Background: Family-9 glycoside hydrolases displaying various organizations are plethoric in cellulosome-producing bacteria.
Results: The 13 family-9 enzymes synthesized by Clostridium cellulolyticum exhibit different substrate specificities, activities, and relationships with key cellulosomal cellulases in artificial cellulosomes.
Conclusion: This variety is required for efficient degradation of crystalline cellulose.
Significance: This first global picture provides insights on the multiplicity and diversity of family-9 enzymes in bacterial cellulosomes.
Keywords: Carbohydrate, Cellulase, Enzyme Kinetics, Enzymes, Protein Complexes, GH9, Cellulose, Cellulosome, Clostridium cellulolyticum, Synergy
Abstract
The genome of Clostridium cellulolyticum encodes 13 GH9 enzymes that display seven distinct domain organizations. All but one contain a dockerin module and were formerly detected in the cellulosomes, but only three of them were previously studied (Cel9E, Cel9G, and Cel9M). In this study, the 10 uncharacterized GH9 enzymes were overproduced in Escherichia coli and purified, and their activity pattern was investigated in the free state or in cellulosome chimeras with key cellulosomal cellulases. The newly purified GH9 enzymes, including those that share similar organization, all exhibited distinct activity patterns, various binding capacities on cellulosic substrates, and different synergies with pivotal cellulases in mini-cellulosomes. Furthermore, one enzyme (Cel9X) was characterized as the first genuine endoxyloglucanase belonging to this family, with no activity on soluble and insoluble celluloses. Another GH9 enzyme (Cel9V), whose sequence is 78% identical to the cellulosomal cellulase Cel9E, was found inactive in the free and complexed states on all tested substrates. The sole noncellulosomal GH9 (Cel9W) is a cellulase displaying a broad substrate specificity, whose engineered form bearing a dockerin can act synergistically in minicomplexes. Finally, incorporation of all GH9 cellulases in trivalent cellulosome chimera containing Cel48F and Cel9G generated a mixture of heterogeneous mini-cellulosomes that exhibit more activity on crystalline cellulose than the best homogeneous tri-functional complex. Altogether, our data emphasize the importance of GH9 diversity in bacterial cellulosomes, confirm that Cel9G is the most synergistic GH9 with the major endoprocessive cellulase Cel48F, but also identify Cel9U as an important cellulosomal component during cellulose depolymerization.
Introduction
Glycoside hydrolases (GH)2 catalyze the hydrolysis of glycosidic bonds in oligo- or polysaccharides. To date, these enzymes have been classified into 133 families on the basis of sequence homologies, but new families arise continuously, as new enzyme sequences and activity patterns become available (1). Some families are rather heterogeneous in terms of enzyme specificities. Family-5, for instance (∼3,800 sequences), contains a large variety of specificities as reflected by the 18 different experimentally determined activities denoted with a specific Enzyme Commission number (1). The GH5 sequences have recently been assigned into 51 subfamilies (2).
Enzymes characterized as cellulases are found in 17 families of the classification initiated in 1991 by Henrissat et al. (3). In contrast to family-5, some families like family-9 and -48 are more homogeneous in terms of specificities because they almost exclusively contain cellulases (EC 3.2.1.4, EC 3.2.1.91, and EC 3.2.1.176). Indeed, 13 of the 14 GH48 enzymes characterized to date were described as cellulases, and only one as a chitinase. Similarly, the 132 characterized GH9 enzymes (out of the 1,525 sequences classified in family 9) were identified as cellulases except for one enzyme from Photobacterium profundum that was found to be more active on chito-oligosaccharides than on cellodextrins (4). Naturally, many GH9 cellulases also display side activities on related polysaccharides such as β1,3–1,4-glycans (5), xylan (6), or xyloglucan (7), but their favorite substrate is either soluble (carboxymethylcellulose and cellodextrins) or insoluble (amorphous) cellulose. In addition to their catalytic domains, a large proportion of known GH9 enzymes contain ancillary modules/domains like carbohydrate-binding modules (CBM) (8–10) or modules whose functions have not yet been fully elucidated such as Ig-like (11) and X2 domains (12). GH9-encoding genes are widespread among cellulolytic microorganisms (except aerobic fungi) and plants, but the genes coding this family of enzymes are particularly abundant in anaerobic bacteria producing cellulosomes. The cellulosomes are heterogeneous large extracellular complexes that efficiently degrade cellulose and related plant cell wall polysaccharides. The simplest cellulosomes such as those produced by mesophilic and cellulolytic clostridia are composed of a single major scaffolding protein hosting a CBM displaying affinity for cellulose and a series of cohesin modules that strongly interact with a complementary module, the dockerin, borne by the catalytic subunits. In other cellulolytic bacteria, such as Clostridium thermocellum and rumen bacteria, several interacting scaffoldins are synthesized, and different types of specific cohesin/dockerin devices are used to assemble and attach to the cell surface these intricate cellulosomes (13). The genome of cellulosome-producing bacteria usually contains only one or two genes coding for a cellulosomal GH48 enzyme, whereas GH9-encoding genes are plethoric. Thus, 15, 13, 8, and 12 genes that putatively encode cellulosomal (i.e. appended with a dockerin) GH9 were discovered in the sequenced genomes of C. thermocellum (DSM 1313) (14), Clostridium papyrosolvens (DSM 2782), Clostridium cellulovorans (743B) (15), and C. cellulolyticum (ATCC 35319) (16), respectively. Furthermore, one or few genes encoding noncellulosomal GH9 were also found in these bacterial genomes. In the case of C. cellulolyticum, the 12 GH9-encoding genes would account for 70% of the genes predicted to code for cellulosomal cellulases. Moreover, former studies (16, 17) have shown using proteomic analyses that their products are systematically detected in the cellulosomes whatever cellulosic substrate (pure crystalline cellulose or hatched straw) is present. This observation indicates that all dockerin-bearing GH9 enzymes are recurrent components of the cellulosomes, in contrast to other cellulosomal catalytic subunits whose synthesis and participation to the cellulosomes are substrate-dependent (16, 17). As for other cellulolytic clostridia, only few C. cellulolyticum GH9 enzymes displaying very different organizations were selected to date for characterization. Thus, Cel9G (8) and Cel9M (18) were described as endoglucanases, whereas Cel9E (9) was found to be an endoprocessive cellulase.
In this study, the multiplicity and the diversity of GH9 enzymes in bacterial cellulosomes were investigated by overproducing, purifying, and establishing the catalytic properties of all uncharacterized GH9 enzymes from C. cellulolyticum. Their contribution to cellulosome efficiency was evaluated using the cellulosome chimera technology by complexation of the GH9 enzymes with pivotal cellulosomal cellulases onto hybrid scaffoldins. The sole noncellulosomal GH9 of C. cellulolyticum was also studied, as well as an engineered form of this enzyme displaying a dockerin module.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
Genomic DNA from C. cellulolyticum ATCC 35319 served as template for amplification by PCR of the DNA encoding the mature form of the various GH9s. A list of primers used in this study is provided in supplemental Table 1. The amplicons were cloned in pET28a(+) (Novagen, Madison, WI) at NcoI/XhoI sites, except for the gene encoding Cel9R which was cloned in pET23(+) at NheI/XhoI sites. In all cases, six His codons were introduced at the 3′ extremity of the coding sequence. Positive clones were verified by DNA sequencing. The BL21(DE3) Escherichia coli strain (Novagen) was used as production strain.
Protein Production and Purification
Purification of Cel48Fc, Cel48Ft, Cel9Gc, Cel9Gf, Cel9Ec, Cel9Et, Cel9Mc, Scaf2, Scaf4, and Scaf6 were formerly described (8, 9, 18–22). The BL21(DE3) overproducing the various GH9 enzymes were grown in 2.5-liter toxin flasks at 37 °C in Luria-Bertani medium supplemented with glycerol (12 g/liter) and the appropriate antibiotic until A600 = 1.5–2. To prevent the formation of inclusion bodies, the cultures were then cooled down, and induction of the expression was performed overnight at 16 °C with 50 μm isopropyl thio-β-d-galactoside for the strains BL21(DE3) carrying pET28a-Cel9H, pET28a-Cel9J, pET28a-Cel9Q, pET28a-Cel9T, pET23a-Cel9R, pET28a-Cel9X, and pET28a-Cel9V and 80 μm isopropyl thio-β-d-galactoside for the strains containing the vectors pET28a-Cel9W and pET28a-Cel9Wc, and 100 μm isopropyl thio-β-d-galactoside for the strain carrying pET28a-Cel9P and pET28a-Cel9U. After 16 h of induction, the cells were harvested by centrifugation (3,000 × g, 15 min) and resuspended in 30 mm Tris-HCl, pH 8.0, 1 mm CaCl2, supplemented with a few milligrams of DNase I (Roche Applied Science), and broken in a French press. The crude extract was centrifuged 15 min at 10,000 × g and loaded on 2–5 ml of nickel-nitrilotriacetic acid resin (Qiagen, Vanloo, The Netherlands) equilibrated in the same buffer. The proteins of interest were then eluted with 100 mm imidazole in 30 mm Tris-HCl, pH 8.0, 1 mm CaCl2. The purification of the recombinant proteins was achieved on Q-Sepharose fast flow (GE Healthcare) equilibrated in 30 mm Tris-HCl, pH 8.0, 1 mm CaCl2. The proteins of interest were eluted by a linear gradient of 0–500 mm NaCl in 30 mm Tris-HCl, pH 8.0, 1 mm CaCl2. In the case of Cel9Vc, an alternative purification procedure was also used. After the first purification step on nickel-nitrilotriacetic acid resin, the fraction containing the protein of interest was concentrated by ultrafiltration to 2 ml, and purification was achieved by gel filtration on a Superdex 200 10/300 GL resin (GE Healthcare) equilibrated in 30 mm Tris-HCl, pH 8.0, 1 mm CaCl2, 150 mm NaCl.
The purified proteins were dialyzed by ultrafiltration against 10 mm Tris-HCl, pH 8.0, 1 mm CaCl2, and stored at −80 °C. The concentration of the proteins was estimated by absorbance at 280 nm in 25 mm sodium phosphate, pH 6.5, using the program ProtParam tool.
Verification of Complex Formation
Scaf2-, Scaf4-, and Scaf6-based complexes were verified by nondenaturing PAGE (19, 20). Interacting protein components (enzymes bearing a dockerin and scaffoldin) were mixed at a final concentration of 10 μm at room temperature in 20 mm Tris maleate, pH 6.0, 1 mm CaCl2, and 4 μl were subjected to PAGE (4–15% gradient) using a Phastsystem apparatus (GE Healthcare). In the case of Scaf6-based heterogeneous mixtures with variable GH9 enzyme occupying the C. cellulolyticum cohesin, each of the 11 GH9 enzymes (bearing a C. cellulolyticum dockerin) was at final concentration of 0.91 μm, whereas Scaf6, the cellulase appended with a C. thermocellum dockerin (Cel48Ft), and the cellulase hosting a Ruminococcus flavefaciens dockerin (Cel9Gf) were at 10 μm.
