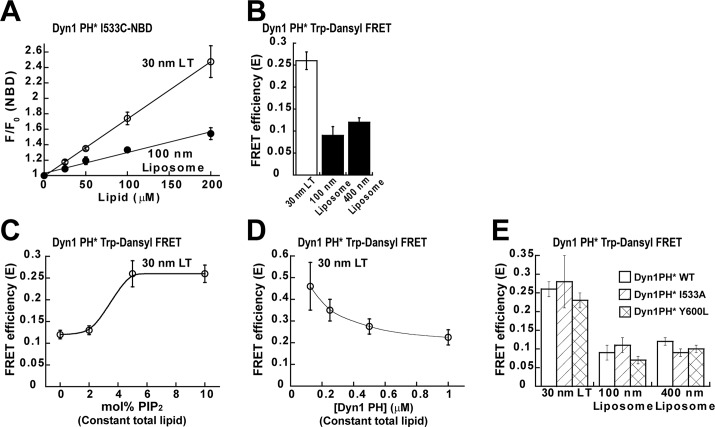
We first explored the possibility that the isolated, monomeric Dyn1 PH domain (Dyn1PH) can sense high membrane curvature and/or tubular membrane geometry on its own. To this end, we generated lipid templates of varying membrane curvature or geometry and used a previously characterized NBD-labeled Dyn1PH VL1 fluorescent derivative (Dyn1PH* I533C-NBD) to directly probe for curvature-sensitive membrane interactions in Dyn1PH (Ramachandran and Schmid, 2008; Ramachandran et al., 2009). Briefly, the water-sensitive NBD dye conjugated to the tip of PH-domain VL1 experiences a dramatic increase in emission intensity upon VL1 insertion into the membrane bilayer (Ramachandran and Schmid, 2008; Ramachandran et al., 2009; Liu et al., 2011). This NBD fluorescence change was therefore used as a sensitive reporter of VL1–membrane interactions in Dyn1PH. The relative size distribution and the average diameters of the various lipid templates generated were ascertained using a combination of electron microscopy (EM), atomic force microscopy (AFM), and dynamic light scattering (DLS) techniques (Supplemental Figure S1).
Dyn1PH, similar to full-length Dyn1 (Ramachandran and Schmid, 2008; Ramachandran et al., 2009), preferentially inserted into highly curved, cylindrical lipid nanotubes (LTs; ∼30 nm in diameter) compared with relatively planar, spherical liposomes (∼100 nm in diameter; Figure 2A). However, significant membrane insertion of Dyn1PH into either template was detected only at high lipid concentrations, indicating a lower membrane-binding affinity for the isolated Dyn1 PH domain relative to the full-length molecule (Ramachandran and Schmid, 2008). On the basis of this assay, we conclude that Dyn1PH can independently insert into membranes and function as an assembly-independent sensor of high membrane curvature and/or tubular membrane geometry.
FIGURE 2:
Dyn1PH is an assembly-independent sensor of membrane curvature. (A) VL1 membrane insertion of the isolated Dyn1PH monomer (0.5 μM Dyn1PH* I533C-NBD) into increasing concentrations of LT and liposomes containing 10 mol% PIP2. The average membrane-dependent NBD emission intensity increase is indicated as F/F0, where F0 is the emission intensity at time 0 before lipid addition and F is the intensity after 30 min of incubation. (B) Binding of 0.5 μM Dyn1PH* to lipid templates of varying membrane curvature (150 μM total lipid) detected by Trp-Dansyl FRET after 30 min of incubation. Lipid templates were labeled with 10 mol% Dansyl-PE. FRET efficiency (E) was calculated according to Materials and Methods. (C) Binding of 0.5 μM Dyn1PH* to LT (150 μM constant total lipid) containing increasing mol% of PIP2 detected by Trp-Dansyl FRET. (D) FRET-detected binding of increasing concentrations of Dyn1PH* to LT (150 μM constant total lipid). (E) Binding of 0.5 μM Dyn1PH I533A (VL1) or Dyn1PH Y600L (VL3) to lipid templates of varying membrane curvature (150 μM total lipid) measured by Trp-Dansyl FRET relative to Dyn1PH WT as shown in B. All steady-state data here and in subsequent figures are averages ± SD (n ≥ 3).
To probe independently for curvature-sensitive Dyn1PH–membrane interactions, we developed an alternative FRET-based approach. We used intrinsic tryptophan (Trp) residues in Dyn1PH as FRET donors for Dansyl-labeled acceptor lipid molecules (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(5-dimethylamino-1-naphthalenesulfonyl) [Dansyl-PE]) distributed randomly on the target membrane surface. This atypical variation of the FRET technique, commonly referred to as plane-to-plane FRET, has been used extensively to determine protein association and topography on a membrane surface (Wang et al., 2003; Ramachandran et al., 2005). Trp-Dansyl FRET measurements confirmed that Dyn1PH is indeed a sensor of membrane curvature and/or geometry, independent of dynamin self-assembly (Figure 2B). Dyn1PH association was positively cooperative with respect to PIP2 density at the membrane surface, similar to full-length Dyn1 (Figure 2C; Ramachandran and Schmid, 2008). However, Dyn1PH membrane association was competitive with respect to protein concentration (Figure 2D). These results, taken together, suggest that PH-domain monomers compete for binding to a relatively small fraction of PIP2 molecules (or negative charges) dynamically clustered at the membrane surface. This competition for membrane binding may explain why, when placed in the crowded confines of a membrane-associated dynamin polymer, the neighboring PH domains of a dimeric repeating dynamin subunit appear asymmetrically oriented on the membrane surface. EM of Dyn1PH-incubated LT did not reveal any ordering of the PH-domain monomers on the membrane, in contrast to the helical morphology of the full-length Dyn1 polymer (Supplemental Figure S2, A and B).
We then determined whether critical VL1 residue I533 or VL3 residue Y600 plays a role in the curvature-sensitive membrane interactions of the Dyn1 PH domain. To this end, we used Trp-Dansyl FRET to compare the behavior of Dyn1PH WT with that of mutants Dyn1PH I533A and Dyn1PH Y600L. Unexpectedly, neither mutation altered the extent or the curvature sensitivity of Dyn1PH–membrane interactions (Figure 2E). Trp-Dansyl FRET measures the extent of PH domain–membrane association but not that of PH domain–membrane insertion. Thus these data indicate that the foregoing mutations do not perturb PH domain–membrane association. These results also suggested that the individual mutations do not effectively suppress the curvature-sensitive membrane interactions of the isolated Dyn1 PH domain. Coupled size exclusion chromatography (SEC)–multiangle light scattering (MALS) measurements confirmed that both Dyn1PH WT and mutants formed monomers in solution (Supplemental Figure S3A). Thus these mutations also do not cause additional structural defects in the PH domain that may lead to protein aggregation.
Dyn1PH is not a membrane-curvature generator
We next determined whether the Dyn1PH monomer could generate curvature from relatively planar membrane surfaces on its own through membrane insertion. To this end, we generated giant unilamellar vesicles (GUVs; ∼5–20 μm in diameter) and visualized membrane remodeling (membrane tubulation) by confocal light microscopy. Despite Dyn1PH enrichment on the membrane surface at very high bulk protein concentrations (20–35 μM), no membrane remodeling was observed (Figure 3, A–C). In contrast, full-length Dyn1 at a much lower bulk protein concentration (1–2 μM) extracted protein-decorated membrane tubules (Figure 3D). EM visualization of liposomes incubated with Dyn1PH (Supplemental Figure S2, C and D) presented no evidence of membrane remodeling, corroborating our conclusion that whereas Dyn1PH is an exquisite membrane-curvature sensor, it is not a membrane-curvature generator in the absence of concerted dynamin self-assembly.
FIGURE 3.
