Hypoxia-inducible factors (HIF) are essential for the adaptive response of cells to low-oxygen conditions. Transcription of HIF-α subunits and HIF activity are repressed by the arginine methyltransferase PRMT1. Therefore PRMT1 is a novel regulator of hypoxic cell responses.
Abstract
Hypoxia-inducible factors (HIF-1 and HIF-2) are essential mediators for the adaptive transcriptional response of cells and tissues to low-oxygen conditions. Under hypoxia or when cells are treated with various nonhypoxic stimuli, the active HIF-α subunits are mainly regulated through increased protein stabilization. For HIF-1α, it is clear that further transcriptional, translational, and posttranslational regulations are important for complete HIF-1 activity. Novel evidence links hypoxia and HIF-1 to arginine methylation, an important protein modification. These studies suggest that arginine methyltransferases may be important for hypoxic responses. Protein arginine methyltransferase 1 (PRMT1), the predominant arginine methyltransferase, can act as a transcriptional activator or repressor by modifying a diverse set of substrates. In this work, we show that PRMT1 is a repressor of both HIF-1 and HIF-2. The cellular depletion of PRMT1 by small interference RNA targeting leads to increased HIF transcriptional activity. This activation is the result of enhanced HIF-α subunit transcription, which allows increased HIF-α subunit availability. We provide evidence that PRMT1-dependent HIF-1α regulation is mediated through the activities of both specificity protein 1 (Sp1) and Sp3, two transcription factors known to control HIF-1α expression. This study therefore identifies PRMT1 as a novel regulator of HIF-1– and HIF-2–mediated responses.
INTRODUCTION
The maintenance of oxygen homeostasis is essential for most organisms. Transient or prolonged imbalance between oxygen availability and requirement can result in oxygen deficiency (hypoxia), requiring rapid adaptive mechanisms. The hypoxia-signaling pathway, mediated through hypoxia-inducible transcription factors (HIFs), is responsible for triggering cellular adaptive responses to this physiological stress. HIFs activate the primary transcriptional response to hypoxia, regulating processes that include cell metabolism, survival, and proliferation (Majmundar et al., 2010) by recognizing and binding to hypoxic response elements (HREs) on the promoters of numerous target genes (Wenger et al., 2005). HIFs belong to the basic helix-loop-helix-Per/ARNT/Sim (bHLH-PAS) transcription factor family, which includes the two main hypoxia-responsive complexes, HIF-1 and HIF-2 (Loboda et al., 2010). Both complexes are heterodimers formed from a distinct oxygen-sensitive HIF-α subunit and a constitutive HIF-1β subunit (Wang et al., 1995). Although it is usually expressed in a constitutive manner, HIF-α is rapidly degraded under normal oxygen tension (Kaelin and Ratcliffe, 2008). HIF-α protein stability is controlled through its oxygen-dependent degradation domain, which contains two key proline residues hydroxylated by specific HIF prolyl-hydroxylases (PHDs) when oxygen is present (Bruick and McKnight, 2001; Epstein et al., 2001). HIF-α prolyl-hydroxylation allows for recognition by von Hippel–Lindau tumor suppressor protein (pVHL), the product of the von Hippel–Lindau tumor suppressor gene (Ivan et al., 2001; Jaakkola et al., 2001; Masson et al., 2001; Yu et al., 2001). As the recognition component of an E3 ubiquitin ligase complex, pVHL allows for HIF-α polyubiquitination and subsequent proteasomal degradation (Maxwell et al., 1999). Hence, after hydroxylation, HIF-α is rapidly targeted for proteasomal degradation (Kaelin and Ratcliffe, 2008). This regulatory mechanism allows for HIF-α to be quickly and efficiently stabilized in hypoxic conditions.
Contributing to HIF-α regulation are diverse factors acting at the level of transcription and translation and through posttranslational modifications (Kuschel et al., 2012). These factors allow precise modulation of HIF activity in response to oxygen-dependent and -independent stimuli. An important posttranslational modification in eukaryotic cells is arginine methylation. This modification interferes with protein interactions, activity, stability, and localization and affects processes such as transcription, RNA processing, translation, signal transduction, and DNA repair (Bedford and Clarke, 2009). Arginine methylation is catalyzed by the protein arginine methyltransferase (PRMT) family, the members of which are responsible for the direct transfer of a methyl group from S-adenosyl-l-methionine to the guanidine nitrogen atom of an arginine residue frequently located in arginine/glycine–rich sequences termed RGG/RG motifs (Bedford and Clarke, 2009; Thandapani et al., 2013). PRMTs are able to catalyze the formation of monomethylarginines and either asymmetric (type I PRMTs) or symmetric dimethylarginines (type II PRMTs). PRMT1, a type I PRMT, is the predominant arginine methyltransferase in mammalian cells, responsible for >85% of all arginine methylation (Bedford and Clarke, 2009; Yu et al., 2009). PRMT1 has a diverse set of substrates and is implicated in the regulation of various cellular processes (Boisvert et al., 2005; Yang and Bedford, 2013). Among these, PRMT1 acts to regulate transcription by methylating histones, transcription factors, transcriptional coactivators and corepressors, RNA-binding proteins, and transcription elongation factors (Yang and Bedford, 2013).
Of interest, hypoxia has been linked to changes in arginine methylation. S-Adenosyl-l-methionine is reduced under hypoxia due to decreased methionine adenosyltransferase expression (Chawla and Jones, 1994; Chawla et al., 1996; Avila et al., 1998). In addition, mice kept under chronic hypoxia had increased PRMT2 expression and increased asymmetric dimethyl-arginine (ADMA) levels, the residues of arginine-methylated protein proteolysis (Yildirim et al., 2006). Finally, a recent study directly linked HIF-1 to arginine methylation, demonstrating that PRMT5 promotes HIF-1α translation via its 5′-untranslated region sequence (Lim et al., 2012). Together these studies suggest a role for arginine methylation in the hypoxic response and potential cross-talk between HIFs and PRMTs.
In the present study, we set out to further evaluate the role of arginine methylation in HIF-dependent hypoxic responses. We were specifically interested in PRMT1’s potential role in regulating HIF activity. We demonstrate that the specific depletion of PRMT1 leads to increased HIF-α transcription. Increased HIF-α mRNA expression during PRMT1 depletion correlates with augmented HIF-α protein levels and HIF-dependent transcriptional activity. We show that HIF repression by PRMT1 is mediated through the activity of specificity protein 1 (Sp1) and Sp3 transcription factors, known regulators of HIF-α expression. Taken together, these results identify PRMT1 as a novel regulator of HIF-mediated transcriptional responses.
RESULTS
PRMT1 represses HIF-1 complex accumulation
To examine the role of arginine methylation in regulating HIF-1α subunit accumulation, we depleted the major type I enzyme, PRMT1, using RNA interference in HeLa cells. Of interest, as shown in Figure 1, siPRMT1-transfected cells showed significantly higher HIF-1α protein levels under hypoxic conditions (1% O2) than did siCtrl-transfected cells (2.1 ± 0.4–fold). A similar increase was also observed under CoCl2 treatment, a known hypoxia mimetic and activator of HIF complexes (Supplemental Figure SIA). In addition, increased HIF-1α accumulation was observed at all time points studied under hypoxic conditions (from 15 min to 48 h; Supplemental Figure SIB). HIF-1α’s sister protein, HIF-2α, was also increased in PRMT1-depleted cells under both hypoxic and CoCl2 treatments (Supplemental Figure SIIA). These results identify PRMT1 as a repressor of HIF-1/2α subunit accumulation. Under our conditions, we consistently observed a PRMT1 protein depletion of >80%. To control for siPRMT1 specificity, we also assayed a second siPRMT1 sequence (siPRMT1*). As seen in Supplemental Figure SIIIA, siPRMT1* also increased HIF-1α protein accumulation. We then determined whether siPRMT1 could modify the expression of other PRMTs. As seen in Supplemental Figure SIIIB, siPRMT1 had no effect on PRMT5 protein levels. In addition, although it decreased the expression of PRMT1 mRNA, siPRMT1 and siPRMT1* did not decrease the expression of PRMT2 through to PRMT9 mRNA (unpublished data). Finally, the depletion of PRMT5, previously identified as a positive regulator of HIF-1α (Lim et al., 2012), did not increase HIF-1α protein levels under our experimental conditions (Supplemental Figure SIIIC).