Enzyme and Complex Activity
Hydrolytic activity on soluble polysaccharides like carboxymethylcellulose (CMC) medium viscosity, laminarin (Sigma), arabinan (sugar beet), and xyloglucan (Megazyme, Wiclow, Ireland) was determined by mixing 4 ml of substrate solution at 10 g/liter (CMC) or 3.5 g/liter (laminarin, arabinan, and xyloglucan) in 20 mm Tris maleate, pH 6.0, 1 mm CaCl2, 0.01% (w/v) azide with 40 μl of an appropriate dilution of enzyme (final enzyme concentration ranging from 1 to 100 nm) at 37 °C. At specific intervals, 0.5-ml aliquots were cooled down in ice and analyzed for reducing sugar contents using the Park and Johnson method (23) and glucose as the standard. Released sugars were in some cases also analyzed by high performance anion exchange chromatography coupled with pulsed amperometric detection (HPAEC-PAD) (see below). Phosphoric acid swollen cellulose (PASC) was prepared as described previously (24). Activity on insoluble substrates like Avicel (PH101, Fluka, Buchs, Switzerland), PASC, barley glucan (Megazyme), and oat spelled xylan (Sigma) was performed similarly under mild shaking (70 rpm), except that 0.8-ml aliquots were pipetted at specific intervals and centrifuged at 4 °C for 10 min at 10,000 × g. The supernatants were analyzed for soluble released sugars by the Park and Johnson method (23) and/or HPAEC-PAD (25). The determination of insoluble reducing extremities during the hydrolysis of PASC was performed according to Ref. 26.
Activity on p-nitrophenyl (pNP) β-d-glucoside, pNP β-d-cellobioside, pNP α-l-arabinoside, pNP β-d-xyloside (Sigma) at 1 g/liter in 50 mm potassium phosphate buffer, pH 7.0, 0.01% (w/v) azide was assayed by incubating at 37 °C 1 ml of substrate solution with 10 μl of enzyme at 10 μm and monitoring the pNP release at 400 nm. One IU corresponds to 1 μmol of pNP (using pNP from Sigma as the standard) released per min. Kinetics on cellodextrins ranging from cellobiose to cellohexaose were performed by incubating at 37 °C 10 μl of enzyme (1 μm) with 90 μl of substrate at 1.11 mm in 20 mm Tris maleate, pH 6.0, 1 mm CaCl2, 0.01% (w/v) sodium azide. Samples (20 μl) were extracted at specific intervals and analyzed by HPAEC-PAD.
HPAEC-PAD analyses of the released soluble sugars were performed in a Dionex ICS 3000 (Sunnyvale, CA) equipped with a pulsed amperometric detector. 200 μl of sample (or 10 times diluted sample in distilled water) were mixed with 50 μl of 0.5 m NaOH, and 25 μl were applied to a Dionex CarboPac PA1 column (4 × 250 mm) preceded by the corresponding guard column (4 × 50 mm). Sugars were eluted with the buffers 0.1 m NaOH and 0.5 m sodium acetate + 0.1 m NaOH as the eluants A and B, respectively, using for cellodextrins the multistep procedure as follows: isochratic separation (5 min, 95% A + 5% B), separation gradient (8 min, 10–37% B), column wash (2 min, 99% B), and subsequent column equilibration (2.5 min, 95% A + 5% B). For analysis of xyloglucan dextrins, the same A and B buffers were used, but a multistep procedure was as follows: isochratic separation (5 min, 95% A + 5% B), separation gradient (25.5 min, 10–99% B), column wash (2 min, 99% B), and subsequent column equilibration (2.5 min, 95% A + 5% B). The flow rate was kept at 1 ml/min in all cases. Injection of samples containing glucose, cellobiose, cellotriose, cellotetraose, cellopentaose, and cellohexaose (Sigma) or the xyloglucan-derived oligosaccharides (Megazyme) isoprimeverose, heptasaccharide (XXXG type), octasaccharide (XLXG type), and nonasaccharides (XLLG type) (27) at known concentrations (ranging from 5 to 100 μm) were used to identify and quantify the released sugars.
Viscosimetric Assays
Viscosimetric assays were performed by monitoring the flow time of 3.5 g/liter CMC or xyloglucan solutions (in 20 mm Tris maleate, pH 6.0, 1 mm CaCl2, 0.01% (w/v) sodium azide) mixed at 37 °C with appropriate quantities of enzyme for different incubation times. 1-ml samples were boiled for 15 min, and the fluidity was measured at room temperature. The relative fluidity ΔF was determined as (To/(T − To)) − (To/(To′ − To)), where To, To′, and T correspond to the flow time of buffer, the flow time of CMC or xyloglucan solutions without enzyme, and the flow time of substrate with enzyme, respectively. In parallel, the reducing sugars content of the samples was determined using the Park and Johnson method (23).
Substrate Binding Assays
The binding of the enzymes to insoluble Avicel and PASC was investigated essentially as described previously (28) by incubating at 4 °C for 1 h under mild shaking, 200 μl of cellulose at 7 g/liter in 50 mm potassium phosphate, pH 7.0, 0.01% (w/v) sodium azide with 1.5 μm of protein. The suspension was subsequently centrifuged for 10 min at 4 °C (10,000 × g), and the supernatant was collected. Ten μl of supernatant were mixed with 5 μl of SDS-PAGE loading buffer and boiled for 5 min. The cellulose-containing pellet was subsequently washed twice with 200 μl of 50 mm potassium phosphate, pH 7.0, resuspended in 200 μl of diluted loading buffer, and boiled for 5 min. The sample was again centrifuged 10 min at 4 °C (10,000 × g), and the supernatant was collected. Samples (first and last supernatants) were subjected to SDS-PAGE using precast gels 4–15% (Bio-Rad). The binding to soluble xyloglucan was monitored by loading the 3 μg of protein of interest in 8% acrylamide gel ProSieve (Lonza, Basel, Switzerland) containing 0 or 0.1% (w/v) xyloglucan. In some cases, the binding to PASC was monitored as described above, except that xyloglucan (7 g/liter) was added to the cellulose suspension before the 1-h incubation at 4 °C.
Far-UV CD Spectroscopy
Far-UV CD spectra were acquired for Cel9Ec and two forms of Cel9Vc obtained by different purification procedures in a quartz cell with 1-mm path length at 25 °C using a Jasco-815 spectropolarimeter (Oklahoma City, OK). The protein concentration was 1 μm in 20 mm potassium phosphate, pH 7.0, buffer. All spectra were first buffer-corrected for background contributions and normalized for slight variations in protein concentrations.
RESULTS
Sequence Homologies and Domain Organization of GH9Enzymes from C. cellulolyticum
The annotation of the genome of C. cellulolyticum revealed the presence of 13 genes that putatively encode GH9 enzymes. These enzymes were all predicted to be cellulases, but this activity was only confirmed for three of them which were formerly characterized; Cel9M and Cel9G were thus reported to be endoglucanases (8, 18), and an endoprocessive cellulase activity was demonstrated for Cel9E (9). All these GH9s were detected by mass spectrometry in the cellulosomes, except the product (called Cel9W) of the gene at locus Ccel_2226. This protein exhibits a typical signal sequence but is devoid of a dockerin module (16) and would thus be secreted as a free enzyme in the external medium.
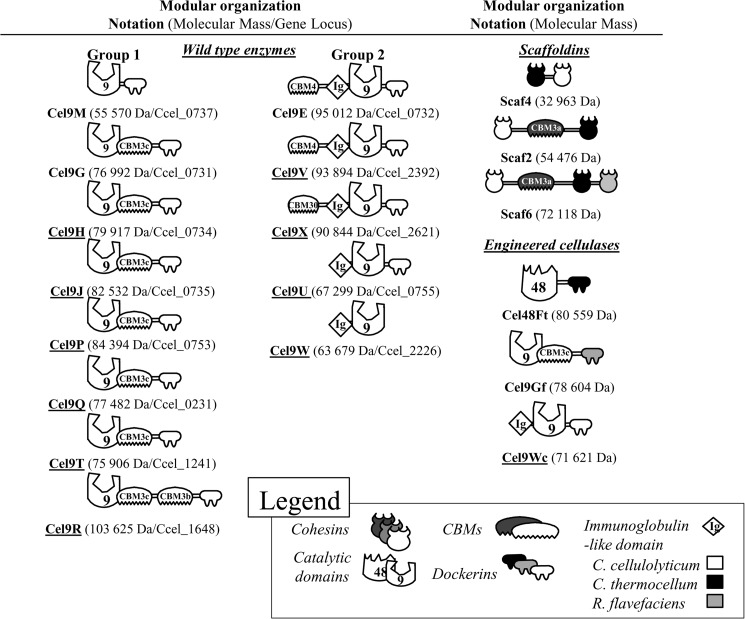
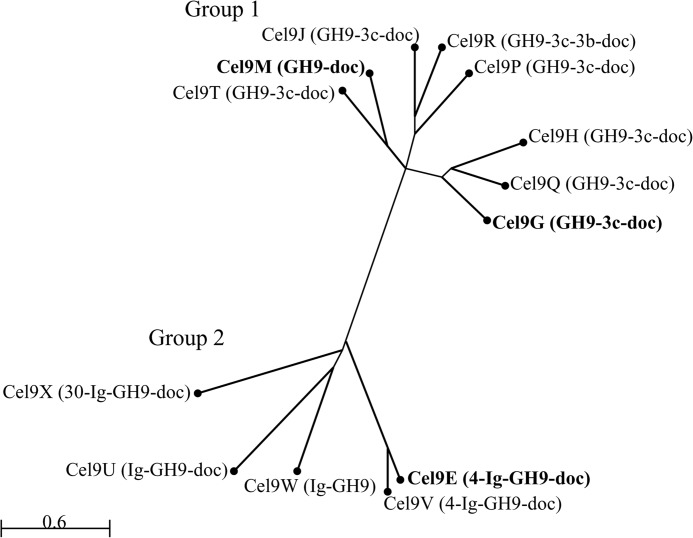
Although the 13 proteins contain a catalytic domain classified in the GH9 family, the sequences of these catalytic domains vary significantly, as well as the presence and nature of ancillary domains (Fig. 1). A phylogenetic tree based on the homologies between catalytic domains distinguishes two groups of GH9 (Fig. 2). The first group includes eight GH9 enzymes. Six of them, including the formerly characterized endoglucanase Cel9G, possess a family-3c CBM located at the C terminus of the catalytic domain followed by the dockerin module. Within this organization formerly termed Theme B1 (29), the CBM3c is considered as a “helper” CBM or a catalytic CBM because its deletion generates an inactive form of the enzyme (8). Cel9R is similar to Cel9G but contains a second CBM that belongs to family 3b′, located between the CBM3c′ and the C-terminal dockerin (organization termed Theme B2 (29)). Interestingly, the formerly characterized endoglucanase Cel9M (18), which only hosts a C-terminal dockerin module (organization termed Theme A (29)), clearly belongs to this group of GH9 as shown in Fig. 2, despite the absence of a CBM3c. It should be noted that sequence similarity between the catalytic domains within this group is not high, even among the six enzymes (Cel9G, Cel9H, Cel9J, Cel9P, Cel9Q, and Cel9T) sharing the same organization (GH9-CBM3c-dockerin), because their catalytic domains only exhibit 30–44% sequence identity (45–61% homology).
FIGURE 1.
Schematic representation of the recombinant proteins used in this study. The GH and CBM families are indicated. Underlined names correspond to the enzymes produced, purified, and characterized in this study. Cel48Ft designates Cel48F from C. cellulolyticum bearing a C. thermocellum dockerin. Cel9Gf designates Cel9G from C. cellulolyticum appended with a R. flavefaciens dockerin. Cel9Wc refers to an engineered form of Cel9W appended with a C. cellulolyticum dockerin.
FIGURE 2.
Phylogenic analysis of the catalytic domains of the GH9 enzymes from C. cellulolyticum. The phylogenetic tree was generated using neighbor joining analyses based on COBALT alignment of the GH9 catalytic domain sequences. Proteins in bold correspond to enzymes that were formerly characterized. The modular organization of each enzyme is indicated as follows: GH9, glycoside hydrolase family 9 catalytic domain; doc, dockerin; Ig, immunoglobulin like module; 3c, 3b, 4, and 30, family 3c, 3b, 4, and 30 carbohydrate binding module, respectively. Scale bar corresponds to 0.6% amino acid substitution.
