The VRK1 protein kinase regulates the phosphorylation of BAF, which binds to dsDNA and LEM domain–containing proteins. VRK1 depletion increases the immobile fraction of BAF at the nuclear periphery and disturbs nuclear envelope architecture. It also leads to the retention of BAF on chromosomes as cells enter and progress through mitosis.
Abstract
Barrier to autointegration factor (BAF), which is encoded by the BANF1 gene, binds with high-affinity to double-stranded DNA and LEM domain–containing proteins at the nuclear periphery. A BANF1 mutation has recently been associated with a novel human progeria syndrome, and cells from these patients have aberrant nuclear envelopes. The interactions of BAF with its DNA- and protein-binding partners are known to be regulated by phosphorylation, and previously we validated BAF as a highly efficient substrate for the VRK1 protein kinase. Here we show that depletion of VRK1 in MCF10a and MDA-MB-231 cells results in aberrant nuclear architecture. The immobile fraction of green fluorescent protein (GFP)–BAF at the nuclear envelope (NE) is elevated, suggesting that prolonged interactions of BAF with its binding partners is likely responsible for the aberrant NE architecture. Because detachment of BAF from its binding partners is associated with NE disassembly, we performed live-imaging analysis of control and VRK1-depleted cells to visualize GFP-BAF dynamics during mitosis. In the absence of VRK1, BAF does not disperse but instead remains chromosome bound from the onset of mitosis. VRK1 depletion also increases the number of anaphase bridges and multipolar spindles. Thus phosphorylation of BAF by VRK1 is essential both for normal NE architecture and proper dynamics of BAF–chromosome interactions during mitosis. These results are consistent with previous studies of the VRK/BAF signaling axis in Caenorhabditis elegans and Drosophila melanogaster and validate VRK1 as a key regulator of NE architecture and mitotic chromosome dynamics in mammalian cells.
INTRODUCTION
Mammalian cells undergo an open mitosis; therefore the faithful segregation of chromosomes first requires disassembly of the nuclear envelope (nuclear envelope breakdown [NEBD]) to allow the replicated chromosomes to interact with the microtubules of the mitotic spindle. The disassembly and subsequent reassembly of the nuclear envelope reflect dramatic intracellular reorganization of proteins and membranes that requires precise orchestration. NEBD is achieved by phosphorylation of the nuclear pore structures and nuclear lamina, which leads to disassembly of protein complexes, as well as by mechanical disruption of the membrane by the mitotic spindle (Burke and Ellenberg, 2002). This process leaves the mitotic chromatin free of membranes while the transmembrane proteins of the NE (NETs) are absorbed into the contiguous endoplasmic reticulum (ER; Anderson and Hetzer, 2007; Puhka et al., 2007). At the end of mitosis, this process is reversed through a wave of dephosphorylation, and two new NEs reassemble around the segregated chromosomes. NE reformation appears to require the chromatin-binding abilities of several protein constituents of the inner nuclear membrane (INM), which function to direct the association of ER tubules with chromatin (Anderson and Hetzer, 2007, 2008).
It is now recognized that the NE is far more than a barrier between the nucleus and the cytoplasm (Gruenbaum et al., 2005; Shimi et al., 2012). Interactions between the lamina, proteins of the NE, and the cytoskeleton position and anchor the nucleus within the cell. Moreover, the arrangement of chromatin within the interphase nucleus and the interaction between chromatin and the lamina have major implications for gene expression and cell function. Inherited mutations in lamina components and NETs, such as lamin A/C, lamin B receptor, and emerin, lead to serious cellular pathologies known as envelopathies (Bengtsson and Wilson, 2004; Gruenbaum et al., 2005; Burke and Stewart, 2013). Patients inheriting these mutations display a clinical spectrum that includes cardiomyopathy, lipodystrophy, muscle pathology, and premature aging (progeria).
Recently a rare autosomal recessive progeria syndrome was linked to a nucleotide substitution in the BANF1 gene, which encodes the barrier to autointegration factor (BAF) protein (Puente et al., 2011). BAF binds with high affinity to double-stranded DNA in a sequence-independent manner (Zheng et al., 2000) and to a subset of NETs that share a common LEM domain (Lap-2, emerin, MAN1; Lee et al., 2001; Lin et al., 2000). These interactions underlie the key role that BAF plays in both nuclear organization and nuclear envelope assembly (Haraguchi et al., 2001, 2007, 2008; Furukawa et al., 2003; Dechat et al., 2004; Margalit et al., 2005). Given the dynamic changes that the NE undergoes during the cell cycle, one would hypothesize that the proposed role of BAF as a bridge between the lamina and the NE must be reversible. Indeed, even in interphase cells, fluorescence recovery after photobleaching (FRAP) analysis shows that BAF is highly mobile (Shimi et al., 2004), suggesting that interactions of BAF with chromatin and NETs may be intermittent. Posttranslational modification of BAF by kinases and phosphatases is a likely mechanism for regulating BAF's interactions with its protein and DNA partners, and BAF has been known for some time to undergo phosphorylation in vivo (Bengtsson and Wilson, 2006).
We determined some years ago that BAF is a high-affinity substrate for the VRK1 protein kinase (Nichols et al., 2006). Phosphorylation of BAF on ser4 abrogates its ability to bind DNA and reduces its affinity for the LEM domain (Nichols et al., 2006). Recently the Caenorhabditis elegans and Drosophila melanogaster VRK1 protein kinases have been identified as important participants in mitotic and meiotic regulation, and the VRK/BAF signaling axis has been shown to be evolutionarily conserved (Gorjanacz et al., 2007; Lancaster et al., 2007). VRK-mediated BAF phosphorylation is important for disassociation of chromosomes from the NE at the onset of mitosis and/or meiosis, a key step in ensuring accurate progression to and through metaphase. When VRK was depleted in these model organisms, BAF (and associated NETs and, perhaps membrane fragments), remained chromatin associated through the early stages of mitosis, jeopardizing faithful chromosome segregation. Moreover, NE assembly at the end of mitosis was incomplete (Gorjanacz et al., 2007; Lancaster et al., 2007).
It is clear that deregulation of the VRK1/BAF signaling axis in these model organisms has devastating effects on NEBD, chromatin release from the NE, faithful progression through mitosis, and NE reassembly. Our goal was to investigate the effect of the VRK1/BAF signaling axis in mammalian cells, which, unlike Drosophila and C. elegans, encode three VRK paralogues. VRK1, a nuclear and enzymatically active kinase, most closely resembles the single VRK paralogue in lower eukaryotes; VRK2 is an active kinase that localizes to the ER (and NE) membranes; and VRK3 is an inactive pseudokinase that localizes to the nucleus (Nichols and Traktman, 2004). Because VRK1 overexpression and signaling has been associated with a variety of cancers and its overexpression is associated with poor clinical outcome in breast cancer patients (Fournier et al., 2006; Martin et al., 2008), we chose to perform our studies on normal (MCF10a) and malignant (MDA-MB-231) mammary epithelial cells. Our approach was to deplete VRK1 using lentiviral-mediated expression of short hairpin RNA (shRNA) and investigate the effect on the morphology of the nuclear envelope during interphase and on NE and chromosome dynamics during mitosis. The work presented here confirms a key role for the mammalian VRK1 kinase in regulating the phosphorylation of BAF and its mobility within the NE and also demonstrates that VRK1 depletion leads to mislocalization of BAF during mitosis and increased frequency of mitotic errors.
RESULTS
Assessment of the intracellular stoichiometry of VRK:BAF
Kinase/substrate interactions can be transient, and our biochemical analyses indicated that VRK1 can phosphorylate a 1000-fold molar excess of BAF to completion in 10 min (Nichols et al., 2006). BAF is known to form complexes with DNA, histones, and NET proteins within the nucleus (Margalit et al., 2007; Berk et al., 2013), and yet FRAP analyses show that the majority of the protein is highly mobile (Shimi et al., 2004; Haraguchi et al., 2008). On the other hand, evidence that BAF and VRK1 can form stable interactions comes from yeast two-hybrid and coimmunoprecipitation analyses (Giot et al., 2003; Gorjanacz et al., 2007). Therefore we wanted to determine the stoichiometric relationship of the two proteins to inform our models of VRK1-mediated regulation of BAF. Lysates of MCF-10a and MDA-MB-231 cells were resolved electrophoretically alongside serially diluted preparations of purified HIS-BAF and 3xFLAG-VRK1. Immunoblot analysis was then performed with antisera to VRK1 and BAF to determine the endogenous levels of both proteins within the cellular lysates (Figure 1). The molar ratio of BAF to VRK1 was calculated to be 5.93:1 in MCF-10a cells and 13.22:1 in MDA-MB-231 cells (we also found a ratio of 9.04:1 in HEK-293 cells; unpublished data). These data indicate that the BAF substrate is only in ∼10-fold excess over the kinase VRK1 in these cell lines.
FIGURE 1:

Determination of the stoichiometric ratio of VRK1 and its substrate BAF in MCF10a and MDA-MB-231 cells. Serial dilutions of purified FL-VRK1 (MW 46K) and His-BAF (MW 10K) were subjected to immunoblot analysis in parallel with whole-cell lysates prepared from MCF-10a and MDA-MB-231 cells. Right, molecular masses (in kilodaltons) of relevant molecular protein standards. We calculated the abundance of each protein within the lysates prepared from each cell type (micrograms of protein/milligram of lysate), as well as the molar ratio of the amount of the two proteins present within the lysates.