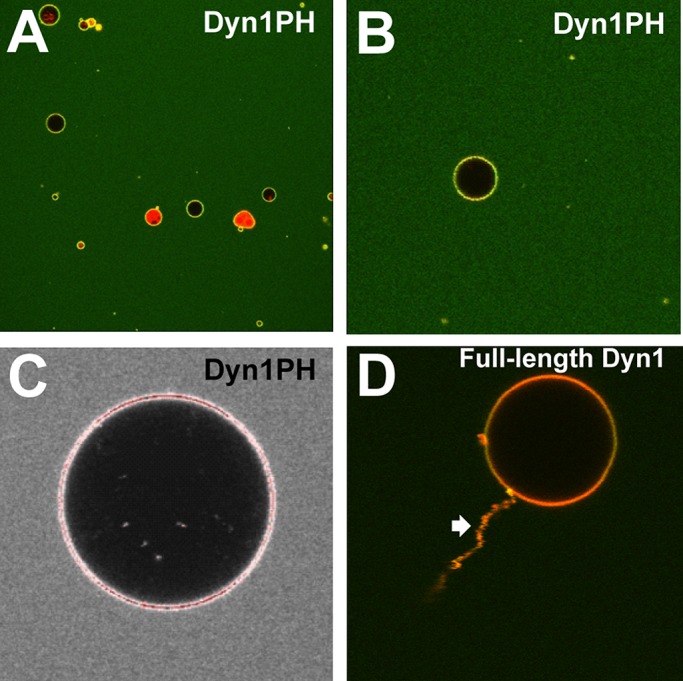
Dyn1PH is not a membrane-curvature generator. (A) Low-magnification confocal image of a field of PIP2-containing, RhPE-labeled GUVs (∼5–20 μm in diameter) incubated with 20 μM Dyn1PH* I533C-BODIPY in solution. (B) Magnified view of a single GUV in A, showing uniform membrane adsorption of BODIPY-Dyn1PH*. (C) Saturated intensity profile (red pixels) of a BODIPY-Dyn1PH*-enriched GUV (35 μM protein in solution) with no detectable membrane-remodeling activity at its surface. (D) High-magnification confocal image of a PIP2-containing, RhPE-labeled GUV with detectable membrane-remodeling activity (tubulation; block arrow) upon incubation with 1 μM BODIPY-labeled, full-length Dyn1 in solution. Representative images.
VL mutations variably affect PH domain–membrane interactions
We then asked how PH-domain mutations I533A (VL1) and Y600L (VL3) affect function in full-length Dyn1. Because we were unable to detect the effects of the I533A and Y600L mutations in the context of the weaker membrane interactions of the isolated PH domain, we reasoned that their effects may be manifested in the stronger, high-avidity membrane interactions of the full-length molecule.
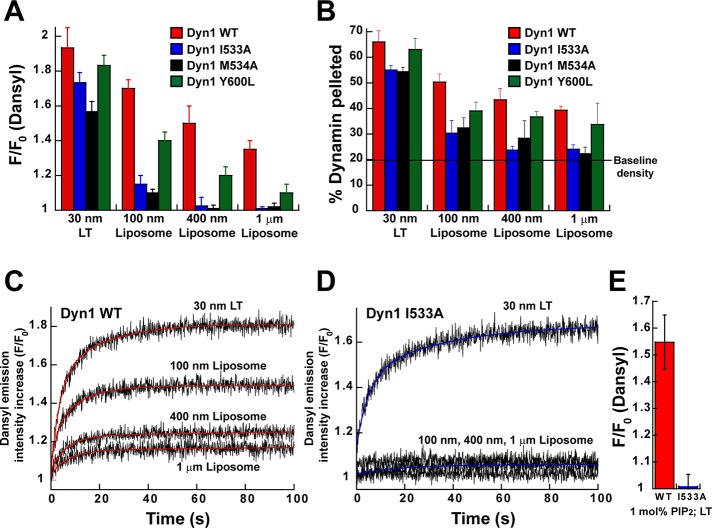
Four of the five Trp residues in Dyn1 are located in the PH domain, and the remaining one is present in the PRD located far above the membrane surface and outside the short-distance range for Trp-Dansyl FRET (R0 ≈ 21 Å; 1.5R0 ≈ 32 Å; see Materials and Methods for explanation; Gustiananda et al., 2004; Piston and Kremers, 2007). We therefore used Trp-Dansyl FRET as a measure (Supplemental Figure S4A) to probe the effects of the I533A and Y600L mutations on the curvature-sensitive PH domain–membrane interactions of full-length Dyn1. In addition to I533A, we also characterized the effect of another VL1 hydrophobic character–disrupting mutation, M534A.
Although FRET was not dramatically altered on LT for either mutant, indicating a more stable interaction with highly curved membranes (Figure 4A), VL1 mutants Dyn1 I533A and Dyn1 M534A experienced a significant loss of FRET on increasingly planar liposomes relative to Dyn1 WT (Figure 4A). In comparison, only a partial loss of FRET was observed for the VL3 mutant Dyn1 Y600L (Figure 4A). These data indicate that the VL1 and VL3 mutations, when placed in the context of a full-length Dyn1 molecule, differentially affect curvature-sensitive PH domain–membrane interactions. Using SEC-MALS, we ascertained that the full-length Dyn1 PH-domain mutants formed tetramers in solution similar to Dyn1 WT (Supplemental Figure S3B). Spin-sedimentation profiles for wild type (WT) and mutants mirrored the FRET data, indicating that the in-equilibrium solution FRET assay directly probes for stable PH domain–membrane interactions (Figure 4B).
FIGURE 4:
VL mutations variably affect PH domain–membrane interactions in full-length Dyn1. (A) Binding of full-length Dyn1 WT and mutants (0.1 μM protein) to lipid templates of varying membrane curvature (5 μM total lipid; 10 mol% PIP2; 10 mol% Dansyl-PE) monitored by the FRET-sensitized increase in Dansyl (acceptor) emission intensity upon Trp (donor) excitation. The average increase in Dansyl emission intensity monitored at 530 nm is indicated as F/F0, where F0 is the background emission intensity of Dansyl (caused by direct excitation at λex = 280 nm) at time 0 before protein addition and F is the intensity after 15 min of protein incubation. (B) Stable membrane association of Dyn1WT or mutants (2 μM protein; 100 μM total lipid; 10 mol% PIP2) was examined by spin-sedimentation followed by SDS–PAGE and densitometric ratio analyses of the supernatant (S) and pellet (P) fractions (see Materials and Methods). Plot of percentage dynamin pelleted of total upon incubation with lipid templates of varying membrane curvature. Baseline density is defined in Materials and Methods. (C, D) Stopped-flow kinetic measurements of Trp-Dansyl FRET in full-length Dyn1 WT (C) or Dyn1 I533A (D). The sensitized increase in Dansyl emission intensity upon Trp excitation is plotted as a function of time. The final protein and lipid concentrations upon mixing were 0.1 and 5 μM, respectively. PIP2 was present at 10 mol% in each lipid template. Representative traces. Kinetic traces were best fitted (colored traces) to a biexponential kinetic equation (Materials and Methods). Measured kinetic rate constants are summarized in Materials and Methods. (E) Stable PH domain–membrane association in Dyn1 WT and Dyn1 I533A at 1 mol% PIP2 in LT detected by Trp-Dansyl FRET. Average FRET-sensitized increase in Dansyl emission intensity is plotted as F/F0 as in A.
To determine the mechanistic bases of Dyn1 impairment by VL1 mutations, we initiated a rigorous examination of Dyn1 I533A in comparison to Dyn1 WT by a variety of biochemical and biophysical assays. Rapid kinetics measurements of Trp-Dansyl FRET first revealed that the differential curvature sensitivities of Dyn1 WT and Dyn1 I533A relate more to the extent of membrane binding than to the rate of PH domain–membrane interactions (Figure 4, C and D; see Materials and Methods for observed kinetic rate constants). We next compared the extent of PH domain–membrane association in Dyn1 I533A relative to Dyn1 WT at limiting concentrations of PIP2 (1 mol%) in LT. Under these particular conditions, Dyn1 I533A has been shown to similarly constrict but, in contrast, to fail in mediating the fission of tethered membrane tubules relative to Dyn1 WT in vitro (Shnyrova et al., 2013). Intriguingly, at 1 mol% PIP2 in LT, Trp-Dansyl FRET was virtually absent for Dyn1 I533A, indicating that the mutant PH domain does not stably engage with highly curved membranes (Figure 4E). Although supporting the fission defect, these results did not explain the unaffected constriction behavior of Dyn1 I533A on membrane tubules (Shnyrova et al., 2013). We therefore explored the various caveats of Trp-Dansyl FRET.
Evidence for an alternate PH-domain membrane orientation
The four Trp residues present in the Dyn1 PH domain are clustered at one end of the roughly globular domain (Figure 1A), and all are located within Trp-Trp homotransfer distance (R0 ≈ 6–12 Å) to collectively serve as energy donors for Trp-Dansyl FRET (Moens et al., 2004). We therefore explored the possibility that in the absence of stable, hydrophobic VL1–membrane interactions, the PH domain (∼40 Å in globular diameter) might adopt an alternate orientation on the membrane surface that positions the Trp cluster outside the effective distance for Trp-Dansyl FRET (i.e., distance >1.5R0 or >∼32 Å; Piston and Kremers, 2007; Supplemental Figure S5). This alternate orientation of the PH domain may involve strong electrostatic adsorptive forces from other regions of the molecule, possibly from the distinct electrostatic lobe of VL3 that helps retain dynamin on the membrane surface in the absence of stable, hydrophobic VL1–membrane insertion. Such a scenario is supported by two different modeled orientations of the PH domain in recent cryo-EM maps of the Dyn1 polymer, which alternately position VL1 or VL3 proximal to the membrane surface (Chappie et al., 2011).