The second group includes five GH9 enzymes that all display one or several ancillary modules at the N terminus, like the endoprocessive cellulase Cel9E that contains a family-4 CBM and an Ig-like domain that precede the catalytic domain. As for Cel9G, deletion of the CBM of Cel9E was found to abolish the activity (9). A second enzyme, Cel9V, displays the same organization as Cel9E (also known as Theme D (29)) and shares 78% sequence identity (95% homology) throughout the entire primary sequence. In contrast, Cel9U and Cel9W only host an Ig-like module at the N terminus (Theme C (29)), and as mentioned above, the Cel9W lacks a dockerin module. Finally Cel9X displays an organization that resembles that of Cel9E, except that the CBM4 is replaced by a CBM30. Such organization, which is found in some GH9 enzymes produced by other cellulolytic bacteria (5), is proposed to be termed Theme E. Aside from the enzyme pair Cel9E and Cel9V, the overall similarity among the GH9 catalytic domains of group 2 enzymes is, as for group 1 enzymes, relatively low.
Characterization of New GH9 Enzymes in the Free State
The specific activity of each enzyme on CMC, PASC, Avicel, barley glucan, xylan, and pNP-cellobioside was determined together with that of already characterized Cel9G and Cel9E, which were used as examples of C. cellulolyticum GH9 enzymes belonging to groups 1 and 2, respectively. The data (Table 1) indicate large variations in terms of specific activities and substrate specificities. Two enzymes, Cel9V and Cel9X, were found completely inactive on all substrates mentioned above. In contrast, Cel9U and Cel9W (as well as the engineered form Cel9Wc displaying a C-terminal dockerin) exhibit the broadest substrate specificity because they were found to degrade all tested substrates. Besides, Cel9U was identified as the most efficient cellulase of C. cellulolyticum characterized to date on CMC, PASC, barley glucan, and pNP-cellobioside. The activity of the other GH9 enzymes is more restricted to cellulose and the closely related polysaccharide barley glucan. The specific activities of the enzymes active on CMC vary considerably from 17 to 6.6 103 IU/μmol, even among the six enzymes displaying the same organization (Cel9G, -H, -J, -P, -Q, and -T). Indeed, such discrepancy is also reflected by the Km and kcat values determined on CMC (Table 2). Variations in the same range (162-fold, Table 1) were also observed between the most and least active cellulases on barley glucan. On PASC, the differences in specific activities are less drastic but still remain very significant because Cel9U is 16-fold more efficient than Cel9G, the least active enzyme on this substrate. On the most recalcitrant cellulose Avicel, however, the least active enzyme Cel9W is “only” 4.5 times less active than Cel9E, which remains the most efficient cellulase of C. cellulolyticum on the crystalline substrate. The data reported in Table 1 also indicate that grafting a C. cellulolyticum dockerin at the C terminus of Cel9W to generate Cel9Wc had a moderate impact on the activity on most tested substrates. Such differences are probably due to minor conformational changes of the catalytic domain induced by the introduction of the dockerin module. It should be noted that the removal of the dockerin in the case of some cellulosomal cellulases was formerly reported to modify the specific activity on cellulosic substrates, although the dockerin module does not participate in catalysis (30, 31).
TABLE 1.
Specific activities of C. cellulolyticum GH9 enzymes on a range of plant cell wall polysaccharides
| GH9 Enzyme | CMCa | PASC | Avicel | Barley glucan | Xylan | Xyloglucan | pNP-cellobioside |
|---|---|---|---|---|---|---|---|
| Cel9G | 711 ± 97b | 47 ± 1.3 | 54.7 ± 9.1 | 417 ± 87 | 0 | 0 | 0 |
| Cel9H | 1,777 ± 199 | 61 ± 1 | 68.2 ± 4.6 | 636 ± 95 | 0 | 0 | 0 |
| Cel9J | 2,365 ± 100 | 124 ± 6.3 | 48.3 ± 5.2 | 96 ± 2.0 | 0 | 0 | 0 |
| Cel9P | 2,131 ± 221 | 285 ± 10 | 64.5 ± 3.8 | 77 ± 9.0 | 0 | 0 | 0 |
| Cel9Q | 1,927 ± 171 | 48 ± 1 | 28.5 ± 5.0 | 46 ± 1.0 | 0 | 0 | 0 |
| Cel9T | 17 ± 8 | 102 ± 4.7 | 51.1 ± 4.1 | 91 ± 1.2 | 0 | 0 | 0 |
| Cel9R | 103 ± 28 | 99 ± 1.9 | 31.5 ± 1.0 | 34 ± 1.7 | 0 | 0 | 0 |
| Cel9E | 33 ± 3 | 106 ± 7.6 | 72.9 ± 1.9 | 6.6 ± 1.0 | 2.3 | 0 | 1.9 ± 0.1 |
| Cel9U | 6,666 ± 310 | 736 ± 6.5 | 44.9 ± 0.4 | 1 069 ± 243 | 10.9 ± 3.1 | 931 ± 39 | 17.4 ± 0.7 |
| Cel9V | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cel9W | 3,079 ± 717 | 49 ± 1 | 16.5 ± 0.4 | 438 ± 62 | 9.7 ± 0.4 | 40 ± 1.3 | 4.1 ± 0.3 |
| Cel9Wc | 1,942 ± 228 | 87 ± 3 | 24.2 ± 1.0 | 253 ± 37 | 10.4 ± 0.1 | 74 ± 12.5 | 4.4 ± 1.1 |
| Cel9X | 0 | 0 | 0 | 0 | 0 | 1 250 ± 85 | 0 |
a Assays for determining specific activities were performed in triplicate at 37 °C with 0.35% (w/v) of substrate except for CMC and pNP-cellobioside, where 1 and 0.1% were used, respectively.
b Values are given in micromoles of product released per μmol of enzyme min−1, except for Avicel where the reported values are in micromoles of released products after 24 h of incubation with 0.1 μmol of enzyme.
TABLE 2.
Kinetic parameters of GH9 cellulases on CMC
| Enzyme | kcat | Km |
|---|---|---|
| min−1 | g/liter | |
| Cel9G | 776 ± 13 | 0.8 ± 0.04 |
| Cel9H | 1810 ± 47 | 1.2 ± 0.2 |
| Cel9J | 6012 ± 137 | 15.4 ± 0.6 |
| Cel9P | 2276 ± 59 | 1 ± 0.07 |
| Cel9Q | 3091 ± 350 | 5.3 ± 0.7 |
| Cel9T | NDa | ND |
| Cel9R | 142 ± 1.05 | 0.6 ± 0.02 |
| Cel9E | 28 ± 0.1 | 2.5 ± 0.02 |
| Cel9U | 7495 ± 953 | 2 ± 0.6 |
| Cel9V | ND | ND |
| Cel9W | 3759 ± 656 | 8.3 ± 1.67 |
| Cel9Wc | 2412 ± 10 | 5 ± 0.46 |
| Cel9X | ND | ND |
a ND means not determined due to insufficient activity or lack of detectable activity.
The influence of pH (ranging from 4 to 11) on the hydrolytic activity on CMC (Cel9H, -J, -Q, -U, -W, and -Wc) or PASC (Cel9P, -R, and -T) indicated that the newly purified GH9 enzymes have similar pH optimum at 6.0 ± 0.5 (data not shown), in the same range as those formerly determined for other C. cellulolyticum cellulases (8, 9, 18, 22, 30, 31).
The soluble sugars released from amorphous and crystalline celluloses by GH9 cellulases were also analyzed by HPAEC-PAD (Fig. 3, A and B). After 30 min of incubation with PASC, the GH9 enzymes classified in group 1 released a complex mixture of cellodextrins ranging from glucose, cellobiose, cellotriose, and cellotetraose to cellopentaose, but three distinct types of patterns were observed. The first type corresponds to Cel9G and Cel9H, which produce primarily cellotetraose, very little cellopentaose, and identical quantities of the other cellodextrins. The second type of released cellodextrins pattern is produced by Cel9J and Cel9P and is characterized by a high proportion of cellobiose (46%). Finally, Cel9Q, -T, and -R exhibit a similar pattern, with both cellobiose and cellotetraose being preferentially released. For GH9 enzymes active on PASC and classified in group 2 (Cel9E, -U, -W, and -Wc), the produced cellodextrins are shorter. No cellopentaose was detected, and cellotetraose is only present in trace amounts, whereas cellobiose is the main product. The proportions of released cellodextrins by these enzymes, however, are different from that of Cel9E, which produces almost exclusively cellobiose (94%). Cel9W and Cel9Wc released identical proportions of cellodextrins, thereby indicating that the introduction of the dockerin affected only quantitatively the activity on PASC.
FIGURE 3.
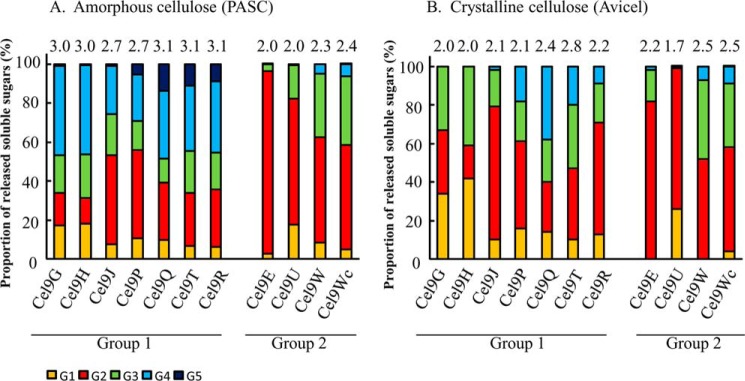
Analysis of the cellodextrins released by the various GH9 cellulases on insoluble celluloses. Soluble sugars generated by 0.1 μm of enzyme after 30 min of incubation on amorphous cellulose (A) or 24 h on crystalline cellulose (B) were identified and quantified by HPAEC-PAD. Numbers on top of the bars designate the average degree of polymerization of the released cellodextrins.
The soluble oligosaccharides released from crystalline cellulose after 24 h of incubation by GH9 cellulases of group 1 were shorter than those produced on PASC by the same enzyme. Thus, the proportion of released glucose increased, whereas that of cellotetraose was reduced, and no cellopentaose was detected. In contrast, similar cellodextrins were released by the GH9 enzymes classified in group 2 on both substrates. The reduction of the average degree of polymerization of the released cellodextrins from Avicel compared with PASC is probably due to lower amounts of reactive sites in the crystalline substrate for group 1 enzymes and to further degradation of the long cellodextrins initially released from Avicel. Despite this general trend, the released cellodextrin patterns were found to be more diverse among the various tested GH9 than in the case of PASC hydrolysis. For instance, the proportions of the cellodextrins released by Cel9Q, -T, and -R diverge significantly, whereas they were highly similar on PASC.
Activity of GH9 Enzymes on Cellodextrins
The activity of newly purified GH9 enzymes together with the previously characterized cellulases Cel9G and Cel9E was tested on cellodextrins ranging from cellobiose to cellohexaose using HPAEC-PAD (Table 3). None of the GH9 enzymes was found to be active on cellobiose. Furthermore, no activity was detected for Cel9V and Cel9X on any cellodextrin, whereas cellotriose was not hydrolyzed by Cel9P, -Q, and -T. As observed for insoluble celluloses, the specific activities varied tremendously among GH9 enzymes active on cellodextrins, but Cel9U was found much more active on all oligosaccharides than any other tested cellulase. In particular cellotriose is degraded 23 times faster by Cel9U than the second most active enzyme on the trisaccharide, Cel9W. In contrast, the least active enzymes on cellodextrins were Cel9Q and Cel9R. Except in the case of Cel9E, the specific activity of all GH9 cellulases increased with the degree of polymerization of the oligosaccharide, suggesting that the active site of these enzymes can accommodate at least 6 glucosyl residues. This hypothesis is supported by the fact that these enzymes exhibit two or more degradation patterns for cellodextrins larger than cellotriose. The main degradation patterns of cellotetraose and cellopentaose were also found to differ, even among GH9 enzymes sharing the same organization (Cel9G, -H, -J, -P, -Q, and -T) suggesting that the preferred positioning of these cellodextrins in the active site differs with respect to the catalytic residues.
