Background: Tryptase is a serglycin proteoglycan-dependent protease localized in mast cell granules.
Results: Tryptase was found to degrade core histones, both during apoptosis and in viable cells.
Conclusion: A serglycin-tryptase axis is identified as a novel mechanism for histone modification.
Significance: Secretory granule compounds are implicated in the regulation of nuclear events.
Keywords: Apoptosis, Histone Modification, Mast Cell, Protease, Proteoglycan, Proteolytic Enzymes, mMCP-6, Secretory Granules, Serglycin, Tryptase
Abstract
A hallmark feature of mast cells is their high content of cytoplasmic secretory granules filled with various preformed compounds, including proteases of tryptase-, chymase-, and carboxypeptidase A3 type that are electrostatically bound to serglycin proteoglycan. Apart from participating in extracellular processes, serglycin proteoglycan and one of its associated proteases, tryptase, are known to regulate cell death by promoting apoptosis over necrosis. Here we sought to outline the underlying mechanism and identify core histones as primary proteolytic targets for the serglycin-tryptase axis. During the cell death process, tryptase was found to relocalize from granules into the cytosol and nucleus, and it was found that the absence of tryptase was associated with a pronounced accumulation of core histones both in the cytosol and in the nucleus. Intriguingly, tryptase deficiency resulted in defective proteolytic modification of core histones even at baseline conditions, i.e. in the absence of cytotoxic agent, suggesting that tryptase has a homeostatic impact on nuclear events. Indeed, tryptase was found in the nucleus of viable cells and was shown to cleave core histones in their N-terminal tail. Moreover, it was shown that the absence of the serglycin-tryptase axis resulted in altered chromatin composition. Together, these findings implicate histone proteolysis through a secretory granule-derived serglycin-tryptase axis as a novel principle for histone modification, during both cell homeostasis and cell death.
Introduction
Mast cells (MCs)3 are hematopoietic cells of the immune system, contributing to both the innate and the adaptive arms of immunity (1). A hallmark feature of MCs is their high content of cytoplasmic secretory granules densely packed with numerous different preformed compounds, including bioactive amines (e.g. histamine, serotonin), preformed cytokines (e.g. TNF), and anionic proteoglycans of serglycin type and various MC-specific proteases, the latter including tryptases, chymases, and carboxypeptidase A3 (2–4). Previous studies have shown that serglycin proteoglycan has a key regulatory role in maintaining the homeostasis of the secretory granules by forming complexes with the various granule proteases and bioactive amines, thereby promoting their storage within granules (5, 6).
The general notion is that the preformed compounds present in immune cell granules predominantly carry out pro-inflammatory functions after their exocytosis in the context of various pathological situations, in particular during allergic conditions (1, 7, 8). However, we have recently shown that secretory granules and their contained serglycin-bound proteases in addition can influence cell death. When assessing the mechanism of cell death in MCs, we found that although wild type (WT) MCs predominantly die by apoptosis in response to various cytotoxic agents, cells lacking serglycin predominantly undergo necrotic cell death (9). This could potentially have a large impact on MC-mediated events, considering that apoptosis occurs with minimal damage to the surrounding milieu, whereas necrotic cell death is characterized by extensive leakage of pro-inflammatory alarmins from the interior of necrotic cells to the cell surroundings (10–12).
When dissecting this process further, we found that granule compounds leaked into the cytosol in conjunction with the cell death process. Moreover, we found that the apoptosis-promoting effect of serglycin was attributed to tryptase, i.e. one of the proteases that are complex-bound to serglycin and dependent on serglycin for storage within granules (9). However, the mechanism by which the serglycin-tryptase axis regulates cell death is not known, one essential aspect being to identify the downstream intracellular targets(s) for the serglycin-tryptase axis. Here we address this issue and identify core histones as primary proteolytic targets for tryptase during cell death. Intriguingly, we show that tryptase also causes proteolysis of core histones during cell homeostasis. In line with this finding, we show that tryptase, in addition to being localized in secretory granules, is found within the nucleus of viable cells and cleaves off N-terminal tails of core histones. Together, these findings introduce a novel principle for histone modification, mediated by a secretory granule protease.
EXPERIMENTAL PROCEDURES
Reagents
Cycloheximide (CHX), digitonin, Pefabloc SC and E-64d were from Sigma-Aldrich (Steinheim, Germany). Rabbit anti-histone H2A, H2B, H3, and H4 polyclonal antibodies were from Abcam (Cambridge, UK). Goat polyclonal antibody to actin was from Santa Cruz Biotechnology (Santa Cruz, CA). Recombinant human skin β-tryptase was from Promega (Madison, WI). Recombinant human histone H2A, H2B, H3.1, and H4 were from New England Biolabs (Ipswich, MA). LysoTracker Red DND-99 was from Life Technologies.
Animals
Mice (8–18 weeks old) deficient in serglycin (serglycin−/−) (6) and mMCP-6−/− mice (13) deficient in tryptase as well as WT controls were on C57BL/6J genetic background. All animal experiments were approved by the local ethical committee.
Bone Marrow-derived MCs (BMMCs)
Mice were sacrificed by CO2 asphyxiation. Femurs and tibiae were removed, and marrow was flushed from the bones with PBS. BMMCs were obtained by culturing bone marrow cells in Dulbecco's modified Eagle's medium (SVA, Uppsala, Sweden), supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen), 50 μg/ml streptomycin sulfate, 60 μg/ml penicillin G, 2 mm l-glutamine (National Veterinary Institute (SVA)), 1 μg/ml mouse recombinant IL-3, and 30% WEHI-3B conditioned medium. The cells were kept at a concentration of 0.5 × 106 cells/ml, at 37 °C in 5% CO2,and the medium was changed every 3rd day. Cell viability in mature BMMCs was >95%.
Induction/Inhibition of Cell Death
Triplicates of 1 ml BMMCs (0.5 × 106 cells/ml) were transferred into individual wells of a 24-well flat-bottomed plate and were either left untreated or treated with 5 μg/ml CHX in complete culture medium. Inhibitory assays were performed using the same methodology. However, before the addition of CHX, cells were previously incubated for 30 min with 0.1 mm Pefabloc SC.
Confocal Microscopy
Aliquots of 100 μl from 0.5 × 106 cells/ml suspensions were dropped into round areas (prepared using liquid-repellent slide marker pen) on microscopic glasses. After 15 min, liquid was removed carefully with filter paper. The remaining cells were left to dry for 15 min. Samples were fixed with 4% paraformaldehyde in PBS for 30 min followed by a 15-min drying step. 100 μl of 50 μg/ml digitonin solution in PBS was added to each individual glass and incubated for 10 min at room temperature. Next, 50 μl of mMCP-6 antibody (1:500) in TBS, 1% BSA or isotype control antibody at the same concentration was left in contact with cells for 1 h followed by washing three times with TBS-T (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 0.01% Tween 20). 50 μl of Alexa Fluor 488-conjugated antibody against rabbit Ig in TBS, 1% BSA was added to each slide. The slides were kept in the dark for 1 h and washed three times with TBS-T. Finally, 50 μl of 1 μg/ml DAPI in TBS, 1% BSA was added for 2 min followed by washing three times with TBS-T. The slides were mounted with Fluoromount-G (Southern Biotech, Birmingham, AL) and cover glass. Samples were analyzed using a laser-scanning microscope equipped with ZEN 2009 software (LSM 710; Carl Zeiss, Berlin, Germany).
