Abstract
The non-structural protein 1 (NS1) of human parvovirus B19 plays a critical role in viral DNA replication. Previous studies identified the origin of replication in the viral DNA, which contains four DNA elements, namely NSBE1 to NSBE4, that are required for optimal viral replication (Guan et al, 2009, J. Virology, 83, 9541-9553). Here we have demonstrated in vitro that the NS1 N-terminal domain (NS1N) binds to the origin of replication in a sequence-specific, length-dependent manner that requires NSBE1 and NSBE2, while NSBE3 and NSBE4 are dispensable. Mutagenesis analysis has identified nucleotides in NSBE1 and NSBE2 that are critical for NS1N binding. These results suggest that NS1 binds to the NSBE1-NSBE2 region in the origin of replication, while NSBE3 and NSBE4 may provide binding sites for potential cellular factors. Such a specialized nucleoprotein complex may enable NS1 to nick the terminal resolution site and separate DNA strands during replication.
Keywords: parvovirus, B19, non-structural protein 1, origin of replication, protein:DNA interaction
Introduction
Human parvovirus B19 (B19V) belongs to the erythrovirus genus in the family of Parvoviridae, a group of single-stranded DNA (ssDNA) virus (Berns and Parrish, 2007; Siegl et al., 1985). B19V causes several diseases including erythema infectiosum in children, acute arthropathy commonly in women and hydrops-fetalis in pregnant women (Young and Brown, 2004). It also causes destruction of erythroid precursors, which are the main targets for B19V replication, resulting in anemia and occasionally leading to hematopoietic disorders, e.g., transient aplastic crisis, transient red cell aplasia, thrombocytopenia and pancytopenia (Anderson et al., 1986; Moffatt et al., 1998; Zhi et al., 2006). B19V shows a cytotoxic effect on human primary erythroid progenitor cells in bone marrow (Ozawa et al., 1986) and fetal liver (Yaegashi et al., 1989).
The B19V virion contains a linear, ssDNA genome of 5.6 kb encapsidated in a non-enveloped, icosahedrally symmetric capsid (Cotmore, 2005; Fauquet, 2004). The B19V genome encodes an essential 74-kDa non-structural protein 1 (NS1), two capsid proteins VP1 and VP2 of 86 and 61 kDa respectively, and two smaller proteins of 7.5 kDa and 11 kDa. The 11kDa protein was shown to be involved in viral replication (Zhi et al., 2006) and to induce apoptosis during B19V infection of primary erythroid progenitor cells (Chen et al., 2010) and to specifically interact with the host growth factor receptor-binding protein 2 (Grb2) in vitro in a Src homology 3 dependent manner (Fan et al., 2001), while the function of the 7.5 kDa protein is not known. NS1 consists of an N-terminal DNA-binding/nickase domain, a central domain displaying sequence motifs for helicase/ATPase, and a C-terminal domain whose function remains unknown (Astell CR, 1997; Doerig et al., 1990; Raab et al., 2002). NS1 is a multi-functional protein that plays a pivotal role in viral DNA replication (Cotmore et al., 2007; Han et al., 2013; Li and Rhode, 1990; Zhi et al., 2006), and has also been implicated in transactivation of viral (Gareus et al., 1998; Raab et al., 2002) and cellular genes (Fu et al., 2002; Moffatt et al., 1996; Nakashima et al., 2004), cell cycle arrest (Luo et al., 2013; Morita et al., 2003; Wan et al., 2010), apoptosis (Moffatt et al., 1998), DNA damage response (Lou et al., 2012; Luo et al., 2011), modulation of host innate immunity (Hsu et al., 2011), and packaging of viral DNA into capsid (Bleker et al., 2006; Cotmore and Tattersall, 2005a). It has been shown that null mutants of B19V NS1 protein in which the translational initiation codon was substituted to termination codon completely abolished the viral infectivity (Zhi et al., 2006). Transcription of the B19V genome is controlled by NS1 operating on the viral p6 promoter (Blundell et al., 1987; Ozawa et al., 1987). Protein-protein interactions between NS1 and the cellular transcription factors Sp1/Sp3 are required to bind NS1 to the p6 promoter (Krady and Ward, 1995; Raab et al., 2002). NS1 might also be responsible for packaging of viral DNA into empty capsid, potentially through a channel in the 5-fold vertex, based on data on the adeno-associate virus Rep proteins which bear similar domain structure and sequence homology (Bleker et al., 2006; King et al., 2001) as well as structural data on the minute virus of mice capsid (Plevka et al., 2011).
The left and right ends of the B19V genome have identical inverted terminal repeats (ITRs) and these ITRs fold back onto themselves to form hairpin structures (Deiss et al., 1990; Zhi et al., 2004). Previous studies identified a 67-bp region in the ITR as the minimal origin of DNA replication (Ori), which constitutes the terminal resolution site (trs) and four GC-rich motifs, namely NSBE1 to NSBE4, that are required for optimal virus replication and could form the potential NS1-binding site (Fig 1A) (Guan et al., 2009). NSBEs 1 and 2 are identical 8-bp sequences with a 2-bp interval, while NSBEs 3 and 4 display degenerative sequences. NSBEs 1 to 3 are essential for virus replication, while NSBE4 is required for maximal virus replication (Guan et al., 2009). The organization of NSBEs in the B19V Ori is distinct from the five tandem tetranucleotide repeats of the Rep-binding site in dependovirus (Ward, 2005) and is also different from that identified in minute virus of mice (MVM), a member of the parvovirus genus (Astell et al., 1983; Cotmore and Tattersall, 2005b), and that predicted in minute virus of canine, a member of the bocavirus genus (Sun et al., 2009). Based on studies on MVM, AAV and other parvoviruses, it is thought that binding of NS1 to a sequence-specific site within the double-stranded Ori DNA leads to nicking of a single strand at an adjacent site termed the terminal resolution site (trs), which generates a free 3′-OH end that primes extension of DNA synthesis (Cotmore, 2005).
Figure 1.
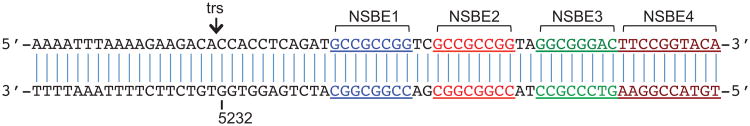
The 67-bp B19V Ori. The four DNA elements (NSBE; shown in blue, red, green and brown respectively) that were predicted to be NS1-binding sites are indicated. The terminal resolution site (trs) is also shown. The nucleotide 5232 in the B19V genome is labeled. There are two identical GC-rich repeats called NSBE1 and NSBE2 and one degenerative element NSBE3, which are essential for virus replication. The fourth element, NSBE4, is required to support maximal virus replication.
