Background: The role of Notch signaling in regulation of cell metabolism remains to be elucidated.
Results: A two-dimensional DIGE study shows Notch signaling activation alters the expression of proteins of various metabolic pathways.
Conclusion: Notch signaling activation leads to the remodeling of cellular metabolic pathways. Glutamine catabolism pathway proteins deregulated via canonical Notch pathway.
Significance: This study connects an important signaling pathway to cellular metabolism.
Keywords: Glutamine, Mitochondria, Notch Pathway, Proteomics, Respiratory Chain, Complex 1
Abstract
The Notch signaling pathway, a known regulator of cell fate decisions, proliferation, and apoptosis, has recently been implicated in the regulation of glycolysis, which affects tumor progression. However, the impact of Notch on other metabolic pathways remains to be elucidated. To gain more insights into the Notch signaling and its role in regulation of metabolism, we studied the mitochondrial proteome in Notch1-activated K562 cells using a comparative proteomics approach. The proteomic study led to the identification of 10 unique proteins that were altered due to Notch1 activation. Eight of these proteins belonged to mitochondria-localized metabolic pathways like oxidative phosphorylation, glutamine metabolism, Krebs cycle, and fatty acid oxidation. Validation of some of these findings showed that constitutive activation of Notch1 deregulated glutamine metabolism and Complex 1 of the respiratory chain. Furthermore, the deregulation of glutamine metabolism involved the canonical Notch signaling and its downstream effectors. The study also reports the effect of Notch signaling on mitochondrial function and status of high energy intermediates ATP, NADH, and NADPH. Thus our study shows the effect of Notch signaling on mitochondrial proteome, which in turn affects the functioning of key metabolic pathways, thereby connecting an important signaling pathway to the regulation of cellular metabolism.
Introduction
Mitochondria are one of the most important subcellular organelles that plays a key role in many cellular functions. Two main functions of mitochondria are energy production in the form of ATP via the oxidative phosphorylation system (OXPHOS)3 and regulation of the apoptosis pathway. Apart from these mitochondria are hubs where most of the metabolic pathways occur completely or partially, like the Krebs cycle, fatty acid metabolism, amino acid metabolism, pyrimidine biosynthesis, urea cycle, heme biosynthesis, etc. The size of the mammalian mitochondrial proteome predicted by computational studies is ∼1500–2000 proteins (1) of which the mitochondrial DNA encodes only for 13 OXPHOS proteins and the rest are encoded by the nuclear DNA. Nuclear DNA-coded proteins are translated in the cytoplasm and are imported into the mitochondria by a specialized protein import system that recognizes a mitochondria localizing signal (2). In this study we aim to find the alteration of the mitochondrial proteome due to activation of Notch1 pathway.
The Notch signaling constitutes an evolutionarily conserved developmental pathway that can control cell fate decisions, proliferation, and apoptosis (3). It consists of single-pass transmembrane receptors Notch 1–4 that are activated by direct contact with the membrane-bound ligands Delta 1–4 and jagged1 and -2 (4). Notch upon interaction with its ligands undergoes successive cleavage of the Notch receptor by ADAM family metalloproteases followed by γ-secretase, resulting in the production of Notch intracellular domain, which then enters the nucleus. In the nucleus, this interacts with the DNA-binding protein recombinant signal-binding protein for immunoglobulin-κJ region (RBP-Jκ, also known as CBF-1), displaces the co-repressor from of RBP-Jκ complex, and converts it into a transcriptional activator that includes other co activators (5). This constitutes the canonical Notch pathway. Apart from acting as a transcriptional activator, downstream signaling of Notch pathway may also act as a transcriptional repressor (6). Notch1 has been found to be deregulated in many types of cancers (7, 8), and most patients with T-cell acute lymphoblastic leukemia harbor activating Notch1 mutations (9). There is a growing interest in understanding the role of Notch signaling in the regulation of cellular metabolism. Earlier studies showed that during T-cell development Notch signaling activation could up-regulate glycolysis via Akt (10); recently it was found that both activation and deactivation of Notch pathway lead to increased glycolysis affecting the progression of tumor (11), but very little has been explored regarding the role of Notch pathway on the other metabolic pathways.
In this paper we therefore aimed to decipher the effect of the activation of Notch1 pathway on mitochondrial proteome. We compared the mitochondrial proteome of K562 and K562 with constitutively activated Notch1 pathway by two-dimensional differential in-gel electrophoresis (DIGE) analyses. The study showed that Notch1 pathway activation alters expression of key mitochondria-localized metabolic pathway proteins. Further validation of some of the protein expression by other methods showed that glutamine metabolism and Complex 1 of respiratory chain are deregulated, but the mitochondrial integrity is maintained. The deregulation of glutamine metabolism has been shown to be regulated by canonical Notch signaling pathway and establish the cross-talk between Notch signaling and glutamine utilization. These data provide new insight into the effect of Notch pathway in the regulation of cellular metabolism.
EXPERIMENTAL PROCEDURES
Cell Culture
K562, HEL, and Jurkat cells were cultured in RPMI1640 (Invitrogen). MCF7 and NIH 3T3 cells were cultured in DMEM (Invitrogen). Cells were stably transfected with Notch1 intracellular domain, pcDNA-Myc-HisN1IC (N1ICD), and control vector by lipofection (Lipofectamine 2000, Invitrogen). For a metabolic dependence study, RPMI without glutamine/glucose were used along with 10% FBS. NIH-3T3 cells expressing either empty vector or jagged1, a ligand for Notch receptor, was used in co-culture experiments to induce endogenous Notch signaling pathway (12). On the other hand, γ-secretase inhibitor (Calbiochem) as well as dominant negative CBF-1 (DN-CBF-1) were used to inhibit the cellular Notch signaling.
Mitochondria Isolation
Mitochondria were isolated as described previously (13); briefly, cells were collected by centrifugation at 400 × g for 10 min at 4 °C. The cell pellets were washed twice with ice-cold phosphate buffer saline (PBS) and resuspended with isolation buffer (IB: 20 mm HEPES-KOH, pH 7.5, 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, 0.25 m sucrose, and a mixture of protease inhibitors), 1.5 ml per ∼107 cells. After 30 min of incubation on ice, the cells were homogenized by using a needle and a syringe. The homogenates were centrifuged twice at 1000 × g for 10 min at 4 °C to remove nuclei and unbroken cells (Fraction 1). The postnuclear supernatants were centrifuged at 6000 × g for 25 min at 4 °C. The pellet is the mitochondrial fraction (Fraction 3), and the supernatant is the cytosolic fraction (Fraction 2). The pellet was again resuspended in IB and washed by centrifuging at 6000 × g for 25 min at 4 °C. This mitochondrial fraction was then resuspended in isotonic sucrose buffer (0.25 m sucrose, 1 mm EDTA, and 10 mm Tris-HCl, pH 7.4), layered on a 1.0/1.5 m discontinuous sucrose gradient in isotonic sucrose buffer, and centrifuged at 30,000 rpm for 40 min at 4 °C in OptimaTM TLX, Beckman Coulter. The mitochondria were collected from the interphase of 1.0 and 1.5 m sucrose, diluted in the isotonic sucrose buffer, and centrifuged again at 12,000 × g for 20 min to pellet mitochondria (enriched mitochondria). Enriched mitochondrial pellets were washed with isolation buffer and then preserved at −80 °C until further analysis. The enriched mitochondria fraction contained mitochondria along with mitochondria-associated membrane (MAM) fraction. For further removal of the MAM fraction from enriched mitochondria, the method reported previously was used (14) with modifications. Briefly, the enriched mitochondria was resuspended in 350 μl of isolation buffer 2 (IB2: 250 mm sucrose, 10 mm Hepes, pH 7.5, 1 mm EDTA, protease inhibitor mixture) and layered over 2.0 ml of 30% (v/v) Percoll (GE Healthcare) in IB2 and centrifuged (95,000 × g, 30 min, 4 °C) in Optima™ TLX, Beckman Coulter. After centrifugation two distinct bands separated by a clear zone were isolated and diluted 4-fold in cold IB2 buffer to remove the Percoll. The lower band constituted the MAM free mitochondria, and the upper band constituted the MAM fraction with incomplete removal of mitochondria (Fraction 4).
Protein Extraction and Two-dimensional DIGE
The enriched mitochondrial pellet was lysed by incubating in DIGE labeling buffer (4% w/v CHAPS, 7 m urea, 2 m thiourea, 30 mm Tris-HCl, pH 8.3) for 1 h on ice with vortexing at regular intervals and centrifuged at 15,000 × g for 15 min at 4 °C. Protein concentrations were determined by the Bradford assay reagent (Bio-Rad). CyDye (GE Healthcare) labeling was performed according to the manufacturer's instructions. Briefly, 50 μg of mitochondrial protein from K562 or KN1ICD was minimally labeled with 400 pmol of either Cy3 or Cy5 for comparison on the same two-dimensional gel. Internal standard aliquots of each sample were pooled and labeled with Cy2. The labeled samples were incubated for 30 min on ice in the dark and then 1 μl of 10 mm lysine was added to stop the reaction. The samples were diluted in equal volume of 2×-DIGE labeling buffer (4% w/v CHAPS, 7 m urea, 2 m thiourea, 30 mm Tris-HCl, pH 8.3, 130 mm DTT, 4% 3/10 ampholytes), and a total volume of 150 μl was made up with rehydration buffer (2% w/v CHAPS, 7 m urea, 2 m thiourea, 50 mm DTT, and 2% 3/10 ampholytes). Samples were loaded via cup loading on overnight rehydrated immobilized pH gradient strips (17 cm, pH 5–8, Bio-Rad). Focusing was performed at a maximum of 20 °C for 100,000 volt hours. After focusing, the immobilized pH gradient strips were equilibrated for 30 min with 50 mm Tris-HCl, pH 8.8, 6 m urea, 30% glycerol, 2% SDS, and 1% of DTT for reduction followed by alkylation with 2.5% iodoacetamide. The second-dimension separations were carried out in 12% SDS-polyacrylamide gels run at 20 mA/gel for 30 min and then 30 mA/gel until the dye reached the bottom of the gels. All samples and their dye-swapped counterparts were run in triplicate generating six replicates.