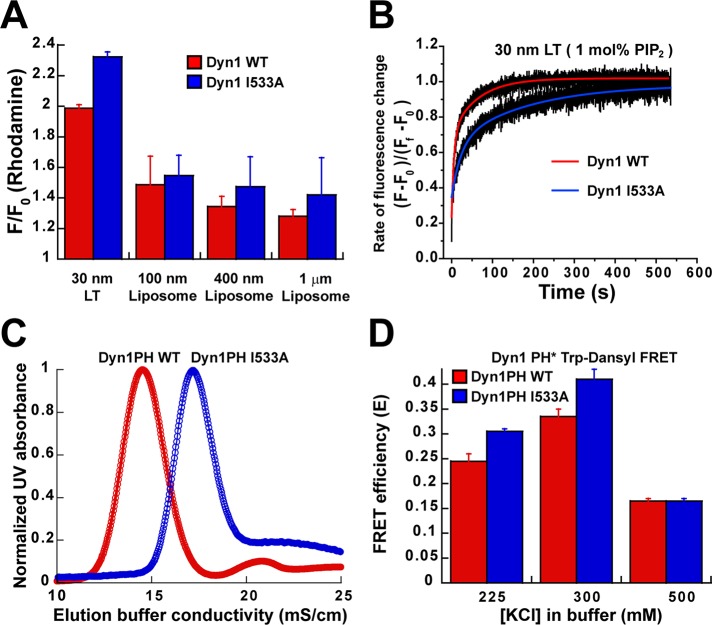
We therefore developed a secondary but complementary FRET approach to probe for dynamin–membrane interactions that would be unaffected by PH-domain orientation on the membrane surface. An extrinsic BODIPY-Fl (BODIPY) dye was coupled to a surface-exposed native Cys residue in the GED (C708; Figure 1A and Supplemental Figure S4B) located far above the base of the molecule (∼70 Å). This dye was then used as a FRET donor for rhodamine-labeled acceptor lipid molecules (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(Lissamine rhodamine B sulfonyl) [rhodamine-PE or RhPE]) distributed randomly on the membrane surface (Supplemental Figure S4C), similar in principle to Trp-Dansyl FRET. The BODIPY-rhodamine pair, however, has a longer distance range for FRET (R0 ≈ 57 vs. 21 Å for Trp-Dansyl) and a higher donor quantum yield, ideal to probe for dynamin–membrane interactions independent of PH-domain membrane orientation (Ramachandran et al., 2005). BODIPY-labeled Dyn1 WT and Dyn1 I533A behaved identically to their unlabeled counterparts in spin-sedimentation assays (Supplemental Figure S6A).
Despite the dramatic loss of Trp-Dansyl FRET in Dyn1 I533A on relatively planar liposomes (Figure 4A), BODIPY-rhodamine FRET in Dyn1 I533A was equivalent to Dyn1 WT on all lipid templates tested (Figure 5A). These data indicate that the lack of short-range Trp-Dansyl FRET in Dyn1 I533A on planar membrane templates is not due to a loss of membrane binding but instead to an alternate orientation of the PH domain that retains dynamin on the membrane surface. Consistent with this interpretation, at 1 mol% PIP2 in LT, the magnitude of BODIPY-rhodamine FRET in Dyn1 I533A was identical to that of Dyn1 WT (F/F0 ≈ 1.3 ± 0.1; n = 3), in stark contrast to the lack of Trp-Dansyl FRET observed for the mutant earlier (Figure 4E). The electrostatic dependence of this alternate PH domain–membrane interaction was clearly evident when at 1 mol% PIP2, Dyn1 I533A bound to LT at a considerably slower rate than Dyn1 WT (Figure 5B). On the basis of these results, we attribute the loss of membrane fission, but not membrane constriction, in Dyn1 I533A to an alternate PH-domain orientation on the membrane surface that does not perturb stalk-dependent dynamin self-assembly. Membrane interactions effected by this alternate PH-domain membrane orientation, however, were considerably weaker, as spin-sedimentation profiles mirrored Trp-Dansyl, and not BODIPY-rhodamine, FRET (Supplemental Figure S6A).
FIGURE 5:
Evidence for an alternate PH-domain membrane orientation. (A) FRET-sensitized increase in rhodamine emission intensity upon incubation of various lipid templates containing 1 mol% RhPE (5 μM total lipid) with BODIPY-Dyn1 WT or -Dyn1 I533A (0.1 μM) monitored at 590 nm and plotted as F/F0, where F0 is the background emission intensity of rhodamine (caused by direct excitation at λex = 470 nm) at time 0 before protein addition and F is the intensity after 15 min of protein incubation. (B) Direct comparison of the FRET-detected rate of dynamin–membrane association in BODIPY-Dyn1 WT and -Dyn1 I533A at limiting concentrations of PIP2 (1 mol%) in LT. Representative kinetic profiles for the FRET-sensitized rhodamine emission intensity increase are normalized and plotted as total fractional intensity change as a function of time. Kinetic traces were best fitted to the biexponential kinetic equation (Materials and Methods). (C) Representative SP-Sepharose cation-exchange chromatography elution profiles for isolated PH domains of Dyn1PH WT and Dyn1PH I533A plotted as a function of increasing ionic strength (salt) measured by buffer conductivity in millisiemens/centimeter. (D) Binding of 0.5 μM Dyn1PH WT or Dyn1PH I533A to LT (150 μM total lipid) at varying buffer ionic strengths measured by Trp-Dansyl FRET as earlier.
Under the same conditions, Dyn1 M534A and Dyn1 Y600L experienced a significant loss of BODIPY-rhodamine FRET relative to Dyn1 WT (Supplemental Figure S6B). However, remarkably, their spin-sedimentation profiles were not considerably different from that of wild type (Supplemental Figure S6C). Because the spin-sedimentation assay does not distinguish between membrane-associated and membrane-dissociated polymeric dynamin species, whereas FRET does, we attribute the loss of function in Dyn1 M534A and Dyn1 Y600L to an overall instability of the dynamin polymer on the membrane surface. This finding is in general agreement with the established roles of VL1 and VL3 in stable phosphoinositide binding (Lee et al., 1999; Vallis et al., 1999).
To gain further insight into the alternate mode of PH domain–membrane association stabilized by Dyn1 I533A, we tested for membrane interactions of a largely electrostatic nature in isolated Dyn1PH I533A relative to Dyn1PH WT. To address such a possibility experimentally, we first took advantage of an empirically established positive correlation between Dyn1PH membrane-binding affinity and the affinity of the PH domain for cation-exchange chromatography resins that putatively mimic the surface of a negatively charged phospholipid bilayer (Kenniston and Lemmon, 2010). To determine whether Dyn1PH I533A binds to negatively charged membranes in an alternate manner to Dyn1PH WT, we tested their relative binding strengths on a sulfopropyl (SP) cation-exchange resin under a linear gradient of increasing ionic strength (see Materials and Methods). Remarkably, Dyn1PH I533A was displaced from the column at a far greater ionic strength than Dyn1PH WT (Figure 5C), indicating a stronger electrostatic interaction for the mutant PH domain on the resin. Weakening of a strong, hydrophobic interaction in Dyn1 I533A therefore appears to considerably enhance the relative contribution of electrostatic interactions in PH domain–membrane association. These results suggest that a balance of counteracting electrostatic and hydrophobic forces engage and orient the dynamin PH domain on the membrane surface. Interestingly, Dyn1PH Y600L exhibited a broad elution profile (Supplemental Figure S7), indicating a mixed population of PH-domain orientations on the membrane surface. This may very well explain the overall instability of Dyn1 Y600L polymer on the membrane surface observed earlier (Supplemental Figure S6, B and C).