Western Blot Analysis
0.5 × 106 BMMCs were collected by centrifugation and immediately solubilized in SDS-PAGE sample buffer followed by boiling for 10 min. Control experiments showed that tryptase activity was completely abolished immediately after the addition of SDS (without boiling). Samples corresponding to equal numbers of cells were subjected to Western blot analysis as described previously (14). Membranes were scanned using an Odyssey Infrared Imager (LI-COR, Lincoln, NE).
Cytosolic Extract Preparation
BMMCs (106 cells) were collected by centrifugation (150 × g, 8 min, 4 °C) in 1.5-ml Eppendorf tubes and then resuspended in 300 μl of ice-cold digitonin extraction buffer (10 μg/ml digitonin, 250 mm sucrose, 20 mm HEPES, pH 7.5, 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA). After 10 min of incubation on ice, cells were centrifuged (11,500 × g, 5 min, 4 °C), and the supernatant (cytosolic fraction) was quickly removed and frozen.
Chromatin Extraction and Analysis
BMMCs (2 × 106 cells) were collected by centrifugation, washed with PBS, and resuspended in 200 μl of 10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 1 mm DTT, 1:100 protease inhibitor cocktail (Sigma-Aldrich), and 0.1% Triton X-100. Samples were incubated for 8 min on ice followed by centrifugation (1,300 × g, 5 min, 4 °C). The supernatant was discharged, and pellets were lysed for 30 min with 100 μl of 3 mm EDTA, 0.2 mm EGTA, 1 mm DTT, and 1:100 protease inhibitor cocktail followed by centrifugation (1,700 × g, 5 min, 4 °C). The supernatant was discharged, and pellets (chromatin-enriched sample) were resuspended each in 100 μl of 10 mm HEPES, pH 7.5, 0.5 mm MgCl2, and 0.05 mm CaCl2. 1 unit of micrococcal nuclease from Staphylococcus aureus (Sigma-Aldrich) was added followed by incubation for 5 min. 10 μl of 0.1 m EDTA/tube was added to stop the reaction. Samples were analyzed by agarose gel (2%) electrophoresis and visualized under UV transillumination.
Electron Microscopy
BMMCs were pelleted and fixed in 1% glutaraldehyde in 0.1 sodium cacodylate buffer (pH 7.4) for 30 min. The samples were washed three times in 0.1 sodium cacodylate buffer (pH 7.4) and then postfixed with 1% osmium tetroxide for 1 h at 4 °C. Postfixed cells were dehydrated with an ascending ethanol series ending with 100% ethanol and then embedded in Dow epoxy resin (DER332; Unione Chimica Europea, Milan, Italy) and DER732 (Serva, Heidelberg, Germany). Ultrathin sections were prepared with an Ultrathome III (GE Healthcare, Uppsala, Sweden). The immunogold postembedding technique was performed as described previously (15). Ultrathin sections were mounted on nickel grids etched for 1 min with 1% periodic acid and rinsed three times in distilled water (2 min each time). The sections were incubated in 20 mm Tris-HCl, pH 8.2, containing 2% BSA, 225 mm NaCl, 20 mm NaN3, 1% Tween 20, and 0.05% Triton X-100, for 15 min at room temperature, and exposed overnight at 4 °C to primary antibodies: rabbit anti-mMCP-6 (diluted 1:30) or nonimmune rabbit serum (diluted 1:10) as control. The grids were washed three times (over 15 min) in the same buffer with 0.5% BSA and were thereafter incubated with secondary antibodies (goat anti-rabbit IgG-20 nm gold; British Biocell International (Cardiff, Wales, UK)) at 1:50 final dilution, for 1 h at room temperature. The grids were finally rinsed five times (over 15 min) with buffer and distilled water (last washing), double-stained with uranyl acetate and lead citrate for 5 min each, and observed with a transmission electron microscope (EM208; Philips, Eindhoven, The Netherlands). Micrographs were taken with a Morada camera (Olympus Soft Imaging Solutions (OSIS), Munster, Germany). Control experiments using nonimmune rabbit serum showed no significant immunogold labeling. Many sections were obtained from two experiments. In each experiment at least 100 sections were analyzed, and the images displayed were representative. The amount of gold particles scored on ultrathin sections depends on many factors, but also on the amount of antigen in a specific subcellular site (16). Accordingly, we enumerated the gold particles in the various subcellular sites by scoring at least 25 micrographs taken at the same magnification, showing 20 secretory granules on average and about one-third of the nuclear area.
Identification of Cleavage Sites
Histone fragments generated after tryptase cleavage were separated by SDS-PAGE followed by extraction of fragments from gels and N-terminal sequencing by the Edman degradation procedure, using a Procise 494 protein sequencer from Applied Biosystems (Stockholm, Sweden).
RESULTS
Core Histones Accumulate in the Cytosol of Dying Serglycin- and Tryptase-deficient MCs
To monitor the effect of the serglycin-tryptase axis on cell death, we treated MCs from WT, serglycin-deficient (serglycin−/−), and tryptase (mouse mast cell protease 6 (mMCP-6))-deficient mice with cytotoxic agent (cycloheximide) followed by annexin V/propidium iodide staining. In agreement with our previous findings (9), WT MCs showed clear signs of apoptotic cell death, whereas MCs lacking either serglycin or tryptase underwent necrosis (data not shown). To approach the mechanism by which the serglycin-tryptase axis regulates cell death, we next performed experiments to identify proteolytic substrates for tryptase during the apoptotic process. To this end, cytosolic extracts were prepared from MCs subjected to cytotoxic agent followed by proteomic analysis using one-dimensional SDS-PAGE. As seen in Fig. 1A, prominent protein bands at ∼17, ∼15, and ∼13 kDa were observed in cytosolic extracts from both serglycin−/− and mMCP-6−/− cells, but were absent in cytosolic extracts from WT cells. Mass spectrometry analysis identified these bands as histone 2B (H2B; most abundant), H2A, and H4, i.e. all representing core histones involved in nucleosome organization. By immunoblot analysis, a profound accumulation of H2B was confirmed in cytosolic extracts from dying cells lacking either serglycin or tryptase (Fig. 1B).
FIGURE 1.
