Abstract
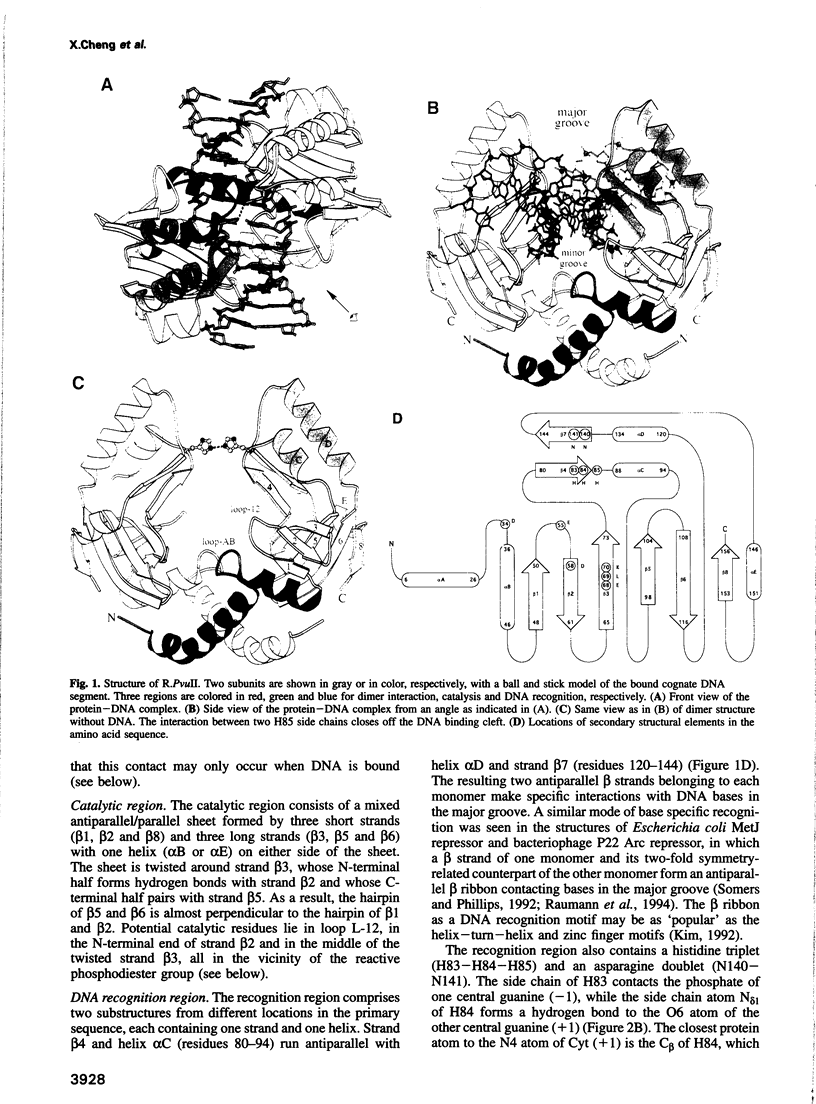
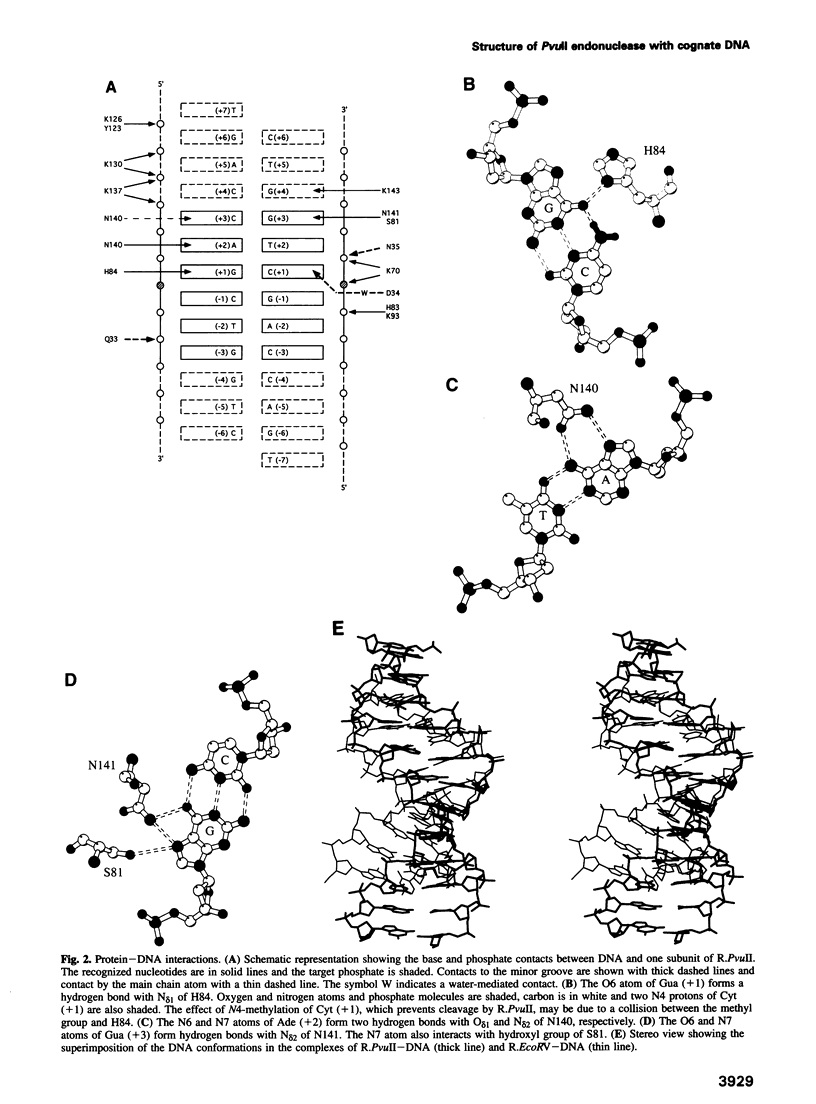
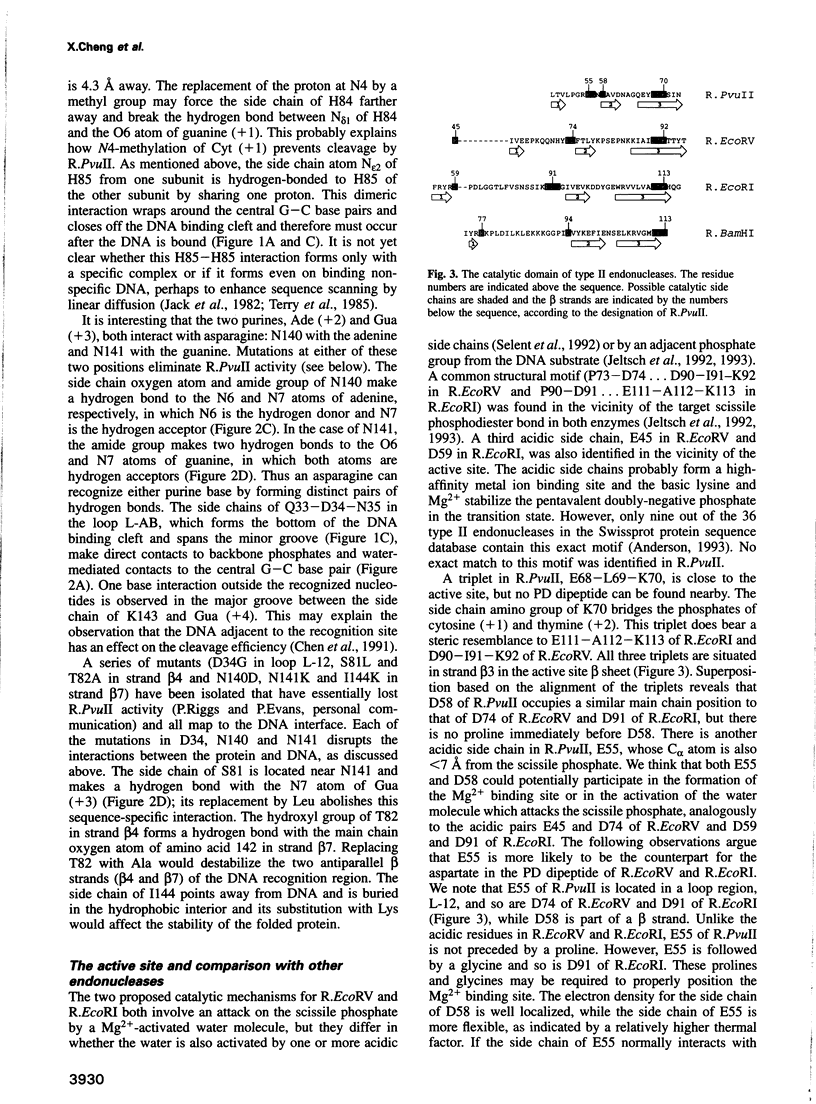
We have determined the structure of PvuII endonuclease complexed with cognate DNA by X-ray crystallography. The DNA substrate is bound with a single homodimeric protein, each subunit of which reveals three structural regions. The catalytic region strongly resembles structures of other restriction endonucleases, even though these regions have dissimilar primary sequences. Comparison of the active site with those of EcoRV and EcoRI endonucleases reveals a conserved triplet sequence close to the reactive phosphodiester group and a conserved acidic pair that may represent the ligands for the catalytic cofactor Mg2+. The DNA duplex is not significantly bent and maintains a B-DNA-like conformation. The subunit interface region of the homodimeric protein consists of a pseudo-three-helix bundle. Direct contacts between the protein and the base pairs of the PvuII recognition site occur exclusively in the major groove through two antiparallel beta strands from the sequence recognition region of the protein. Water-mediated contacts are made in the minor grooves to central bases of the site. If restriction enzymes do share a common ancestor, as has been proposed, their catalytic regions have been very strongly conserved, while their subunit interfaces and DNA sequence recognition regions have undergone remarkable structural variation.
Full text