Image Analysis, In-gel Digestion, and Mass Spectrometry (MS)
Fluorescence images of the gels were acquired on a Typhoon TRIO scanner (GE Healthcare) with a resolution of 100 μm. Spot detection was performed on the gel images using the DeCyder software (Version 6.5). A differential in-gel analysis module was done by setting the target spot number to 1000. All six gels were added to the appropriate workspace and group and matched to a master gel sequentially using the DeCyder module BVA (Version 6.5.14). For grouped comparison, significance was calculated using two-tailed t tests, and differentially expressed proteins with an absolute ratio of at least 1.4-fold and p ≤ 0.05 were selected as proteins of interest. For the preparative gel, >500 μg of mitochondrial protein was resolved in a two-dimensional gel and stained with Blue Silver (15). Proteins of interest were excised. The excised spots were destained, and then tryptic digestion was performed overnight using Trypsin Gold (Promega, Madison, WI). The digestion mixture was lyophilized in a Heto vacuum centrifuge (Thermo) and resuspended in 50% acetonitrile and 0.1% trifluoroacetic acid solution. The dissolved samples were spotted on the MALDI plate along with matrix α-cyano-4-hydroxycinnamic acid. MALDI analyses were performed with a 4700 Proteomic Analyzer MALDI-TOF/TOF (Applied Biosystems) for identification of the protein spots. Peptide mass fingerprinting data were acquired in positive MS reflector mode, and for tandem mass spectrometry the peptides were selected according to their intensity and subjected to further fragmentation and MS identification. Spectral data were analyzed with GPS ExplorerTM Version 3.6 (Applied Biosystems). Peptide identification was based on the MASCOT database-scoring algorithm from MSDB protein databases using search settings of single missed tryptic cut, fixed modification of carbamidomethylation, precursor tolerance of 300 ppm, and MS/MS fragment tolerance of 1.2 Da. Autotryptic peaks were excluded in the MASCOT search parameter, and p < 0.05 was considered significant during identification along with at least two MS/MS hits were considered a true “hit.” Because we had taken a gel-based proteomics approach, we could identify specific proteins from a particular spot according to the molecular weight and pI from the protein list given by MASCOT server.
Quantitative RT-PCR
RNA was isolated using TRIzol (Roche Applied Science). The expression level of genes was assessed by quantitative RT-PCR using SYBR Green core PCR reagents (Applied Biosystems). HPRT1 was used to normalize the expression of the genes. The reactions were carried out in 7500 Sequence Detection Systems (Applied Biosystems). The primers used HPRT1 5′-GACACTGGCAAAACAATGCAGAC-3′ and 5′-TGGCTTATATCCAACACTTCGTGG-3′; GLS 5′-TGGGCAACAGTGTTAAGGGAA-3′ and 5′- GGAATGCCTTTGATCACCACC-3′; OAT 5′-GCACATTTGCAGAGGTTTGCT-3′ and 5′-GTCATCAGAGGTTGGAGGGC-3′; NDUFV2 5′-CCTGTGTGAACGCACCAATG-3′ and 5′-TGGCCCTGGTTTTGGGATTT-3′; NDUFS1 5′-CGTCGTGGTCCAGACAGTTT-3′ and 5′-TCAAGTTGCTTGCTGCTGTG-3′; ECHS1 5′-GCCTCGGGTGCTAACTTTGA-3′ and 5′-GCCATCGCAAAGTGCATTGA-3′; Hey1 5′-CTGCAGATGACCGTGGATCA-3′ and 5′-CAACTTCTGCCAGGCATTCC-3′; Hes1 5′-GAAGGCGGACATTCTGGAAA-3′ and 5′GTTCATGCACTCGCTGAAGC3′. Experiments were repeated at least three times.
Western Blotting
For Western blotting cells or mitochondria were lysed in cold radioimmune precipitation assay buffer. Equal amounts of protein was run on SDS-PAGE and blotted on PVDF membranes (GE Healthcare). After incubation with primary and HRP-conjugated secondary antibodies, the blots were developed using Enhanced Chemiluminescent substrate (Thermo Fischer Scientific). For whole cell lysate GAPDH (Cell Signaling Technology, Denver, CO) or β-actin (Abcam) was used as the loading control. For mitochondria fraction prohibitin (Thermo Fischer Scientific) was used as the loading control as the two-dimensional DIGE Decyder based analysis showed that it remains constant across the six replicates. The other antibodies used for the study are OAT, NUDFS1, GLS, ECHS1, RBP-Jκ, ERp57, and c-myc (Santa Cruz Biotechnology), VDAC (Cell Signaling Technology), TIM-23 (Pharmingen), cleaved Notch1, Lamin, Hey1 (Abcam), and calreticulin (Stressgen, Victoria, BC, Canada).
RBP-Jκ Reporter Assay
We used the RBP-Jκ Cignal kit (Qiagen, Valencia, CA) for the quantification of Notch canonical pathway signaling. The expression of the firefly luciferase was quantified with a Dual Luciferase Reporter assay system (Promega) according to the manufacturer's protocol. All transfections were performed in triplicate. The level of the firefly luciferase activity was normalized by the corresponding level of the Renilla luciferase activity, and the values for the negative control were normalized. Luminescence was measured using a Sirius Luminometer, Berthold Detection Systems.
Cell Survival (through MTT Assay) Measurement
MTT assay was performed with the Cell Proliferation Kit I (MTT) (Roche Applied Science) for understanding the metabolic dependence on glutamine and glucose. Briefly, an equal number of cells (1 × 104) was plated in specific media (RPMI, glutamine-free, or glucose free RPMI) for 48 h. Then they were incubated with MTT reagent (final concentration, 0.5 mg/ml) for 2 h. At the end of incubation period the dye was solubilized with acidic isopropyl alcohol (0.04 m HCl in absolute isopropyl alcohol). The absorbance of the dye was measured at a 570-nm wavelength with background subtraction at 650 nm. The readings at 48 h were normalized with the reading at 0 h.
ADP/ATP Quantification, NAD+/NADH, Cellular NADPH, and Oxygen Consumption Rate Measurement
ADP/ATP, NAD+/NADH, and cellular NADPH were measured according to the manufacturer's instructions (Abcam). NADPH was expressed as pmol of NADPH per μg of protein. To measure oxygen consumption rate cells were suspended in equal concentration, and 500 μl of the suspension was added to the oxygraph chamber (Hansatech) in air-saturated PBS + 20 mm glucose.
Assay for Complex 1 of the Respiratory Chain
The rotenone-sensitive NADH:ubiquinone oxidoreductase activity of Complex 1 was performed spectrophotometrically. Complex I oxidizes NADH, and the electrons produced reduce the artificial substrate decylubiquinone, which subsequently delivers the electrons to dichloroindophenol. The reduction of dichloroindophenol can be followed spectrophotometrically at 600 nm as reported previously by others (16) and normalized to the protein amount.
Pyruvate Dehydrogenase Complex (PDH) Activity
A Dipstick assay kit for PDH activity from Mitosciences was employed. The assay was performed according to the manufacturer's protocol for isolated mitochondria. Samples containing equal amounts of protein were added to a 96-well plate followed by adsorption onto dipsticks containing immobilized antibody against the PDH complex and the addition of the activity buffer, which contained substrates for the PDH reaction. PDH activity was determined densitometrically from the bands with the target protein-antibody complexes formed on dipsticks in images taken with Versa Doc (Bio-Rad) scanner in colorimetric setting.
Glutamine Consumption Assay
Cells (1 × 104/well) were cultured in a 24-well plate for 48 h in specific medium. Medium was collected, and cells were lysed. Concentrations of glutamine in the medium were determined with the glutamine/glutamate determination kit (Sigma). The experiment was performed according to the manufactures protocol. Briefly, each sample was divided into two; one was measured with glutaminase for transferring the glutamine into glutamate, and the other was measured directly. The glutamate present was then converted to α-ketoglutarate by glutamate dehydrogenase accompanied by reduction of NAD+ to NADH. The amount of NADH is proportional to the amount of glutamate and was measured using a spectrophotometer at 340 nm. Glutamine levels were calculated by subtracting the second reading from total glutamate obtained from the first reading. The glutamine level of their culture medium was measured similarly, and the glutamine consumption was calculated (glutamine in culture medium-glutamine in medium after culturing cells for 48 h), normalized to protein level, and then the -fold change with respect to the control cell was calculated.
Mitochondrial Membrane Potential and Mitochondrial Mass
The mitochondrial membrane potential sensitive dye, 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethyl benzimidazolyl carbocyanine iodide (JC1; Invitrogen) was used in flow cytometry analysis on a BD Biosciences FACS Calibur platform using FL1 channel (unbound JC1) and FL2 channel (mitochondrial membrane bound JC-1). Carbonyl cyanide m-chlorophenyl hydrazone-treated cells had their membrane potential disrupted. Cells were stained with 50 nm nonyl acridine orange (Invitrogen) for 45 min at 37 °C in dark and were then subjected to flow cytometry.
Statistical Analysis
Statistical significance of the difference between the different conditions was assessed using Student's two-tailed t test. All calculations were performed using Microsoft Office Excel unless otherwise mentioned.