Core histones accumulate in the cytosol of tryptase- and serglycin-deficient mast cells during cell death. A, WT, tryptase-deficient (mMCP-6−/−) and serglycin−/− mast cells were treated with cytotoxic agent (CHX). At the time points indicated, cytosolic extracts were prepared and were analyzed by SDS-PAGE followed by staining with Coomassie Brilliant Blue. The indicated protein bands were excised and identified by mass spectrometry analysis. B, immunoblot analysis of cytosolic extracts, showing the preservation of H2B in cytosolic extracts from serglycin−/− and mMCP-6−/− mast cells after their exposure to cytotoxic agent.
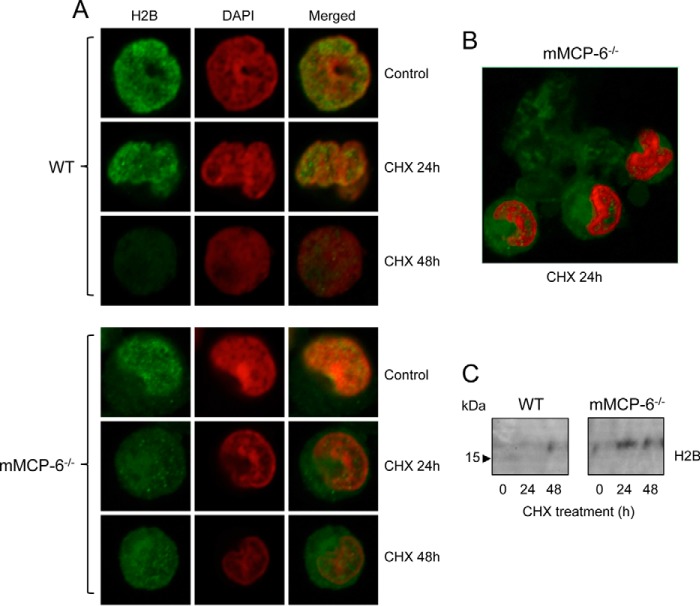
To further substantiate the effect of the serglycin-tryptase axis on core histone accumulation during cell death, we performed confocal microscopy analysis. As expected, H2B co-localized with DAPI in viable cells, both of WT and of mMCP-6−/− genotype (Fig. 2A). When WT cells were treated with cytotoxic agent, H2B staining was gradually reduced, suggesting degradation of H2B during the apoptotic cell death, and it was also notable that the core histone showed a nuclear localization in viable WT cells as well as in all stages of cell death. In contrast, preserved H2B staining was apparent in mMCP-6−/− MCs even at 48 h after the addition of cytotoxic agent, suggesting defective degradation of the core histone. Moreover, in mMCP-6−/− cells treated with cytotoxic agent, strong H2B staining was seen also in the cytosol. Hence, these findings indicate that the absence of tryptase is associated with profound defects in core histone degradation during cell death, leading to defective histone degradation in the nucleus accompanied by an accumulation of core histones in the cytosolic compartment.
FIGURE 2.
Effect of tryptase on histone 2B in mast cells undergoing cell death. WT and tryptase-deficient (mMCP-6−/−) mast cells were exposed to cytotoxic agent (CHX). A, at the time points indicated, cells were stained for histone 2B (H2B) and with DAPI followed by confocal microscopy analysis. Note the preservation of and cytosolic location of H2B in mMCP-6−/− as opposed to WT cells. B, lower magnification showing extracellular release of H2B from mMCP-6−/− cells undergoing cell death. C, immunoblot analysis confirming the release of H2B from mMCP-6−/− cells exposed to cytotoxic agent. Note that the release of H2B is minimal from WT cells.
Core Histones Are Released from Tryptase-deficient MCs Undergoing Cell Death
Necrotic cell death is associated with permeabilization of cell membranes and leakage of various intracellular compounds to the extracellular space (12). Based on the predominant necrotic cell death and accumulation of cytosolic histones in tryptase-deficient MCs, we reasoned that histones might be released from mMCP-6−/− MCs undergoing cell death. Indeed, confocal microscopy analysis revealed strong extracellular H2B staining after treatment of tryptase-deficient MCs with cytotoxic agent (Fig. 2B), whereas this was not observed in WT MCs (data not shown). Moreover, release of H2B from dying mMCP-6−/−, but not WT, cells was supported by immunoblot analysis (Fig. 2C).
The Serglycin-Tryptase Axis Affects Core Histones during Both Cell Death and Homeostasis
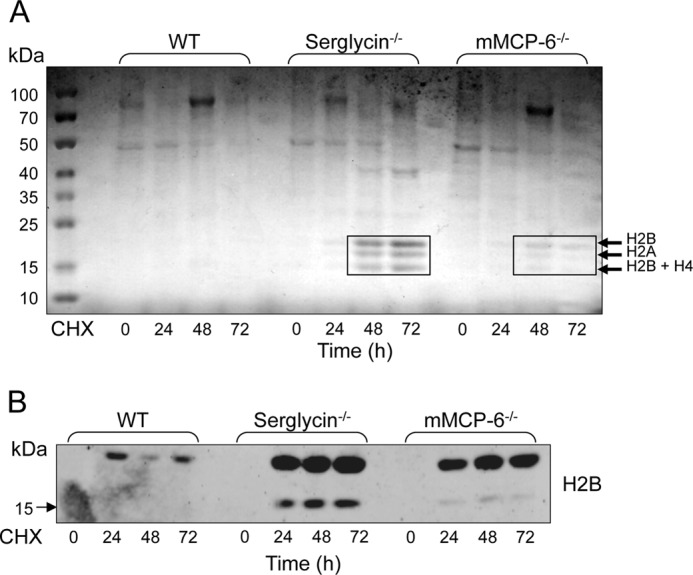
To provide further insight into the impact of the serglycin-tryptase axis on histones, we performed immunoblot analysis for all core histones (H2A, H2B, H3, and H4) in total cell extracts, i.e. including the nuclear compartment. As expected, full-length forms of the core histones were readily detected in extracts from viable MCs of all genotypes (Fig. 3). When subjecting the cells to cytotoxic agent, a time-dependent reduction of core histone levels was clearly seen in WT MCs, all core histones being virtually undetectable after 72 h of treatment. In contrast, the reduction in core histone levels was substantially delayed in serglycin−/− and mMCP-6−/− MCs, supporting a role for the serglycin-tryptase axis in regulating core histone levels during apoptotic cell death (Fig. 3). Most likely, the effect of the serglycin-tryptase axis on core histones is explained by a proteolytic mechanism where tryptase, complex-bound to serglycin, degrades the respective histones. In support of this, the addition of a general inhibitor of serine proteases (Pefabloc; tryptase belongs to the serine protease family) to WT MCs caused a profound blockade of the reduction of core histone levels in response to cytotoxic agent (Fig. 3). Notably, after the addition of serine protease inhibitor to WT cells, the pattern of core histone reduction in response to cytotoxic agent was highly similar to those of serglycin−/− and mMCP-6−/− cells. After the addition of cytotoxic stimulus, it is notable that actin degradation is observed.