FIGURE 1:
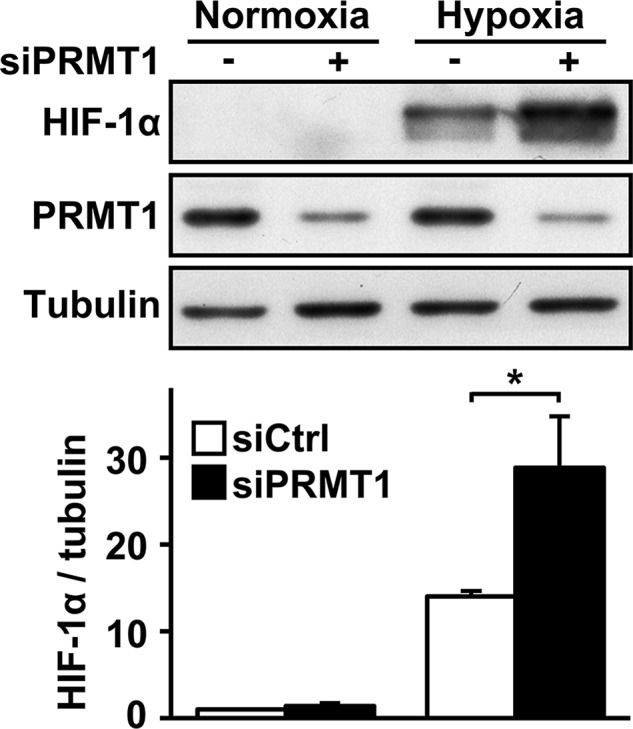
PRMT1 depletion increases HIF-1α protein accumulation. HeLa cells were transfected with a control siRNA or a siRNA against PRMT1 mRNA (20 nM). At 48 h posttransfection, cells were maintained under control conditions or under 1% oxygen for 2 h. HIF-1α, PRMT1, and α-tubulin levels were analyzed by Western blot (top) and quantified (bottom) with the Odyssey Infrared Imaging System using α-tubulin as a loading control. *p < 0.05 as compared with siCtrl cells under hypoxia.
PRMT1 does not modify HIF-1α protein stability
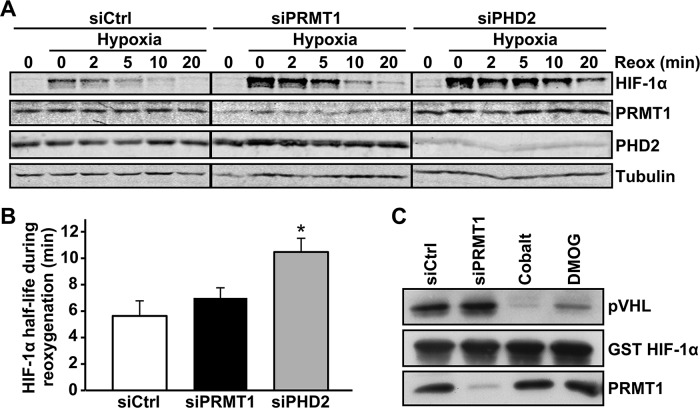
Because protein stabilization is the major regulatory mechanism for HIF-1α induction, we investigated whether PRMT1 could regulate HIF-1α stabilization or degradation. First, HIF-1α half-life was determined under reoxygenation conditions using cycloheximide, a general protein synthesis inhibitor. Under these conditions, changes in HIF-1α half-life reflect changes in HIF-1α stability (Laughner et al., 2001; Page et al., 2008; Michaud et al., 2009; Patten et al., 2010; Labbe et al., 2012). siPRMT1- or siCtrl-transfected cells were maintained under hypoxic conditions for 1 h to induce HIF-1α. Cells were then treated with cycloheximide and transferred to normoxic conditions (20.9% O2; reoxygenation) for different periods of time (Figure 2A). HIF-1α half-life was determined by quantifying HIF-1α protein levels over the time of reoxygenation. Surprisingly, HIF-1α half-life was not significantly modified by PRMT1 depletion (5.6 ± 1.1 and 7.0 ± 0.8 min for siCtrl- and siPRMT1-transfected cells, respectively; Figure 2B). As expected, increased HIF-1α half-life was observed in siPHD2-transfected cells (10.5 ± 1.0 min). Second, we determined whether PRMT1 modulates HIF-1α binding to pVHL. The binding of HIF-α subunits to pVHL is a key step for targeted proteasomal degradation and is a direct consequence of PHD-dependent HIF-α hydroxylation (Ivan et al., 2001). To determine the effect of PRMT1 depletion on pVHL binding, we performed a pVHL-capture assay. A glutathione S-transferase (GST)–HIF-1α fusion protein comprising amino acids 344–582 from human HIF-1α was subjected to modification by extracts from siPRMT1- or siCtrl-transfected HeLa cells. This was followed by interaction with in vitro–translated pVHL and GST pull down. As demonstrated in Figure 2C, the binding of HIF-1α to pVHL was not modified by PRMT1 depletion, consistent with the results obtained for HIF-1α half-life. As expected, incubation of GST-HIF-1α fusion protein with extracts containing CoCl2 or dimethyloxalylglycine, two inhibitors of HIF-1α hydroxylation, blocked the binding of HIF-1α to pVHL. Taken together, these results indicate that the regulation of HIF-1α levels by PRMT1 is not mediated through changes in HIF-1α protein stability.
FIGURE 2:
PRMT1 depletion does not modify HIF-1α protein stability. (A) HeLa cells were transfected with a control siRNA or siRNAs against PRMT1 or PHD2 mRNA (20 nM). At 48 h posttransfection, cells were maintained under control conditions or under 1% oxygen for 1 h. The HIF-1α degradation rate was determined by treating cells with cycloheximide (30 μg/ml) for 5 min to block all de novo protein synthesis, followed by reoxygenation for the indicated times. HIF-1α, PRMT1, PHD2, and α-tubulin protein levels were evaluated by Western blot and quantified with the Odyssey Infrared Imaging System. (B) HIF-1α half-life was determined by plotting data as HIF-1α/α-tubulin ratio vs. time under reoxygenation, as quantified from A. Results are an average ± SEM of four independent experiments. *p < 0.05 as compared with siCtrl cells. (C) HeLa cells were transfected with a control siRNA or a siRNA against PRMT1 mRNA (20 nM). At 48 h posttransfection, cells were lysed, and cytoplasmic extracts were incubated with GST-HIF-1α coupled to Sepharose beads for 1 h in the presence or not of 200 μM CoCl2 or 1 mM dimethyloxalylglycine (DMOG). Samples were then incubated in the presence of in vitro–translated pVHL and resolved by SDS–PAGE (12%). Immunoblotting was performed using anti-HA (pVHL) and anti-GST antibodies.
PRMT1 represses HIF-1α mRNA expression
The regulation of HIF-1α gene transcription can be an important mechanism to modify HIF-1 complex formation, accumulation, and activity (Kuschel et al., 2012). Furthermore, previous studies showed that PRMT1 can regulate gene transcription (Bedford and Clarke, 2009; Nicholson et al., 2009). Therefore we next determined whether PRMT1 could regulate HIF-1α mRNA expression. We first evaluated the effect of PRMT1 depletion on HIF-1α mRNA levels by real time quantitative reverse transcription (RT) PCR experiments. Of interest, siPRMT1-transfected cells showed a striking increase of HIF-1α transcript levels (2.3 ± 0.1–fold over siCtrl-transfected cells; Figure 3A). In addition, increased HIF-1α mRNA levels during PRMT1 depletion occurred under both normoxic and hypoxic conditions (Supplemental Figure SIC). A similar increase in HIF-2α mRNA levels was observed in siPRMT1-transfected cells (2.1 ± 0.3–fold over siCtrl-transfected cells; Supplemental Figure SIIB). It is important to note that hypoxia did not increase basal HIF-1α mRNA levels under these conditions. To further evaluate the role of PRMT1 in HIF-1α mRNA expression, we used a luciferase reporter plasmid driven by the human HIF-1α gene promoter region (pHIF1A-571/+32Luc). When cells were transiently cotransfected with this reporter along with siPRMT1, a significant increase in reporter activity was observed (1.5 ± 0.1–fold over siCtrl-transfected cells; Figure 3B). We also assessed HIF-1α mRNA stability after PRMT1 depletion by determining the rate of mRNA degradation. After treatment of cells with actinomycin D during different periods of time to block transcription, the level of HIF-1α mRNA remained high, demonstrating its high stability (Figure 3C). More important, PRMT1 depletion did not significantly modify HIF-1α mRNA stability. As a control for actinomycin D effectiveness, increased 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (PFKFB4) mRNA expression under CoCl2 treatment was blocked by pretreating cells with actinomycin D (Supplemental Figure SIV). These results indicate that increased mRNA stability is not responsible for augmented HIF-1α mRNA levels, confirming a role for enhanced transcription in HIF-1α regulation during PRMT1 depletion. Taken together, these results demonstrate that PRMT1 is a transcriptional repressor of HIF-1α expression.