RESULTS
Activation of the Notch1 Signaling Pathway Alters the Mitochondrial Proteome
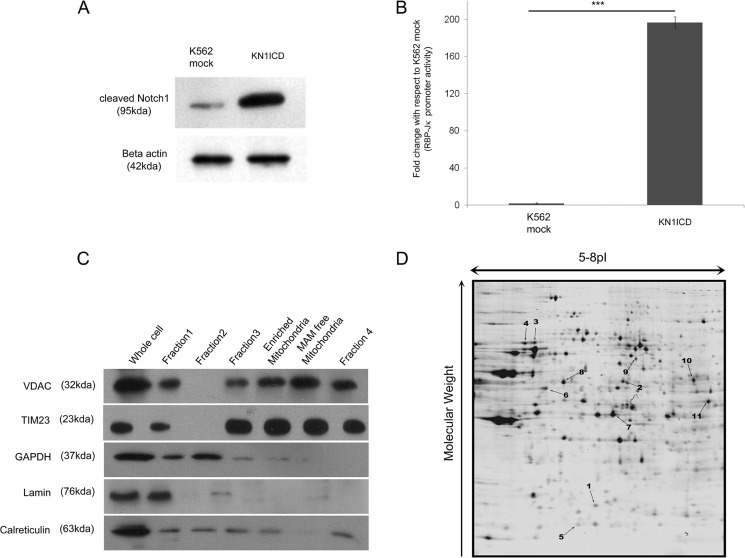
For examining the effect of Notch1 signaling on the mitochondrial proteome, we stably expressed the constitutively active form of Notch1, i.e. the Notch1 intracellular domain in human cell line K562 (this is referred as KN1ICD). K562 cells express Notch1 receptors, but the pathway is not activated (17). Western blot analysis showed that there was a high expression of cleaved Notch1 in KN1ICD cells compared with the mock-transfected K562 cells (K562 mock) (Fig. 1A). An increase in Notch activity was identified from RBP-Jκ luciferase reporter activity (Fig. 1B). An earlier effect of Notch1 activation in K562 cells has been studied to understand its influence on differentiation or cell growth (18–20). Here the effect of Notch signaling on the expression of mitochondrial proteins and the cellular metabolism has been investigated. For this, the two-dimensional DIGE-based comparative study of the mitochondrial proteome was performed. Mitochondria were isolated from K562 mock and KN1ICD, and its quality was judged by Western blotting using organelle specific markers, nuclear marker lamin, cytoplasm marker GAPDH, mitochondrial markers VDAC, TIM23, and MAM fraction marker calreticulin, as depicted in Fig. 1C. For a further study the enriched mitochondrial fraction containing mitochondria and MAM was used. The Decyder-based analysis of the two-dimensional DIGE gels revealed a set of 11 protein spots that showed significant (absolute -fold change ≥1.4, p ≤ 0.05) alteration and was represented in all the six replicates. Fig. 1D shows a representative gel with spot numbers indicating the altered proteins. This indicates that Notch1 activation can alter the mitochondrial proteome and affect the expression of mitochondrial proteins.
FIGURE 1.
Notch1 activation alters mitochondrial proteome (A)-cleaved Notch1 levels in K562 mock and KN1ICD cells. B, Notch signaling activity from RBP-Jκ luciferase reporter activity in K562 mock and KN1ICD. C, Western blot analysis mitochondrial purity isolated by differential centrifugation. D, two-dimensional gel of mitochondrial protein separated in 5–8 pI and 12% polyacrylamide gel. The spot numbers of the altered proteins are indicated.
Mass Spectrometry-based Identification of Protein Spots
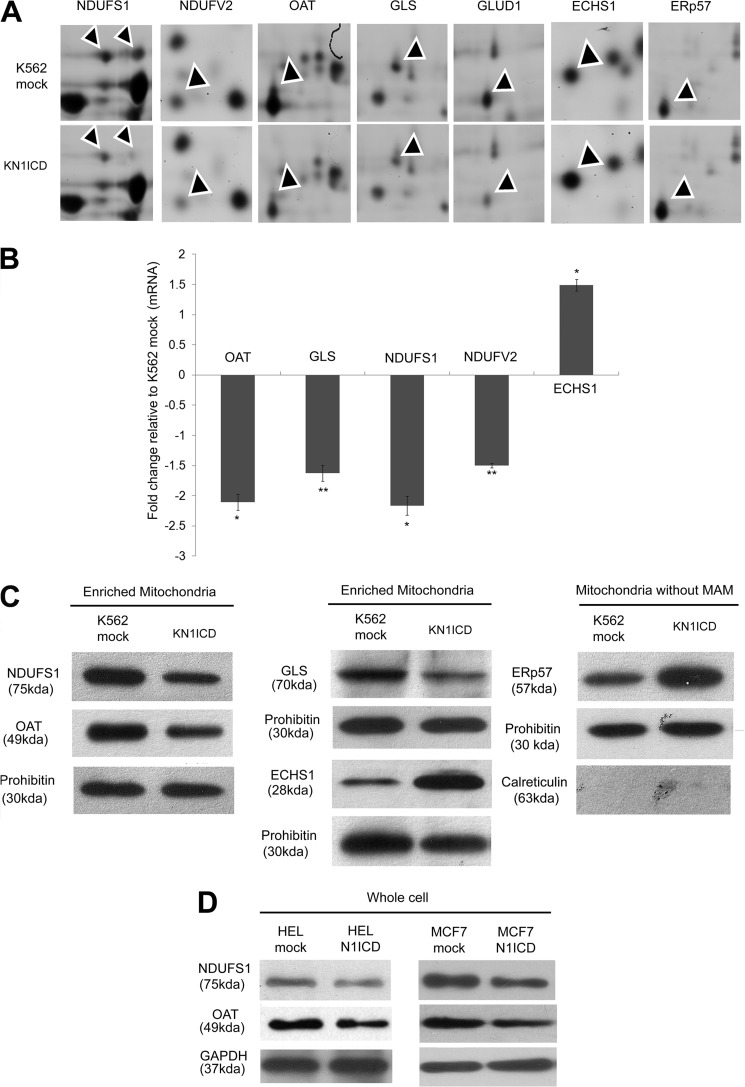
The 11 protein spots that altered significantly were subjected to MALDI TOF/TOF-based tandem MS analysis. 10 unique proteins were identified based on mass spectrometry. All the altered proteins that were identified matched with more than one peptide and gave at least two MS/MS hits with p < 0.05. Among these, 10 proteins, 8 proteins were known mitochondrial protein, and the pathways to which these proteins belonged were analyzed from the gene ontology (GO) provided in UniProtKB/Swiss-Prot database. The eight proteins are enoyl-CoA hydratase, mitochondrial (ECHS1, 1.8 ± 0.074) (1), glutaminase kidney isoform, mitochondrial (GLS, −1.44 ± 0.0580) (2), NADH-ubiquinone oxidoreductase 75-kDa subunit, mitochondrial (NDUFS1, −2.63 ± 0.0427, −1.95 ± 0.0316) (3 and 4), NADH dehydrogenase (ubiquinone) flavoprotein 2, mitochondrial (NDUFV2, −1.53 ± 0.0583 (5), ornithine aminotransferase, mitochondrial (OAT, −2.07 ± 0.1077 (7), succinate dehydrogenase (ubiquinone) flavoprotein subunit, mitochondrial (SDHA, −1.56 ± 0.0419) (9), dihydrolipoyl dehydrogenase, mitochondrial (DLD, 1.49 ± 0.1189) (10), and glutamate dehydrogenase 1, mitochondrial (GLUD1, −1.44 ± 0.1046) (11), and two proteins were non-mitochondrial keratin, type II cytoskeletal 8 (KRT8, 3.55 ± 0.5745) (6) and protein disulfide isomerase A3 (ERP57, 1.68 ± 0.126) (8). The eight mitochondrial proteins identified all belonged to mitochondria-localized metabolic pathways like oxidative phosphorylation (OXPHOS), glutamine metabolism, Krebs cycle, and fatty acid oxidation. The metabolic pathways to which these proteins belong along with their mass spectrometry identification details are provided in Table 1, and the details of the mass spectrometry spectra are provided in the supplemental material. Fig. 2A shows the two-dimensional DIGE segment of some of the altered proteins. Validation of some of the proteins at mRNA and protein levels was also performed (Fig. 2, B and C). Alteration of OAT and NDUFS1 in other cell lines (HEL and MCF7) due to Notch1 activation has also been checked (Fig. 2D). Thereby, further studies address how Notch1 signaling effects the expression of these proteins, the pathways to which they belong, and to the overall cellular metabolic state.
TABLE 1.
Decyder-based alterations in mitochondrial protein expression of KN1ICD in comparison with K562 mock along with their mass spectrometry-based identification and GO annotation for biological functions
Mr, molecular weight.
| Spot no. | Protein name (gene name) | Uniprot accession ID | GO annotation | Average-fold change±S.E. | t Test p value | Theoretical Mr/pI | Mascot score | No of peptides matched | Ion score | % Sequence coverage |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Enoyl-CoA hydratase, mitochondrial (ECHS1) | ECHM_HUMAN | Fatty acid β-oxidation | 1.8 ± 0.074 | 8.40E-06 | 28.32445/5.88 | 116 | 12 | 84 | 37 |
| 2 | Glutaminase kidney isoform, mitochondrial (GLS) | GLSK_HUMAN | Glutamine catabolic process | −1.44 ± 0.0580 | 6.40E-05 | 71.51485/7.01 | 270 | 18 | 207 | 42 |
| 3 | NADH-ubiquinone oxidoreductase 75-kDa subunit, mitochondrial (NDUFS1) | NDUS1_HUMAN | Mitochondrial electron transport, NADH to ubiquinone | −2.63 ± 0.0427 | 0.00018 | 76.92606/5.42 | 273 | 30 | 177 | 38 |
| 4 | NADH-ubiquinone oxidoreductase 75-kDa subunit, mitochondrial (NDUFS1) | NDUS1_HUMAN | Mitochondrial electron transport, NADH to ubiquinone | −1.95 ± 0.0316 | 0.00046 | 76.92606/5.42 | 205 | 27 | 115 | 41 |
| 5 | NADH dehydrogenase (ubiquinone) flavoprotein 2, mitochondrial (NDUFV2) | NDUV2_HUMAN | Mitochondrial electron transport, NADH to ubiquinone | −1.53 ± 0.0583 | 0.00089 | 23.74512/5.71 | 72 | 10 | 25 | 44 |
| 6 | Keratin, type II cytoskeletal 8 (KRT8) | K2C8_HUMAN | Cytoskeleton organization | 3.55 ± 0.5745 | 9.90E-08 | 53.67113/5.52 | 235 | 29 | 83 | 44 |
| 7 | Ornithine aminotransferase, mitochondrial (OAT) | OAT_HUMAN | Cellular amino acid biosynthetic process | −2.07 ± 0.1077 | 0.0014 | 44.78017/5.72 | 454 | 29 | 389 | 52 |
| 8 | Protein disulfide-isomerase A3 (ERP57) | PDIA3_HUMAN | Cell redox homeostasis | 1.68 ± 0.126 | 4.70E-05 | 54.23121/5.61 | 443 | 30 | 310 | 48 |
| 9 | Succinate dehydrogenase (ubiquinone) flavoprotein subunit, mitochondrial (SDHA) | DHSA_HUMAN | Respiratory electron transport chain | −1.56 ± 0.0419 | 3.30E-06 | 67.96871/6.25 | 317 | 33 | 186 | 47 |
| 10 | Dihydrolipoyl dehydrogenase, mitochondrial (DLD) | DLDH_HUMAN | 2-Oxoglutarate metabolic process | 1.49 ± 0.1189 | 0.0004 | 54142.97/7.95 | 378 | 24 | 303 | 39 |
| 11 | Glutamate dehydrogenase 1, mitochondrial (GLUD1) | DHE3_HUMAN | Glutamate catabolic process | −1.44 ± 0.1046 | 0.038 | 61359.19/7.66 | 436 | 24 | 355 | 35 |
FIGURE 2.
