Key Points
We found the defective ALDH2 variant is associated with accelerated progression of BMF in Japanese FA patients.
The data support the view that aldehydes are an important source of genotoxicity in the human hematopoietic system.
Abstract
Fanconi anemia (FA) is a severe hereditary disorder with defective DNA damage response and repair. It is characterized by phenotypes including progressive bone marrow failure (BMF), developmental abnormalities, and increased occurrence of leukemia and cancer. Recent studies in mice have suggested that the FA proteins might counteract aldehyde-induced genotoxicity in hematopoietic stem cells. Nearly half of the Japanese population carries a dominant-negative allele (rs671) of the aldehyde-catalyzing enzyme ALDH2 (acetaldehyde dehydrogenase 2), providing an opportunity to test this hypothesis in humans. We examined 64 Japanese FA patients, and found that the ALDH2 variant is associated with accelerated progression of BMF, while birth weight or the number of physical abnormalities was not affected. Moreover, malformations at some specific anatomic locations were observed more frequently in ALDH2-deficient patients. Our current data indicate that the level of ALDH2 activity impacts pathogenesis in FA, suggesting the possibility of a novel therapeutic approach.
Introduction
Fanconi anemia (FA) is a genomic instability disorder with phenotypes including progressive bone marrow failure (BMF), developmental abnormalities, and increased occurrence of leukemia and cancer.1 To date, 16 genes have been implicated in FA, and their products form a common DNA repair network (“FA pathway”).2,3 Because FA cells are hypersensitive to DNA interstrand cross-links (ICLs), the FA pathway has been considered to be involved in the repair of ICLs.2,3 However, it remains unclear what type of endogenous DNA damage is repaired through the FA pathway. Recent studies have suggested that FA cells are also sensitive to aldehydes,4 which may create DNA adducts including ICLs or DNA-protein crosslinks. Furthermore, double knockout mice deficient in Fancd2 and Aldh2, but neither of the single mutant mice, display an accelerated development of leukemia and BMF.5,6 On the other hand, Fanc-deficient mice in general do not fully recapitulate the human FA phenotype, including overt BMF.7 Thus, the role of aldehydes in the pathogenesis of human FA is still uncertain.
ALDH2 deficiency resulting from a Glu504Lys substitution (rs671, hereinafter referred to as the A allele) is highly prevalent in East Asian populations. The A allele (Lys504) acts as a dominant negative, since the variant form can suppress the activity of the Glu504 form (G allele) in GA heterozygotes by the formation of heterotetramers.8 Individuals with the A variant experience flushing when drinking alcohol, and have an elevated risk of esophageal cancer with habitual drinking.9 Because the frequency of the A allele is close to 50% in the Japanese population at large, some Japanese FA patients are expected to be deficient in ALDH2. We thus set out to determine the ALDH2 status in a collection of Japanese FA patients.
Study design
The onset of BMF was defined according to the criteria used in the International Fanconi Anemia Registry (IFAR) study.10 Criteria for diagnosis of aplastic anemia and other conditions are described in supplemental Methods (available on the Blood Web site). We observed physical abnormalities characteristic of FA, including skin abnormalities (hyperpigmentation and café au lait spots), low birth weight, growth defects, and malformations affecting skeletal systems and deep organs. Extensive malformation was defined as the involvement of at least 3 sites including at least 1 deep organ.11 Mutation analysis of FANCA/FANCC/FANCG genes,12 ALDH2 genotyping,13 multiplex ligation-mediated probe amplification (MLPA) test for FANCA (Falco), and whole-exome sequencing (WES)14 were done as previously described. Details are provided as supplemental Methods. Development of BMF or acute myeloid leukemia (AML)/myelodysplasia (MDS) was analyzed by the Kaplan-Meier method or the cumulative incidence method,15,16 respectively, since competing events (eg, death and stem cell transplantation [SCT]) existed in AML/MDS but not in BMF. This study was approved by the Research Ethics Committee of the Tokai University Hospital and Kyoto University. We obtained family informed consent from all subjects involved in this work in accordance with the Declaration of Helsinki.