TABLE 3.
Degradation patterns and specific activities of the various GH9 enzymes on cellotriose (G3), cellotetratose (G4), cellopentaose (G5), and cellohexaose (G6)
| Substrate | Cel9G | Released products and specific activitiesa |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cel9H | Cel9J | Cel9P | Cel9Q | Cel9T | Cel9R | Cel9E | Cel9U | Cel9V | Cel9W | Cel9Wc | Cel9X | ||
| G3b | G2 + G1c | G2 + G1 | G2 + G1 | G2 + G1 | G2 + G1 | G2 + G1 | G2 + G1 | G2 + G1 | |||||
| 2 | 4 | 14 | 0 | 0 | 0 | 8 | 1.4 | 664 | 0 | 28 | 13 | 0 | |
| G4 | G3 + G1d | G3 + G1 | G2 + G2 | G2 + G2 | G2 + G2 | G2 + G2 | G2 + G2 | G2 + G2 | G2 + G2 | G2 + G2 | G2 + G2 | ||
| G2 + G2 | G2 + G2 | G3 + G1 | G3 + G1 | G3 + G1 | G3 + G1 | G3 + G1 | G3 + G1 | G3 + G1 | G3 + G1 | ||||
| 27 | 17 | 108 | 59 | 17 | 63 | 64 | 84 | 3 652 | 0 | 206 | 180 | 0 | |
| G5 | G4 + G1 | G4 + G1 | G3 + G2 | G3 + G2 | G4 + G1 | G3 + G2 | G3 + G2 | G3 + G2 | G3 + G2 | G3 + G2 | G3 + G2 | ||
| G3 + G2 | G3 + G2 | G4 + G1 | G4 + G1 | G3 + G2 | G4 + G1 | G4 + G1 | G4 + G1 | G4 + G1 | G4 + G1 | ||||
| 91 | 171 | 579 | 109 | 14 | 151 | 51 | 75 | 8 658 | 0 | 1 069 | 916 | 0 | |
| G6 | G4 + G2 | G4 + G2 | G4 + G2 | G4 + G2 | G4 + G2 | G4 + G2 | G4 + G2 | G4 + G2 | G4 + G2 | G4 + G2 | G4 + G2 | ||
| G3 + G3 | G3 + G3 | G3 + G3 | G3 + G3 | G3 + G3 | G3 + G3 | G3 + G3 | G3 + G3 | G3 + G3 | G3 + G3 | G3 + G3 | |||
| G5 + G1 | G5 + G1 | G5 + G1 | G5 + G1 | G5 + G1 | G5 + G1 | G5 + G1 | G5 + G1 | G5 + G1 | G5 + G1 | ||||
| 1 170 | 729 | 876 | 1 231 | 284 | 607 | 260 | 52 | 11 546 | 0 | 2 796 | 3 455 | 0 | |
a Values are in micromoles of consumed substrate per μmol of enzyme min−1.
b Assays were performed at 37 °C with 1 mm substrate. The identification and quantification of the cellodextrins were performed by HPAEC-PAD.
c G1 and G2 designate glucose and cellobiose, respectively.
d Underlined oligosaccharides correspond to the major degradation pattern when two or three degradation patterns are observed.
Mode of Action
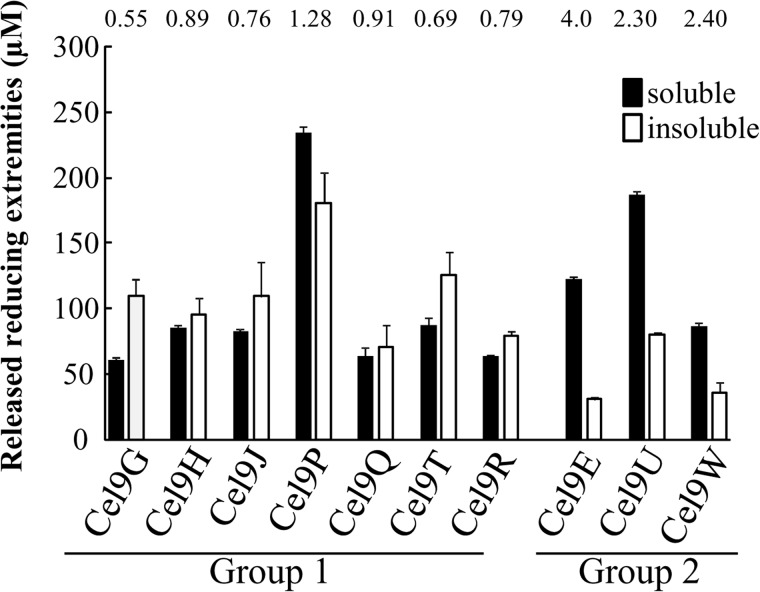
The activity profile of the various GH9 active on cellulosic substrates was explored by quantifying the amounts of soluble and insoluble reducing extremities generated on PASC (26). The endoglucanase Cel9G, which produces slightly more insoluble reducing extremities, and the endoprocessive cellulase Cel9E, which generates four times more soluble reducing extremities, were used as references (Fig. 4). Most of the newly purified GH9 enzymes display an endoglucanase profile, with similar proportions of soluble and insoluble reducing extremities. In contrast, Cel9U and Cel9W were found to be rather processive cellulases, but their processive mode of action is less pronounced than in the case of Cel9E.
FIGURE 4.
Mode of action of the various GH9 cellulases on amorphous cellulose. The amount of soluble and insoluble reducing extremities generated by each enzyme on 3.5 g/liter amorphous cellulose is reported. Numbers on top of the bars indicate the soluble-to-insoluble ratios. The data show the mean and standard deviation of three independent experiments.
Cases of Cel9X and Cel9V
Although Cel9X and Cel9V belong to a GH family whose characterized members were almost all described as cellulases, none of them displayed any activity on the oligo- and polysaccharides mentioned above. Their activity was therefore assayed on other sugars, some of them being more distantly related to cellulose. Cel9X and Cel9V did not exhibit any activity on laminarin, arabinan, pNP β-d-glucoside, pNP α-l-arabinoside, pNP β-d-galactoside, pNP β-d-xyloside, and pNP β-d-lactoside. When assayed on straw, both Cel9X and Cel9V failed to release any detectable soluble sugar. Nevertheless, Cel9X was found to be highly active on xyloglucan (Table 1), and determination of the kcat and Km provided values of 6 560 min−1 and 8.4 g/liter, respectively, and its optimum pH was found to be 6.0. As regards its mode of action on xyloglucan, the change in relative fluidity versus the production of reducing sugars was monitored and indicates an endo mode of action (data not shown). The endo-xyloglucanase activity was further confirmed by HPAEC-PAD analysis because Cel9X essentially releases a range of xyloglucan oligosaccharides (XGO) with glucan backbones greater than four at the beginning of the kinetics (Fig. 5). Thus, Cel9X is a GH9 enzyme whose activity seems to be restricted to xyloglucan. Xyloglucanase activity (Table 1) was also observed for Cel9U with kcat and Km values of 2 302 min−1 and 7.5 g/liter, respectively. Cel9U also displays an endo mode of action on this substrate. Nevertheless, careful examination of the data obtained by HPAEC-PAD indicates that in contrast to Cel9X, which does not seem to be selective, Cel9U preferentially releases XGOs from regions of the substrate where the side chains are only composed of a single α-xylosyl residue (Fig. 5). Among the other GH9 enzymes, only Cel9W and Cel9Wc were found to degrade the highly decorated polysaccharide but with reduced specific activities compared with that of Cel9X and Cel9U (Table 1).
FIGURE 5.

Xyloglucan degradation patterns by Cel9U and Cel9X. The samples were analyzed by HPAEC-PAD. Black line, xyloglucan at 3.5 g/liter incubated for 2 h at 37 °C; red line, xyloglucan (3.5 g/liter) incubated for 2 h at 37 °C with 10 nm of Cel9U; blue line, xyloglucan (3.5 g/liter) incubated for 30 min at 37 °C with 10 nm of Cel9X. XGOGlc4, XGOGlc8, and XGOGlc24) refer to xyloglucan oligosaccharides displaying 4, 8, and 24 glucosyl residues backbone, respectively. G designates unbranched β(1→4)-linked backbone glucosyl residue. The X unit represents a (Xylα(1→6))Glc β(1→4) moiety. The L unit refers to ([Galβ(1→2))Xylα(1→6)]Glc β(1→4) moiety (27).
The lack of activity of Cel9V on PASC at pH values ranging from 4 to 9 and on all other tested substrates (at pH 6), despite its high sequence identity with the active cellulase Cel9E, could be due to a loss of activity during the purification. The binding to an anion exchanger resin was formerly found to induce a total inactivation of the cellulase Cel8C from C. cellulolyticum.3 To rule out this hypothesis, purification of Cel9V was carried out using a different two-step procedure in which the final anion exchanger chromatography was replaced by a gel filtration step. Nevertheless, the newly purified Cel9V was also completely inactive toward all tested substrates. The two samples of purified Cel9V were subjected to CD spectroscopy. In both cases, the same far-UV CD spectrum was obtained. The spectra were found to be very similar to that of Cel9E (Fig. 6) and characteristic of α-helix-rich proteins, thus indicating that the two purified Cel9V are likely to be folded and to display a similar secondary structure than the active processive endocellulase Cel9E.
FIGURE 6.
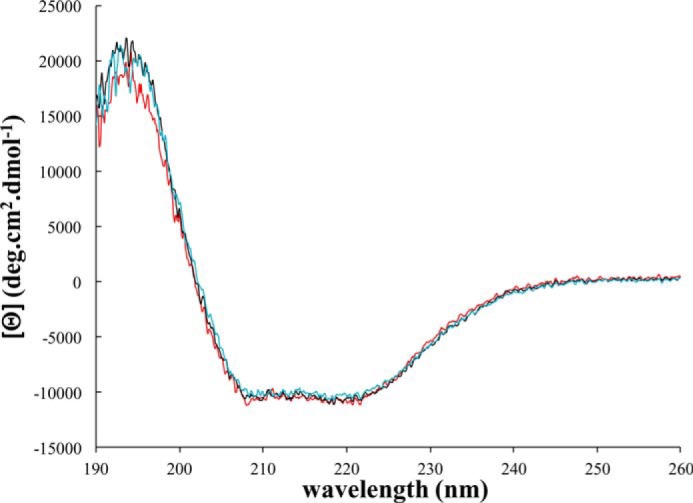
Far-UV CD spectra of Cel9E and Cel9V. Spectra were recorded at 25 °C in 1-mm path length quartz cell. Protein concentration was 1 μm. Red line, Cel9E; black line, Cel9V purified by anion exchange chromatography (final purification step); blue line, Cel9V purified using gel filtration chromatography (final purification step).