Although the essentiality of NSBEs for B19V replication has been documented and the NSBEs were predicted to be the potential NS1-binding site (Guan et al., 2009), it hasn't been clear if and how NS1 physically binds to Ori. In this study we have heterologously expressed and purified the N-terminal DNA-binding/nickase domain of the B19V NS1 (NS1N) and characterized in vitro molecular interaction with Ori. We demonstrate that the NS1N specifically binds to the B19V NSBE region. The four NSBEs exhibit differential roles in NS1N binding. Mutagenesis analysis has identified nts in NSBE1 and NSBE2 that are critical for NS1N binding. The results suggest a mode of interaction between NS1 and Ori, in which NS1N binds to an extended region encompassing NSBE1 and NSBE2, while NSBE3 and NSBE4 may provide a site for binding of potential cellular factors. Such molecular interaction may lead to assembly of a specialized nucleoprotein complex, enabling NS1 to nick the terminal resolution site and separate DNA strands during replication.
Results
Expression and purification of NS1N
Expression of the full-length NS1 in E. coli was not successful. Therefore, we designed a construct that covered the putative N-terminal Ori-recognition and nickase domain (residues 4-180), namely NS1N. NS1N was expressed in E. coli, and purified with a Ni-NTA affinity followed by size-exclusion chromatography (SEC). The purified protein was >95% pure (Figure 2 inset). The homogeneity of the protein was demonstrated by a single peak on SEC at an elution volume of 18.59 ml corresponding to a molecular weight of approximately 28 kDa, indicating that the NS1N existed as a monomer in solution (Figure 2).
Figure 2.
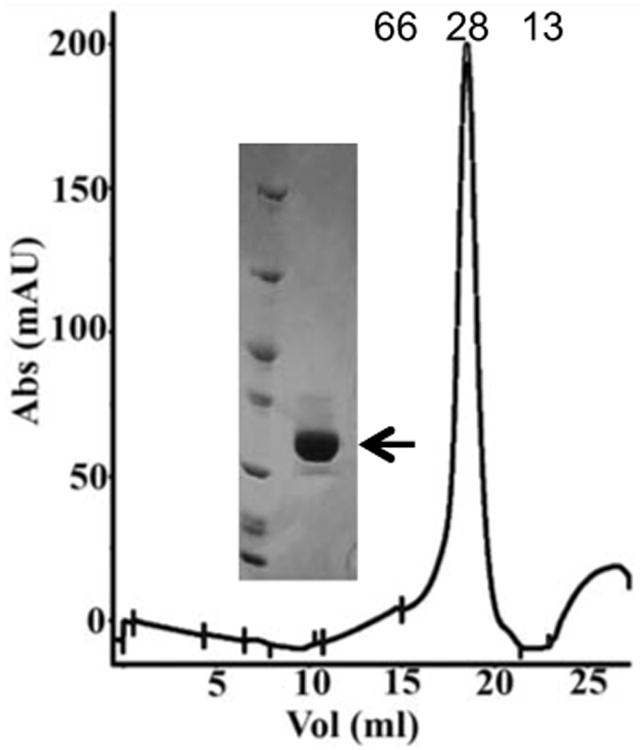
Analytical gel filtration chromatography of purified B19V NS1N showing a peak at an elution volume of 18.6 ml that corresponds to a molecular weight of ∼28 kDa. Positions of the molecular weight standards at 66 and 13 kDa, bovine serum albumin and cytochrome-C respectively, are indicated. Inset, SDS polyacrylamide gel electrophoresis of the purified NS1N (arrow in the right lane) and unstained protein molecular weight marker (Thermo Scientific™) (left lane).
NS1N specifically binds to the B19V Ori
Previous studies showed that NSBEs were essential for B19V replication, suggesting that this region may serve as the potential NS1-binding site (Guan et al., 2009). However, whether and how NS1 physically binds to NSBEs hasn't been clear. We characterized in vitro binding of NS1N and the B19V Ori. Synthesized plus and minus strands of NSBE were annealed to generate double-stranded DNA (dsDNA) molecules as described in Materials and Methods. DNA molecules were incubated with the purified NS1N. It is observed that NS1N strongly and specifically bound to B19V 67-bp Ori, a 38-bp wt DNA encompassing the four NSBEs, and a 28-bp wt DNA encompassing NSBE1 to NSBE3 (Figure 3; Table 1). A single clear band was observed that migrated differently from the free DNA, indicating that NS1N and DNA formed a single species of nucleoprotein complex (Figure 3A, lanes 1-3; S1A, lanes 1-7 and S1B, lanes 2-5 in the supplemental materials). The putative nucleoprotein complexes of NS1N with the 38-bp and 28-bp DNA molecules both showed a sharp, single band, suggesting formation of stable protein:DNA complexes. The putative nucleoprotein complex between NS1N and the 67-bp DNA showed a diffusive band (Figure 3A, lane 1), which suggests dynamics of the NS1N:DNA interaction and/or reflects sequential binding of multiple copies of NS1N to the DNA resulting in continuous bands that were not resolved in the gel. Smears were observed between the unbound DNA bands and the bands for the putative NS1N:DNA complexes (Figure 3A, lanes 1-3), reflecting dynamics of NS1N:DNA binding and/or the presence of non-specific NS1N:DNA interaction. No binding was observed for a 21-bp non-specific DNA (Figure 3A, lane 7). These data demonstrate that the NSBE region is indeed the NS1N-binding site, and NS1N specifically binds to that region to form a stable nucleoprotein complex.
Figure 3.
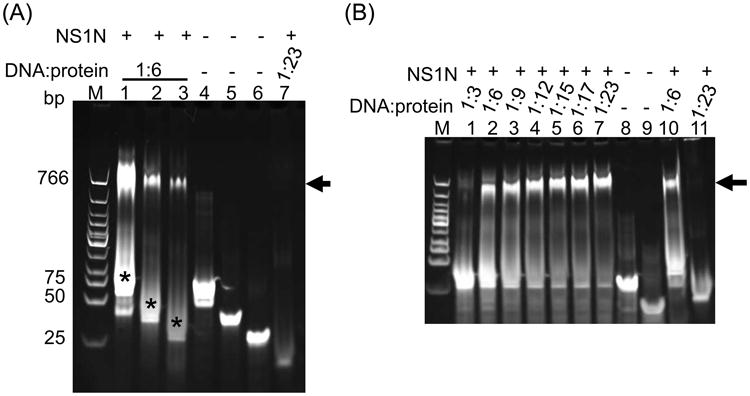
B19V NS1N specifically binds to Ori. (A) Lanes 1-3, NS1N incubated with the 67-bp Ori, the 38-bp NSBE1+2+3+4 wt and the 28-bp NSBE1+2+3 wt, respectively (for nomenclature of DNA molecules, refer to Table 1). Lanes 4-6, the 67-bp, 38-bp and 28-bp DNA alone, respectively. Lane 7, NS1N incubated with 21-bp non-specific DNA. Smears in Lanes 1-3 representing dynamic and/or non-specific DNA:protein interactions are indicated by asterisks. (B) Lanes 1-7, NS1N binding with the 28-bp NSBE1+2+3 wt at various DNA:protein molar ratios as indicated above the gel. Lane 8 and 9, the 28-bp NSBE1+2+3 wt DNA and its upper strand alone, respectively. Lane 10, NS1N binding with the 38-bp NSBE1+2+3+4 wt as a comparison. Lane 11, NS1N incubated with the 21-bp non-specific DNA. The potential nucleoprotein complex is indicated with arrow. Lane M, DNA marker (New England Biolabs).
