Background: Productive epithelial interactions require actin filament assembly at E-cadherin adhesions.
Results: Cortactin localizes to the zonula adherens through interactions with E-cadherin and N-WASP; there it recruits Arp2/3 and WAVE2 by separate mechanisms to support actin nucleation.
Conclusion: Cortactin acts as a coincident scaffold.
Significance: Cortactin can regulate the dynamic integration of cadherin adhesion with the actin cytoskeleton.
Keywords: Actin, Cell Adhesion, Cell Junctions, Cytoskeleton, E-cadherin, Arp2/3, Cortactin, WAVE2
Abstract
Cadherin junctions arise from the integrated action of cell adhesion, signaling, and the cytoskeleton. At the zonula adherens (ZA), a WAVE2-Arp2/3 actin nucleation apparatus is necessary for junctional tension and integrity. But how this is coordinated with cadherin adhesion is not known. We now identify cortactin as a key scaffold for actin regulation at the ZA, which localizes to the ZA through influences from both E-cadherin and N-WASP. Using cell-free protein expression and fluorescent single molecule coincidence assays, we demonstrate that cortactin binds directly to the cadherin cytoplasmic tail. However, its concentration with cadherin at the apical ZA also requires N-WASP. Cortactin is known to bind Arp2/3 directly (Weed, S. A., Karginov, A. V., Schafer, D. A., Weaver, A. M., Kinley, A. W., Cooper, J. A., and Parsons, J. T. (2000) J. Cell Biol. 151, 29–40). We further show that cortactin can directly bind WAVE2, as well as Arp2/3, and both these interactions are necessary for actin assembly at the ZA. We propose that cortactin serves as a platform that integrates regulators of junctional actin assembly at the ZA.
Introduction
The epithelial zonula adherens (ZA)2 is a specialized adhesive junction located at the apical-lateral interfaces of cell-cell contacts. The ZA supports epithelial organization and junctional tension (2, 3) by integrating E-cadherin-mediated adhesion and the dynamic actomyosin cytoskeleton (4). In particular, the ZA cortex is a site of dynamic actin assembly, where Arp2/3 mediates actin filament nucleation (5, 6) and thereby promotes the accumulation of myosin II to generate junctional tension (6–8).
Control of actin nucleation at the ZA exemplifies a broader issue in the cell biology of actin regulation. Arp2/3 has little activity in isolation and must, instead, be stimulated by nucleation-promoting co-factors (9). This presents two challenges for the cell. First, multiple molecules must be co-recruited into functional complexes to activate nucleation. Of the Arp2/3 activators (9), both N-WASP and WAVE2 are found at the ZA (6, 10, 11). However, N-WASP does not support nucleation but rather contributes to junctional actin homeostasis by stabilizing nascent filaments (10). Instead, WAVE2 promotes actin nucleation at the ZA and, like Arp2/3, supports F-actin homeostasis and junctional tension (6). But how WAVE2 might be co-localized with Arp2/3 at the ZA is not known.
The second challenge for regulation of Arp2/3 is to ensure that these ensembles of molecules are co-localized with subcellular spatial fidelity. Potentially, actin nucleation at the ZA entails a form of coincident regulation (12, 13), where multiple molecules must coincide with high spatial and temporal fidelity for actin assembly to occur. Here E-cadherin itself can play a central role. Cadherin adhesion exerts an instructive role to mark sites for cortical recruitment of Arp2/3 and actin assembly (14, 15) and E-cadherin associates with both Arp2/3 (14) and WAVE2 (6). However, these interactions do not appear to be constitutive, as they are lost when cadherins are not engaged in homophilic adhesion (14). This implies that other mechanisms may exist to coordinate the elements of the actin nucleation apparatus with E-cadherin at the ZA.
In this study, we examine the potential role for the scaffolding protein, cortactin, to support coincident regulation of Arp2/3 at the ZA. Cortactin is a multidomain protein whose acidic N terminus can directly bind to, and activate, Arp2/3 (1). Cortactin also contributes to other steps associated with Arp2/3-mediated actin assembly (16), including association with other regulatory proteins, such as N-WASP (17) and WIP (18), and stabilizing nascent branched actin networks (19). Previously, we reported that cortactin can form a complex with cadherin and contribute to cadherin-based actin regulation (15). We now demonstrate that cortactin integrates inputs from E-cadherin and N-WASP to localize selectively to the ZA, where it promotes actin nucleation by scaffolding both Arp2/3 and WAVE2 to the ZA cortex, through distinct binding interactions.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfections
Caco-2 cells were cultured in RPMI complete growth media and grown at 37 °C in 5% CO2. Cells were transfected with Lipofectamine 2000 or Lipofectamine RNAiMAX (both from Invitrogen) according to the manufacturer's instructions for plasmids or siRNAs, respectively. N-WASP KD cells were generated using lentiviral transduction and have been previously described (10).
Plasmids and siRNA
Cortactin-GFP was expressed in cells using a previously described lentivirus-based system (10, 20). Briefly, shRNA against human cortactin (5′-GAGAAGCACGAGTCACAGA-3′) and human cortactin containing four silent mutations within the target sequence were cloned into a pLL5.0 vector carrying an enhanced GFP reporter gene. The shRNA was cloned downstream of a U6 promoter, whereas the RNAi-resistant cortactin sequence was cloned downstream of an LTR promoter and upstream of the enhanced GFP gene. This resulted in simultaneous knockdown of endogenous cortactin and expression of RNAi-resistant cortactin-GFP at levels suitable for imaging. Cortactin mutant constructs were generated by site-directed mutagenesis using the Stratagene kit or by cloning of truncated cortactin sequences into the vector.
WAVE2-GFP was described previously (6, 10). Arp3-GFP was a kind gift from Matt Welch. mRFP-PAGFP-Arp3 was made from the Arp3-GFP plasmid by replacing GFP with the mRFP-PAGFP fragment from mRFP-PAGFP-actin. Plasmids for mammalian expression of human β-catenin-GFP and human cortactin-GFP were obtained by cloning the corresponding cDNA into pEGFP-N1 vectors (Clontech). Similarly, murine E-cadherin-Cherry was obtained by cloning the murine Ecad cDNA (20) into pmCherry-N1.
siRNAs used in this study were Dharmacon SMARTpool RNAi duplexes against WAVE2 (L-012141-00-0010), E-cadherin (L-003877-00), or control scrambled siRNA (D-001810-01-05). siRNAs were used at a final concentration of 100 nm.
Antibodies and Immunoprecipitation
Primary antibodies used in this study were as follows: mouse monoclonal against cortactin (4F11, a kind gift from S. Weed, West Virginia University; 1:1000 for immunofluorescence (IF) and 1:1000 for Western blot (WB)); rabbit polyclonal raised against E-cadherin (1:1000 IF and 1:2000 WB); mouse monoclonal HECD-1 against E-cadherin (1:50 IF and 1:100 WB, a kind gift from P. Wheelock, University of Nebraska, with permission of M. Takeichi); mouse monoclonal against Arp3 subunit of Arp2/3 complex (Sigma, 1:50 IF and 1:400 WB); rabbit polyclonal raised against p34 subunit of Arp2/3 complex (1:1000 IF); rabbit polyclonal against GAPDH (R&D Systems, 1:2000 WB); rabbit polyclonal against GFP (Molecular Probes, 1:1000 WB); rabbit polyclonal against WAVE2 (Cell Signaling, 1:50 IF and 1:2000 WB); rabbit polyclonal against N-WASP (Cell Signaling, 1:50 IF and 1:1000 WB); rabbit polyclonal against dsRED/mCherry (Clontech, 1:1000 WB); rabbit polyclonal against Nap1 (Sigma, 1:50 IF and 1:1000 WB); and mouse monoclonal against Abi1 (Abcam, 1:100 IF, 1:1000 WB). Secondary antibodies were species-specific antibodies conjugated with AlexaFluor 488, 594, or 647 (Invitrogen) for immunofluorescence or with horseradish peroxidase (Bio-Rad) for immunoblotting. F-actin was stained using Alexa488- or 594-phalloidin (Invitrogen, 1:500).
Cells were lysed in 1 ml of lysis buffer (1% Nonidet P-40, 150 mm NaCl, 50 mm Tris-HCl, pH 7.4, 1 mm EDTA, 50 mm sodium fluoride, 2 mm sodium vanadate, 0.1% bovine serum albumin, and Complete protease inhibitors (Roche Applied Science)). Immunoprecipitations were performed using ∼1 μg of total protein, to which 2 μg of antibody and 20 μl of packed slurry of protein A-Sepharose beads (GE Healthcare) or 15 μl of GFP-Trap coupled to agarose beads (Chromotek) were added. Protein complexes were dissociated from beads, and samples were run on SDS-polyacrylamide gels.