Results and discussion
All of the patients in this study (n = 64; supplemental Table 1) were referred to the Tokai University Hospital because of pancytopenia, in some cases with MDS or leukemia. The clinical diagnosis of FA was made based on clinical presentation and diepoxybutane (DEB)-induced chromosome fragility tests in peripheral blood lymphocytes,17 except for 3 cases in which the DEB test was negative due to FANCA reversion mosaicism (supplemental Tables 1-2). Most of the patients underwent allogeneic SCT, indicating that our patients probably represent an FA population with relatively severe hematologic symptoms.
To determine which FA gene was mutated in each of these patients, we applied combinations of polymerase chain reaction–based methods (n = 26), the MLPA test for FANCA mutations (n = 44), and WES (n = 29). In our WES analysis, >90% of the 50-Mb target sequences were analyzed by >10 independent reads (data not shown). Fifty-nine patients were found to have a mutation in FA genes in at least 1 allele, but 5 of them were mutation-free in the known 16 FA genes, even after WES (Table 1; supplemental Table 1). These unclassified cases might be caused by large deletions or intronic mutations that are difficult to detect with these methods,18 or possibly mutations in a novel FA gene.
Table 1.
Summary of genotypes and clinical characteristics of the patients studied
| Total | ALDH2 genotype | |||
|---|---|---|---|---|
| GG | GA | AA | ||
| No. of cases | 64 | 36 | 25 | 3 |
| Mutated FA gene* | ||||
| FANCA | 39 | 26† | 11 | 2 |
| FANCG | 15 | 7 | 8 | — |
| FANCI | 2 | — | 2 | — |
| FANCM | 1 | — | 1 | — |
| FANCP | 2 | — | 1 | 1 |
| Unknown | 5 | 3 | 2 | — |
| Disease | ||||
| Aplastic anemia | 2 | 2 | — | — |
| Severe aplastic anemia | 40 | 21 | 19 | — |
| MDS/AML | 22 | 13 | 6 | 3‡ |
| Tongue cancer | 2 | 1 | 1 | — |
| Median months of onset (range) | ||||
| BMF | 52 (0-297) | 72 (27-297) | 28 (7-87) | 0 (0-7) |
| MDS/AML | 118 (4-384) | 156 (61-384) | 85 (41-192) | 4 (4-12) |
| No. of cases with SCT (%) | 58 (91) | 33 (92) | 23 (92) | 2 (67) |
| Median months at SCT (range) | 118 (12-448) | 130 (52-448) | 86 (28-248) | 25 (13-36) |
–, no case was found.
Mutations found in the patients were listed in supplemental Table 1. Some of them were presumptive because their functional significance has not been determined.
Somatic mosaicism due to reversion was confirmed in 2 cases and suspected in 1 case.
In these cases, onset of severe aplastic anemia and MDS was essentially simultaneous.
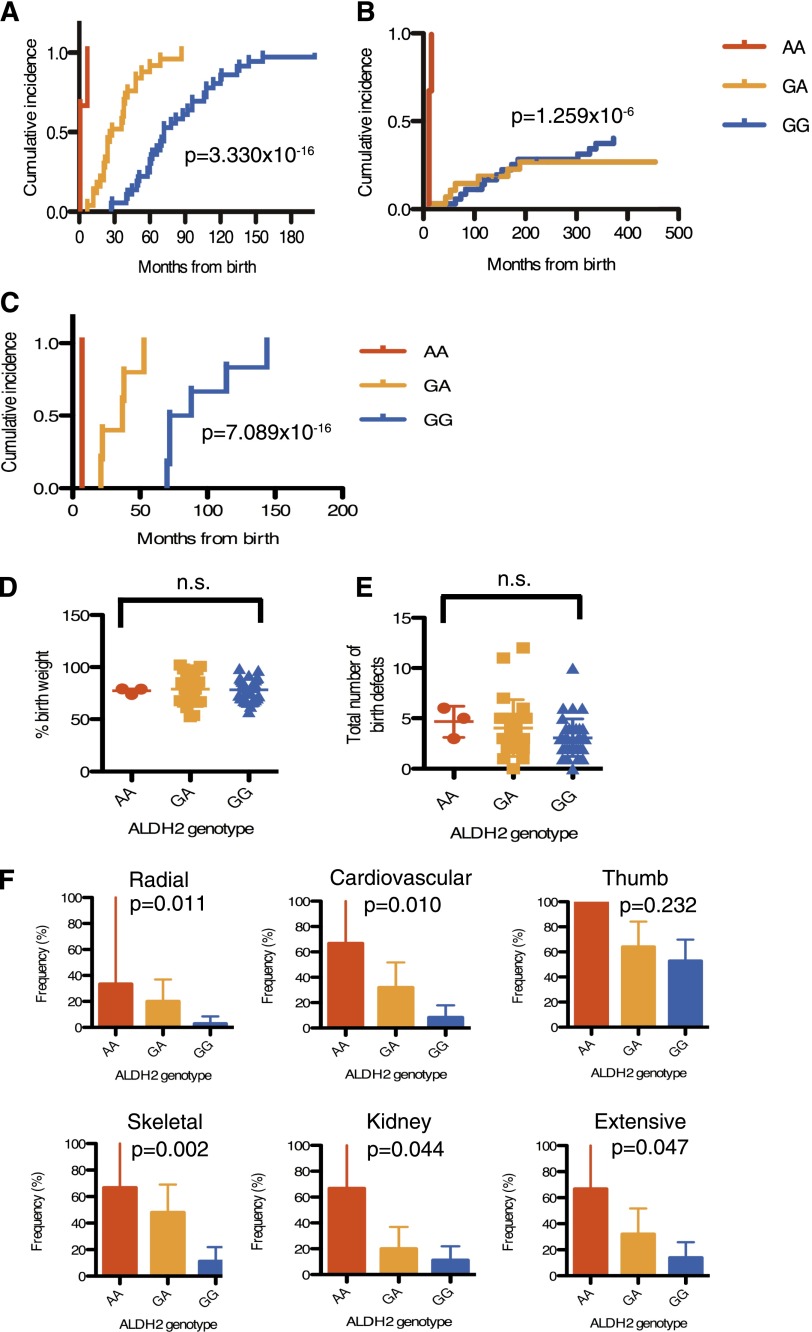