FIGURE 3.
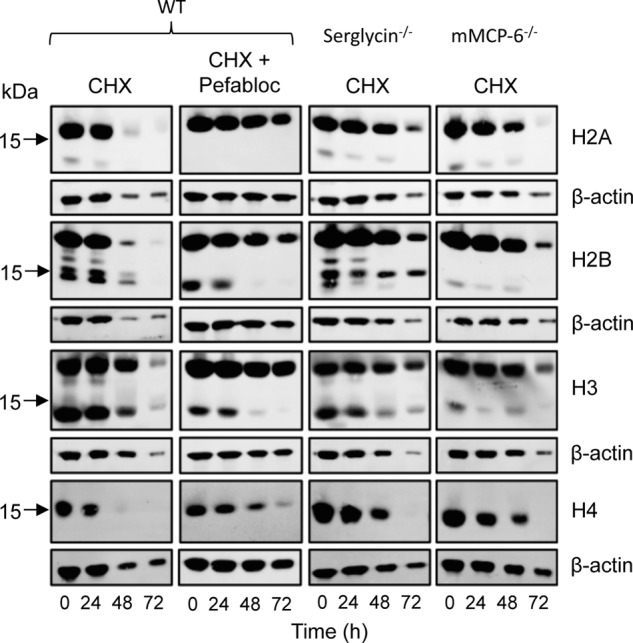
The processing of histones in viable and apoptotic mast cells is dependent on tryptase and serglycin. WT, serglycin−/−, and mMCP-6−/− mast cells were treated with cytotoxic agent (CHX) or left untreated. In addition, WT cells were incubated in the presence of both cytotoxic agent and a general serine protease inhibitor (Pefabloc) as indicated. At the time points indicated, total cell extracts (corresponding to 0.5 × 106 cells/sample) were prepared and subjected to immunoblot analysis for core histones H2A, H2B, H3, and H4. β-Actin was used as loading control.
In addition to their full-length variants, truncated forms of core histones, in particular H2B and H3, were seen in WT MCs. Strikingly, truncation of H2B and H3 was clearly observed in cells that had not been subjected to cytotoxic agent, suggesting that core histone truncation occurs as a normal process in viable MCs. Even more strikingly, the truncation of H2B and H3 at baseline conditions was strongly reduced in MCs lacking tryptase, and partly reduced in serglycin−/− MCs (Fig. 3). These findings suggest that tryptase has an impact on core histones even under normal conditions, i.e. outside of a cell death context. This was highly unexpected considering the general view that tryptase is confined to the secretory granules, being a well established marker for this cellular compartment (3, 4).
Although H2B as expected showed an apparent nuclear localization in WT and tryptase-deficient MCs at baseline conditions (Fig. 2), we reasoned that defective histone processing in tryptase-deficient cells might lead to low levels of histone accumulation in the cytosol. Indeed, although cytosolic H2B staining was not apparent using low contrast images (Fig. 2), when increasing the contrast of the confocal images, we noted H2B accumulation in the cytosol of tryptase-deficient MCs (Fig. 4). In contrast, no cytosolic H2B staining was seen in high contrast images from WT cells (Fig. 4). These findings support a role for tryptase in regulating the levels of core histones during normal MC homeostasis.
FIGURE 4.
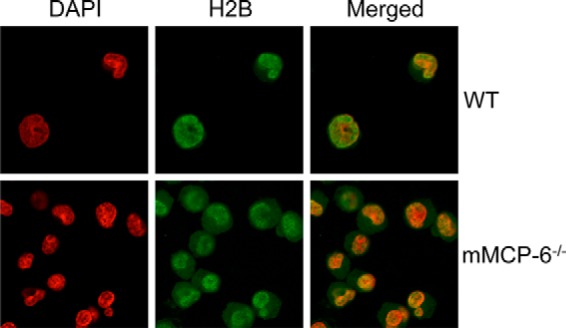
Histones accumulate in the cytosol of tryptase-deficient mast cells at baseline conditions. Viable WT and tryptase-deficient mast cells were stained for histone 2B (H2B; green) and with DAPI (red) followed by confocal microscopy analysis. The images were generated using high contrast. Note that cytosolic H2B is seen in tryptase-deficient cells, whereas H2B is exclusively localized to the nucleus of WT cells.
Tryptase Is Found in the Nucleus in Both Viable and Apoptotic Cells
The findings above introduce the intriguing possibility that tryptase, a protein that hitherto has been recognized as being localized exclusively to secretory granules, in fact may enter the nucleus. To directly address this possibility, we stained MCs for mMCP-6 followed by confocal microscopy analysis. As seen in Fig. 5A, strong granular staining for mMCP-6 was as expected seen in viable WT MCs. As controls, negligible staining was seen when preimmune serum was used (Fig. 5A). When MCs were subjected to cytotoxic agent, the granular staining was lost, accompanied by a more diffuse staining throughout the cells, suggesting release of tryptase from the granules into the cytosol (Fig. 5B). We also noted nuclear staining for mMCP-6, both in untreated cells and in cells treated with cytotoxic agent (Fig. 5B). mMCP-6 positivity was mostly evident in the periphery of the nuclei, but staining was also seen in internal regions of the nucleus (Fig. 5B and data not shown). Upon treatment with cytotoxic agent, a time-dependent gradual increase in nuclear mMCP-6 staining was seen, indicating increased migration of tryptase from granule compartments to the nucleus during apoptotic cell death (Fig. 5B). As an additional sign of lost integrity of acidic compartments during cell death, granular staining with LysoTracker was evident in viable cells but was gradually lost as MCs were exposed to cytotoxic agent (Fig. 5C).
FIGURE 5.