Immunofluorescence Microscopy and Image Analysis
Cells were fixed with 4% paraformaldehyde in cytoskeletal stabilization buffer (10 mm PIPES at pH 6.8, 100 mm KCl, 300 mm sucrose, 2 mm EGTA, and 2 mm MgCl2) on ice for 10 min. Fixed coverslips were then permeabilized with 0.25% Triton X-100 in PBS for 5 min at room temperature. Confocal images were acquired on a Zeiss LSM710 microscope and processed using ImageJ (National Institutes of Health), Imaris (Bitplane), or Photoshop (Adobe).
Quantitative analysis of staining intensity at contacts was performed in ImageJ with the line scan function (7). Briefly, a line of 40 pixels in length was selected centered on and perpendicular to a contact. The PlotProfile feature was used to record the pixel intensities along the selected line, which were corrected for background and used to calculate the mean and standard deviation of the maximum pixel intensity.
G-actin Incorporation Assay
Barbed ends were labeled by G-actin incorporation (10). Confluent monolayers of Caco-2 cells were permeabilized with saponin for 7 min in the presence of 0.45 μm Alexa594-tagged G-actin to favor barbed end incorporation. Cells were then fixed in 4% paraformaldehyde in cytoskeletal stabilization buffer containing 2% Triton X-100 and Alexa488-phalloidin (1:500).
Protein Preparation
In vitro translation was performed as described previously (21). Proteins of interest were cloned into a derivative of the pLTE vector carrying T7 promoter and species-independent translation initiation sequences as well as His or fluorescent protein tags (22). The vectors included recombination sites compatible with Gateway cloning system (Clontech).3 This allowed rapid cloning of genes of interest and in vitro synthesis of tagged gene products used for subsequent single molecule coincidence analysis, AlphaScreen, or pulldown experiments. Expressed proteins were fluorescently labeled during synthesis by random incorporation of BODIPY-Lys (1:250, Promega), then resolved on a 4–25% Tris/glycine gel, and visualized using a fluorescent scanner (Typhoon, GE Healthcare).
Protein Interaction Analysis
Single molecule fluorescence analysis was performed as described previously (23, 24). Briefly, GFP- and mCherry-tagged proteins were co-expressed in vitro for 3 h and diluted to a single molecule concentration (∼100 pm) immediately before measurement. For each experiment, 20 μl of sample were placed into a custom-made silicone 192-well plate equipped with a 70 × 80-mm glass coverslip (ProSciTech). Plates were analyzed on a Zeiss LSM710 microscope with a Confocor3 module at room temperature. For coincidence experiments, two lasers (488 and 561 nm) were focused in solution using a 40 × 1.2 NA water immersion objective. Fluorescence was collected and separated using a 565-nm dichroic mirror; the signal from GFP was filtered by a 505–540 bandpass filter, and fluorescence from mCherry was filtered by a 580-nm long pass filter. The fluorescence of the two channels was recorded simultaneously and separately, adding the number of photons collected in 1-ms time bins. A single molecule event was detected when the total intensity of the two channels was above a threshold of 80 photons.
For each event, the intensities of the GFP and mCherry bursts were corrected for background and GFP fluorescence bleed through (10% of the GFP signal into the mCherry channel). The coincidence ratio C was then measured ratiometrically as the corrected mCherry signal, divided by the total intensity of the burst. In the absence of mCherry fluorescence, C is close to zero, and in the absence of GFP, C tends toward 1. Events with 0.25 < C < 0.75 are considered coincident events.
Single molecule coincidence histograms were plotted by measuring >1000 events per interaction and were fitted by gaussian peaks for GFP-only, coincidence, and mCherry-only contributions. The bound fraction was calculated as the proportion of coincidence to total events.
FCS was performed using the Zeiss710 microscope and the Confocor3 software. The 488-nm laser was used at low power to reduce bleaching and triplet contributions. The FCS measurements were calibrated using Alexa488 dye, and the setup was aligned until the structural parameter S was strictly less than 10. This ensured accurate fitting of the FCS curves by purely diffusive components. The calibration was further performed with GFP and GFP-mCherry tandem proteins, and the increases in diffusion times were consistent with the increase of physical size of the diffusing object. To perform these measurements, the individually expressed proteins were diluted to 10 nm concentration in cell-free lysate to take into account possible viscosity effects. We found that the diffusions of individually expressed GFP-EcadTail, GFP-β-catenin, and GFP-cortactin were consistent with the calibration of diffusion times, with GFP-Ecadtail diffusing faster than the tandem but slower than GFP. To measure the potential effect of binding, GFP-EcadTail was co-expressed with unlabeled β-catenin or cortactin, and the co-expression was diluted in lysate expressing unlabeled β-catenin or cortactin. This ensures that the proteins have undiluted binding partners to maximize association. We estimate that the cell-free system produces low micromolar concentrations of the proteins.
AlphaScreen was performed using a c-Myc detection kit (PerkinElmer Life Sciences). Mixtures of GFP- and mCherry-Myc-tagged in vitro translated proteins were serially diluted in 0.5 m PBS containing 200 mm NaCl and 1 mm DTT. 1.5 μl of sample was incubated with 8.5 μl of water, 2 μl of acceptor beads, and 1.5 μl of biotinylated GFP-nanotrap for 30 min, after which 2 μl of donor beads was added. After another 30-min incubation, samples were analyzed on an EnVision Multilabel plate reader (PerkinElmer Life Sciences), and the signal was corrected for background noise. For competition assays, a 10-fold excess of His-tagged in vitro translated proteins was added prior to dilution.
RESULTS
Cortactin Recruits Arp2/3 to Regulate Actin Dynamics at the ZA
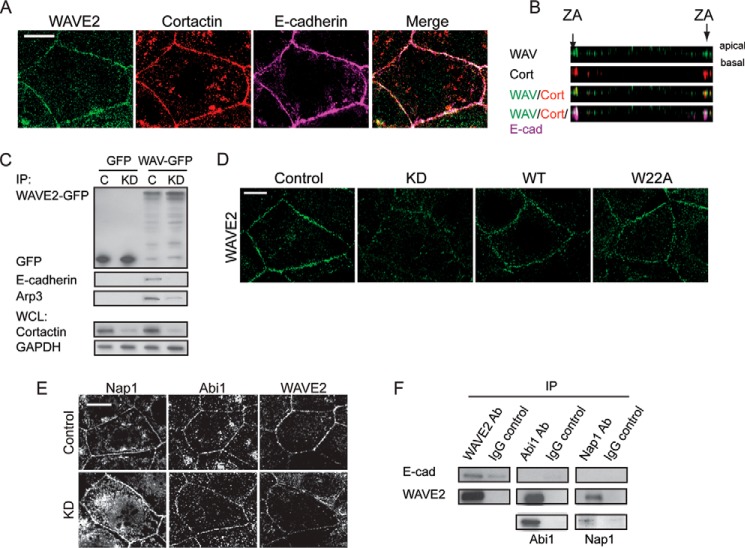
We used Caco-2 cells, which assemble prominent zonulae adherentes within their cell-cell contacts (25, 26), as a model in which to analyze how cortactin might influence Arp2/3 at junctions. In this system, Arp2/3 is found at cell-cell junctions where it supports actin filament nucleation for the junctional actin cytoskeleton (6). Although E-cadherin adhesion can recruit Arp2/3 (14, 26), the molecular intermediates responsible are not well understood. We first used immunofluorescence and co-immunoprecipitation analysis to assess whether cortactin was a junctional protein in Caco-2 cells, as it is in other cell lines (15, 27). Indeed, we found that cortactin co-localized with Arp2/3 at cadherin junctions in Caco-2 cells (Fig. 1, A and B) and co-immunoprecipitated with E-cadherin in these cells (Fig. 1C). Furthermore, junctional cortactin was reduced when E-cadherin was depleted by RNAi (Fig. 1D); the remaining cortactin staining persisted principally where residual E-cadherin was found, although we cannot exclude a contribution from a nonjunctional cortical pool. Because cortactin can interact directly with Arp2/3 (1, 16), it therefore appeared an attractive candidate to mediate the recruitment of Arp2/3 to E-cadherin.
FIGURE 1.