Table 1.
| DNA | Length/bp | Sequence* | Binding affinity** |
|---|---|---|---|
| Ori | 67 | 5′-AAAATTTAAAAGAAGACACCAAATCAGATGCCGCCGGTCGCC GCCGGTAGGCGGGACTTCCGGTACA -3′ | ++++ |
| NSBE1+2+3+4 wt | 38 | 5′- TGCCGCCGGTCGCCGCCGGTAGGCGGGACTTCCGGTACA -3 | ++++ |
| NSBE1+2+3 wt | 28 | 5 - TGCCGCCGGTCGCCGCCGGTAGGCGGGAC -3′ | ++++ |
| NSBE2+3+4 | 28 | 5′-AGCCGCCGGTAGGCGGGACTTTCCGGTACA-3 | + |
| NSBE1-3456 | 28 | 5 - AGCTATTGGTCGCCGCCGGTAGGCGGGAC -3′ | + |
| NSBE2-3456 | 28 | 5′-AGCCGCCGGTCGCTATTGGTAGGCGGGAC-3 | + |
| NSBE3-3456 | 28 | 5 -AGCCGCCGGTCGCCGCCGGTAGGTAAAA C-3 | ++++ |
| NSBE1+2-3456 | 28 | 5′-AGCTATTGGTCGCTATTGGTAGGCGGGAC-3′ | - |
| NSBE3-1to8 | 28 | 5 - AGCCGCCGGTCGCCGCCGGTATTATTTCA -3′ | ++++ |
| NSBE1+2 | 18 | 5′- TGCCGCCGGTCGCCGCCGG -3 | - |
| NSBE2+3 | 18 | 5′- TGCCGCCGGTAGGCGGGAC -3′ | - |
| NSBE3+4 | 18 | 5′- AGGCGGGACTTCCGGTACA -3 | - |
| NSBE/19 | 19 | 5′-AGCCGCCGGTCGCCGCCGGT-3 | - |
| NSBE/20 | 20 | 5 -AGCCGCCGGTCGCCGCCGGTA-3′ | - |
| NSBE/21 | 21 | 5′-AGCCGCCGGTCGCCGCCGGTAG-3′ | ++ |
| NSBE/23 | 23 | 5′- AGCCGCCGGTCGCCGCCGGTAGGC -3′ | +++ |
| NSBE/25 | 25 | 5′- AGCCGCCGGTCGCCGCCGGTAGGCGG-3′ | ++++ |
| NSBE1+2-1AA | 28 | 5′- AACCGCCGGTCACCGCCGGTAGGCGGGAC-3′ | +++ |
| NSBE1+2-2TT | 28 | 5′-AGTCGCCGGTCGTCGCCGGTAGGCGGGAC-3′ | ++++ |
| NSBE1+2-3TT | 28 | 5′-AGCTGCCGGTCGCTGCCGGTAGGCGGGAC-3′ | ++++ |
| NSBE1+2-4AA | 28 | 5′-AGCCACCGGTCGCCACCGGTAGGCGGGAC-3′ | +++ |
| NSBE1+2-5TT | 28 | 5′-AGCCGTCGGTCGCCGTCGGTAGGCGGGAC-3′ | ++++ |
| NSBE1+2-6TT | 28 | 5′-AGCCGCTGGTCGCCGCTGGTAGGCGGGAC-3′ | + |
| NSBE1+2-7AA | 28 | 5′-AGCCGCCAGTCGCCGCCAGTAGGCGGGAC-3′ | + |
| NSBE1+2-8AA | 28 | 5′-AGCCGCCGATCGCCGCCGATAGGCGGGAC-3′ | + |
Double-stranded DNA molecules were used in EMSA, but only plus strands of those DNA molecules are listed here. All dsDNA molecules except for the Ori contain a single 5′ overhang on both strands. The bold underlined letters indicate positions of mutation. wt, wild type DNA.
The binding affinity of NS1N is shown by plus signs, and the minus sings mean no observed binding.
NSBEs 1 and 2 are required for NS1N binding in a sequence-dependent manner, while NSBEs 3 and 4 are dispensable
To examine the roles of the four NSBEs with NS1N interaction, we constructed 28-bp dsDNA molecules that encompass NSBEs 1 to 3 designated as NSBE1+2+3 wt (Figure 1A; Table 1). EMSA shows that 28-bp wt DNA binds to NS1N at an affinity similar to that of the 38-bp wt DNA (Figure 3A, lanes 2-3; 3B, lanes 2-7; S1B, lanes 2-5), suggesting that NSBE4 is dispensable for NS1N binding. We then tested the sequence dependence of the NSBEs for NS1N binding by mutagenesis analysis using a variety of mutated DNA molecules (Table 1). The nts at the 3rd, 4th, 5th and 6th positions (that is, those in the center of the NSBE segments) in either NSBE1, NSBE2 or NSBE3 were mutated concomitantly. The four concomitant mutations in NSBE3 caused no apparent change for NS1N binding (Table 1; Figure 4, lane 3; Figure S2C, lanes 2-7). Furthermore, concomitant mutation of all eight nts in NSBE3 also showed no apparent effect on NS1N binding (Table 1; Figure 4, lane 6; Figure S2F, lanes 2-7). These data indicate that NSBE3 is dispensable for NS1N binding and the sequence of NSBE3 plays a minimal, if any, role in specific interaction with NS1N.
Figure 4.
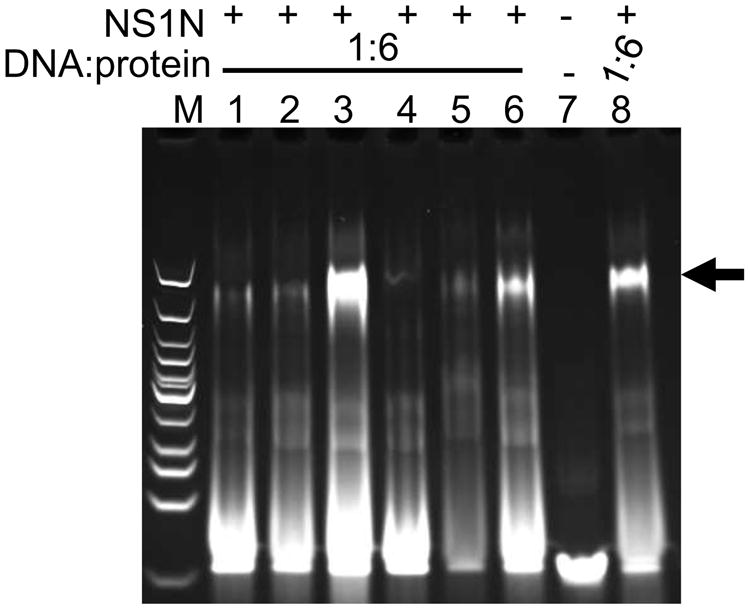
NS1N binding to DNA with mutations in NSBEs. Lanes 1-6, NS1N incubated with the 28-bp NSBE1-3456, NSBE2-3456, NSBE3-3456, NSBE1+2-3456, NSBE2+3+4 and NSBE3-1to8, DNA, respectively. Lane 7, the 28-bp NSBE1+2+3 wt DNA alone. Lane 8, NS1N incubated with the 28-bp NSBE1+2+3 wt DNA. The DNA:protein molar ratios are shown. The potential nucleoprotein complexes are indicated with arrow. Lane M, DNA marker. See Table 1 for nomenclature of DNA molecules.