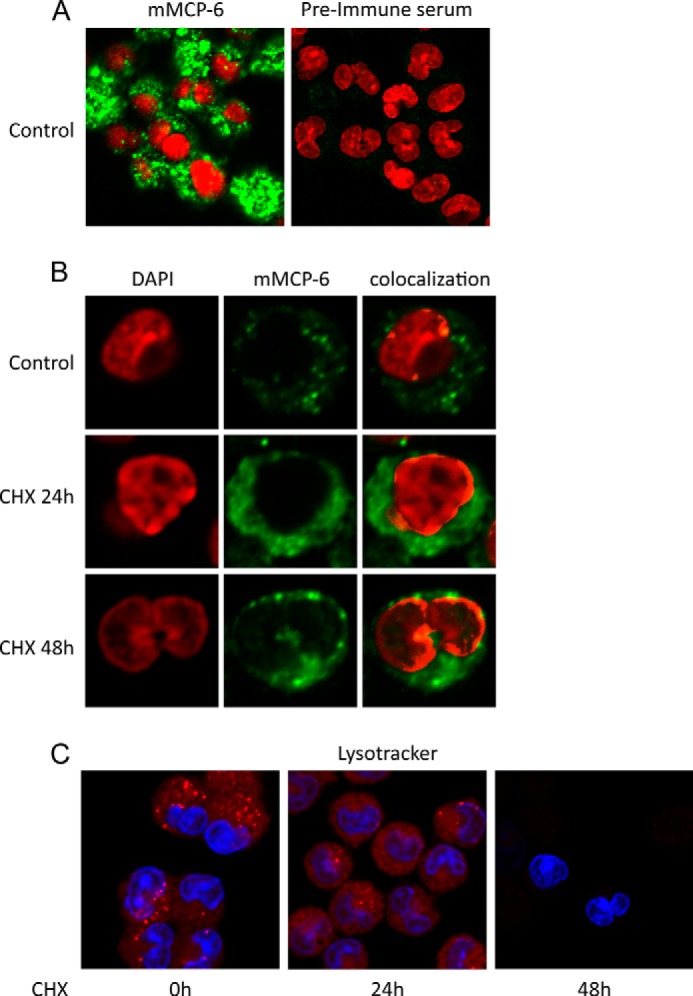
Tryptase mMCP-6 is found in the nucleus of viable and apoptotic mast cells. A and B, control cells (0 h) or mast cells treated with pro-apoptotic agent (CHX) for various time periods were stained for tryptase (mMCP-6; green) and with DAPI (red). Note that mMCP-6 is found both within granular structures and in the nucleus of mast cells and that the nuclear fraction of mMCP-6 increases in apoptotic cells. C, control cells (0 h) and mast cells treated with pro-apoptotic agent (CHX) were stained with LysoTracker (red) to visualize acidic mast cell compartments and with DAPI (blue). Note that the staining with LysoTracker is reduced in apoptotic cells.
To provide more detailed insight into the subcellular localization of tryptase, we performed immunogold transmission electron microscopy analysis. As displayed in Fig. 6A, strong mMCP-6 positivity was as expected seen in many secretory granules, although heterogeneous staining among individual granules was evident. In agreement with the confocal analysis, mMCP-6 staining was also evident in the nuclear compartment (Fig. 6, A–D). Approximately 20% of the total tryptase positivity was seen in the nucleus. In particular, heterochromatin areas were positive for mMCP-6, whereas weaker staining of euchromatin was seen (Fig. 6, B and D). We also observed mMCP-6 positivity in the cytosol, and higher magnification of these signals indicated that mMCP-6 was found within membrane-enclosed small vesicles (Fig. 6A, lower right panel). In contrast, nuclear mMCP-6 was membrane-free. Strikingly, mMCP-6 was seen protruding through nuclear clefts (Fig. 6C, arrow). Together, these findings indicate that tryptase, in addition to its granule localization, is found within the nucleus of viable MCs.
FIGURE 6.
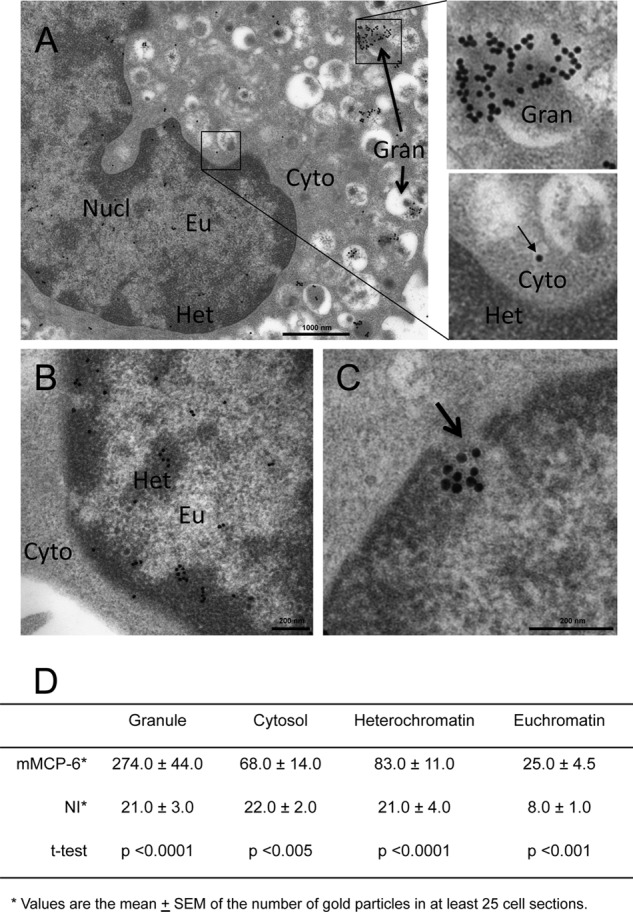
Electron microscopy analysis of tryptase location in mast cells. Mast cells were stained with anti-tryptase (mMCP-6) antibody followed by incubation with gold-labeled secondary antibody and transmission electron microscopy analysis. A, general distribution of tryptase, showing extensive positivity in granules but also positivity in the nucleus and in the cytosol. The right panels show enlargements of the indicated areas, the upper panel highlights the strong positivity of granules, and the lower panel shows membrane-enclosed tryptase in the cytosol. Nucl, nucleus; Eu, euchromatin; Cyto, cytoplasm; Gran, granule; Het, heterochromatin. B, enlarged view of the nucleus, showing tryptase positivity mainly of the heterochromatin. C, view of tryptase protruding through a nuclear cleft (arrow). D, quantification of the tryptase staining, showing the distribution of tryptase in cellular compartments. NI, nonimmune control serum.
Tryptase-dependent Core Histone Processing Accompanies MC Differentiation
Tryptase is a well established MC marker, being expressed in differentiated but not in immature MCs (17). We therefore reasoned that tryptase-dependent core histone processing might accompany the process of MC differentiation. To investigate this possibility, we developed MCs from bone marrow precursor cells and assessed H2B processing and mMCP-6 levels in MCs at different stages of differentiation. In agreement with mMCP-6 being a marker of MC differentiation (17), mMCP-6 was not detected in bone marrow cells (day 0) of either genotype but was strongly induced in WT cells as they acquired MC phenotype (high expression from day ∼16 of culture)(Fig. 7). As expected, mMCP-6 was absent in mMCP-6−/− cells and strongly diminished in serglycin−/− cells (Fig. 7). Processed H2B was detected in day 0 bone marrow cells of all genotypes. From day ∼4, H2B processing was less pronounced in cells of all three genotypes, but became again apparent starting from day ∼16. However, although pronounced H2B processing was seen in maturing WT MCs, H2B processing was minimal in mMCP-6−/− cells and reduced in serglycin−/− MCs (Fig. 6). It is also notable that the onset of H2B processing in maturing cells coincided with the onset of high mMCP-6 expression (Fig. 7). Together, these findings indicate that tryptase-dependent core histone processing accompanies the process of MC maturation.