PDF

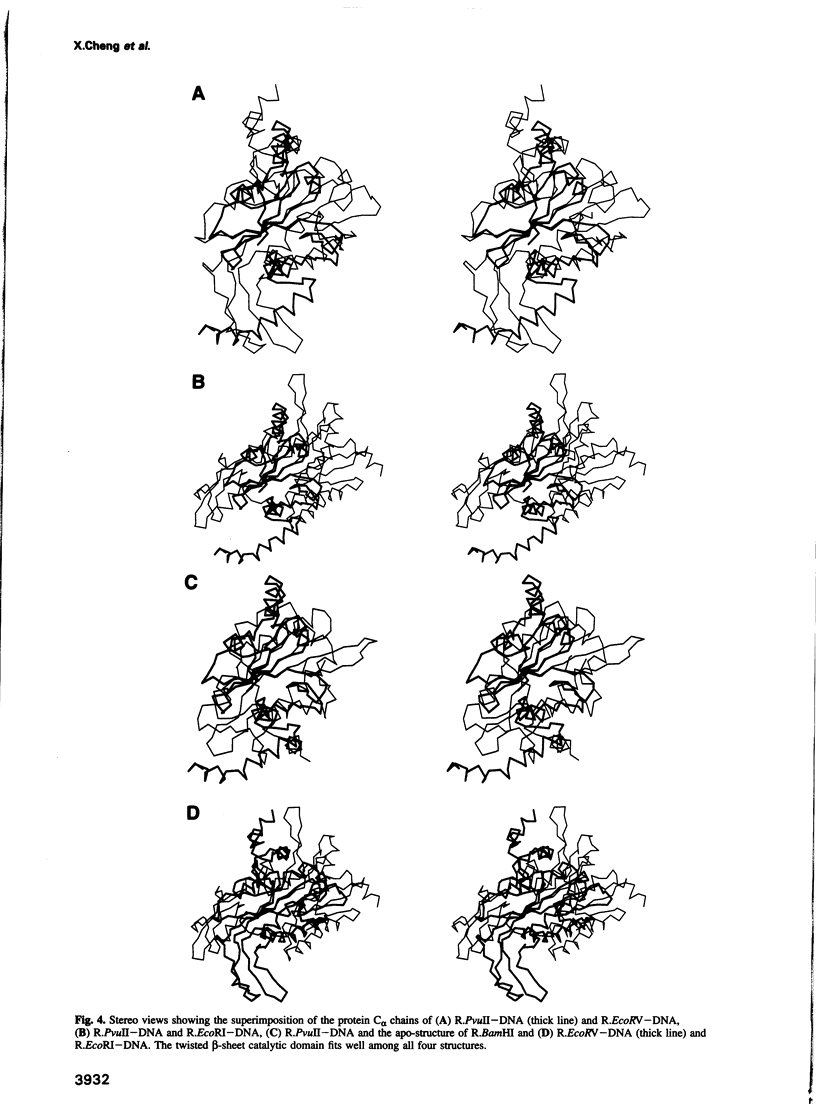


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Athanasiadis A., Gregoriu M., Thanos D., Kokkinidis M., Papamatheakis J. Complete nucleotide sequence of the PvuII restriction enzyme gene from Proteus vulgaris. Nucleic Acids Res. 1990 Nov 11;18(21):6434–6434. doi: 10.1093/nar/18.21.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis A., Kokkinidis M. Purification, crystallization and preliminary X-ray diffraction studies of the PvuII endonuclease. J Mol Biol. 1991 Dec 5;222(3):451–453. doi: 10.1016/0022-2836(91)90486-p. [DOI] [PubMed] [Google Scholar]
- Balendiran K., Bonventre J., Knott R., Jack W., Benner J., Schildkraut I., Anderson J. E. Expression, purification, and crystallization of restriction endonuclease PvuII with DNA containing its recognition site. Proteins. 1994 May;19(1):77–79. doi: 10.1002/prot.340190110. [DOI] [PubMed] [Google Scholar]
- Blumenthal R. M., Gregory S. A., Cooperider J. S. Cloning of a restriction-modification system from Proteus vulgaris and its use in analyzing a methylase-sensitive phenotype in Escherichia coli. J Bacteriol. 1985 Nov;164(2):501–509. doi: 10.1128/jb.164.2.501-509.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkus V., Klimasauskas S., Petrauskiene L., Maneliene Z., Lebionka A., Janulaitis A. Interaction of AluI, Cfr6I and PvuII restriction-modification enzymes with substrates containing either N4-methylcytosine or 5-methylcytosine. Biochim Biophys Acta. 1987 Aug 25;909(3):201–207. doi: 10.1016/0167-4781(87)90078-9. [DOI] [PubMed] [Google Scholar]