To probe directly for this alternate, electrostatic mode of Dyn1PH membrane association, we resorted to the Trp-Dansyl FRET assay and measured the extents of membrane binding in Dyn1PH I533A relative to Dyn1PH WT at varying buffer ionic strengths. At high ionic strength (500 mM KCl), which effectively screens electrostatic PH domain–membrane interactions, the binding of Dyn1PH I533A to LT was equivalent to that of Dyn1PH WT (Figure 5D). As expected, this binding was significantly lower than that observed earlier at physiologically relevant ionic strength (150 mM KCl; Figure 2E). However, at intermediate ionic strengths (225 and 300 mM KCl), Dyn1PH I533A exhibited significantly increased binding to LT relative to Dyn1PH WT (Figure 5D). These data reflected the greater resistance of the mutant PH domain to displacement from the chromatographic resin observed earlier (Figure 5C). We therefore conclude on the basis of the above data that VL1-insertion-independent, alternate membrane interactions of the Dyn1 PH domain are predominantly electrostatic in nature.
VL1 senses high membrane curvature exclusively via hydrophobic interactions
To reveal the mechanism of membrane-curvature sensing in the dynamin PH domain and discern the role(s) of hydrophobic and electrostatic interactions, we introduced and characterized the effect of two additional mutations, I533D and K535A, in Dyn1 PH domain VL1. Introduced one at a time, these mutations effectively render the membrane-inserting VL1 electrically neutral, that is, with a net charge equivalent to zero. However, the K535A mutation renders the tip of VL1 entirely hydrophobic, whereas the loop bearing the I533D mutation is drastically reduced in hydrophobicity and also harbors charged residues of opposite polarity.
Remarkably, when assayed on curved LT, the extents of both Trp-Dansyl and BODIPY-rhodamine FRET in Dyn1 K535A were almost identical to that of Dyn1 WT (Figure 6A), indicating that electrostatic interactions of VL1 are not involved in LT curvature sensing. This unexpected finding was further corroborated by the spin-sedimentation assay (Figure 6B). However, binding of Dyn1 K535A to liposomes was significantly impaired (Supplemental Figure S8), suggesting an important role for the positively charged residue in the electrostatic adsorption of dynamin to negatively charged, planar membrane surfaces. In contrast, both the curvature-sensing and membrane-binding properties of the charge-bearing yet electrically neutral Dyn1 I533D were dramatically reduced on LT (Figure 6, A and B). These data conclusively demonstrate that VL1 senses high membrane curvature exclusively via hydrophobic membrane interactions. Consistent with the foregoing data, the LT-stimulated GTPase activity of Dyn1 K535A was comparable to that of Dyn1 WT, whereas the corresponding activities of VL1 mutants Dyn1 I533D and Dyn1 M534A were significantly reduced (Figure 6C). As previously shown, the stimulated GTPase activities of Dyn1 I533A and Dyn1 Y600L on LT were not considerably altered (Ramachandran et al., 2009; Liu et al., 2011; Figure 6C). The inability of Dyn1 K535A to self-assemble efficiently and in proper register at low protein concentrations (≤0.1 μM; Ramachandran and Schmid, 2008) might explain the loss of membrane stability and stimulated GTPase activity observed previously for mutants of K535 (Achiriloaie et al., 1999; Vallis et al., 1999).
FIGURE 6:
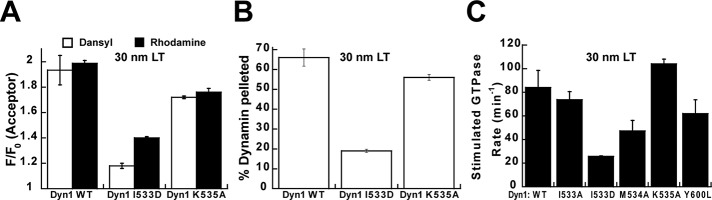
VL1 is a hydrophobic sensor of high membrane curvature. (A) Binding of Dyn1 I533D or Dyn1 K535A (0.1 μM) to LT (5 μM total lipid) measured by Trp-Dansyl and BODIPY-rhodamine FRET. (B) Stable association of Dyn1 I533D or Dyn1 K535A (2 μM) to LT (100 μM total lipid) analyzed by spin sedimentation. (C) Stimulated GTPase activity of Dyn1 WT or mutants (0.5 μM) preincubated on LT (150 μM total lipid) measured as described in Materials and Methods.
PH-domain orientation regulates dynamin polymer dynamics
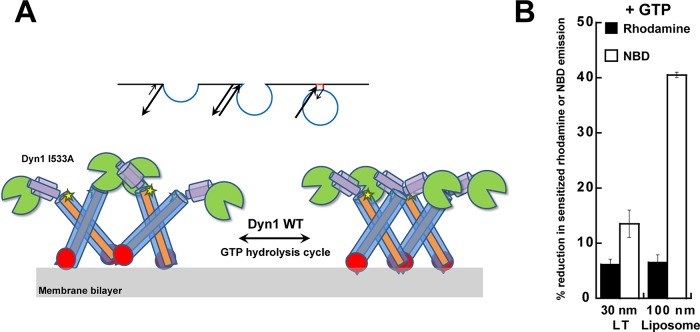
To discern the influence of the alternate PH-domain orientation on dynamin morphology, we compared the membrane-bound polymers of Dyn1 I533A to those of Dyn1 WT by EM. Because possible structural variations in polymer morphology between the two were not readily apparent on LT of preset membrane curvature (Ramachandran et al., 2009), we alternatively examined Dyn1 I533A polymers that were stabilized on the sparse membrane tubules generated from liposomes. EM data revealed that Dyn1 I533A polymers were extremely variable in diameter, fluctuating between highly constricted and relaxed conformations, in contrast to the near-uniform diameter of Dyn1 WT (Figure 7A). The variable and nonuniform polymerization of Dyn1 I533A was further evidenced in real time using a previously established BODIPY self-quenching assay for dynamin self-assembly in proper register (Ramachandran et al., 2007). Imperfect register of the assembly interface in Dyn1 I533A coincided with a distinct loss of BODIPY self-quenching relative to Dyn1 WT even on LT (Figure 7B). From these data, we conclude that PH-domain membrane insertion primarily functions to stabilize uniform, narrow membrane curvature through the concerted control of dynamin polymer morphology. We suggest that the cyclical weakening of VL1–membrane interactions during the GTP hydrolysis cycle (as observed by Ramachandran and Schmid, 2008) likely results in dynamic fluctuations of the dynamin polymer between uniformly constricted (VL1-inserted) and variably relaxed (VL1-retracted) conformational states (Figure 8A). The latter conformational state is stabilized in Dyn1 I533A. Supporting this model, BODIPY-rhodamine FRET-detected alternate, electrostatic PH domain–membrane interactions were largely insensitive to GTP hydrolysis, in striking contrast to NBD-detected hydrophobic, VL1-membrane insertion (Figure 8B; Ramachandran and Schmid, 2008). Thus the dynamin PH domain likely functions as a regulator, as early studies suggested (Muhlberg et al., 1997), rather than as a direct effector of membrane fission.