FIGURE 7.
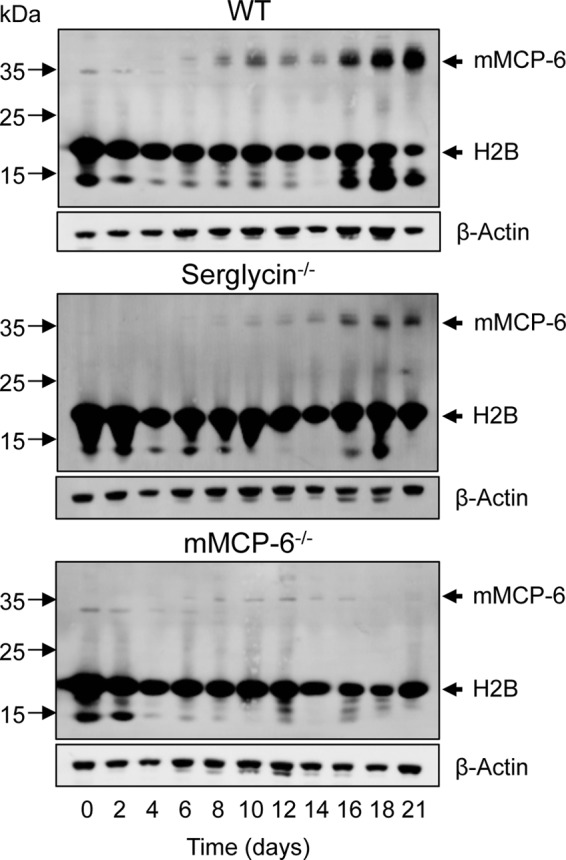
Tryptase-dependent core histone processing accompanies mast cell differentiation. Mast cells were developed by differentiation of bone marrow cells from WT, serglycin−/−-deficient, and tryptase-deficient (mMCP-6−/−) mice. At the time points indicated, cells were harvested and subjected to immunoblot analysis for histone 2B and, as a control for mast cell differentiation, for tryptase mMCP-6. β-Actin was used as loading control.
Core Histones Are Substrates for Tryptase
Next, we employed a purified system to verify that purified core histones are proteolytic substrates for tryptase. Indeed, all core histones were substrates for recombinant tryptase, although the efficiency of cleavage and pattern of degradation products varied among the different core histones. As displayed in Fig. 8A, H2A was rapidly and extensively degraded into fragments too small to be visualized on SDS-PAGE. H2B was also rapidly degraded, into distinct fragments of ∼15, ∼14, and ∼13 kDa followed by further degradation. Incubation of H3 with tryptase resulted in rapid formation of one dominant fragment of ∼15 kDa followed by further degradation into a somewhat smaller fragment (∼11 kDa). Degradation was also apparent for H4, leading to the generation of a fragment of ∼14 kDa. However, the degradation of H4 was not as efficient as for the other core histones.
FIGURE 8.
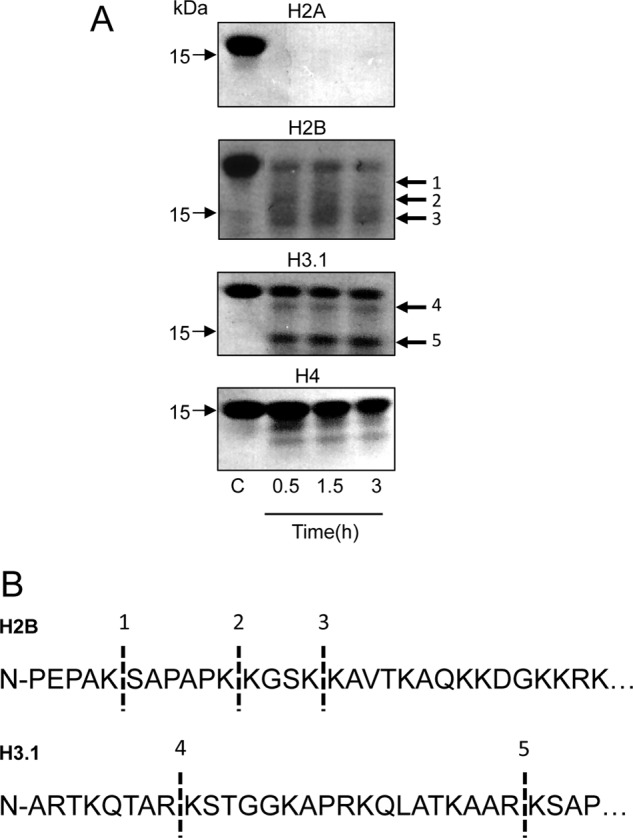
Tryptase cleaves core histones in their N terminus. A, purified tryptase (2.5 μg; 2.6 μm) was incubated with 5 μg of either purified histone 2B (H2B) or purified histone 3 (H3.1)(12 and 11 μm for H2B and H3.1, respectively). At the time points indicated, incubations were interrupted, and the samples were analyzed by SDS-PAGE (12% gels) followed by Coomassie Brilliant Blue staining. Fragments indicated by arrows were excised and subjected to N-terminal sequencing. B, sequence of H2B and H3.1. Dashed lines indicate the sites that were cleaved by tryptase. The numbering of fragments in A corresponds to the numbering of identified cleavage sites in B.
Tryptase Cleaves Off Core Histone N-terminal Portions
To identify the sites in the core histones that are targets for tryptase, N-terminal sequencing of cleavage products was carried out. N-terminal sequencing of H2B cleavage products revealed tryptase-catalyzed cleavage at Lys-5, Lys-11, and Lys-15, whereas H3 (H3.1 variant) was cleaved at Lys-9 and Arg-26 (Fig. 8B). This indicates that the N-terminal portions are truncated in cleaved H2B and H3. Importantly, the N-terminal portions of the core histones, in particular those of H2B and H3, are major sites for extensive post-translational modification (18, 19). Notably, the cleavage patterns seen after incubating purified tryptase with purified histones closely resemble those seen in cultured cells (Fig. 3). For example, highly similar H3 cleavage patterns are seen in cultured cells and after incubation of purified tryptase with purified H3.1 (compare Figs. 3 and 8), and there is a corresponding agreement regarding H2B processing in the respective contexts. Hence, it is likely that the cleavage sites identified through analysis of purified components correspond to the cleavages observed in intact cells.
