Background: GLUT1 is the main glucose transporter in certain immune cells.
Results: Overexpressing GLUT1 in macrophages results in increased glucose uptake and glucose utilization.
Conclusion: Driving glucose uptake and metabolism through GLUT1 induces a proinflammatory response that is dependent upon glycolysis and reactive oxygen species.
Significance: Understanding how macrophage substrate metabolism impacts inflammation is crucial to develop novel therapeutics for obesity and diabetes.
Keywords: Glucose Transport, Glycolysis, Inflammation, Macrophages, Metabolomics, Mitochondrial Metabolism, Obesity, Pentose Phosphate Pathway, Crown-like Structure
Abstract
Glucose is a critical component in the proinflammatory response of macrophages (MΦs). However, the contribution of glucose transporters (GLUTs) and the mechanisms regulating subsequent glucose metabolism in the inflammatory response are not well understood. Because MΦs contribute to obesity-induced inflammation, it is important to understand how substrate metabolism may alter inflammatory function. We report that GLUT1 (SLC2A1) is the primary rate-limiting glucose transporter on proinflammatory-polarized MΦs. Furthermore, in high fat diet-fed rodents, MΦs in crown-like structures and inflammatory loci in adipose and liver, respectively, stain positively for GLUT1. We hypothesized that metabolic reprogramming via increased glucose availability could modulate the MΦ inflammatory response. To increase glucose uptake, we stably overexpressed the GLUT1 transporter in RAW264.7 MΦs (GLUT1-OE MΦs). Cellular bioenergetics analysis, metabolomics, and radiotracer studies demonstrated that GLUT1 overexpression resulted in elevated glucose uptake and metabolism, increased pentose phosphate pathway intermediates, with a complimentary reduction in cellular oxygen consumption rates. Gene expression and proteome profiling analysis revealed that GLUT1-OE MΦs demonstrated a hyperinflammatory state characterized by elevated secretion of inflammatory mediators and that this effect could be blunted by pharmacologic inhibition of glycolysis. Finally, reactive oxygen species production and evidence of oxidative stress were significantly enhanced in GLUT1-OE MΦs; antioxidant treatment blunted the expression of inflammatory mediators such as PAI-1 (plasminogen activator inhibitor 1), suggesting that glucose-mediated oxidative stress was driving the proinflammatory response. Our results indicate that increased utilization of glucose induced a ROS-driven proinflammatory phenotype in MΦs, which may play an integral role in the promotion of obesity-associated insulin resistance.
Introduction
Immune cells infiltrate adipose and other tissues at the onset of weight gain; these cells directly contribute to continued weight gain, adipose inflammation, and systemic insulin resistance associated with obesity (1–3). In this microenvironment, it is likely that particular substrate availability and utilization can alter MΦ2 phenotype. Indeed, fatty acids, lipid metabolism, and fatty acid-binding proteins play an important role in regulating MΦ-mediated inflammation in obesity and diabetes (4–7). However, little work has been completed to date on the metabolic fate of other substrates, such as glucose, in metabolism-dependent reprogramming of MΦ phenotypes.
A range of MΦ phenotypes, or subtypes, exist in which “M1,” or classically activated, proinflammatory MΦs, occupy one extreme of the polarization spectrum, whereas “M2,” or alternatively activated anti-inflammatory MΦs, exist at the opposite end (2). M1 MΦs are activated through the action of classical type 1 T-helper (Th1) inflammatory cytokines interferon-γ (IFNγ) and tumor necrosis factor α (TNFα); these cytokines are produced in response to agents such as saturated fatty acids, cytokines, and Gram-negative bacterial endotoxin (lipopolysaccharide (LPS)). Upon activation, M1 MΦs secrete inflammatory cytokines including TNFα, IL-1β, IL-6, and others. In contrast, M2 MΦs are activated by prototypical type 2 T-helper (Th2) cytokines, IL-4, and IL-13 (2).
Importantly, M1 MΦs preferentially metabolize glucose as an energy substrate (8, 9). Similarly, dendritic cells (10) and T-cells (11–13) exhibit a Warburg-like shift in cellular metabolism upon activation. However, alternatively activated M2 MΦs primarily utilize fatty acids to fuel cellular behavior and activity (8, 9). Taken together, glucose metabolism is central to the function of classically activated MΦs and thus is a potential target for modifying inflammatory responses.
MΦ subtypes modulate inflammation-associated insulin resistance (reviewed in detail in Ref. 2). Broadly, M2 MΦs are immunoregulatory and maintain insulin sensitivity in adipose and liver, whereas M1 MΦs disrupt insulin sensitivity in these tissues. In lean individuals, M2 MΦs reside in adipose tissue and are involved in tissue remodeling and repair (14). At the onset of obesity and as weight gain continues, M1 MΦs infiltrate the adipose tissue contributing to the low grade, chronic inflammation associated with excess adiposity (14). Based upon existing evidence for fuel preferences of M1 versus M2 MΦs, we posited that manipulating MΦ substrate metabolism may serve as a novel approach for controlling macrophage polarization and inflammatory capacity.
We sought to manipulate the MΦ inflammatory response through the modulation of the primary glucose transporter in MΦs, GLUT1 (15–18). To this end, we overexpressed GLUT1 in the RAW264.7 murine MΦ cell line to test the hypothesis that increased intracellular glucose availability and subsequently enhanced glucose metabolism can independently drive a hyperinflammatory response. Through radiotracer, metabolomics, bioenergetics, and expression analysis studies, we have demonstrated that elevated GLUT1-driven glucose metabolism drives reactive oxygen species (ROS) production and expression of proinflammatory mediators. Inhibiting glycolysis or treating cells with an antioxidant reversed GLUT1-mediated proinflammatory elevations. Finally, in vivo detection of GLUT1 in MΦ-laden crown-like structures in adipose tissue and inflammatory loci in livers of obese animals demonstrated important evidence for GLUT1-mediated glucose metabolism in tissue inflammation. Through understanding mechanisms of metabolic reprogramming driven by substrate availability, we provide important insights into the control of inflammation.
EXPERIMENTAL PROCEDURES
Cell Culture and Chemicals
For the GLUT1 construct, Rat Glut1 with a tandem FLAG epitope (DYKDDDDKDYKDDDDK, inserted between amino acids 66 and 67) was cloned into the pEF6 vector (Invitrogen) (11, 19, 20). RAW264.7 (RAW) mouse MΦs were transfected with either the empty vector or Glut1 construct using the AMAXA nucleofector V kit (Lonza, Cologne, Germany). Stable cell lines were established by selecting in 10 μg/ml Blasticidin S (Invitrogen) for 2 weeks. Empty vector MΦs (GLUT1-EV) were diluted serially to obtain clonal isolates. Glut1-transfected MΦs were labeled with a fluorescent anti-FLAG antibody, and Glut1-overexpressing cells (GLUT1-OE) were purified by sterile cell sorting. This population of cells was diluted serially to obtain clonal isolates. RAW cells were maintained in 10 μg/ml Blasticidin S. Studies reported herein were repeated in a second set of clones and the parent heterologous population of GLUT1-EV and GLUT1-OE from which the clones were isolated (data not shown). Lipopolysaccharide (Sigma) was diluted in sterile PBS at 1 mg/ml. Stock reagents were stored at −80 °C under light-free conditions and diluted to appropriate final concentrations in the media. For ROS assays, N-acetylcysteine (Enzo Life Sciences, Inc., Farmingdale, NY) and 2-deoxyglucose (Sigma) were prepared in sterile PBS just before use.
Cells
All cultures were maintained under a fully humidified atmosphere of 95% air and 5% CO2 at 37 °C. RAW264.7 cells (ATCC, Manassas, VA (ATCC# TIB-71)) were grown in Dulbecco's minimal essential media (DMEM) 4.5g/liter (25 mm) glucose supplemented with 10% FBS, 1× l-glutamine, and 1× penicillin/streptomycin antibiotic mix (Sigma). Cell densities were determined by hematocytometer and trypan blue exclusion. For experimental incubations, cells in log-phase growth were plated at a density of 1–5 × 105 cells/ml and allowed to attach overnight. Cells at 80% confluency were then treated as described below. Where indicated, cells were treated with 100 ng/ml LPS for 24 h. Experimental incubations were terminated by directly lysing cells in the appropriate lysis buffer. Cell size, volume, and viability were determined using a SceptorTM handheld automated cell counter (Millipore, Billerica, MA). Cell proliferation was determined by 3-(4, 5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) assay (ATCC) over 24–96 h of culture.