Validation of two-dimensional DIGE data. A, segments of two-dimensional DIGE gels after Decyder analysis. The arrowheads point at the protein of interest. B, relative gene expression in K562 with constitutively activated N1ICD compared with K562 mock. The mRNA expression of the gene was normalized to the expression of HPRT1 mRNA. Data represent the mean ± S.E. *, p < 0.05; **, p < 0.01; n = 3. C, Western blot of enriched mitochondria fraction of K562 mock and KNICD showing relative expression of the altered proteins. D, Western blot of whole cell lysates from mock- and N1ICD-transfected HEL and MCF-7 cell lines showing the decrease in OAT and NDUFS1 expression.
Notch1 Activation Deregulates Complex 1 and the Glutamine Metabolism Pathway
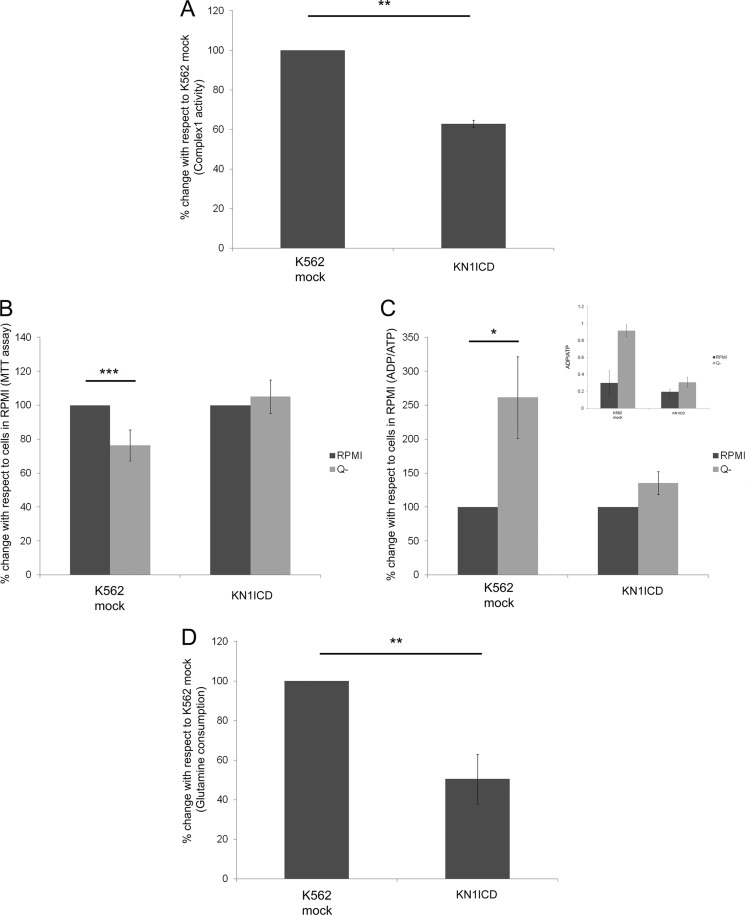
Among the altered proteins, two subunits of Complex 1 of the respiratory chain and three proteins that are directly related to glutamine catabolism process were found. The two-dimensional DIGE analysis showed that the NADH-ubiquinone oxidoreductase 75-kDa subunit (NDUFS1) and NADH dehydrogenase (ubiquinone) flavoprotein 2 (NDUFV2), two subunits of Complex 1, were down-regulated. NDUFS1 is the largest subunit of Complex 1 containing three iron sulfur clusters and plays an important role in electron transfer. Several mutations of this gene are known, and these are reported to cause Complex 1 deficiency and a decrease in its activity (21). NDUFV2 subunit of Complex 1 is one of its core subunits that is very much conserved from bacteria to mammals (22). The down-regulated proteins related to glutamine catabolism were GLS, OAT, and GLUD1. GLS performs the first step of conversion of glutamine to glutamate and is often the rate-limiting step of glutamine metabolism in many types of cells and is often found to be up-regulated in highly proliferating cells (23). Ornithine aminotransferase (OAT) is the intermediate enzyme for the synthesis of arginine from glutamate. Glutamate dehydrogenase (GLUD1) converts glutamate in to α-ketoglutarate, which is an intermediate of the Krebs cycle. The comparison of the rotenone-sensitive NADH:ubiquinone oxidoreductase activity in K562 mock and KN1ICD showed that the activity decreases by ∼40% in KN1ICD (Fig. 3A). Glutamine starvation of K562 cells for 48 h leads to a decrease in cell survival and increased ADP/ATP ratio, but KN1ICD cells were unaffected by the glutamine starvation (Fig. 3, B and C). The glutamine consumption assay showed that KN1ICD cells consumed less glutamine than K562 mock cells (Fig. 3D), supporting our glutamine dependence study.
FIGURE 3.
Effect of Notch signaling activation on the Complex 1 activity and glutamine dependence in K562 cells. A, rotenone-sensitive NADH:ubiquinone oxidoreductase activity of Complex 1 of the respiratory chain normalized by the protein amount. The data represent the percentage of Complex 1 activity in KN1ICD with respect to K562 mock. B, K562 mock and KN1ICD were plated in equal amounts in RPMI and RPMI without glutamine media (Q-) for 48 h. The cell survival was assessed by MTT assay. The readings were normalized by 0 h reading. The data give the MTT assay readings with respect to readings of cells in RPMI. Data show the mean reading of at least four separate experiments (***, p < 0.001). C, ADP/ATP ratio was determined by luciferase activity for cells growing in RPMI or RPMI without glutamine media. The data are represented as the % change, and the inset shows the actual ADP/ATP ratio (**, p < 0.01). D, percentage change in glutamine consumption/amount of protein for cells cultured in complete media for 48 h (**, p < 0.01; ***, p < 0.001).
Activation of Notch in K562 by Fibroblast-Expressing jagged1
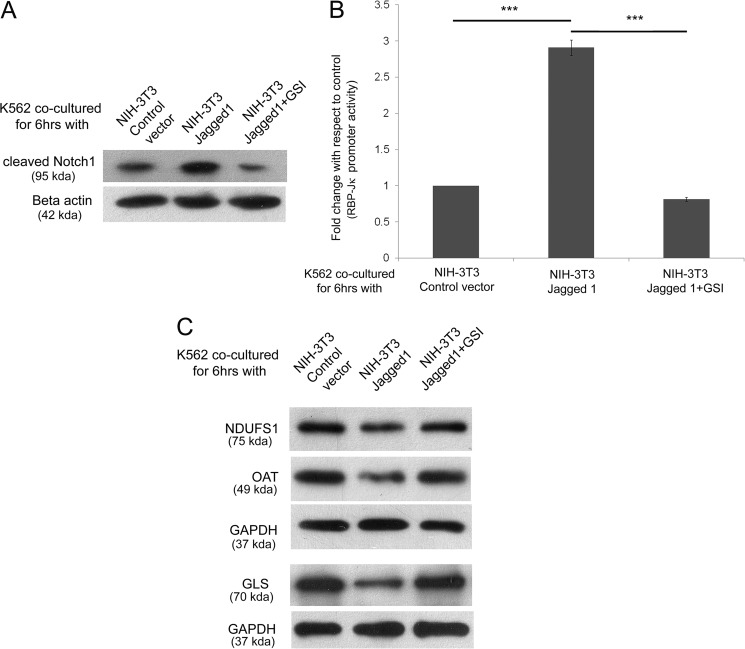
These data indicated that an increase of Notch1 intracellular domain expression in K562 cells resulted in an alteration of NDUFS1, OAT, and GLS among others. Next we asked whether ligand-mediated activation of Notch is sufficient for bringing about these alterations. For this, K562 cells were cultured on a monolayer of NIH-3T3 fibroblasts transfected with either control vector or Notch ligand jagged1 expression vector in the presence or absence of 1 μm GSI for 6 h. The effect of these conditions on Notch activation was determined first. The cleaved Notch1 expression was checked under the three conditions, and it was found that jagged1 induced cleaved Notch1 formation that was inhibited by GSI (Fig. 4A). Similarly, a RBP-Jκ reporter assay also showed the increase in Notch activity due to jagged1 ligand binding. (Fig. 4B). This jagged1 ligand-mediated activation of the Notch pathway decreased the expression of NDUFS1, OAT, and GLS, which could be inhibited by GSI (Fig. 4C). Thus, our results indicate that Notch activation through cell-associated jagged1 ligation can also alter the protein expression.
FIGURE 4.
Activation of Notch in K562 by fibroblast expressing jagged1. A, Western blot of whole cell lysate of K562 after co-culture with NIH-3T3-empty vector, jagged1, and jagged1 + 1 μm GSI showing relative expression of cleaved Notch1. B, Notch signaling activity from RBP-Jκ luciferase reporter activity after co-culture with NIH-3T3-empty vector, jagged1, and jagged1 + 1 μm GSI. C, Western blot for the expression change of NDUFS1, OAT, and GLS in K562 after co-culture.