VRK1 depletion diminishes BAF phosphorylation, especially at the onset of mitosis
The increase in BAF phosphorylation at mitosis affects its interactions with DNA and LEM domain–containing proteins and can influence its subcellular localization (Hirano et al., 2005; Bengtsson and Wilson, 2006; Nichols et al., 2006; Margalit et al., 2007; Capanni et al., 2010). There is definitive evidence for the phosphorylation of BAF by VRK1 in vitro, and although BAF phosphorylation can be identified by altered electrophoretic mobility (Nichols et al., 2006), the small size of the protein can make quantifying the phosphorylation of endogenous BAF by immunoblot difficult. Fortunately, green fluorescent protein (GFP)- and 3xFLAG-tagged versions of BAF have been used successfully to characterize the dynamic properties of BAF within the cell (Haraguchi et al., 2001, 2007, 2008; Dechat et al., 2004; Margalit et al., 2005; Bengtsson and Wilson, 2006; Nichols et al., 2006). Therefore we used a lentiviral delivery system to stably integrate a 3xFLAG-BAF allele into the target cell genome. Stable integration of the transgene also imparted HYGROR, allowing us to select a pure population of FL-BAF–expressing MCF10a and MDA-MB-231 cells. Using a complementary lentiviral delivery system, we then introduced constructs that imparted PUROR and led to the stable expression of control or VRK1-depleting shRNAs.
BAF is hyperphosphorylated during mitosis, and mitotic phosphorylation on ser4 of BAF, putatively by VRK1, negatively regulates BAF–DNA and BAF–LEM-domain binding (Bengtsson and Wilson, 2006; Nichols et al., 2006; Gorjanacz et al., 2007). To examine the importance of VRK1 to the mitotic phosphorylation of BAF, we analyzed VRK1-depleted MCF-10a FL-BAF and MDA-MB-231 FL-BAF cells not only in asynchronously growing populations, but also after arrest in early mitosis by nocodazole treatment (Figure 2, A and B, lanes 1–3 and 4–6, respectively). Whole-cell lysates were prepared and subjected to immunoblot analysis; phosphorylation of BAF was identified by an electrophoretic mobility shift in the FL-BAF protein (Figure 2, A and B, c), as well as by a validated anti–pBAF-ser4 antibody developed in our lab (Figure 2, A and B, d; Wiebe and Traktman, 2007). As shown in Figure 2Aa, both shVRK1-01 and -02 were very effective at depleting VRK1 (gray arrowhead); on average, >92% depletion was obtained. The effectiveness of the nocodazole treatment (lanes 4–6) is evident by the greatly increased levels of the mitotic marker pH3-S10 (Figure 2Ab). When asynchronous MCF-10a cells were depleted of VRK1, a modest reduction in the levels of the slowly migrating p-FL-BAF was seen when using the anti-FLAG antibody (Figure 2Ac, open diamond; compare lane 1 with lanes 2 and 3); the reduction of phosphorylation was far more evident when the phospho-specific anti-pBAFser4 antibody was used (Figure 2Ad, gray diamond), with a greater reduction seen in cells expressing shVRK1-02 than -01.
FIGURE 2:
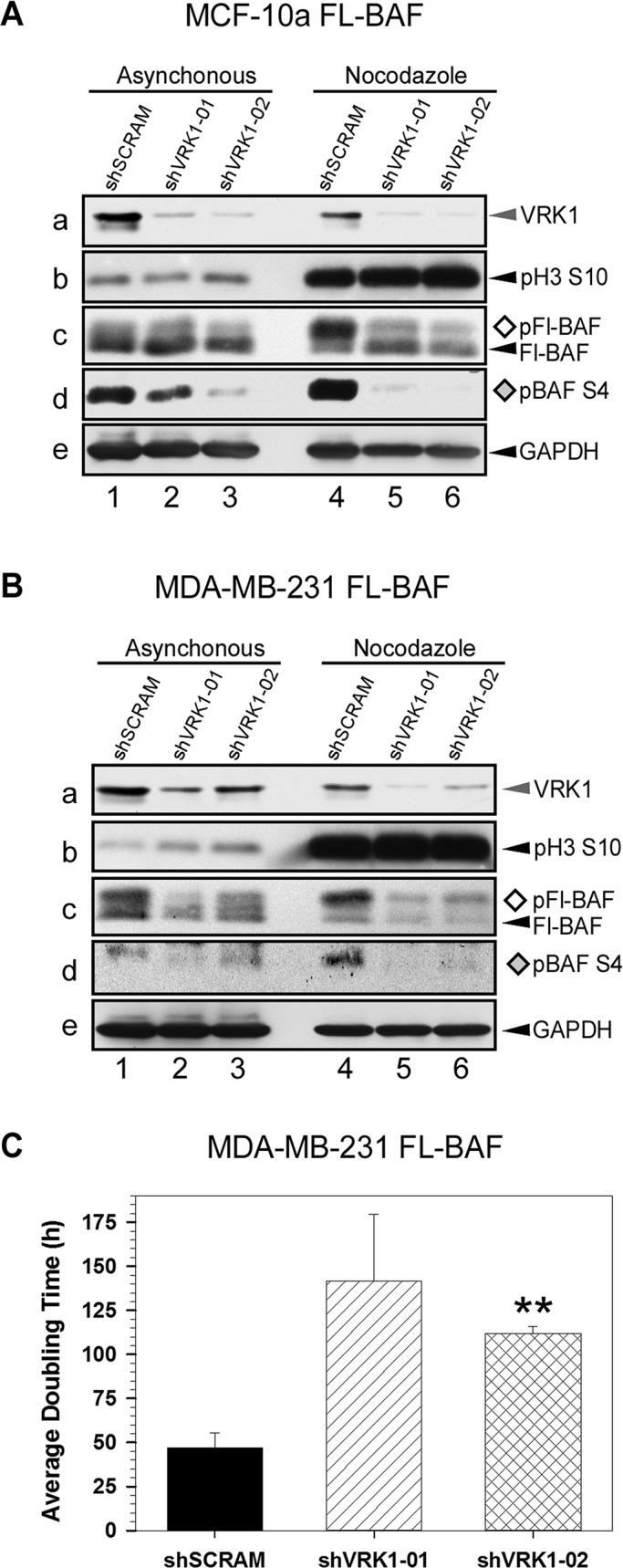
Mitotic BAF phosphorylation is abolished by VRK1-depletion. (A, B) Immunoblot analyses of asynchronous and nocodazole-arrested populations of control and VRK1-depleted cells. MCF-10a (A) and MDA-MB-231 (B) cells were engineered to stably express FL-BAF and subsequently transduced to stably express control (shSCRAM) or VRK1-depleting (shVRK1-01 or -02) shRNAs. Cell lysates were prepared from asynchronous cultures (lanes 1–3) or cells arrested in mitosis by a 16-h treatment with nocodazole (lanes 4–6). Immunoblot analysis was performed with antisera directed against VRK1 (a), phosphohistone H3 (pH3S10) (b), FL-BAF (c), phosphorylated BAF (pBAF S4) (d), and glyceraldehyde-3-phosphate dehydrogenase (e; as loading control). The phosphorylated species of BAF are indicated by ◊ and ♦. (C) Comparison of doubling time of control and VRK1-depleted MDA-MB-231 FL-BAF cells. MDA-MB-231 FL-BAF cells were seeded at equal densities and incubated under normal growth conditions for 170 h. Cell number and viability were determined by flow cytometry, and apparent doubling time was calculated as previously described (n = 3; **p < 0.01; Molitor and Traktman, 2013).
In control cells arrested in mitosis as a result of nocodazole treatment, ∼70% of the total FL-BAF is present in the slower-mobility band (open diamond; lane 4); in cells expressing shVRK1-01 or -02, only ∼40% of the BAF migrates at this position (lanes 5 and 6). The influence of VRK1 depletion on BAF phosphorylation in mitotic cells was seen most dramatically with the phosphospecific antibody (Figure 2Ad; compare lane 4 with lanes 5 and 6). From these data we conclude that VRK1 is a major kinase responsible for BAF phosphorylation during the onset of mitosis in “normal” mammary epithelial cells.
As shown in Figure 2B, MDA-MB-231 FL-BAF cells displayed a similar phenotype upon VRK1 depletion. A modest effect on the levels of phosphorylated BAF was seen in the asynchronous population (lanes 1–3, Figure 2B, c and d), but loss of BAF phosphorylation upon VRK1 depletion was dramatic in nocadazole-treated cells (lanes 4–6, Figure 2B, c and d).