Rodents and Bone Marrow-derived MΦs (BMDM)
All animals were housed in a temperature controlled facility with a 12-h light/dark cycle in compliance with the Duke and University of North Carolina at Chapel Hill Institutional Animal Care and Use Committee protocols. C57BL/6 male mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and bred at the University of North Carolina. To generate C57BL/6-derived BMDM, bone marrow was collected from femur and tibia bones (5, 6). BMDMs were generated by culturing marrow cells in 10-cm non-tissue culture dishes for 6 days with 30% L929 fibroblast-conditioned media and 70% RPMI supplemented with 10% FBS, 1× l-glutamine, and 1× penicillin/streptomycin antibiotic mix. BMDMs were left untreated as naïve unstimulated MΦs (M0) or polarized using 5 ng/ml LPS and 10 ng/ml IFNγ (M1) or 10 ng/ml IL-4 (M2) for 24 h as in Vats et al. (8). To examine adipose and liver in lean and obese rodents, rats were given ad libitum access to either a purified diet containing 10% kcal from fat (low fat diet, Research Diets D07010502, New Brunswick, NJ) or a diet containing 45% kcal from fat (high fat diet, Research Diets, D06011802) as reported in Sampey et al. (4, 5). For tissue collection, animals were anesthetized with tribromoethanol (0.02 ml/g of a 1.25% solution) and euthanized by cervical dislocation. Epididymal white adipose tissue and liver were fixed in 10% formalin and paraffin-embedded for immunohistochemical analysis. To examine in vivo GLUT1 activation and inflammatory gene expression, Myc-epitope-tagged Glut1 knock-in mice (Glut1 myc) mice that express exofacially Myc-tagged GLUT1 at endogenous levels (12) were used (n = 3, 12-week-old females). Mice were sacrificed, and an adipose tissue single cell suspension was generated using a GentleMACS tissue dissociator (Miltenyi Biotec Inc., Cambridge, MA) according to the manufacturer's recommended protocol. Cells were stained with anti-F4/80-APC (eBioscience, San Diego, CA), mouse-anti-Myc tag (Millipore clone 4A6), and V450-anti mouse IgG, then fixed in 1% paraformaldehyde and permeabilized with methanol. After permeabilization, cells were stained with anti-IL-6-phycoerythrin (PE) and anti-TNFα-FITC antibodies (eBioscience) and analyzed using a MacsQuant flow cytometer (Miltenyi Biotec). Data show the mean ± S.E. of the mean fluorescence intensity.
Immunocytochemistry and Immunohistochemistry
GLUT1 localization was determined in RAW264.7 GLUT1-OE cells. Cells were washed in PBS, 2% FBS and blocked with Fc block (1:100 in 5% BSA for 10 min in 5% rat serum). Primary rabbit anti-FLAG (Sigma #F7425), R-PE donkey anti-rabbit (Jackson Immuno catalog 711-116-152), and DAPI were used, and images were captured under a Nikon Microscope (Melville, NY). 5-μm sections of liver and epididymal white adipose from lean and obese rats were used to examine GLUT1 expression in vivo by immunohistochemistry (4).
Western Immunoblots
Cells were washed twice with PBS then lysed in radioimmunoprecipitation assay buffer (50 mm Tris, 150 mm NaCl, 0.1% SDS, 0.5% deoxycholate, 1% Triton X-100) with 1× protease and phosphatase inhibitors (Sigma). Protein concentration of cleared lysates was determined using the BCA assay (Thermo Fisher Scientific, Rockford, IL). Samples were treated with 4× Laemmli (4% SDS, 40% glycerol, 10% β-mercaptoethanol, 125 mm Tris-Cl, pH 6.8) and incubated for 15 min at room temperature. Anti-Glut1 (ab40084, Abcam, Cambridge, MA) primary antibody or anti-actin antibody (MAB1501) (Millipore), followed by peroxidase-conjugated goat anti-mouse secondary antibody (NXA931, GE Healthcare) were used to probe membranes and developed by chemiluminescence (Amersham Biosciences) and quantitated using a VersaDoc 4000 instrument and software (Bio-Rad). Blots were quantitated from three separate experiments. All lanes were normalized to actin for loading, and on each blot, GLUT1 expression in the GLUT1-EV cells was set to 1.
qPCR and qPCR Arrays
mRNA was isolated (Qiagen RNeasy mini kit, Qiagen, Valencia, CA) and reverse-transcribed into cDNA using the iScript cDNA synthesis kit (Bio-Rad). The expression of specific genes was quantitated by qPCR using Assay-On-Demands (ABI) on an ABI 7900HT machine (Invitrogen) (21). Mouse Innate and Adaptive Immune Responses Array qPCR arrays were purchased from SABiosciences (Qiagen). n = 3 for GLUT1-OE and GLUT1-EV MΦs.
Microarray Expression Analysis
Gene expression analysis was performed as previously reported (22) using Agilent microarrays (G4846A) as per the manufacturer instructions. A two-color labeling was performed using Low Input Quick Amp labeling kits (Agilent, Santa Clara, CA) with GLUT1 overexpressing cells labeled with Cy3 and a reference channel (Universal Mouse Reference, Stratagene, Santa Clara, CA) labeled with Cy5. Data analyses were performed using R (Version 2.9.2). Genes were filtered with only genes where >70% of microarrays had a signal >10 dots per inch in both channels included. Data were Lowess-normalized, and missing data were imputed using k-nearest neighbor imputation (with k = 6). Data were analyzed by 2-class significance analysis of microarrays (SAM) to identify genes that significantly changed by GLUT1 overexpression (i.e. GLUT1-EV versus GLUT1-OE). Significant probes were collapsed by gene and visualized using Java Treeview. Significant genes were evaluated for ontological enrichment using Ingenuity Pathway Analysis (Ingenuity® Systems), with Benjamini-Hochberg (B-H) multiple testing correction. Significant functions and pathways were defined as those with B-H p values less than 0.05.
Proteome Profiler Cytokine Array
GLUT1-EV and GLUT1-OE RAW264.7 MΦs were plated into 24-well plates and either treated with media alone or 100 ng/ml LPS for 24 h. Conditioned media was assayed using Proteome Profiler Array Mouse Cytokine array panel A (ARY006, R and D Systems, Inc., Minneapolis, MN) following the manufacturer's protocol. Digital images were quantified using the Versadoc 4000, and relative abundance with background subtracted was normalized to internal standards, as in Sampey et al. (5).
Cellular Bioenergetics-Seahorse Analyses
60,000 GLUT1-EV or GLUT1-OE MΦs were seeded into each well of a Seahorse Bioscience (Billerica, MA) tissue culture plate and incubated overnight at 37 °C in an atmosphere of 5% CO2. Before analysis, cells were washed twice with DMEM assay media (sodium bicarbonate- and glucose-free DMEM supplemented with glutamine and penicillin/streptomycin, pH 7.4) and incubated for 1 h at 37 °C without CO2. Measures of glycolytic rate and glycolytic capacity were determined by recording extracellular acidification rates (ECAR, milli pH/min) and oxygen consumption rates (OCR, pmol/min) on a Seahorse Bioscience XF24–3 Extracellular Flux Analyzer. The injection of glucose was used to measure glycolytic rate (final concentration 25 mm; Sigma), the injection of oligomycin was used to measure glycolytic capacity (final concentration 2.5 μm; Sigma), and the injection of 2-deoxyglucose was used to blunt glycolysis (final concentration 20 mm). Background from cell-free wells was subtracted. Cells were lysed, and protein concentrations were determined using BCA protein assay. n = 6 replicates per treatment for ECAR and OCR. Data were normalized to protein in each well. Experiments were repeated n = 3 times.
Radiotracer Glucose Uptake Assay
GLUT1-EV and GLUT1-OE RAW264.7 MΦs were plated at 1.5 × 105 cells/well into 24-well plates and allowed to attach overnight. Cells were treated with vehicle or 100 ng/ml LPS for 24 h. The cells were washed twice with 0.5 ml of 37 °C 1× PBS and then incubated for 10 min at 37 °C in 0.5 ml/well DMEM (136 mm NaCl, 4.7 mm KCl, 1.25 mm CaCl2, 1.25 mm MgSO4, 10 mm HEPES, pH 7.4) with 0.5 μCi/ml deoxy-d-[2-3H]glucose (PerkinElmer Life Sciences). Cells were washed twice with 0.5 ml of cold 1× PBS with 20 mm d-glucose and then lysed in 0.5 ml of lysis buffer (0.025% SDS, 1% Triton X-100 in 1× PBS). Lysates were centrifuged at 20,000 × g for 5 min at 4 °C, and the protein concentration was determined with a BCA assay. Remaining lysate was counted by scintillation. The data are expressed as dpm/mg of protein.
Radiotracer Glucose Oxidation and Glycogen Synthesis
GLUT1-EV and GLUT1-OE RAW264.7 MΦs were plated as above. Glucose oxidation to CO2 was measured by incubating cells with ubiquitously labeled [14C]glucose for 2 h and then measuring [14C]CO2 in the media. Briefly, the cells were washed twice with 0.5 ml of 37 °C 1× PBS and then incubated for 120 min at 37 °C in 0.5 ml/well DMEM with 2 μCi/ml [14C]uniformly labeled d-glucose (PerkinElmer Life Sciences). [14C]CO2 released from acidified media was captured and subjected to scintillation counting. The data are expressed as dpm/mg of protein/minute. Cells were then washed twice with 0.5 ml of iced 1× PBS with 20 mm d-glucose and lysed in 0.5 ml of lysis buffer (30% KOH saturated with Na2SO4) to ethanol-precipitate glycogen, which was measured by scintillation counting. The data are expressed as dpm/mg of protein/min.
Lactate Assay
Media from experiments was saved in aliquots at −80 °C and assayed for lactate content. Briefly, fresh NAD+ was added to assay buffer (175 mm hydrazine sulfate, 68 mm glycine, 2.9 mm EDTA, 11.3 mm NAD+, pH 9.5) just before assay. In a 96-well plate, 200 μl of assay buffer and 40 μl of lactate standard or media were combined in each well. The plate was then pre-read at 340 nm followed by the addition of 1 unit of lactate dehydrogenase in a 10-μl volume of water. The plate was mixed and read every minute for 30 min at 340 nm. The standard curve measured concentrations of 0, 1, 2, 3, 4, 5, and 10 μm lactate. The lactate concentration in each sample was determined from the standard curve.