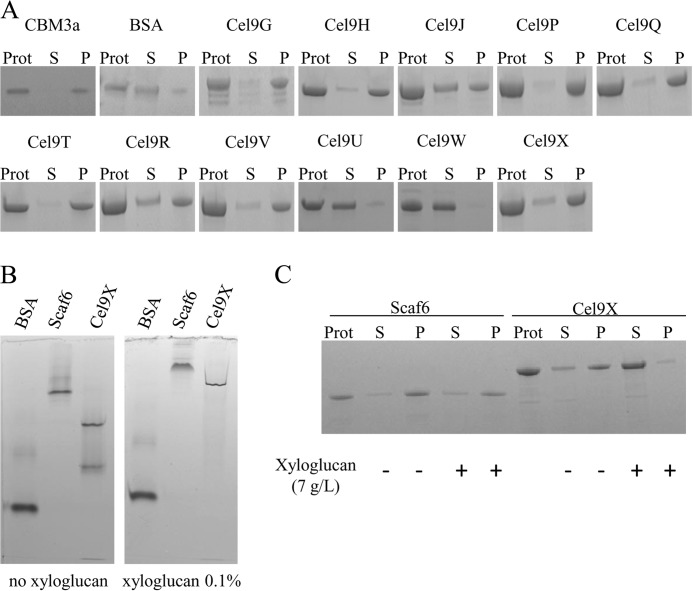
Binding Capacities of GH9 Enzymes
The ability of the newly purified GH9 enzymes to bind to insoluble celluloses was investigated using a simple binding assay and separation by centrifugation of the unbound (supernatant) and cellulose-bound (pellet) proteins prior to SDS-PAGE analysis. When microcrystalline cellulose Avicel was used, none of the new GH9 enzymes was found to significantly bind to the crystalline cellulose, because in all cases the GH9 enzyme was totally recovered in the supernatant (data not shown). In contrast, the CBM-bearing enzymes Cel9H, -J, -P, -Q, -R, -T, -V, and -X were found to bind to amorphous cellulose PASC as a large fraction of the protein is detected in the cellulose-containing pellet (Fig. 7A). Cel9U and Cel9W, however, which do not host a known CBM, displayed no affinity for PASC. Among the cellulose-binding GH9 enzymes, an important proportion of unbound protein is observed for Cel9J and Cel9R, thereby reflecting a weaker affinity for the insoluble cellulose, although the latter contains two CBMs (CBM3c′ and -b′). Interestingly, the inactive Cel9V strongly binds to PASC, thereby suggesting its CBM4 is properly folded and functional. The endo-xyloglucanase Cel9X was also found to significantly bind to amorphous cellulose. The binding capacity of Cel9X to soluble xyloglucan was explored by subjecting the GH9 enzyme with the BSA and a CBM3a-containing scaffoldin to nondenaturing electrophoresis on polyacrylamide gels containing no or 0.1% xyloglucan. As shown in Fig. 7B, the migration of Cel9X is considerably altered in the presence of xyloglucan, whereas that of the scaffoldin Scaf6 is also retarded to some extent, thereby indicating that both proteins display affinity for xyloglucan. Nevertheless, Cel9X exhibits a net preference for xyloglucan because the binding to PASC is abolished in the presence of the decorated polysaccharide (Fig. 7C), whereas the binding of the scaffoldin to amorphous cellulose is maintained in the same experimental conditions.
FIGURE 7.
Binding of GH9 enzymes to cellulosic substrates. The binding to amorphous cellulose (A), xyloglucan (B), and both (C) were investigated. A, proteins (1.5 μm) were mixed with amorphous cellulose (7 g/liter) and incubated for 1 h at 4 °C. The suspension was centrifuged; the pellet (P, bound proteins) was washed twice, and the supernatant fluids (S, unbound protein) were collected, mixed with sample buffer, and subjected to SDS-PAGE, together with an aliquot of the protein solution prior incubation with the substrate (Prot). CBM3a designates the CBM3a of the scaffoldin CipC from C. cellulolyticum produced in E. coli. B, nondenaturing electrophoresis of BSA, Scaf6, and Cel9X on polyacrylamide gel containing no or 0.1% (w/v) xyloglucan. C, binding assays of Scaf6 and Cel9X onto amorphous cellulose in the presence or absence of xyloglucan (7 g/liter). Prot, S, and P same as in Fig. 6A.
Activities of Divalent Cellulosome Chimeras Containing the Novel GH9 Enzymes
Former studies have shown that among the previously characterized cellulases (Cel5A, Cel8C, Cel9E, Cel9G, Cel48F, and Cel9M) from C. cellulolyticum, the enzyme pair Cel48F/Cel9G generated the most efficient cellulosome chimeras on Avicel (19, 21), suggesting this enzyme pair plays a pivotal role in cellulosome efficiency toward pure crystalline cellulose. The superior activity compared with other incorporated enzyme pairs was observed when Cel48F appended with a C. thermocellum dockerin (termed Cel48Ft, Fig. 1), and Cel9G bearing its native C. cellulolyticum dockerin was bound onto hybrid scaffoldins displaying the cognate cohesins and a CBM3a (like Scaf2, Fig. 1), as well as Scaf4, which lacks a CBM (Fig. 1) (19). Indeed, the Scaf4-based complex containing these two cellulases was less efficient than the corresponding Scaf2-based complex, because both enzyme proximity and substrate-targeting effects occurred within the latter complex, whereas complexation onto Scaf4 only induced the proximity effect.
In this study, the activities of the newly purified family-9 glycoside hydrolases appended with their native dockerin from C. cellulolyticum and the engineered enzyme Cel9Wc were assayed on crystalline cellulose in combination with Cel48Ft (Fig. 1). Each enzyme pair was bound onto the corresponding free cohesins, Scaf4 or Scaf2. Cel48Ft was also combined with the previously characterized Cel9G and Cel9E. The xyloglucanase Cel9X, which exhibits no activity on crystalline cellulose, was omitted in these experiments. The released soluble sugars after various incubation times with Avicel were analyzed by HPAEC-PAD, and the total amount of released sugars obtained at the end of the kinetics (24 h) is reported in Table 4. The best Scaf4- and Scaf2-based cellulosome chimeras are those containing Cel48Ft and Cel9G, because none of the new GH9 enzymes combined with Cel48Ft lead to chimeras with higher activity. This phenomenon is more obvious in the case of Scaf4-based complexes because the chimera containing Cel9G proved to be 30% more efficient than the second most active complex that contains Cel9H. This is also reflected by the “stimulation factor,” which we defined as the ratio between the activity of the Scaf4-based complex over the activity of the free cohesin system for a given enzyme pair. In the case of Cel9G and Cel48Ft, the binding onto Scaf4 induced a 2.3-fold improvement of the Avicelase activity, thereby indicating a strong proximity effect, whereas the enzyme pairs containing another GH9 enzyme exhibit an activity enhancement on Scaf4 that varies from 1 to 1.4. No stimulation on Scaf4 was observed either for Cel9R or the engineered form of the noncellulosomal enzyme Cel9W displaying a dockerin. Concerning the Scaf2-based complexes, the most active chimera also contains Cel9G, followed by the Scaf2-based complex hosting Cel9U. This result indicates that a strong substrate targeting effect compensates the weak proximity effect for the enzyme pair Cel48Ft + Cel9U. Other enzyme pairs such as Cel48Ft/Cel9J and Cel48Ft/Cel9Wc are also strongly stimulated when bound onto Scaf2 compared with Scaf4. Thus, in contrast to Cel9G, when the newly purified GH9 cellulases are combined with Cel48Ft in cellulosome chimeras, the substrate targeting effect induced by the CBM3a of the scaffoldin seems predominant over the proximity effect. Clearly, Cel9V remains inactive when combined with Cel48Ft because the enzyme pair displayed the same activity as the corresponding control (Cel48Ft alone) in all configurations (free cohesins and Scaf4- and Scaf2-based complexes).
TABLE 4.
Activity on crystalline cellulose Avicel of the various GH9 enzymes combined with Cel48Ft in divalent chimeras
| Enzyme paira | Cohesins and hybrid scaffoldins |
||||
|---|---|---|---|---|---|
| Free cohesinsb | Scaf4 | SFScaf4c | Scaf2 | SFScaf2d | |
| Cel48Ft+none | 45 ± 2e,f | 48.5 ± 3.4 | 1.1 | 57.1 ± 1 | 1.3 |
| Cel48Ft+Cel9G | 86 ± 5.8 | 200 ± 2.6 | 2.3 | 323 ± 13.6 | 3.8 |
| Cel48Ft+Cel9H | 116 ± 11.4 | 155 ± 1 | 1.3 | 245 ± 2.1 | 2.1 |
| Cel48Ft+Cel9J | 71 ± 1.5 | 87 ± 1.4 | 1.2 | 168 ± 5.4 | 2.4 |
| Cel48Ft+Cel9P | 76 ± 3.2 | 107 ± 4.7 | 1.4 | 179 ± 1.6 | 2.4 |
| Cel48Ft+Cel9Q | 93 ± 1.4 | 107 ± 6.1 | 1.1 | 172 ± 3.1 | 1.8 |
| Cel48Ft+Cel9T | 92 ± 4.6 | 126 ± 7.4 | 1.4 | 165 ± 4.0 | 1.8 |
| Cel48Ft+Cel9R | 82 ± 1.0 | 85.8 ± 3.6 | 1.0 | 155 ± 3.8 | 1.9 |
| Cel48Ft+Cel9E | 87 ± 1.3 | 120 ± 2.9 | 1.4 | 126 ± 1.9 | 1.5 |
| Cel48Ft+Cel9U | 130 ± 3.6 | 140 ± 1 | 1.1 | 256 ± 9.7 | 2.0 |
| Cel48Ft+Cel9V | 45.7 ± 1.2 | 49 ± 5.6 | 1.0 | 60.1 ± 2.2 | 1.3 |
| Cel48Ft+Cel9Wc | 93 ± 5.7 | 91 ± 2.7 | 1.0 | 172 ± 12.7 | 1.8 |
a Enzymes, free cohesins, and complexes were at 0.1 μm.
b Cel48Ft was bound onto free cohesin 2 from CipA of C. thermocellum, and the GH9 enzyme was bound onto free cohesin 1 of CipC from C. cellulolyticum.
c SFScaf4 is the impact of complexation on Scaf4: SFScaf4 = (released soluble sugars by Scaf4-based complex)/(released soluble sugars by the corresponding enzyme pair bound onto free cohesins).
d SFScaf2 is the impact of complexation on Scaf2: SFScaf2 = (released soluble sugars by Scaf2-based complex)/(released soluble sugars by the corresponding enzyme pair bound onto free cohesins).
e Released soluble sugars in μm were measured by HPAEC-PAD after 24 h of incubation at 37 °C with 3.5 g/liter Avicel.
f Average (and standard deviation) of three experiments is shown.
Activities of Trivalent Cellulosome Chimeras Containing the Novel GH9 Enzymes
The GH9 cellulases isolated in this study were also incorporated in Scaf6-based chimeras (Fig. 1). This hybrid scaffoldin displays three divergent cohesins from C. cellulolyticum, C. thermocellum, and R. flavefaciens and one CBM3a and can therefore incorporate three distinct enzymes appended with the cognate dockerins (21). The GH9 enzymes bearing their native C. cellulolyticum dockerins were bound to Scaf6 together with Cel48Ft and Cel9Gf that host dockerin modules from C. thermocellum and R. flavefaciens, respectively. The resulting complexes were assayed on Avicel, and their activity was compared with the calculated sum of activities displayed by the complex Scaf6-(Cel48Ft/Cel9Gf) and by the corresponding GH9 bound onto a cohesin from C. cellulolyticum. Trivalent chimeras with either Cel9G or Cel9E bound onto the C. cellulolyticum cohesin of Scaf6 were also tested. Except in the case of Cel9V, after 24 h of incubation all trivalent cellulosome chimeras released significantly more soluble sugars than the divalent complex Scaf6-(Cel48Ft/Cel9Gf) (Table 5). Apart from the complex containing Cel9E, this activity appears to be superior to the calculated sum of soluble sugars released by the divalent cellulosome chimera and by the corresponding GH9 bound onto the free cohesin. The complex containing Cel9U, however, is clearly the most active, although in the free state this cellulase displays a moderate activity on crystalline cellulose compared with other GH9s (Table 1). Furthermore, the activity of the trivalent chimera that contains Cel9U is 1.5-fold higher than the calculated sum of activities determined for the complexes Scaf6-(Cel48Ft/Cel9Gf) and Cel9U-cohesin (stimulation factor (SF) reported in Table 5). Quite unexpectedly, incorporation of the engineered enzyme Cel9Wc led to the second highest stimulation factor, thereby indicating it can significantly contribute to the overall activity of the minicellulosome, whereas the trivalent complex displaying the major cellulosomal enzyme Cel9E is one of the least.