VRK1 depletion significantly increases apparent doubling time of MDA-MBA-231 FL-BAF cells
We previously reported that depletion of VRK1 slowed the growth of both MCF10a and MDA-MB-231 cells. In the latter cell line, we observed a consistent increase in apparent doubling time from ∼46 to ∼ 57 h (∼25% increase) using shVRK-01 or -02 (Molitor and Traktman, 2013). We therefore reexamined the effect of VRK1 depletion on MDA-MB-231-FL-BAF cells, which overexpress BAF approximately fivefold but retain an apparent doubling time of ∼47 h (Figure 2C). Depletion of VRK1 with shVRK-01 or -02 increased the apparent doubling time to 143 and 110 h, respectively (∼169% increase). These data support a key role for the VRK1/BAF axis as a regulator of cell proliferation and suggest that excessive amounts of unphosphorylated BAF are deleterious to cell growth. Our difficulties in generating a cell line that constitutively expresses an unphosphorylatable allele of BAF (BAF-MAAQ: M1TTSQ → M1AAAQ; Nichols et al., 2006) lends further support to this argument. The extreme inflation in apparent doubling time upon VRK1 depletion in BAF-overexpressing cells (Figure 2C) might solely represent the increased duration of the cell cycle, but we cannot rule out the possibility that the number of quiescent cells might also be increased.
Depletion of VRK1 leads to aberrant nuclear envelope morphology in interphase cells
Genetic lesions in lamin A, lamin B, and emerin that disrupt the architecture of the nuclear lamina can result in alterations in gene expression and increased genomic instability. In affected human patients, these laminopathies manifest as accelerated aging (progeria), muscular and skeletal dystrophies, and cardiomyopathy (Broers et al., 2006; Capell and Collins, 2006; Holaska and Wilson, 2006). More recently, a mutation in the BANF1 gene (encoding BAF) was also associated with a progeroid syndrome (Puente et al., 2011). Fibroblasts biopsied from patients carrying this mutation displayed irregular nuclear envelope morphology. We hypothesized that the depletion of VRK1 and the attendant hypophosphorylation of BAF might disrupt the equilibrium of BAF's interactions with chromatin, the lamina, and the INM and thereby alter the architecture of the nuclear periphery. We therefore performed a comparative analysis of the nuclear envelope morphology in MCF-10a and MDA-MB-231 cell lines stably engineered to express either a control shRNA (shSCRAM) or VRK1-targeting shRNAs (shVRK1-01, shVRK1-02).
Asynchronous cultures of these derivatives of MCF-10a and MDA-MB-231 were stained with antibodies to emerin or lamin A/C and fluorescently labeled secondary antibodies. Cultures were examined by indirect immunofluorescence microscopy, and the nuclear morphology was evaluated with scores from 1 to 6: 1, normal; 2, invaginated; 3, highly invaginated; 4, pinched off; 5, blebbed; and 6, multinucleated (Figures 3B and 4B). We scored 300 cells/cell line and determined the statistical significance of changes in distribution among the six categories by the Kolmogorov–Smirnov test. Figure 3A shows representative images from the analysis of envelope morphology in MCF-10a cells. MCF-10a shSCRAM cells displayed a smooth and even envelope morphology when stained for either emerin (Figure 3A) or lamin A/C (Supplemental Figure S1A). In the MCF-10a VRK1-depleted cells, numerous abnormalities in the profile of the nuclear envelope were seen. The red arrowheads indicate aberrant features such as invaginations. A bubble graph of the envelope morphology scoring is shown in Figure 3C. Whereas 60% of the control cells had a normal envelope profile, this value was reduced to 24 and 26% in the two VRK1-depleted cell lines. In contrast, the fraction of cells with highly invaginated envelopes increased from 13% in the control cells to 35 and 34% in the VRK1-depleted cell lines. A similar increase in aberrant envelopes was seen when the cells were stained for lamin A rather than emerin (Supplemental Figure S1). Note that emerin and lamin A/C continue to localize correctly to the envelope upon VRK1 depletion. These data strongly support the conclusion that in interphase cells, depletion of VRK1 with an attendant decrease in BAF phosphorylation has a profound effect on the architecture of the nuclear envelope but does not impair localization of emerin or lamin A/C.
FIGURE 3:
Depletion of VRK1 in MCF-10a cells leads to an increase in aberrant nuclear envelope morphology as assessed by emerin staining. (A) Analysis of nuclear envelope morphology as assessed by immunofluorescent detection of emerin. Asynchronous populations of MCF10a cells expressing control (shSCRAM) or VRK1-depleting (shVRK1-01 or -02) shRNAs were fixed and subject to immunofluorescence microscopy after labeling with an antibody directed against the nuclear envelope constituent emerin as well the DNA-binding dye DAPI. Red arrowheads indicate sites of nuclear envelope aberrations. Scale bars, 50 μm. (B) Scoring system for nuclear envelope morphology. Nuclear envelope morphology was evaluated according to the following criteria: 1, normal; 2, invaginated; 3, highly invaginated; 4, pinched off; 5, blebbed; and 6, multinucleated. Representative images for each category. (C) Quantification of nuclear envelope morphology. The nuclear morphology of 300 cells per cell line was assessed using the scoring guide described in B. Results are presented by bubble graph for emerin; a parallel analysis for lamin A/C staining is shown in Supplemental Figure S1. Statistical significance was determined by the Kolmogorov–Smirnov test (**p < 0.01).
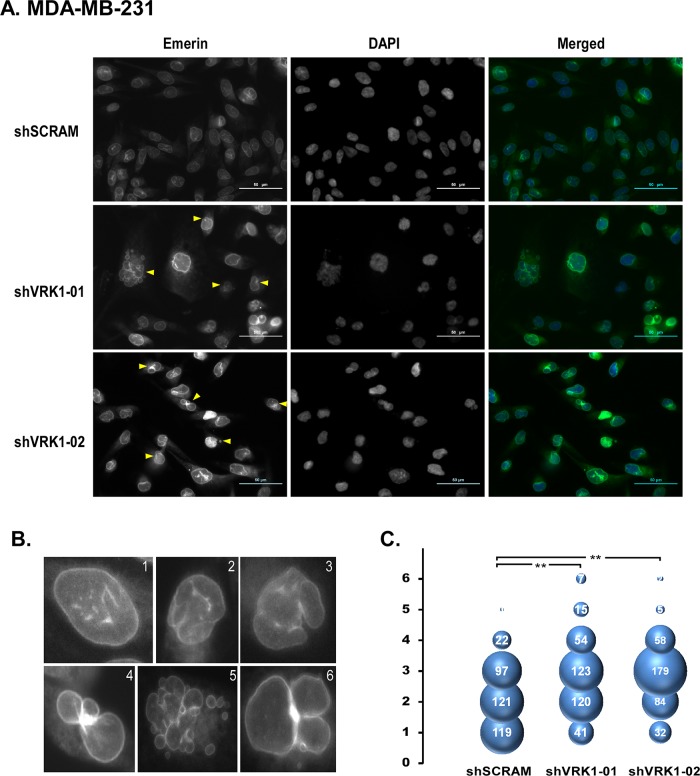
FIGURE 4:
Depletion of VRK1 in MDA-MB-231 cells exacerbates abnormal nuclear envelope morphology as assessed by emerin staining. (A–C) MDA-MB-231 cells expressing control (shSCRAM) or VRK1-depleting (shVRK1-01 or -02) shRNAs were stained with anti-emerin and DAPI, and the nuclear envelope morphologies of 360 cells within each cell line were scored and graphed as described in Figure 3.
Control MDA-MB-231 cells have a more abnormal nuclear envelope phenotype, consistent with the association of irregular envelopes with the highly transformed and malignant state (Pienta and Coffey, 1991; Nickerson, 1998). To determine whether depletion of VRK1 was capable of exacerbating the already abnormal nuclear envelope morphology seen in MDA-MB-231 cells, we performed the comparable analysis described for MCF10a cells, except that we counted 360 cells for each of the three cell lines. Nuclear envelope invaginations were commonplace in MDA-MB-231 shSCRAM cells (Figure 4A), but 33% of the cells exhibited a smooth nuclear envelope profile (bubble plot, Figure 4C). This population was reduced to 11 and 9% in the VRK1-depleted cells, and the frequency of severe aberrations also increased upon VRK1 depletion. More cells displayed nuclear blebbing and multinucleation (Figure 4A, red arrows). Comparable results were obtained when the cells were stained for lamin A/C (Supplemental Figure S1B). Once again, note that both emerin and lamin A/C were still localized at the nuclear envelope after VRK1 depletion, despite the increase in aberrant nuclear morphology (Figure 4 and Supplemental Figure S1).
VRK1 depletion increases the immobile fraction of GFP-BAF in interphase cells
The nuclear pool of BAF interacts with LEM domain–containing proteins, which are stably embedded in the INM, as well as with core histone H3 and DNA (Shimi et al., 2004; Montes de Oca et al., 2005; Margalit et al., 2007). Nevertheless, FRAP analysis shows that the nuclear pool of GFP–BAF is highly mobile during interphase (Shimi et al., 2004; Haraguchi et al., 2008), suggesting that BAF's interactions with its many partners are likely to be transient and reversible. Posttranslational modification of BAF on its amino terminus, such as is regulated by VRK1, can be presumed to play a key role in regulating the duration of these interactions (Bengtsson and Wilson, 2006; Nichols et al., 2006). We used FRAP analysis to assess the effect of VRK1 depletion on the mobility of BAF at the nuclear periphery in interphase cells. MCF-10a cells were engineered to stably express a GFP-BAF fusion protein that was used in previous studies (Haraguchi et al., 2008). As expected, GFP-BAF was present in both nuclear and cytoplasmic pools but was enriched at the nuclear envelope (Shimi et al., 2004; Haraguchi et al., 2008). Transduction of these cells with control (shSCRAM-expressing) or VRK1-depleting (shVRK1-01 and -02 expressing) lentiviruses led to the generation of three cell lines that were then used in FRAP analysis. For each population we photobleached 15 randomly chosen cells within a 2.2-μm2 strip that spanned the nuclear envelope lengthwise, as depicted by the white box in Figure 5A, and measured the rate of recovery of the GFP-BAF signal.