Metabolomics
GLUT1-EV and GLUT1-OE MΦs were plated at 5 × 105 cells/ml and allowed to attach overnight. Fresh medium was added, and cells were grown for an additional 24 h. PBS-washed cells were detached by scraping and pelleted for analysis by Metabolon, Inc. (Durham, NC) as in Bhatt et al. (23). Data are presented as relative measures of “scaled intensity” after normalization to protein and median scaling to 1. Missing values were imputed with the minimum.
Reactive Oxygen Species Production
1.5 × 106 GLUT1-EV or GLUT1-OE MΦs were seeded into 6-well plates. Cells were either untreated or incubated with 5 mm N-acetyl-cysteine (NAC) or 1 mm 2-deoxyglucose (2-DG) and incubated overnight in DMEM at 37 °C in an atmosphere of 5% CO2. Total ROS and superoxide levels in live, unfixed cells were measured by a total ROS/superoxide kit per manufacturer's instructions (Enzo Life Sciences) and analyzed using a Beckman Coulter CyAn ADP flow cytometer (Brea, CA). Flow cytometry data were analyzed using FlowJo software (TreeStar, Ashland, OR), and median fluorescence intensity was used to compare total ROS and superoxide produced by GLUT1-EV or GLUT1-OE MΦs. At least 5 × 103 cellular events were analyzed.
Statistics
Statistical differences between experimental groups were determined using statistics software within GraphPad Prism (GraphPad Software, Inc., La Jolla, CA). 1-Way analysis of variance was used to determine the existence of statistically significant differences between RAW, GLUT1-EV, and GLUT1-OE MΦs. Non-paired Student's t test was then used to determine p values comparing GLUT1-OE MΦs to the two control cell lines. For all experiments, representative results are reported for at least n = 3. Experiments were conducted with n = 3–6 replicates per assay. All data are shown as means ± S.E. For metabolomics study, two-way analysis of variance with p < 0.05 was considered significant.
RESULTS
GLUT1 Was the Predominant Glucose Transporter in MΦs
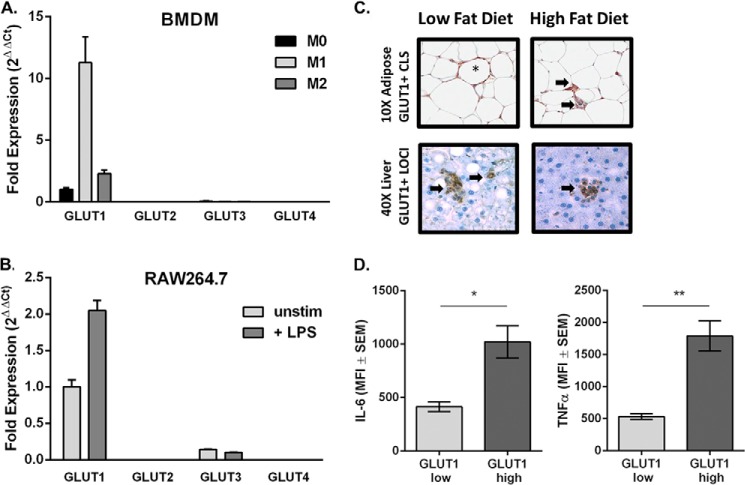
BMDM from C57BL/6J mice were plated and left untreated as naïve unstimulated MΦs (M0) or polarized to M1 or M2 as described under “Experimental Procedures.” qPCR analysis of the glucose transporters GLUTs 1–4 revealed that GLUT1 mRNA was up-regulated 10-fold by M1 polarization compared with M0 BMDMs (Fig. 1A). M2-polarized MΦs up-regulated GLUT1 expression by 2-fold compared with M0. GLUT3 was minimally detected in M0 and down-regulated with M1 polarization. GLUT2 and -4 were undetected (Fig. 1A). Similar to BMDMs, the most abundant glucose transporter in the RAW MΦ cell line was GLUT1 followed by a low level of expression of GLUT3. Expression of GLUT2 or GLUT4 was not detected (Fig. 1B). RAW MΦs up-regulated GLUT1 mRNA with no change in GLUTs 2–4 in response to 24-h LPS stimulation (Fig. 1B).
FIGURE 1.
GLUT1 is the primary transporter on proinflammatory MΦ and is detected in obesity-associated crown-like structures. qPCR expression analysis of GLUT1–4 in BMDM (A) and RAW MΦs (B). A, bone marrow was isolated from wild type C57BL/6 mice. BMDMs were left as unstimulated naïve cells (M0) or polarized toward M1 or M2 phenotype. Fold expression from M0 GLUT1 was set to 1. B, RAW MΦs were plated and left unstimulated or stimulated with 100 ng/ml LPS for 24 h. Fold expression from untreated GLUT1 was set to 1. C, GLUT1+ CLS (crown-like structure) (*) and inflammatory loci (→) are indicated in low fat diet-fed lean and high fat diet-fed obese adipose (top images) and livers (bottom images). D, adipose tissue macrophages derived from mice were stained for macrophages (F4/80+), GLUT1 (myc), IL-6 (*, p = 018), and TNFα (*, p = 0.0062). n = 3. Data show the mean ± S.E. of the mean fluorescence intensity (MFI).
GLUT1 Was Up-regulated in Inflamed Obese Adipose and Liver
To determine the relevance of MΦ GLUT1 in vivo, we stained for GLUT1 expression in adipose and liver of lean and diet-induced obese rats. GLUT1-positive staining was associated with inflammatory MΦ infiltrates in epididymal white adipose tissue and liver from obese animals fed a high fat diet compared with low fat diet-fed lean controls. GLUT1-positive staining was co-localized with MΦs in crown-like structure (CLS) and inflammatory loci (Fig. 1C) (4, 5). In addition, to confirm in vivo expression of GLUT1 and cytokine expression, we isolated F4/80+ macrophages from adipose tissue of mice (Fig. 1D). Macrophages that expressed high levels of surface GLUT1 contained significantly greater levels of IL-6 (p = 0.018) and TNFα (p = 0.0062) compared with macrophages expressing low levels of GLUT1.
Generation of GLUT1 Overexpression MΦ Model
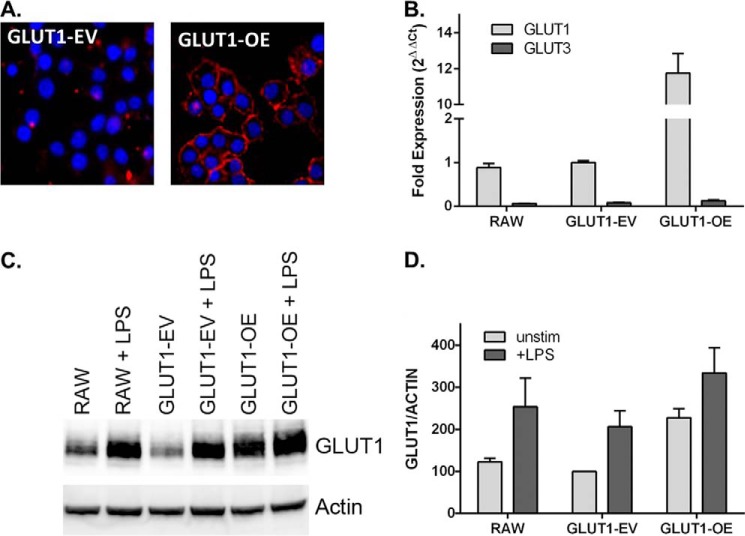
To investigate the role of glucose metabolism in the inflammatory state of MΦs, GLUT1 was stably transfected into RAW MΦs. Clonal lines stably expressing either pEF6 vector alone (GLUT1-empty vector (GLUT1-EV) or pEF6 overexpressing GLUT1 (GLUT1-OE) were generated and grown according to ATCC recommendations in 4.5 g/liter glucose containing DMEM. Immunocytochemical staining of GLUT1-EV and GLUT1-OE clones demonstrated successful overexpression of GLUT1 and correct localization in the GLUT1-OE cell line, as seen in the red punctate and primarily membrane-localized areas (Fig. 2A). qPCR of GLUT1 in untreated RAW, GLUT1-EV, and GLUT1-OE MΦs revealed a nearly 12-fold overexpression of GLUT1 mRNA in the GLUT1-OE cells compared with the other MΦ groups (Fig. 2B). GLUT1-OE MΦs expressed 2.28-fold greater GLUT1 protein levels relative to GLUT1-EV and RAW MΦs (Fig. 2, C and D). LPS stimulation resulted in a 2-fold increase in GLUT1 protein in RAW and GLUT1-EV cells and a 1.5-fold increase in GLUT1-OE cells. The overexpression of GLUT1 at the protein level in this model was at physiologic levels and was comparable to a 1.64-fold induction in GLUT1 protein on adipose derived macrophages after a 3-h intraperitoneal LPS injection (data not shown) as well as 1.8-fold LPS-induced elevations in glucose uptake in BMDMs. The viability and growth characteristics of the MΦs were not altered by the overexpression of GLUT1 as measured by trypan blue exclusion and MTT assay, respectively (data not shown). Using flow cytometry, neither cell size nor cell volume was altered by GLUT1: RAW (10.48 μm, 0.60 pl), GLUT1-EV (9.97 μm, 0.52 pl), and GLUT1-OE (10.53 μm, 0.61 pl).