Tryptase Regulates Chromatin Structure
It is well established that chromatin organization is highly regulated through histone modifications (20). Based on our observed impact of tryptase deficiency on histone modification, combined with our finding that tryptase predominantly localized to heterochromatin, we decided to investigate whether the serglycin-tryptase axis might influence chromatin structure. To address this possibility, we extracted chromatin from WT, serglycin−/−, and mMCP-6−/− cells followed by degradation with micrococcal nuclease and size separation on agarose gels. As shown in Fig. 9, chromatin from WT cells was extensively degraded, whereas a large fraction of the DNA from both mMCP-6−/− and serglycin−/− cells was resistant to treatment. As resistance of chromatin to micrococcal nuclease treatment is an indication of high heterochromatin content (21), these findings indicate that tryptase regulates chromatin structure by promoting the formation of euchromatin over heterochromatin.
FIGURE 9.
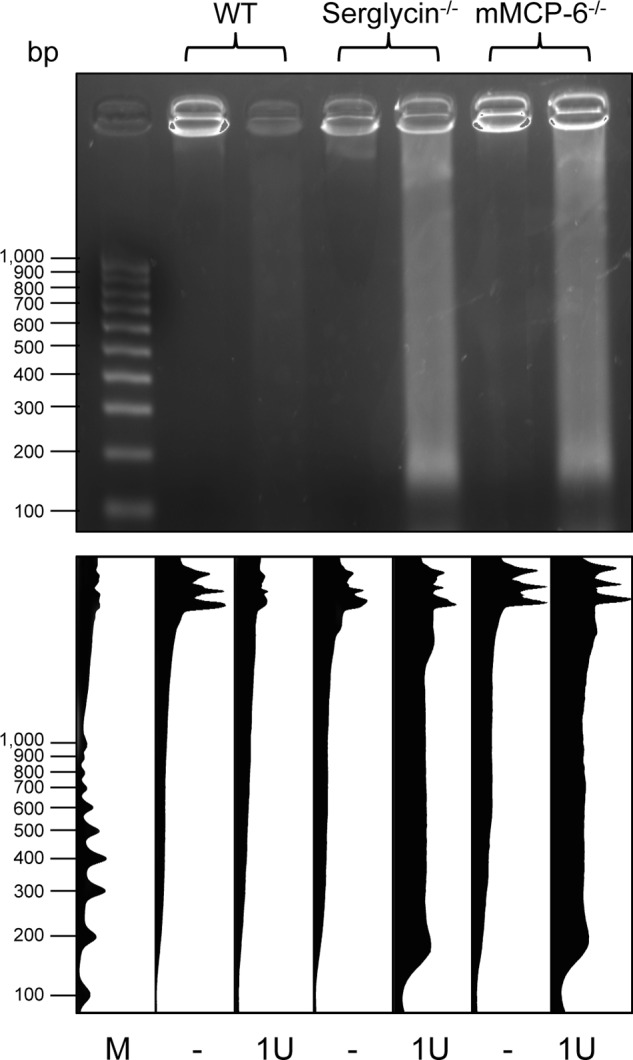
Tryptase regulates chromatin structure. Chromatin was extracted from WT, serglycin−/−-deficient, and tryptase (mMCP-6)-deficient mast cells followed by digestion with one unit (1U) of micrococcal nuclease and agarose gel electrophoresis. The lower panel shows quantification of the data using the ImageJ software. Note that the chromatin from serglycin−/− and mMCP-6−/− cells is largely resistant to digestion, indicative of a high content of heterochromatin. M indicates molecular size markers.
DISCUSSION
A unique feature of MCs is their remarkably high content of granules aimed for regulated secretion, the granules being densely packed with various preformed compounds. Among these, serglycin-protease complexes occupy a dominant position, accounting for a major part of the protein content of the granules. The general view is that the major function of serglycin-protease complexes is to exert extracellular functions, after their release in conjunction with MC degranulation. Indeed, MC proteases including tryptases, chymases, and carboxypeptidase A3 have been implicated in a variety of conditions involving extracellular proteolysis of various substrates, in particular in the context of inflammatory events (3).
Here we introduce a new dimension to the granule-localized proteases by showing that a serglycin-tryptase axis, in addition to impacting on the extracellular environment, can regulate cell-intrinsic processes. Intriguingly, we show that tryptase, during the apoptotic process, is released from the granules into the cytosol and nucleus and that this is accompanied by extensive proteolysis of core histones. As evidence for this scenario, extensive accumulation of core histones was seen both in the cytosol and in the nucleus of tryptase-deficient cells undergoing cell death. In this context it is notable that WT and tryptase-deficient cells differ profoundly in their mechanism of cell death, with WT cells predominantly undergoing apoptotic cell death, whereas tryptase-deficient MCs die by necrosis. A hallmark feature of apoptotic as opposed to necrotic cell death is extensive degradation of DNA mediated by DNA-degrading enzymes such as caspase-activated DNase (12). Conceivably, the DNA degradation seen in WT cells can be facilitated by tryptase-mediated histone proteolysis that may result in unmasking of DNA, thus rendering it sensitive to DNA-degrading enzymes. We may thus propose that the apoptotic mode of cell death seen in WT cells is dependent on tryptase-mediated core histone degradation and, conversely, that the necrotic cell death of tryptase-deficient cells may be linked to defective histone proteolysis.
A highly unexpected finding was that tryptase-mediated histone proteolysis, in addition to occurring in the context of apoptosis, was also evident in viable cells. In agreement with this, tryptase was found in the nucleus of live cells, mostly localizing to the heterochromatin. We also noted that the absence of either tryptase or serglycin resulted in a higher content of heterochromatin versus euchromatin. Plausibly, serglycin-tryptase axis-dependent histone proteolysis could thus affect the general organization of the chromatin. It should be noted that although protease appears to be the major histone-processing tryptase in mast cells, limited histone processing with similar cleavage patterns as those dependent on tryptase is also seen in cells lacking tryptase expression (Fig. 7). Hence, proteases other than tryptase can also contribute to the histone processing, although to a minor extent.
Tryptase has a highly unusual structure, being a tetramer with all of its active sites facing a narrow central pore (22), thus having a macromolecular organization reminiscent of the proteasome. Due to this organization, tryptase is resistant to all known macromolecular protease inhibitors found in mammals (23). The enzyme is thereby well suited for carrying out proteolytic processes in the cellular environment, unhindered by the multitude of protease inhibitors of serpin type that are abundant within the cytosol and also in the nucleus (24, 25). The ability of tryptase to degrade histones intracellularly is thus compatible with its biochemical organization. It is also important to note that tryptase is highly dependent on serglycin proteoglycan, both for tetramerization/activation and stability, as well as for storage within granules (4). In agreement with a central role of serglycin in regulating tryptase, we show that serglycin−/− MCs display similar alterations in histone proteolysis as those seen in tryptase-deficient cells. However, it is important to emphasize that the lack of serglycin does not result in complete tryptase deficiency (Fig. 7). In agreement with the latter notion, serglycin−/− cells showed effects on histone proteolysis that were intermediate to those of WT and tryptase-deficient cells.