Effect of Notch1 Activation on Mitochondrial Function, Mass, and Other Cellular Metabolic Parameters
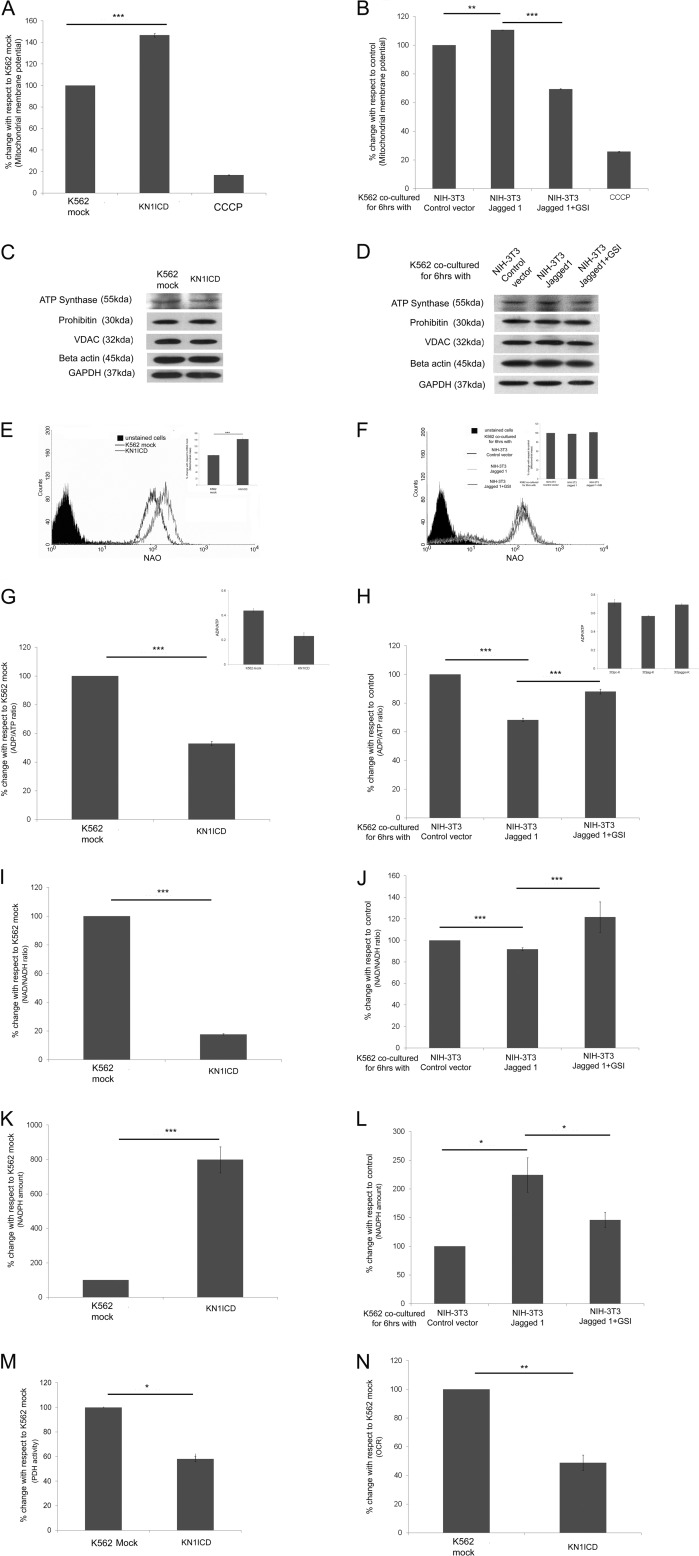
The proteomic study and its subsequent validation show that Notch1 activation down-regulates some mitochondrial protein expression and deregulates the pathways to which they belong. Next the effect of Notch activation on the mitochondrial integrity was determined. In KN1ICD, the membrane potential was found to increase (Fig. 5A). The mitochondrial protein expression from various compartments (VDAC, outer membrane; ATP synthase, complex 5 subunit, inner membrane; prohibitin, matrix) remained unaltered with respect to cytoplasmic proteins GAPDH and β-actin in whole cell lysate (Fig. 5C); however, the mitochondrial content measured using nonyl acridine orange showed an increase in mitochondrial mass (Fig. 5E). It may be noted that activation of Notch signaling by co-culturing with fibroblast expressing jagged1 also showed an increase in membrane potential that could be inhibited in the presence of GSI, whereas the expression of proteins from various mitochondrial compartments remained unaltered with respect to whole cell control (Fig. 5, B and D). No significant change in mitochondrial mass was observed in this case (Fig. 5F). All these indicate that Notch1 activation alters the mitochondrial proteome and can maintain the mitochondrial integrity. Next the status of the high energy intermediates ATP, NADH, and NADPH due to activation of Notch1 was measured. The ADP/ATP ratio decreases in KN1ICD cells and also upon Notch activation by jagged1 ligand interaction (Fig. 5, G and H). The cellular NAD+/NADH ratio decreases in KN1ICD and in K562 cells co-cultured with fibroblasts expressing jagged1. (Fig. 5, I and J). NADPH, the other pyridine nucleotide whose homeostasis is vital for cell survival and proliferation, is found to increase in KN1ICD cells and in jagged1 ligand-mediated Notch-activated K562 cells in comparison to their respective controls (Fig. 5, K and L). The PDH activity was measured in isolated mitochondria from K562 mock and KN1ICD cells, which showed that in KN1ICD cells there was a significant decrease. This indicates a decrease in pyruvate entry in mitochondria for further oxidation (Fig. 5 m). Even the oxygen consumption rate in KN1ICD decreased in comparison to K562 mock (Fig. 5N). The alteration of these major high energy intermediates in favor of cell survival accompanied by a decrease in PDH activity and decreased oxygen consumption rate only indicates that Notch pathway activation decreases the cellular dependence on mitochondria for reduction of oxygen, but the mitochondrial integrity is maintained.
FIGURE 5.
Notch1 activation and mitochondrial function, mass, and other cellular metabolic parameters. Shown are mitochondrial membrane potential in KN1ICD (A) and in Notch induction by NIH-3T3 jagged1 (B). CCCP, carbonyl cyanide m-chlorophenylhydrazone. Shown are mitochondrial proteins from various compartments that were found to be unaltered by a two-dimensional DIGE study compared with cytoplasmic controls β-actin and GAPDH in K562 mock and KN1ICD (C) and in K562 after co-culture with NIH-3T3-empty vector, jagged1, and jagged1 + 1 μm GSI (D). Shown are mitochondrial mass measured by membrane potential independent dye nonyl acridine orange (NAO) in K562 mock (E) and KN1ICD and in K562 after co-culture with NIH-3T3-empty vector, jagged1, and jagged1 + 1 μm GSI (the inset gives the percentage change n = 3; ***, p < 0.001) (F). G, ADP/ATP ratio in K562 mock and KN1ICD (H) and in K562 after co-culture with NIH-3T3-empty vector, jagged1, and jagged1 + 1 μm GSI. The data are represented as percentage change, and the insets show the actual ADP/ATP ratio. Shown are NAD+/NADH ratios in K562 mock (I) and KN1ICD (J) and in K562 after co-culture with NIH-3T3-empty vector, jagged1, and jagged1 + 1 μm GSI. Shown are NADPH amounts per μg of protein in K562 mock (K) and KN1ICD (L) and in K562 after co-culture with NIH-3T3-empty vector, jagged1, and jagged1 + 1 μm GSI. M, percentage PDH activity of KNICD with respect to K562 mock (*, p < 0.05). N, -fold change in oxygen consumption rate (OCR) in KN1ICD measured in nmol/min/cell with respect to K562 mock.
Alteration of the Expression of Proteins Involved in Glutamine Metabolism Is via the Canonical Notch Pathway
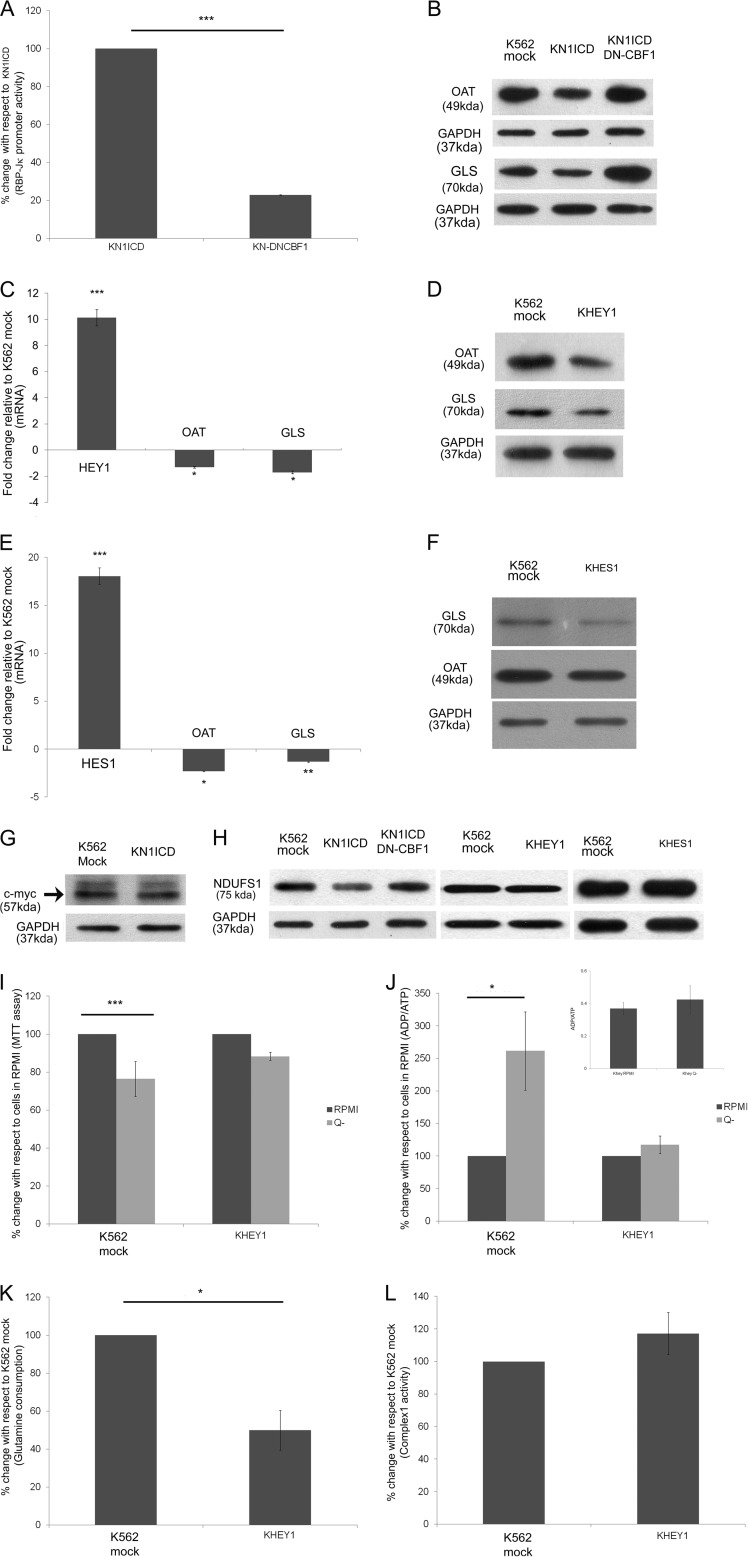
In the canonical pathway NICD interacts with the DNA binding factor, CBF-1/RBP-Jκ, to form a ternary complex acting as a transcriptional activator. Co-expressing DN-CBF1, which binds NICD but cannot interact with DNA, abrogated N1ICD-mediated RBP-Jκ promoter activity in KN1ICD cells (Fig. 6A). This also increased the expression of OAT and GLS in KN1ICD cells co-expressing DN-CBF1 (Fig. 6B). Our previous study (19) showed that Hey1, a direct target of the canonical Notch pathway, is up-regulated in KN1ICD among others. In the stably expressing Hey1 K562 cells (KHEY1), we observed a decrease of OAT and GLS gene and protein expression (Fig. 6, C and D). The canonical Notch pathway increases the expression of many genes, among which two independent primary target genes are Hes1 and Hey1, which are members of the basic helix-loop-helix family of transcriptional repressors (24). Each works either individually or cooperatively to repress target gene expression through its specific DNA-binding sites (25). The effect of Hes1 expression on these two proteins was checked, and they also showed a decrease in the expression of OAT and GLS in K562 cells (Fig. 6, E and F). Again, c-Myc another direct target of Notch signaling pathway is known for up-regulating GLS expression both transcriptionally and post transcriptionally (26, 27). But K562 cells, which inherently have high c-Myc expression, do not show a significant increase in its protein expression in KN1ICD cells (Fig. 6G). On the other hand, due to DN-CBF-1, the NDUFS1 protein expression increased to a lesser extent in comparison to glutamine metabolism proteins (Fig. 6H). Again, Hey1- or Hes1-transfected K562 did not show any alteration in the expression of NDUFS1 protein (Fig. 6H). It may be noted that the UCSC genome browser database (46) analyzed the Hey1 binding by Chip-Seq assay in K562 cells and showed high probability of Hey1 binding upstream of OAT and GLS but a low probability of binding upstream to NDUFS1 and NDUFV2. The glutamine dependence studies with KHEY1 cells showed that similar to KN1ICD cells these cells lose dependence on exogenous glutamine for cell survival and ADP/ATP ratio is not affected significantly by the absence of exogenous glutamine in the growth medium (Fig. 6, I and J). The glutamine consumption also decreased in KHEY1 cells (Fig. 6K). But the rotenone-sensitive NADH:ubiquinone oxidoreductase activity in KHEY1 cells remained unaltered in comparison to K562 mock (Fig. 6L). Hence we clearly show that the canonical Notch signaling pathway is directly linked to the regulation of the glutamine dependence of the cells.