Mutations in either NSBE1 or NSBE2 considerably reduced the NS1N binding affinity, by at least 4-fold (Table 1; Figure 4, lanes 1-2; Figure S2A, lane 2; Figure S2B, lanes 2-3). A DNA molecule encompassing NSBEs 2, 3 and 4 showed a reduction of at least 4-fold in NS1N binding affinity (Table 1; Figure 4, lane 5; S2E, lanes 2-7). Concomitant mutations of the nts at the 3rd, 4th, 5th and 6th positions in both NSBE1 and NSBE2 lead to more significant reduction in NS1N binding and essentially abolished NS1N binding at a protein:DNA molar ratio of up to 12:1 (Figure 4, lane 4; S2D, lanes 2-4; Table 1). These data indicate that NSBEs 1 and 2 are both required for NS1N binding in a sequence-dependent manner, while NSBEs 3 and 4 are dispensable.
NSBEs 1 and 2 are not sufficient for NS1N binding and an extension in DNA length is required for optimal NS1N:DNA interaction
DNA molecules of 18-bp length that encompasses two consecutive NSBEs, designated NSBE1+2, NSBE2+3 and NSBE3+4, were designed as shown in Table1. These DNA molecules showed little binding to NS1N in EMSA (Figure 5, A-C, lanes 2-7). A 21-bp non-specific DNA showed no interactions with excess of the NS1N protein (Figure 5, A-C, lane 11). While the observation of little binding of NSBE3+4 and NSBE2+3 is consistent with the notes in the last section that the NSBEs 3 and 4 are dispensable for NS1N binding and that both NSBE1 and NSBE2 are essential for NS1N binding, virtually no binding of the 18-bp NSBE1+2 molecule suggests that the NS1N:DNA interaction is dependent on DNA length. To further test this, EMSA analysis was carried out with DNA molecules consisting of the NSBEs 1 and 2 but with shorter lengths, that is, 19, 20, 21, 23 and 25 bp, respectively (Table 1). EMSA showed that the NS1N binding affinity correlated well with the length of DNA (Figure 6; Figure S3, A-E). Very weak NS1N binding was observed with the 19-bp DNA (Figure S3A, lanes 2-7). With 20-bp DNA, only weak binding of NS1N was observed at a protein:DNA molar ratio of 17:1 (Figure 6, lane 1 and Figure S3B, lanes 2-5). The minimum length of the NSBE that showed NS1N binding affinity comparable to that of the 28-bp DNA is 21-bp (Figure 6, lane 2; S3C, lanes 2-7). These data suggest that NS1N binding requires a minimal length of DNA that extends beyond the sole NSBE1 and NSBE2 portions of the DNA, which reflects a spatial requirement for accommodating NS1N molecules onto the bound DNA.
Figure 5.
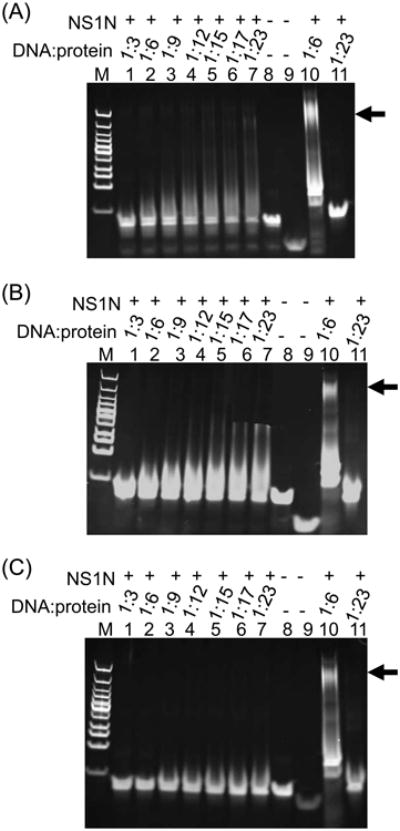
EMSA of NS1N with 18-bp DNA molecules encompassing two NSBEs. (A) Lanes 1-7, NS1N incubated with NSBE1+2 in various DNA:protein molar ratios. Lane 8 and 9, the 18-bp NSBE1+2 DNA and its upper strand alone, respectively. Lane 10, NS1N incubated with the 28-bp NSBE1+2+3 wt DNA. Lane 11, NS1N incubated with 21-bp non-specific DNA. (B) and (C), are same as (A) except that the 18-bp NSBE2+3 and NSBE3+4 were used in lanes 1-7 respectively. The DNA:protein molar ratios are indicated. The potential nucleoprotein complexes in lane 10 in all panels are indicated with an arrow. Lane M, DNA marker.
Figure 6.
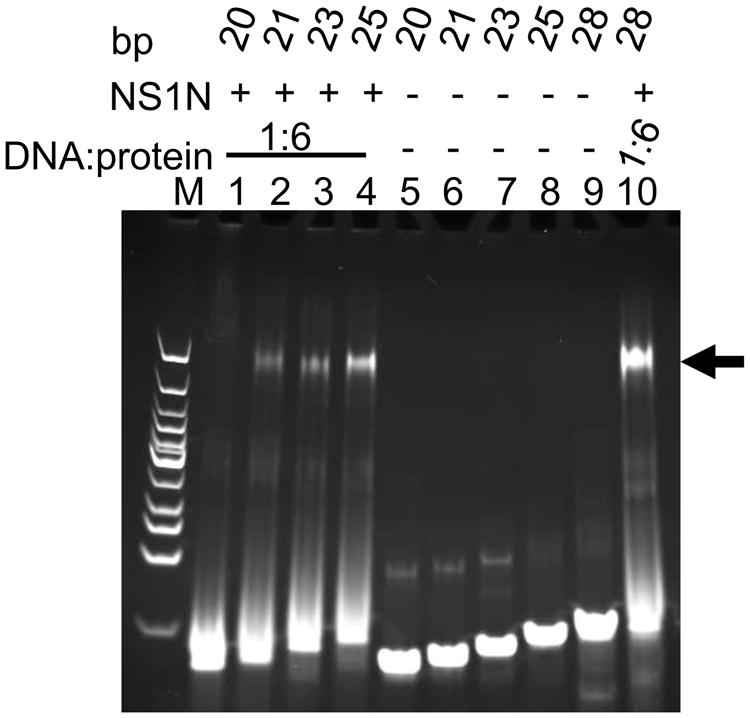
Length dependence of DNA binding by NS1N. Lanes 1-4, NS1N incubated with 20-, 21-, 23- and 25-bp DNA (NSBE/20, NSBE/21, NSBE/23 and NSBE/25 in Table 1), respectively. Lanes 5-8, the 20-, 21-, 23- and 25-bp DNA alone, respectively. Lane 9, the 28-bp NSBE1+2+3 wt DNA. Lane 10, NS1N incubated with the 28-bp NSBE1+2+3 wt DNA. The nucleoprotein complexes are indicated with an arrow. The DNA:protein molar ratios are indicated. The numbers of base pairs for DNA molecules are labeled on the top. Lane M, DNA marker.