Another consequence of the unique macromolecular organization of tryptase is that relatively few proteins have the ability to enter the active site-containing central pore. Accordingly, although the actual cleavage specificity of tryptase is broad, relatively few substrates for tryptase have been identified (4), and in most cases it has not been verified whether the respective identified substrates are of physiological relevance. The present study thus identifies core histones as major substrates for tryptase. In agreement with the trypsin-like cleavage specificity of MC tryptase, histones were exclusively cleaved after Lys and Arg residues. The core histones, in particular those that were shown to be cleaved by tryptase, all possess structurally flexible N-terminal portions that extend outside of the nucleosomes (18). These N-terminal ends would thus be highly susceptible to proteolysis, and it is also likely that the N-terminal ends, due to their flexible structure, can readily enter the central pore of tryptase. In some analogy with the present findings, it has been shown that cathepsin L, a cysteine protease, can modify core histones in embryonic stem cells (26).
In summary, the present study introduces a novel principle for histone modification, mediated by a secretory granule-serglycin-tryptase axis.
Acknowledgment
We are highly grateful to David M. Lee (Harvard Medical School) for providing mMCP-6−/− mice.
This work was supported by grants from The Swedish Research Council (to G. P.) and The Swedish Cancer Foundation (to G. P.).
- MC
- mast cell
- mMCP
- mouse mast cell protease
- BMMC
- bone marrow-derived mast cell
- H
- histone
- CHX
- cycloheximide.
REFERENCES
- 1. Galli S. J., Grimbaldeston M., Tsai M. (2008) Immunomodulatory mast cells: negative, as well as positive, regulators of immunity. Nat. Rev. Immunol. 8, 478–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lundequist A., Pejler G. (2011) Biological implications of preformed mast cell mediators. Cell Mol. Life Sci. 68, 965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pejler G., Rönnberg E., Waern I., Wernersson S. (2010) Mast cell proteases: multifaceted regulators of inflammatory disease. Blood 115, 4981–4990 [DOI] [PubMed] [Google Scholar]
- 4. Pejler G., Abrink M., Ringvall M., Wernersson S. (2007) Mast cell proteases. Adv. Immunol. 95, 167–255 [DOI] [PubMed] [Google Scholar]
- 5. Kolset S. O., Pejler G. (2011) Serglycin – A structural and functional chameleon with wide impact on immune cells. J. Immunol. 187, 4927–4933 [DOI] [PubMed] [Google Scholar]
- 6. Abrink M., Grujic M., Pejler G. (2004) Serglycin is essential for maturation of mast cell secretory granule. J. Biol. Chem. 279, 40897–40905 [DOI] [PubMed] [Google Scholar]
- 7. Rivera J., Fierro N. A., Olivera A., Suzuki R. (2008) New insights on mast cell activation via the high affinity receptor for IgE. Adv. Immunol. 98, 85–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Metcalfe D. D., Baram D., Mekori Y. A. (1997) Mast cells. Physiol. Rev. 77, 1033–1079 [DOI] [PubMed] [Google Scholar]
- 9. Melo F. R., Grujic M., Spirkoski J., Calounova G., Pejler G. (2012) Serglycin proteoglycan promotes apoptotic versus necrotic cell death in mast cells. J. Biol. Chem. 287, 18142–18152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edinger A. L., Thompson C. B. (2004) Death by design: apoptosis, necrosis and autophagy. Curr. Opin Cell Biol. 16, 663–669 [DOI] [PubMed] [Google Scholar]
- 11. Elmore S. (2007) Apoptosis: a review of programmed cell death. Toxicol. Pathol. 35, 495–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Taylor R. C., Cullen S. P., Martin S. J. (2008) Apoptosis: controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 9, 231–241 [DOI] [PubMed] [Google Scholar]
- 13. Shin K., Watts G. F., Oettgen H. C., Friend D. S., Pemberton A. D., Gurish M. F., Lee D. M. (2008) Mouse mast cell tryptase mMCP-6 is a critical link between adaptive and innate immunity in the chronic phase of Trichinella spiralis infection. J. Immunol. 180, 4885–4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rönnberg E., Pejler G. (2012) Serglycin: the master of the mast cell. Methods Mol. Biol. 836, 201–217 [DOI] [PubMed] [Google Scholar]
- 15. Zabucchi G., Soranzo M. R., Menegazzi R., Vecchio M., Knowles A., Piccinini C., Spessotto P., Patriarca P. (1992) Eosinophil peroxidase deficiency: morphological and immunocytochemical studies of the eosinophil-specific granules. Blood 80, 2903–2910 [PubMed] [Google Scholar]
- 16. Griffiths G., Hoppeler H. (1986) Quantitation in immunocytochemistry: correlation of immunogold labeling to absolute number of membrane antigens. J. Histochem. Cytochem. 34, 1389–1398 [DOI] [PubMed] [Google Scholar]
- 17. Gurish M. F., Ghildyal N., McNeil H. P., Austen K. F., Gillis S., Stevens R. L. (1992) Differential expression of secretory granule proteases in mouse mast cells exposed to interleukin 3 and c-kit ligand. J. Exp. Med. 175, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 19. Strahl B. D., Allis C. D. (2000) The language of covalent histone modifications. Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 20. Wolffe A. P., Hayes J. J. (1999) Chromatin disruption and modification. Nucleic Acids Res. 27, 711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim J. A., Hsu J. Y., Smith M. M., Allis C. D. (2012) Mutagenesis of pairwise combinations of histone amino-terminal tails reveals functional redundancy in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 109, 5779–5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pereira P. J., Bergner A., Macedo-Ribeiro S., Huber R., Matschiner G., Fritz H., Sommerhoff C. P., Bode W. (1998) Human β-tryptase is a ring-like tetramer with active sites facing a central pore. Nature 392, 306–311 [DOI] [PubMed] [Google Scholar]
- 23. Sommerhoff C. P., Bode W., Pereira P. J., Stubbs M. T., Stürzebecher J., Piechottka G. P., Matschiner G., Bergner A. (1999) The structure of the human βII-tryptase tetramer: fo(u)r better or worse. Proc. Natl. Acad. Sci. U.S.A. 96, 10984–10991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bird P. I., Trapani J. A., Villadangos J. A. (2009) Endolysosomal proteases and their inhibitors in immunity. Nat. Rev. Immunol. 9, 871–882 [DOI] [PubMed] [Google Scholar]
- 25. Teoh S. S., Whisstock J. C., Bird P. I. (2010) Maspin (SERPINB5) is an obligate intracellular serpin. J. Biol. Chem. 285, 10862–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Duncan E. M., Muratore-Schroeder T. L., Cook R. G., Garcia B. A., Shabanowitz J., Hunt D. F., Allis C. D. (2008) Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell 135, 284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]