FIGURE 6.
Alteration of the expression of proteins involved in glutamine metabolism is via canonical Notch pathway. A, Notch signaling activity from RBP-Jκ luciferase reporter activity in KN1ICD and KN-DNCBF-1. B, Western blot for the expression change of OAT and GLS in KN-DNCBF-1. C, relative gene expression in K562 with constitutively expressing Hey1 compared with K562 mock. The mRNA expression of the gene was normalized to the expression of HPRT1 mRNA. Data represent the mean ± S.E. (*, p < 0.05; ***, p < 0.001, n = 3). D, Western blot of whole cell lysate of K562 mock- and K562-transfected with Hey1showing relative expression of OAT and GLS. E, relative gene expression in K562 with constitutively expressing Hes1 compared with K562 mock. The mRNA expression of the gene was normalized to the expression of HPRT1 mRNA. Data represent the mean ± S.E. (*p < 0.05, **p < 0.01, ***p < 0.001, n = 3). F, Western blot of whole cell lysate of K562 mock- and K562-transfected with Hes1 showing relative expression of OAT and GLS. G, Western blot for c-myc expression in K562 mock and KN1ICD. H, Western blot of whole cell lysate of KN1ICD and KN1ICD transfected with DN-CBF-1, K562 mock, and K562 transfected with Hey1 or Hes1 showing relative expression of NDUFS1. I, K562 mock and KHEY1 were plated in equal amounts in RPMI and RPMI without glutamine for 48 h. The cell survival was assessed by MTT assay. The readings were normalized by 0 h reading. The data give the MTT assay readings with respect to readings of cells in RPMI. Data show the mean reading of at least four separate experiments (***, p < 0.001). J, ADP/ATP ratio was determined by luciferase activity for cells growing in RPMI or RPMI without glutamine (Q-) media. The data are represented as percentage change, and the inset shows the actual ADP/ATP ratio (*, p < 0.05). K, percentage change in glutamine consumption/amount of protein for cells cultured in complete media for 48 h (*, p < 0.05). L, rotenone-sensitive NADH:ubiquinone oxidoreductase activity of Complex 1 of the respiratory chain normalized by the protein amount. The data represent the percentage of Complex 1 activity in KHEY1 with respect to K562 mock.
The Cross-talk between Notch Signaling Pathway and Glutamine Utilization in Jurkat Cells
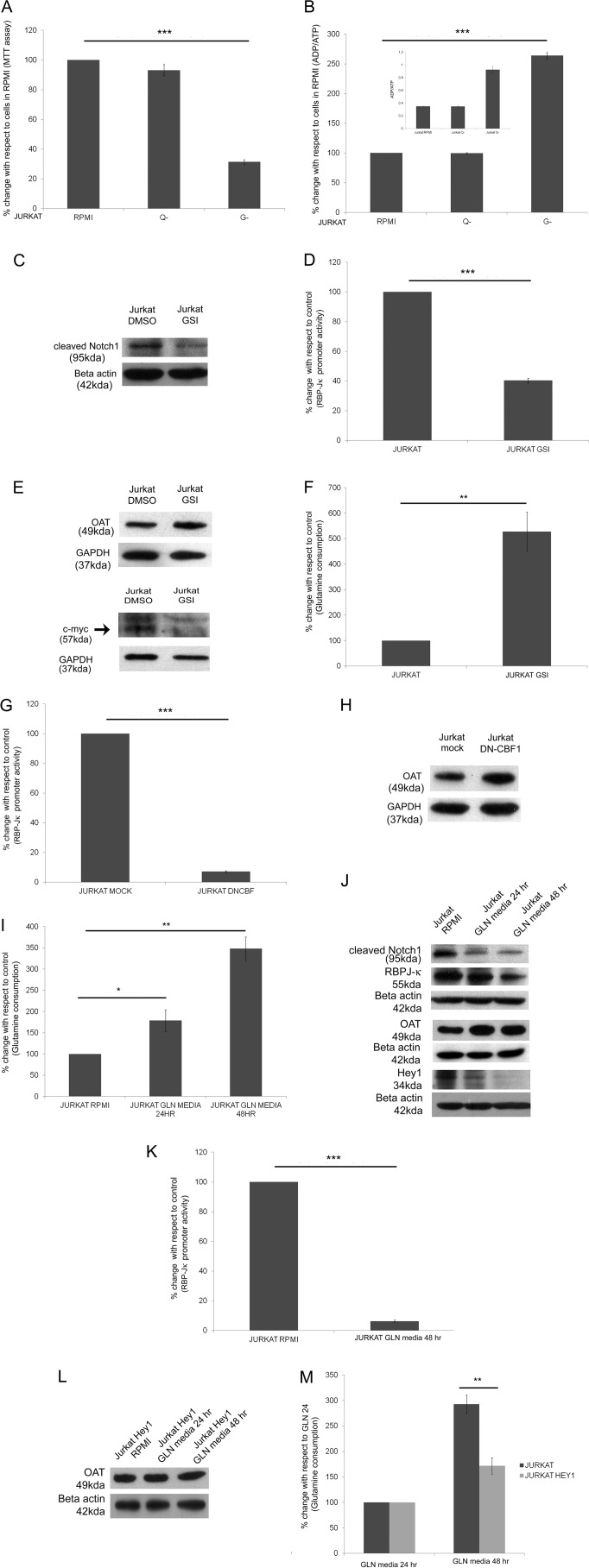
Jurkat, a T-cell acute lymphoblastic leukemia cell line that expresses wild type Notch1 with high expression of N1ICD, and its growth is inhibited by a high dose of GSI (28). This cell line has been reported earlier to have an impaired glutamine utilization capability (29) and has been used for studying change in glutamine dependence for various functions (30). Hence, first the exogenous glutamine dependence of Jurkat cells for cell survival and ADP/ATP ratio maintenance was studied. This showed that Jurkat cells are not affected by the removal of glutamine from the media, but these cells are highly dependent on glucose (Fig. 7, A and B). To understand the cross-talk between Notch signaling and glutamine dependence and utilization, the Notch signaling pathway was inhibited by treating cells with GSI. GSI treatment decreased cleaved Notch1 expression and abrogated RBP-Jκ-mediated Notch pathway activity (Fig. 7, C and D). This inhibition increased the expression of OAT and the glutamine consumption by the cells (Fig. 7, E and F), although c-Myc expression decreased (Fig. 7E), as previously shown by others (31). Again expressing DN-CBF1 in Jurkat cells decreased Notch activity and increased the expression of OAT (Fig. 7, G and H). Thus inhibition of Notch signaling in Jurkat cells affected the glutamine utilization.
FIGURE 7.