FIGURE 2.
Stable overexpression of GLUT1 in RAW264.7 MΦs. A, immunolocalization of GLUT1 to the cell surface (red) in GLUT1-OE compared with GLUT1-EV cells using an anti-FLAG antibody (nuclei are stained blue with DAPI). B, qPCR of GLUT1 and GLUT3 in RAW, GLUT1-EV, and GLUT1-OE. Fold expression is normalized to GLUT1-EV, set to 1. GLUT2 and GLUT4 were not detected. n = 5 separate experiments ± S.E. C, representative Western immunoblot of GLUT1 and loading control actin in RAW, GLUT1- EV, and GLUT1-OE cell lysates ± LPS. D, quantification of immunoblots from three separate experiments ± S.E. For each immunoblot, all lanes are normalized to actin. Unstimulated GLUT1-EV was set to 1.
GLUT1 Overexpression Increased Glucose Metabolism and Metabolites in the Pentose Phosphate Pathway (PPP)
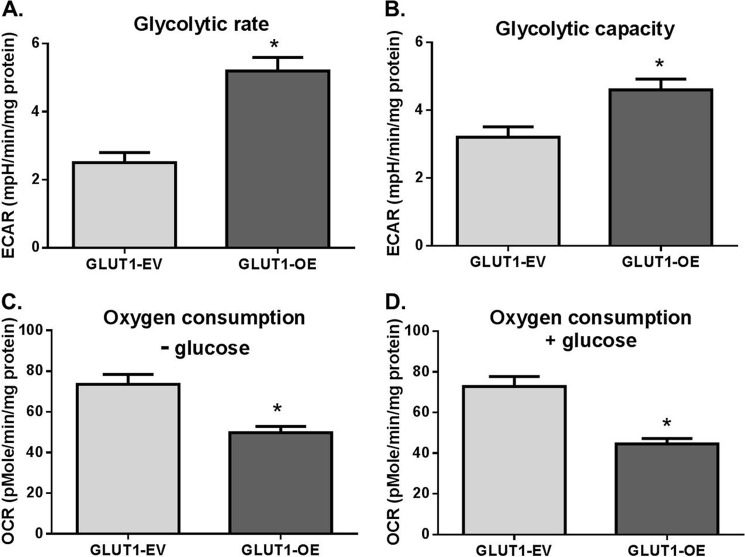
We first analyzed GLUT1-EV and GLUT1-OE cellular bioenergetics through examining ECARs (measure of glycolysis) and OCR (measure of mitochondrial respiration) using the Seahorse Bioanalyzer. ECAR was more than doubled in GLUT1-OE compared with GLUT1-EV MΦs (p = 0.0003, Fig. 3A). The addition of oligomycin (an inhibitor of ATP synthase) to test glycolytic capacity induced a greater increase in GLUT1-OE MΦs ECAR compared with GLUT1-EV (p = 0.01, Fig. 3B). Inhibiting glycolysis with 2-DG reduced ECARs equivalently in GLUT1-OE and GLUT1-EV MΦs, indicating no difference in glycolytic reserve (not shown). Interestingly, GLUT1-OE MΦs demonstrated significantly reduced OCR compared with GLUT1-EV MΦs throughout the experiment regardless of whether glucose was present in the assay media (Fig. 3, C and D, p < 0.002 and p < 0.0005, respectively).
FIGURE 3.
GLUT1 up-regulated glycolytic rates and capacity and blunted respiratory capacity. ECAR and OCR were measured using the Seahorse Bioanalyzer. Glycolytic rate (A; *, p = 0.0003) and glycolytic capacity (B; *, p = 0.01) were measured. Glycolytic reserve was equal in GLUT1-EV and GLUT1-OE (not shown). Oxygen consumption was measured using the Seahorse before glucose addition to the media (C, − glucose, *, p = 0.002) and after glucose injection into the assay (D, + glucose, *, p = 0.0005). n = 3 experiments with n = 6 replicates were conducted, and data were normalized to protein in each well.
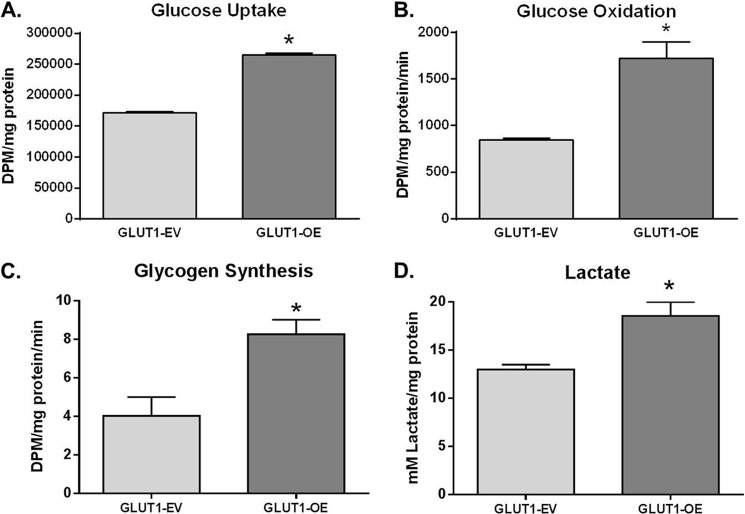
Radiotracer studies were then utilized to directly quantitate glucose uptake and metabolism. GLUT1-OE MΦs took up 1.45-fold more [2-3H]DG compared with GLUT1-EV MΦs (p < 0.0001 GLUT1-OE versus GLUT1-EV, Fig. 4A). GLUT1-mediated [14C]-glucose oxidation was significantly increased in GLUT1-OE compared with RAW or GLUT1-EV MΦs (p = 0.001 GLUT1-OE versus GLUT1-EV, Fig. 4B). [14C]Glycogen synthesis was doubled compared with GLUT1-EV MΦs (p = 0.03 GLUT1-OE versus GLUT1-EV, Fig. 4C). Based on increased glycolytic flux as measured by ECAR, we measured lactate production directly. Lactate concentration in the media demonstrated that GLUT1-OE MΦs displayed a 1.43-fold higher rate of glycolysis compared with controls (p = 0.005 GLUT1-OE versus GLUT1-EV; Fig. 4D).
FIGURE 4.
GLUT1 gain of function up-regulates glucose metabolism in GLUT1-OE MΦs compared with GLUT1-EV. A, [2-3H]DG uptake was measured in GLUT1-EV and GLUT1-OE cells. *, p < 0.0001 GLUT1-OE versus GLUT1-EV. B, [14C] ubiquitously labeled glucose oxidation was measured. *, p = 0.001 GLUT1-OE versus GLUT1-EV. C, [14C] ubiquitously labeled glucose incorporation into glycogen was measured as above. *, p = 0.03 GLUT1-OE versus GLUT1-EV. D, lactate was measured in conditioned media of GLUT-EV and GLUT1-OE MΦs cultured for 24 h. *, p = 0.005 GLUT1-OE versus GLUT1-EV.
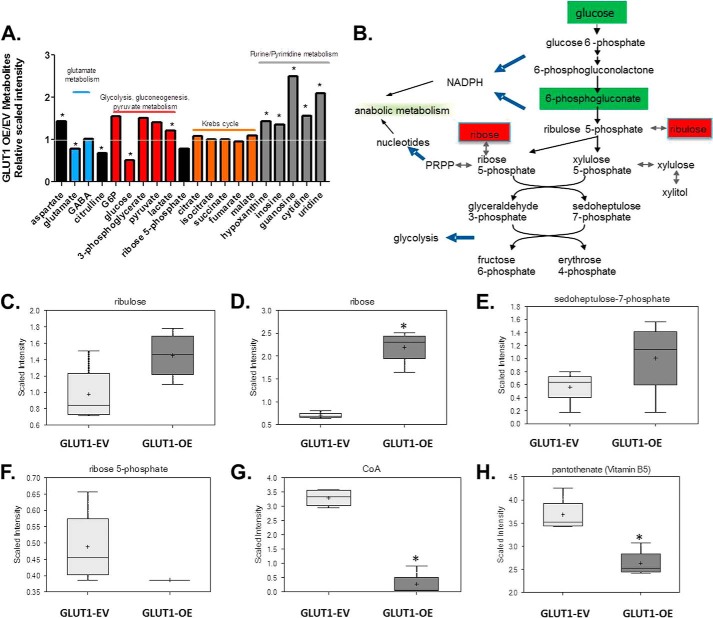
Comprehensive metabolomic analysis was performed on GLUT1-EV and GLUT1-OE cell lysates to examine alterations in glycolytic and other pathway metabolites. 152 biochemicals (45 up-regulated and 107 down-regulated) were significantly different (p < 0.05) between unstimulated GLUT1-OE compared with GLUT1-EV supplemental Table 1). Several glycolytic intermediates were significantly up-regulated (Fig. 5A and supplemental Table 1). GLUT1-OE MΦs contained significantly less intracellular glucose (p = 0.033), likely due to increased glycolysis, as evidenced by elevated intracellular lactate concentrations (p = 0.049) (Fig. 5A and supplemental Table 1). In addition, greater amounts of metabolites in the PPP were detected (Fig. 5B). Specifically, ribulose and sedoheptulose 7-phosphate were elevated by 1.5- and 1.8-fold, respectively, with significantly 3.12-fold greater levels of ribose (p = 4.55 × 10−06) detected in GLUT1-OE MΦ lysates compared with the GLUT1-EV MΦs (Fig. 5, B–E, and supplemental Table 1), whereas ribose 5-phosphate concentration was blunted by 0.79-fold in GLUT1-OE compared with GLUT1-EV (Fig. 5F). There were no significant alterations in steady state measures of TCA cycle intermediates (Fig. 5A). However, content of purine and pyrimidine metabolites was significantly enhanced (supplemental Table 1). Furthermore, cholesterol content was significantly increased by 1.22-fold (p = 0.02) (supplemental Table 1). There was also a significant blunting of coenzyme A concentration by 0.8-fold (CoA, p = 0.0002; Fig. 5G and supplemental Table 1) as well as pantothenate (vitamin B5), a necessary component of functional CoA (p = 0.00007) (Fig. 5H and supplemental Table 1).