FIGURE 5:
Depletion of VRK1 increases the immobile fraction of GFP-BAF in interphase cells. (A) MCF-10a cells stably expressing GFP-BAF were transduced to express control (shSCRAM) or VRK1-depleting (shVRK1-01 or -02) shRNAs. FRAP analysis was performed on 15 randomly selected cells from each population. Whole-cell images are shown over the enlarged version of the bleached area, which is indicated by the white box. Cells were imaged before bleaching (prebleach) and at 0.1-s intervals after photobleaching for 25 frames, then at 0.5-s intervals for 6 frames, and then at 2-s intervals for 15 frames. (B, C) Fluorescence intensity was measured at the nuclear periphery, where the BAF-interacting LEM domain–containing proteins reside. For each cell line, the relative fluorescence intensity of the 15 cells at each time point was averaged and plotted over time. Right, a closer view of the initial 5-s postbleach. Error bars, SEM. The red line represents empirical modeling of recovery kinetics as described by Ellenberg et al. (1997).
Representative images taken before bleaching and at 0.125, 0.5, 1, 5, 10 and 20 s postbleaching are shown for each population. Whereas the GFP-BAF signal within the bleached area in the control cells appeared to recover fully within <10 s, recovery was impaired in the VRK1-depleted cells. Figure 5B, left, is a graphical representation of recovery kinetics during 35 s of postbleach analysis; Figure 5B, right, presents a closer view of the first 5 s. In the control (shSCRAM-expressing) cells, recovery was rapid and the immobile fraction was very low (<5%), confirming that GFP-BAF at the nuclear envelope is highly mobile. Although the bleached area within both VRK1-depleted populations appeared to recover with similar kinetics, the immobile fraction of GFP-BAF was increased dramatically (∼20 and ∼30% for shVRK1-01 and -02, respectively). Table 1 contains a quantitative comparison of the t½ of recovery (in seconds), the immobile fraction (%), and diffusion coefficients for each population. Again, the diffusion coefficients calculated using two distinct methods indicate similar kinetics of recovery for GFP-BAF in all three cell populations, whereas the fraction of GFP-BAF that is immobile is clearly elevated significantly in the VRK1-depleted populations. These data provide strong evidence that VRK1-mediated phosphorylation is an important determinant of the fraction of BAF that is mobile at the nuclear envelope of interphase cells.
TABLE 1:
Comparison of FRAP recovery halftimes, immobile fractions, and diffusion coefficients for GFP-BAF in VRK1-depleted and control cells.
| shSCRAM | shVRK1-01 | shVRK1-02 | |
|---|---|---|---|
| Half-time (t1/2; s) | 1.15 ± 0.02 | 1.13 ± 0.05 | 1.392 ± 0.07 |
| Immobile fraction (%) | 5.89 ± 0.51 | 19.89 ± 0.81 | 31.13 ± 1.28 |
| Diffusion coefficient, Axelrod (μm2/s) | 0.17 ± 0.02 | 0.17 ± 0.05 | 0.14 ± 0.07 |
| Diffusion coefficient, Ellenberg (μm2/s) | 0.56 ± 0.01 | 0.75 ± 0.01 | 0.45 ± 0.02 |
VRK1 depletion leads to atypical association of BAF with mitotic chromosomes
We have shown that VRK1 depletion leads to a reduction in BAF phosphorylation during interphase that is associated with aberrant nuclear envelope morphology and an increase in the immobile fraction of BAF (Figures 2–5). In control cells, the level of BAF phosphorylation increases acutely and dramatically at the onset of mitosis, coincident with NEBD and the association of chromosomes with the mitotic spindle. This mitotic wave of BAF phosphorylation is strongly diminished by VRK1 depletion (Figure 2). To investigate how VRK1 depletion and the persistence of unphosphorylated BAF might affect the progression of mitosis, we used live-cell imaging of MCF-10a GFP-BAF cells. Monolayers of cells engineered to express a control (shSCRAM) or VRK1-depleting (shVRK1-02) shRNA were arrested in mitosis by nocodazole treatment, released into standard media, and analyzed by live-cell imaging at 2-min intervals for 2 h. For each cell population, 10 fields were imaged at each time point, and 40 cells that transited mitosis were examined in detail. Representative still images are shown in Figure 6; in each case, metaphase is set as 0 s, and times before and after metaphase are indicated appropriately.
FIGURE 6:
GFP-BAF remains chromatin associated throughout mitosis in VRK1-depleted populations. MCF-10a cells stably expressing GFP-BAF were transduced to express control (shSCRAM) or VRK1-depleting (shVRK1-01 or -02) shRNAs. Live-cell imaging was performed on cells synchronized by treatment with nocodazole and then released from mitotic arrest. Metaphase is represented by time zero in all images. (A) Representative images from recordings of mitoses in the control MCF-10a GFP-BAF shSCRAM cells demonstrate the exclusion of GFP-BAF from chromatin during prophase, metaphase, and early anaphase and recruitment to chromatin during late anaphase/telophase. (B) Additional images of MCF-10a GFP-BAF shSCRAM at metaphase, showing exclusion of BAF from the metaphase plate. (C, D) MCF-10a GFP-BAF cells expressing shVRK1-01 (C) or -02 (D) cells demonstrate the localization of GFP-BAF to mitotic chromosomes from prophase through telophase. (E) Forty mitoses were scored for each cell line, and the percentage of cells demonstrating chromosome-localized GFP-BAF chromatin is represented graphically. Statistical significance was determined by the χ2 test; a statistically significant difference between the control cells and the VRK1-depleted cells (***p < 0.001) was found for prophase, metaphase, and anaphase.
Figure 6A illustrates the dynamics of GFP-BAF localization in control cells (shSCRAM). After nuclear envelope dissolution (between −600 and −400 s), the GFP-BAF signal became diffuse and was clearly excluded from the chromatin at the metaphase plate (0, 200 s) and during early anaphase (400 s). Additional images of cells in metaphase, showing lack of BAF staining at the metaphase plate, are shown in Figure 6B. BAF rapidly localized to chromatin at late anaphase/early telophase (600 s). The dynamics we observed for these control MCF-10a cells parallels what was previously shown in HeLa cells using similar techniques (Haraguchi et al., 2008), in which BAF was seen to associate with mitotic chromosomes 5 min after the onset of anaphase.
The dynamics of GFP-BAF localization was quite different in both VRK1-depleted populations (Figures 6, C and D). In a significant fraction of the cells, GFP-BAF remained chromatin associated throughout mitosis. After NEBD, although some of the GFP-BAF was spread diffusely throughout the cell, much of it coalesced on the condensed chromatin and was clearly visible on the chromosomes during prophase (Figure 6C, shVRK1-01, −1200 s onward; Figure 6D, shVRK1-02, −600 s onward). BAF-GFP was clearly associated with the metaphase plate (0 s) in both shVRK1-01 and shVRK1-02 populations (Figure 6, C and D, respectively) and remained there during anaphase. At telophase, all of the cellular BAF was found on the chromosomes, as was true in the control cells. Also note that many, but not all, of the VRK1-depleted cells exhibited a slower transit from prophase to cytokinesis (e.g., compare the timing of Figure 6, A and C).
To quantify BAF localization, mitotic cells in telophase were identified by the exclusive localization of BAF on the chromatin. The live-imaging movies for each of these cells were then reviewed, and the association of BAF with chromatin was monitored during all phases of mitosis. The dynamics of BAF localization on 40 cells within each population is represented graphically in Figure 6E. In control cells, GFP-BAF was never observed on chromatin until anaphase, and even then was detected in only ∼20% of the cells at that stage of mitosis. However, in VRK1-depleted populations, GFP-BAF was associated with chromosomes in ∼40% of cells in prophase, ∼50–60% of cells in metaphase, and 70–80% of cells in anaphase. GFP-BAF was bound to telophase chromosomes in 100% of both control and VRK1-depleted cells. In sum, these data uncover a profound difference in the dynamics of BAF localization when VRK1 is depleted and indicate that, after NEBD, BAF remains bound to the chromosomes as they interact with the mitotic spindle and undergo congression and sister chromatid separation.
It would be reasonable to propose that the retention of BAF on mitotic chromosomes might confound their ability to undergo proper interactions with the spindle. Indeed, lagging chromosomes and anaphase bridges were frequently observed in VRK1-depleted populations (55 and 30% of mitoses for shVRK-01 and -02, respectively). Because DNA dyes designed for live-cell imaging were toxic in our system, we were unable to accurately quantify the number of lagging chromosomes in the control cells, in which the metaphase and anaphase chromosomes were not made visible by bound GFP-BAF. We were, however, able to score and quantify cells at telophase that exhibited tripolar (triplet) spindles and found that this frequency increased from 0.25% of control cells to 12.5 and 17.5% for shVRK1-01 and -02 cells, respectively (unpublished data).