The cross-talk between Notch signaling pathway and glutamine utilization in Jurkat cells. A, Jurkat cells were plated in equal amounts in RPMI, RPMI without glutamine (Q-), and RPMI without glucose (G-) media for 48 h. The cell survival was assessed by MTT assay. The readings were normalized by 0-h reading. The data give the MTT assay readings with respect to readings of cells in RPMI. Data show the mean reading of at least four separate experiments (***, p < 0.001). B, ADP/ATP ratio was determined by luciferase activity for cells growing in RPMI, RPMI without glutamine, and RPMI without glucose media. The data are represented as percentage change, and the inset shows the actual ADP/ATP ratio (*, p < 0.001). C, Western blot of whole cell lysate of Jurkat and Jurkat GSI showing relative expression of cleaved Notch1. D, Notch signaling activity from RBP-Jκ luciferase reporter activity in Jurkat and Jurkat GSI. E, Western blot of whole cell lysate of Jurkat and Jurkat GSI showing relative expression of OAT and c-Myc. F, percentage change in glutamine consumption/amount of protein for cells cultured in complete media for 48 h (**, p < 0.01). G, Notch signaling activity from RBP-Jκ luciferase reporter activity in Jurkat and Jurkat DN-CBF1. H, Western blot of whole cell lysate of Jurkat and Jurkat DN-CBF showing relative expression of OAT. I, percentage change in glutamine consumption/amount of protein for cells cultured in complete media or GLN media for 24 or 48 h (*, p < 0.05; **p < 0.01). J, Western blot of whole cell lysates of Jurkat RPMI and Jurkat growing in GLN media for 24 and 48 h showing relative expression of cleaved Notch, RBP-Jκ, Hey1, and OAT. K, Notch signaling activity from RBP-Jκ luciferase reporter activity in Jurkat and Jurkat in GLN media for 48 h. L, Western blot of whole cell lysate of Jurkat expressing Hey1 growing in RPMI and GLN media for 24 or 48 h showing relative expression of OAT. M, percentage change in glutamine consumption/amount of protein for Jurkat Hey1 cells cultured in GLN media for 48 h with respect to cells cultured in GLN media for 24 h (**, p < 0.01).
Next, the effect on Notch signaling due to increased glutamine utilization in Jurkat cells was examined. For this, the glutamine consumption was artificially increased in Jurkat cells by growing the cells in “GLN media,” which is glucose-free media supplemented with glutamine (4 mm) and galactose (10 mm) as previously described (29, 32). Fig. 7I shows that glutamine consumption increases after 48 h of growing the Jurkat cells in GLN media. Under this condition we monitored the expression of cleaved Notch1, RBP-Jκ, Hey1, and OAT (Fig. 7J). As expected, the OAT expression increased in Jurkat cells growing in GLN media, and as per our hypothesis we found there was a decrease in cleaved Notch1, RBP-Jκ, and Hey1 expression. The Notch activity was also down-regulated in cells growing in GLN media for 48 h (Fig. 7K). Next, we stably expressed Hey1 in Jurkat cells (JHEY1). OAT expression in JHEY1 cells remained unaltered when grown in GLN media, and an increase in glutamine consumption in GLN media was significantly less in comparison to Jurkat cells (Fig. 7, L and M), thus verifying further the presence of cross-talk between the Notch signaling pathway and glutamine utilization pathway.
DISCUSSION
In this study we show that Notch1 activation alters the mitochondrial proteome. It was found that key proteins of Complex1 and the glutamine catabolic pathway are down-regulated. These down-regulations also affect the activity and functioning of the pathway in which they are involved, as they lead to the decrease of the rotenone-sensitive NADH:ubiquinone oxidoreductase activity and dependence on exogenous glutamine for cell survival. Our study also shows that this deregulation of glutamine catabolism is regulated by a canonical Notch pathway. The study establishes cross-talk between Notch signaling pathway and glutamine utilization by the cells. Apart from this, our study highlights the effect of Notch activation on the high energy intermediates ATP, NADH, and NADPH.
Earlier studies have shown that T-cell development mediated by Notch signaling pathway is dependent on PI3K/Akt-mediated up-regulation of glycolysis (10). An extension to this regulation of glycolysis by Notch signaling has been shown to cause the formation of aggressive tumor (11). The rearrangement of the metabolic network in cancer cells is being widely explored. A salient feature of cancer cells is that it frequently relies on glycolysis rather than oxidative phosphorylation (OXPHOS) for energy generation. This phenomenon was first observed by Otto Warburg, who more than 80 years ago found that cancer cells undergo aerobic glycolysis (33). Further studies on cancer metabolism have shown that although cancer cells are not dependent on OXPHOS, mitochondrial integrity is not usually compromised (34). The cells have abundance of ATP and tumor cells maintain a low NAD+/NADH ratio (35), and cancer cells possess high amounts of NADPH (36). These apart from its regular functions as high energy units and redox regulation are important for de novo synthesis of metabolic intermediates, which are required by highly proliferating tumor cells (37). From our study we find that Notch signaling up-regulation down-regulates OXPHOS proteins, oxygen consumption rate, and PDH activity but maintains mitochondrial membrane potential and mass. Upon Notch activation the ADP/ATP and NAD+/NADH decreases, and the amounts of NADPH increases. These patterns of alteration of the metabolic intermediates are similar to the metabolic adaptations during tumor progression. This further supports the role of Notch signaling in modifying the metabolic networks of tumor cells.
The first step of glutamine catabolism is glutaminolysis mediated by mitochondrial glutaminase to convert it into glutamate. Glutamate is a very important metabolic intermediate as it is involved in a variety of important biological processes such as conversion to α-ketoglutarate, which is a substrate for the Kreb cycle, amino acid metabolism, formation of glutathione, etc. Highly proliferating cells are addicted to glutamine, and glutamine catabolism is often up-regulated in tumor cells (23). It is interesting to note that from our two-dimensional DIGE study we found that up-regulation of Notch1 signaling in K562 cells decreases expression of GLS, OAT, and GLUD1. The activation of Notch1 not only alters protein expression but also decreases glutamine consumption. Moreover, Notch1 activation alters the cellular metabolism such that the cells lose their dependence on glutamine for survival, and the ADP/ATP ratio becomes independent of the presence of exogenous glutamine. Thus, on one hand increased Notch activity decreases glutamine utilization and on the other we show that increased glutamine utilization disrupts Notch signaling pathway. A literature survey shows that metastasis and growth of brain tumor is inhibited by targeting glutamine metabolism (38) and increased expression of Notch1 (39), respectively. Again, Jurkat, a T-cell acute lymphoblastic leukemia cell line that harbors activated Notch1 pathway, has a lower growth rate in high glutamine media, which increases upon inducing glutamine utilization ability (29). The process of naïve T-cell activation is a highly glutamine-dependent process (40) and is inhibited by the activation of Notch pathway (41). Furthermore, during T-cell activation, CBF1, a key effector of the Notch signaling pathway, is among the few genes that are down-regulated (42). It was shown nearly a decade earlier that Notch3 mutation is related to mitochondrial dysfunction (43). Recently it has been shown that increased Notch activity decreases rotenone sensitivity for cell death, whereas decreasing Notch activity by DN-CBF1 decreases Complex 1 activity and rotenone sensitivity (11). Our study shows that the expression of NDUFS1 (Complex 1 subunit) was reduced, accompanied with a decrease in the rotenone-sensitive NADH:ubiquinone oxidoreductase activity due to Notch activation. In contrast, inhibition of the canonical Notch pathways or expression of its downstream effectors did not result in a significant alteration of NDUFS1 and rotenone-sensitive NADH:ubiquinone oxidoreductase activity. This apparent inconsistency suggests that the regulation of rotenone-sensitive NADH:ubiquinone oxidoreductase activity might follow a non-canonical pathway as observed in various regulation of cellular functions by Notch signaling (44, 45). The study thus confirms that Notch signaling is related to mitochondrial OXPHOS regulation and that Notch signaling and glutamine utilization are inversely dependent and regulated by the canonical Notch pathway.
In summary, this study underlines that Notch1 pathway activation affects the mitochondrial proteome and causes alteration of common metabolic pathway proteins. It decreases Complex 1 activity and makes the cells independent of exogenous glutamine. Notch signaling inhibition or endogenous activation also impacts the protein expression. The canonical Notch pathway regulates the glutamine metabolism pathway proteins. Expression of Hey1 alone can make the cells independent of exogenous glutamine. Last, our study put forward the fact that Notch signaling pathway causes a major rearrangement of the metabolic network partly mimicking the cancer cells.
Acknowledgments
We acknowledge Dr. J. Aster, Dr. S. Krishna, Dr. A. Sarin, Dr. S. Stifani, and Dr. K. Helin for the plasmid constructs of pcDNA-(His)-NICD, pcDNA-jagged1, DN-CBF-1, Hes1, and Hey1, respectively. We acknowledge the laboratory of Dr. S. Adhya (of Council of Scientific and Industrial Research IICB, Kolkata) for providing the Oxygraph instrument.
The work was supported by the Integrative Biology on Omics Platform project of Dept. of Atomic Energy, Government of India.

This article contains supplemental Spectra 1–11.