FIGURE 5.
GLUT1-OE drives pentose phosphate pathway. A, metabolomic analysis of cell lysates reveals increases in glycolysis, the pentose phosphate pathway, and purine/pyrimidine metabolism. B, diagram of pentose phosphate pathway (PRPP, phosphoribosyl pyrophosphate). Red and green-shaded cells indicate p ≤ 0.05; red indicates that the mean values are significantly higher, and green values significantly lower for GLUT1-OE versus GLUT1-EV. Ribulose (C) and ribose (*, p = 4.55 × 10−6) (D), sedoheptulose-7-phosphate (E), ribose-5-phosphate (F), coenzyme A (*, p = 0.0002) (G), and pantothenate (*, p = 0.00007) (H) were modulated by GLUT1 expression.
GLUT1 Drives a Proinflammatory Phenotype and Alters Redox Status of MΦs
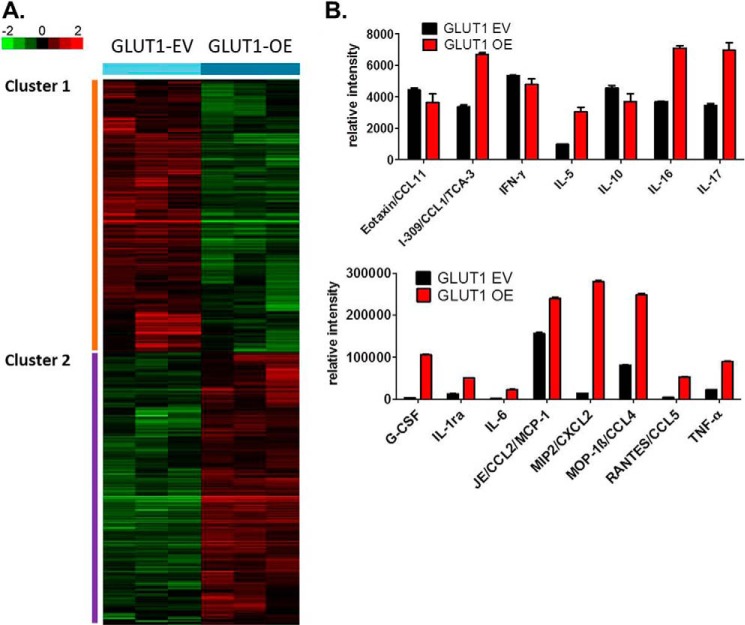
Having established that GLUT1-OE MΦs take up and metabolize more glucose compared with the GLUT1-EV MΦs, we next set out to test our central hypothesis that increasing glucose metabolism in MΦs renders these cells more proinflammatory. Supervised two-class analysis comparing GLUT1-OE to GLUT1-EV revealed that 1547 genes were differentially regulated by GLUT1 (775 genes down-regulated, 772 genes up-regulated) using a false discovery rate of 3.2% (supplemental Table 2). Significantly regulated genes were visualized using hierarchical clustering (Fig. 6A). Using Ingenuity Pathway Analysis, the genes in cluster 1 (Fig. 6A) were defined by expression of genes involved in the cellular function and maintenance, protein synthesis, cellular development, cellular growth and proliferation, and cell morphology functional pathways. Down-regulated canonical pathways included cross-talk between dendritic cells and Natural Killer cells, antigen presentation pathway, communication between innate and adaptive immune cells, allograft rejection signaling, and role of pattern recognition signaling in recognition of bacteria and viruses (Table 1). Cluster 2 (Fig. 6A) was associated with a GLUT1-mediated increase in expression of genes involved in cell cycle, DNA replication, recombination, and repair, cellular assembly, and organization, cell-to-cell signaling and interaction, and cellular function and maintenance. Up-regulated canonical pathways in cluster 2 included cholesterol biosynthesis I, II, III and caveolar-mediated endocytosis signaling (Table 1).
FIGURE 6.
GLUT1 gain of function drives increased expression of inflammatory mediators. A, microarray analysis of untreated GLUT1-EV and GLUT1-OE MΦs was conducted. Supervised two-class significance analysis of microarray analysis identified 1,547 genes differentially regulated by GLUT1 at a false discovery rate of 3.2% (supplemental Table 2). 775 were down-regulated and 772 were up-regulated in a GLUT1-dependent manner. Cluster analysis revealed two clusters of genes regulated by GLUT1. B, media from GLUT1-EV and GLUT1-OE MΦs were examined for secretion of inflammatory mediators using a proteome profiler array and normalized to protein lysates. Significantly (p < 0.05) regulated proteins are shown with the exception of eotaxin and IL-10.
TABLE 1.
Top canonical pathways regulated by GLUT1 expression
mRNA levels were quantified in GLUT1-EV and GLUT1-OE macrophages by microarray. Pathways were identified by Ingenuity Pathway Analysis.
| Ingenuity canonical pathways | p Value | Genes in pathway |
|---|---|---|
| Down-regulated | ||
| Cross-talk between dendritic cells and natural killer cells | 6.13E-07 | IL2RG, HLA-B, CD83, NFKB2, TREM2, ITGAL, HLA-G, CD28, CSF2RB, CD80, TNFRSF1B, TNF, HLA-E, PVRL2 |
| Antigen presentation pathway | 7.11E-07 | HLA-G, B2M, NLRC5, HLA-B, PSMB8, TAP1, TAPBP, HLA-E, MR1 |
| Communication between innate and adaptive immune cells | 8.11E-06 | HLA-G, B2M, CD28, CD80, IL1RN, HLA-B, TLR1, CD83, CCL5, TNFRSF13B, TNF, HLA-E |
| Role of pattern or recognition receptors in recognition of bacteria and viruses | 1.62E-05 | OAS1, C5AR1, MAPK9, CCL5, NFKB2, PRKCG, IFIH1, CLEC7A, PRKCD, SYK, TLR1, DDX58, TNF, C3AR1 |
| Allograft rejection signaling | 4.17E-05 | HLA-G, B2M, CD28, H2-T10, CD80, H2-K2/H2-Q9, HLA-B, TNF, HLA-E |
| Up-regulated | ||
| Cholesterol biosynthesis I | 1.36E-03 | SQLE, DHCR24, HSD17B7, TM7SF2 |
| Cholesterol biosynthesis II (via 24,25-dihydrolanosterol) | 1.36E-03 | SQLE, DHCR24, HSD17B7, TM7SF2 |
| Cholesterol biosynthesis III (via desmosterol) | 1.36E-03 | SQLE, DHCR24, HSD17B7, TM7SF2 |
| Superpathway of cholesterol biosynthesis | 3.93E-03 | SQLE, DHCR24, HSD17B7, TM7SF2, HMGCS1 |
| Noradrenaline and adrenaline degradation | 5.41E-03 | ADH7, Aldh1a7, PECR, SMOX, ALDH7A1 |
Because GLUT1 overexpression and subsequent enhanced glucose metabolism regulated important signaling and metabolic pathways that could affect MΦ inflammatory function, we next investigated the effect of GLUT1 overexpression on inflammatory activation using quantitative gene and protein expression analysis. First, using a targeted qPCR array for 84 genes involved in the innate and adaptive immune response, GLUT1-OE was found to significantly regulate 12 genes including a 5-fold increase in serpine1, also known as plasminogen activator inhibitor 1 (PAI-1, p < 0.05, Table 2). We next examined secreted cytokines using a proteome profiling array. GLUT1-OE cells significantly increased secretion of 11 cytokines and growth factors (Fig. 6B and Table 3). Notably, G-CSF, IL-6, CXCL2, and RANTES all show a >10-fold increase compared with GLUT1-EV MΦs, whereas CXCL1, TNF-α, IL-1 receptor antagonist, IL-5, CCL4, IL-17, and CCL1 were up-regulated by at least 2-fold (Fig. 6B). Three cytokines (IL-2, IL-27, and CXCL10) were significantly reduced by GLUT1-OE (Table 3).
TABLE 2.
GLUT1-mediated regulation of inflammatory genes
mRNA levels were quantified in GLUT1-EV and GLUT1-OE macrophages by qPCR array. Using a confidence interval of 95% and a p value < 0.05, 4 genes were up-regulated (boldface type), and 6 were down-regulated >2-fold (italic type) when comparing GLUT1-OE to GLUT1-EV.
| PCR array (OE/EV) | -Fold | p value |
|---|---|---|
| SERPINE1 | 5.36 | 0.014 |
| CD55 | 3.06 | 0.001 |
| IL1R2 | 2.30 | 0.005 |
| TREM1 | 2.20 | 0.015 |
| DEFB4 | 5.17 | 0.041 |
| CASP1 | 4.19 | 0.003 |
| CCR3 | 3.50 | 0.005 |
| COLEC12 | 3.17 | 0.033 |
| TLR10 | 2.60 | 0.004 |
| IL1F10 | 2.46 | 0.001 |
TABLE 3.