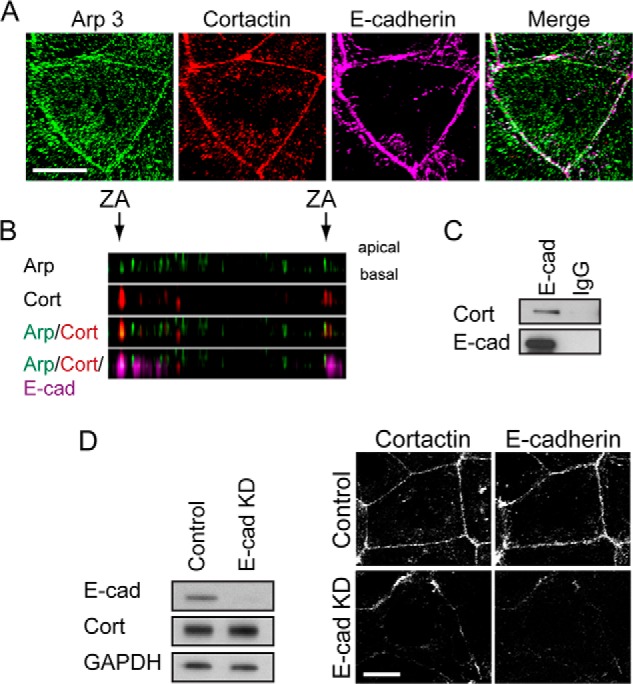
Cortactin accumulates with Arp2/3 at apical E-cadherin junctions. A, maximum projection view from a confocal stack of Arp3, cortactin, and E-cadherin immunostaining in Caco-2 cells. Scale bar, 10 μm. B, Z-section of image in A showing that Arp3 (Arp, green) and cortactin (cort, red) co-localize at the apical regions (ZA) of cell-cell contacts, as marked by E-cadherin (E-cad, magenta). C, E-cadherin or control IgG immunoprecipitates from Caco-2 cells immunoblotted for cortactin (Cort) or E-cadherin (E-cad). D, impact of E-cadherin on cortactin localization. Cells treated with E-cadherin siRNA to KD E-cadherin were analyzed by Western blotting (left panel) and immunofluorescence (right panel) for cortactin and E-cadherin; GAPDH was used as a loading control. Junctional cortactin was lost, despite stable cellular expression, in E-cadherin KD cells. Scale bar, 10 μm.
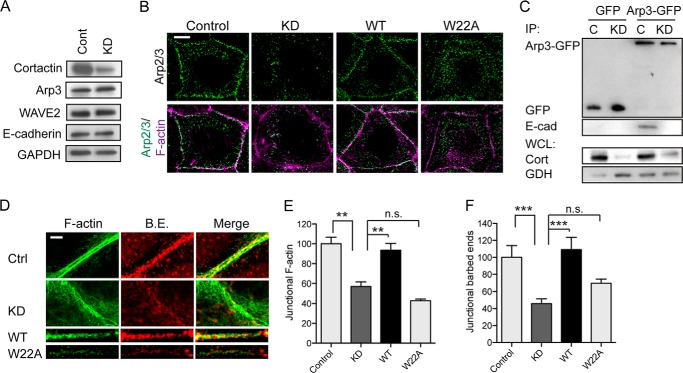
To test this hypothesis, we infected Caco-2 cells with lentiviral shRNA that reduced cortactin levels by 70–90% (Fig. 2A). Junctional Arp3 staining, used as a marker of the Arp2/3 complex, was substantially reduced in cortactin shRNA (KD) cells (Fig. 2B), without any change in its overall cellular expression (Fig. 2A). This was restored by expression of an shRNA-resistant cortactin transgene (Fig. 2B), confirming that the effects were attributable to cortactin depletion. However, junctional Arp3 was not restored when cortactin KD cells were transfected with a cortactin mutant (W22A) that bears a point mutation in the N-terminal DDW motif (28) necessary for cortactin to directly bind Arp2/3 (Figs. 2B and 7C).
FIGURE 2.
Cortactin recruits Arp2/3 to regulate the junctional actin cytoskeleton. A, Western analysis of control (Cont) and cortactin KD cell lysates probed for cortactin, Arp3, WAVE2, E-cadherin, and GAPDH (loading control). B, representative confocal images of control cells, cortactin KD cells, and KD cells reconstituted with wild-type (WT) cortactin, or a mutant (W22A) incapable of binding Arp2/3, stained for Arp3 and F-actin (phalloidin). Junctional Arp3 was substantially reduced at junctions (marked by F-actin staining) in KD cells and was restored by expression of WT but not W22A cortactin. C, impact of cortactin on the E-cadherin-Arp2/3 interaction. Control (C) and cortactin KD cells were transfected with either pEGFP (GFP) or Arp3-GFP constructs and immunoprecipitated (IP) with anti-GFP antibody. Immunocomplexes were immunoblotted for GFP or E-cadherin; whole cell lysates (WCL) were probed for cortactin (cort) to confirm depletion and GAPDH (GDH) as a loading control. Arp3-GFP could immunoprecipitate E-cadherin in control cells but not in cortactin KD cells. D–F, junctional actin cytoskeleton was analyzed in control (Ctrl) cells, cortactin shRNA (KD) cells, and cortactin shRNA cells reconstituted with wild-type cortactin (WT) or cortactin W22A (W22A). D, representative confocal images of apical intercellular contacts. Cells were incubated with Alexa594-G-actin prior to fixation to label barbed ends (B.E.) and stained for F-actin after fixation. In control cells, barbed ends localized between the parallel bundles of F-actin filaments adjacent to the intercellular membrane. In KD cells, barbed ends were reduced at the ZA and F-actin appeared more dispersed on either side of the membrane. Expression of WT, but not W22A, restored F-actin and barbed end labeling at the ZA. E and F, quantitative line scan analysis of junctional F-actin (E) and barbed end labeling (F). Scale bar in B and D are 10 and 2 μm, respectively; data are means ± S.E.; **, p < 0.01; ***, p < 0.001; n.s., not significant.
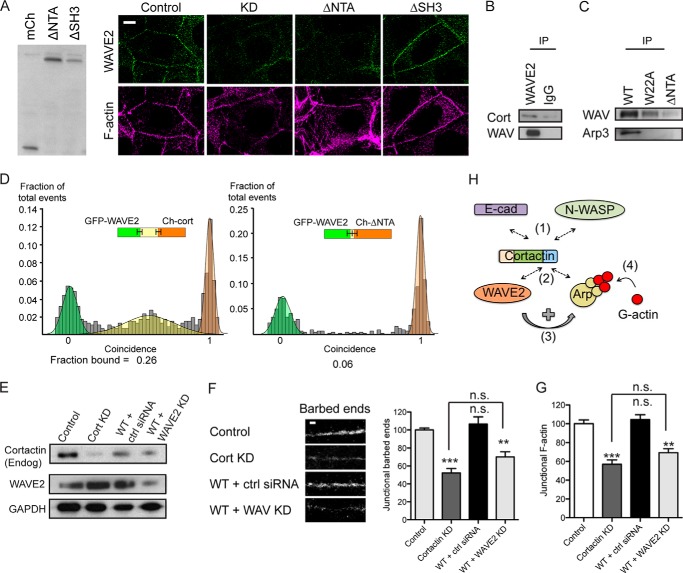
FIGURE 7.
WAVE2 can interact with the N terminus of cortactin. A, characterization of control cells, cortactin KD cells, and KD cells reconstituted with cortactin ΔNTA or ΔSH3 mutants. Immunoblots of cell lysates were probed for mCherry (left panel); all cortactin mutants ran at the expected molecular mass. Representative confocal images were stained for WAVE2 and F-actin (right panel). Junctional WAVE2 was lost from the contacts (marked by F-actin staining) in cortactin KD and ΔNTA cells but restored by ΔSH3 cortactin. B, WAVE2 immunoprecipitates (IP) from Caco-2 cell lysates probed for cortactin (Cort) and WAVE2. C, cell lysates from cortactin KD cells expressing GFP-tagged wild-type cortactin (WT), W22A mutant, or ΔNTA mutant were immunoprecipitated with anti-GFP antibody and immunoblotted for WAVE2 and Arp3. The W22A mutant could co-immunoprecipitate WAVE2 but not Arp3, whereas the ΔNTA mutant could pull down neither protein. D, direct interactions between in vitro translated WAVE2 and cortactin assessed by single molecule coincidence analysis. WAVE2 interacted with full-length cortactin but not with the ΔNTA mutant. E–G, cortactin knockdown cells reconstituted with wild type (WT) cortactin were transfected with nontargeting siRNA (+ ctrl siRNA) or siRNA targeted against WAVE2 (+ WAVE2 KD). E, Western analysis of whole cell lysates immunoprobed for cortactin (endogenous (Endog)), WAVE2, and GAPDH as a loading control (left panel). F, representative images of barbed end labeling at junctions in cell lines (left panel) and quantitation of junctional barbed end labeling (right panel). G, quantitation of junctional F-actin. (n = 48–60 contacts pooled from 4 to 5 independent experiments). Scale bar is 10 μm in A and 2 μm in F; data are means ± S.E.; **, p < 0.01; ***, p < 0.001; n.s., not significant. H, conceptual model of cortactin as a coincident scaffold that regulates actin nucleation at the ZA. Cortactin binds directly to β-catenin-bound E-cadherin at the ZA via an N-WASP-dependent mechanism (bar 1). The N-terminal acidic domain of cortactin recruits both Arp2/3 and WAVE2 (bar 2), whereupon WAVE2 activates Arp2/3 (bar 3) to promote actin nucleation and polymerization at the ZA (bar 4).