FIGURE 3:
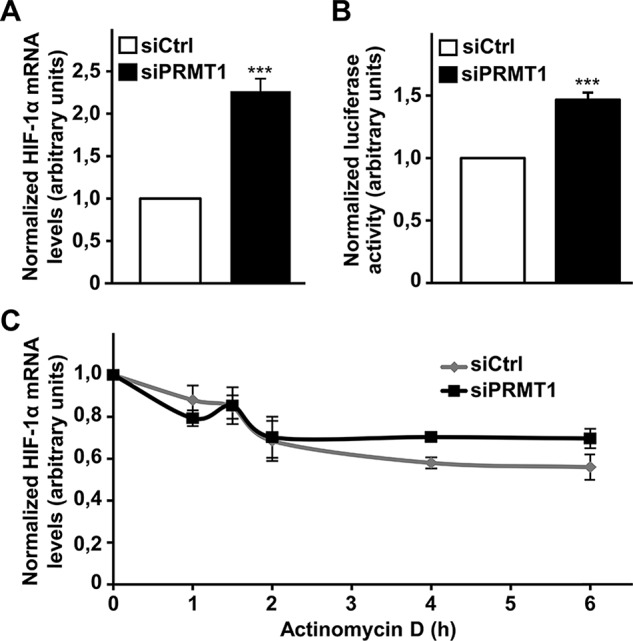
Cellular PRMT1 depletion increases HIF-1α mRNA expression. (A) HeLa cells were transfected with a control siRNA or a siRNA against PRMT1 mRNA (20 nM). At 48 h posttransfection, cells were lysed, followed by RNA extraction and cDNA synthesis. Real-time PCR was performed using HIF-1α and HPRT (internal control) oligonucleotides. Results are an average ± SEM of 10 independent experiments. ***p < 0.001 (B) HEK 293T cells were transfected with a control siRNA or a siRNA against PRMT1 mRNA (20 nM) along with pGL2 HIF-1α -572/+32 and an expression vector coding for R. reniformis luciferase. At 48 h posttransfection, cells were lysed and luciferase activity was measured. Results are expressed as a ratio of luciferase activity to R. reniformis luciferase activity and are an average ± SEM of seven independent experiments performed in triplicate. ***p < 0.001 (C) HeLa cells were transfected with a control siRNA or a siRNA against PRMT1 mRNA (20 nM). At 48 h posttransfection, cells were treated with actinomycin D (1 μg/ml) for the indicated times to block transcription, followed by RNA extraction and cDNA synthesis. Real-time PCR was performed using HIF-1α and HPRT (reference gene) oligonucleotides. Results are an average ± SEM of at least three independent experiments.
PRMT1 regulates HIF-1α levels in a methyltransferase-dependent manner
To evaluate the requirement of PRMT1’s methyltransferase activity in regulating HIF-1α accumulation, we studied the effect of an inactive form of PRMT1 on HIF-1α protein levels. Small interfering RNA (siRNA)–insensitive and catalytically inactive PRMT1 was generated by inserting mutations into the region recognized by siPRMT1 and the methyl-donor-binding region (residues 68–70; VLD to AAA). The VLD mutation was reported to cause complete loss of methyltransferase activity and act as a dominant-negative protein by forming hetero-oligomers with endogenous PRMT1 (Herrmann and Fackelmayer, 2009). To confirm the effect of the VLD mutation on PRMT1 activity, we evaluated total asymmetric protein arginine methylation levels using an anti–methyl-arginine antibody, ASYM25 (Boisvert et al., 2003). As expected, siPRMT1-transfected cells showed decreased asymmetric arginine methylation (Supplemental Figure SV). This decrease was reversed by expressing an siRNA-insensitive form of wild-type PRMT1 (PRMT1siR) but not the catalytically inactive PRMT1 VLD mutant (PRMT1siR-VLD-A). These mutants were then used to “rescue” the changes in HIF-1α protein expression caused by PRMT1 depletion. The expression of PRMT1siR at endogenous levels in siPRMT1-transfected cells blocked increased HIF-1α protein levels during PRMT1 depletion (Figure 4). These results indicate that PRMT1 is indeed a repressor of HIF-1α protein levels. In addition, the expression of inactive PRMT1siR-VLD-A was not able to reestablish HIF-1α protein levels caused by PRMT1 depletion. These results indicate that PRMT1’s methyltransferase activity is required to repress HIF-1α expression and protein levels.
FIGURE 4:
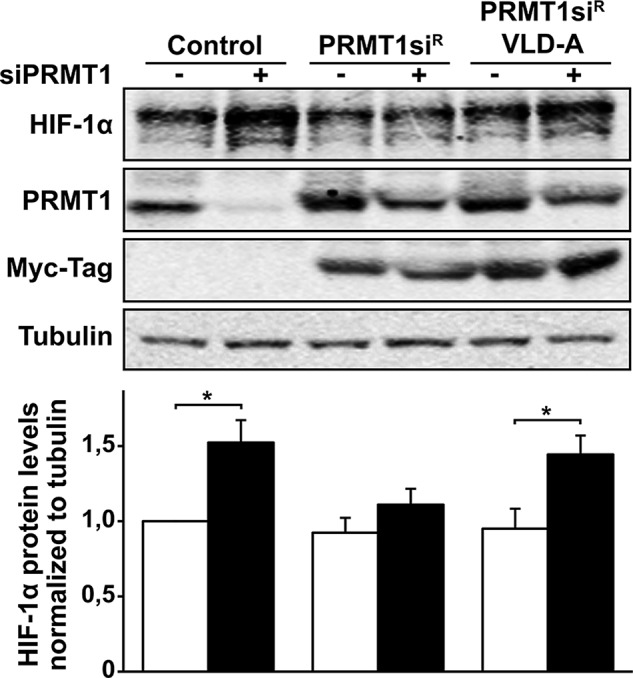
PRMT1’s methyltransferase activity is required for HIF-1α repression. Cells were transfected with a control siRNA or a siRNA against PRMT1 mRNA (20 nM), along with pcDNA3 empty vector, pCDNA3-PRMT1siR (siRNA-resistant mutant), or pCDNA3-PRMT1siR VLD-A (siRNA-resistant and catalytically inactive mutant). At 48 h posttransfection, cells were maintained under 1% oxygen for 2 h. HIF-1α, PRMT1, myc-Tag, and α-tubulin levels were analyzed by Western blot (top) and quantified (bottom) with the Odyssey Infrared Imaging System using α-tubulin as a loading control. *p < 0.05 as compared with siCtrl cells.
PRMT1 represses HIF transcriptional activity
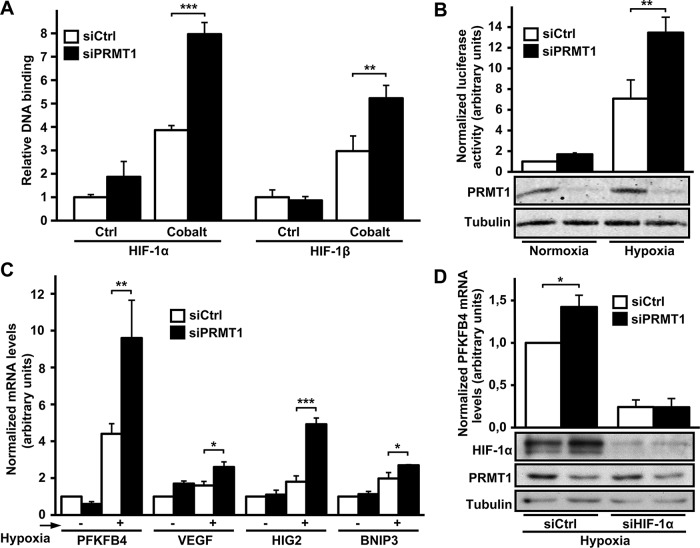
To examine the consequence of HIF-α regulation by PRMT1 on HIF transcriptional activity, we first investigated the effect of PRMT1 on the binding of HIF complexes to a DNA sequence containing a hypoxic response element. Active HIF complexes bind to specific HRE DNA sequences upon dimerization of HIF-α with HIF-1β (Wang et al., 1995). To undertake these studies, we performed HIF transcription factor enzyme-linked immunoassays using specific HIF-binding (HRE-containing) double-stranded DNA (dsDNA) oligonucleotides (Blouin et al., 2004). Nuclear extracts from HeLa cells treated with CoCl2 showed increased DNA binding of HIF-1 complexes, identified through detection of HIF-1α and HIF-1β bound to HRE oligonucleotides (Figure 5A). More important, siPRMT1-transfected cells showed increased HIF-1 DNA binding as compared with siCtrl-transfected cells (2.1 ± 0.1– and 1.7 ± 0.2–fold for HIF-1α and HIF-1β, respectively), which parallels increased HIF-1α protein levels observed in Figure 1. To determine binding specificity, we used a dsDNA oligonucleotide substituted on two residues essential for HIF-complex binding. As expected, no increased HIF-1 binding to these oligonucleotides was observed under hypoxia-mimetic conditions or after PRMT1 depletion (unpublished data). To further examine the role of PRMT1 in regulating HIF activity, we performed gene reporter assays using a luciferase reporter plasmid driven by three HRE sequences (pGL3 [R2.2] 3HRE-TK). Exposure of siCtrl-transfected cells to hypoxic conditions increased HRE reporter activity (7.1 ± 1.8–fold) as compared with normoxic conditions (Figure 5B). Of interest, siPRMT1-transfected cells showed enhanced HRE reporter activity in hypoxic conditions (1.9 ± 0.2–fold) over siCtrl-transfected cells. It is important to note that increased HRE reporter activity in siPRMT1-transfected cells was also consistently observed in normoxic conditions (1.7 ± 0.1–fold over siCtrl-transfected cells), although not statistically significantly. The repressive role of PRMT1 on HIF-mediated transcriptional activity was further demonstrated by evaluating the expression of known HIF target genes. Among others, the exposure of HeLa cells to hypoxic conditions increased the expression of PFKFB4, vascular endothelial growth factor (VEGF), hypoxia-inducible gene 2 (HIG2), and BCL2/adenovirus E1B 19-kDa interacting protein 3 (BNIP3) mRNA in an HIF-dependent manner (Supplemental Figure SVI). As seen in Figure 5C, the hypoxic inductions of PFKFB4, VEGF, HIG2, and BNIP3 were all enhanced by PRMT1 depletion (2.2 ± 0.5–, 1.6 ± 0.2–, 2.7 ± 0.2x–, and 1.4 ± 0.02–fold over hypoxic siCtrl-transfected cells, respectively). To confirm that increased HIF-responsive gene expression by PRMT1 depletion was dependent on HIF activity, we targeted HIF-1α expression with a specific small interfering RNA (siHIF-1α). Indeed, as seen in Figure 5D, HIF-1α depletion abolished increased PFKFB4 expression in siPRMT1-transfected cells. Taken together, these results indicate that PRMT1 depletion induces the accumulation of HIF-1/2α subunits, allowing the formation of HIF-1/2 complexes, which can bind to specific HRE DNA sequences and increase HIF-dependent gene transcription.