Identification of the nucleotides in NSBEs 1 and 2 that are critical for sequence-specific NS1N binding
The NSBE1 and NSBE2 are identical in DNA sequences (Figure 1A). We systematically analyzed the dependence of NS1N binding on the DNA sequence. Single mutations on either NSBE1 or NSBE2 did not confer significant influence on NS1N binding (data not shown), in agreement with a cooperative binding mode between the two NSBEs and NS1N. Thus, a series of 28-bp DNA molecules encompassing NSBEs 1, 2 and 3 were designed, each with an identical point mutation in both NSBE1 and NSBE2 to facilitate assessment of the effect of mutation on NS1N binding (Table 1). Those double mutations exhibited differential effects on NS1N:DNA interaction (Figure 7, lanes 1-8; S4 A-H, lanes 2-7). Little or minimal effects on NS1N binding were observed for mutations on the 2nd, 3rd and 5th positions (Figure 7A, lanes 2, 3 and 5; S4 B, C and E, lanes 2-7; 7B). The mutations at the 1st and 4th positions showed very weak NS1N binding at a DNA:protein molar ratio of 1:6 but showed clear binding at 1:9 (Figure 7A, lane 1 and 4; S4 A and D, lane 2; 7B). Mutations at positions 6, 7 and 8 showed significantly weaker NS1N binding than mutations at other positions (Figure 7A, Lanes 6-8; S4 F-H; 7B). Among them, mutations at positions 7 and 8 appeared to have the maximal effect on NS1N binding (Figure 7A, Lanes 7-8; Figure S3 G and H; 7B). Taken together, these data suggest that the nts at positions 2, 3 and 5 in the NSBEs 1 and 2 sequence may be not involved in sequence-specific NS1N:DNA recognition, although they may participate in NS1N:DNA interaction in a non-sequence-specific manner. The nts at positions 1 and 4 may be involved in sequence-specific NS1N:DNA recognition, and the nts at positions 6-8 are critical for sequence-specific NS1N:DNA recognition.
Figure 7.
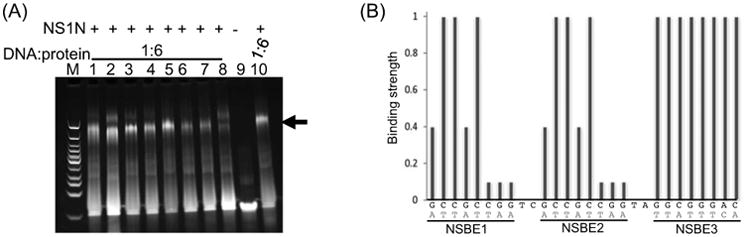
Identification of nucleotides in NSBEs 1 and 2 critical for NS1N binding. (A) Lanes 1-8, NS1N incubated with the 28-bp NSBE1+2-1AA, NSBE-1+2-2TT, NSBE1+2-3TT, NSBE1+2-4AA, NSBE1+2-5TT, NSBE1+2-6TT, NSBE1+2-7AA and NSBE1+2-8AA DNA, respectively (for nomenclature of DNA molecules, refer to Table 1). Lane 9, the 28-bp NSBE1+2+3 wt DNA alone. Lane 10, NS1N incubated with the 28-bp NSBE1+2+3 wt DNA. The potential nucleoprotein complexes are indicated with an arrow. Lane M, DNA marker. (B) A plot showing effects of point mutations in NSBEs on NS1N binding, which combines EMSA results for the eight DNA molecules in (A) (see Table 1 for actual sequences of DNA molecules used in the assay). The vertical axis is semi-quantified binding strength based on EMSA results. The horizontal axis shows the nucleotide sequence of the region of NSBEs 1 to 3 (upper row in black) as well as the nucleotides they were mutated to (bottom row in red). The effect of mutation in NSBE3 was based on a single EMSA result using the 28-bp NSBE3-1to8 DNA, in which all the eight nucleotides were mutated (Table 1; Figure 4, lane 6).
Discussion
The minimal origin of DNA replication in B19V was previously identified, and four DNA fragments located therein were shown to be essential for optimal virus replication and could serve as the potential NS1-binding sites (Guan et al., 2009). However, whether and how those DNA fragments physically bind to NS1 hasn't been addressed. In this work, we have characterized in vitro molecular interaction of NS1N with Ori. We have demonstrated that the NS1N binds to Ori to form a nucleoprotein complex, and this binding is sequence-specific. NSBE1 and NSBE2 are both essential for NS1 binding, whereas NSBE3 and NSBE4 are dispensable. These data indicate that NSBEs 1 and 2 may be the binding sites for NS1, whereas NSBEs 3 and 4 may not directly interact with NS1. The differential roles of the NSBEs 1 to 4 in NS1N:DNA interactions observed here are in good agreement with previous results showing that NSBEs 1 to 3 were essential for virus replication and NSBE4 was not essential but was needed for maximal virus replication (Guan et al., 2009). In MVM, two copies of the ACGT motif near the NS1 binding sites were demonstrated to be the binding sites for host glucocorticoid modulating element binding proteins that were dubbed parvovirus initiation factor (PIF), and NS1 and PIF must form a ternary complex with Ori for nicking to occur (Christensen et al., 1997, 1999, 2001). Thus, it is likely that NSBEs 3 and 4 may serve as the binding sites for potential host factor(s). This is in agreement with the fact that the B19V NSBEs 1 and 2 are identical but NSBEs 3 and 4 show distinct sequences, making it unlikely for NS1 to bind to the four NSBEs in a same sequence-specific manner. Nevertheless, only the N-terminal domain of NS1 was used in the present study, and it couldn't be ruled out that other portions of the NS1 protein might interact with NSBEs 3 and/or 4. Participation of cellular factors was also observed during NS1 interaction with the p6 promoter, which involves the cellular Sp1/Sp3 transcription factors (Doerig et al., 1990; Gareus et al., 1998; Raab et al., 2002). Thus, NS1 functions appear to be highly orchestrated with cellular factors. B19V has a strict tropism for human erythroid progenitor cells, and B19V infection causes cell cycle arrest in the S phase in which cellular factors for viral DNA replication are available (Luo et al., 2013). In MVM, it was shown in a reconstituted DNA replication system that NS1 bound to single-stranded DNA binding protein RPA to form a functional replication fork (Christensen and Tattersall, 2002). It would be interesting to address if and how the potential host factors that bind to NSBEs 3 and 4 are involved in B19V host tropism and B19V-induced cell cycle arrest.