FIGURE 7:
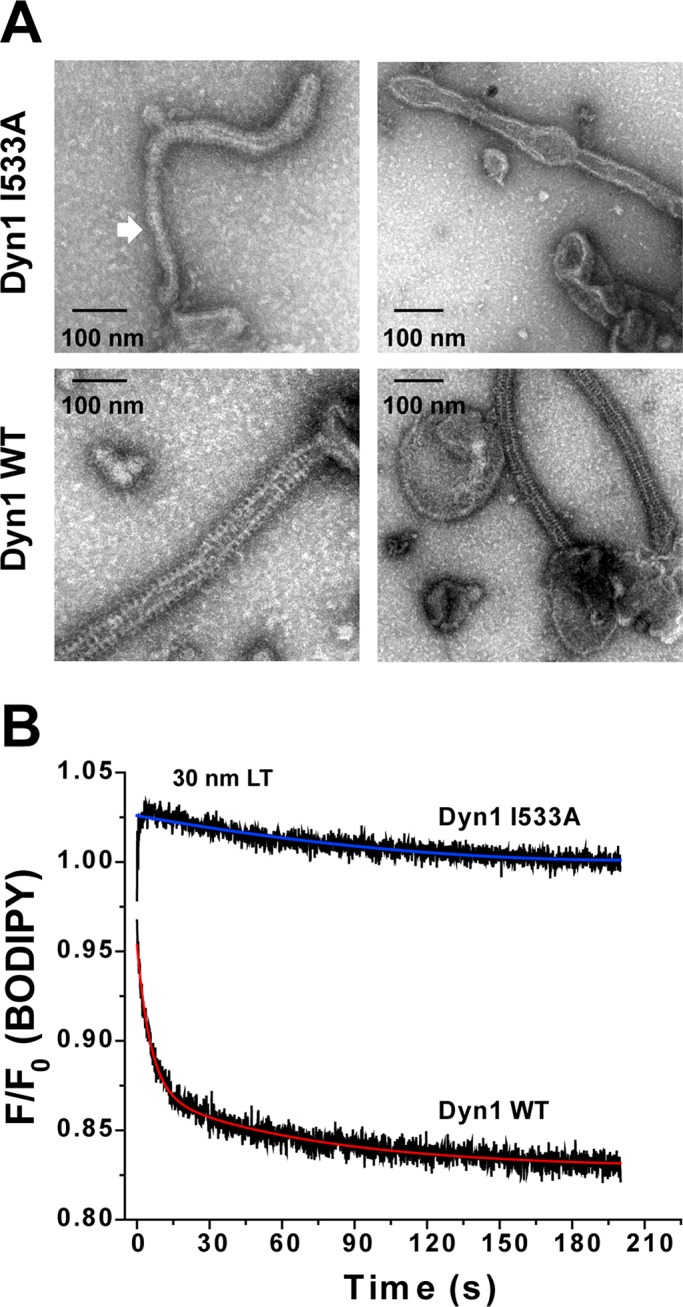
Alternate PH-domain orientations affect dynamin polymer morphology. (A) Representative EM images of Dyn1 WT and Dyn1 I533A self-assembled on 400-nm-diameter PIP2-containing liposomes. Block arrow points to a highly constricted polymer conformation sampled by Dyn1 I533A relative to the uniform curvature of Dyn1 WT. (B) BODIPY emission intensity change at 510 nm (λex = 490 nm) upon self-assembly of BODIPY-Dyn1 WT or -Dyn1 I533A (0.1 μM) on unlabeled LT (5 μM total lipid; 10 mol% PIP2) was monitored by stopped-flow kinetics. F0 is the emission intensity at time 0 before LT addition and F is the intensity at time t. Representative kinetic traces are shown. Kinetic traces were best fitted (colored traces) to a biexponential kinetic equation (Materials and Methods).
FIGURE 8:
Model for dynamin polymer dynamics and the role of GTP hydrolysis. (A) Proposed model for alternate PH-domain orientations in the regulation of dynamin polymer morphology and membrane constriction. The Dyn1 WT polymer oscillates between VL1-inserted, uniformly constricted (right) and VL1-retracted, variably relaxed (left) states during the dynamin GTP hydrolysis cycle. Dyn1 I533A is stabilized in the latter state, causing nonuniform membrane tubule constriction. The short spike in the PH-domain illustration represents VL1, and the star represents the BODIPY dye coupled to C708 in GED, which detects dynamin self-assembly in the proper register. Curvature-sensitive association of the dynamin PH domain to the progressively narrowing endocytic pit membrane neck is illustrated (inset) as a ratio of on- and off-rates (arrows). (B) Percentage reduction in BODIPY-rhodamine FRET or NBD emission intensity upon addition of GTP (1 mM) to BODIPY-Dyn1 WT or RCLDyn1 G532C-NBD (0.1 μM protein), respectively, in the presence of either LT or liposomes. BODIPY-rhodamine FRET detects alternate PH domain–membrane interactions, whereas NBD detects VL1 membrane insertion (Ramachandran et al., 2009). Note that in the constant presence of GTP, highly curved LT stabilizes VL1 membrane insertion far greater than relatively planar liposomes, underscoring the critical role of preacquired membrane curvature in the mechanism of dynamin-mediated membrane fission.
DISCUSSION
Previous work (Roux et al., 2010) had suggested that the ability of dynamin to sense high membrane curvature relies on its inherent propensity to form short polymeric arcs and rings whose internal diameters closely approximated the outer diameter of a highly curved membrane tube such as the narrow membrane neck of an endocytic coated pit. Here we demonstrate, using the isolated monomeric Dyn1 PH domain, that the mechanism of curvature sensing in dynamin exclusively involves hydrophobic interactions effected by VL1 membrane insertion and is not critically reliant on helical dynamin self-assembly. Furthermore, our FRET measurements reveal the existence of an alternate PH-domain orientation on the membrane surface that is stabilized predominantly by electrostatic interactions and is unaffected by VL1 membrane insertion. Stabilization of this alternate orientation, as observed for Dyn1 I533A, resulted in variable dynamin polymer morphology and nonuniform membrane tubule constriction, indicating that the PH domain regulates membrane fission through the control of dynamin polymer dynamics. Our results therefore suggest that the dynamin PH-domain VL1 primarily functions 1) as a hydrophobic membrane anchor to constrain dynamin conformational dynamics and 2) as a fulcrum to direct dynamin polymerization to the uniform, narrow tubular membrane diameters required for membrane fission. Through reversible and regulated membrane penetration of its PH-domain VL1 during the GTP hydrolysis cycle, dynamin is expected to oscillate between uniformly narrow and variably relaxed polymer conformations to stochastically catalyze membrane fission.
An unexpected finding from our high-sensitivity fluorescence measurements is that the tip of VL1, when rendered entirely hydrophobic by mutagenic substitution (K535A), was still able to recognize high membrane curvature and stabilize dynamin–membrane association. Electrostatic interactions of K535 necessary for dynamin adsorption onto relatively planar, negatively charged membrane surfaces therefore appear to be dispensable on a highly curved membrane that permits hydrophobic protein–membrane interactions. These results lend further support to the growing realization that curvature sensing is a generic property of the membrane manifesting curvature-induced defects in lipid packing (e.g., LT) to attract hydrophobic elements (e.g., VL1) from large protein densities rather than a high protein affinity for highly curved membranes (Bhatia et al., 2010). We speculate that K535, initially adsorbed onto relatively planar membranes through electrostatic surface interactions, subsequently “snorkels” upon VL1 insertion into highly curved membranes. This is presumably accomplished via the insertion of the hydrophobic part of the K535 side chain into the hydrocarbon membrane core, while the residue's positively charged tip is positioned at the more-polar lipid–water interface. Such a “snorkeling” behavior for K residues positioned at the lipid–water interface has been described in a number of membrane proteins (Strandberg et al., 2002; Strandberg and Killian, 2003). Thus VL1, through the hydrophobic membrane interactions of I533, M534, and K535, functions as an exquisite sensor of high membrane curvature.
On the basis of our observation that the isolated Dyn1PH monomer can insert into relatively planar membrane templates, albeit less efficiently, but not generate membrane tubules, we argue that the membrane-inserting VL1 does not function as a lipid-splaying, membrane-bending “wedge.” Instead, it acts as a hydrophobic “anchor” to stabilize the PH domain on the membrane surface. This is consistent with the small footprint of membrane insertion effected by the membrane-proximal residues, I533, M534, and K535. A membrane-anchoring role for similarly positioned hydrophobic residues has been observed for many other PH domains that are unrelated to membrane remodeling (Manna et al., 2007). We concur that dynamin-mediated membrane remodeling requires concerted dynamin self-assembly in addition to PH domain membrane insertion.
Although we contend that the isolated, low-affinity Dyn1 PH domain is not a membrane-curvature generator in its own right, a structurally related high-affinity PH domain from the Golgi remodeling protein FAPP that inserts its hydrophobic β1-β2 loop similarly into the membrane has been demonstrated to remodel (tubulate) membranes independently and efficiently (Lenoir et al., 2010). It is thus plausible that at very high bulk protein concentrations not attained in our experiments, the Dyn1 PH-domain monomer may generate membrane curvature on its own. Yet, under our experimental conditions, we were unable to correlate detectable PH domain membrane insertion with any corresponding membrane remodeling. We further speculate that the closely quartered Y600 residue in Dyn1 VL3, whose mutation (Dyn1 Y600L) impairs membrane fission in vitro (Liu et al., 2011), may function to orient the PH domain favorably at the membrane surface for efficient VL1 membrane penetration. A role for aromatic residues in the partitioning of membrane-interacting protein elements to the lipid–water interface has been described for a number of proteins (Wimley and White, 1996). Indeed, aromatic Trp residues present in the β1-β2 loop of the structurally related FAPP PH domain appear to function as “buoys” at the lipid–water interface to support optimal membrane insertion (Lenoir et al., 2010).