FIGURE 5:
HIF activity is increased in PRMT1-depleted cells. (A) HeLa cells were transfected with a control siRNA or a siRNA against PRMT1 mRNA (20 nM). At 48 h posttransfection, cells were exposed or not to 200 μM CoCl2 for 2 h. Nuclear extract proteins (10 μg) were incubated in a 96-well plate coated with oligonucleotides containing the HIF-binding site. Presence of HIF transcription complexes was evaluated with antibodies to either HIF-1α or HIF-1β. Results are expressed as the fold increase of absorbance at 450 nM over control conditions and are an average ± SEM of three independent experiments performed in triplicate. **p < 0.01 and ***p < 0.001 as compared with CoCl2-treated siCtrl cells. (B) HEK293T cells were transfected with a control siRNA or a siRNA against PRMT1 mRNA (20 nM), along with pGL3 (R2.2) 3HRE-tk-LUC and an expression vector coding for R. reniformis luciferase. At 48 h posttransfection, cells were maintained under control conditions or under 1% oxygen for 6 h. Cells were lysed, and luciferase activity was measured. Results are expressed as a ratio of firefly luciferase activity to R. reniformis luciferase activity and are an average ± SEM of three independent experiments performed in triplicate. **p < 0.01 as compared with siCtrl cells under hypoxia. PRMT1 and α-tubulin protein levels were evaluated by Western blot. (C) HeLa cells were transfected with a control siRNA or a siRNA against PRMT1 mRNA (20 nM). At 48 h posttransfection, cells were maintained under control conditions or under 1% oxygen for 4 h, followed by RNA extraction and cDNA synthesis. Real-time PCR was performed using PFKFB4, VEGF, HIG2, BNIP3, and HPRT (reference gene) oligonucleotides. Results are an average ± SEM of at least three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001 as compared with siCtrl cells under hypoxia. (D) HeLa cells were transfected with a control siRNA or siRNAs against PRMT1 mRNA (20 nM) and/or HIF-1α mRNA (10 nM) as indicated. At 48 h posttransfection, cells were maintained under control conditions or under 1% oxygen for 4 h, followed by RNA extraction and cDNA synthesis. Real-time PCR was performed using PFKFB4 and HPRT (reference gene) oligonucleotides. Results are an average ± SEM of three independent experiments. *p < 0.05 as compared with siCtrl cells. HIF-1α, PRMT1, and α-tubulin protein levels were evaluated by Western blot.
Sp1 and Sp3 mediate HIF-1α repression by PRMT1
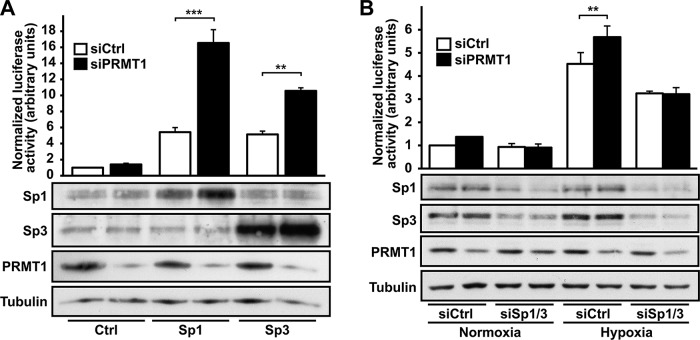
The Sp family of transcription factors are important activators of HIF-1α expression, and their specific DNA-binding motifs (GC-rich sequences) are present on the HIF-1α promoter (Iyer et al., 1998; Minet et al., 1999). As seen in Supplemental Figure SVIIA, the transient transfection of Sp1 or Sp3 constructs (Sapetschnig et al., 2004), the two main Sp family members, both significantly increased HIF-1α gene promoter (pHIF1A-571/+32Luc) activity by 1.8 ± 0.1– and 1.6 ± 0.1–fold over control levels (empty expression vector), respectively. The transfection of both Sp1 and Sp3 did not produce an additive effect on HIF-1α promoter activity (1.7 ± 0.2–fold), indicating competition for the same DNA-binding sites. We therefore investigated whether Sp1 and/or Sp3 are involved in the transcriptional repression of HIF by PRMT1. We first evaluated whether Sp1 and/or Sp3 activity could be repressed by PRMT1. For these experiments, we performed a luciferase assay using a reporter construct driven by a promoter containing six Sp-specific GC-rich elements (Dynan and Tjian, 1983; Supplemental Figure SVIIB). Transient transfection of either Sp1 or Sp3 increased promoter activity by 5.4 ± 0.6– and 5.1 ± 0.4–fold, respectively (Figure 6A). Of greater interest, increased Sp-specific promoter activity was further enhanced in siPRMT1-transfected cells (3.1 ± 0.3 and 2.1 ± 0.07 over siCtrl-transfected cells for Sp1 and Sp3, respectively). These results identify PRMT1 as a repressor of Sp1/3 transcriptional activity, which would therefore have a direct effect on HIF-1α expression. We then evaluated the importance of Sp1/3 in the regulation of HIF activity by PRMT1, using Sp1- and Sp3-specific siRNAs (Supplemental Figure SVIIC). As demonstrated in Figure 6B, Sp1/3 depletion caused a complete inhibition of the siPRMT1-dependent increase of HIF transcriptional activity, as demonstrated with the HIF-sensitive HRE luciferase reporter. Taken together, these results indicate that the repression of Sp1/3 by PRMT1 is an essential step in blocking HIF-α expression and decreasing HIF activity.
FIGURE 6:
Sp1 and Sp3 transcriptional activity is increased after PRMT1 depletion and required for HIF induction. (A) HEK 293T cells were transfected with a control siRNA or a siRNA against PRMT1 mRNA (20 nM), along with pN3-Control, pN3-Sp1, or pN3-Sp3 constructs and pGL3 promoter vector. At 48 h posttransfection, cells were lysed and luciferase activity was measured (top). Results are expressed as a ratio of luciferase activity to total protein levels and are an average ± SEM of three independent experiments performed in triplicate. ***p < 0.001 and **p < 0.01 as compared with Sp1/siCtrl or Sp3/siCtrl cells, respectively. Sp1, Sp3, PRMT1 and α-tubulin protein levels were analyzed by Western blot (bottom). (B) HEK 293T cells were transfected with a control siRNA or siRNAs against PRMT1 (20 nM), Sp1 (10 nM), or Sp3 mRNA (10 nM), as indicated, along with pGL3 (R2.2) 3HRE-tk-LUC and an expression vector coding for R. reniformis luciferase. At 48 h posttransfection, cells were maintained under control conditions or under 1% oxygen for 6 h. Cells were lysed, and luciferase activity was measured (top). Results are expressed as a ratio of firefly luciferase activity to R. reniformis luciferase activity and are an average ± SEM of at least three independent experiments performed in triplicate. **p < 0.01 as compared with siCtrl cells under low oxygen. Sp1, Sp3, PRMT1, and α-tubulin protein levels were analyzed be Western blot (bottom).