We then asked whether cortactin might support the biochemical interaction between E-cadherin and Arp2/3. To test this, we expressed Arp3-GFP in Caco-2 cells and isolated protein complexes with GFP-Trap (Fig. 2C). E-cadherin was detected in the Arp3-GFP precipitates isolated from control cells. However, the amount of E-cadherin that associated with Arp3-GFP was significantly reduced in cortactin KD cells (Fig. 2C). Altogether, these findings identified cortactin as necessary for the junctional localization of Arp2/3, presumably by mediating recruitment of Arp2/3 to the E-cadherin molecular complex.
To characterize how this molecular action of cortactin might affect junctional actin homeostasis, we quantitated phalloidin staining to assess junctional F-actin content and labeled free barbed ends with fluorescently tagged G-actin as an index of actin nucleation (10). Whereas control cells showed prominent perijunctional F-actin cables at the ZA, junctional F-actin was reduced, and appeared more loosely organized, in cortactin KD cells (Fig. 2, D and E). Furthermore, cortactin KD significantly reduced barbed end labeling at the ZA (Fig. 2, D and F). Therefore, cortactin was necessary for filament nucleation at the ZA, which was consistent with our observation that cortactin KD depleted junctions of Arp2/3 (Fig. 2B). Both barbed end labeling and F-actin content at the ZA were restored to cortactin KD cells by expression of WT cortactin (Fig. 2, D–F), confirming that the junctional changes observed were due to loss of cortactin. However, neither of these indices of junctional actin homeostasis were restored when cortactin KD cells were reconstituted with W22A-cortactin (Fig. 2, D–F). Overall, these implied that cortactin contributes to junctional actin homeostasis by its ability to recruit Arp2/3.
Cortactin Interacts Directly with E-cadherin
Given that cortactin can interact with E-cadherin in a variety of epithelial cells (Fig. 1C) (15), we then used in vitro approaches to further characterize the biochemical interaction between recombinant cortactin and the cytoplasmic tail of E-cadherin (Ecad-Tail). For these experiments, we chose to use a eukaryotic cell-free translation system based on Leishmania tarentolae extract (LTE). LTE is well suited for the analysis of protein interactions as it supports co-expression of multiple cDNAs and allows production of large multidomain proteins in their active form (22, 29). We were able to express full-length polypeptides of Ecad-Tail, cortactin, or β-catenin tagged with either GFP or mCherry in the LTE system. We confirmed with SDS-PAGE that untagged and tagged protein products were of the expected molecular weight (Fig. 3A). To analyze interactions, we applied single molecule fluorescence methods that require only minimal amounts of sample and directly give quantitative information on the oligomerization state of the proteins (30).
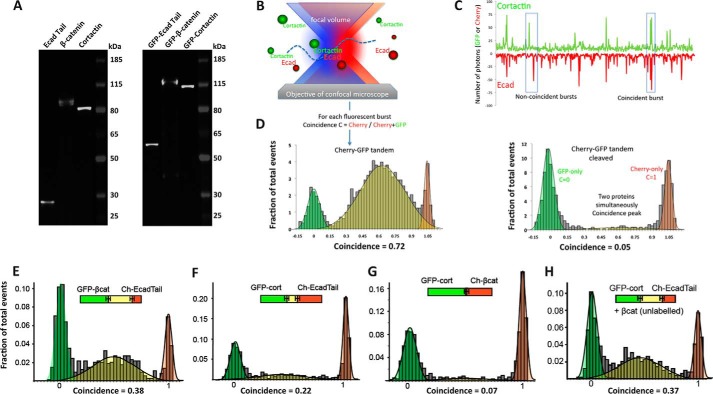
FIGURE 3.
Single molecular coincidence analysis of E-cadherin-cortactin interaction. A, characterization of in vitro translated proteins. Untagged (left panel) and GFP-tagged (right panel) in vitro translated and BODIPY-labeled proteins were separated on a 4–25% Tris/glycine gel and visualized on a Typhoon scanner. Proteins ran at expected sizes. Note that cortactin has a predicted mass of 55 kDa but usually runs at 80 kDa. B–D, illustration and validation of experimental approach. B, diagram illustrating single molecule coincidence method. Leishmania extracts expressing GFP- or mCherry-tagged proteins are diluted to concentrations at which only single molecules or molecular complexes are moved through the focal volume by Brownian motion. C, representative trace showing that binding partners diffuse together through the focal volume, generating coincident bursts of red and green signals, whereas nonbinding partners that diffuse independently generate uncorrelated bursts. Coincidence was measured as the ratio of mCherry signal to the total fluorescent signal (GFP + mCherry) for each signal peak. D, single molecule coincidence analysis of in vitro translated GFP-Cherry fusion protein containing a TEV protease cleavage site between the GFP and Cherry tags. Analysis was performed before (left panel) and after (right panel) treatment with TEV protease. Events were binned into GFP-only (green, coincidence C <0.25), coincidence (yellow, 0.25 < C < 0.75) and mCherry-only (red, C > 0.75). The fraction bound was calculated based on the proportion of coincidence events to total events. E–H, single molecule coincidence analysis of LTE co-expressed GFP-β-catenin and Cherry-Ecad-Tail proteins (E), GFP-cortactin and Cherry-Ecad-Tail (F), GFP-cortactin and Cherry-β-catenin (G), and GFP-cortactin and Cherry-Ecad-Tail co-expressed with unlabeled β-catenin (H). Events were binned into GFP-only (green, coincidence C < 0.25), coincidence (yellow, 0.25 < C < 0.75), and mCherry-only (red, C > 0.75). The fraction bound was calculated based on the proportion of coincidence events to total events.
As we wanted to observe the interaction between multiple proteins, we adapted the single molecule two-color coincidence (SMC) technique that was used earlier to directly visualize binding interactions between DNA fragments (31–34). Fig. 3B shows the principle of the experiment. On a confocal microscope, two lasers are focused within the same excitation volume, and samples bearing appropriate fluorophores are diluted to concentrations below 100 pm. When a molecule diffuses in the focal volume, the fluorophore emits photons that are recorded as bursts in the fluorescence time trace. The two-color experiment enables direct observation of co-diffusion of two interacting proteins when the bursts of fluorescence in the two channels coincide (Fig. 3C). As a test of principle, we expressed an mCherry-GFP fusion protein containing a TEV protease site between the two fluorophores (Fig. 3D). A high coincidence between mCherry and GFP signals was detected from the intact fusion protein (0.72), but coincidence was essentially eliminated (0.05) when the fusion protein was cleaved into its constituents.
We next applied SMC to analyze the interaction between GFP-β-catenin and mCherry-E-Cad-Tail in the LTE protein expression reaction (Fig. 3E). We observed clear coincidence (0.38), consistent with the well established capacity of these proteins to interact directly (35). GFP-cortactin and mCherry-E-Cad-Tail were then co-expressed along with unlabeled β-catenin (Fig. 3H). SMC further revealed that cortactin and the cadherin tail interacted under these circumstances (coincidence 0.37), evidence that the three proteins formed a complex. We then examined the pairwise interactions between these proteins. Strikingly, we found that cortactin bound Ecad-Tail with a high coincidence value (0.22), even in the absence of β-catenin (Fig. 3F). This suggested that Ecad-tail and cortactin could interact directly.