Our complementary FRET-based approaches provide the first experimental evidence for the functional role of alternate PH-domain orientations in the mechanism of dynamin-catalyzed membrane fission. However, suggestions of alternate dynamin PH domain orientations in the regulation of dynamin function are not unprecedented. Alternate orientations of the dynamin PH domain were found in the recently solved crystal structures of Dyn1 variants (Faelber et al., 2011; Ford et al., 2011). In the upright dynamin criss-cross dimer, the stalk-interfaced PH domain orientation (as shown in Figure 1A; Faelber et al., 2011) restricts access to a key assembly interface (interface 3) located at the base of the molecule. In this orientation, the PH-domain positions VL3, and not VL1, proximally to the membrane surface. On the other hand, a flexibly tethered orientation of the PH domain (as shown in Ford et al., 2011) provides unrestricted access to interface 3 while repositioning VL1 for membrane insertion. Alternate PH-domain orientations were also modeled in cryo-EM maps of the membrane-bound dynamin polymer (Chappie et al., 2011). Our FRET-detected measurements of alternate PH-domain orientations in dynamin function correlate well with structural data from these techniques.
The presence of alternate modes of membrane recognition is not unique to the dynamin PH domain and is found in others, including the membrane-inserting PH domain of general receptor of phosphoinositides 1 (GRP1; Manna et al., 2007; Lai et al., 2013). Weak electrostatic membrane interactions rotationally steer the GRP1 PH domain on the membrane surface for effective recognition and capture of the target lipid, phosphatidylinositol (3,4,5)-trisphosphate (PIP3). Coincidence detection of PIP3 and local lipid-packing defects subsequently facilitates the insertion of a PH-domain hydrophobic residue that functions to anchor the molecule onto the membrane surface. We speculate that the dynamin PH domain functions similarly in using electrostatic and hydrophobic interactions for the targeting and subsequent anchoring of the molecule onto the membrane surface, respectively. On the basis of the character-defining VL sequence variations in Dyn1 and Dyn2 (Liu et al., 2011), we further speculate that the differential membrane recognition properties of the two dynamin isoforms may arise from an altered balance of electrostatic and hydrophobic forces in their respective PH domain–membrane interactions. Further studies are necessary to elucidate these differences.
Finally, BAR domain–containing partner proteins such as SNX9 and amphiphysin I have been shown to differentially regulate dynamin VL1–membrane interactions (Ramachandran and Schmid, 2008). Based on our current understanding of PH-domain function, it is plausible that these effectors differentially stabilize either the relaxed or the uniformly constricted conformations of the membrane-bound dynamin polymer to control membrane fission kinetics and efficiency.
On the basis of the above data, we propose the following order of events in the recruitment and regulated polymerization of dynamin at the endocytic pit neck before membrane fission: 1) Cytosolic dynamin prevalent as a dimer of dimers (Ramachandran et al., 2007) probes for high membrane curvature at the progressively narrowing membrane neck of an endocytic pit. This interaction uses weak ionic interactions mediated by the electrostatic lobes of PH-domain VL1 and VL3, with both PIP2 and other negatively charged lipids (e.g., phosphatidylserine) present at the membrane surface. 2) Coincidence detection of high membrane curvature, sequestered PIP2 (Bethoney et al., 2009), and SH3 domain–containing binding partners (BAR proteins) engages VL1 stably in the membrane through strong hydrophobic interactions. Stabilization of PH domain–membrane interactions concomitantly exposes key assembly interfaces in dynamin for helical self-assembly around the BAR-stabilized membrane neck. 3) At the onset of stimulated GTP hydrolysis, VL1 cyclically inserts into and retracts from the membrane, while the PH domain is retained on the membrane surface through alternate, electrostatic interactions. The resultant fluctuations of the dynamin polymer between uniformly constricted and variably relaxed states stochastically then leads to the formation of a hemifission membrane intermediate (Bashkirov et al., 2008), which ultimately precipitates membrane fission.
MATERIALS AND METHODS
Preparation of isolated Dyn1PH and mutants
DNA encoding Cys-less Dyn1PH* (Dyn1PH C607S) subcloned in pET11a (Ferguson et al., 1994) was used as the template for PCR-based mutagenesis using the QuikChange protocol (Agilent Technologies, Santa Clara, CA). All mutations were confirmed by automated DNA sequencing. Recombinant Dyn1PH* and mutants were expressed in BL21 Star (DE3) Escherichia coli and purified as previously described (Ferguson et al., 1994), except that a sulfopropyl (SP)–Sepharose resin was used for cation-exchange chromatography under a linear ionic gradient of 25–250 mM NaCl. Dyn1PH* I533C was fluorescently labeled with either NBD or BODIPY-Fl as described previously (Ramachandran and Schmid, 2008). The labeling efficiency for either dye was determined to be >80%. Expression of Dyn1PH M534A surprisingly did not yield soluble protein.
Preparation of full-length Dyn1 and derivatives
Unless noted otherwise, DNA encoding full-length human Dyn1 WT or mutants subcloned in pRSET C (Invitrogen, Carlsbad, CA) was used for protein expression in E. coli BL21 Star (DE3) following published protocols (Kenniston and Lemmon, 2010). Full-length Dyn1 was purified using an AmphII-SH3-domain affinity protocol as described previously (Leonard et al., 2005) and stored in buffer containing 10% vol/vol glycerol. Dyn1 I533D was expressed and purified from insect cells as described earlier (Ramachandran et al., 2009). Dyn1 K535A was expressed and purified from both bacterial and insect cells. Dyn1 WT and mutants were labeled with the fluorophore BODIPY-Fl as described earlier (Ramachandran and Schmid, 2008). The efficiency of labeling determined using a molar absorptivity coefficient of 76,000 M−1 cm−1 for BODIPY-Fl at 502 nm was 1:1 dye-to-protein in all cases. RCLDyn1 G532C-NBD was prepared as described previously (Ramachandran et al., 2009). The efficiency of NBD labeling was determined to be ∼90% using a molar absorptivity coefficient of 25,000 M−1 cm−1 for NBD at 478 nm.
SEC-MALS
Dyn1PH (42–75 μM) and full-length Dyn1 samples (5–7 μM) were fractionated on Superdex 75 10/300 GL or Superose 6 10/300 GL SEC columns (GE Healthcare, Piscataway, NJ), respectively, and analyzed online using miniDAWN Treos MALS and Optilab rEX differential refractive index (dRI) detectors (Wyatt Technologies, Santa Barbara, CA) as described earlier (Ramachandran et al., 2007). Data analysis was accomplished using the ASTRA 6.1 software package (Wyatt Technologies).
Preparation of liposomes and LT
All phospholipids were purchased from Avanti Polar Lipids (Alabaster, AL). Liposomes of 100-, 400-, and 1000-nm average diameter composed of 90 mol% 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 10 mol% porcine PIP2, and preformed lipid nanotubes composed of 45 mol% DOPC, 45 mol% C24:1 β-d-galactosyl ceramide and 10 mol% PIP2 were generated according to procedures described previously (Leonard et al., 2005). One mol% of RhPE or 10 mol% of Dansyl-PE replacing an equivalent mole fraction of DOPC was introduced for dynamin–membrane FRET measurements. LT containing 1 mol% PIP2 were prepared likewise.
Atomic force microscopy
LT samples (50 μM total lipid) were incubated on a freshly cleaved mica surface for ∼10 min and subsequently rinsed with water to remove buffer and salts. After the mica surface was allowed to dry, the samples were imaged using the tapping mode on a Digital Instruments MultiMode atomic force microscope equipped with a Nanoscope IV controller and a type E scanner (Bruker, Billerica, MA). All images were acquired using single-beam silicon probes with nominal spring constant of 40 N/m and nominal tip radius of 10 nm.
Electron microscopy
Negative-stain EM imaging of Dyn1 WT and Dyn1 I533A polymers stabilized on 400-nm-diameter liposomes was performed as described previously (Ramachandran et al., 2007, 2009). Dynamin (1.0 μM) and liposomes (25 μM total lipid) were incubated together for 15 min at room temperature before deposition on carbon-coated EM grids. EM of Dyn1 WT on LT was performed under similar conditions. For Dyn1PH imaging, the final concentrations of protein and lipid were both 50 μM.
Dynamic light scattering
DLS measurements of liposome size were performed in a Dynapro Nanostar (Wyatt Technologies) instrument according to previously published methods (Drin et al., 2008). Samples (50 μl) of liposomes (0.5 mM total lipid) prepared by extrusion were analyzed in an Eppendorf UVette at room temperature. Autocorrelation curves from a set of 10 acquisitions (10-s integration time each) were analyzed using Dynamics version 7.1.3 software (Wyatt Technologies) to resolve the relative size distribution of liposomes within each sample.