DISCUSSION
HIF activity is under the control of a wide variety of factors. This diversity ensures fine tuning of HIF-mediated responses under different conditions. Although HIF-1α subunit stability is the principal pathway involved in HIF-1 activation, other regulatory mechanisms have also gained importance. HIF-1α has been found to be transcriptionally regulated by Sp1/3, NFκB, Egr-1, and Stat3 transcription factors (Frede et al., 2006; Belaiba et al., 2007; Bonello et al., 2007; Niu et al., 2008; Oh et al., 2008; Rius et al., 2008; van Uden et al., 2008; Koshikawa et al., 2009; Patel and Kalra, 2010; Biswas et al., 2013). Translation of HIF-1α mRNA was also found to be regulated by the PI3K/AKT/mTOR and Ras/MEK/ERK pathways, by activation of the ribosomal 6S protein, and the eukaryotic translation initiation factor 4E (eIF-4E), as well as by IRES-mediated translation (Laughner et al., 2001; Fukuda et al., 2002, 2003; Lang et al., 2002; Zhou et al., 2004; Schepens et al., 2005; Young et al., 2008). Studies provide clear evidence that increased HIF-1α mRNA expression is important for enhanced HIF-1 activity after cell activation by several stimuli. Previous studies with angiotensin II, lipopolysaccharide, growth factors, and oncogenes all demonstrated that increased HIF-1α transcription can be essential for HIF-1 induction and activation (Page et al., 2002; Blouin et al., 2004; Gorlach, 2009; Kuschel et al., 2012). Even under oxygenated conditions, when the HIF-1α degradation pathway is fully active, increased HIF-1α expression can maintain sufficient protein synthesis to overcome the rate of degradation and allow normoxic HIF-1α accumulation. This has been observed in numerous studies during HIF-1α overexpression experiments using expression plasmids under normoxic conditions.
The transcription factors Sp1 and Sp3 have putative binding sites on the HIF-1α gene promoter (Iyer et al., 1998). The Sp-binding sites were suggested to be involved in the maintenance of constitutive HIF-1α gene transcription (Minet et al., 1999). Of interest, although less studied, HIF-2α gene expression has also been linked to Sp1 (Wada et al., 2006). Sp1 and Sp3 are the main members of the Sp family of transcription factors that bind to GC-rich elements on promoter regions of target genes. They are constitutively expressed, share >90% sequence homology, and are implicated in activation or repression of a wide variety of genes. Sp1 and Sp3 are able to either synergize or compete for the regulation of target genes, depending on the particular promoter context (Wierstra, 2008; Li and Davie, 2010). Here we confirm the role of Sp1 as a transcriptional activator of HIF-1α and further demonstrate a similar role of Sp3 for HIF-1α expression. Our results suggest that Sp1 and Sp3 compete for the same GC sites on the HIF-1α promoter and act as equivalent activators, since overexpression of both factors was not additive for HIF-1α promoter activity. Even though the HIF-1α promoter contains several Sp-binding sites, lipopolysaccharide stimulates significant Sp1 binding at only one of the three proximal sites (Oh et al., 2008). Therefore we speculate that in this context, only one Sp site is fully active on the HIF-1α promoter, allowing both Sp1 and Sp3 to act as competitive and equivalent transcriptional activators.
We report here that the protein arginine methyltransferase PRMT1 represses HIF-1α/2α mRNA expression by blocking Sp1/3 activity. This effect may provide a positive feedback loop in order to regulate HIF activity. Indeed, the production of S-adenosyl-l-methionine, the principal biological methyl donor, was reduced under hypoxic conditions through changes in methionine adenosyltransferase expression (MAT; Chawla and Jones, 1994; Chawla et al., 1996; Avila et al., 1998; Liu et al., 2011). Reduced S-adenosyl-l-methionine levels were associated with decreased DNA methylation after hypoxic exposure (Chawla et al., 1996; Liu et al., 2011). Furthermore, MAT2A expression is regulated in an HIF-1–dependent manner in hepatoma cells under hypoxic conditions (Liu et al., 2011). We believe that during hypoxia and HIF activation, PRMT1 activity could be reduced in order to decrease its repressive effects on HIF and maintain elevated HIF-1α levels. Even if we did not observe this phenomenon under our hypoxic conditions in HeLa cells, it is important to consider that this feedback regulation may be observed only under longer periods of hypoxic exposure and vary among cell types. We are investigating this possibility in various cell lines sensitive to oxygen level variations.
A recent report indicated that PRMT5 depletion positively regulates HIF-1α translation in A549 lung adenocarcinoma cells but that depletion of other PRMTs had no apparent effect on HIF-1α levels after 8 h of hypoxia (Lim et al., 2012). We investigated the effect of our specific siPRMT1 in A549 cells at different hypoxic time points. Of interest, after hypoxic exposure of A549 cells from 15 min to 6 h, we observed an increase in HIF-1α protein levels during siPRMT1 depletion (unpublished results). However, after prolonged periods of hypoxia, increased HIF-1α protein induction gradually decreased. These observations support our hypothesis of down-regulation of PRMT1 activity through reduced S-adenosyl-l-methionine availability under longer hypoxic exposure, ensuring maintenance of elevated HIF-1α levels. Further supporting our results and hypothesis is a recent observation that PRMT1 depletion slightly modulates HIF-1α protein levels in A549 cells after 24 h of hypoxia (Lim et al., 2013). We also believe that short-term HIF regulation, even if not sustained for longer periods, leads to significant changes in gene regulation and biological responses, as observed here through specific target gene expression. Nonetheless, different feedback loop mechanisms in HIF regulation have been reported and shown to be important to fine tune responses and adaptations to prolonged hypoxia. Finally, our results indicate that HIF-1α mRNA repression also occurs under normoxic conditions, indicating that the effect of PRMT1 on HIF-1α mRNA levels can occur regardless of changes in oxygen levels. In future studies, it will be interesting to determine the effect of PRMT1 on different nonhypoxic activators of HIF-1 (Dery et al., 2005).
Our study establishes a novel and interesting link between PRMT1, Sp1/3 activity, and HIF regulation. The mechanism underlying the direct regulation of Sp1/3 activity by PRMT1 remains to be determined and is under investigation. It will be interesting to evaluate the modulation of Sp1/3 DNA binding by PRMT1 through chromatin immunoprecipitation assays on various promoters and identify the target of PRMT1’s methyltransferase activity in this regulation. Finally, our work provides a better understanding of how protein arginine methylation can control transcriptional regulation in diverse cell activation conditions and provides an interesting connection between this important posttranslational modification and hypoxic signaling.
MATERIALS AND METHODS
Materials
Cobalt chloride, cycloheximide, and actinomycin D were all from Sigma-Aldrich (St. Louis, MO). Dimethyloxalylglycine was from Frontier Scientific (Logan, UT). Polyclonal anti–HIF-1α and anti–HIF-2α antibodies were raised in our laboratory in rabbits immunized against the last 20 amino acids of the C-terminus of each human protein (Richard, 1999; Lauzier et al., 2008). Anti–HIF-1α, anti–HIF-2α, and anti–HIF-1β used in DNA-binding assay and anti-GST and anti-PHD2 antibodies were from Novus Biologicals (Littleton, CO). Anti–α-tubulin antibody was from Sigma-Aldrich. Polyclonal anti-PRMT1 antibody was from Cell Signaling Technology (Danvers, MA), and polyclonal anti-PRMT5 antibody was obtained from Millipore (Billerica, MA). Polyclonal anti-ASYM25 was raised as previously described (Boisvert et al., 2003). Anti-Sp1 and anti-Sp3 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal HA.11 antibody was from Covance (Emeryville, CA). Horseradish peroxidase–coupled anti-mouse and anti-rabbit antibodies were from Promega (Madison, WI). A GST–HIF-1α fusion protein comprising amino acids 344–582 from human HIF-1α and pVHL-hemagglutinin (HA) constructs were kind gifts from Jacques Pouysségur (Université de Nice, France) and Peter Ratcliffe (University of Oxford, UK), respectively. pCDNA3-PRMT1-myc expression vector was previously described (Cote et al., 2003). pCDNA3-PRMT1siR (siRNA-resistant mutant) and pCDNA3-PRMT1siR VLD-A (catalytically inactive mutant) were constructed by site-directed mutagenesis by inserting mutations into 1) the region recognized by siPRMT1 (nucleotide 650; T to C) and 2) the methyl-donor-binding region (residues 68–70; VLD to AAA). pN3-Control, pN3-Sp1FL, and pN3-Sp3FL expression vectors were obtained from Guntram Suske (Philipps-Universität, Marburg, Germany) (plasmids 24544, 24543, and 24541, respectively [Addgene, Cambridge, MA]; Sapetschnig et al., 2004). pGL3 (R2.2) 3HRE-tk-LUC luciferase reporter vector was generated in our laboratory (Lauzier et al., 2007). pHIF-1A-572/+32Luc reporter vector was graciously provided by Gregg Semenza (Johns Hopkins University, Baltimore, MD). pGL3 promoter vector and Renilla reniformis luciferase expression vectors were from Promega (Madison, WI).