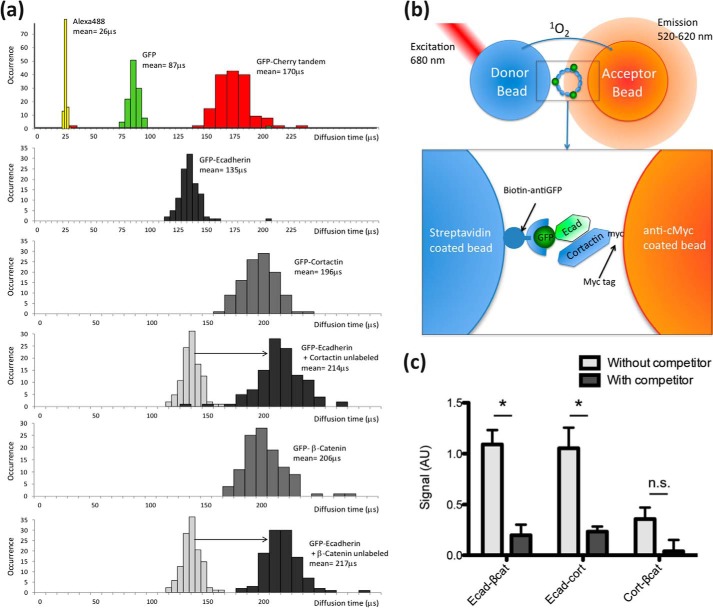
To confirm this result, we used FCS to measure the diffusion rates of GFP-tagged-Ecad-Tail co-expressed with unlabeled β-catenin or cortactin (Fig. 4a) (36). The diffusion time of Ecad-tail was greatly increased when it was co-expressed with β-catenin, consistent with an interaction between these two proteins (35). Similarly, the diffusion time of Ecad-Tail was increased in the presence of cortactin (Fig. 4a), suggesting a direct interaction. FCS measurements also enabled us to further characterize the individually expressed proteins. We found that all the diffusion times of these proteins were consistent with their monomeric forms, as calibrated by a small organic dye (Alexa488), GFP alone, and a GFP-Cherry tandem protein (Fig. 4a).
FIGURE 4.
Analysis of E-cadherin-cortactin protein interactions by fluorescence correlation spectroscopy and AlphaScreen. a, frequency histograms of diffusion times of AlexaFluor 488 dye and in vitro translated GFP and GFP-Cherry fusion protein measured by FCS. The data were used to calibrate the size dependence of the diffusion times and demonstrate that the diffusion times of individually expressed proteins were within the expected range for their monomeric sizes. The diffusion times of GFP-E-cadherin were significantly increased when co-expressed with cortactin or β-catenin, consistent with an increase in size of the resulting complex. b, schematic representation of the AlphaScreen assay. A donor bead is excited by a 680-nm laser pulse and emits singlet oxygen that can react with the acceptor bead. A luminescence signal is only observed if the beads are in close proximity and brought into contact by interacting proteins (inset). A GFP-tagged protein is recognized by a biotinylated GFP-nanotrap, bound to the streptavidin donor bead. The other protein is expressed as a C-terminal Cherry-myc fusion and binds the anti-Myc-coated acceptor bead. c, AlphaScreen analysis of the interactions among in vitro translated Ecad-Tail, β-catenin, and cortactin. The specificity of interactions was validated by competition assays with untagged proteins as follows: unlabeled Ecad-Tail for the interactions between GFP-Ecad-Tail and either β-catenin or cortactin, and unlabeled β-catenin for the interaction of GFP-β-catenin and cortactin. The interaction between Ecad-Tail and β-catenin was used as a positive control. Cortactin could bind the E-cadherin cytoplasmic tail but not β-catenin. *, p < 0.05; n.s., nonsignificant.
To verify the observed interactions using an independent method, we employed the amplified luminescent proximity homogeneous assay (AlphaScreen). This assay measures interactions between protein-coated beads using proximity-dependent singlet oxygen transfer (30, 37) and is able to detect interactions over a broad range of affinities (picomolar to millimolar). To enhance the compatibility of the method with commonly used cell biological probes, we adapted the assay to analyze GFP- and Myc-tagged proteins (Fig. 4b). The AlphaScreen assay was performed directly on the cell-free co-expression mixtures, without purification of the recombinant protein. As a positive control, we tested the interaction between Ecad-Tail and β-catenin, which was substantially reduced by incubation in the presence of unlabeled Ecad-Tail, confirming the specificity of this association (Fig. 4c). An interaction was also detected between Ecad-Tail and cortactin, which was also largely abolished in the presence of unlabeled Ecad-Tail (Fig. 4c), thus corroborating the SMC and FCS assay results.
Together, these studies suggested that cortactin can bind directly to the E-cadherin cytoplasmic tail. However, binding in the SMC analysis was enhanced in the presence of β-catenin. This implies that incorporation of β-catenin into the complex enhanced the association of cadherin and cortactin (Fig. 3H). One potential explanation was that an independent binding site for cortactin existed in β-catenin. However, we detected little coincidence (0.07) between cortactin and β-catenin in pairwise SMC analysis (Fig. 3G), suggesting that these two proteins do not bind directly in the experimental concentrations that we used. Similarly, no specific interaction between cortactin and β-catenin was detected in the AlphaScreen assay (Fig. 4c). This suggested that the presence of β-catenin might enhance a direct interaction between cortactin and the E-cadherin cytoplasmic tail.
N-WASP Refines Cortactin Localization to the ZA
In Caco-2 cells, E-cadherin is found both in the apical ring of the ZA and as clusters distributed throughout the lateral contact surfaces, below the ZA (26). To further characterize the spatial distribution of cortactin at cell-cell junctions, we then compared its localization with that of E-cadherin in these two regions (Fig. 5A). For this purpose, we took advantage of the fact that mature, polarized Caco-2 monolayers often display en face cell-cell junctions that are tilted relative to the optical axis, allowing closer examination of protein localization throughout the apico-lateral axis of the contact zones (26). Deconvolution imaging of these en face contacts confirmed that cortactin is concentrated at the ZA where it strongly co-localizes with E-cadherin, but this co-localization was less obvious throughout the rest of the contact despite the extensive presence of E-cadherin clusters (Fig. 5A). Thus, cortactin is highly enriched at the ZA relative to the rest of the cell-cell contact.
FIGURE 5.
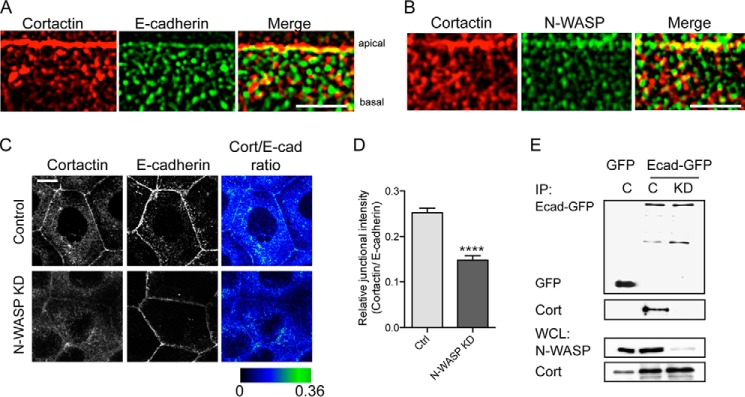
N-WASP promotes the preferential localization of cortactin to the apical zonula adherens. A and B, deconvolved z-stacks of cell-cell junctions immunostained for cortactin (red) and E-cadherin (green) (A) or cortactin (red) and N-WASP (green) (B). Bars are 5 μm. C and D, representative confocal images of control and N-WASP KD cells immunostained for cortactin and E-cadherin (C). Ratiometric analysis of staining intensities revealed a significant reduction in the ratio of cortactin to E-cadherin at junctions in N-WASP KD cells (D). Ctrl, control. ****, p < 0.0001 (Student's t test). E, impact of N-WASP on the E-cadherin-cortactin interaction. Anti-GFP immunoprecipitates (IP) from control (C) and N-WASP KD cells stably expressing either pEGFP (GFP) or E-cadherin-GFP were immunoblotted for GFP or cortactin (Cort). Whole cell lysates (WCL) were probled for N-WASP and cortactin. E-cadherin-GFP could immunoprecipitate cortactin in control cells but not in N-WASP KD cells.
A direct association with E-cadherin did not readily explain how cortactin might concentrate with E-cadherin at the ZA but not with cadherin clusters at the lateral junctions. This discrepancy suggested that another molecule at the ZA might participate in defining cortactin's localization within cadherin-based contacts. We assessed the potential contribution of N-WASP, which can bind cortactin (17, 38) and localize to the ZA (10, 26) where it interacts with the cadherin molecular complex and supports junctional integrity (10, 39). Indeed, N-WASP co-accumulated with cortactin at the ZA in Caco-2 cells (Fig. 5B). We then tested how depletion of N-WASP by lentiviral shRNA (10) affected the localization of cortactin at cell-cell junctions. Junctional cortactin immunofluorescence was substantially reduced at the apical junctions in N-WASP KD cells (Fig. 5C), without total cellular cortactin being affected (Fig. 5E). This suggested that N-WASP might influence the cortical accumulation of cortactin at the ZA. However, because N-WASP depletion also reduces the amount of E-cadherin that concentrates at the ZA (10), it was possible that the reduced cortactin reflected changes in the quantity of the junctional cadherin·β-catenin complex, rather than an additional effect of N-WASP on intra-junctional localization. To pursue this, we performed a ratiometric analysis of cortactin and E-cadherin immunostaining at cell-cell junctions. This revealed that junctional cortactin was significantly reduced by N-WASP depletion, over and above changes in E-cadherin (Fig. 5, C and D).