Our results provided further details regarding NS1:Ori interaction. A DNA molecule solely consisting of NSBEs 1 and 2 did not support NS1N binding, and any DNA molecules consisting of two consecutive NSBEs did not support NS1 binding either. Moreover, a 18-bp DNA fragment encompassing NSBE1 and NSBE2 failed to bind to NS1N, and the minimal length of DNA allowing NS1N binding requires at least a 3-bp extension to the 3′ end of that fragment, indicating a requirement for sufficient space on DNA to accommodate bound NS1. Our mutagenesis analysis has identified nucleotides in NSBE1 and NSBE2 that are critical for NS1N binding. The nucleotides at positions 6, 7 and 8 in the NSBEs 1 and 2 sequence motif are critical for sequence-specific NS1N:DNA recognition, whereas those at positions 1 and 4 may also be involved in NS1N:DNA recognition but may be less critical, and those at positions 2, 3 and 5 may not be involved in sequence-specific NS1N:DNA recognition, although they may participate in NS1N:DNA interaction in a non-sequence-specific manner. These data suggest that the nucleotides at positions 6-8 may be involved in direct interaction with NS1N by, for example, insertion of amino acid residues of the NS1 protein into the grooves in the DNA, thus interacting with DNA bases. Such interactions can confer sequence specificity for the NS1:DNA recognition. Such protein:DNA interactions were observed in AAV Rep N-terminal nickase domain (Hickman et al., 2004). The Rep nickase domain interacts with tetra-nucleotide repeats in AAV Ori by providing a loop to one repeat and an α helix to an adjacent repeat, where long side chains of Arg and Lys residues insert into the DNA major and minor grooves respectively (Hickman et al., 2004).
The presence of two identical NSBEs (1 and 2) suggests that multiple NS1 molecules participate in binding to the Ori region. In AAV, five copies of the Rep nuclease domain bound to the Rep-binding site, consistent with the presence of five tetranucleotide repeats (Hickman et al., 2004). It was proposed that binding of multiple copies of Rep would lead to oligomerization of Rep to form a ring-like hexamer, which would then generate ssDNA around the trs region, enabling nicking and DNA strand separation (Hickman et al., 2004). In the B19V Ori, the NSBEs 1 and 2 can accommodate at least two NS1 molecules, assuming each of the two NSBEs is bound by a single copy of NS1. Interestingly, the sequence of the NSBEs 1 and 2 region in Ori (5′- GCCGCCGG tc GCCGCCGG -3′ where the two-nucleotide spacer are shown in lower case; Figure 1) can be considered as five consecutive repeats of a single tetranucleotide motif GCCG, in which the first and second repeats share a nucleotide G (the same for the fourth and fifth repeats) and the third repeat possesses a degenerative sequence of GTCG. It is tempting to speculate that such a DNA structure can allow multiple copies of NS1 to bind to the DNA in a tandem and approximately helical arrangement bearing similarity to AAV Rep, which promotes subsequent assembly of a nucleoprotein complex between NS1 and Ori in which NS1 is activated for DNA nicking. Nevertheless, the detailed modes of Ori binding in B19V NS1 and AAV Rep must be different as evidenced by the different DNA sequence motifs and lack of apparent amino acid sequence identity between B19V NS1 and AAV Rep.
Taken together, these results suggest a mode of interaction between NS1 and Ori, in which NS1 physically binds to the region encompassing NSBE1 and NSBE2, while NSBE3 and NSBE4 may provide sites for binding of potential cellular factors (Figure 8). Such molecular interactions lead to assembly of multiple copies of NS1 with Ori into a specialized nucleoprotein complex, enabling nicking at the terminal resolution site and displacement of DNA strands during DNA replication. Further studies are needed to address what the exact NS1-binding motif is, how NS1 assembles with Ori, how the NS1 nickase and helicase domains are coordinated during viral DNA replication, and the identity of potential cellular factors that are involved in NS1:Ori interaction.
Figure 8.
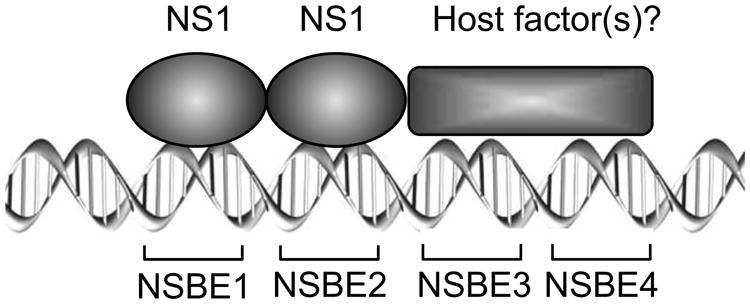
A model for the binding of NS1 to the B19V Ori. The NSBE1 and NSBE2 are the binding sites for NS1 (ovals). It is likely that multiple NS1 molecules participate in the binding, but only two NS1 molecules are shown schematically. NSBE3 and MSBE4 may provide the binding sites for potential host factors (rectangle). See text for details.
Materials and Methods
Protein expression and purification
The fragment encoding non-structural protein 1 (NS1N; residues 4-180) was PCR amplified from the B19V genomic DNA (Zhi et al., 2004) with forward and reverse primers containing NdeI and BamHI restriction sites respectively with a stop codon. Digested PCR product was ligated into the expression vector pDZ1, resulting in the NS1N protein carrying a N-terminal hexa-histidine tag followed by an 8-kDa GB1 tag (Streptococcal protein G B1 domain). NS1N was expressed in E. coli strain BL21(DE834) cells in LB media supplemented with 30 μg/ml carbenicillin. After growth at 37°C to an absorbance at 600 nm of 0.6, NS1N expression was induced with 1 mM IPTG (isopropyl-β -D-thiogalactopyranoside) and the culture was further grown at 28°C for 16 h. The overnight grown cells were collected and lysed on a French press in a lysis buffer containing 20 mM Tris-HCl pH 8.0, 500 mM NaCl and 5mM β-mercaptoethanol. NS1N was purified with the N-terminal hexa-histidine tag by metal-chelating chromatography using Ni-NTA beads (Qiagen). The elution from the Ni-NTA beads was further purified by gel filtration chromatography on a sephacryl S-300 26/60 column (GE Healthcare) pre-equilibrated with a gel filtration buffer containing 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EDTA and 1mM DTT. Typically, one liter of E. coli culture yielded about 3 mg of purified protein, which was concentrated to approximately 10 mg/ml using a Millipore concentrator (Molecular weight cut off 10 kDa). The homogeneity of the purified protein was analyzed by the analytical gel filtration column Superose 6 10/300 (GE Healthcare) (Fig. 2).