GLUT1-mediated regulation of inflammatory protein secretion
Protein levels were quantified in conditioned media from GLUT1-EV and GLUT1-OE macrophages by proteome profiling. Proteins up (boldface type)- or down (italic type)-regulated in the GLUT1-OE cells compared to the GLUT1-EV cells are indicated. Keratinocyte-derived chemokine, KC; interleukin-1 receptor antagonist, IL-1ra.
| Cytokine (OE/EV) | -Fold |
|---|---|
| G-CSF | 31.90 |
| MIP2/CXCL2 | 19.87 |
| IL-6 | 13.42 |
| RANTES/CCL5 | 12.64 |
| KC/CXCL1 | 7.32 |
| TNF-α | 4.05 |
| IL-1ra | 3.98 |
| IL-5 | 3.13 |
| MOP-1β/CCL4 | 3.06 |
| IL-17 | 2.02 |
| I-309/CCL1/TCA-3 | 2.00 |
| IL-2 | −1.45 |
| IP-10/CXCL10/CRG-2 | −1.30 |
| IL-27 | −1.28 |
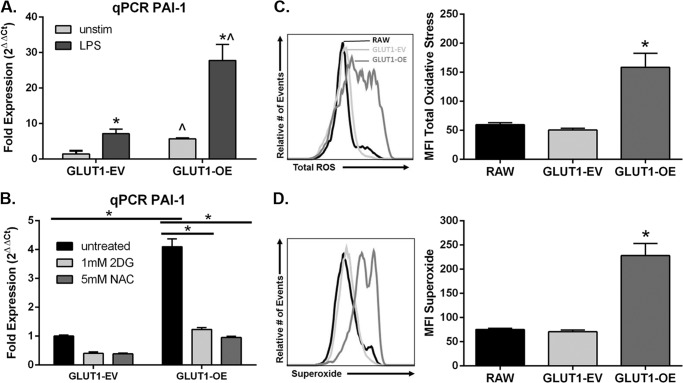
PAI-1 is a proinflammatory mediator and is established to be glucose-regulated (24). Single gene qPCR validated that PAI-1 was up-regulated (p = 0.009, Fig. 7A; p = 0.0001, Fig. 7B) in the GLUT1-OE MΦs compared with the GLUT1-EV MΦs, confirming findings from microarray (supplemental Table 2) and the qPCR array (Table 2). LPS stimulation for 24 h increased PAI-1 expression similarly between GLUT1-EV and GLUT1-OE by 5.0 (p = 0.02) and 4.9-fold (p = 0.008), respectively. LPS treatment did not modify the GLUT1-mediated increase in PAI-1 (4.0-fold in unstimulated (p = 0.009) and 3.9-fold (p = 0.012) in LPS-stimulated macrophages; Fig. 7A). To determine whether the increased PAI-1 expression was dependent upon GLUT1-mediated glycolysis, cells were exposed to 1 mm 2-DG for 24 h. Inhibition of glycolysis by 2-DG significantly decreased expression of PAI-1 in GLUT1-OE MΦs, restoring expression to levels detected in untreated GLUT1-EV MΦs (p = 0.0001, Fig. 7B).
FIGURE 7.
GLUT1 overexpression inflammatory mediator production is dependent upon glycolysis and ROS production. A, qPCR of PAI-1 was measured in GLUT1-EV and GLUT1-OE MΦs left unstimulated or treated with 100 ng/ml LPS for 24 h. Graphs are of combined data from three independent experiments ± S.E. *, p < 0.05 versus unstimulated; ^, p < 0.05 versus GLUT1-EV). B, qPCR of PAI-1 was measured in GLUT1-EV and GLUT1-OE MΦs left untreated or co-treated with 1 mm 2-DG to inhibit glycolysis or 5 mm NAC to quench ROS for 24 h. Graphs are of combined data from three independent experiments ± S.E. GLUT1-EV is set to 1. *, p = 0.0001. C, ROS production was measured in unstimulated RAW, GLUT1-EV, and GLUT1-OE cells. MFI, mean fluorescence intensity. Total ROS (C) and superoxide (D) in RAW, GLUT1-EV, and GLUT1-OE were detected. Graphs shown are representative of three separate experiments done in triplicate. Means ± S.E. are shown. *, p < 0.0005 versus RAW and GLUT1-EV.
Finally, ROS production is a hallmark of M1 MΦ polarization (2). Based on the increased glycolytic rate of the GLUT1-OE MΦs, we hypothesized that elevated ROS may be the mechanism linking increased glucose metabolism to proinflammatory cytokine production. Metabolomic analysis indicated a shift in intracellular redox balance with GLUT1 overexpression, such that that GLUT1-OE had a greater GSSG/GSH ratio (0.764) than GLUT1-EV (0.337) (supplemental Table 1). To examine ROS production directly, RAW, GLUT1-EV, and GLUT1-OE MΦs were labeled with fluorescent probes to measure total ROS and superoxide. Indeed, mean fluorescent intensity of GLUT1-OE MΦs was 3-fold greater than RAW cells and GLUT1-EV for both total ROS (p < 0.0005, Fig. 7C) and superoxide (p < 0.0005, Fig. 7D). To test the dependence of PAI-1 expression on ROS signaling, GLUT1-EV, and GLUT1-OE, MΦs were incubated with 5 mm NAC for 24 h to quench ROS, and PAI-1 expression was again assessed by qPCR. NAC treatment resulted in a complete reversal in the up-regulation of PAI-1 in the GLUT1-OE MΦs (p = 0.0001, Fig. 7B).
DISCUSSION
Immune cell activation in response to infection or other stressors is a metabolically expensive event (2). To date there is evidence that MΦ glucose metabolism plays a key role in mounting the robust, proinflammatory response to bacterial infection. This proinflammatory response includes increased expression of GLUT1 (SLC2A1), glucose-6-phosphate dehydrogenase (the rate-limiting enzyme in the PPP), hexokinase, and increased glucose uptake (9, 25–28). Other stresses including alcohol exposure, burn, and sepsis also up-regulate GLUT1 expression on MΦs (data not shown and Refs. 29 and 30). These data suggest that glucose is the primary fuel metabolized in proinflammatory MΦs.
However, little is known about the metabolic underpinnings controlling how MΦ substrate metabolism regulates MΦ polarization during states of chronic stress like obesity. One potential modifier of MΦ polarization is the availability of fuel substrates within the tissue microenvironment, such as high glucose in diabetes or elevated fatty acids with hypertriglyceridemia and with elevated lipolysis (2). Indeed, Chait and co-workers (31–33) demonstrated that elevated glucose exposure drives inflammation in cultured adipocytes as well as adipose tissue MΦ infiltration.
Adipose tissue MΦs displaying a proinflammatory M1-like phenotype interfere with insulin sensitivity and perpetuate obesity-induced inflammation (2). Strategies that modulate substrate metabolism in MΦs would be innovative approaches to address insulin resistance and diabetes. To this end we investigated whether manipulating the amount of glucose substrate available to MΦs for metabolism by modulating the expression of the GLUT1 transporter would alter the inflammatory characteristics of MΦs. Herein, we report that GLUT1 is the primary glucose transporter in murine BMDM and RAW264.7 MΦs and that proinflammatory activation of MΦs either through M1 polarization of BMDM or LPS stimulation of RAW MΦs increased GLUT1 expression. When GLUT1 was overexpressed in MΦs, elevations in glycolytic rates and lactate production in conjunction with increased glucose oxidation and glycogen storage were detected, suggesting that there was an overall increased flux though glucose-dependent metabolic pathways within the cell. Metabolomic analysis also demonstrated activation of the PPP. The PPP utilizes glucose to generate NADPH for use in biosynthetic pathways, ROS production, and ribose sugars for nucleotide biosynthesis. Increased levels of PPP metabolic intermediates in GLUT1-OE indicate glucose availability in excess of energetic requirements of the cell. Bioenergetic analysis further demonstrated that GLUT1-overexpressing cells are highly glycolytic, with blunted overall oxidative capacity compared with controls. GLUT1-overexpressing cells also contained less CoA and B vitamin pantothenate (necessary for CoA generation). The lack of CoA in GLUT1-OE cells may limit excess pyruvate or fatty acids from entering the TCA cycle for oxidative metabolism. Taken together, GLUT1-mediated glucose metabolism drives metabolic programming of macrophages by switching fuel metabolism from oxidative to glycolytic.