To test whether these changes in localization patterns corresponded to a biochemical alteration, we then asked whether N-WASP shRNA affected the capacity of cortactin and E-cadherin to co-immunoprecipitate. We isolated E-cadherin-GFP expressed in cadherin shRNA cells (20). The amount of cortactin that co-precipitated with E-cadherin-GFP was reduced by N-WASP RNAi (Fig. 5E). We therefore conclude that the selective localization of cortactin to the ZA involves coordinated contributions from both E-cadherin and N-WASP.
Cortactin Co-scaffolds WAVE2 to Arp2/3 at the ZA
In isolation, Arp2/3 is essentially inert; its biological action is strictly regulated by association with nucleation-promoting factors, such as members of the WASP/WAVE family (9). Recently, we found that one such protein, WAVE2, was a major factor in activating actin nucleation at the ZA in Caco-2 cells (6). We therefore asked whether cortactin might also influence WAVE2.
WAVE2 co-accumulated with cortactin at the ZA (Fig. 6, A and B), and WAVE2-GFP co-immunoprecipitated both Arp3 and E-cadherin (Fig. 6C), evidence that their junctional co-localization might reflect a molecular complex. Of note, both junctional WAVE2 (Fig. 6D) and the association between E-cadherin with WAVE2-GFP (Fig. 6C) were reduced by cortactin KD. As cellular WAVE2 levels were not affected by cortactin KD (Fig. 2A), this implied that cortactin supported the accumulation of WAVE2 at the ZA by recruiting it to E-cadherin. However, because WAVE2 can bind to Arp2/3 (40), it was possible that cortactin supported junctional WAVE2 indirectly, through its action on Arp2/3. However, we found that expression of the cortactin W22A mutant, which does not restore junctional Arp2/3 (Fig. 2B), effectively rescued junctional WAVE2 (Fig. 6D). Similarly, the W22A cortactin mutant could still co-immunoprecipitate WAVE2 (Fig. 7C), despite its inability to bind Arp2/3. Thus, the ability of cortactin to control the junctional localization of WAVE2 was mechanistically distinguishable from its ability to localize Arp2/3.
FIGURE 6.
Cortactin controls the junctional localization of WAVE2. A, maximum projection view of a confocal stack of Caco-2 cells immunostained for WAVE2, cortactin, and E-cadherin. B, Z-section of image in A shows that WAVE2 (WAV, green) and cortactin (Cort, red) co-localize (arrows, 3rd panel) at the ZA, as marked by E-cadherin (E-cad, magenta). C, anti-GFP immunoprecipitates from control (C) and cortactin KD cells transfected with either pEGFP (GFP) or WAVE2-GFP were immunoblotted for GFP, E-cadherin or Arp3. WAVE2-GFP could co-immunoprecipitate (IP) E-cadherin in control cells but not in cortactin KD cells. Whole cell lysates (WCL) were immunoblotted with cortactin to confirm depletion and GAPDH as a loading control. D, representative confocal images of WAVE2 staining in control, cortactin KD, and cortactin KD cells reconstituted with wild-type (WT) or mutant (W22A) cortactin. E, representative confocal images of control and cortactin KD cells immunostained for Nap1, Abi1, and WAVE2. Junctional WAVE2 was reduced in KD cells, without concomitant reduction in Nap1 or Abi1. F, Caco-2 lysates were immunoprecipitated (IP) with antibodies directed against WAVE2, Abi1, or Nap1 or control IgG and Western-blotted for E-cadherin, WAVE2, Abi1, or Nap1. E-cadherin was detected in WAVE2 immunoprecipitates but not in Abi1 or Nap1 immune complexes. All scale bars are 10 μm.
In cells, WAVE proteins also interacts with four other proteins (Sra1, Nap1, Abi, and HSPC300) to form the WAVE regulatory complex (WRC). WAVE2 is trans-inhibited by this complex, and inhibition can be released by Rac signaling (41, 42). In Caco-2 cells, both Nap1 and Abi1 localize with WAVE2 at cell-cell junctions (Fig. 6E) (6). However, Nap1 and Abi1 persisted at junctions of cortactin KD cells, despite WAVE2 being substantially reduced at these same sites (Fig. 6E). This suggested that the interactions of the WRC might be altered at the ZA. Consistent with this, E-cadherin co-immunoprecipitated with WAVE2 but not with either Nap1 or Abi1 (Fig. 6F). Thus, junctional localization of WAVE2 and the WRC may be regulated via distinct mechanisms.
These observations suggested that cortactin might influence the junctional localization of WAVE2 by a novel mechanism. To pursue this, we expressed further cortactin mutants in cortactin KD cells (Fig. 7A). Interestingly, junctional WAVE staining was restored by a cortactin mutant lacking the SH3 domain (ΔSH3) but not by one lacking the N-terminal acidic domain (ΔNTA) (Fig. 7A). This suggested that the N terminus of cortactin might mediate binding to WAVE. Indeed, whereas WAVE and full-length cortactin co-immunoprecipitated (Fig. 7, B and C), WAVE2 did not interact with the ΔNTA cortactin mutant (Fig. 7C). Furthermore, SMC analysis of recombinant proteins demonstrated that WAVE2 can bind directly to full-length cortactin (Fig. 7D). However, recombinant WAVE2 did not bind the ΔNTA mutant (Fig. 7D). These finding therefore suggest that the cortactin N terminus can bind WAVE2 independently of Arp2/3.
Together, these findings suggested that cortactin served to scaffold Arp2/3 and WAVE2 to the ZA by distinguishable molecular mechanisms. We postulated that cortactin acted to coordinately stabilize Arp2/3 and WAVE2 at the junction, thereby co-localizing these two elements of the nucleation apparatus to promote actin assembly. However, cortactin can also directly activate Arp2/3 (1). To further assess how cortactin supported junctional actin nucleation, we therefore asked if exogenous WT-cortactin alone could rescue the junctional cytoskeleton in cortactin KD cells or whether it required WAVE2 to be present. Accordingly, we tested whether WT cortactin could rescue cortactin KD cells if WAVE2 was also depleted (Fig. 7E). We found that reconstitution with WT-cortactin did not rescue junctional actin nucleation (Fig. 7F) or steady-state F-actin content (Fig. 7G) in cortactin KD cells that were also deficient in WAVE2. This implies that the intrinsic capacity of cortactin to activate Arp2/3 (1, 19, 28) cannot by itself support actin assembly at the ZA.
DISCUSSION
The apical zonula adherens in simple epithelial cells is a specialized adhesive junction where actomyosin contractility is coupled to E-cadherin adhesion (4). The prominent myosin-decorated actin rings that lie adjacent to the ZA reflect the interaction of myosin II with actin networks that are nucleated by Arp2/3 at the ZA itself (6). Building and maintaining the junctional cytoskeleton at the ZA thus requires mechanisms that localize the Arp2/3 nucleator to apical cadherin junctions. Furthermore, as Arp2/3 is essentially inactive in isolation, it must also be localized to interact with upstream-activating proteins, notably WAVE2, a major nucleation promotor at the ZA (6). Our present findings identify a key role for cortactin in scaffolding both Arp2/3 and WAVE2 to the apical cadherin pool. Cortactin was necessary for Arp2/3 to localize at the ZA, a process that is attributable to the known capacity for these two proteins to associate directly (1). Consistent with this, actin nucleation and steady-state F-actin content were reduced at apical cadherin junctions when cortactin was depleted, and this was not rescued by a cortactin mutant that could not bind Arp2/3.
Although the N terminus of cortactin is best understood for its capacity to interact with Arp2/3, we now present evidence that it can also interact with WAVE2. Thus, WAVE2 failed to recruit to junctions and did not interact with E-cadherin in cortactin knockdown cells. This impact of cortactin mapped to its N terminus but could be experimentally distinguished from the ability of cortactin to bind Arp2/3, as the W22A cortactin mutant restored junctional WAVE2 but not Arp2/3. Similarly, W22A cortactin retained the ability to co-immunoprecipitate WAVE2 but not Arp2/3. Therefore, although WAVE2 can associate with Arp2/3, our data suggest that another interaction mapping to the N terminus of cortactin is required for the stable localization of WAVE2 at the ZA. Although we earlier found WAVE2 to be a major contributor to junctional actin assembly, cortactin can itself activate Arp2/3 by direct association (1), suggesting that it might provide an alternative pathway for Arp2/3 activation at cadherin junctions. However, this did not appear to be the case, as we found that cortactin was not sufficient to restore junctional actin assembly when WAVE2 was also depleted. This suggests that an ability to coordinately scaffold both Arp2/3 and WAVE2 is a major mechanism for cortactin to support actin assembly at the ZA. Such coordinate scaffolding would provide an attractive mechanism to ensure that the fully functional nucleation apparatus is efficiently localized to cadherin junctions.