DISCUSSION
Of the three VRK paralogues encoded within the mammalian genome, the nuclear VRK-1 protein is most similar to the single VRK gene present in the genomes of D. melanogaster (NHK-1) and C. elegans (Ce-VRK). Several substrates have been reported for VRK-1, but the VRK1-BAF signaling axis appears to be the most highly conserved among these diverse eukaryotes. BAF plays key roles in regulating the architecture of the nuclear lamina and the disassembly and reassembly of the NE at mitosis (Nichols et al., 2006; Gorjanacz et al., 2007; Lancaster et al., 2007). Phosphorylation of BAF regulates its interaction with both DNA and LEM domain–containing proteins such as emerin (Bengtsson and Wilson, 2006; Nichols et al., 2006). Here we show conclusively that VRK1 is the principal, if not the only, kinase responsible for the wave of BAF phosphorylation that accompanies the onset of mitosis in MCF10a and MDA-MB-231 cells (Figure 2). The levels of VRK1 do not appear to vary during the cell cycle (unpublished data), but the dramatic increase in VRK1-mediated phosphorylation of BAF at the onset of mitosis may reflect release from inhibitory proteins, such as the histone variant macroH2A1.2 (Kim et al., 2012a), or reduced competition from counteracting phosphatases (Asencio et al., 2012).
Mutations that affect components of the nuclear lamina cause syndromes known as laminopathies or envelopathies (Bengtsson and Wilson, 2004; Gruenbaum et al., 2005; Burke and Stewart, 2013). At a cellular level, these defects cause aberrant nuclear architecture and altered patterns of gene expression; at an organismal level, the defects range from muscular dystrophies to premature aging (progeria). Given that BAF is one of the proteins whose mutation can cause an envelopathy (Cabanillas et al., 2011; Puente et al., 2011), and since its function is known to be regulated by posttranslatonal modification, we examined whether the phosphorylation of BAF would affect the architecture of the nuclear envelope (Figures 3 and 4). Depletion of VRK1 and the consequent reduction in BAF phosphorylation caused abnormalities in the nuclear envelope of interphase cells (Figures 2 and 3 and Supplemental Figure S1). Moreover, depletion of VRK1 from these cells, which express only endogenous BAF, has a deleterious effect on cell proliferation, increasing the apparent doubling time from ∼46 to 57 h (Molitor and Traktman, 2013).
A clue to the cause of these abnormalities came from our FRAP analyses of GFP-BAF–expressing cells (Figure 5). Consistent with previous findings (Shimi et al., 2004; Haraguchi et al., 2008), we found that BAF is highly mobile within the nuclear envelope. Their observed t1/2 of recovery of 270 ms prompted the “touch and go” model, in which BAF forms frequent but short-lived interactions with its DNA- and protein-binding partners during interphase. We showed that depletion of VRK1 did not change the recovery rate of the mobile fraction of BAF (t1/2 = 1.15 s for control MCF10a/GFP-BAF cells and 1.13 and 1.39 s for VRK-1 depleted cell lines) but raised the immobile fraction from ∼5% of the total BAF pool in control cells to 20–30% in VRK1-depleted cells. We propose that, during interphase, BAF phosphorylation is dynamic due to the action of VRK1 and other kinases, as well as of opposing phosphatases, and that this dynamic phosphorylation allows BAF to make transient contacts with its binding partners. When VRK1, one of the kinases responsible for BAF phosphorylation in interphase, is depleted, this equilibrium is disturbed. A significantly increased fraction of BAF remains unphosphorylated, extending the duration of the interactions between BAF and its DNA or protein partners. The altered duration of these interactions may be largely responsible for the distorted structure of the nuclear envelopes seen in VRK1-dpeleted MCF10a and MDA-MB-231 cells (Figures 2 and 3 and Supplemental Figure S1). It is of course possible that VRK1 is also responsible for phosphorylating other proteins at the nuclear periphery and that reducing their phosphorylation may also disturb the architecture of the nuclear envelope. In this regard, a fuller appreciation of VRK1’s substrates is an important goal.
The aberrant NE profile seen upon depletion of VRK1 resembles that seen in cells bearing certain lamin A/C or emerin mutations. In addition to the altered appearance of the NE, cells carrying these mutations have altered distributions of heterochromatin and altered profiles of gene expression (Wilson, 2000; Wilson et al., 2005; Somech et al., 2005; Capell and Collins, 2006; Chi et al., 2009; Taimen et al., 2009; Cox et al., 2011; Burke and Stewart, 2013). BAF interacts with several transcription factors, as well as with chromatin, and its mutation or absence can have a serious effect on gene expression (Wang et al., 2002; Holaska et al., 2003; Holaska and Wilson, 2006; Cox et al., 2011). Therefore investigations of whether reduced VRK1-mediated phosphorylation of BAF also alters heterochromatin disposition within the nucleus and leads to modified profiles of gene expression will be important for future study. It is also to be expected that the aberrant prolongation of BAF-DNA and BAF-LEM interactions at the onset of mitosis will cause defects in NEBD. Future thorough study of NEBD is therefore also important.
Our analyses of mitotic cells uncovered another important component of the phenotype engendered by VRK1 depletion (see schematic in Figure 7). In a normal mitosis (Figure 7, top), the NE with its transmembrane proteins is absorbed into the ER, and BAF remains dispersed throughout the cell until after the start of anaphase. On VRK1 depletion, however, BAF remains on the chromosomes from NEBD all the way through mitosis (Figure 7, bottom). This is a profound change and is highly analogous to the observed phenotype in a C. elegans strain carrying a mutated VRK allele (Gorjanacz et al., 2007). The retention of BAF on mitotic chromosomes raises the question of whether the LEM domain–containing proteins, and even fragments of the NE, are also bound to the periphery of these chromosomes. In C. elegans, small interfering RNA–mediated VRK depletion led to the retention of emerin and LEM-2 on mitotic chromosomes along with BAF (Gorjanacz et al., 2007). Recently the natural product obtusilactone B was shown to bind to BAF and prevent its phosphorylation by VRK1; when A549 cells were treated with this compound, lamin A/C was aberrantly retained on mitotic chromosomes (Kim et al., 2012b). These data are provocative and suggest that upon VRK1 depletion, mitotic chromosomes may in fact be covered with fragments of the lamina and the nuclear membranes (Figure 7, bottom). Determining whether NE reassembly at the end of mitosis is therefore aberrant and whether the mitotic defects contribute to the altered NE structure seen in interphase cells are important questions for future study.
FIGURE 7:
Schematic model of the phenotypic effect of VRK1 depletion on mitosis. The localization of BAF and VRK1 for wild-type (WT) and VRK1-depleted chromosomes is shown schematically. Note the absence of BAF from prophase and metaphase chromosomes in WT cells and the presence of chromosome-bound BAF during these stages in VRK1-depleted cells. We hypothesize that BAF will remain bound to LEM/NET proteins in VRK1-deficient mitotic cells and that these proteins, as well as NE remnants or ER membranes, may also remain with mitotic chromosomes. This scenario could lead to difficulties in separating these membranes during late anaphase/telophase and during reassembly of the NE. Note that during interphase, the NE of VRK1-deficient cells has an irregular profile, and we have drawn an increased number of LEM/NET- BAF-NE bridges to reflect a greater immobile fraction of BAF. The legend is given at the bottom of the figure.
The altered chromosome dynamics during mitosis that we observed upon VRK1 depletion raises the question of whether VRK1-mediated BAF phosphorylation is a determinant of genome fidelity. It seems intuitive that retention of BAF, LEM domain–containing proteins, lamins, and perhaps membrane fragments on the periphery of mitotic chromosomes might impair congression at the metaphase plate and separation at anaphase. Indeed, our live-imaging analyses of control and VRK1-depleted MCF10-GFP/BAF cells revealed an increased number of lagging chromosomes and anaphase bridges. We performed karyotypic analysis of our control and VRK1-depleted cells (20 d post lentiviral transduction), and in one study we observed the expansion of a subpopulation of VRK1-depleted cells that exhibited an altered genotype (Supplemental Figure S2). These data suggest that VRK1 deficiency might promote aneuploidy and chromosome breakage/rejoining. However, because we examined the cultures several weeks after VRK1 depletion, these analyses are likely to underestimate the effect on genomic instability, since only viable cells will survive and expand sufficiently for identification in karyotype analysis. Additional studies that can uncover ongoing chromosome instability should be undertaken. We also noted that the frequency of triplet spindles was increased in VRK1-depleted cells. Previously, mitotic defects, including multipolar spindles, chromosomes with unattached kinetochores, and lagging chromosomes, were associated with null alleles of the Drosophila VRK (Cullen et al., 2005). It will therefore be of interest to determine whether VRK1 phosphorylates components of the centrosome or the kinetochore and whether its absence causes an activation of the spindle assembly checkpoint due to impaired interactions with centromeres and kinetochores.
In sum, the results presented here and summarized schematically in Figure 7 solidify the importance of VRK1 in regulating several aspects of nuclear structure and function and validate the conclusion that the C. elegans and D. melanogaster VRK enzymes and the mammalian VRK1 enzyme share many of the same functions. VRK1 depletion and the attendant reduction in BAF phosphorylation cause a NE phenotype that is reminiscent of that seen in several nuclear envelopathies that result from mutations in lamin A/C and emerin. Moreover, VRK1 depletion leads to the retention of BAF (and perhaps other NE components) on mitotic chromosomes, which has significant implications for genomic fidelity.