Electrophoretic mobility shift assay of NS1N:DNA interaction
The wild type (wt) B19V Ori DNA as well as mutated ssDNA oligos were synthesized (Integrated DNA Technologies) as described in Table 1. The synthesized DNA oligos were annealed to complementary strands in a buffer containing 20 mM Tris-HCl pH 7.0, 10 mM KCl, 3 mM MgCl2 and 50 mM NaCl at 95°C for 10 min followed by rapid cooling on ice. Annealed DNA molecules were used in subsequent electrophoretic mobility shift assay (EMSA). Briefly, increasing amounts of NS1N protein were mixed with a constant amount of DNA at various DNA:protein molar ratios, followed by incubation at 25°C for 1 h. Samples were loaded onto 10% polyacrylamide Tris-borate EDTA (TBE) gels in a TBE buffer containing 45 mM Tris-HCl pH 8.30, 45 mM boric acid and 1 mM EDTA, which were run at 200 Volts for 1 h. TBE gels were stained with ethidium bromide for 10 min on a rocker before recording the images on a trans-illuminator.
Supplementary Material
Research Highlights.
The NS1 protein of human parvovirus B19 plays critical roles in viral replication.
The NS1 N-terminal domain specifically binds to the origin of replication.
NSBE1 and 2 are essential while NSBE3 and 4 are dispensable for NS1 binding.
NSBE3 and 4 may provide binding sites for potential cellular factors.
Acknowledgments
This work was supported by the grant R01AI070723 from the National Institute of Allergy and Infectious Diseases, the National Institutes of Health to J.Q. We thank Dr. Roberto De Guzman for sharing the pDZ1 vector.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson LJ, Tsou C, Parker RA, Chorba TL, Wulff H, Tattersall P, Mortimer PP. Detection of antibodies and antigens of human parvovirus B19 by enzyme-linked immunosorbent assay. Journal of clinical microbiology. 1986;24:522–526. doi: 10.1128/jcm.24.4.522-526.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astell CR, L W, Brunstein J, St Amannd J. B19 Parvovirus: biochemical and molecular features. In: Anderson LJ, Young N, editors. Human parvovirus B19. Karger; Basel, Switzerland: 1997. pp. 16–41. [Google Scholar]
- Astell CR, Thomson M, Chow MB, Ward DC. Structure and replication of minute virus of mice DNA. Cold Spring Harbor symposia on quantitative biology. 1983;47 Pt 2:751–762. doi: 10.1101/sqb.1983.047.01.086. [DOI] [PubMed] [Google Scholar]
- Berns K, Parrish CR. Parvoviridae. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields' virology. 5. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 2437–2477. [Google Scholar]
- Bleker S, Pawlita M, Kleinschmidt JA. Impact of capsid conformation and Rep-capsid interactions on adeno-associated virus type 2 genome packaging. J Virol. 2006;80:810–820. doi: 10.1128/JVI.80.2.810-820.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell MC, Beard C, Astell CR. In vitro identification of a B19 parvovirus promoter. Virology. 1987;157:534–538. doi: 10.1016/0042-6822(87)90296-0. [DOI] [PubMed] [Google Scholar]
- Chen AY, Zhang EY, Guan W, Cheng F, Kleiboeker S, Yankee TM, Qiu J. The small 11 kDa nonstructural protein of human parvovirus B19 plays a key role in inducing apoptosis during B19 virus infection of primary erythroid progenitor cells. Blood. 2010;115:1070–1080. doi: 10.1182/blood-2009-04-215756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Cotmore SF, Tattersall P. Parvovirus initiation factor PIF: a novel human DNA-binding factor which coordinately recognizes two ACGT motifs. J Virol. 1997;71:5733–5741. doi: 10.1128/jvi.71.8.5733-5741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Cotmore SF, Tattersall P. Two new members of the emerging KDWK family of combinatorial transcription modulators bind as a heterodimer to flexibly spaced PuCGPy half-sites. Molecular and cellular biology. 1999;19:7741–7750. doi: 10.1128/mcb.19.11.7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Cotmore SF, Tattersall P. Minute virus of mice initiator protein NS1 and a host KDWK family transcription factor must form a precise ternary complex with origin DNA for nicking to occur. J Virol. 2001;75:7009–7017. doi: 10.1128/JVI.75.15.7009-7017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Tattersall P. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J Virol. 2002;76:6518–6531. doi: 10.1128/JVI.76.13.6518-6531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore SF, Tattersall P. Structure and organization of the viral genome. In: Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. Hodder Arnold; London, United Kingdom: 2005. pp. 73–94. [Google Scholar]
- Cotmore SF, Gottlieb RL, Tattersall P. Replication initiator protein NS1 of the parvovirus minute virus of mice binds to modular divergent sites distributed throughout duplex viral DNA. Journal of virology. 2007;81:13015–13027. doi: 10.1128/JVI.01703-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore SF, Tattersall P. Encapsidation of minute virus of mice DNA: aspects of the translocation mechanism revealed by the structure of partially packaged genomes. Virology. 2005a;336:100–112. doi: 10.1016/j.virol.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Cotmore SF, Tattersall P. Structure and organization of the viral genome. In: Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. Hodder Arnold; London, United Kindom: 2005b. pp. 73–94. [Google Scholar]
- Deiss V, Tratschin JD, Weitz M, Siegl G. Cloning of the human parvovirus B19 genome and structural analysis of its palindromic termini. Virology. 1990;175:247–254. doi: 10.1016/0042-6822(90)90205-6. [DOI] [PubMed] [Google Scholar]
- Doerig C, Hirt B, Antonietti JP, Beard P. Nonstructural protein of parvoviruses B19 and minute virus of mice controls transcription. J Virol. 1990;64:387–396. doi: 10.1128/jvi.64.1.387-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan MM, Tamburic L, Shippam-Brett C, Zagrodney DB, Astell CR. The small 11-kDa protein from B19 parvovirus binds growth factor receptor-binding protein 2 in vitro in a Src homology 3 domain/ligand-dependent manner. Virology. 2001;291:285–291. doi: 10.1006/viro.2001.1217. [DOI] [PubMed] [Google Scholar]
- Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. Virus Taxonomy, VIIIth Report of the ICTV. Elsevier/Academic Press; London: 2004. pp. 277–370. [Google Scholar]