A number of actin regulators that contribute to the junctional cortex are recruited to junctions in an E-cadherin-dependent fashion. Some of these, such as cortactin and N-WASP (10, 26), preferentially localize to the apical ZA. This is somewhat surprising because, although apical zonulae adherentes have been equated with cadherin (or adherens) junctions (43), they actually represent a subpopulation of E-cadherin clusters (25) that are stabilized at the apical-lateral interface (44). Many, albeit less stable (44), E-cadherin clusters can be found throughout the lateral junctions below the ZA (26). This raises the question of how proteins can be recruited to the cortex in a cadherin-dependent fashion, yet selectively accumulate with a subpopulation of cadherins within junctions. We propose that for cortactin this involves the integration of two molecular processes.
The first is a direct association with E-cadherin, which accounts for the ability of these two proteins to co-immunoprecipitate within cells (15, 27) and aligns with the observation that cadherin is necessary for cortactin to localize to the junctional cortex. Our protein interaction analysis further indicates that cortactin can bind directly to the cytoplasmic tail of E-cadherin. Thus, associations between these two polypeptides were identified using both single molecule coincidence analysis and a quantitative proximity assay (AlphaScreen). It was noteworthy that, even though an interaction was detected between cortactin and Ecad-Tail alone, SMC analysis suggested that the strength of this association was increased when β-catenin was co-translated. This suggests that the presence of β-catenin, which is an obligate component of the native cadherin·catenin complex (3), might enhance the association with cortactin. Potentially, this might be due to the presence of an independent binding site for cortactin in β-catenin itself. However, this seems unlikely, as a significant association between these two proteins alone was not detected by either SMC or AlphaScreen. An alternative explanation is that the binding of cortactin to E-cadherin is allosterically enhanced by the presence of β-catenin. This notion is consistent with evidence that the isolated cadherin cytoplasmic tail is an unstructured polypeptide, but it becomes structured upon binding β-catenin (35), potentially revealing binding sites for other proteins, such as cortactin.
The second interaction that is necessary for cortactin to concentrate at the ZA involves the actin regulator, N-WASP. Thus, cortactin concentrated with N-WASP at the apical ZA, but its junctional localization was reduced when N-WASP was depleted. How then might interactions with both cadherin and N-WASP contribute to localizing cortactin at the ZA? One possibility is that the intrinsic interaction between cortactin and E-cadherin is relatively weak in cells, and it must be stabilized by an additional binding interaction for cortactin to concentrate at junctions. Indeed, cortactin can potentially interact with N-WASP through both direct and indirect mechanisms. Earlier studies reported that N-WASP can bind to the SH3 domain of cortactin (17). However, as the ΔSH3 cortactin mutant could restore junctional organization in our experiments, this interaction may not be critical at the ZA. N-WASP could use other molecular mechanisms, such as members of the WIP family (18), to recruit cortactin to the ZA. The stringent localization of N-WASP to the apical junctions is, in turn, likely to reflect upstream signals, such as the Cdc42 exchange factor Tuba (45, 46). Thus the specific localization of cortactin to apical junctions may reflect a convergence of multiple biochemical pathways and intermolecular interactions that ultimately polarize the cell cortex of epithelial cells.
In summary, we postulate that cortactin may serve as a node for coincident regulation of actin assembly at the ZA (Fig. 7H). It would integrate upstream inputs from both E-cadherin and N-WASP that define its spatial accumulation at the apical-lateral zone of contact between cells. There it would serve to coordinately scaffold both Arp2/3 and WAVE2 to promote actin nucleation and filament assembly. It is interesting to note that, although in vitro WAVE forms a very stable complex with other members of the WRC (42), cortactin depletion appeared to delocalize WAVE2 from junctions without affecting other members of the WRC. This suggests that the interaction between WAVE and the WRC may be altered by association with cortactin within the context of the intact ZA. Further research will be needed to test this hypothesis. Nonetheless, the wealth of actin regulators that are regulated to co-accumulate at cadherin junctions may provide opportunities for novel protein-protein interactions.
Acknowledgments
We thank Dr. Sergey Mureev for the Gateway vectors, and our many other laboratory colleagues for their unstinting support and advice. Imaging was performed at the Australian Cancer Research Foundation Cancer Biology Imaging Facility at the Institute for Molecular Bioscience, established with the generous support of the Australian Cancer Research Foundation.
This work was supported by the National Health and Medical Research Council of Australia Project Grants 631377, APP1010489, and Research Fellowship 631383 (to A. Y.), Program Grant 511005 (to K. A. and R. G. P.), Program Grant 1037320 (to A. Y., K. A., and R. G. P.), and Australia Fellowship 569542 (to R. G. P.), by Australian Research Council Future Fellowship FT0991611 and Project Grant DP120101423 (to K. A.) and Future Fellowship FT110100478 (to Y. G.), and by the Kid's Cancer Project of the Oncology Children's Foundation.
This article was selected as a Paper of the Week.
D. Gagoski, S. Mureev, Y. Gambin, N. Giles, J. Johnston, M. Dahmer-Heath, D. Škalamera, T. J. Gonda, and K. Alexandrov, manuscript in preparation.
- ZA
- zonula adherens
- LTE
- L. tarentolae extract
- SMC
- single molecule coincidence
- FCS
- fluorescence correlation spectroscopy
- mRFP
- monomeric red fluorescent protein
- KD
- knockdown
- WRC
- WAVE regulatory complex
- IF
- immunofluorescence
- WB
- Western blot
- SH3
- Src homology 3 domain
- TEV
- tobacco etch virus.
REFERENCES
- 1. Weed S. A., Karginov A. V., Schafer D. A., Weaver A. M., Kinley A. W., Cooper J. A., Parsons J. T. (2000) Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J. Cell Biol. 151, 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sawyer J. M., Harrell J. R., Shemer G., Sullivan-Brown J., Roh-Johnson M., Goldstein B. (2010) Apical constriction: a cell shape change that can drive morphogenesis. Dev. Biol. 341, 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Niessen C. M., Leckband D., Yap A. S. (2011) Tissue organization by classical cadherin adhesion molecules: dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol. Rev. 91, 691–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ratheesh A., Yap A. S. (2012) A bigger picture: classical cadherins and the dynamic actin cytoskeleton. Nat. Rev. Mol. Cell Biol. 13, 673–679 [DOI] [PubMed] [Google Scholar]
- 5. Tang V. W., Brieher W. M. (2012) α-Actinin-4/FSGS1 is required for Arp2/3-dependent actin assembly at the adherens junction. J. Cell Biol. 196, 115–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Verma S., Han S. P., Michael M., Gomez G. A., Yang Z., Teasdale R. D., Ratheesh A., Kovacs E. M., Ali R. G., Yap A. S. (2012) A WAVE2-Arp2/3 actin nucleator apparatus supports junctional tension at the epithelial zonula adherens. Mol. Biol. Cell 23, 4601–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smutny M., Cox H. L., Leerberg J. M., Kovacs E. M., Conti M. A., Ferguson C., Hamilton N. A., Parton R. G., Adelstein R. S., Yap A. S. (2010) Myosin II isoforms identify distinct functional modules that support integrity of the epithelial zonula adherens. Nat. Cell Biol. 12, 696–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ratheesh A., Gomez G. A., Priya R., Verma S., Kovacs E. M., Jiang K., Brown N. H., Akhmanova A., Stehbens S. J., Yap A. S. (2012) Central spindlin and α-catenin regulate Rho signalling at the epithelial zonula adherens. Nat. Cell Biol. 14, 818–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Insall R. H., Machesky L. M. (2009) Actin dynamics at the leading edge: from simple machinery to complex networks. Dev. Cell 17, 310–322 [DOI] [PubMed] [Google Scholar]
- 10. Kovacs E. M., Verma S., Ali R. G., Ratheesh A., Hamilton N. A., Akhmanova A., Yap A. S. (2011) N-WASP regulates the epithelial junctional actin cytoskeleton through a non-canonical post-nucleation pathway. Nat. Cell Biol. 13, 934–943 [DOI] [PubMed] [Google Scholar]