Preparation of GUVs and confocal light microscopy
GUVs were prepared under physiological salt conditions (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.5, 150 mM KCl) using a modified electroformation protocol and laboratory-built platinum wire electrode chambers as described previously (Montes et al., 2007). The lipid mixture contained, in addition to DOPC (∼75 mol%), 5 mol% PIP2, 20 mol% 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS), 0.1 mol% RhPE for fluorescence imaging, and 0.5 mol% biotin-PE for GUV adhesion to streptavidin (Sigma-Aldrich, St. Louis, MO)–coupled, biotin-LC-BSA (Thermo Scientific, Rockford, IL)–coated LabTek II chambered cover slides (Nalge Nunc International, Rochester, NY) prepared according to published protocols (Yildiz et al., 2003). Confocal images were captured using a 60× oil-immersion objective and an Olympus FV1000 IX81 confocal microscope (Olympus USA, Melville, NY).
Fluorescence spectroscopy and stopped-flow kinetics
All fluorescence measurements were carried out at 25°C on a Fluorolog 3-22 photon-counting spectrofluorimeter (Horiba Jobin Yvon, Edison, NJ) equipped with a 450-W xenon lamp, double-excitation and double-emission monochromators, a cooled PMT housing, a temperature-controlled sample compartment, and a rapid kinetics accessory (Hi-Tech Scientific, Bradford-on-Avon, United Kingdom). Fluorescence intensities were corrected for background emission and lamp fluctuations. To enhance Tyr-Trp hetero- and Trp-Trp homo-energy transfer for efficient Trp-Dansyl FRET, samples were excited at 280 nm (2-nm bandpass) and emission was monitored between 315 and 550 nm (4-nm bandpass; Weber and Shinitzky, 1970). FRET efficiency (E) was calculated using the equation E = 1 − (FDA/FD), where FDA is the emission intensity of Trp monitored at 330 nm in the presence of acceptor Dansyl-labeled lipids and FD is the corresponding emission intensity in the absence of acceptors. The Förster radius (R0) is the characteristic distance for a FRET pair at which E = 50%. Owing to the inverse-sixth-power dependence of FRET on distance, E rapidly approaches zero as the distance separating the FRET pair increases to 1.5R0 (Piston and Kremers, 2007). FRET-sensitized Dansyl emission intensity increase was monitored at 530 nm (4 nm). Likewise, for BODIPY-rhodamine FRET, samples were excited at 470 nm (2 nm) and emission was monitored between 500–700 nm (4 nm). FRET-sensitized rhodamine emission intensity increase was monitored at 590 nm (4 nm). The fractional emission intensity contribution of BODIPY at 590 nm was taken into account. Unless noted otherwise, for both steady-state and rapid kinetics measurements, 0.1 μM labeled or unlabeled full-length Dyn1 was mixed with 5 μM total lipid to maintain pseudo-first-order conditions. The protein and lipid concentrations noted correspond to those after mixing. For isolated Dyn1PH* and derivatives, 0.5 μM protein was used at a protein-to-lipid ratio of 1:300 (150 μM total lipid). Excitation and emission wavelengths were 490 and 510 nm, respectively, for monitoring the self-quenching of BODIPY-labeled Dyn1 on unlabeled lipid templates. The kinetics in each case was fast enough to monitor the fluorescence intensities continuously (0.1-s integration time) without photobleaching. The apparent rate constant kobs and the amplitudes were determined from the kinetic traces by including a dead-time correction t0 that takes into account the delay between the mixing of reactants and the start of the measurements. All fitting procedures were carried out with Origin 8.5 statistical software based on nonlinear, least-square methods and the Levenberg–Marquardt algorithm using the equation
where F0, Ff, and F(t) are initial, final, and time-t fluorescence intensities, respectively; kobs1 and kobs2 are apparent kinetic rate constants; t0 is the dead time; and a is the relative amplitude of the fast component. The kobs1 corresponding to the fast component measured for Dyn1 WT by Trp-Dansyl FRET on LT, 100-, 400-, and 1000-nm-diameter liposome populations (Figure 4C) was, respectively, 0.33 ± 0.13, 0.28 ± 0.11, 0.14 ± 0.01, and 0.14 ± 0.01 s−1. The kobs1 for Dyn1 I533A measured by Trp-Dansyl FRET on LT was 0.23 ± 0.10 s−1 (Figure 4D). Similarly, kobs1 measured for Dyn1 WT and Dyn1 I533A on 1 mol% PIP2–containing LT by BODIPY-rhodamine FRET (Figure 5B) was 0.16 ± 0.01 and 0.06 ± 0.01 s−1, respectively. The kobs2 was lower than 0.1 s−1 in all cases.
Steady-state end-point measurements of NBD emission intensity were made as reported previously (Ramachandran et al., 2009). All experiments were performed in buffer containing 20 mM HEPES, pH 7.5, 150 mM KCl, and 2 mM MgCl2.
Spin-sedimentation assay
Supernatant (S) and pellet (P) fractions of Dyn1 WT and mutants (2 μM protein final) incubated with various lipid templates (100 μM lipid final) at room temperature for 30 min were obtained by high-speed centrifugation at 20,800 × g in a refrigerated microcentrifuge maintained at 4°C according to published protocols (Ramachandran et al., 2009). Densitometry of S and P fractions was performed using ImageJ (National Institutes of Health, Bethesda, MD). Regardless of the presence of membrane or protein aggregates, ∼20% of protein intensity was always found associated with the pellet fraction. We attribute this to the inefficient retrieval of protein adsorbed onto the microfuge tube wall from the S fraction. We therefore defined a “baseline density” to describe this constant fraction.
GTPase assay
GTP hydrolysis rates of full-length Dyn1 and mutants (0.5 μM final) preincubated on LT (150 μM final) for 15 min at room temperature before GTP addition were monitored at 37°C as a function of time using a malachite green–based GTPase assay as described earlier (Leonard et al., 2005). GTP (Sigma-Aldrich) was used at a final concentration of 1 mM.
Supplementary Material
Acknowledgments
We thank Corey Smith for critical comments on the manuscript, Marcin Apostol for atomic force microscopy, Jason Mears for electron microscopy, and Patrick Macdonald for GTPase assays. R.R. was a Special Fellow (3344-09) of the Leukemia and Lymphoma society.
Abbreviations used:
- Dyn
dynamin
- EM
electron microscopy
- FRET
Förster resonance energy transfer
- GUVs
giant unilamellar vesicles
- LT
lipid nanotubes
- MALS
multiangle light scattering
- PH
pleckstrin homology
- PIP2
phosphatidylinositol-4,5-bisphosphate
- SEC
size exclusion chromatography
- VL
variable loop
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-09-0548) on January 29, 2014.