Cell culture
Human cervical carcinoma (HeLa) and human embryonic kidney (HEK 293T) cells were from the American Type Culture Collection (Manassas, VA). Cells were cultured in DMEM containing 10% heat-inactivated fetal bovine serum. Culture medium was also supplemented with antibiotics (50 U/ml penicillin and 50 μg/ml streptomycin) and 2 mM glutamine (Life Technologies, Grand Island, NY). Cells were cultured in a humid atmosphere (5% CO2, 95% air) and serially passaged upon reaching confluence. Hypoxic conditions were obtained by placing cells in a sealed hypoxic workstation (Ruskinn Technology, Bridgend, United Kingdom). The oxygen level in this workstation was maintained at 1%, with the residual gas mixture containing 94% nitrogen and 5% CO2.
Western blot analysis
Cells were lysed in a 2× Laemmli buffer. The protein concentration was determined by Lowry assay. Samples were resolved on SDS–polyacrylamide gels and electrophoretically transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore) or nitrocellulose membranes (Hybond C Extra; GE Healthcare Life Sciences, Piscataway, NJ). Proteins were analyzed using indicated antibodies and visualized with enhanced chemiluminescence system (GE Healthcare Life Sciences) or with the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE). Western blots were quantitated using LI-COR Image Studio software 2.0 (LI-COR Biosciences). For Western blot studies, results are representative of at least three independent experiments.
RNA silencing
Cells were seeded in 60-mm plates. At 24 h after plating, siRNA oligonucleotides were transfected by calcium phosphate precipitation. At 16 h after transfection, the medium was changed and the cells were allowed to recover. At 48 h posttransfection, cells were treated and harvested. Ambion siRNA duplexes were obtained from Life Technologies/Applied Biosystems (Carlsbad, CA). The specific sequences used were as follows: human PRMT1 (accession no. NM_001536; predesigned siRNA 110967; sense, 5′-GCCAACAAGUUAGACCACGtt-3′), human PRMT1* (validated siRNA s6917), human HIF-1α (accession no. NM_001530; sense, 5′-AGGACAAGUCACAACAGGAuu-3′), human HIF-2α (accession no. NM_001430; sense, 5′-GGGUCAGGUAGUAAGUGGCuu-3′), human EGLN1/PHD2 (accession no. NM_022051; validated siRNA s29231), human Sp1 (accession no. NM_138473; sense, 5′-GGUAGCUCUAAGUUUUGAUtt-3′), and human Sp3 (accession no. NM_003111; predesigned siRNA 115336; sense, 5′-CCUUCUGCUAACAUCCAGAtt-3′). As a control, Ambion Silencer Negative Control #2 siRNA (Life Technologies/Applied Biosystems) was used.
Luciferase assay
HEK 293T cells seeded in six-well plates were transfected by calcium phosphate precipitation with pGL3 (R2.2) 3HRE-tk-LUC, pHIF-1A-572/+32Luc, or pGL3 promoter vector (500 ng/well). R. reniformis luciferase expression vector (25 ng/well) was used as a control for transfection efficiency. In experiments with Sp1 and/or Sp3 overexpression, total protein levels determined by Lowry assay were used to normalize samples. At 48 h posttransfection, cells were exposed to hypoxia for 6 h, and luciferase assays were performed using the Dual Luciferase Reporter Assay System (Promega). Results were quantified with a Luminoskan Ascent microplate reader with integrated injectors (Thermo Fisher Scientific, Waltham, MA). Results are expressed as a ratio of firefly luciferase activity over R. reniformis luciferase activity or total protein level. Experiments are an average ± SEM of at least three independent experiments performed in triplicate.
Real time quantitative RT PCR
HeLa cells were lysed with TRIzol reagent and RNA was extracted according to the manufacturer's protocol (Life Technologies). RT was performed with the qScript cDNA SuperMix kit, followed by quantitative PCR analysis with the Perfecta SYBR Green SuperMix, Low ROX kit according to the manufacturer's protocol (Quanta BioSciences, Gaithersburg, MD). Quantitative RT-PCR primers used in this study are outlined in Table 1. The expression of the gene of interest relative to HPRT, a reference gene, was calculated based on the threshold cycle (Ct) using the Pfaffl formula (Pfaffl, 2001). HPRT1 stability was assessed using geNorm (www.biogazelle.com). Experiments are an average ± SEM of at least three independent experiments.
TABLE 1:
Primers used for real time quantitative RT PCR.
| Gene | Sequence |
|---|---|
| BNIP3 forward | 5′-GATGATATTGAAAGAAGGAAAGAAGTTG-3′ |
| BNIP3 reverse | 5′-GTTCCTCATGCTGAGGG-3′ |
| HIF1A forward | 5′-GAGATGTTAGCTCCCTATATCCCA-3′ |
| HIF1A reverse | 5′-TAGGTTCTTGTATTTGAGTCTGCTG-3′ |
| HIF2A forward | 5′-AAGCATCCCTGCCACCGTGC-3′ |
| HIF2A reverse | 5′-CAACGGCGCTGCTCCCAAGA-3′ |
| HIG2 forward | 5′-GTTGTGTGGGTGGGCTGT-3′ |
| HIG2 reverse | 5′-GGTGGCCACAATGTCCATA-3′ |
| HPRT forward | 5′-CAGTCCCAGCGTCGTGATTAGT-3′ |
| HPRT reverse | 5′-ATCCAGCAGGTCAGCAAAGAAC-3′ |
| PFKFB4 forward PFKFB4 reverse | 5′-ATGGGCTGAGGTGTAGCTGA-3′ 5′-TATTTATTGCAAAGAATGCTGGA-3′ |
| PRMT1 forward | 5′-CCAGTGGAGAAGGTGGACAT-3′ |
| PRMT1 reverse | 5′-GTCATTCCGCTTCACTTGCA-3′ |
| PRMT2 forward | 5′-TGAATCGCAGGGAGAAGAGC-3′ |
| PRMT2 reverse | 5′-CGATGGCCACAAACTCCTCT-3′ |
| PRMT3 forward | 5′-CAGGGTCGTGTTCTCTACGG-3′ |
| PRMT3 reverse | 5′-CACGGTGAGAGAACGTGGAT-3′ |
| PRMT4 forward | 5′-TAAGTGCTCAGTGTCCCGAG-3′ |
| PRMT4 reverse | 5′-CGGCAGGTTTTCAGGATGTT-3′ |
| PRMT5 forward | 5′-ATGAATTGTCGCCTGAGTGC-3′ |
| PRMT5 reverse | 5′-AAAGGAAGTGTACTCCCCGG-3′ |
| PRMT6 forward | 5′-GTCGCAGCCCAAGAAAAGAA-3′ |
| PRMT6 reverse | 5′-GTAGTACAGCTGGTCCCGTT-3′ |
| PRMT7 forward | 5′-GCTGTGGTTGTGGAGTTCAG-3′ |
| PRMT7 reverse | 5′-GGCACGCTTAATCATGTCGT-3′ |
| PRMT8 forward | 5′-GGCAGCTTTGTACGTGGTAG-3′ |
| PRMT8 reverse | 5′-GCTTTGGATCCACGATGTCC-3′ |
| PRMT9 forward | 5′-CTCTAGCTGGAGTCTGGGTG-3′ |
| PRMT9 reverse | 5′-TAAACACCAACCTGCTTGCC-3′ |
| VEGFA forward | 5′-TACTGCCATCCAATCGAGAC-3′ |
| VEGFA reverse | 5′-GCATGGTGATGTTGGACT-3′ |
Transcription factor enzyme-linked immunoassay
Experiments were performed as described previously (Blouin et al., 2004). In brief, high-bind NeutrAvidin-coated 96-well strip plates (Thermo Fisher Scientific) were incubated with a 5′-biotinylated 26–base pair dsDNA oligonucleotide for 1 h at room temperature. This sequence contains the wild-type or mutant (underlined) HIF-1–binding motif described previously (Semenza and Wang, 1992). We used 5′-GATCGCCCTACGTGCTGTCTCAGATC-3′ for W26 wild-type sequence and 5′-GATCGCCCTAAAAGCTGTCTCAGATC-3′ for M26 mutant sequence. Nuclear extracts from HeLa cells were incubated with oligonucleotides, and HIF-1 complexes bound to DNA were detected using anti–HIF-1α or anti–HIF-1β antibodies (Novus Biologicals), horseradish peroxidase–conjugated secondary antibodies, and TMB-ONE solution (Promega). Results are expressed as an average ± SEM of at least three independent experiments performed in triplicate.