- Chen D. F., Liu Q. A., Chen X. W., Zhao X. L., Chen Y. W. The inhibition of restriction endonuclease PvuII cleavage activity by methylation outside its recognition sequence. Nucleic Acids Res. 1991 Oct 25;19(20):5703–5705. doi: 10.1093/nar/19.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T. R., Greenough L., Schildkraut I., Roberts R. J. Two new restriction endonucleases from Proteus vulgaris. Nucleic Acids Res. 1981 Sep 25;9(18):4525–4536. doi: 10.1093/nar/9.18.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack W. E., Terry B. J., Modrich P. Involvement of outside DNA sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4010–4014. doi: 10.1073/pnas.79.13.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A., Alves J., Maass G., Pingoud A. On the catalytic mechanism of EcoRI and EcoRV. A detailed proposal based on biochemical results, structural data and molecular modelling. FEBS Lett. 1992 Jun 8;304(1):4–8. doi: 10.1016/0014-5793(92)80576-3. [DOI] [PubMed] [Google Scholar]
- Jeltsch A., Alves J., Wolfes H., Maass G., Pingoud A. Substrate-assisted catalysis in the cleavage of DNA by the EcoRI and EcoRV restriction enzymes. Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8499–8503. doi: 10.1073/pnas.90.18.8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kim S. H. Beta ribbon: a new DNA recognition motif. Science. 1992 Mar 6;255(5049):1217–1218. doi: 10.1126/science.1546321. [DOI] [PubMed] [Google Scholar]