We determined the ALDH2 genotype in our series of 64 patients (Table 1; supplemental Table 1). The distribution of the ALDH2 variant alleles appeared not significantly different from the reported allele frequencies in the healthy Japanese population.13 The occurrence of leukemia and/or MDS was also not significantly different between patients with GA and GG genotypes. Strikingly, however, we found that progression of BMF was accelerated in heterozygous carriers of the variant A allele compared with homozygous GG patients (Figure 1A-B). Moreover, the 3 individuals carrying AA alleles developed MDS with BMF at a very young age (Figure 1A-B). None of these 3 patients belonged to FA-D1 or FA-N, the FA subgroups with severe symptoms.19,20 Patient number 3 had biallelic frameshift mutations (S115AfsX11) in FANCP/SLX4. By contrast, of the FA-P patients that have previously been reported, none have displayed particularly severe symptoms.21-23
Figure 1.
Effects of the ALDH2 deficiency on Japanese FA patients. (A-B) Cumulative incidence of BMF (A) or MDS/AML (B) were analyzed in 64 FA subjects. Numbers of AA, GA, and GG patients were 3, 25, and 36, respectively. (C) Cumulative incidence of BMF was analyzed in patients with confirmed biallelic FANCA mutations having protein truncations and/or large deletions (n = 12). Numbers of AA, GA, and GG patients were 1, 5, and 6, respectively. P values shown were calculated by the Gray test. In panel A, P values between genotypes were 8.625 × 10−7 (GG vs GA), 2.107 × 10−10 (GG vs AA), 1.259 × 10−6 (GA vs AA), respectively. In (B), the difference between GG and GA subjects was not significant (P = .4564479), whereas other statistical comparisons were highly significant (GG vs AA, 2.911 × 10−10; GA vs AA, 8.813 × 10−8). In panel C, the P values between GG and GA, GG and AA, or GA and AA were calculated as 0.001228433, 0.01430588, 0.02534732, respectively. (D) Percentage of birth weight or (E) total number of physical abnormalities (shown in supplemental Table 1) in 64 FA patients with 3 ALDH2 genotypes. Birth weight was normalized to mean weight at gestational age in Japan. Mean and SEM are indicated. Birth weight records were missing for 3 patients (supplemental Table 1). There was no significant difference between the ALDH2 genotypes (Kruskal-Wallis test). (F) Frequency (percentage) of cardiovascular, radial, thumb, skeletal, kidney, and extensive malformations in each ALDH2 genotype. P values were calculated by the Cochran-Armitage test for trend, which detects statistical significance of effects across the genotypes. The error bars represent 95% confidence intervals.