MΦs, which display elevated GLUT1-mediated glucose uptake and metabolism, are forced into a hyperinflammatory state with increased production of multiple inflammatory pathways and protein mediators. Our findings support those of Haschemi et al. (34) that demonstrated classical, M1 MΦ activation is dependent upon increased flux through the glycolytic and pentose phosphate pathways and extend those findings to show that increasing glucose uptake through GLUT1 is, by itself, sufficient to impart an M1-like phenotype in MΦs. Furthermore, Tannahill (9) elegantly demonstrated glycolytic dependence of LPS-induced IL-1β release in BMDMs; however, alterations in succinate or GABA were not detected in our cell system, suggesting an alternate mechanism linking glucose metabolism to inflammation. One mechanism by which GLUT1-mediated metabolism may drive inflammation is through ROS production, which we demonstrated was highly increased with GLUT1 overexpression. Upon activation, classically activated M1 MΦs undergo a “respiratory burst,” referred to as the “glycolytic burst,” driven by significantly enhanced NADPH oxidase activity, the purpose of which is to generate large amounts of ROS to destroy invading bacteria (35). To support redox balance, the glycolysis-PPP axis is activated in response to M1 polarization, presumably to sustain enhanced production of NADPH for use by NADPH oxidase in addition to production of glutathione used by the MΦs to protect from the excessive amounts of superoxide being produced (35). We report that overexpression of GLUT1 alone leads to increased overall ROS burden and increased intracellular oxidative stress as evidenced by an increased GSSG/GSH ratio. GLUT1-mediated metabolism ties increased cellular glucose to oxidative stress; the role of oxidative stress in obesity-induced inflammation is well established (2).
PAI-1 is an important proinflammatory mediator linked to the pathophysiology of obesity and vascular diseases such as atherosclerosis (2). It was robustly up-regulated in GLUT1-OE MΦs regardless of LPS stimulation or not. Therefore, we tested the dependence of PAI-1 expression on GLUT1-mediated glycolysis. Pharmacologic inhibition of glycolysis using 2-DG blunted expression PAI-1. The fact that PAI-1 is an established NFκB target gene and NFκB is regulated by redox status (36) could be a mechanism linking GLUT1, enhanced glucose metabolism, and increased proinflammatory mediators, including ROS. We have reported that treatment of GLUT1-OE MΦs with the antioxidant NAC also blunted GLUT1-driven PAI-1 expression. Taken together, because the M1 MΦ subtype has been shown to modulate inflammation-associated insulin resistance in tissues such as adipose and liver (14, 37, 38), it is likely that GLUT1 may contribute to maintenance of the proinflammatory M1 phenotype present in obese tissue. Indeed, in vivo evidence for the role of GLUT1 in proinflammatory MΦs was demonstrated; GLUT1 positive MΦs co-localized with crown-like structures and inflammatory loci in white adipose and livers, respectively, of obese rats versus lean controls (4, 5). In addition, adipose tissue macrophages displaying high surface GLUT1 contained greater cytokines, suggesting that GLUT1 correlates with in vivo inflammation. The dependence of these findings upon GLUT1 remains to be determined in future studies.
Elevated lipogenesis and cholesterol accumulation are other potential mechanisms by which glucose metabolism can lead to inflammation (39–45). GLUT1-regulated pathways identified by Ingenuity Pathway Analysis included elevated cholesterol synthesis. Cholesterol is a known activator of inflammatory pathways (46, 47). Importantly, we and others have shown that lipids such as free cholesterol can induce endoplasmic reticulum stress and promote MΦ-mediated inflammation via elevations in inflammatory kinase activity such as JNK and IκK, which promote insulin resistance, obesity, and atherosclerosis (46). Hence, the link between GLUT1-mediated elevated glucose metabolism and elevated cholesterol production in MΦs is currently under investigation.
Taken together our work demonstrates that MΦ metabolic reprogramming, which is critical to MΦ inflammatory capacity, is driven by GLUT1-mediated glucose uptake and subsequent enhanced glucose metabolism through the PPP. This phenomenon was detected in the immortalized RAW macrophage cell line, which are inherently highly glycolytic. The relevance of GLUT1 to in vivo inflammatory states is currently under investigation. It is likely that glucose metabolism may aid in fine-tuning the chronic, obesity-associated MΦ-mediated inflammation in insulin resistance. As obesity has reached globally epidemic proportions (48), the ability to better understand obesity-induced inflammation and identify putative targets that result in the manipulation of substrate metabolism in MΦs will aid in the discovery of novel pathways that regulate MΦ-mediated inflammation and the promotion of obesity and diabetes.
This work was supported, in whole or in part, by National Institutes of Health Grants AA017376 (UNC University Cancer Research Fund), ES019472, and P30DK034987; to L. M.), P30DK056350 (UNC Nutrition Obesity Research Consortium (to L. M. and M. T.), and R01HL108006 (J. C. R.).

This article contains supplemental Tables 1 and 2.
- MΦ
- macrophage
- GLUT
- glucose transporter
- RAW
- RAW264.7
- EV
- empty vector
- OE
- overexpresser
- BMDM
- bone marrow-derived macrophage
- ECAR
- extracellular acidification rate
- OCR
- oxygen consumption rate
- NAC
- N-acetyl-cysteine
- ROS
- reactive oxygen species
- PPP
- pentose phosphate pathway
- DMEM
- Dulbecco's minimal essential media
- MTT
- 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide
- qPCR
- quantitative PCR
- 2-DG
- 2-deoxyglucose
- RANTES
- regulated on activation normal T cell expressed and secreted
- PAI-1
- plasminogen activator inhibitor 1.
REFERENCES
- 1. Xu H., Barnes G. T., Yang Q., Tan G., Yang D., Chou C. J., Sole J., Nichols A., Ross J. S., Tartaglia L. A., Chen H. (2003) Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest. 112, 1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Johnson A. R., Milner J. J., Makowski L. (2012) The inflammation highway. Metabolism accelerates inflammatory traffic in obesity. Immunol. Rev. 249, 218–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weisberg S. P., McCann D., Desai M., Rosenbaum M., Leibel R. L., Ferrante A. W., Jr. (2003) Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 112, 1796–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sampey B. P., Vanhoose A. M., Winfield H. M., Freemerman A. J., Muehlbauer M. J., Fueger P. T., Newgard C. B., Makowski L. (2011) Cafeteria diet is a robust model of human metabolic syndrome with liver and adipose inflammation. Comparison to high fat diet. Obesity 19, 1109–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sampey B. P., Freemerman A. J., Zhang J., Kuan P. F., Galanko J. A., O'Connell T. M., Ilkayeva O. R., Muehlbauer M. J., Stevens R. D., Newgard C. B., Brauer H. A., Troester M. A., Makowski L. (2012) Metabolomic profiling reveals mitochondrial-derived lipid biomarkers that drive obesity-associated inflammation. PLoS ONE 7, e38812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Makowski L., Boord J. B., Maeda K., Babaev V. R., Uysal K. T., Morgan M. A., Parker R. A., Suttles J., Fazio S., Hotamisligil G. S., Linton M. F. (2001) Lack of macrophage fatty acid-binding protein aP2 protects mice deficient in apolipoprotein E against atherosclerosis. Nat. Med. 7, 699–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Makowski L., Brittingham K. C., Reynolds J. M., Suttles J., Hotamisligil G. S. (2005) The fatty acid-binding protein, aP2, coordinates macrophage cholesterol trafficking and inflammatory activity. Macrophage expression of aP2 impacts peroxisome proliferator-activated receptor γ and IκB kinase activities. J. Biol. Chem. 280, 12888–12895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vats D., Mukundan L., Odegaard J. I., Zhang L., Smith K. L., Morel C. R., Wagner R. A., Greaves D. R., Murray P. J., Chawla A. (2006) Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab. 4, 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tannahill G. M., Curtis A. M., Adamik J., Palsson-McDermott E. M., McGettrick A. F., Goel G., Frezza C., Bernard N. J., Kelly B., Foley N. H., Zheng L., Gardet A., Tong Z., Jany S. S., Corr S. C., Haneklaus M., Caffrey B. E., Pierce K., Walmsley S., Beasley F. C., Cummins E., Nizet V., Whyte M., Taylor C. T., Lin H., Masters S. L., Gottlieb E., Kelly V. P., Clish C., Auron P. E., Xavier R. J., O'Neill L. A. (2013) Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krawczyk C. M., Holowka T., Sun J., Blagih J., Amiel E., DeBerardinis R. J., Cross J. R., Jung E., Thompson C. B., Jones R. G., Pearce E. J. (2010) Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 115, 4742–4749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maciver N. J., Jacobs S. R., Wieman H. L., Wofford J. A., Coloff J. L., Rathmell J. C. (2008) Glucose metabolism in lymphocytes is a regulated process with significant effects on immune cell function and survival. J. Leukoc. Biol. 84, 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Michalek R. D., Gerriets V. A., Jacobs S. R., Macintyre A. N., MacIver N. J., Mason E. F., Sullivan S. A., Nichols A. G., Rathmell J. C. (2011) Cutting edge. distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186, 3299–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi L. Z., Wang R., Huang G., Vogel P., Neale G., Green D. R., Chi H. (2011) HIF1α-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 208, 1367–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lumeng C. N., Bodzin J. L., Saltiel A. R. (2007) Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 117, 175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fu Y., Maianu L., Melbert B. R., Garvey W. T. (2004) Facilitative glucose transporter gene expression in human lymphocytes, monocytes, and macrophages. A role for GLUT isoforms 1, 3, and 5 in the immune response and foam cell formation. Blood Cells Mol. Dis. 32, 182–190 [DOI] [PubMed] [Google Scholar]