- Kim Y. C., Grable J. C., Love R., Greene P. J., Rosenberg J. M. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science. 1990 Sep 14;249(4974):1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- Newman M., Strzelecka T., Dorner L. F., Schildkraut I., Aggarwal A. K. Structure of restriction endonuclease BamHI and its relationship to EcoRI. Nature. 1994 Apr 14;368(6472):660–664. doi: 10.1038/368660a0. [DOI] [PubMed] [Google Scholar]
- Newman M., Strzelecka T., Dorner L. F., Schildkraut I., Aggarwal A. K. Structure of restriction endonuclease bamhi phased at 1.95 A resolution by MAD analysis. Structure. 1994 May 15;2(5):439–452. doi: 10.1016/s0969-2126(00)00045-9. [DOI] [PubMed] [Google Scholar]
- Park Y. W., Breslauer K. J. A spectroscopic and calorimetric study of the melting behaviors of a "bent" and a "normal" DNA duplex: [d(GA4T4C)]2 versus [d(GT4A4C)]2. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1551–1555. doi: 10.1073/pnas.88.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raumann B. E., Rould M. A., Pabo C. O., Sauer R. T. DNA recognition by beta-sheets in the Arc repressor-operator crystal structure. Nature. 1994 Feb 24;367(6465):754–757. doi: 10.1038/367754a0. [DOI] [PubMed] [Google Scholar]