- OXPHOS
- oxidative phosphorylation system
- RBP-Jκ
- recombinant signal-binding protein for immunoglobulin-κJ region
- DIGE
- differential in gel electrophoresis
- NICD
- Notch intracellular domain
- N1ICD
- Notch1 intracellular domain
- GSI
- γ-secretase inhibitor
- DN
- dominant negative CBF-1
- MAM
- mitochondria-associated membrane
- IB
- isolation buffer
- IB2
- isolation buffer 2
- VDAC
- voltage-dependent anion channel protein
- PDH
- pyruvate dehydrogenase complex
- GLUD1
- glutamate dehydrogenase 1
- OAT
- ornithine aminotransferase
- GLS
- glutaminase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
REFERENCES
- 1. Taylor S. W., Fahy E., Zhang B., Glenn G. M., Warnock D. E., Wiley S., Murphy A. N., Gaucher S. P., Capaldi R. A., Gibson B. W., Ghosh S. S. (2003) Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 21, 281–286 [DOI] [PubMed] [Google Scholar]
- 2. Neupert W., Herrmann J. M. (2007) Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 3. Artavanis-Tsakonas S., Rand M. D., Lake R. J. (1999) Notch signaling. Cell fate control and signal integration in development. Science 284, 770–776 [DOI] [PubMed] [Google Scholar]
- 4. Kopan R., Ilagan M. X. (2009) The canonical Notch signaling pathway. Unfolding the activation mechanism. Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schroeter E. H., Kisslinger J. A., Kopan R. (1998) Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 [DOI] [PubMed] [Google Scholar]
- 6. Nakagawa O., McFadden D. G., Nakagawa M., Yanagisawa H., Hu T., Srivastava D., Olson E. N. (2000) Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signaling. Proc. Natl. Acad. Sci. U.S.A. 97, 13655–13660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Radtke F., Raj K. (2003) The role of Notch in tumorigenesis. Oncogene or tumour suppressor? Nat. Rev. Cancer 3, 756–767 [DOI] [PubMed] [Google Scholar]
- 8. Callahan R., Egan S. E. (2004) Notch signaling in mammary development and oncogenesis. J. Mammary Gland Biol. Neoplasia 9, 145–163 [DOI] [PubMed] [Google Scholar]
- 9. Weng A. P., Ferrando A. A., Lee W., Morris J. P., 4th, Silverman L. B., Sanchez-Irizarry C., Blacklow S. C., Look A. T., Aster J. C. (2004) Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271 [DOI] [PubMed] [Google Scholar]
- 10. Ciofani M., Zúñiga-Pflücker J. C. (2005) Notch promotes survival of pre-T cells at the β-selection checkpoint by regulating cellular metabolism. Nat. Immunol. 6, 881–888 [DOI] [PubMed] [Google Scholar]
- 11. Landor S. K., Mutvei A. P., Mamaeva V., Jin S., Busk M., Borra R., Grönroos T. J., Kronqvist P., Lendahl U., Sahlgren C. M. (2011) Hypo- and hyperactivated Notch signaling induce a glycolytic switch through distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 108, 18814–18819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng P., Gabrilovich D. (2008) Notch signaling in differentiation and function of dendritic cells. Immunol. Res. 41, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rezaul K., Wu L., Mayya V., Hwang S. I., Han D. (2005) A systematic characterization of mitochondrial proteome from human T leukemia cells. Mol. Cell. Proteomics 4, 169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoffstrom B. G., Kaplan A., Letso R., Schmid R. S., Turmel G. J., Lo D. C., Stockwell B. R. (2010) Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat. Chem. Biol. 6, 900–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G. M., Carnemolla B., Orecchia P., Zardi L., Righetti P. G. (2004) Blue silver. A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25, 1327–1333 [DOI] [PubMed] [Google Scholar]
- 16. Janssen A. J., Trijbels F. J., Sengers R. C., Smeitink J. A., van den Heuvel L. P., Wintjes L. T., Stoltenborg-Hogenkamp B. J., Rodenburg R. J. (2007) Spectrophotometric assay for complex I of the respiratory chain in tissue samples and cultured fibroblasts. Clin. Chem. 53, 729–734 [DOI] [PubMed] [Google Scholar]
- 17. Kannan S., Sutphin R. M., Hall M. G., Golfman L. S., Fang W., Nolo R. M., Akers L. J., Hammitt R. A., McMurray J. S., Kornblau S. M., Melnick A. M., Figueroa M. E., Zweidler-McKay P. A. (2013) Notch activation inhibits AML growth and survival. A potential therapeutic approach. J. Exp. Med. 210, 321–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ishiko E., Matsumura I., Ezoe S., Gale K., Ishiko J., Satoh Y., Tanaka H., Shibayama H., Mizuki M., Era T., Enver T., Kanakura Y. (2005) Notch signals inhibit the development of erythroid/megakaryocytic cells by suppressing GATA-1 activity through the induction of HES1. J. Biol. Chem. 280, 4929–4939 [DOI] [PubMed] [Google Scholar]
- 19. Roy A., Basak N. P., Banerjee S. (2012) Notch1 intracellular domain increases cytoplasmic EZH2 levels during early megakaryopoiesis. Cell Death Dis. 3, e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yin D. D., Fan F. Y., Hu X. B., Hou L. H., Zhang X. P., Liu L., Liang Y. M., Han H. (2009) Notch signaling inhibits the growth of the human chronic myeloid leukemia cell line K562. Leuk Res. 33, 109–114 [DOI] [PubMed] [Google Scholar]
- 21. Hoefs S. J., Skjeldal O. H., Rodenburg R. J., Nedregaard B., van Kaauwen E. P., Spiekerkötter U., von Kleist-Retzow J. C., Smeitink J. A., Nijtmans L. G., van den Heuvel L. P. (2010) Novel mutations in the NDUFS1 gene cause low residual activities in human complex I deficiencies. Mol. Genet. Metab. 100, 251–256 [DOI] [PubMed] [Google Scholar]
- 22. Liu H. Y., Liao P. C., Chuang K. T., Kao M. C. (2011) Mitochondrial targeting of human NADH dehydrogenase (ubiquinone) flavoprotein 2 (NDUFV2) and its association with early-onset hypertrophic cardiomyopathy and encephalopathy. J. Biomed. Sci. 18, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wise D. R., Thompson C. B. (2010) Glutamine addiction. A new therapeutic target in cancer. Trends Biochem. Sci. 35, 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leal M. C., Surace E. I., Holgado M. P., Ferrari C. C., Tarelli R., Pitossi F., Wisniewski T., Castaño E. M., Morelli L. (2012) Notch signaling proteins HES-1 and Hey-1 bind to insulin degrading enzyme (IDE) proximal promoter and repress its transcription and activity. Implications for cellular Aβ metabolism. Biochim. Biophys. Acta 1823, 227–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iso T., Sartorelli V., Poizat C., Iezzi S., Wu H. Y., Chung G., Kedes L., Hamamori Y. (2001) HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol. Cell Biol. 21, 6080–6089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wise D. R., DeBerardinis R. J., Mancuso A., Sayed N., Zhang X. Y., Pfeiffer H. K., Nissim I., Daikhin E., Yudkoff M., McMahon S. B., Thompson C. B. (2008) Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. U.S.A. 105, 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gao P., Tchernyshyov I., Chang T. C., Lee Y. S., Kita K., Ochi T., Zeller K. I., De Marzo A. M., Van Eyk J. E., Mendell J. T., Dang C. V. (2009) c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458, 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rao S. S., O'Neil J., Liberator C. D., Hardwick J. S., Dai X., Zhang T., Tyminski E., Yuan J., Kohl N. E., Richon V. M., Van der Ploeg L. H., Carroll P. M., Draetta G. F., Look A. T., Strack P. R., Winter C. G. (2009) Inhibition of NOTCH signaling by γ-secretase inhibitor engages the RB pathway and elicits cell cycle exit in T-cell acute lymphoblastic leukemia cells. Cancer Res. 69, 3060–3068 [DOI] [PubMed] [Google Scholar]
- 29. Rathore M. G., Saumet A., Rossi J. F., de Bettignies C., Tempé D., Lecellier C. H., Villalba M. (2012) The NF-κB member p65 controls glutamine metabolism through miR-23a. Int. J. Biochem. Cell Biol. 44, 1448–1456 [DOI] [PubMed] [Google Scholar]
- 30. Chang W. K., Yang K. D., Chuang H., Jan J. T., Shaio M. F. (2002) Glutamine protects activated human T cells from apoptosis by up-regulating glutathione and Bcl-2 levels. Clin. Immunol. 104, 151–160 [DOI] [PubMed] [Google Scholar]
- 31. Chadwick N., Zeef L., Portillo V., Fennessy C., Warrander F., Hoyle S., Buckle A. M. (2009) Identification of novel Notch target genes in T cell leukaemia. Mol. Cancer 8, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rossignol R., Gilkerson R., Aggeler R., Yamagata K., Remington S. J., Capaldi R. A. (2004) Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 64, 985–993 [DOI] [PubMed] [Google Scholar]
- 33. Warburg O. (1956) On the origin of cancer cells. Science 123, 309–314 [DOI] [PubMed] [Google Scholar]
- 34. Wallace D. C. (2012) Mitochondria and cancer. Nat. Rev. Cancer 12, 685–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chiarugi A., Dölle C., Felici R., Ziegler M. (2012) The NAD metabolome. A key determinant of cancer cell biology. Nat. Rev. Cancer 12, 741–752 [DOI] [PubMed] [Google Scholar]
- 36. Jeon S. M., Chandel N. S., Hay N. (2012) AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature 485, 661–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oka S., Hsu C. P., Sadoshima J. (2012) Regulation of cell survival and death by pyridine nucleotides. Circ. Res. 111, 611–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fan X., Mikolaenko I., Elhassan I., Ni X., Wang Y., Ball D., Brat D. J., Perry A., Eberhart C. G. (2004) Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 64, 7787–7793 [DOI] [PubMed] [Google Scholar]
- 39. Shelton L. M., Huysentruyt L. C., Seyfried T. N. (2010) Glutamine targeting inhibits systemic metastasis in the VM-M3 murine tumor model. Int. J. Cancer 127, 2478–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carr E. L., Kelman A., Wu G. S., Gopaul R., Senkevitch E., Aghvanyan A., Turay A. M., Frauwirth K. A. (2010) Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol. 185, 1037–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eagar T. N., Tang Q., Wolfe M., He Y., Pear W. S., Bluestone J. A. (2004) Notch 1 signaling regulates peripheral T cell activation. Immunity 20, 407–415 [DOI] [PubMed] [Google Scholar]
- 42. Tyagi M., Karn J. (2007) CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 26, 4985–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. de la Peña P., Bornstein B., del Hoyo P., Fernández-Moreno M. A., Martín M. A., Campos Y., Gómez-Escalonilla C., Molina J. A., Cabello A., Arenas J., Garesse R. (2001) Mitochondrial dysfunction associated with a mutation in the Notch3 gene in a CADASIL family. Neurology 57, 1235–1238 [DOI] [PubMed] [Google Scholar]
- 44. Perumalsamy L. R., Nagala M., Banerjee P., Sarin A. (2009) A hierarchical cascade activated by non-canonical Notch signaling and the mTOR-Rictor complex regulates neglect-induced death in mammalian cells. Cell Death Differ. 16, 879–889 [DOI] [PubMed] [Google Scholar]
- 45. Lee S. F., Srinivasan B., Sephton C. F., Dries D. R., Wang B., Yu C., Wang Y., Dewey C. M., Shah S., Jiang J., Yu G. (2011) γ-Secretase-regulated proteolysis of the Notch receptor by mitochondrial intermediate peptidase. J. Biol. Chem. 286, 27447–27453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Meyer L. R., Zweig A. S., Hinrichs A. S., Karolchik D., Kuhn R. M., Wong M., Sloan C. A., Rosenbloom K. R., Roe G., Rhead B., Raney B. J., Pohl A., Malladi V. S., Li C. H., Lee B. T., Learned K., Kirkup V., Hsu F., Heitner S., Harte R. A., Haeussler M., Guruvadoo L., Goldman M., Giardine B. M., Fujita P. A., Dreszer T. R., Diekhans M., Cline M. S., Clawson H., Barber G. P., Haussler D., Kent W. J. (2013) The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 41, D64–D69 [DOI] [PMC free article] [PubMed] [Google Scholar]