TABLE 5.
Activity on crystalline cellulose Avicel of trivalent chimeras containing the various GH9 enzymes combined with Cel48Ft and Cel9Gf
| Incorporated enzymes | Scaf6 | SFa |
|---|---|---|
| Cel48Ft+Cel9Gf | 310 ± 6.4b,c | |
| Cel48Ft+Cel9Gf+Cel9G | 459 ± 5.5 | 1.26 |
| Cel48Ft+Cel9Gf+Cel9H | 443 ± 14 | 1.20 |
| Cel48Ft+Cel9Gf+Cel9J | 419 ± 2.7 | 1.17 |
| Cel48Ft+Cel9Gf+Cel9P | 429 ± 9.7 | 1.15 |
| Cel48Ft+Cel9Gf+Cel9Q | 395 ± 8.7 | 1.17 |
| Cel48Ft+Cel9Gf+Cel9T | 402 ± 7.8 | 1.10 |
| Cel48Ft+Cel9Gf+Cel9R | 380 ± 5.2 | 1.11 |
| Cel48Ft+Cel9Gf+Cel9E | 362 ± 7.1 | 0.95 |
| Cel48Ft+Cel9Gf+Cel9U | 528 ± 5.6 | 1.49 |
| Cel48Ft+Cel9Gf+Cel9V | 308 ± 4.1 | 1 |
| Cel48Ft+Cel9Gf+Cel9Wc | 447 ± 9.1 | 1.34 |
a Soluble sugars (in μm) released by 0.1 μm of complex measured by HPAEC-PAD after 24 h of incubation at 37 °C with 3.5 g/liter Avicel.
b Average and standard deviation of two independent experiments are shown.
c SF = (released soluble sugars by trivalent chimeras)/(sum of released soluble sugars by Scaf6(Cel48Ft/Cel9Gf) + released soluble sugars by corresponding GH9 enzyme bound onto free cohesin).
Avicelase Activity of a Heterogeneous Mixture of Trivalent Scaf6-based Chimeras
To evaluate the role of the functional diversity of GH9 cellulases as cellulosome components, the divalent complex Scaf6-(Cel48Ft/Cel9Gf) was mixed with the 11 C. cellulolyticum dockerin-bearing GH9 enzymes active on crystalline cellulose (Cel9E, -G, -H, -J, -M, -P, -Q, -T, -R, -U and W-c). Because of their lack of activity on cellulose, the enzymes Cel9V and Cel9X were omitted in this experiment. The concentration of each GH9 cellulase was 1/11 that of the divalent complex Scaf6-(Cel48Ft/Cel9Gf) so that all the GH9s bearing a C. cellulolyticum dockerin could be bound onto Scaf6. The resulting sample contained 11 different trivalent Scaf6-based complexes systematically composed of Cel48Ft and Cel9Gf but displaying enzyme heterogeneity at the C. cellulolyticum cohesin module. The resulting mixture was assayed on crystalline cellulose Avicel. As shown in Fig. 8, the degradation of the crystalline cellulose by the heterogeneous Scaf6-based complexes was considerably more important than in the case of the divalent complex. The soluble sugars released after 24 h of incubation by the heterogeneous complexes were 1.85-fold that generated by the most efficient homogeneous trivalent Scaf6-based complex (containing Cel9U, Cel48Ft, and Cel9Gf). Furthermore, the activity of the heterogeneous mixture of trivalent complexes was found 2.3-fold higher than the calculated mean of all homogeneous chimeras (419 ± 52 μm after 24 h of incubation). The comparison with the mixture of free cellulases (supplemented with free Cel48Ft and Cel9Gf) also indicates that the complexation of all catalytic subunits onto Scaf6 induced a 4.5-fold improvement of the overall activity.
FIGURE 8.
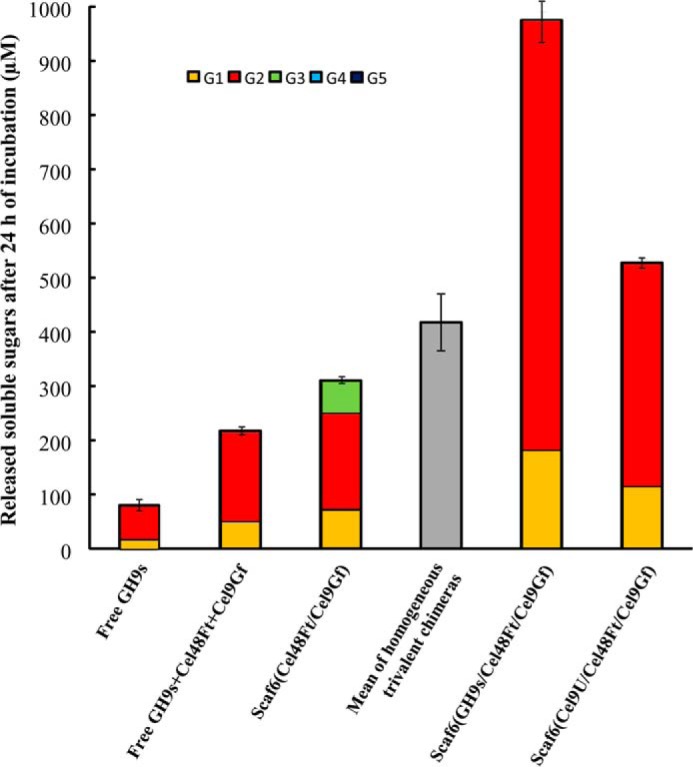
Degradation of crystalline cellulose by a mixture of 11 Scaf6-based trivalent chimeras containing Cel48Ft, Cel9Gf, and one GH9 cellulase bearing a C. cellulolyticum dockerin. The amount of released soluble cellodextrins and their proportion after 24 h of incubation at 37 °C with 3.5 g/liter Avicel are shown. Free GH9s refers to a mixture of free Cel9E + Cel9G + Cel9H + Cel9J + Cel9M + Cel9P + Cel9Q + Cel9R + Cel9T + Cel9U + Cel9Wc, each enzyme being at a final concentration of 0.0091 μm. Free GH9s + Cel48Ft + Cel9Gf refers to free GH9s (each enzyme at 0.0091 μm) supplemented with free Cel48Ft and Cel9Gf at 0.1 μm. Scaf6(Cel48Ft/Cel9Gf) refers to the Scaf6-based divalent complex at 0.1 μm with no enzyme bound onto the C. cellulolyticum cohesin of the scaffoldin. Mean of homogeneous trivalent chimeras corresponds to the average activity on Avicel of all homogenous trivalent chimeras at 0.1 μm containing Cel48Ft, Cel9Gf, and one GH9 cellulase, which was calculated from the data reported in Table 5. Scaf6(GH9s/Cel48Ft/Cel9Gf) designates a mixture of the Scaf6-based complexes that systematically contain Cel48Ft and Cel9Gf and either Cel9E, Cel9G, Cel9H, Cel9J, Cel9M, Cel9P, Cel9Q, Cel9R, Cel9T, Cel9U, or Cel9Wc bound onto the C. cellulolyticum cohesin of the scaffoldin. Each distinct trivalent complex in the mixture was at a concentration of 0.0091 μm. Scaf6(Cel9U/Cel48Ft/Cel9Gf) refers to the best homogeneous trivalent chimera (see Table 5), which was at a concentration of 0.1 μm. The data show the mean of at least three independent experiments, and bars indicate the standard deviation.
DISCUSSION
The profusion of GH9 enzyme-encoding genes in cellulosome-producing microorganisms suggests a pivotal role for this family of enzymes during plant cell wall degradation by the cellulolytic complexes. Moreover, former studies using cellulosome chimeras have also demonstrated that the synergy between GH48 and GH9 cellulases within the complex plays a critical role during cellulose hydrolysis (19, 21). In C. cellulolyticum, only three GH9s (Cel9E, Cel9G, and Cel9M) were characterized to date, but cellulosomes were shown to contain 12 different GH9 enzymes, and the bacterium is likely to secrete an additional GH9 that lacks a dockerin (16, 17). Among the GH9s synthesized by the mesophilic Clostridium, a large variety of domain organizations (or Themes) is found. Some organizations are displayed by only one GH9 enzyme, whereas one specific organization, previously named Theme B1 (29) (GH9-CBM3c-dockerin), is common to six different enzymes. The diversity but also paradoxically the redundancy of the GH9 raises several questions with respect to this category of enzymes, which is the most prominent in bacterial cellulosomes. Are they all authentic cellulases as suggested by their classification in a GH family almost exclusively composed of cellulose hydrolyzing enzymes? Do they share similar or distinct activity profiles, especially in the case of the six GH9 displaying the same organization? Do they equally contribute to the activity of the cellulosomes? Do they display the same functional relationships with the major endoprocessive cellulase Cel48F in the cellulosomes?
These open questions prompted us to overproduce, purify, and characterize in the free and complexed states the 10 C. cellulolyticum GH9 enzymes whose activity patterns had not yet been explored, thereby leading to the first global enzymatic picture of the arsenal of GH9 enzymes synthesized by a cellulosome-producing bacterium.
The characterization of all GH9 enzymes not only showed that the novel GH9 cellulases display different activity patterns but identified Cel9X as a genuine endoxyloglucanase. Some other GH9 cellulases were formerly described to display side activity on xyloglucan, and this study has shown that both Cel9U and Cel9W have significant activities on this substrate. Nevertheless, to our knowledge Cel9X is the first GH9 enzyme shown to be an authentic xyloglucanase with no activity on soluble (cellodextrins, CMC) and insoluble (PASC, Avicel) celluloses. Cel9X harbors a CBM30 that can bind to both cellulose and xyloglucan, but it exhibits a preference for xyloglucan. Cel9X had no detectable activity on the hemicellulosic polysaccharides barley glucan, xylan, arabinan, and laminarin, thus suggesting this enzyme is highly specific of xyloglucan. This activity pattern is very unusual for a GH9 enzyme and also differs from typical xyloglucanases belonging to family-44 like CelB from R. flavefaciens FD1 (32) or family 74 like Xgh74 from C. thermocellum (33), which are described to exhibit some activity on barley glucan and/or CMC. Interestingly, Cel9X resembles (53% sequence identity) the N terminus part of CelJ from C. thermocellum made of CBM30-Ig-GH9 (also called Cel9D), which was shown to be an endocellulase (5) having significant activities on barley glucan, lichenan, and xyloglucan (7). This broad substrate specificity of Cel9D moiety is shared by the C-terminal part of CelJ, made of GH44-dockerin-PKD-CBM44 and termed Cel44A, which was found to be active on CMC, xyloglucan, lichenan, glucomannan, and crystalline cellulose (10). The domain organization of Cel44A moiety of CelJ matches the enzyme Cel44O (gene locus Ccel_0429) from C. cellulolyticum (59% sequence identity), suggesting that cel9X and cel44O are phylogenetically connected to celJ. One could hypothesize that the celJ gene was transferred to the mesophilic Clostridium and subsequently split into cel9X and cel44O followed by a restriction of the specificity of Cel9X to xyloglucan. Nevertheless, the unusual dockerin of CelJ, whose first segment displays the typical C. cellulolyticum dyad AV and was shown to bind to cohesins from both C. thermocellum and Clostridium josui (34), a bacterium closely related to C. cellulolyticum, rather suggests a horizontal gene transfer in the opposite way. The combination of both genes would have subsequently occurred in C. thermocellum leading to celJ and followed by a broadening of the substrate specificity of the N-terminal part (Cel9D) of CelJ.