- 16. Fukuzumi M., Shinomiya H., Shimizu Y., Ohishi K., Utsumi S. (1996) Endotoxin-induced enhancement of glucose influx into murine peritoneal macrophages via GLUT1. Infect. Immun. 64, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bentley J., Itchayanan D., Barnes K., McIntosh E., Tang X., Downes C. P., Holman G. D., Whetton A. D., Owen-Lynch P. J., Baldwin S. A. (2003) Interleukin-3-mediated cell survival signals include phosphatidylinositol 3-kinase-dependent translocation of the glucose transporter GLUT1 to the cell surface. J. Biol. Chem. 278, 39337–39348 [DOI] [PubMed] [Google Scholar]
- 18. Buller C. L., Loberg R. D., Fan M. H., Zhu Q., Park J. L., Vesely E., Inoki K., Guan K. L., Brosius F. C., 3rd. (2008) A GSK-3/TSC2/mTOR pathway regulates glucose uptake and GLUT1 glucose transporter expression. Am. J. Physiol. Cell Physiol. 295, C836–C843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wofford J. A., Wieman H. L., Jacobs S. R., Zhao Y., Rathmell J. C. (2008) IL-7 promotes Glut1 trafficking and glucose uptake via STAT5-mediated activation of Akt to support T-cell survival. Blood 111, 2101–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wieman H. L., Wofford J. A., Rathmell J. C. (2007) Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Mol. Biol. Cell 18, 1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qin Y., Hamilton J. L., Bird M. D., Chen M. M., Ramirez L., Zahs A., Kovacs E. J., Makowski L. (2014) Adipose inflammation and macrophage infiltration after binge ethanol and burn injury. Alcohol. Clin. Exp. Res. 38, 204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stewart D. A., Yang Y., Makowski L., Troester M. A. (2012) Basal-like breast cancer cells induce phenotypic and genomic changes in macrophages. Mol. Cancer Res. 10, 727–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bhatt A. P., Jacobs S. R., Freemerman A. J., Makowski L., Rathmell J. C., Dittmer D. P., Damania B. (2012) Dysregulation of fatty acid synthesis and glycolysis in non-Hodgkin lymphoma. Proc. Natl. Acad. Sci. U.S.A. 109, 11818–11823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tada H., Tsukamoto M., Ishii H., Isogai S. (1994) A high concentration of glucose alters the production of tPA, uPA, and PAI-1 antigens from human mesangial cells. Diabetes Res. Clin. Pract. 24, 33–39 [DOI] [PubMed] [Google Scholar]
- 25. Salh B., Wagey R., Marotta A., Tao J. S., Pelech S. (1998) Activation of phosphatidylinositol 3-kinase, protein kinase B, and p70 S6 kinases in lipopolysaccharide-stimulated Raw 264.7 cells. Differential effects of rapamycin, Ly294002, and wortmannin on nitric oxide production. J. Immunol. 161, 6947–6954 [PubMed] [Google Scholar]
- 26. Spolarics Z., Navarro L. (1994) Endotoxin stimulates the expression of glucose-6-phosphate dehydrogenase in Kupffer and hepatic endothelial cells. J. Leukoc. Biol. 56, 453–457 [DOI] [PubMed] [Google Scholar]
- 27. Malide D., Davies-Hill T. M., Levine M., Simpson I. A. (1998) Distinct localization of GLUT-1, -3, and -5 in human monocyte-derived macrophages. Effects of cell activation. Am. J. Physiol. 274, E516–E526 [DOI] [PubMed] [Google Scholar]
- 28. Chang M., Hamilton J. A., Scholz G. M., Masendycz P., Macaulay S. L., Elsegood C. L. (2009) Phosphatidylinostitol-3 kinase and phospholipase C enhance CSF-1-dependent macrophage survival by controlling glucose uptake. Cell Signal 21, 1361–1369 [DOI] [PubMed] [Google Scholar]
- 29. Gamelli R. L., Liu H., He L. K., Hofmann C. A. (1996) Augmentations of glucose uptake and glucose transporter-1 in macrophages following thermal injury and sepsis in mice. J. Leukoc. Biol. 59, 639–647 [DOI] [PubMed] [Google Scholar]
- 30. Newsholme P., Curi R., Gordon S., Newsholme E. A. (1986) Metabolism of glucose, glutamine, long-chain fatty acids and ketone bodies by murine macrophages. Biochem. J. 239, 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yeop Han C., Kargi A. Y., Omer M., Chan C. K., Wabitsch M., O'Brien K. D., Wight T. N., Chait A. (2010) Differential effect of saturated and unsaturated free fatty acids on the generation of monocyte adhesion and chemotactic factors by adipocytes. Dissociation of adipocyte hypertrophy from inflammation. Diabetes 59, 386–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Subramanian S., Chait A. (2009) The effect of dietary cholesterol on macrophage accumulation in adipose tissue. Implications for systemic inflammation and atherosclerosis. Curr. Opin. Lipidol. 20, 39–44 [DOI] [PubMed] [Google Scholar]
- 33. Subramanian S., Han C. Y., Chiba T., McMillen T. S., Wang S. A., Haw A., 3rd, Kirk E. A., O'Brien K. D., Chait A. (2008) Dietary cholesterol worsens adipose tissue macrophage accumulation and atherosclerosis in obese LDL receptor-deficient mice. Arterioscler. Thromb. Vasc. Biol. 28, 685–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haschemi A., Kosma P., Gille L., Evans C. R., Burant C. F., Starkl P., Knapp B., Haas R., Schmid J. A., Jandl C., Amir S., Lubec G., Park J., Esterbauer H., Bilban M., Brizuela L., Pospisilik J. A., Otterbein L. E., Wagner O. (2012) The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 15, 813–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sheppard F. R., Kelher M. R., Moore E. E., McLaughlin N. J., Banerjee A., Silliman C. C. (2005) Structural organization of the neutrophil NADPH oxidase. Phosphorylation and translocation during priming and activation. J. Leukoc. Biol. 78, 1025–1042 [DOI] [PubMed] [Google Scholar]
- 36. Nishi T., Shimizu N., Hiramoto M., Sato I., Yamaguchi Y., Hasegawa M., Aizawa S., Tanaka H., Kataoka K., Watanabe H., Handa H. (2002) Spatial redox regulation of a critical cysteine residue of NF-κB in vivo. J. Biol. Chem. 277, 44548–44556 [DOI] [PubMed] [Google Scholar]
- 37. Lumeng C. N., DelProposto J. B., Westcott D. J., Saltiel A. R. (2008) Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes 57, 3239–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Odegaard J. I., Ricardo-Gonzalez R. R., Goforth M. H., Morel C. R., Subramanian V., Mukundan L., Red Eagle A., Vats D., Brombacher F., Ferrante A. W., Chawla A. (2007) Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature 447, 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Griffin E., Re A., Hamel N., Fu C., Bush H., McCaffrey T., Asch A. S. (2001) A link between diabetes and atherosclerosis. Glucose regulates expression of CD36 at the level of translation. Nat. Med. 7, 840–846 [DOI] [PubMed] [Google Scholar]
- 40. Mauerer R., Ebert S., Langmann T. (2009) High glucose, unsaturated, and saturated fatty acids differentially regulate expression of ATP-binding cassette transporters ABCA1 and ABCG1 in human macrophages. Exp. Mol. Med. 41, 126–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mauldin J. P., Nagelin M. H., Wojcik A. J., Srinivasan S., Skaflen M. D., Ayers C. R., McNamara C. A., Hedrick C. C. (2008) Reduced expression of ATP-binding cassette transporter G1 increases cholesterol accumulation in macrophages of patients with type 2 diabetes mellitus. Circulation 117, 2785–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hayek T., Kaplan M., Kerry R., Aviram M. (2007) Macrophage NADPH oxidase activation, impaired cholesterol fluxes, and increased cholesterol biosynthesis in diabetic mice. A stimulatory role for d-glucose. Atherosclerosis 195, 277–286 [DOI] [PubMed] [Google Scholar]
- 43. Kaplan M., Kerry R., Aviram M., Hayek T. (2008) High glucose concentration increases macrophage cholesterol biosynthesis in diabetes through activation of the sterol regulatory element binding protein 1 (SREBP1). Inhibitory effect of insulin. J. Cardiovasc. Pharmacol. 52, 324–332 [DOI] [PubMed] [Google Scholar]
- 44. O'Rourke L., Gronning L. M., Yeaman S. J., Shepherd P. R. (2002) Glucose-dependent regulation of cholesterol ester metabolism in macrophages by insulin and leptin. J. Biol. Chem. 277, 42557–42562 [DOI] [PubMed] [Google Scholar]
- 45. Porstmann T., Griffiths B., Chung Y. L., Delpuech O., Griffiths J. R., Downward J., Schulze A. (2005) PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene 24, 6465–6481 [DOI] [PubMed] [Google Scholar]
- 46. Erbay E., Babaev V. R., Mayers J. R., Makowski L., Charles K. N., Snitow M. E., Fazio S., Wiest M. M., Watkins S. M., Linton M. F., Hotamisligil G. S. (2009) Reducing endoplasmic reticulum stress through a macrophage lipid chaperone alleviates atherosclerosis. Nat. Med. 15, 1383–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Feng B., Yao P. M., Li Y., Devlin C. M., Zhang D., Harding H. P., Sweeney M., Rong J. X., Kuriakose G., Fisher E. A., Marks A. R., Ron D., Tabas I. (2003) The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat. Cell Biol. 5, 781–792 [DOI] [PubMed] [Google Scholar]
- 48. Ogden C. L., Carroll M. D., Kit B. K., Flegal K. M. (2013) Prevalence of obesity among adults. NCHS Data Brief 131, 1–8 [PubMed] [Google Scholar]