FA is a heterogeneous disorder, and our cohort of patients is quite heterogeneous in terms of complementation groups and types of mutations (Table 1). To reduce some of the variability, we selected only the FANCA patients having nonsense, frameshift, or large deletion mutations identified at both alleles, (n = 12; supplemental Table 1), and repeated the analysis. A patient with probable FANCA reversion (patient number 55) was excluded. In this subset of patients, a highly significant statistical difference was reproduced in BMF progression (Figure 1C) but not in AML/MDS development (data not shown).
We could not detect any significant difference in terms of the percentage of birth weight (Figure 1D) or number of physical abnormalities (Figure 1E) that correlated with the ALDH2 genotypes. However, a significant difference was observed in the incidence of each class of malformations in the case of radial, cardiovascular, skeletal, or kidney anomalies, and in the incidence of extensive malformation (Figure 1F).
In conclusion, our current data indicate that endogenous aldehydes are an important source of genotoxicity in the human hematopoietic system, and the FA pathway counteracts them. If the FA pathway is compromised, hematopoietic stem cells (HSCs) likely accumulate aldehyde-induced DNA damage, resulting in BMF due to p53/p21-mediated cell death or senescence.6,24 Consistent with this model, a recent study showed that the HSCs in aldh2/fancd2 double knockout mice accumulate more DNA damage than HSCs in either of the single knockout mice.6 Because some ALDH2-proficient FA patients developed BMF early, other modifier genes or environmental factors might affect levels of aldehydes or other genotoxic substances. Interestingly, our data predict that Japanese FA patients in general develop BMF at an earlier age compared with patients of other ethnic origins. We need to establish a Japanese FA registry similar to IFAR to test whether this is true or not. Finally, it seems worth considering ALDH2 agonists such as Alda-1 as protective drugs against BMF in FA patients. Alda-1 can stimulate the enzymatic activity of both the normal and variant ALDH2,25 suggesting that Alda-1 or a similar drug could be beneficial even for ALDH2-proficient FA cases.
Supplementary Material
Acknowledgments
The authors thank the individual patients and families in the study who made this work possible, Dr K. J. Patel (University of Cambridge) for communicating unpublished results, Dr James Hejna (Graduate School of Biostudies, Kyoto University) for critical reading of the manuscript and English editing, Mr Naoya Suzuki and Drs Akira Niwa and Megumu Saito (CiRA, Kyoto University) for discussion, and Ms Fumiko Tsuchida, Emi Uchida, Sumiyo Ariga, Chinatsu Ohki, and Mao Hisano for expert technical assistance.
This work was supported by grants from the Ministry of Health, Labor and Welfare, and grants from the Ministry of Education, Culture, Sports, Science and Technology (MEXT).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.Y. and H.Y. examined DEB-induced chromosome aberrations, carried out MLPA testing, and analyzed clinical records; K.Y., Y.O., Y.S., K.C., H.T., S.M., S.K., and S.O. performed WES and analyzed sequence data; A.H. validated exome data and carried out genotyping; A.H., M.Y., H.Y., K.M., J.N., and M.T. analyzed data; and M.Y., M.T., and K.M. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Minoru Takata, Laboratory of DNA Damage Signaling, Department of Late Effects Studies, Radiation Biology Center, Kyoto University, Kyoto 606-8501, Japan; e-mail: mtakata@house.rbc.kyoto-u.ac.jp; and Miharu Yabe, Department of Cell Transplantation and Regenerative Medicine, Tokai University School of Medicine, Isehara 259-1193, Japan; e-mail: miharu@is.icc.u-tokai.ac.jp.