- 11. Ivanov A. I., Bachar M., Babbin B. A., Adelstein R. S., Nusrat A., Parkos C. A. (2007) A unique role for nonmuscle myosin heavy chain IIA in regulation of epithelial apical junctions. PLoS ONE 2, e658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carlton J. G., Cullen P. J. (2005) Coincidence detection in phosphoinositide signaling. Trends Cell Biol. 15, 540–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lebensohn A. M., Kirschner M. W. (2009) Activation of the WAVE complex by coincident signals controls actin assembly. Mol. Cell 36, 512–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kovacs E. M., Goodwin M., Ali R. G., Paterson A. D., Yap A. S. (2002) Cadherin-directed actin assembly: E-cadherin physically associates with the Arp2/3 complex to direct actin assembly in nascent adhesive contacts. Curr. Biol. 12, 379–382 [DOI] [PubMed] [Google Scholar]
- 15. Helwani F. M., Kovacs E. M., Paterson A. D., Verma S., Ali R. G., Fanning A. S., Weed S. A., Yap A. S. (2004) Cortactin is necessary for E-cadherin-mediated contact formation and actin reorganization. J. Cell Biol. 164, 899–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ammer A. G., Weed S. A. (2008) Cortactin branches out: roles in regulating protrusive actin dynamics. Cell Motil. Cytoskeleton 65, 687–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martinez-Quiles N., Ho H.-Y., Kirschner M. W., Ramesh N., Geha R. S. (2004) Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP. Mol. Cell. Biol. 24, 5269–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kinley A. W., Weed S. A., Weaver A. M., Karginov A. V., Bissonette E., Cooper J. A., Parsons J. T. (2003) Cortactin interacts with WIP in regulating Arp2/3 activation and membrane protrusion. Curr. Biol. 13, 384–393 [DOI] [PubMed] [Google Scholar]
- 19. Weaver A. M., Karginov A. V., Kinley A. W., Weed S. A., Li Y., Parsons J. T., Cooper J. A. (2001) Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 11, 370–374 [DOI] [PubMed] [Google Scholar]
- 20. Smutny M., Wu S. K., Gomez G. A., Mangold S., Yap A. S., Hamilton N. A. (2011) Multicomponent analysis of junctional movements regulated by myosin II isoforms at the epithelial zonula adherens. PLoS ONE 6, e22458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kovtun O., Mureev S., Jung W., Kubala M. H., Johnston W., Alexandrov K. (2011) Leishmania cell-free protein expression system. Methods 55, 58–64 [DOI] [PubMed] [Google Scholar]
- 22. Mureev S., Kovtun O., Nguyen U. T., Alexandrov K. (2009) Species-independent translational leaders facilitate cell-free expression. Nat. Biotechnol. 27, 747–752 [DOI] [PubMed] [Google Scholar]
- 23. Gambin Y., Schug A., Lemke E. A., Lavinder J. J., Ferreon A. C., Magliery T. J., Onuchic J. N., Deniz A. A. (2009) Direct single-molecule observation of a protein living in two opposed native structures. Proc. Natl. Acad. Sci. U.S.A. 106, 10153–10158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gambin Y., VanDelinder V., Ferreon A. C., Lemke E. A., Groisman A., Deniz A. A. (2011) Visualizing a one-way protein encounter complex by ultrafast single-molecule mixing. Nat. Methods 8, 239–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meng W., Mushika Y., Ichii T., Takeichi M. (2008) Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell 135, 948–959 [DOI] [PubMed] [Google Scholar]
- 26. Wu S. K., Gomez G. A., Michael M., Verma S., Cox H. L., Lefevre J. G., Parton R. G., Hamilton N. A., Neufeld Z., Yap A. S. (2014) Cortical F-actin stabilization generates apical-lateral patterns of junctional contractility that integrate cells into epithelia. Nat. Cell Biol. 16, 167–178 [DOI] [PubMed] [Google Scholar]
- 27. Ren G., Helwani F. M., Verma S., McLachlan R. W., Weed S. A., Yap A. S. (2009) Cortactin is a functional target of E-cadherin-activated Src family kinases in MCF7 epithelial monolayers. J. Biol. Chem. 284, 18913–18922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Uruno T., Liu J., Zhang P., Fan Y.-X., Egile C., Li R., Mueller S. C., Zhan X. (2001) Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat. Cell Biol. 3, 259–266 [DOI] [PubMed] [Google Scholar]
- 29. Guo Z., Johnston W., Kovtun O., Mureev S., Bröcker C., Ungermann C., Alexandrov K. (2013) Subunit organisation of in vitro reconstituted HOPS and CORVET multisubunit membrane tethering complexes. PLoS ONE 8, e81534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sierecki E., Giles N., Polinkovsky M. E., Moustaqil M., Alexandrov K. A., Gambin Y. (2013) A cell-free approach to accelerate the study of protein-protein interaction in vitro. Interface Focus 10.1098/rsfs.2013.0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Orte A., Clarke R., Balasubramanian S., Klenerman D. (2006) Determination of the fraction and stoichiometry of femtomolar levels of biomolecular complexes in an excess of monomer using single-molecule, two-color coincidence detection. Anal. Chem. 78, 7707–7715 [DOI] [PubMed] [Google Scholar]
- 32. Orte A., Clarke R., Klenerman D. (2010) Single-molecule two-colour coincidence detection to probe biomolecular associations. Biochem. Soc. Trans. 38, 914–918 [DOI] [PubMed] [Google Scholar]
- 33. Orte A., Clarke R. W., Klenerman D. (2011) Single-molecule fluorescence coincidence spectroscopy and its application to resonance energy transfer. Chemphyschem 12, 491–499 [DOI] [PubMed] [Google Scholar]
- 34. Yahiatène I., Doose S., Huser T., Sauer M. (2012) Correlation-matrix analysis of two-color coincidence events in single-molecule fluorescence experiments. Anal. Chem. 84, 2729–2736 [DOI] [PubMed] [Google Scholar]
- 35. Huber A. H., Stewart D. B., Laurents D. V., Nelson W. J., Weis W. I. (2001) The cadherin cytoplasmic domain is unstructured in the absence of β-catenin. A possible mechanism for regulating cadherin turnover. J. Biol. Chem. 276, 12301–12309 [DOI] [PubMed] [Google Scholar]
- 36. Ferreon A. C., Gambin Y., Lemke E. A., Deniz A. A. (2009) Interplay of α-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc. Natl. Acad. Sci. U.S.A. 106, 5645–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ullman E. F., Kirakossian H., Switchenko A. C., Ishkanian J., Ericson M., Wartchow C. A., Pirio M., Pease J., Irvin B. R., Singh S., Singh R., Patel R., Dafforn A., Davalian D., Skold C., Kurn N., Wagner D. B. (1996) Luminescent oxygen channeling assay (LOCI): sensitive, broadly applicable homogeneous immunoassay method. Clin. Chem. 42, 1518–1526 [PubMed] [Google Scholar]
- 38. Kowalski J. R., Egile C., Gil S., Snapper S. B., Li R., Thomas S. M. (2005) Cortactin regulates cell migration through activation of N-WASP. J. Cell Sci. 118, 79–87 [DOI] [PubMed] [Google Scholar]
- 39. Rajput C., Kini V., Smith M., Yazbeck P., Chavez A., Schmidt T., Zhang W., Knezevic N., Komarova Y., Mehta D. (2013) Neural Wiskott-Aldrich syndrome protein (N-WASP)-mediated p120-catenin interaction with Arp2-Actin complex stabilizes endothelial adherens junctions. J. Biol. Chem. 288, 4241–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu X. P., Rouiller I., Slaughter B. D., Egile C., Kim E., Unruh J. R., Fan X., Pollard T. D., Li R., Hanein D., Volkmann N. (2012) Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J. 31, 236–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ismail A. M., Padrick S. B., Chen B., Umetani J., Rosen M. K. (2009) The WAVE regulatory complex is inhibited. Nat. Struct. Mol. Biol. 16, 561–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen Z., Borek D., Padrick S. B., Gomez T. S., Metlagel Z., Ismail A. M., Umetani J., Billadeau D. D., Otwinowski Z., Rosen M. K. (2010) Structure and control of the actin regulatory WAVE complex. Nature 468, 533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Farhadifar R., Röper J. C., Aigouy B., Eaton S., Jülicher F. (2007) The influence of cell mechanics, cell-cell interactions, and proliferation on epithelial packing. Curr. Biol. 17, 2095–2104 [DOI] [PubMed] [Google Scholar]
- 44. Priya R., Yap A. S., Gomez G. A. (2013) E-cadherin supports steady-state Rho signaling at the epithelial zonula adherens. Differentiation 86, 133–140 [DOI] [PubMed] [Google Scholar]
- 45. Kovacs E. M., Verma S., Thomas S. G., Yap A. S. (2011) Tuba and N-WASP function cooperatively to position the central lumen during epithelial cyst morphogenesis. Cell Adh. Migr. 5, 344–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Otani T., Ichii T., Aono S., Takeichi M. (2006) Cdc42 GEF Tuba regulates the junctional configuration of simple epithelial cells. J. Cell Biol. 175, 135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]