- Fu Y, Ishii KK, Munakata Y, Saitoh T, Kaku M, Sasaki T. Regulation of tumor necrosis factor alpha promoter by human parvovirus B19 NS1 through activation of AP-1 and AP-2. J Virol. 2002;76:5395–5403. doi: 10.1128/JVI.76.11.5395-5403.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareus R, Gigler A, Hemauer A, Leruez-Ville M, Morinet F, Wolf H, Modrow S. Characterization of cis-acting and NS1 protein-responsive elements in the p6 promoter of parvovirus B19. J Virol. 1998;72:609–616. doi: 10.1128/jvi.72.1.609-616.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W, Wong S, Zhi N, Qiu J. The genome of human parvovirus b19 can replicate in nonpermissive cells with the help of adenovirus genes and produces infectious virus. J Virol. 2009;83:9541–9553. doi: 10.1128/JVI.00702-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Wang Q, Qiu Y, Wu W, He H, Zhang J, Hu Y, Zhou X. Periplaneta fuliginosa densovirus nonstructural protein NS1 contains an endonuclease activity that is regulated by its phosphorylation. Virology. 2013;437:1–11. doi: 10.1016/j.virol.2012.12.006. [DOI] [PubMed] [Google Scholar]
- Hickman AB, Ronning DR, Perez ZN, Kotin RM, Dyda F. The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces. Mol Cell. 2004;13:403–414. doi: 10.1016/s1097-2765(04)00023-1. [DOI] [PubMed] [Google Scholar]
- Hsu GJ, Tzang BS, Tsai CC, Chiu CC, Huang CY, Hsu TC. Effects of human parvovirus B19 on expression of defensins and Toll-like receptors. The Chinese journal of physiology. 2011;54:367–376. [PubMed] [Google Scholar]
- King JA, Dubielzig R, Grimm D, Kleinschmidt JA. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 2001;20:3282–3291. doi: 10.1093/emboj/20.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krady JK, Ward DC. Transcriptional activation by the parvoviral nonstructural protein NS-1 is mediated via a direct interaction with Sp1. Molecular and cellular biology. 1995;15:524–533. doi: 10.1128/mcb.15.1.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Rhode SL., 3rd Mutation of lysine 405 to serine in the parvovirus H-1 NS1 abolishes its functions for viral DNA replication, late promoter trans activation, and cytotoxicity. Journal of virology. 1990;64:4654–4660. doi: 10.1128/jvi.64.10.4654-4660.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou S, Luo Y, Cheng F, Huang Q, Shen W, Kleiboeker S, Tisdale JF, Liu Z, Qiu J. Human parvovirus B19 DNA replication induces a DNA damage response that is dispensable for cell cycle arrest at phase G2/M. J Virol. 2012;86:10748–10758. doi: 10.1128/JVI.01007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Kleiboeker S, Deng X, Qiu J. Human parvovirus B19 infection causes cell cycle arrest of human erythroid progenitors at late S phase that favors viral DNA replication. J Virol. 2013 doi: 10.1128/JVI.02333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Lou S, Deng X, Liu Z, Li Y, Kleiboeker S, Qiu J. Parvovirus B19 infection of human primary erythroid progenitor cells triggers ATR-Chk1 signaling, which promotes B19 virus replication. J Virol. 2011;85:8046–8055. doi: 10.1128/JVI.00831-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt S, Tanaka N, Tada K, Nose M, Nakamura M, Muraoka O, Hirano T, Sugamura K. A cytotoxic nonstructural protein, NS1, of human parvovirus B19 induces activation of interleukin-6 gene expression. Journal of virology. 1996;70:8485–8491. doi: 10.1128/jvi.70.12.8485-8491.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt S, Yaegashi N, Tada K, Tanaka N, Sugamura K. Human parvovirus B19 nonstructural (NS1) protein induces apoptosis in erythroid lineage cells. J Virol. 1998;72:3018–3028. doi: 10.1128/jvi.72.4.3018-3028.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Nakashima A, Asao H, Sato H, Sugamura K. Human parvovirus B19 nonstructural protein (NS1) induces cell cycle arrest at G(1) phase. J Virol. 2003;77:2915–2921. doi: 10.1128/JVI.77.5.2915-2921.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A, Morita E, Saito S, Sugamura K. Human Parvovirus B19 nonstructural protein transactivates the p21/WAF1 through Sp1. Virology. 2004;329:493–504. doi: 10.1016/j.virol.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Ozawa K, Ayub J, Hao YS, Kurtzman G, Shimada T, Young N. Novel transcription map for the B19 (human) pathogenic parvovirus. J Virol. 1987;61:2395–2406. doi: 10.1128/jvi.61.8.2395-2406.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa K, Kurtzman G, Young N. Replication of the B19 parvovirus in human bone marrow cell cultures. Science. 1986;233:883–886. doi: 10.1126/science.3738514. [DOI] [PubMed] [Google Scholar]
- Plevka P, Hafenstein S, Li L, D'Abrgamo A, Jr, Cotmore SF, Rossmann MG, Tattersall P. Structure of a packaging-defective mutant of minute virus of mice indicates that the genome is packaged via a pore at a 5-fold axis. J Virol. 2011;85:4822–4827. doi: 10.1128/JVI.02598-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab U, Beckenlehner K, Lowin T, Niller HH, Doyle S, Modrow S. NS1 protein of parvovirus B19 interacts directly with DNA sequences of the p6 promoter and with the cellular transcription factors Sp1/Sp3. Virology. 2002;293:86–93. doi: 10.1006/viro.2001.1285. [DOI] [PubMed] [Google Scholar]
- Siegl G, Bates RC, Berns KI, Carter BJ, Kelly DC, Kurstak E, Tattersall P. Characteristics and taxonomy of Parvoviridae. Intervirology. 1985;23:61–73. doi: 10.1159/000149587. [DOI] [PubMed] [Google Scholar]
- Sun Y, Chen AY, Cheng F, Guan W, Johnson FB, Qiu J. Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J Virol. 2009;83:3956–3967. doi: 10.1128/JVI.02569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Z, Zhi N, Wong S, Keyvanfar K, Liu D, Raghavachari N, Munson PJ, Su S, Malide D, Kajigaya S, Young NS. Human parvovirus B19 causes cell cycle arrest of human erythroid progenitors via deregulation of the E2F family of transcription factors. The Journal of clinical investigation. 2010;120:3530–3544. doi: 10.1172/JCI41805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P. Replication of adeno-associated virus DNA. In: Kerr J, Cotmore SF, Bloom ME, Linden ME, Parrish CR, editors. Parvoviruses. Hodder Arnold; London, United Kingdom: 2005. pp. 189–211. [Google Scholar]
- Yaegashi N, Shiraishi H, Takeshita T, Nakamura M, Yajima A, Sugamura K. Propagation of human parvovirus B19 in primary culture of erythroid lineage cells derived from fetal liver. Journal of virology. 1989;63:2422–2426. doi: 10.1128/jvi.63.6.2422-2426.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NS, Brown KE. Parvovirus B19. N Engl J Med. 2004;350:586–597. doi: 10.1056/NEJMra030840. [DOI] [PubMed] [Google Scholar]
- Zhi N, Mills IP, Lu J, Wong S, Filippone C, Brown KE. Molecular and functional analyses of a human parvovirus B19 infectious clone demonstrates essential roles for NS1, VP1, and the 11-kilodalton protein in virus replication and infectivity. J Virol. 2006;80:5941–5950. doi: 10.1128/JVI.02430-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi N, Zadori Z, Brown KE, Tijssen P. Construction and sequencing of an infectious clone of the human parvovirus B19. Virology. 2004;318:142–152. doi: 10.1016/j.virol.2003.09.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