References
- 1.Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res. 2009;668(1-2):4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim H, D’Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26(13):1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493(7432):356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ridpath JR, Nakamura A, Tano K, et al. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007;67(23):11117–11122. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- 5.Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475(7354):53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- 6.Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489(7417):571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- 7.Parmar K, D’Andrea A, Niedernhofer LJ. Mouse models of Fanconi anemia. Mutat Res. 2009;668(1-2):133–140. doi: 10.1016/j.mrfmmm.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crabb DW, Edenberg HJ, Bosron WF, Li TK. Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. J Clin Invest. 1989;83(1):314–316. doi: 10.1172/JCI113875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuo K, Hamajima N, Shinoda M, et al. Gene-environment interaction between an aldehyde dehydrogenase-2 (ALDH2) polymorphism and alcohol consumption for the risk of esophageal cancer. Carcinogenesis. 2001;22(6):913–916. doi: 10.1093/carcin/22.6.913. [DOI] [PubMed] [Google Scholar]
- 10.Butturini A, Gale RP, Verlander PC, Adler-Brecher B, Gillio AP, Auerbach AD. Hematologic abnormalities in Fanconi anemia: an International Fanconi Anemia Registry study. Blood. 1994;84(5):1650–1655. [PubMed] [Google Scholar]
- 11.Guardiola P, Pasquini R, Dokal I, et al. Outcome of 69 allogeneic stem cell transplantations for Fanconi anemia using HLA-matched unrelated donors: a study on behalf of the European Group for Blood and Marrow Transplantation. Blood. 2000;95(2):422–429. [PubMed] [Google Scholar]
- 12.Tachibana A, Kato T, Ejima Y, et al. The FANCA gene in Japanese Fanconi anemia: reports of eight novel mutations and analysis of sequence variability. Hum Mutat. 1999;13(3):237–244. doi: 10.1002/(SICI)1098-1004(1999)13:3<237::AID-HUMU8>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 13.Matsuo K, Wakai K, Hirose K, Ito H, Saito T, Tajima K. Alcohol dehydrogenase 2 His47Arg polymorphism influences drinking habit independently of aldehyde dehydrogenase 2 Glu487Lys polymorphism: analysis of 2,299 Japanese subjects. Cancer Epidemiol Biomarkers Prev. 2006;15(5):1009–1013. doi: 10.1158/1055-9965.EPI-05-0911. [DOI] [PubMed] [Google Scholar]
- 14.Kunishima S, Okuno Y, Yoshida K, et al. ACTN1 mutations cause congenital macrothrombocytopenia. Am J Hum Genet. 2013;92(3):431–438. doi: 10.1016/j.ajhg.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat. 1988;16(3):1141–1154. [Google Scholar]
- 16.Klein JP, Rizzo JD, Zhang MJ, Keiding N. Statistical methods for the analysis and presentation of the results of bone marrow transplants. Part I: unadjusted analysis. Bone Marrow Transplant. 2001;28(10):909–915. doi: 10.1038/sj.bmt.1703260. [DOI] [PubMed] [Google Scholar]
- 17.Auerbach AD, Rogatko A, Schroeder-Kurth TM. International Fanconi Anemia Registry: relation of clinical symptoms to diepoxybutane sensitivity. Blood. 1989;73(2):391–396. [PubMed] [Google Scholar]
- 18.Chandrasekharappa SC, Lach FP, Kimble DC, et al. NISC Comparative Sequencing Program. Massively parallel sequencing, aCGH, and RNA-Seq technologies provide a comprehensive molecular diagnosis of Fanconi anemia. Blood. 2013;121(22):e138–e148. doi: 10.1182/blood-2012-12-474585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner JE, Tolar J, Levran O, et al. Germline mutations in BRCA2: shared genetic susceptibility to breast cancer, early onset leukemia, and Fanconi anemia. Blood. 2004;103(8):3226–3229. doi: 10.1182/blood-2003-09-3138. [DOI] [PubMed] [Google Scholar]
- 20.Reid S, Schindler D, Hanenberg H, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39(2):162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43(2):142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoepker C, Hain K, Schuster B, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43(2):138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- 23.Schuster B, Knies K, Stoepker C, et al. Whole exome sequencing reveals uncommon mutations in the recently identified Fanconi anemia gene SLX4/FANCP. Hum Mutat. 2013;34(1):93–96. doi: 10.1002/humu.22221. [DOI] [PubMed] [Google Scholar]
- 24.Ceccaldi R, Parmar K, Mouly E, et al. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11(1):36–49. doi: 10.1016/j.stem.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CH, Budas GR, Churchill EN, Disatnik MH, Hurley TD, Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321(5895):1493–1495. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.