MATERIALS AND METHODS
Cell culture
All cells were cultured at 37°C in the presence of 5% CO2. Human mammary epithelial cell lines MCF10a (CRL-10317) and MDA-MB 231 (HTB-26) were obtained from the American Tissue Culture Collection (Rockville, MD). MCF-10a cells were maintained in DMEM/F12 (1:1) plus l-glutamine (Gibco, Life Technologies, Carlsbad, CA) supplemented with 5% horse serum, hydrocortisone (500 ng/ml), cholera toxin (100 ng/ml), insulin (10 μg/ml), and epidermal growth factor (10 ng/ml). Stable cell lines generated from these parental lines by lentiviral transduction were maintained as described with the addition of either puromycin (7.5 μg/ml) or hygromycin (50 μg/ml). Cell viability and enumeration was performed as previously described (Molitor and Traktman, 2013). We used the equation 2x = nfinal/nstart to determine the number of doublings that occurred during incubation, where x equals the number of doublings. The number of hours of growth (h) was then divided by the number of doublings (x) to give the apparent doubling time of the population. For synchronization experiments, cells were arrested at the G2/M border by incubation with 400 nm nocodazole (Sigma-Aldrich, St. Louis, MO) for 16 h. For immunoblot analyses, nocodazole-treated cells were subjected to a mitotic shakeoff to retrieve the nonadherent mitotic cells and leave any nonsynchronized cells on the dish.
BAF and VRK1 purification
Recombinant WT BAF was prepared from the insoluble fraction of induced cultures of Escherichia coli essentially as described (Lee and Craigie, 1998; Umland et al., 2000), except that, after denaturing, Ni2+-agarose purification, and removal of the amino-terminal hexahistidine tag with thrombin, BAF dimers were purified by gel filtration on Sephacryl S100 (Amersham, Piscataway, NJ) and concentrated with Centriprep-10 filtration units (Millipore, Bedford, MA; Nichols et al., 2006). For 3xFLAG-VRK1 purification, 293TRex-Fl VRK1 cells (Nichols et al., 2006) were incubated for 48 h with 1 μg/ml tetracycline. Cells were scraped and Dounce homogenized on ice in FLAG lysis buffer (FLB; 20 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 2 mM MgCl2, 25 U/ml Benzonase, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg/ml leupeptin, 1 μg/ml pepstatin). The lysate was brought to 500 mM NaCl, clarified (600 × g, 10 min), and incubated with anti-FLAG M2 Affinity gel (A2220; Sigma-Aldrich) at 4°C overnight with end-over-end mixing. Bound Fl-VRK1 was eluted from the beads by incubation with 3xFLAG peptide (F4799; Sigma-Aldrich) for 1 h with end-over-end mixing at 4°C. Eluted Fl-VRK1 was stored in FLB supplemented with 5 mM EDTA, 5 mM dithiothreitol, and 15% glycerol. Quantification of purified wild-type BAF and Fl-VRK1 was performed by imaging and quantifying silver-stained gels containing the purified protein preparations and a dilution series of bovine serum albumin (BSA) on the FluorChemHD2 Imaging System (Protein Simple, Santa Clara, CA).
VRK1-BAF stoichiometry
To determine the stoichiometric ratio of VRK1 and its substrate BAF, purified preparations of each protein were subjected to immunoblot analysis in parallel with whole-cell lysates. Serial dilutions of HIS-BAF and FL-VRK1 (10–100 ng) were resolved along with 50 and 100 μg of whole-cell lysate prepared from asynchronous cultures of MCF10a and MDA-MB-231 cells. Immunoblot analysis was performed with the α-BAF and α-VRK1 antibodies described. Quantification of the signals was obtained on the FluorChemHD2 Imaging System, and micrograms of protein/milligram of lysate was calculated based on a comparison of the immunoblot signal obtained from the cell lysates and the titration of the purified standards. The molar ratio of BAF to VRK1 was derived from the mass ratio using the molecular weights 10,059 and 45,476 g/mol for BAF and VRK1, respectively.
Lentiviral construction and transduction for shRNA expression
The pLL 3.7 LentiLox system (Addgene, Cambridge, MA) was used to generate replication-deficient lentiviral particles for stable transduction of shRNA. shRNA sequences inserted into the transfer vector pLL 3.7-PUROR were as follows, with uppercase letters signifying target sequence within the VRK1 transcript: VRK1-01 sense, 5′-tGAAAGAGAGTCCAGAAGTAttcaagagaTACTTCTGGACTCTCTTTCtttttggaa ac-3′; VRK1-02 sense, 5′-tGCAAGGAACCTGG TGTTGAttcaagagaTCAACACCAGGTTCCTTGCtttttggaaac-3′; and SCRAM sense, 5′-tCAGTCGCGTTTGCGACTGGttcaagagaCCAGTCGCAAACGCGACTGtttttggaaac-3′.
For construction of the viral particles, 2 μg of the helper plasmids pRRE and pRSV-REV and 4 μg of pVSVG along with 12 μg of the pLL 3.7-PUROR transfer vector were transfected into low-passage 293T cells. The viral inoculum was collected from the cells 48 h posttransfection, and target cells were transduced with 1 ml of the viral inoculum in the presence of Polybrene (8 μg/ml; Sigma-Aldrich) for 3–6 h at 37°C. The standard medium was replaced, and cells were selected with puromycin (7.5–10 μg/ml) 36–48 h later. Assays were performed on transduced lines from 5–10 d postinfection.
Construction of lentivirus for stable transduction of transgenes
A variant of the pHAGE lentiviral system (Mostoslavsky et al., 2006; Schneider and Hudson, 2011; generous gift of Amy Hudson, Medical College of Wisconsin, Milwaukee, WI) containing a multiple cloning site (MCS) was used to generate replication-deficient viral particles for stable transduction of cDNA. Transfer vectors included either a puromycin resistance cassette, pHAGE-PUROR-MCS (PPM), or a hygromycin-resistance cassette, pHAGE-HYGR-MCS (PHM). The helper plasmids HIV Tat (0.52 μg), HIV Rev (0.52 μg), MLV gag/pol (0.52 μg), and VSVG (1.04 μg) were transfected (Lipofectamine 2000; Invitrogen, Carlsbad, CA) into low-passage 293T cells along with 10 μg of PPM or PHM. Viral harvest, transduction, and selection were performed as described for the LentiLox system. PHM-transduced cells were selected with 250–500 μg/ml hygromycin. This system was used to generate cells that express N′-terminally 3xFLAG-tagged BAF or a GFP-BAF fusion protein as previously described (Bengtsson and Wilson, 2006; Nichols et al., 2006).
Cell lysis and immunoblot analysis
Cell lysates were prepared by homogenizing cell pellets in lysis buffer (100 mM NaPO4, 100 mM NaCl, 1% [vol/vol] Triton X-100, 1.0% [vol/vol] SDS, 0.5% deoxycholate, Benzonase nuclease [25 U/ml; Sigma-Aldrich]) supplemented with protease inhibitors (1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin) and phosphatase inhibitors (10 mM sodium fluoride, 1 mM sodium orthovanadate, 5 nm calyculin A). Protein quantification was performed using the BCA Protein Assay (Pierce, Thermo Scientific, Rockford, IL), and equal protein quantities were resolved by SDS–PAGE for immunoblot analysis. Commercial antibodies used in this study include pH3-S10 (06-570; Upstate, Millipore), glyceraldehyde-3-phosphate dehydrogenase (5019A-2; Imgenex, San Diego, CA), and VRK1 (HPA000660; Sigma-Aldrich). In addition, a rabbit polyclonal antiserum was generated against a peptide corresponding to amino acids 4–20 of human BAF, as well as an antibody specific for BAF modified by phosphorylation on Ser-4 (produced for us by Bethyl Laboratories, Montgomery, TX; Wiebe and Traktman, 2007). Secondary antibodies were conjugated to horseradish peroxidase, and blots were developed with the SuperSignal West Pico Chemiluminescence Substrate (Pierce) and quantitated on a FluorChemHD2 Imaging System.
Assessment of nuclear envelope morphology
MCF-10a and MDA-MB-231 cell lines were engineered to express a control shRNA (shSCRAM) or one of two shVRK1s directed against VRK1 (shVRK1-01 or -02) as described. The cells were cultured under normal conditions until ∼70% confluence was reached. The cells were then fixed for 15 min with 4% paraformaldehyde and 4% sucrose and permeabilized with 0.2% Triton-X for 5 min on ice. Samples were incubated with antibodies directed against the nuclear envelope constituent emerin (SC15378; Santa Cruz Biotechnology, Santa Cruz, CA) or lamin A/C (MAB3211; Millipore) in a 3% BSA/phosphate-buffered saline (PBS) solution. After overnight incubation the samples were washed three times with BSA/PBS and incubated with secondary antibodies conjugated to Alexa Fluor 488 or 596. DNA was visualized by staining with 4′,6′-diamidino-2-phenylindole (DAPI). In each population of cells the envelope morphology was scored from 1 to 6: 1, normal; 2, invaginated; 3, highly invaginated; 4, pinched off; 5, blebbed; 6, multinucleated. From 300 to 400 cells were scored per line in a blinded manner, and statistical significance was determined by the Kolmogorov–Smirnov test.