HIF-1α half-life analysis
HeLa cells were exposed to hypoxia for 1 h, after which cycloheximide (30 μg/ml) was added for 5 min. Cells were then exposed to normal oxygen conditions for different periods of time (up to 20 min) and lysed in a 2× Laemmli buffer. HIF-1α, and α-tubulin protein levels were evaluated by Western blotting, followed by quantification, and the ratio of HIF-1α to α-tubulin was determined. HIF-1α half-life under experimental conditions was estimated by plotting data as the HIF-1α/α-tubulin ratio versus time under cycloheximide treatment.
pVHL capture assay
pVHL capture assay was performed as previously described (Lauzier et al., 2008). HeLa cells were grown to confluence and stimulated as indicated. Cells were washed once in phosphate-buffered saline and twice in HEB buffer (20 nM Tris, pH 7.5, 5 mM KCl, 1.5 mM MgCl2, and 1 mM dithiothreitol). Cells were then lysed using a Dounce homogenizer, and cytoplasmic extracts were cleared by centrifugation (20,000 × g). Cytoplasmic extracts (250 μg) were incubated with Sepharose-bound GST–HIF-1α 344–582 (30 μg) for 1 h at room temperature, with or without specific inhibitors. The Sepharose-bound GST–HIF-1α was washed with NETN buffer (150 nM NaCl, 0.5 mM EDTA, 20 mM Tris, pH 8.0, 0.5% Igepal, and 100 μM deferoxamine) and incubated overnight with in vitro–translated pVHL-HA in NETN at 4°C. Samples were then washed with NETN, denatured with 2× Laemmli buffer, resolved in SDS–polyacrylamide gels (12%), and revealed by Western blotting with anti-HA and anti-GST antibodies.
Statistical analysis
Statistical analyses of different experiments were performed using InStat 3 (GraphPad, La Jolla, CA). Unless otherwise noted, one-way analysis of variance tests were performed. Results were deemed significant if they attained a 95% confidence level (p < 0.05).
Supplementary Material
Acknowledgments
We thank J. Pouysségur and P. Ratcliffe for the GST–HIF-1α and pVHL-HA constructs and Wagner Welber Arrais-Silva and Marc-André C. Déry for their perspective and insightful advice. This work was supported by research grants from the Canadian Institutes of Health Research (MOP-93811 to S.R. and MOP-102760 to D.E.R.) and the Heart and Stroke Foundation of Québec (to D.E.R.). V.N.L. is the recipient of a Frederick Banting and Charles Best Canada Graduate Scholarship. D.E.R. is a senior research scholar from the Fonds de recherche du Québec-Santé.
Abbreviations used:
- BNIP3
BCL2/adenovirus E1B 19-kDa interacting protein 3
- HIF
hypoxia-inducible factor
- HIG
hypoxia-inducible gene
- HRE
hypoxic response element
- PFKFB4
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4
- PHD
HIF prolyl-hydroxylase
- PRMT
protein arginine methyltransferase
- pVHL
von Hippel–Lindau tumor suppressor protein
- Sp
specificity protein
- VEGF
vascular endothelial growth factor
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-07-0423) on January 22, 2014.
REFERENCES
- Avila MA, Carretero MV, Rodriguez EN, Mato JM. Regulation by hypoxia of methionine adenosyltransferase activity and gene expression in rat hepatocytes. Gastroenterology. 1998;114:364–371. doi: 10.1016/s0016-5085(98)70489-5. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaiba RS, Bonello S, Zahringer C, Schmidt S, Hess J, Kietzmann T, Gorlach A. Hypoxia up-regulates hypoxia-inducible factor-1alpha transcription by involving phosphatidylinositol 3-kinase and nuclear factor kappaB in pulmonary artery smooth muscle cells. Mol Biol Cell. 2007;18:4691–4697. doi: 10.1091/mbc.E07-04-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Mukherjee R, Tapryal N, Singh AK, Mukhopadhyay CK. Insulin regulates hypoxia-inducible factor-1alpha transcription by reactive oxygen species sensitive activation of Sp1 in 3T3-L1 preadipocyte. PLoS One. 2013;8:e62128. doi: 10.1371/journal.pone.0062128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin CC, Page EL, Soucy GM, Richard DE. Hypoxic gene activation by lipopolysaccharide in macrophages: implication of hypoxia-inducible factor 1alpha. Blood. 2004;103:1124–1130. doi: 10.1182/blood-2003-07-2427. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Chenard CA, Richard S. Protein interfaces in signaling regulated by arginine methylation. Sci STKE. 2005;2005:re2. doi: 10.1126/stke.2712005re2. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Cote J, Boulanger MC, Richard S. A proteomic analysis of arginine-methylated protein complexes. Mol Cell Proteomics. 2003;2:1319–1330. doi: 10.1074/mcp.M300088-MCP200. [DOI] [PubMed] [Google Scholar]
- Bonello S, Zahringer C, BelAiba RS, Djordjevic T, Hess J, Michiels C, Kietzmann T, Gorlach A. Reactive oxygen species activate the HIF-1alpha promoter via a functional NFkappaB site. Arterioscler Thromb Vasc Biol. 2007;27:755–761. doi: 10.1161/01.ATV.0000258979.92828.bc. [DOI] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- Chawla RK, Jones DP. Abnormal metabolism of S-adenosyl-L-methionine in hypoxic rat liver. Similarities to its abnormal metabolism in alcoholic cirrhosis. Biochim Biophys Acta. 1994;1199:45–51. doi: 10.1016/0304-4165(94)90094-9. [DOI] [PubMed] [Google Scholar]
- Chawla RK, Watson WH, Jones DP. Effect of hypoxia on hepatic DNA methylation and tRNA methyltransferase in rat: similarities to effects of methyl-deficient diets. J Cell Biochem. 1996;61:72–80. doi: 10.1002/(SICI)1097-4644(19960401)61:1%3C72::AID-JCB9%3E3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Cote J, Boisvert FM, Boulanger MC, Bedford MT, Richard S. Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1. Mol Biol Cell. 2003;14:274–287. doi: 10.1091/mbc.E02-08-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dery MA, Michaud MD, Richard DE. Hypoxia-inducible factor 1: regulation by hypoxic and non-hypoxic activators. Int J Biochem Cell Biol. 2005;37:535–540. doi: 10.1016/j.biocel.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Dynan WS, Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Epstein ACR, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Frede S, Stockmann C, Freitag P, Fandrey J. Bacterial lipopolysaccharide induces HIF-1 activation in human monocytes via p44/42 MAPK and NF-kappaB. Biochem J. 2006;396:517–527. doi: 10.1042/BJ20051839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R, Hirota K, Fan F, Jung YD, Ellis LM, Semenza GL. Insulin-like growth factor 1 induces hypoxia-inducible factor 1-mediated vascular endothelial growth factor expression, which is dependent on MAP kinase and phosphatidylinositol 3-kinase signaling in colon cancer cells. J Biol Chem. 2002;277:38205–38211. doi: 10.1074/jbc.M203781200. [DOI] [PubMed] [Google Scholar]
- Fukuda R, Kelly B, Semenza GL. Vascular endothelial growth factor gene expression in colon cancer cells exposed to prostaglandin E2 is mediated by hypoxia-inducible factor 1. Cancer Res. 2003;63:2330–2334. [PubMed] [Google Scholar]
- Gorlach A. Regulation of HIF-1alpha at the transcriptional level. Curr Pharm Des. 2009;15:3844–3852. doi: 10.2174/138161209789649420. [DOI] [PubMed] [Google Scholar]
- Herrmann F, Fackelmayer FO. Nucleo-cytoplasmic shuttling of protein arginine methyltransferase 1 (PRMT1) requires enzymatic activity. Genes Cells. 2009;14:309–317. doi: 10.1111/j.1365-2443.2008.01266.x. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Iyer NV, Leung SW, Semenza GL. The human hypoxia-inducible factor 1alpha gene: HIF1A structure and evolutionary conservation. Genomics. 1998;52:159–165. doi: 10.1006/geno.1998.5416. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Koshikawa N, Hayashi J, Nakagawara A, Takenaga K. Reactive oxygen species-generating mitochondrial DNA mutation up-regulates hypoxia-inducible factor-1alpha gene transcription via phosphatidylinositol 3-kinase-Akt/protein kinase C/histone deacetylase pathway. J Biol Chem. 2009;284:33185–33194. doi: 10.1074/jbc.M109.054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuschel A, Simon P, Tug S. Functional regulation of HIF-1alpha under normoxia–is there more than post-translational regulation. J Cell Physiol. 2012;227:514–524. doi: 10.1002/jcp.22798. [DOI] [PubMed] [Google Scholar]
- Labbe A, Lafleur VN, Patten DA, Robitaille GA, Garand C, Lamalice L, Lebel M, Richard DE. The Werner syndrome gene product (WRN): a repressor of hypoxia-inducible factor-1 activity. Exp Cell Res. 2012;318:1620–1632. doi: 10.1016/j.yexcr.2012.04.010. [DOI] [PubMed] [Google Scholar]