- Selent U., Rüter T., Köhler E., Liedtke M., Thielking V., Alves J., Oelgeschläger T., Wolfes H., Peters F., Pingoud A. A site-directed mutagenesis study to identify amino acid residues involved in the catalytic function of the restriction endonuclease EcoRV. Biochemistry. 1992 May 26;31(20):4808–4815. doi: 10.1021/bi00135a010. [DOI] [PubMed] [Google Scholar]
- Somers W. S., Phillips S. E. Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands. Nature. 1992 Oct 1;359(6394):387–393. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]
- Tao T., Blumenthal R. M. Sequence and characterization of pvuIIR, the PvuII endonuclease gene, and of pvuIIC, its regulatory gene. J Bacteriol. 1992 May;174(10):3395–3398. doi: 10.1128/jb.174.10.3395-3398.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao T., Walter J., Brennan K. J., Cotterman M. M., Blumenthal R. M. Sequence, internal homology and high-level expression of the gene for a DNA-(cytosine N4)-methyltransferase, M.Pvu II. Nucleic Acids Res. 1989 Jun 12;17(11):4161–4175. doi: 10.1093/nar/17.11.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry B. J., Jack W. E., Modrich P. Facilitated diffusion during catalysis by EcoRI endonuclease. Nonspecific interactions in EcoRI catalysis. J Biol Chem. 1985 Oct 25;260(24):13130–13137. [PubMed] [Google Scholar]
- Wang B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- Winkler F. K., Banner D. W., Oefner C., Tsernoglou D., Brown R. S., Heathman S. P., Bryan R. K., Martin P. D., Petratos K., Wilson K. S. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993 May;12(5):1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]