Fluorescence recovery after photobleaching
For FRAP experiments, MCF10a stable cell lines expressing GFP-BAF under HYGROR selection were transduced with lentiviruses that imparted PUROR and expressed either control (shSCRAM) or VRK1-targeting shRNAs (shVRK1-01 or -02). Interphase cells were imaged on the Leica TCS SP5 broadband confocal microscope system using the Leica HC PL APO 63×/1.4 oil immersion lens (Leica, Wetzlar, Germany). Cells were bleached by 488-nm light from an argon laser in a 2.2-μm2 strip localized at the nuclear periphery of the cell. Images were scanned before and after photobleach (bidirectional scanning, 1400 Hz, 256 × 256 resolution) every 0.1 s for 25 frames, then every 0.5 s for 6 frames, and then every 2 s for 15 frames. To correct for bleaching during monitoring, relative intensity (Irel) was calculated for each time point according to Dundr and Misteli (2003) and plotted over time. Recovery kinetics was modeled by single-exponential association, and diffusion coefficient (D) was calculated according to D = 0.88(r2/4t1/2) (Axelrod et al., 1976). Alternatively, D was calculated empirically according to I(t) = Ifinal {1 – [w2(w2 + 4πDt)−1]1/2}, where w (0.95 μm) is the width of the bleached area (Ellenberg et al., 1997). Kinetic modeling was performed on the mean of 15 cells per shRNA-expressing population.
Live-cell imaging
MCF10a stable cell lines expressing GFP-BAF under HYGROR selection were transduced with lentivirus that imparted PUROR and expressed shSCRAM, shVRK1-01, or shVRK1-02. We plated 1 × 105 cells on glass-bottom 35-mm dishes with a 20-mm microwell (MatTek, Ashland, MA) and incubated them for 24 h. Cells were then treated with 400 ng/ml nocodazole for 16 h and released for imaging. Live cells were imaged every 2 min for a total of 2 h using the Tokai Hit INU Stage Top Incubator (Fujinomiya-shi, Shizuoka-ken, Japan) for a Nikon Eclipse Ti microscope (Kawasaki, Kanagawa, Japan) with perfect focus. The Nikon Ti-S-EJOY motorized staging system permitted parallel imaging of control and VRK1-knockdown lines with 10 separate fields for each cell line. Mitotic cells were first identified by rounded morphology under phase optics and subsequently evaluated for GFP-BAF localization throughout mitosis. GFP-BAF was scored for chromatin association in all mitotic stages in 40 cells/line. GFP-BAF cells were also examined for proper segregation of chromosomes and presence of anaphase bridges or irregular spindle formation. Each cell line was scored for number of irregular divisions per mitotic event observed. MCF10a H2B-GFP cells (generated with pHM-H2-GFP), engineered to express control or VRK1-depleting shRNAs (shSCRAM and shVRK1–02, respectively), were also examined as a control for proper chromosome dynamics after nocodazole arrest and release. The H2B-GFP cDNA was a generous gift of Joseph Besharse (Medical College of Wisconsin).
Karyotype analysis
MCF10a cells were transduced with lentiviruses encoding control (shSCRAM) or VRK1-depleting (shVRK1-01 or -02) shRNAs and passaged for 20 d. The cells were then transferred to the Dyancare Clinical Cytogenetics Lab (Milwaukee, WI) for karyotypic analysis. The karyotype of five cells per population was determined there by Peter Van Tuinen (Department of Pathology, Medical College of Wisconsin).
Supplementary Material
Acknowledgments
This work was supported by grants to P.T. from the Medical College of Wisconsin Cancer Center (Breast Cancer Showhouse). We thank Jeremy Nichols and Matthew Wiebe for helpful discussions, all of the members of the Traktman laboratory for their insights, Noah Zimmerman for his guidance in setting up the live-cell imaging, the Medical College of Wisconsin Biotechnology and Bioengineering Center for the use of the confocal microscope for the FRAP analysis, Jason Pokorny for help with software, and the Medical College of Wisconsin Biostatistics drop-in center for advice on biostatistical analysis.
Abbreviations used:
- BAF
barrier to autointegration factor
- ER
endoplasmic reticulum
- FL-BAF
3XFLAG-BAF
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- INM
inner nuclear membrane
- NE
nuclear envelope
- NEBD
nuclear envelope breakdown
- NETs
nuclear envelope transmembrane proteins
- VRK
vaccinia-related kinase
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-10-0603) on January 15, 2014.
*Present address: The Parkinson's Institute, Sunnyvale, CA 94085.
REFERENCES
- Anderson DJ, Hetzer MW. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat Cell Biol. 2007;9:1160–1166. doi: 10.1038/ncb1636. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW. Shaping the endoplasmic reticulum into the nuclear envelope. J Cell Sci. 2008;121:137–142. doi: 10.1242/jcs.005777. [DOI] [PubMed] [Google Scholar]
- Asencio C, Davidson IF, Santarella-Mellwig R, Ly-Hartig TB, Mall M, Wallenfang MR, Mattaj IW, Gorjanacz M. Coordination of kinase and phosphatase activities by Lem4 enables nuclear envelope reassembly during mitosis. Cell. 2012;150:122–135. doi: 10.1016/j.cell.2012.04.043. [DOI] [PubMed] [Google Scholar]
- Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson L, Wilson KL. Multiple and surprising new functions for emerin, a nuclear membrane protein. Curr Opin Cell Biol. 2004;16:73–79. doi: 10.1016/j.ceb.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Bengtsson L, Wilson KL. Barrier-to-autointegration factor phosphorylation on Ser-4 regulates emerin binding to lamin A in vitro and emerin localization in vivo. Mol Biol Cell. 2006;17:1154–1163. doi: 10.1091/mbc.E05-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk JM, Maitra S, Dawdy AW, Shabanowitz J, Hunt DF, Wilson KL. O-linked beta-N-acetylglucosamine (O-GlcNAc) regulates emerin binding to barrier to autointegration factor (BAF) in a chromatin- and lamin B-enriched “Niche.”. J Biol Chem. 2013;288:30192–30209. doi: 10.1074/jbc.M113.503060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broers JL, Ramaekers FC, Bonne G, Yaou RB, Hutchison CJ. Nuclear lamins: laminopathies and their role in premature ageing. Physiol Rev. 2006;86:967–1008. doi: 10.1152/physrev.00047.2005. [DOI] [PubMed] [Google Scholar]
- Burke B, Ellenberg J. Remodelling the walls of the nucleus. Nat Rev Mol Cell Biol. 2002;3:487–497. doi: 10.1038/nrm860. [DOI] [PubMed] [Google Scholar]
- Burke B, Stewart CL. The nuclear lamins: flexibility in function. Nat Rev Mol Cell Biol. 2013;14:13–24. doi: 10.1038/nrm3488. [DOI] [PubMed] [Google Scholar]
- Cabanillas R, et al. Nestor-Guillermo progeria syndrome: a novel premature aging condition with early onset and chronic development caused by BANF1 mutations. Am J Med Genet A. 2011;155A:2617–2625. doi: 10.1002/ajmg.a.34249. [DOI] [PubMed] [Google Scholar]
- Capanni C, Cenni V, Haraguchi T, Squarzoni S, Schuchner S, Ogris E, Novelli G, Maraldi NM, Lattanzi G. Lamin A precursor induces barrier-to-autointegration factor nuclear localization. Cell Cycle. 2010;9:2600–2610. doi: 10.4161/cc.9.13.12080. [DOI] [PubMed] [Google Scholar]
- Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- Chi YH, Chen ZJ, Jeang KT. The nuclear envelopathies and human diseases. J Biomed Sci. 2009;16:96. doi: 10.1186/1423-0127-16-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JL, Mallanna SK, Ormsbee BD, Desler M, Wiebe MS, Rizzino A. Banf1 is required to maintain the self-renewal of both mouse and human embryonic stem cells. J Cell Sci. 2011;124:2654–2665. doi: 10.1242/jcs.083238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen CF, Brittle AL, Ito T, Ohkura H. The conserved kinase NHK-1 is essential for mitotic progression and unifying acentrosomal meiotic spindles in Drosophila melanogaster. J Cell Biol. 2005;171:593–602. doi: 10.1083/jcb.200508127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Gajewski A, Korbei B, Gerlich D, Daigle N, Haraguchi T, Furukawa K, Ellenberg J, Foisner R. LAP2alpha and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J Cell Sci. 2004;117:6117–6128. doi: 10.1242/jcs.01529. [DOI] [PubMed] [Google Scholar]
- Dundr M, Misteli T. Measuring dynamics of nuclear proteins by photobleaching. Curr Protoc Cell Biol. 2003. Chapter 13, Units 13.5.1–13.5.5. [DOI] [PubMed]
- Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, Lippincott-Schwartz J. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier MV, Martin KJ, Kenny PA, Xhaja K, Bosch I, Yaswen P, Bissell MJ. Gene expression signature in organized and growth-arrested mammary acini predicts good outcome in breast cancer. Cancer Res. 2006;66:7095–7102. doi: 10.1158/0008-5472.CAN-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Sugiyama S, Osouda S, Goto H, Inagaki M, Horigome T, Omata S, McConnell M, Fisher PA, Nishida Y. Barrier-to-autointegration factor plays crucial roles in cell cycle progression and nuclear organization in Drosophila. J Cell Sci. 2003;116:3811–3823. doi: 10.1242/jcs.00682. [DOI] [PubMed] [Google Scholar]
- Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Gorjanacz M, Klerkx EP, Galy V, Santarella R, Lopez-Iglesias C, Askjaer P, Mattaj IW. Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly. EMBO J. 2007;26:132–143. doi: 10.1038/sj.emboj.7601470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- Haraguchi T, Kojidani T, Koujin T, Shimi T, Osakada H, Mori C, Yamamoto A, Hiraoka Y. Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J Cell Sci. 2008;121:2540–2554. doi: 10.1242/jcs.033597. [DOI] [PubMed] [Google Scholar]
- Haraguchi T, Koujin T, Osakada H, Kojidani T, Mori C, Masuda H, Hiraoka Y. Nuclear localization of barrier-to-autointegration factor is correlated with progression of S phase in human cells. J Cell Sci. 2007;120:1967–1977. doi: 10.1242/jcs.03461. [DOI] [PubMed] [Google Scholar]
- Haraguchi T, Koujin T, Segura-Totten M, Lee KK, Matsuoka Y, Yoneda Y, Wilson KL, Hiraoka Y. BAF is required for emerin assembly into the reforming nuclear envelope. J Cell Sci. 2001;114:4575–4585. doi: 10.1242/jcs.114.24.4575. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Segawa M, Ouchi FS, Yamakawa Y, Furukawa K, Takeyasu K, Horigome T. Dissociation of emerin from barrier-to-autointegration factor is regulated through mitotic phosphorylation of emerin in a Xenopus egg cell-free system. J Biol Chem. 2005;280:39925–39933. doi: 10.1074/jbc.M503214200. [DOI] [PubMed] [Google Scholar]
- Holaska JM, Lee KK, Kowalski AK, Wilson KL. Transcriptional repressor germ cell-less (GCL) and barrier to autointegration factor (BAF) compete for binding to emerin in vitro. J Biol Chem. 2003;278:6969–6975. doi: 10.1074/jbc.M208811200. [DOI] [PubMed] [Google Scholar]
- Holaska JM, Wilson KL. Multiple roles for emerin: implications for Emery-Dreifuss muscular dystrophy. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:676–680. doi: 10.1002/ar.a.20334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Chakraborty G, Kim S, Shin J, Park CH, Jeong MW, Bharatham N, Yoon HS, Kim KT. Macro histone H2A1.2 (macroH2A1) protein suppresses mitotic kinase VRK1 during interphase. J Biol Chem. 2012a;287:5278–5289. doi: 10.1074/jbc.M111.281709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Lyu HN, Kwon HS, Kim YS, Lee KH, Kim DY, Chakraborty G, Choi KY, Yoon HS, Kim KT. Obtusilactone B from Machilus thunbergii targets barrier-to-autointegration factor to treat cancer. Mol Pharmacol. 2012b;83:367–376. doi: 10.1124/mol.112.082578. [DOI] [PubMed] [Google Scholar]
- Lancaster OM, Cullen CF, Ohkura H. NHK-1 phosphorylates BAF to allow karyosome formation in the Drosophila oocyte nucleus. J Cell Biol. 2007;179:817–824. doi: 10.1083/jcb.200706067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Haraguchi T, Lee RS, Koujin T, Hiraoka Y, Wilson KL. Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J Cell Sci. 2001;114:4567–4573. doi: 10.1242/jcs.114.24.4567. [DOI] [PubMed] [Google Scholar]
- Lee MS, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Blake DL, Callebaut I, Skerjanc IS, Holmer L, McBurney MW, Paulin-Levasseur M, Worman HJ. MAN1, an inner nuclear membrane protein that shares the LEM domain with lamina-associated polypeptide 2 and emerin. J Biol Chem. 2000;275:4840–4847. doi: 10.1074/jbc.275.7.4840. [DOI] [PubMed] [Google Scholar]
- Margalit A, Brachner A, Gotzmann J, Foisner R, Gruenbaum Y. Barrier-to-autointegration factor—a BAFfling little protein. Trends Cell Biol. 2007;17:202–208. doi: 10.1016/j.tcb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Margalit A, Segura-Totten M, Gruenbaum Y, Wilson KL. Barrier-to-autointegration factor is required to segregate and enclose chromosomes within the nuclear envelope and assemble the nuclear lamina. Proc Natl Acad Sci USA. 2005;102:3290–3295. doi: 10.1073/pnas.0408364102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KJ, Patrick DR, Bissell MJ, Fournier MV. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS One. 2008;3:e2994. doi: 10.1371/journal.pone.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor TP, Traktman P. Molecular genetic analysis of VRK1 in mammary epithelial cells: depletion slows proliferation in vitro and tumor growth and metastasis in vivo. Oncogenesis. 2013;2:e48. doi: 10.1038/oncsis.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca R, Lee KK, Wilson KL. Binding of barrier to autointegration factor (BAF) to histone H3 and selected linker histones including H1.1. J Biol Chem. 2005;280:42252–42262. doi: 10.1074/jbc.M509917200. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky G, Fabian AJ, Rooney S, Alt FW, Mulligan RC. Complete correction of murine Artemis immunodeficiency by lentiviral vector-mediated gene transfer. Proc Natl Acad Sci USA. 2006;103:16406–16411. doi: 10.1073/pnas.0608130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols RJ, Traktman P. Characterization of three paralogous members of the mammalian vaccinia related kinase family. J Biol Chem. 2004;279:7934–7946. doi: 10.1074/jbc.M310813200. [DOI] [PubMed] [Google Scholar]
- Nichols RJ, Wiebe MS, Traktman P. The vaccinia-related kinases phosphorylate the N’ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol Biol Cell. 2006;17:2451–2464. doi: 10.1091/mbc.E05-12-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson JA. Nuclear dreams: the malignant alteration of nuclear architecture. J Cell Biochem. 1998;70:172–180. [PubMed] [Google Scholar]
- Pienta KJ, Coffey DS. Correlation of nuclear morphometry with progression of breast cancer. Cancer. 1991;68:2012–2016. doi: 10.1002/1097-0142(19911101)68:9<2012::aid-cncr2820680928>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Puente XS, et al. Exome sequencing and functional analysis identifies BANF1 mutation as the cause of a hereditary progeroid syndrome. Am J Hum Genet. 2011;88:650–656. doi: 10.1016/j.ajhg.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhka M, Vihinen H, Joensuu M, Jokitalo E. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J Cell Biol. 2007;179:895–909. doi: 10.1083/jcb.200705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CL, Hudson AW. The human herpesvirus-7 (HHV-7) U21 immunoevasin subverts NK-mediated cytoxicity through modulation of MICA and MICB. PLoS Pathog. 2011;7:e1002362. doi: 10.1371/journal.ppat.1002362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Butin-Israeli V, Goldman RD. The functions of the nuclear envelope in mediating the molecular crosstalk between the nucleus and the cytoplasm. Curr Opin Cell Biol. 2012;24:71–78. doi: 10.1016/j.ceb.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Koujin T, Segura-Totten M, Wilson KL, Haraguchi T, Hiraoka Y. Dynamic interaction between BAF and emerin revealed by FRAP, FLIP, and FRET analyses in living HeLa cells. J Struct Biol. 2004;147:31–41. doi: 10.1016/j.jsb.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Somech R, Shaklai S, Amariglio N, Rechavi G, Simon AJ. Nuclear envelopathies—raising the nuclear veil. Pediatr Res. 2005;57:8R–15R. doi: 10.1203/01.PDR.0000159566.54287.6C. [DOI] [PubMed] [Google Scholar]
- Taimen P, et al. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc Natl Acad Sci USA. 2009;106:20788–20793. doi: 10.1073/pnas.0911895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umland TC, Wei SQ, Craigie R, Davies DR. Structural basis of DNA bridging by barrier-to-autointegration factor. Biochemistry. 2000;39:9130–9138. doi: 10.1021/bi000572w. [DOI] [PubMed] [Google Scholar]
- Wang X, et al. Barrier to autointegration factor interacts with the cone-rod homeobox and represses its transactivation function. J Biol Chem. 2002;277:43288–43300. doi: 10.1074/jbc.M207952200. [DOI] [PubMed] [Google Scholar]
- Wiebe MS, Traktman P. Poxviral B1 kinase overcomes barrier to autointegration factor, a host defense against virus replication. Cell Host Microbe. 2007;1:187–197. doi: 10.1016/j.chom.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KL. The nuclear envelope, muscular dystrophy and gene expression. Trends Cell Biol. 2000;10:125–129. doi: 10.1016/s0962-8924(99)01708-0. [DOI] [PubMed] [Google Scholar]
- Wilson KL, Holaska JM, Montes de Oca R, Tifft K, Zastrow M, Segura-Totten M, Mansharamani M, Bengtsson L. Nuclear membrane protein emerin: roles in gene regulation, actin dynamics and human disease. Novartis Found Symp. 2005;264:51–58. [PubMed] [Google Scholar]
- Zheng R, Ghirlando R, Lee MS, Mizuuchi K, Krause M, Craigie R. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc Natl Acad Sci USA. 2000;97:8997–9002. doi: 10.1073/pnas.150240197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.