- Lang KJ, Kappel A, Goodall GJ. Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol Biol Cell. 2002;13:1792–1801. doi: 10.1091/mbc.02-02-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughner E, Taghavi P, Chiles K, Mahon PC, Semenza GL. HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1alpha (HIF-1alpha) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol Cell Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauzier MC, Page EL, Michaud MD, Richard DE. Differential regulation of hypoxia-inducible factor-1 through receptor tyrosine kinase transactivation in vascular smooth muscle cells. Endocrinology. 2007;148:4023–4031. doi: 10.1210/en.2007-0285. [DOI] [PubMed] [Google Scholar]
- Lauzier MC, Robitaille GA, Chan DA, Giaccia AJ, Richard DE. (2R)-[(4-biphenylylsulfonyl)amino]-N-hydroxy-3-phenylpropionamide (BiPS), a matrix metalloprotease inhibitor, is a novel and potent activator of hypoxia-inducible factors. Mol Pharmacol. 2008;74:282–288. doi: 10.1124/mol.108.045690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Davie JR. The role of Sp1 and Sp3 in normal and cancer cell biology. Ann Anat. 2010;192:275–283. doi: 10.1016/j.aanat.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Lim JH, Choi YJ, Cho CH, Park JW. Protein arginine methyltransferase 5 is an essential component of the hypoxia-inducible factor 1 signaling pathway. Biochem Biophys Res Commun. 2012;418:254–259. doi: 10.1016/j.bbrc.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Lim SK, Jeong YW, Kim DI, Park MJ, Choi JH, Kim SU, Kang SS, Han HJ, Park SH. Activation of PRMT1 and PRMT5 mediates hypoxia- and ischemia-induced apoptosis in human lung epithelial cells and the lung of miniature pigs: the role of p38 and JNK mitogen-activated protein kinases. Biochem Biophys Res Commun. 2013;440:707–713. doi: 10.1016/j.bbrc.2013.09.136. [DOI] [PubMed] [Google Scholar]
- Liu Q, Liu L, Zhao Y, Zhang J, Wang D, Chen J, He Y, Wu J, Zhang Z, Liu Z. Hypoxia induces genomic DNA demethylation through the activation of HIF-1alpha and transcriptional upregulation of MAT2A in hepatoma cells. Mol Cancer Ther. 2011;10:1113–1123. doi: 10.1158/1535-7163.MCT-10-1010. [DOI] [PubMed] [Google Scholar]
- Loboda A, Jozkowicz A, Dulak J. HIF-1 and HIF-2 transcription factors: similar but not identical. Mol Cells. 2010;29:435–442. doi: 10.1007/s10059-010-0067-2. [DOI] [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson N, Willam C, Maxwell PH, Pugh CW, Ratcliffe PJ. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 2001;20:5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Michaud MD, Robitaille GA, Gratton JP, Richard DE. Sphingosine-1-phosphate: a novel nonhypoxic activator of hypoxia-inducible factor-1 in vascular cells. Arterioscler Thromb Vasc Biol. 2009;29:902–908. doi: 10.1161/ATVBAHA.109.185280. [DOI] [PubMed] [Google Scholar]
- Minet E, Ernest I, Michel G, Roland I, Remacle J, Raes M, Michiels C. HIF1A gene transcription is dependent on a core promoter sequence encompassing activating and inhibiting sequences located upstream from the transcription initiation site and cis elements located within the 5′ UTR. Biochem Biophys Res Commun. 1999;261:534–540. doi: 10.1006/bbrc.1999.0995. [DOI] [PubMed] [Google Scholar]
- Nicholson TB, Chen T, Richard S. The physiological and pathophysiological role of PRMT1-mediated protein arginine methylation. Pharmacol Res. 2009;60:466–474. doi: 10.1016/j.phrs.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Niu G, et al. Signal transducer and activator of transcription 3 is required for hypoxia-inducible factor-1alpha RNA expression in both tumor cells and tumor-associated myeloid cells. Mol Cancer Res. 2008;6:1099–1105. doi: 10.1158/1541-7786.MCR-07-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh YT, Lee JY, Yoon H, Lee EH, Baik HH, Kim SS, Ha J, Yoon KS, Choe W, Kang I. Lipopolysaccharide induces hypoxia-inducible factor-1 alpha mRNA expression and activation via NADPH oxidase and Sp1-dependent pathway in BV2 murine microglial cells. Neurosci Lett. 2008;431:155–160. doi: 10.1016/j.neulet.2007.11.033. [DOI] [PubMed] [Google Scholar]
- Page EL, Chan DA, Giaccia AJ, Levine M, Richard DE. Hypoxia-inducible factor-1alpha stabilization in nonhypoxic conditions: role of oxidation and intracellular ascorbate depletion. Mol Biol Cell. 2008;19:86–94. doi: 10.1091/mbc.E07-06-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page EL, Robitaille GA, Pouyssegur J, Richard DE. Induction of hypoxia-inducible factor-1alpha by transcriptional and translational mechanisms. J Biol Chem. 2002;277:48403–48409. doi: 10.1074/jbc.M209114200. [DOI] [PubMed] [Google Scholar]
- Patel N, Kalra VK. Placenta growth factor-induced early growth response 1 (Egr-1) regulates hypoxia-inducible factor-1alpha (HIF-1alpha) in endothelial cells. J Biol Chem. 2010;285:20570–20579. doi: 10.1074/jbc.M110.119495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten DA, Lafleur VN, Robitaille GA, Chan DA, Giaccia AJ, Richard DE. Hypoxia-inducible factor-1 activation in nonhypoxic conditions: the essential role of mitochondrial-derived reactive oxygen species. Mol Biol Cell. 2010;21:3247–3257. doi: 10.1091/mbc.E10-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard DE. p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1alpha (HIF-1alpha) and enhance the transcriptional activity of HIF-1. J Biol Chem. 1999;274:32631–32637. doi: 10.1074/jbc.274.46.32631. [DOI] [PubMed] [Google Scholar]
- Rius J, Guma M, Schachtrup C, Akassoglou K, Zinkernagel AS, Nizet V, Johnson RS, Haddad GG, Karin M. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453:807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapetschnig A, Koch F, Rischitor G, Mennenga T, Suske G. Complexity of translationally controlled transcription factor Sp3 isoform expression. J Biol Chem. 2004;279:42095–42105. doi: 10.1074/jbc.M404989200. [DOI] [PubMed] [Google Scholar]
- Schepens B, Tinton SA, Bruynooghe Y, Beyaert R, Cornelis S. The polypyrimidine tract-binding protein stimulates HIF-1alpha IRES-mediated translation during hypoxia. Nucleic Acids Res. 2005;33:6884–6894. doi: 10.1093/nar/gki1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thandapani P, O'Connor TR, Bailey TL, Richard S. Defining the RGG/RG motif. Mol Cell. 2013;50:613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- van Uden P, Kenneth NS, Rocha S. Regulation of hypoxia-inducible factor-1α by NF-κB. Biochem J. 2008;412:477–484. doi: 10.1042/BJ20080476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Shimba S, Tezuka M. Transcriptional regulation of the hypoxia inducible factor-2a (HIF-2a) gene during adipose differentiation in 3T3-L1 Cells. Biol Pharm Bull. 2006;29:49–54. doi: 10.1248/bpb.29.49. [DOI] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci STKE. 2005;2005:re12. doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- Wierstra I. Sp1: emerging roles—beyond constitutive activation of TATA-less housekeeping genes. Biochem Biophys Res Commun. 2008;372:1–13. doi: 10.1016/j.bbrc.2008.03.074. [DOI] [PubMed] [Google Scholar]
- Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- Yildirim AO, Bulau P, Zakrzewicz D, Kitowska KE, Weissmann N, Grimminger F, Morty RE, Eickelberg O. Increased protein arginine methylation in chronic hypoxia: role of protein arginine methyltransferases. Am J Respir Cell Mol Biol. 2006;35:436–443. doi: 10.1165/rcmb.2006-0097OC. [DOI] [PubMed] [Google Scholar]
- Young RM, Wang SJ, Gordan JD, Ji X, Liebhaber SA, Simon MC. Hypoxia-mediated selective mRNA translation by an internal ribosome entry site-independent mechanism. J Biol Chem. 2008;283:16309–16319. doi: 10.1074/jbc.M710079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Chen T, Hebert J, Li E, Richard S. A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation. Mol Cell Biol. 2009;29:2982–2996. doi: 10.1128/MCB.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, White SB, Zhao Q, Lee FS. HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc Natl Acad Sci USA. 2001;98:9630–9635. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Callapina M, Goodall GJ, Brune B. Functional integrity of nuclear factor kappaB, phosphatidylinositol 3’-kinase, and mitogen-activated protein kinase signaling allows tumor necrosis factor alpha-evoked Bcl-2 expression to provoke internal ribosome entry site-dependent translation of hypoxia-inducible factor 1alpha. Cancer Res. 2004;64:9041–9048. doi: 10.1158/0008-5472.CAN-04-1437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.