REFERENCES
- Achiriloaie M, Barylko B, Albanesi JP. Essential role of the dynamin pleckstrin homology domain in receptor-mediated endocytosis. Mol Cell Biol. 1999;19:1410–1415. doi: 10.1128/mcb.19.2.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barylko B, Wang L, Binns DD, Ross JA, Tassin TC, Collins KA, Jameson DM, Albanesi JP. The proline/arginine-rich domain is a major determinant of dynamin self-activation. Biochemistry. 2010;49:10592–10594. doi: 10.1021/bi101343p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov PV, Akimov SA, Evseev AI, Schmid SL, Zimmerberg J, Frolov VA. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell. 2008;135:1276–1286. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethoney KA, King MC, Hinshaw JE, Ostap EM, Lemmon MA. A possible effector role for the pleckstrin homology (PH) domain of dynamin. Proc Natl Acad Sci USA. 2009;106:13359–13364. doi: 10.1073/pnas.0906945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia VK, Hatzakis NS, Stamou D. A unifying mechanism accounts for sensing of membrane curvature by BAR domains, amphipathic helices and membrane-anchored proteins. Semin Cell Dev Biol. 2010;21:381–390. doi: 10.1016/j.semcdb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Chappie JS, Dyda F. Building a fission machine—structural insights into dynamin assembly and activation. J Cell Sci. 2013;126:2773–2784. doi: 10.1242/jcs.108845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie JS, Mears JA, Fang S, Leonard M, Schmid SL, Milligan RA, Hinshaw JE, Dyda F. A pseudoatomic model of the dynamin polymer identifies a hydrolysis-dependent powerstroke. Cell. 2011;147:209–222. doi: 10.1016/j.cell.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Morello V, Casella JF, Gounon P, Antonny B. Asymmetric tethering of flat and curved lipid membranes by a golgin. Science. 2008;320:670–673. doi: 10.1126/science.1155821. [DOI] [PubMed] [Google Scholar]
- Faelber K, Held M, Gao S, Posor Y, Haucke V, Noe F, Daumke O. Structural insights into dynamin-mediated membrane fission. Structure. 2012;20:1621–1628. doi: 10.1016/j.str.2012.08.028. [DOI] [PubMed] [Google Scholar]
- Faelber K, Posor Y, Gao S, Held M, Roske Y, Schulze D, Haucke V, Noe F, Daumke O. Crystal structure of nucleotide-free dynamin. Nature. 2011;477:556–560. doi: 10.1038/nature10369. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Crystal structure at 2.2 A resolution of the pleckstrin homology domain from human dynamin. Cell. 1994;79:199–209. doi: 10.1016/0092-8674(94)90190-2. [DOI] [PubMed] [Google Scholar]
- Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol. 2012;13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MG, Jenni S, Nunnari J. The crystal structure of dynamin. Nature. 2011;477:561–566. doi: 10.1038/nature10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustiananda M, Liggins JR, Cummins PL, Gready JE. Conformation of prion protein repeat peptides probed by FRET measurements and molecular dynamics simulations. Biophys J. 2004;86:2467–2483. doi: 10.1016/S0006-3495(04)74303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JA, Hinshaw JE. Dynamins at a glance. J Cell Sci. 2009;122:3427–3431. doi: 10.1242/jcs.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenniston JA, Lemmon MA. Dynamin GTPase regulation is altered by PH domain mutations found in centronuclear myopathy patients. EMBO J. 2010;29:3054–3067. doi: 10.1038/emboj.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DE, Lee A, Frank DW, Marks MS, Lemmon MA. The pleckstrin homology domains of dynamin isoforms require oligomerization for high affinity phosphoinositide binding. J Biol Chem. 1998;273:27725–27733. doi: 10.1074/jbc.273.42.27725. [DOI] [PubMed] [Google Scholar]
- Lai CL, Srivastava A, Pilling C, Chase AR, Falke JJ, Voth GA. Molecular mechanism of membrane binding of the GRP1 PH domain. J Mol Biol. 2013;425:3073–3090. doi: 10.1016/j.jmb.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Frank DW, Marks MS, Lemmon MA. Dominant-negative inhibition of receptor-mediated endocytosis by a dynamin-1 mutant with a defective pleckstrin homology domain. Curr Biol. 1999;9:261–264. doi: 10.1016/s0960-9822(99)80115-8. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem J. 2000;350:1–18. [PMC free article] [PubMed] [Google Scholar]
- Lenoir M, Coskun U, Grzybek M, Cao X, Buschhorn SB, James J, Simons K, Overduin M. Structural basis of wedging the Golgi membrane by FAPP pleckstrin homology domains. EMBO Rep. 2010;11:279–284. doi: 10.1038/embor.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard M, Song BD, Ramachandran R, Schmid SL. Robust colorimetric assays for dynamin's basal and stimulated GTPase activities. Methods Enzymol. 2005;404:490–503. doi: 10.1016/S0076-6879(05)04043-7. [DOI] [PubMed] [Google Scholar]
- Liu YW, Neumann S, Ramachandran R, Ferguson SM, Pucadyil TJ, Schmid SL. Differential curvature sensing and generating activities of dynamin isoforms provide opportunities for tissue-specific regulation. Proc Natl Acad Sci USA. 2011;108:E234–E242. doi: 10.1073/pnas.1102710108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna D, Albanese A, Park WS, Cho W. Mechanistic basis of differential cellular responses of phosphatidylinositol 3,4-bisphosphate- and phosphatidylinositol 3,4,5-trisphosphate-binding pleckstrin homology domains. J Biol Chem. 2007;282:32093–32105. doi: 10.1074/jbc.M703517200. [DOI] [PubMed] [Google Scholar]
- Moens PD, Helms MK, Jameson DM. Detection of tryptophan to tryptophan energy transfer in proteins. Protein J. 2004;23:79–83. doi: 10.1023/b:jopc.0000016261.97474.2e. [DOI] [PubMed] [Google Scholar]
- Montes LR, Alonso A, Goni FM, Bagatolli LA. Giant unilamellar vesicles electroformed from native membranes and organic lipid mixtures under physiological conditions. Biophys J. 2007;93:3548–3554. doi: 10.1529/biophysj.107.116228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlberg AB, Warnock DE, Schmid SL. Domain structure and intramolecular regulation of dynamin GTPase. EMBO J. 1997;16:6676–6683. doi: 10.1093/emboj/16.22.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piston DW, Kremers GJ. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci. 2007;32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Ramachandran R. Vesicle scission: dynamin. Semin Cell Dev Biol. 2011;22:10–17. doi: 10.1016/j.semcdb.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Pucadyil TJ, Liu YW, Acharya S, Leonard M, Lukiyanchuk V, Schmid SL. Membrane insertion of the pleckstrin homology domain variable loop 1 is critical for dynamin-catalyzed vesicle scission. Mol Biol Cell. 2009;20:4630–4639. doi: 10.1091/mbc.E09-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Schmid SL. Real-time detection reveals that effectors couple dynamin's GTP-dependent conformational changes to the membrane. EMBO J. 2008;27:27–37. doi: 10.1038/sj.emboj.7601961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Surka M, Chappie JS, Fowler DM, Foss TR, Song BD, Schmid SL. The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J. 2007;26:559–566. doi: 10.1038/sj.emboj.7601491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Tweten RK, Johnson AE. The domains of a cholesterol-dependent cytolysin undergo a major FRET-detected rearrangement during pore formation. Proc Natl Acad Sci USA. 2005;102:7139–7144. doi: 10.1073/pnas.0500556102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebecchi MJ, Scarlata S. Pleckstrin homology domains: a common fold with diverse functions. Annu Rev Biophys Biomol Struct. 1998;27:503–528. doi: 10.1146/annurev.biophys.27.1.503. [DOI] [PubMed] [Google Scholar]
- Roux A, Koster G, Lenz M, Sorre B, Manneville JB, Nassoy P, Bassereau P. Membrane curvature controls dynamin polymerization. Proc Natl Acad Sci USA. 2010;107:4141–4146. doi: 10.1073/pnas.0913734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim K, et al. Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton's tyrosine kinase. EMBO J. 1996;15:6241–6250. [PMC free article] [PubMed] [Google Scholar]
- Schmid SL, Frolov VA. Dynamin: functional design of a membrane fission catalyst. Annu Rev Cell Dev Biol. 2011;27:79–105. doi: 10.1146/annurev-cellbio-100109-104016. [DOI] [PubMed] [Google Scholar]
- Shnyrova AV, Bashkirov PV, Akimov SA, Pucadyil TJ, Zimmerberg J, Schmid SL, Frolov VA. Geometric catalysis of membrane fission driven by flexible dynamin rings. Science. 2013;339:1433–1436. doi: 10.1126/science.1233920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandberg E, Killian JA. Snorkeling of lysine side chains in transmembrane helices: how easy can it get. FEBS Lett. 2003;544:69–73. doi: 10.1016/s0014-5793(03)00475-7. [DOI] [PubMed] [Google Scholar]
- Strandberg E, Morein S, Rijkers DT, Liskamp RM, van der Wel PC, Killian JA. Lipid dependence of membrane anchoring properties and snorkeling behavior of aromatic and charged residues in transmembrane peptides. Biochemistry. 2002;41:7190–7198. doi: 10.1021/bi012047i. [DOI] [PubMed] [Google Scholar]
- Vallis Y, Wigge P, Marks B, Evans PR, McMahon HT. Importance of the pleckstrin homology domain of dynamin in clathrin-mediated endocytosis. Curr Biol. 1999;9:257–260. doi: 10.1016/s0960-9822(99)80114-6. [DOI] [PubMed] [Google Scholar]
- Wang P, Wang CT, Bai J, Jackson MB, Chapman ER. Mutations in the effector binding loops in the C2A and C2B domains of synaptotagmin I disrupt exocytosis in a nonadditive manner. J Biol Chem. 2003;278:47030–47037. doi: 10.1074/jbc.M306728200. [DOI] [PubMed] [Google Scholar]
- Weber G, Shinitzky M. Failure of energy transfer between identical aromatic molecules on excitation at the long wave edge of the absorption spectrum. Proc Natl Acad Sci USA. 1970;65:823–830. doi: 10.1073/pnas.65.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- Zheng J, Cahill SM, Lemmon MA, Fushman D, Schlessinger J, Cowburn D. Identification of the binding site for acidic phospholipids on the PH domain of dynamin: implications for stimulation of GTPase activity. J Mol Biol. 1996;255:14–21. doi: 10.1006/jmbi.1996.0002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.