Another unexpected result concerns Cel9V, which was found totally inactive toward all tested cellulosic and hemicellulosic substrates. This observation is surprising because this protein is detected in the cellulosomes of C. cellulolyticum in various culture conditions and shares 78% sequence identity (95% similarity) with the active endoprocessive cellulase Cel9E. The predicted catalytic acid (Asp-377) and base (Glu-788) are identical to those found in Cel9E and other GH9 enzymes exhibiting the same modular organization (Theme D) (35, 36). Furthermore, the CD spectra of Cel9V and Cel9E can be superimposed, thereby indicating that Cel9V has the same α-helix rich secondary structure than the previously characterized cellulase and suggesting Cel9V is correctly folded. In addition, Cel9V was found to bind to amorphous cellulose, which indicates that the N-terminal CBM4 is functional. Thus, the reasons why Cel9V is completely inactive are not yet elucidated. Cel9V may be specific of a noncellulosic substrate that was not assayed in this study. Another possibility could be that this enzyme, conversely to Cel9E, may require specific post-translational modifications that E. coli cannot perform. An alternative hypothesis would be a duplication of the cel9E gene that would have progressively evolved to encode an inactive form of the enzyme. It is worth noticing that the genomes of two closely related cellulolytic bacteria, Clostridium papyrosolvens and Clostridium sp. BNL1100, also contain two genes that putatively encode highly homologous GH9 enzymes displaying the same modular organization (CBM4-Ig-GH9-dockerin) than Cel9E and Cel9V. Their genetic context is also comparable because one these genes, the cel9E-like gene, is located in a large cluster of genes analogous to the cip-cel cluster in C. cellulolyticum (37), whereas the cel9V-like gene occupies a distant locus on the chromosome (16). The characterization of these enzymes from both C. papyrosolvens and Clostridium sp. BNL1100 would indeed help to assess the hypotheses mentioned above.
The other C. cellulolyticum GH9 enzymes characterized in this study are all genuine cellulases but display distinct activity patterns. The enzymes classified in “group 1,” which lack ancillary domains at the N terminus (Themes B1 and B2), are authentic endoglucanases, whose substrate specificity is restricted to soluble and insoluble celluloses and the closely related polysaccharide barley glucan. Nevertheless, on a given substrate their specific activities vary considerably. For instance, the specific activities on CMC range from 17 IU/μmol for Cel9T to 2,363 IU/μmol for Cel9J, although both enzymes share the same modular organization (Theme B1). Similarly, the released cellodextrins from amorphous and crystalline celluloses by the group 1 cellulases, as well as their proportions, noticeably fluctuate from one enzyme to another. The GH9 cellulases classified in group 2 (Cel9E, Cel9U, and Cel9W) that harbor an Ig-like module and an optional CBM4 exhibit a broader substrate specificity because in addition to barley glucan, soluble and insoluble celluloses, they also hydrolyze xylan. Furthermore, Cel9U and Cel9W are active on xyloglucan. The specific activities determined on each substrate as well as the soluble sugars released from insoluble celluloses also vary significantly among the cellulases of this group. Cel9U, however, was found to be the most active GH9 enzyme on almost all tested substrates, except Avicel and xyloglucan that were faster hydrolyzed by Cel9E and Cel9X, respectively.
Although the previously characterized cellulase Cel9G has rather moderate activities in the free state on cellulosic substrates compared with other GH9 enzymes characterized in this study, its complexation with Cel48Ft in divalent Scaf4- and Scaf2-based mini-cellulosomes systematically generated the most active complexes on crystalline cellulose. Consequently, this enzyme pair is characterized by the highest proximity effect (SFscaf4) and the best stimulation by complexation onto Scaf2, which induces both proximity and substrate targeting effects. This result confirms the special role of this highly synergistic enzyme pair hypothesized in previous reports (19, 21). Quite unexpectedly, the other GH9 cellulases displaying the same modular organization as Cel9G were much less synergistic in divalent complexes with Cel48Ft. This observation suggests that these cellulases are not just additional copies of Cel9G and may, as discussed below, play a specific role in the cellulosomes. The cellulase pair Cel9U/Cel48Ft generated the second most active Scaf2-based complex. The latter pair, however, displayed a modest stimulation factor by complexation onto the scaffoldin Scaf4 that lacks a CBM, probably because the corresponding free cohesin system displays an elevated activity on Avicel. Thus, Cel9U and Cel48Ft exhibit an important synergy (1.5-fold) in the free state that is not significantly improved by physical proximity within the cellulosome chimeras.
Interestingly, the use of homogeneous trivalent cellulosome chimeras that systematically contain Cel48Ft and Cel9Gf revealed more complex relationships within the cellulosomes. The highest Avicelase activity was observed for the trivalent complex, including Cel9U, which proved to be 25% more efficient than the trivalent chimera containing the cellulases Cel9G-Cel48Ft-Cel9Gf and formerly reported as the best complex with characterized cellulases from C. cellulolyticum (21). Thus, Cel9U in complex can act synergistically with the enzyme pair Cel48Ft/Cel9Gf, whereas the physical proximity with Cel48Ft alone does not trigger significant additional synergies in divalent complexes. This observation could be explained by the fact that bacterial major scaffoldins are usually composed of five or more cohesins, thereby allowing a complex network of functional relationships among all the bound catalytic subunits. Such network can only be glimpsed using two- or three-enzyme cellulosome chimeras, even on a “simple” substrate such as pure crystalline cellulose.
This high degree of complexity is also suggested by the data obtained with the heterogeneous mixture of trivalent chimeras, containing Cel48Ft, Cel9Gf, and variable GH9-cellulases bound onto the C. cellulolyticum cohesin of the hybrid scaffoldin (Fig. 8). The combination of the 11 complexes displaying limited heterogeneity proved to be 85% more efficient than the best homogeneous trivalent chimera composed of Cel9U, Cel48Ft, and Cel9Gf. This result demonstrates that, as formerly described, complexes displaying even slightly different compositions can cooperate (25, 38). Indeed the native cellulosomes produced by the bacterium were shown to be highly heterogeneous in terms of enzyme composition (38). Taken together, these data suggest that the enzymatic contribution of cellulases such as Cel9J, Cel9Q, Cel9T, or Cel9R, which seems minor in simple and homogeneous chimeras, may significantly increase in more complex and heterogeneous systems.
It should also be noted that the major processive cellulase Cel48F was selected in this study as the preferred partner of the newly isolated GH9 enzymes in divalent complexes. Nevertheless, the cellulosomes also contain copious amounts of the endoprocessive cellulase Cel9E (9, 39, 40), which may have been a more suitable partner within the mini-cellulosome for some of the GH9 enzymes described in this study. Furthermore, we have recently shown that the initial binding of a particular enzyme onto a cohesin of the scaffoldin induces preferences on the occupation of the adjacent cohesins, thereby inducing enzyme discrimination during cellulosome assembly and optimized enzyme combinations (25). Such property could be exploited to identify the best partner for each GH9 enzyme characterized in this study or alternatively to discover which GH9 enzyme is preferentially incorporated in the neighborhood of the major cellulosomal cellulases Cel48F and Cel9E.
To summarize, our data indicate that the vast repertoire of GH9 enzymes, which was selected by C. cellulolyticum, is characterized by a large variety of substrate specificities and activity patterns in both free and complexed states. Our results also strongly suggest that this diversity is most likely required to achieve complete degradation of cellulose by the cellulolytic macrocomplexes. This conclusion can probably be extended to the other cellulosome-producing bacteria that share similar GH9 collections. Nevertheless, this study does not elucidate why this particular family of enzymes was selected by the cellulolytic clostridia to generate such diversity in modular organizations and activities rather than the GH families 5, 8, or 48, which also contain cellulosomal cellulases. The redundancy of these enzymes may be related to the unusual relationships between the GH9 catalytic domains and their surrounding domains in multimodular GH9 enzymes. Many plant cell wall-degrading enzymes contain auxiliary module(s) connected to the catalytic module by a flexible linker. However, extended linkers are not found in GH9 enzymes between the flanking auxiliary domain(s) such as the Ig-like (11, 35, 36, 41) or CBM3c (42, 43) domains and the catalytic domain. These domains are structurally linked to the catalytic domain and may be considered as part of it. As a consequence, deletion of the ancillary contiguous domain usually generates an inactive form of the GH9 enzyme (44), whereas in other families of glycoside hydrolases the catalytic domain is a module that remains generally active even when it is produced individually. The recurrent physical association of the GH9 catalytic domains and their neighboring ancillary domains may thus have been selected to generate a larger array of activity patterns, further diversified by addition of supplementary modules/domains such as the CBM3b or CBM4, which are not specific of GH9 enzymes as they are found in bacterial scaffoldins and glycoside hydrolases classified in other GH families.
Acknowledgments
We are grateful to Marianne Ilbert from the Laboratoire de Bioénergétique et d'Ingénierie des Protéines (CNRS, UMR7281) for help during CD experiments. We are indebted to Stéphanie Perret and Sandrine Pagès from the Laboratoire de Chimie Bactérienne (CNRS, UMR7283) for fruitful discussions.
This work was supported by a fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche (to J. R.).

This article contains supplemental Table S1.
H.-P. Fierobe, unpublished data.
- GH
- glycoside hydrolase
- CBM
- carbohydrate-binding module
- doc
- dockerin module
- Cel48Ft
- Cel48F from Clostridium cellulolyticum bearing a C. thermocellum dockerin
- Cel9Gf
- Cel9G from C. cellulolyticum bearing a Ruminococcus flavefaciens dockerin
- Cel9Wc
- Cel9W from C. cellulolyticum engineered to host a C. cellulolyticum dockerin
- PASC
- phosphoric acid swollen cellulose
- CMC
- carboxymethyl cellulose
- pNP-cellobioside
- p-nitrophenyl β-d-cellobioside
- XGO
- xyloglucan oligosaccharide
- HPAEC-PAD
- high pressure anion exchange chromatography coupled with pulsed amperometric detection
- SF
- stimulation factor.
REFERENCES
- 1. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aspeborg H., Coutinho P. M., Wang Y., Brumer H., 3rd, Henrissat B. (2012) Evolution, substrate specificity and subfamily classification of glycoside hydrolase family-5 (GH5). BMC Evol. Biol. 12, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Henrissat B., Claeyssens M., Tomme P., Lemesle L., Mornon J. P. (1989) Cellulase families revealed by hydrophobic cluster analysis. Gene 81, 83–95 [DOI] [PubMed] [Google Scholar]
- 4. Honda Y., Shimaya N., Ishisaki K., Ebihara M., Taniguchi H. (2011) Elucidation of exo-β-d-glucosaminidase activity of a family 9 glycoside hydrolase (PBPRA0520) from Photobacterium profundum SS9. Glycobiology 21, 503–511 [DOI] [PubMed] [Google Scholar]
- 5. Arai T., Araki R., Tanaka A., Karita S., Kimura T., Sakka K., Ohmiya K. (2003) Characterization of a cellulase containing a family 30 carbohydrate-binding module (CBM) derived from Clostridium thermocellum CelJ: importance of the CBM to cellulose hydrolysis. J. Bacteriol. 185, 504–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eckert K., Zielinski F., Lo Leggio L., Schneider E. (2002) Gene cloning, sequencing, and characterization of a family 9 endoglucanase (CelA) with an unusual pattern of activity from the thermoacidophile Alicyclobacillus acidocaldarius ATCC27009. Appl. Microbiol. Biotechnol. 60, 428–436 [DOI] [PubMed] [Google Scholar]
- 7. Hirano N., Hasegawa H., Nihei S., Haruki M. (2013) Cell-free protein synthesis and substrate specificity of full-length endoglucanase CelJ (Cel9D-Cel44A), the largest multi-enzyme subunit of the Clostridium thermocellum cellulosome. FEMS Microbiol. Lett. 344, 25–30 [DOI] [PubMed] [Google Scholar]
- 8. Gal L., Gaudin C., Belaich A., Pages S., Tardif C., Belaich J. P. (1997) CelG from Clostridium cellulolyticum: a multidomain endoglucanase acting efficiently on crystalline cellulose. J. Bacteriol. 179, 6595–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaudin C., Belaich A., Champ S., Belaich J. P. (2000) CelE, a multidomain cellulase from Clostridium cellulolyticum: a key enzyme in the cellulosome? J. Bacteriol. 182, 1910–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Najmudin S., Guerreiro C. I., Carvalho A. L., Prates J. A., Correia M. A., Alves V. D., Ferreira L. M., Romão M. J., Gilbert H. J., Bolam D. N., Fontes C. M. (2006) Xyloglucan is recognized by carbohydrate-binding modules that interact with β-glucan chains. J. Biol. Chem. 281, 8815–8828 [DOI] [PubMed] [Google Scholar]
- 11. Kataeva I. A., Uversky V. N., Brewer J. M., Schubot F., Rose J. P., Wang B. C., Ljungdahl L. G. (2004) Interactions between immunoglobulin-like and catalytic modules in Clostridium thermocellum cellulosomal cellobiohydrolase CbhA. Protein Eng. Des. Sel. 17, 759–769 [DOI] [PubMed] [Google Scholar]
- 12. Zhang X. Z., Sathitsuksanoh N., Zhang Y. H. (2010) Glycoside hydrolase family 9 processive endoglucanase from Clostridium phytofermentans: heterologous expression, characterization, and synergy with family-48 cellobiohydrolase. Bioresource Technol. 101, 5534–5538 [DOI] [PubMed] [Google Scholar]
- 13. Fontes C. M., Gilbert H. J. (2010) Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu. Rev. Biochem. 79, 655–681 [DOI] [PubMed] [Google Scholar]
- 14. Feinberg L., Foden J., Barrett T., Davenport K. W., Bruce D., Detter C., Tapia R., Han C., Lapidus A., Lucas S., Cheng J. F., Pitluck S., Woyke T., Ivanova N., Mikhailova N., Land M., Hauser L., Argyros D. A., Goodwin L., Hogsett D., Caiazza N. (2011) Complete genome sequence of the cellulolytic thermophile Clostridium thermocellum DSM1313. J. Bacteriol. 193, 2906–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tamaru Y., Miyake H., Kuroda K., Nakanishi A., Matsushima C., Doi R. H., Ueda M. (2011) Comparison of the mesophilic cellulosome-producing Clostridium cellulovorans genome with other cellulosome-related clostridial genomes, Microb. Biotechnol. 4, 64–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blouzard J. C., Coutinho P. M., Fierobe H. P., Henrissat B., Lignon S., Tardif C., Pagès S., de Philip P. (2010) Modulation of cellulosome composition in Clostridium cellulolyticum: adaptation to the polysaccharide environment revealed by proteomic and carbohydrate-active enzyme analyses. Proteomics 10, 541–554 [DOI] [PubMed] [Google Scholar]
- 17. Celik H., Blouzard J. C., Voigt B., Becher D., Trotter V., Fierobe H. P., Tardif C., Pagès S., de Philip P. (2013) A two-component system (XydS/R) controls the expression of genes encoding CBM6-containing proteins in response to straw in Clostridium cellulolyticum. PLoS One 8, e56063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Belaich A., Parsiegla G., Gal L., Villard C., Haser R., Belaich J. P. (2002) Cel9M, a new family 9 cellulase of the Clostridium cellulolyticum cellulosome. J. Bacteriol. 184, 1378–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fierobe H. P., Bayer E. A., Tardif C., Czjzek M., Mechaly A., Bélaïch A., Lamed R., Shoham Y., Bélaïch J. P. (2002) Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J. Biol. Chem. 277, 49621–49630 [DOI] [PubMed] [Google Scholar]
- 20. Fierobe H. P., Mechaly A., Tardif C., Belaich A., Lamed R., Shoham Y., Belaich J. P., Bayer E. A. (2001) Design and production of active cellulosome chimeras. Selective incorporation of dockerin-containing enzymes into defined functional complexes. J. Biol. Chem. 276, 21257–21261 [DOI] [PubMed] [Google Scholar]
- 21. Fierobe H. P., Mingardon F., Mechaly A., Bélaïch A., Rincon M. T., Pagès S., Lamed R., Tardif C., Bélaïch J. P., Bayer E. A. (2005) Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J. Biol. Chem. 280, 16325–16334 [DOI] [PubMed] [Google Scholar]
- 22. Reverbel-Leroy C., Pages S., Belaich A., Belaich J. P., Tardif C. (1997) The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J. Bacteriol. 179, 46–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park J. T., Johnson M. J. (1949) A submicrodetermination of glucose. J. Biol. Chem. 181, 149–151 [PubMed] [Google Scholar]
- 24. Wood T. M. (1988) Preparation of crystalline, amorphous and dyed cellulase substrates. Methods Enzymol. 160, 19–25 [Google Scholar]
- 25. Borne R., Bayer E. A., Pagès S., Perret S., Fierobe H. P. (2013) Unraveling enzyme discrimination during cellulosome assembly independent of cohesin-dockerin affinity. FEBS J. 280, 5764–5779 [DOI] [PubMed] [Google Scholar]
- 26. Irwin D. C., Spezio M., Walker L. P., Wilson D. B. (1993) Activity studies of eight purified cellulases: Specificity, synergism, and binding domain effects. Biotechnol. Bioeng. 42, 1002–1013 [DOI] [PubMed] [Google Scholar]
- 27. Eklöf J. M., Ruda M. C., Brumer H. (2012) Distinguishing xyloglucanase activity in endo-beta(1→4)glucanases. Methods Enzymol. 510, 97–120 [DOI] [PubMed] [Google Scholar]
- 28. Yaniv O., Jindou S., Frolow F., Lamed R., Bayer E. A. (2012) A simple method for determining specificity of carbohydrate-binding modules for purified and crude insoluble polysaccharide substrates. Methods Mol. Biol. 908, 101–107 [DOI] [PubMed] [Google Scholar]
- 29. Jindou S., Xu Q., Kenig R., Shulman M., Shoham Y., Bayer E. A., Lamed R. (2006) Novel architecture of family-9 glycoside hydrolases identified in cellulosomal enzymes of Acetivibrio cellulolyticus and Clostridium thermocellum. FEMS Microbiol. Lett. 254, 308–316 [DOI] [PubMed] [Google Scholar]
- 30. Fierobe H. P., Bagnara-Tardif C., Gaudin C., Guerlesquin F., Sauve P., Belaich A., Belaich J. P. (1993) Purification and characterization of endoglucanase C from Clostridium cellulolyticum. Catalytic comparison with endoglucanase A. Eur. J. Biochem. 217, 557–565 [DOI] [PubMed] [Google Scholar]
- 31. Fierobe H. P., Gaudin C., Belaich A., Loutfi M., Faure E., Bagnara C., Baty D., Belaich J. P. (1991) Characterization of endoglucanase A from Clostridium cellulolyticum. J. Bacteriol. 173, 7956–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Warner C. D., Go R. M., García-Salinas C., Ford C., Reilly P. J. (2011) Kinetic characterization of a glycoside hydrolase family-44 xyloglucanase/endoglucanase from Ruminococcus flavefaciens FD-1. Enzyme Microb. Technol. 48, 27–32 [DOI] [PubMed] [Google Scholar]
- 33. Zverlov V. V., Schantz N., Schmitt-Kopplin P., Schwarz W. H. (2005) Two new major subunits in the cellulosome of Clostridium thermocellum: xyloglucanase Xgh74A and endoxylanase Xyn10D. Microbiology 151, 3395–3401 [DOI] [PubMed] [Google Scholar]
- 34. Sakka K., Kishino Y., Sugihara Y., Jindou S., Sakka M., Inagaki M., Kimura T. (2009) Unusual binding properties of the dockerin module of Clostridium thermocellum endoglucanase CelJ (Cel9D-Cel44A). FEMS Microbiol. Lett. 300, 249–255 [DOI] [PubMed] [Google Scholar]
- 35. Pereira J. H., Sapra R., Volponi J. V., Kozina C. L., Simmons B., Adams P. D. (2009) Structure of endoglucanase Cel9A from the thermoacidophilic Alicyclobacillus acidocaldarius. Acta Crystallogr. D Biol. Crystallogr. 65, 744–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schubot F. D., Kataeva I. A., Chang J., Shah A. K., Ljungdahl L. G., Rose J. P., Wang B. C. (2004) Structural basis for the exocellulase activity of the cellobiohydrolase CbhA from Clostridium thermocellum. Biochemistry 43, 1163–1170 [DOI] [PubMed] [Google Scholar]
- 37. Abdou L., Boileau C., de Philip P., Pagès S., Fiérobe H. P., Tardif C. (2008) Transcriptional regulation of the Clostridium cellulolyticum cip-cel operon: a complex mechanism involving a catabolite-responsive element. J. Bacteriol. 190, 1499–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fendri I., Tardif C., Fierobe H. P., Lignon S., Valette O., Pagès S., Perret S. (2009) The cellulosomes from Clostridium cellulolyticum: identification of new components and synergies between complexes. FEBS J. 276, 3076–3086 [DOI] [PubMed] [Google Scholar]
- 39. Blouzard J. C., Bourgeois C., de Philip P., Valette O., Bélaïch A., Tardif C., Bélaïch J. P., Pagès S. (2007) Enzyme diversity of the cellulolytic system produced by Clostridium cellulolyticum explored by two-dimensional analysis: identification of seven genes encoding new dockerin-containing proteins. J. Bacteriol. 189, 2300–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gal L., Pages S., Gaudin C., Belaich A., Reverbel-Leroy C., Tardif C., Belaich J. P. (1997) Characterization of the cellulolytic complex (cellulosome) produced by Clostridium cellulolyticum. Appl. Environ. Microbiol. 63, 903–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eckert K., Vigouroux A., Lo Leggio L., Moréra S. (2009) Crystal structures of A. acidocaldarius endoglucanase Cel9A in complex with cello-oligosaccharides: strong-1 and -2 subsites mimic cellobiohydrolase activity. J. Mol. Biol. 394, 61–70 [DOI] [PubMed] [Google Scholar]
- 42. Mandelman D., Belaich A., Belaich J. P., Aghajari N., Driguez H., Haser R. (2003) X-Ray crystal structure of the multidomain endoglucanase Cel9G from Clostridium cellulolyticum complexed with natural and synthetic cello-oligosaccharides. J. Bacteriol. 185, 4127–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sakon J., Irwin D., Wilson D. B., Karplus P. A. (1997) Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat. Struct. Biol. 4, 810–818 [DOI] [PubMed] [Google Scholar]
- 44. Burstein T., Shulman M., Jindou S., Petkun S., Frolow F., Shoham Y., Bayer E. A., Lamed R. (2009) Physical association of the catalytic and helper modules of a family-9 glycoside hydrolase is essential for activity. FEBS Lett. 583, 879–884 [DOI] [PubMed] [Google Scholar]